Abstract
Ribosomal proteins are indispensable components of a living cell, and yet their structures are remarkably diverse in different species. Here we use manually curated structural alignments to provide a comprehensive catalog of structural variations in homologous ribosomal proteins from bacteria, archaea, eukaryotes, and eukaryotic organelles. By resolving numerous ambiguities and errors of automated structural and sequence alignments, we uncover a whole new class of structural variations that reside within seemingly conserved segments of ribosomal proteins. We then illustrate that these variations reflect an apparent adaptation of ribosomal proteins to the specific environments and lifestyles of living species. Finally, we show that most of these structural variations reside within nonglobular extensions of ribosomal proteins—protein segments that are thought to promote ribosome biogenesis by stabilizing the proper folding of ribosomal RNA. We show that although the extensions are thought to be the most ancient peptides on our planet, they are in fact the most rapidly evolving and most structurally and functionally diverse segments of ribosomal proteins. Overall, our work illustrates that, despite being long considered as slowly evolving and highly conserved, ribosomal proteins are more complex and more specialized than is generally recognized.
Keywords: ribosome, ubiquitous proteins, ribosomal proteins, adaptation, protein structure, protein evolution
Introduction
Ribosomes are present in every living cell, but their structures are astonishingly distinct in different species. Even relatively simple ribosomes from bacterial species, whose molecular weights vary around 2.4 MDa, carry ∼0.7 MDa of unique RNA and protein moieties, which are missing in eukaryotic ribosomes (Melnikov et al. 2012). Remarkably, these species-specific moieties decorate every functional center of the ribosome, including the peptidyl-transferase, the peptide exit tunnel, the messenger RNA (mRNA) channel, the decoding site, and the binding sites of translation factors and regulatory proteins (Ban et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001; Yusupova et al. 2001; Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011; Armache et al. 2013). And as we rapidly uncover the structural diversities of ribosomes from all branches of life, we are just beginning to understand the physiological roles behind species-specific variations in ribosome architecture.
This impressive diversity of ribosome composition originates to a great extent from the high variability in ribosomal protein content. To date, more than 200 nonhomologous ribosomal proteins have been found in nature, but only 33 of them are present in all domains of life (Lecompte et al. 2002; Agrawal 2011). Furthermore, these 33 proteins vary in size and sequence to such an extent that when higher eukaryotes are compared to bacteria, or free-living species are compared to obligate intracellular parasites, some of these proteins differ up to five times in length and may have as little as ∼30% of sequence similarity (Wool et al. 1995; Katinka et al. 2001; Ben-Shem et al. 2011; Melnikov et al. 2012).
Our understanding of how and why ribosomal proteins have diverged across species has been greatly advanced by structural studies. Over the past two decades, ribosome structures have been determined in species from all domains of life, including common laboratory models of bacterial, archaeal, and eukaryotic species and organelles of eukaryotic cells (Ban et al., 2000; Wimberly et al., 2000; Yusupov et al., 2001; Harms et al., 2001; Haldar et al., 2006; Ben-Shem et al., 2011; Klinge et al., 2011; Rabl et al., 2011; Greber et al., 2012; Anger et al., 2013; Armache et al. 2013; Hashem et al., 2013; Fernandez et al., 2014; Gogala et al., 2014; Kaushal et al. 2014; Park et al., 2014; Voorhees et al., 2014; Amunts et al. 2015; Aylett et al., 2015; Eyal et al. 2015; Greber et al., 2015; Noeske et al., 2015; Sohmen et al., 2015; Voorhees and Hegde, 2015; Khusainov et al., 2016; Shalev-Benami et al., 2016; Ahmed et al., 2017; Bieri et al., 2017; Desai et al., 2017; Graf et al., 2017; Hentschel et al., 2017; Li et al., 2017; Wong et al., 2017). These studies gave us a unique opportunity to visualize variations in the ribosome structure and therefore shed light on their biological roles.
However, despite the abundance of structural data, comparison of ribosomes from different species remains laborious, and most studies are still relying on multiple sequence alignments to identify structurally conserved and structurally variable segments of ribosomal proteins (Klein et al. 2004; Nakao et al. 2004; Hartman et al. 2006; Teeling and Gloeckner, 2006; Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011). We previously showed that the problem of this traditional approach is that multiple sequence alignments tend to underestimate the structural diversity of ribosomal proteins (Ben-Shem et al. 2011; Melnikov et al. 2012, 2015). First, we found that multiple sequence alignments frequently align structurally dissimilar segments of homologous ribosomal proteins and annotate these segments as structurally conserved (Ben-Shem et al. 2011). We then showed that a more accurate annotation of structurally conserved and structurally variable protein segments could be achieved by automated alignments of secondary structures (Melnikov et al. 2012). However, even these alignments were not fully accurate because many segments of ribosomal proteins lack a well-defined secondary structure (Melnikov et al. 2015).
