Abstract
In modern applications of molecular epidemiology, genetic sequence data are routinely used to identify clusters of transmission in rapidly evolving pathogens, most notably HIV-1. Traditional ‘shoe-leather’ epidemiology infers transmission clusters by tracing chains of partners sharing epidemiological connections (e.g., sexual contact). Here, we present a computational tool for identifying a molecular transmission analog of such clusters: HIV-TRACE (TRAnsmission Cluster Engine). HIV-TRACE implements an approach inspired by traditional epidemiology, by identifying chains of partners whose viral genetic relatedness imply direct or indirect epidemiological connections. Molecular transmission clusters are constructed using codon-aware pairwise alignment to a reference sequence followed by pairwise genetic distance estimation among all sequences. This approach is computationally tractable and is capable of identifying HIV-1 transmission clusters in large surveillance databases comprising tens or hundreds of thousands of sequences in near real time, that is, on the order of minutes to hours. HIV-TRACE is available at www.hivtrace.org and from www.github.com/veg/hivtrace, along with the accompanying result visualization module from www.github.com/veg/hivtrace-viz. Importantly, the approach underlying HIV-TRACE is not limited to the study of HIV-1 and can be applied to study outbreaks and epidemics of other rapidly evolving pathogens.
Keywords: molecular epidemiology, HIV, network, transmission cluster, surveillance
Research into fundamental questions of epidemiology and public health, such as ‘Who infected whom?’ (Volz and Frost 2013; Romero-Severson et al. 2016), ‘How does pathogen X spread through a population’? (Dennis et al. 2014), and ‘Is a particular prevention or treatment effective at slowing or stopping the spread of disease?’ (Little et al. 2014) has greatly benefited from large-scale analyses of molecular sequences obtained during surveillance or through routine diagnostics. For rapidly evolving pathogens, such as HIV-1 or hepatitis C virus, viral isolates from different hosts will typically not be genetically identical, and analyses of these genetic differences via phylogenetic, phylodynamic, or other evolutionary methods have proven tremendously powerful. Phylogenetic analyses have been used in criminal cases involving deliberate HIV-1 transmission (Scaduto et al. 2010), to understand the introduction of HIV-1 into regions and countries (Gilbert et al. 2007), and to define recent clusters of transmission cases (Peters et al. 2016). Recent work in the field of phylodynamics has established a template on how to use sequence data to inform inference of epidemiological transmission parameters, for example, R0 or transmission rates between different risk groups (Frost and Volz 2013; Volz and Frost 2014). The fundamental insight shared by all these methods is that genetic similarity, or relatedness, between pathogen sequences can be used to identify strains that are connected in an epidemiologically meaningful way: as potential source–recipient pairs (Campbell et al. 2011) or members of a distinct transmission cluster (Campbell et al. 2017; Wertheim et al. 2017a).
Real-time or near real-time surveillance of pathogen transmission is an area of great interest to local, national, and global public health agencies (Division of HIV/AIDS Prevention 2017). Real-time surveillance seeks to quickly analyze newly obtained pathogen genetic sequences in the context of large, preexisting reference samples and to deliver actionable inference results: ‘A new rapidly growing HIV-1 transmission cluster has been identified’, or ‘An unusual pattern of transmission between people with different risk factors has been detected’, or ‘An HIV-1 transmission prevention is effectively reducing population level incidence’.
Defining molecular transmission clusters is a challenging problem, and currently there is no consensus in the field of molecular epidemiology on what should or should not constitute a transmission cluster or whether certain definitions are more germane to particular research questions or public health interventions (Grabowski and Redd 2014; Wertheim et al. 2014; Hassan et al. 2017; Novitsky et al. 2017).
Here, we present the algorithmic software implementation, and operational usage details for HIV-TRACE (TRAnsmission Cluster Engine), a platform that has been used extensively for rapid inference of transmission networks from large sets of pathogen genetic sequences to identify potential transmission links and to describe putative transmission clusters. An early version of HIV-TRACE was used to analyze nearly 100,000 HIV-1 sequences sampled worldwide, and this analysis revealed that there was a surprising amount of global (country-to-country) connectivity in this network (Wertheim et al. 2014). Since then, HIV-TRACE has been used to investigate transmission patterns among risk groups (Oster et al., 2015; Whiteside et al. 2015), characterize transmission fitness of HIV drug-resistance-associated mutations (Wertheim et al. 2017 b), and to identify rapidly growing transmission clusters (Campbell et al. 2017; Monterosso et al. 2017).