Here, to address this issue, we perform a manually curated comparison of tertiary structures of ribosomal proteins. By doing so, we provide a comprehensive annotation of unique features of tertiary structure in the conserved ribosomal proteins from eukaryotes, bacteria, archaea, and eukaryotic mitochondria. We uncover a new class of structural variations that are systematically omitted by commonly used tools of multiple sequence alignments and automated alignments of secondary structures. We show that many protein segments that were previously regarded as structurally conserved, have dissimilar structures, and differing contacts within the ribosome structure in different species. We then illustrate that these local variations in the ribosome structure appear to reflect functional specialization of ribosomal proteins across the three domains of life, ultimately suggesting that these unique protein features might be used as markers of evolution and biodiversity and as species-specific therapeutic targets.
Results
Manually Curated Comparison of Ribosomal Proteins Across the Three Domains of Life
To compare homologous ribosomal proteins, we used ribosome structures from bacteria Escherichia coli, Thermus thermophilus, Deinococcus radiodurans, Bacillus subtilis, and Staphylococcus aureus; archaea Methanocaldococcusjannaschii, Haloarculamarismortiu, and Pyrococcus furiosus; and eukaryotes Homo sapiens, Drosophila melanogaster, Sacromyces cerevisiae, Plasmodium falciparum, Trypanosoma brucei, Trypanosoma cruzi, Tetrahymenathermophila, and Triticum aestivum (supplementary table S1, Supplementary Material online). In our approach, we aimed to describe variations in tertiary structure of ribosomal proteins. Therefore, rather than comparing protein sequences, we compared position of Cα atoms in the three-dimensional structures of homologous ribosomal proteins (Materials and Methods section). Because tertiary structures evolve much slower than protein sequence (Illergard et al. 2009), this approach could reveal those changes in protein structure which likely reflect an emergence of new biological activities rather than being functionally neutral variations.
During the analysis, we excluded protein segments that are poorly resolved in the electron density maps of crystallographic or electron microscopy data to avoid coordinate mistakes and refinement artifacts (see Materials and Methods section). We also used local structural alignments for proteins that reside in flexible parts of the ribosome, such as the central protuberance and the stalks in the large ribosomal subunit, ribosomal RNA (rRNA) helix h16, the head-to-body junction in the small ribosomal subunit, the intersubunit bridges, and a few protein segments at the ribosome periphery.
To illustrate major changes in homologous proteins from different domains of life, we used one reference ribosome structure per each domain of life. These ribosome structures were from bacterium E. coli, archaeon M. jannaschii, and eukaryote H. sapiens. In a few cases, we also used ribosome structures from other species to compensate incompleteness of the reference models or exemplify variations in protein structures within single domains of life. To facilitate comparison of ribosomal proteins from different species, we used a unified nomenclature in which conserved proteins are named according to their names in bacterial species (supplementary table S2, Supplementary Material online) (Ban et al. 2014).
By comparing homologous proteins from E. coli, M. jannaschii, and H. sapiens ribosomes, we first defined which protein segments have conserved three-dimensional fold of a polypeptide chain and which segments have dissimilar folds or occurrence in bacteria, archaea, and eukaryotes (supplementary fig. S1, supplementary Data S1, supplementary table S3, Supplementary Material online). Then, we used this structural comparison to correct errors and ambiguities of multiple sequence alignments of ribosomal proteins from the three domains of life (fig. 1; supplementary Data S2).
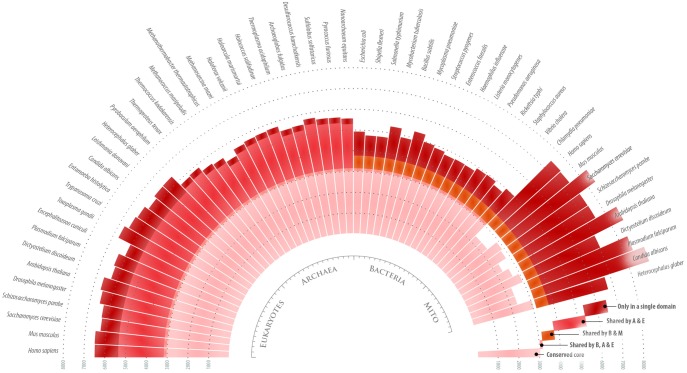
Fig. 1.
Homologous ribosomal proteins have highly diversified structure across the three domains of life. The diagram provides an overview of structural variability in 33 conserved proteins from 45 different species in the three domains of life. Each bar indicates the total number of amino acids in the 33 proteins in each species. The bars are colored to show the number of residues that form either the structurally invariable core (pink) or the variable protein segments (red). As the diagram shows, conserved ribosomal proteins carry nearly as many residues in structurally conserved protein segments as they carry in protein segments with distinct structure in different domains of life, suggesting a high degree of functional specialization of ribosomal proteins across the three domains of life.
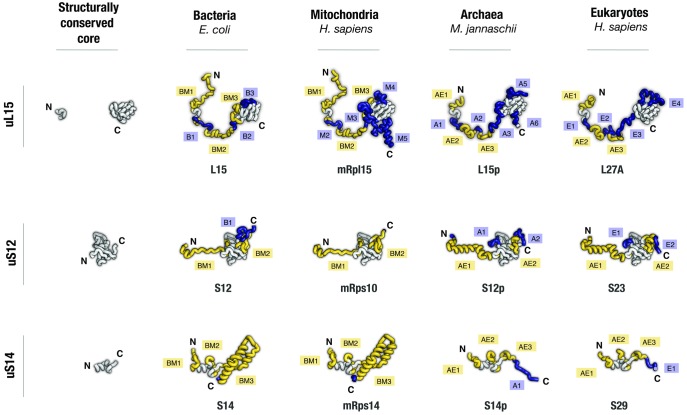
Our analysis shows that the conserved ribosomal proteins comprise a large number of variable segments that are systematically overlooked by multiple sequence alignments. Typically, these variations represent local remodeling of protein structure, such as transformations of α-helices into β-strands or transformations of protein loops into α-helices or into polyproline helices. One striking example of these structural variations is the structure of protein uL15. This protein has a long nonglobular N-terminal extension that has long been considered conserved due to its largely invariable length (∼80 residues), poor secondary structure, and similar position in the ribosome interior in different species (fig. 2). However, we found that only one short segment of this extension (residues 29–35 in E. coli) has invariable structure across species, whereas the other parts have dissimilar folds in bacterial, archaeal, and eukaryotic ribosomes (fig. 2). Similar variations are present nearly every ribosomal protein, most prominently in uL2, uL10, uL13, uS3, and uS4 (fig. 2, supplementary fig. S1, Supplementary Material online). In total, we have identified ∼60 of protein segments (∼600 residues in total), which have seemingly conserved occurrence in the three domains of life but in fact have dissimilar structures in bacteria, archaea, and eukaryotes.
Fig. 2.
Homologous ribosomal proteins have largely conserved globular domains but highly divergent nonglobular extensions. Aligned structures of homologous ribosomal proteins are shown as they appear in bacterial, mitochondrial, archaeal, and eukaryotic ribosomes (pdb ids 4y4b, 3j9m, 4v4n, and 4v6x, respectively). Proteins are colored according to structural conservation: segments that have identical tertiary structure in all four protein homologs are shown in gray; protein segments that have unique tertiary structure or occurrence only in one of the four homologs are shown in blue; protein segments that have unique tertiary structure or occurrence only in bacterial and mitochondrial proteins (labeled as BM) or only in archaeal and eukaryotic proteins are shown in yellow (labeled as AE). The segments are labeled with “B,” “M,” “A,” and “E” to indicate that a protein segment have unique structure or occurrence in bacterial, mitochondrial, archaeal, and eukaryotic proteins, respectively; the segments are numbered as they appear in each protein from its N- to the C-terminus. Apart from common protein names, each protein is named according to its name in bacterial, archaeal, and eukaryotic species. The panel illustrates that, despite homologous ribosomal proteins having comparable size across species, many segments in these proteins have dissimilar secondary and tertiary structure in different domains of life.