The source code, installation instruction (via pip3), and documentation for HIV-TRACE is available at github.com/veg/hivtrace, and the accompanying result visualization module—at github.com/veg/hivtrace-viz. In addition, a public instance of the HIV-TRACE web-application is hosted at www.hivtrace.org, as a part of the Datamonkey family of services (Weaver et al. 2018).
New Approaches
HIV-TRACE does not infer a phylogenetic tree from sequence data because phylogenetic inference is a computational bottleneck and because the phylogenies themselves are typically not directly useful for epidemiological inference. In most applications, phylogenies are converted to summary features (e.g., clades) or summary statistics (e.g., patristic distances) to identify clusters. In lieu of phylogenetic inference, HIV-TRACE identifies groups of putative transmission partners and assembles these partners in transmission clusters. This approach is analogous to the traditional epidemiological definition of an infectious disease transmission cluster: a group of infected people with direct or indirect epidemiological connections. In HIV-TRACE, genetic linkage serves as a proxy for these direct or indirect epidemiological connections, and a cluster is constructed based on these connections. This approach is fundamentally different from phylogenetic-based cluster inference (Grabowski and Redd 2014; Wertheim et al. 2014), which seeks to identify a point in evolutionary history from which all cluster members descend (i.e., a point that gives rise to a clade on a phylogeny). Importantly, several independent studies have shown that in many cases relevant to HIV-1 epidemiology, HIV-TRACE reports very similar sets of clusters to phylogeny-based methods (Poon 2016; Rose et al. 2017 b), although whether or not clusters arise due to increased transmission rate or from increased sampling rates or recent transmission is potentially difficult to identify with this (or alternative) approaches (Le Vu et al. 2017; McCloskey and Poon 2017).
Inference Procedure
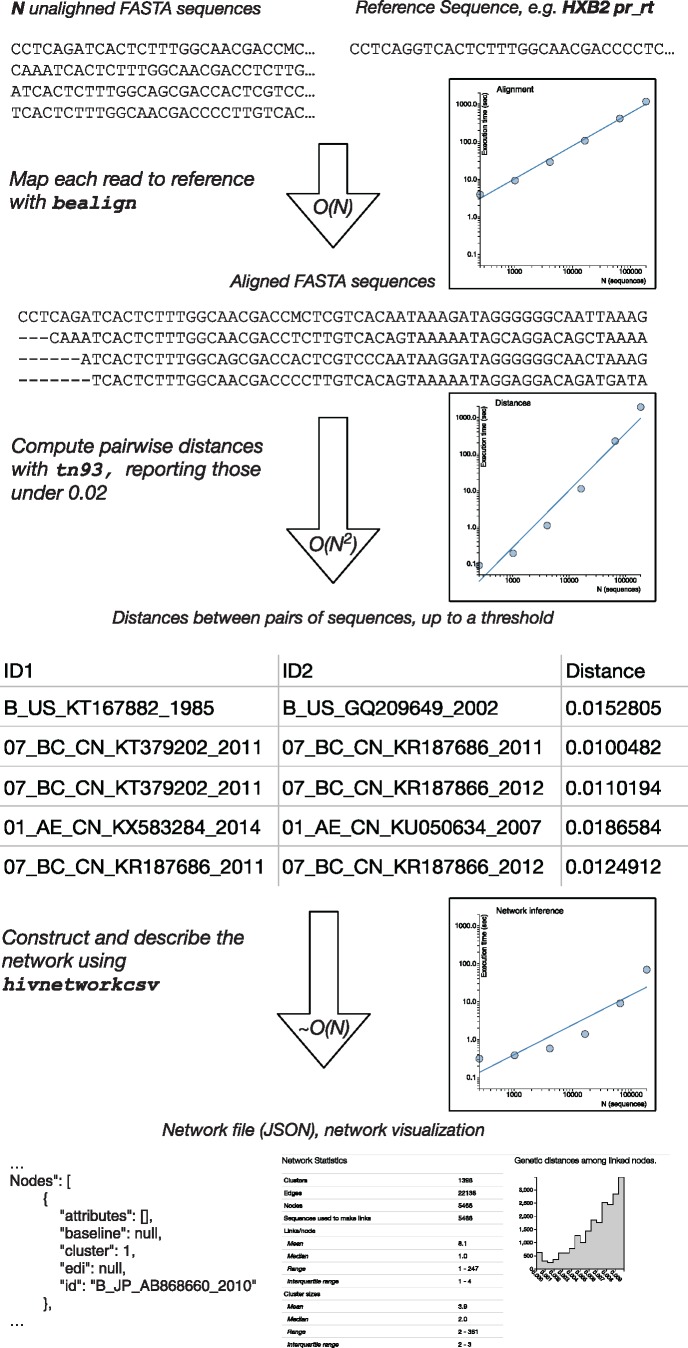
HIV-TRACE takes in a collection of N unaligned coding viral sequences sampled from M ≤ N individuals (multiple sequences per individual are supported) formatted as a FASTA file, and it outputs a JSON file containing the description of the inferred transmission network as nodes (individuals) and links (potential transmission partners). When additional clinical, demographic, or other data are available, they can be included in the network as attributes. Key parameters controlling network inference are summarized in table 1, and the schematic of program flow is depicted in figure 1.