We also found that for the largest proteins, their sequence alignments establish a false amino acid correspondence by aligning a variable segment in one protein to a conserved segment in its homolog. For instance, protein uL3 has two β-strands comprising residues 45YRAIQVTT52 and 78GLWEFR83 in E. coli and 160IRVIAHTQ167 and 179HLMEIQ184 in H. sapiens. Although these β-strands are structurally conserved across the three domains of life, in the sequence alignment, the corresponding sequences are aligned to eukaryote- and bacteria-specific protein segments, respectively, instead of being aligned to each other (supplementary figs. S2 and S3, Supplementary Material online). Furthermore, the multiple sequence alignment of protein uL3 has 155 aligned pairs of residues between E. coli and H. sapiens homologs of uL3, but only 95 of these pairs represent the actual correspondence of the amino acids, whereas the remaining 60 pairs are false (supplementary fig. S2, Supplementary Material online). Similarly, protein uS4 (S9 in eukaryotes) has a conserved globular domain made of five α-helices and four β-strands of which only three α-helices and three β-strands are aligned to each other between bacterial and eukaryotic homologs (supplementary figs. S4 and S5, Supplementary Material online). In total, the multiple sequence alignment of uS4 contains ∼100 falsely paired residues of 146 pairs between E. coli and H. sapiens homologs of protein uS4 (supplementary fig. S4, Supplementary Material online).
A closer examination of structurally conserved cores of ribosomal proteins shows that the errors in multiple sequence alignments stem from two factors. First, the misaligned protein segments have relatively low sequence conservation. For instance, misaligned α-helices 1 and 2 of E. coli and H. sapiens homologs of ribosomal protein uS4 have only 39% sequence similarity, whereas properly aligned segments of uS4 have 55% of sequence similarity (supplementary fig. S6, Supplementary Material online). Second, the structurally conserved core has a discontinuous primary structure: its sequence is typically split into several short segments that are intermingled with bacteria- and eukaryote-specific protein segments (supplementary figs. S2 and S4, Supplementary Material online). Together, these two factors make sequence alignments prone to artifacts in which structurally conserved protein segments of one protein are aligned to bacteria- or eukaryote-specific segments of its homolog.
In summary, the examples above illustrate that ribosomal proteins carry a previously unknown class of structural variations that evade detection by commonly used methods of automated structural and sequence alignments. These local variations in protein structures create a great structural diversity of homologous ribosomal proteins from different domains of life and possibly reflect emergence of new biological functions.
Structural Diversity of Conserved Ribosomal Proteins Between the Three Domains of Life
Our analysis shows that in total the structurally invariable core of ribosomal proteins comprises ∼3,000 amino acid residues. In addition to the core, ∼2,200 residues form protein segments which are found only in bacteria and eukaryotic mitochondria; ∼2,700 of residues form archaea- or archaea/eukaryote-specific protein segments; and ∼1,100 residues form eukaryote-specific protein segments (fig. 1). These numbers illustrate that conserved ribosomal proteins have highly diversified structures and carry many appended segments in the three domains of life. Even relatively simple bacterial species carry nearly as many residues within unique structural protein features as they carry in the conserved core, suggesting high degree of functional specialization of ribosomal proteins in each domain of life (fig. 1).
These unique structural elements represent protein segments, whose lengths vary from a few to a few dozens of residues and which are typically exposed on a protein’s surface. These segments are so abundant that nearly every of the 33 conserved proteins carries at least one variable segment in each domain of life (supplementary fig. S1, Supplementary Material online). The largest ribosomal proteins, such as uL2, uL3 or uL4, harbor more than ten segments that have dissimilar structure or occurrence in bacteria, archaea and eukaryotes (supplementary fig. S1, Supplementary Material online). The only proteins that lack variable features are bacterial proteins uS13 and uS14, although archaeal and eukaryotic homologs of these proteins develop appended protein segments (supplementary fig. S1, Supplementary Material online).
Structural variations in ribosomal proteins have one similarity. Most frequently, they are found in nonglobular protein extensions, whereas invariable segments typically form protein globules. For instance, the invariable structure of protein uS14 is represented by a miniature globular domain—a 30-amino acid-long zinc finger–whereas the variable parts are represented by long N- and C-terminal extensions, which have different folds in bacteria and eukaryotes (supplementary fig. S1, Supplementary Material online). Similarly, variations in the protein extensions can be found in two-thirds of the 33 conserved ribosomal proteins (supplementary fig. S1, Supplementary Material online). It is worth mentioning, however, that some of the nonglobular extensions have invariable structures across three domains of life. These extensions are typically found in the largest ribosomal proteins, such as uL2, uL3, uL4, uS12, and uS13, where they stabilize conserved rRNA junctions or mediate critical contacts between ribosomes and their ligands. However, even these proteins develop additional nonglobular extensions in archaeal and eukaryotic species. Thus, ribosomal proteins typically evolve in such a way that their globular domains remain largely invariable, but the nonglobular extensions change their size and tertiary structure across three domains of life.
Some of the local structural variations in ribosomal proteins reflect their apparent functional specialization. For instance, the previously mentioned N-terminal extension of protein uL15 stabilizes the universally conserved helical junction H26-H27-H32-H36 within 23S/28S rRNA, in all the three domains of life. In eukaryotes, this extension has an additional biological activity: it serves as a nuclear localization signal that directs ribosomal protein delivery to the nucleolus, the site of eukaryotic ribosome biogenesis (Underwood and Fried, 1990). Our structural alignments show that the emergence of the nuclear localization signal is accompanied with local transformations of the secondary structure in which a segment of this extension transforms from being devoid of secondary structure in bacteria, to partly folded as α-helices in archaeal species, and to a fully α-helical protein segment in eukaryotes (supplementary fig. S1, Supplementary Material online).
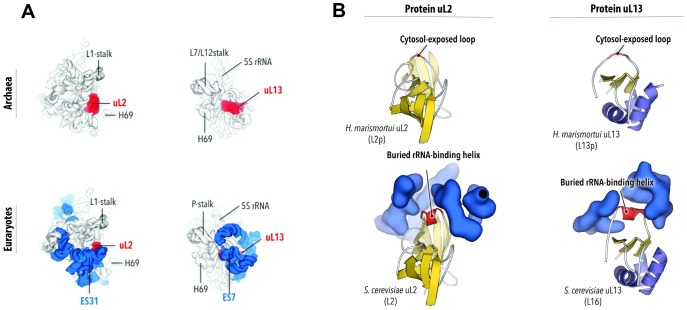
Another notable example of apparent functional innovations through local structural change is present in proteins uL2 and uL13. As the ribosomes grow in size upon transition from bacteria to eukaryotes, these ribosome-exposed proteins in bacterial ribosome are getting buried in the ribosome interior under a layer of additional, eukaryote-specific rRNA expansions (fig. 3A). Our analysis shows that both uL2 and uL13 transform their surface-exposed loops (in bacteria and archaea) into short α-helices or polyproline helices (in eukaryotes) to evolve additional RNA binding sites. These additional helices stabilize interactions between the ribosomal core and eukaryotic rRNA expansions (fig. 3B).
Fig. 3.
Variations in protein globules in an apparent adaptation to ribosomal RNA (rRNA) expansion. The figure compares structures of Haloarcula marismortui and Sacromyces cerevisiae ribosomes. It illustrates how transition from prokaryotes to eukaryotes was accompanied with the formation of a novel secondary structure in cytosol-exposed ribosomal proteins. (A) Views on the large ribosomal subunits illustrate that upon the transition from prokaryotes to eukaryotes, ribosomes have markedly increased in size, and ribosomal proteins uL2 and uL13 (in red) that were exposed on the surface of prokaryotic ribosome became buried in the interior of the eukaryotic ribosome. In eukaryotes, uL2 and uL13 are associated with eukaryote-specific rRNA expansion segments, ES31 and ES7 (in blue). (B) Close-up views on uL13 and uL2 show that the prokaryote-to-eukaryote transition was accompanied with secondary structure transformation in which surface-exposed protein loops of uL2 and uL13 were remodeled into rRNA-binding helices. Names in parenthesis show protein names in H. marismortui and S. cerevisiae.
These examples illustrate that at least some of the local structural changes represent development of new biological activities that adapt ribosomal proteins to new requirements of ribosome and cell architecture.
Structural Variations in Ribosomal Proteins Within a Single Domain of Life
While preparing the catalog of structural variations in homologous ribosomal proteins, we were aware that our analysis is not comprehensive, because some ribosomal proteins have very diverse structures within a single domain of life. Therefore, to show limitations of our catalog, we sought to provide examples illustrating the structural variations of ribosomal proteins within a single domain of life.
Overall, ribosomal proteins have highly conserved size and sequence in species from a single domain of life (Lecompte et al. 2002; Nakao et al. 2004; Wool et al. 1995). For instance, 72 of 81 ribosomal proteins from H. sapiens have nearly identical size and identical tertiary structures as their homologs from S. cerevisiae, and only 8 ribosomal proteins have long extensions in humans compared to yeasts, and one protein (uS5) has a large deletion in humans compared to yeasts (Anger et al. 2013). However, in each domain of life there are distant lineages of species in which ribosomes lose or acquire unique protein features in many ribosomal proteins.