Table 1.
Key Parameters Controlling HIV-TRACE.
| Parameter | Meaning | Phase |
|---|---|---|
| −r, --reference | Reference sequence for mapping | Alignment |
| −m, --minoverlap | Sequences must have at least this many aligned characters | Distance estimation |
| −a, --ambiguities | Sets policy for handling ambiguous nucleotides | Distance estimation |
| −g, --fraction | Sets the maximum fraction of resolvable ambiguous nucleotides | Distance estimation |
| −s, --strip_drams | Mask HIV-1 drug resistance associated sites | Distance estimation |
| −t, --threshold | Distance threshold for drawing a network link | Network construction |
| −u, --curate | Sets policy for handling potential contaminants | Network construction |
Fig. 1.
A schematic of the HIV-TRACE workflow. For each stage, we show example input and output data, indicate computational complexity, and provide empirical run-times as functions of the number of sequences on the example HIV-1 data sets described in the text. Trend lines show linear fits in the log-log space.
To demonstrate method performance, we downloaded all publicly available HIV-1 polymerase sequence (one sequence per patient, minimum length 500 nt) from the Los Alamos National Laboratories HIV database (hiv.lanl.gov), resulting in N = M = 185,849 sequences. We randomly sampled a set of 256, 1,024, 4,096, 16,384, and 65,536 sequences to plot computational time scaling. We ran each step of the pipeline 10 times (to average out computing environment stochasticity) on a 64-core (2× 32 AMD Opteron 6356) system running at 2 GHz clock rate (fig. 1).
Sequence Alignment
HIV-TRACE first aligns each of the input sequences to a single reference sequence using a codon-aware extension of the Smith–Waterman dynamic programming algorithm (Smith and Waterman 1981), previously developed by us in the context of high throughput sequencing read mapping (e.g., Gianella et al. 2011). For standard HIV-1 analyses, the HXB2 sequence (GenBank Accession number: K03455) is used as a reference sequence, although any in-frame coding sequence can be supplied as reference. Both the forward and the reverse-complement versions of each sequence are considered, and the one with the higher alignment score is retained. Codon-aware alignment leverages protein homology to align nucleotide data and is able to identify and correct relatively frequent (i.e., up to 5% of sequences in some data sets) frame-shifting insertions or deletions involving one or two nucleotides. In this case, correction means maintaining the frame relative to the reference. The resulting pairwise alignment is merged into a single multiple-sequence alignment (MSA). As the vast majority of HIV-1 sequence data arise from surveillance screening for drug resistance in a 1497-nucleotide protease and reverse transcriptase genomic region, which only rarely exhibit insertions/deletions relative to the reference sequence, this ‘mapping’ approach is effective and scales linearly in the number of sequences. Traditional progressive alignment methods have superlinear (e.g., up to quadratic) computational cost. In our example, computational complexity scaled linearly as expected, and the alignment of 185,849 sequences to a reference took about 20 min on average. Importantly, HIV-TRACE is also capable of handling previously aligned sequences, which may be desirable for analyses of HIV-1 envelope sequences or other pathogens with low evolutionary conservation, where ‘all-to-one’ alignment is not likely to recover more distant homologies. However, genes or sequence regions that are challenging to align may be suboptimal for molecular epidemiology applications.
Estimation of Genetic Distances
Given an MSA on N sequences, HIV-TRACE computes all N × (N – 1)/2 pairwise genetic distances under the Tamura-Nei 93 (TN93) (Tamura and Nei 1993) nucleotide substitution model, which is the most general nucleotide substitution model for which distances can be estimated directly from counts of nucleotide pairs in aligned sequences. Whereas more complex models substitution models are typically preferable in the context of phylogenetic inference, especially for more distantly related strains (Posada and Crandall 2001), when genetic distances are low (e.g., 0.05), all sensible nucleotide distance measures perform comparably (Wertheim and Kosakovsky Pond 2011). A key option controlling this step in HIV-TRACE is how to handle ambiguous nucleotide characters that represent within-host population polymorphisms or sequencing errors (see Parameterizing genetic distance estimates section). An important example of epidemiological processes that yield sequences with high fractions of ambiguous nucleotides is multiple (super- or dual-) HIV infection (Pacold et al. 2010).
Pairwise distances are reported to a comma separated file, and are typically limited only to those pairs that are below a user-specified threshold (e.g., 0.015 substitutions/site) to retain only pairs of sequences that have an epidemiological link. This step is computationally costly, scaling as N2, but an efficient parallelized implementation of the tool allows rapid processing of 105–106 sequences. For instance, it took approximately 32 min to compute all pairwise distances between 185,849 sequences. Our implementation is also memory efficient, requiring O(NL) space, where L is the sequence length. For data sets of this size, traditional rapid phylogeny reconstruction techniques, such as Neighbor Joining are already infeasible, because they scale as N3 and require the storage of the entire distance matrix (this would require ∼256 GiB of RAM for our example), which HIV-TRACE deliberately avoids. Because most phylogenetic methods for cluster definition require some measure of clade support (e.g., Grabowski and Redd 2014), it is also necessary to perform a version of bootstrapping. Our implementation compares favorably to even the fastest tree construction methods, such as FastTree 2 (Price et al. 2010) or IQ-Tree (Nguyen et al. 2015), which takes at least 10x longer to process these sizes of data; for example, typical run times of FastTree 2 (the fastest tool to our knowledge) on ∼200,000 sequences is on the order of 10–20 h (Price et al. 2010). It is worth noting that FastTree 2 has an asymptotically better run time , but it does considerably more work than needed for our application (resulting in slower run times), and uses heuristics which are not guaranteed to always find all distances below a certain threshold.
Network Construction
The transmission network is inferred from the file of pairwise distances and optionally annotated with data from attribute files. Nodes within the network are all keyed on either the entire sequence name or parts thereof extracted by regular expressions. A link is drawn between two individuals if and only if the pairwise distances between any of the paired sequences from these individuals is below a user specified threshold, D. (for example, D = 0.0015). A cluster is defined as a connected component of the network. Optionally, the network can be screened for contaminants (i.e., any query sequences that link to lab strains or other user-specified contaminant sequences). Global statistics of the network, such as the number of nodes, edges, clusters, cluster sizes, and the degree distribution, are computed and reported. Lastly, the degree distribution is fit to one of four generative models of network growth: random attachment, preferential attachment, preferential attachment mixed with a component of random attachment, and power law, using the methods described by Handcock and Jones (2004). If the best fitting model is from a scale-free family (i.e., preferential attachment), the characteristic exponent ρ of the network is estimated and reported. This step is computationally relatively inexpensive, taking only a few seconds.
Parameterizing Genetic Distance Estimates
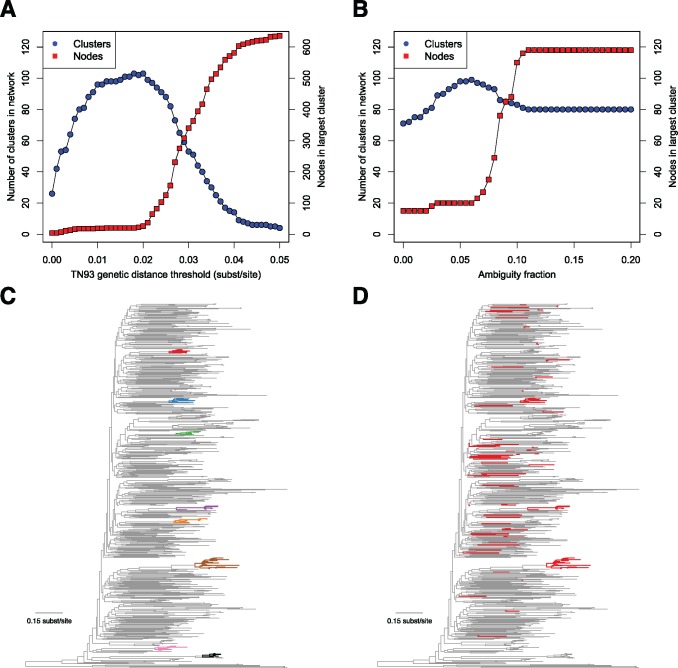
Selecting appropriate parameters governing genetic distance estimation is critical to HIV-TRACE analysis. Investigations in the US, the UK, and Canada have consistently found natural breakpoints in genetic distance between putative transmission partners and ‘random’ cases or within-host and between-host diversity (Lewis et al. 2008; Smith et al. 2009; Poon et al. 2015; Rose et al. 2017a; Wertheim et al. 2017a). In New York city, Wetheim et al (2017a) found that genetic distance thresholds between 0.01 and 0.02 substitutions/site were more strongly associated with probable transmission partners than traditional epidemiological connections (i.e., naming of sexual and injection drug using partners) and that a distance of 0.015 could serve as a use proxy for epidemiological relatedness in a surveillance setting. Moreover, these genetic distance thresholds have been validated by molecular epidemiological studies in U.S. public health surveillance populations (Oster et al. 2015; Whiteside et al. 2015; Wertheim et al. 2016, 2017b), which have reported results that are typically robust to thresholds in this range. Lower distance thresholds (e.g., 0.005 substitutions/site) may be more appropriate for distinguishing rapidly growing clusters (Division of HIV/AIDS Prevention 2017) or populations where faster evolving (i.e., non-B subtypes) predominate. As distance thresholds increase, smaller clusters merge into larger, less informative clusters (fig. 2A). At the extreme, all sequences would belong to a single cluster, which while technically correct, since all HIV-1 sequences are related through a series of transmissions, this finding is unlikely to be of interest in the context of molecular epidemiology. The same principle—that D should separate within-host or epidemiologically recent diversity from between-host diversity has been used successfully for other epidemics, genetic regions, and viruses. For example, Rose et al. (2017b) used D = 0.053 for HIV-1 gp41, Bartlett et al. (2017) selected D = 0.03 for the core gene of Hepatitis C virus. Regional and national epidemics HIV-1 also tend to require larger thresholds due to sparser sampling and the prevalence of chronically infected individuals (Hassan et al. 2017).
Fig. 2.
Effect of genetic distance threshold and ambiguity fraction on network construction. (A) Number of clusters and size of largest cluster across increasing genetic distance thresholds. (B) Number of clusters and size of largest cluster across increasing ambiguity fractions. (C) Largest clusters (≥ 7 nodes) from the San Diego Primary Infection Cohort, inferred with a 0.015 substitutions/site genetic distance threshold and a 0.05 ambiguity fraction on a phylogenetic tree (each cluster has its own color and is shown in bold). (D) Members of the large, artifactual cluster when ambiguity fraction is increased to 1.0 and distances from ambiguities in all sequences are resolved (shown in bold, colored in red). San Diego sequence data are from Little et al. (2014), and phylogeny was inferred using FastTree2 (Price et al. 2010 b).
Nucleotide ambiguities (e.g., Y indicating a mixed population of both C and T at the same genomic position) have the potential to compromise HIV-TRACE analysis, or phylogenetic inference in general. By default, HIV-TRACE will resolve (here, to ‘resolve’ means to choose the value of the ambiguity to match the other nucleotide if possible) the genetic distance between ambiguities (i.e., Y is 0 substitutions from both C and T). However, sequences with a high fraction of nucleotide ambiguities have the tendency to link to distantly related sequences when ambiguities are resolved, resulting in artifactual larger clusters (fig. 2B). When ambiguities are properly accounted for, HIV-TRACE clusters tend to resemble clades on a phylogenetic tree (fig. 2C). However, when distances ambiguities are resolved irrespective of ambiguity fraction, distantly related sequences are connected through these high ambiguity sequences, forming large artifactual clusters (fig. 2D and Aldous et al. 2012). Therefore, HIV-TRACE includes a parameter (ambiguity fraction) that averages the genetic distance from ambiguities (i.e., Y is 0.5 substitutions from both C and T) in sequences with a higher proportion of ambiguities than the indicated ambiguity fraction. In cohorts of fewer than 1,000 individuals (i.e., San Diego Primary Infection Cohort), an ambiguity fraction of 0.05 is appropriate based on empirical network sensitivity analyses. For US surveillance data, an ambiguity fraction above 0.015 produces spurious clusters. As a consequence, sequences with high ambiguity fractions are less likely to cluster using HIV-TRACE.
In HIV-TRACE, excluding sites containing ambiguities has a similar effect on network construction as resolving ambiguities. Many popular phylogenetic packages used for constructing HIV-1 molecular transmission networks (e.g., BEAST—Drummond et al. 2012 and FastTree—Price et al. 2010) exclude sites containing ambiguities from likelihood calculations. It remains unclear how treatment of nucleotide ambiguities will affect phylogenetic inference of HIV transmission clusters (Fearnhill et al. 2017).
Visualization
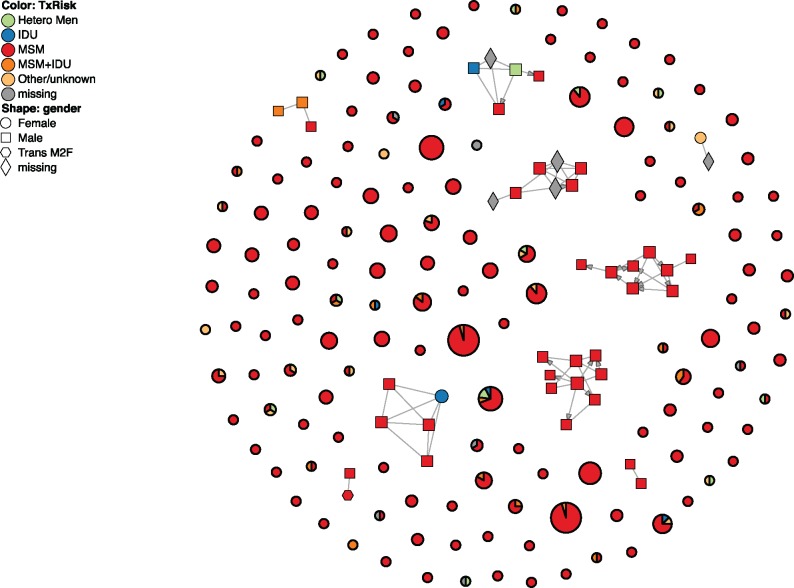
The JSON file output by HIV-TRACE can be explored using an interactive JavaScript application which we call hivtrace-viz. It is based on the open source data visualization library d3.js. This application runs within any modern web-browser and provides means to view the overall structure of the network, explore individual clusters, display network summary, and explore associations among attributes for connected nodes. When clinical and demographic attributes are available, they can be overlaid on the network structure as shown in figure 3.
Fig. 3.
Visualization of the San Diego Primary Infection Cohort cluster (Little et al. 2014) using hivtrace-viz. Circles without connections and darker borders represent clusters, and their area is proportional to cluster size. Nine of the clusters have been expanded, showing individual nodes (individuals) and edges (putative transmission links). Nodes and clusters are colored by risk factor (this is user selectable, and is obtained from network annotation data); for clusters, the distribution of risk factors is shown as a pie chart. The shape of individual nodes indicates the gender of the corresponding individual.
Software Components
Alignment
bealign is implemented in Python 3 as a part of BioExt library (github.com/veg/BioExt) which extends the functionality of the popular BioPython library (Cock et al. 2009). The core alignment routine is implemented in C and incorporated via Cython. When the program is run in a multicore/multiprocessing environment, it will distribute alignment tasks across cores.
Distance Calculation
tn93 is a self-contained C++ program (available from github.com/veg/tn93) which is tuned to allow ∼105–106 distance calculations per second per core on ∼1,000 bp long sequences. It uses OpenMP to distribute distance calculations across multiple CPU cores whenever possible. For example, tn93 achieved parallelized (64 cores) throughput of ∼107 pairwise distance calculations per second when computing distances on the LANL example data set.
Network Inference
hivnetworkcsv is a Python 3 module, which is available from github.com/veg/hivclustering, along with the attendant documentation.
Concluding Remarks
HIV-TRACE is a powerful computational tool for the rapid and automated characterization of molecular transmission clusters in populations of HIV infected individuals. Its applicability for HIV research and public health surveillance and prevention activities is apparent, as first illustrated by the unsupervised recovery of many previously characterized clusters (defined via phylogenetic analyses) in our global-scale analysis of HIV-1 databases (Wertheim et al. 2014). As viral sequence, databases increase in size and transition to using Next Generation Sequencing (NGS) data, scalable tools like HIV-TRACE will be increasingly relevant.
HIV-TRACE can accommodate NGS data in three different ways. First, NGS data can be used to generate a consensus sequence for each individual, which is then handled the same way as Sanger sequences are now. Phylogenetic approaches most commonly use this route, and HIV-TRACE has already been used in this context (Rose et al. 2017b). Second, NGS reads could be converted into a smaller collection of individual haplotypes; HIV-TRACE can directly handle multiple sequences per individual, and supports two mode of drawing links between individuals A and B: single linkage (at least one pair of sequences from A and B are closer than D substitutions per site) or complete linkage (all pairs of sequences are closer than D substitutions per site). Lastly, for NGS amplicon data that have been mapped to the reference, HIV-TRACE can be used to quickly compute the distribution of genetic distances between reads from individuals A and B; links can then be drawn if the distribution meets a particular condition, for example, at least X% of read pairs are closer than D substitutions per site.
In addition to extensive applications in the HIV-1 domain, HIV-TRACE has demonstrated utility for other pathogens including acute hepatitis C virus infection (Bartlett et al. 2017; Rose et al. 2017a) and norovirus (Drumright et al. 2014). As any computational tool, HIV-TRACE has advantages and drawbacks. Speed, easy to understand clusters definitions, persistence of clusters when more sequences are added, robustness to recombination, and systematic handling of mixed bases count among the former. The latter include the difficulty in interpreting what variables drive cluster formation and growth, inability to ascertain that any particular link is a direct transmission (i.e., source attribution), and loss of information contained in the phylogenetic tree, including timing (which can be leveraged by molecular clock methods), and branching (which can be taken advantage of by phylodynamics methods). For most rigorous analyses, clusters identified by HIV-TRACE are further analyzed using compute-intensive molecular clock phylogenetic inference tools (e.g., BEAST; Drummond et al. 2012; (Wertheim et al. 2016, 2017; Chaillon et al. 2017). By using HIV-TRACE first to identify transmission cluster of interest, these more computationally intensive tools can be reserved for smaller, focused analyses.
Acknowledgments
This study was supported in part by grants R01 AI134384 (NIH/NIAID), R01 GM093939 (NIH/NIGMS) and U01 GM110749 (NIH/NIGMS). J.O.W. was funded by an NIH-NIAID Career Development Award (K01AI110181) and the California HIV/AIDS Research Program (ID15-SD-052). We thank N. Lance Hepler for his work on the initial development of HIV-TRACE.
References
- Aldous JL, Pond SK, Poon A, Jain S, Qin H, Kahn JS, Kitahata M, Rodriguez B, Dennis AM, Boswell SL et al. , . 2012. Characterizing HIV transmission networks across the united states. Clin Infect Dis. 558:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett SR, Wertheim JO, Bull RA, Matthews GV, Lamoury FM, Scheffler K, Hellard M, Maher L, Dore GJ, Lloyd AR et al. , . 2017. A molecular transmission network of recent hepatitis C infection in people with and without HIV: implications for targeted treatment strategies. J Viral Hepat. 245:404–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Jia H, Shankar A, Hanson D, Luo W, Masciotra S, Owen SM, Oster AM, Galang RR, Spiller MW et al. , . 2017. Detailed transmission network analysis of a large opiate-driven outbreak of HIV infection in the United States. J Infect Dis. 216:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Mullins JI, Hughes JP, Celum C, Wong KG, Raugi DN, Sorensen S, Stoddard JN, Zhao H, Deng W, Partners in Prevention HSV/HIV Transmission Study Team, et al. 2011. Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One 63:e16986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillon A, Avila-Ríos S, Wertheim JO, Dennis A, García-Morales C, Tapia-Trejo D, Mejía-Villatoro C, Pascale JM, Porras-Cortés G, Quant-Durán CJ, Mesoamerican Project Group, et al. 2017. Identification of major routes of HIV transmission throughout Mesoamerica. Infect Genet Evol. 54:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B et al. , . 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2511:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis AM, Herbeck JT, Brown AL, Kellam P, de Oliveira T, Pillay D, Fraser C, Cohen MS.. 2014. Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest?. J Acquir Immune Defic Syndr. 672:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Division of HIV/AIDS Prevention 2017. Detecting, investigating, and responding to HIV transmission clusters. Technical report, Centers for Disease Control and Prevention.
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 298:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumright LN, Leigh Brown AL, Frost SDW.. 2014. The global circulation of norovoris GII.3 and GII.4. In 21st International HIV Dynamics and Evolution Conference.
- Fearnhill E, Gourlay A, Malyuta R, Simmons R, Ferns RB, Grant P, Nastouli E, Karnets I, Murphy G, Medoeva A, CASCADE Collaboration in EuroCoord, et al. 2017. A phylogenetic analysis of HIV-1 sequences in Kiev: findings among key populations. Clin Infect Dis. 2017 May 29. doi: 10.1093/cid/cix499. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost SDW, Volz EM.. 2013. Modelling tree shape and structure in viral phylodynamics. Philos Trans R Soc Lond B Biol Sci. 3681614:20120208.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianella S, Delport W, Pacold ME, Young JA, Choi JY, Little SJ, Richman DD, Kosakovsky Pond SL, Smith DM.. 2011. Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol. 8516:8359–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M.. 2007. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A. 10447:18566–18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski MK, Redd AD.. 2014. Molecular tools for studying HIV transmission in sexual networks. Curr Opin HIV AIDS 92:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handcock MS, Jones JH.. 2004. Likelihood-based inference for stochastic models of sexual network formation. Theor Popul Biol. 654:413–422. [DOI] [PubMed] [Google Scholar]
- Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J.. 2017. Defining HIV-1 transmission clusters based on sequence data. AIDS 319:1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vu S, Ratmann O, Delpech V, Brown AE, Gill ON, Tostevin A, Fraser C, Volz EM.. 2017. Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics pii: S1755-4365(17): 30115–30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ.. 2008. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 53:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, May S, Smith DM.. 2014. Using HIV networks to inform real time prevention interventions. PLoS One 96:e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey RM, Poon AFY.. 2017. A model-based clustering method to detect infectious disease transmission outbreaks from sequence variation. PLoS Comput Biol. 1311:e1005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monterosso A, Minnerly S, Goings S, Morris A, France AM, Dasgupta S, Oster A, Fanning M.. 2017. Identifying and investigating a rapidly growing HIV transmission cluster in Texas. In Conference on Retroviruses and Opportunistic Infections, page 845LB.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 321:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitsky V, Moyo S, Essex M.. 2017. Phylogenetic inference of hiv transmission clusters. Infect Dis Transl Med. 32:51–59. [Google Scholar]
- Oster AM, Wertheim JO, Hernandez AL, Ocfemia MCB, Saduvala N, Hall HI.. 2015. Using molecular HIV surveillance data to understand transmission between subpopulations in the United States. J Acquir Immune Defic Syndr. 704:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold M, Smith D, Little S, Cheng PM, Jordan P, Ignacio C, Richman D, Pond SK.. 2010. Comparison of methods to detect HIV dual infection. AIDS Res Hum Retroviruses 2612:1291–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Pontones P, Hoover KW, Patel MR, Galang RR, Shields J, Blosser SJ, Spiller MW, Combs B, Switzer WM, Indiana HIV Outbreak Investigation Team, et al. 2016. HIV infection linked to injection use of oxymorphone in Indiana, 2014–2015. N Engl J Med. 3753:229–239. [DOI] [PubMed] [Google Scholar]
- Poon AFY. 2016. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol. 22:vew031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AFY, Joy JB, Woods CK, Shurgold S, Colley G, Brumme CJ, Hogg RS, Montaner JSG, Harrigan PR.. 2015. The impact of clinical, demographic and risk factors on rates of HIV transmission: a population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis. 2116:926–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA.. 2001. Selecting models of nucleotide substitution: an application to human immunodeficiency virus 1 (HIV-1). Mol Biol Evol. 186:897–906. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 53:e9490.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Severson EO, Bulla I, Leitner T.. 2016. Phylogenetically resolving epidemiologic linkage. Proc Natl Acad Sci U S A. 11310:2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Lamers SL, Dollar JJ, Grabowski MK, Hodcroft EB, Ragonnet-Cronin M, Wertheim JO, Redd AD, German D, Laeyendecker O.. 2017b. Identifying transmission clusters with Cluster Picker and HIV-TRACE. AIDS Res Hum Retroviruses 333: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R, Lamers SL, Massaccesi G, Osburn W, Ray SC, Thomas DL, Cox AL, Laeyendecker O.. 2017a. Complex patterns of Hepatitis-C virus longitudinal clustering in a high-risk population. Infect Genet Evol. 58:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML.. 2010. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A. 10750:21242–21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, May SJ, Tweeten S, Drumright L, Pacold ME, Kosakovsky Pond SL, Pesano RL, Lie YS, Richman DD, Frost SDW et al. , . 2009. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS 232:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS.. 1981. Identification of common molecular subsequences. J Mol Biol. 1471:195–197. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M.. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 103:512–526. [DOI] [PubMed] [Google Scholar]
- Volz EM, Frost SDW.. 2013. Inferring the source of transmission with phylogenetic data. PLoS Comput Biol. 912:e1003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz EM, Frost SDW.. 2014. Sampling through time and phylodynamic inference with coalescent and birth-death models. J R Soc Interface 11101:20140945.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 2018 Jan 2. doi: 10.1093/molbev/msx335. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Kosakovsky Pond SL.. 2011. Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol. 2812:3355–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Kosakovsky Pond SL, Forgione LA, Mehta SR, Murrell B, Shah S, Smith DM, Scheffler K, Torian LV.. 2017a. Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog. 131:e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, Kosakovsky Pond SL.. 2014. The global transmission network of HIV-1. J Infect Dis. 2092:304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Oster AM, Hernandez AL, Saduvala N, Bañez Ocfemia MC, Hall HI.. 2016. The international dimension of the U.S. HIV transmission network and onward transmission of HIV recently imported into the United States. AIDS Res Hum Retroviruses 32(10–11):1046–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Oster AM, Johnson JA, Switzer WM, Saduvala N, Hernandez AL, Hall HI, Heneine W.. 2017b. Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol. 31:vex008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside YO, Song R, Wertheim JO, Oster AM.. 2015. Molecular analysis allows inference into HIV transmission among young men who have sex with men in the united states. AIDS 2918:2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]