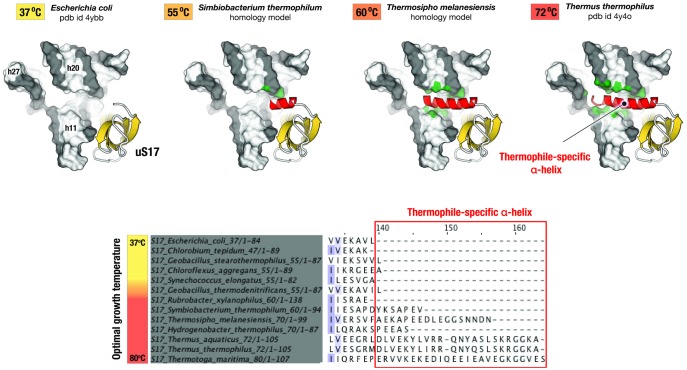
Our preliminary analysis shows that some ribosomal proteins appear to adapt their secondary and tertiary structures to the specific environments. For instance, in thermophilic bacteria, protein uS17 has an additional α-helix, which stabilizes RNA folding in a highly conserved three-way helical junction h12-h13-h8 in the 16S rRNA (fig. 4). Most remarkably, the size of this α-helix correlates with optimal growth temperature and increases from ∼13 residues in bacteria thriving at 55°C, to ∼16 residues in bacteria thriving at 60°C, and to 20 residues in bacteria thriving at 72°C (fig. 4).
Fig. 4.
Variations in protein structures within one domain of life as a possible adaptation to extreme environments. The figure illustrates structural variations in homologous ribosomal proteins upon transition from mesophilic to thermophilic species. The species used for comparison are arranged according to their optimal growth temperature. Fragments of Escherichia coli and Thermus thermophilus ribosome structures and homology models of Symbiobacterium thermophilum and Thermosipho melanesiensis ribosomes illustrate that, in thermophilic species, ribosomal protein uS17 develops an additional C-terminal helix (in red). This helix creates a new RNA–protein interface and stabilizes the RNA fold in the three-way helical junction in the 16S rRNA. Remarkably, as the optimal growth temperature for a given species gets higher, this helix gets progressively longer, and its apparent contacts with rRNA get more extensive (highlighted in green). This example shows that structure of some ribosomal proteins appear to evolutionary respond to higher temperatures by increasing the size of nonglobular extensions to establish new protein–rRNA or protein–protein contacts within the ribosome.
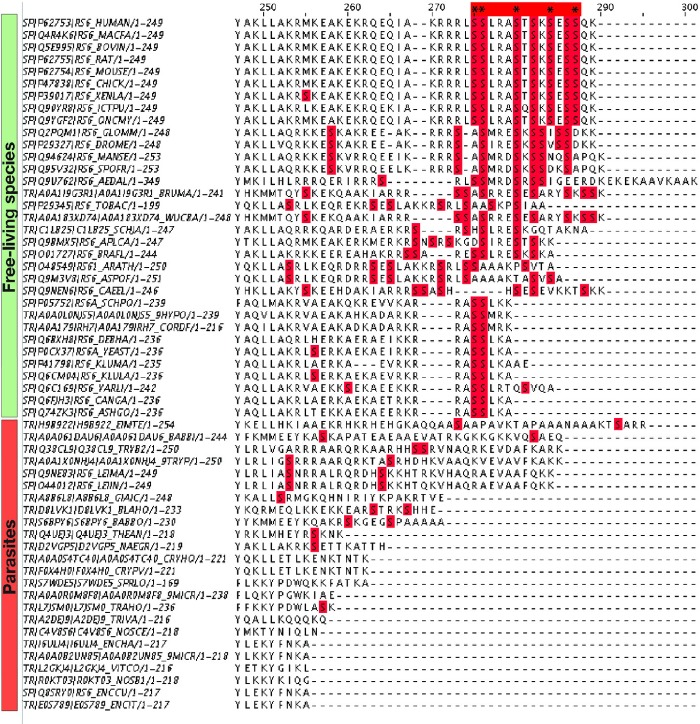
Some other ribosomal proteins have altered secondary and tertiary structure in an apparent adaptation to specific lifestyles. For instance, protein eS6 serves as a build-in nutrient sensor in the eukaryotic ribosome (Ruvinsky and Meyuhas, 2006). In its C-terminal segment, eS6 carries conserved serine residues (Ser235, Ser236, Ser240, Ser244, and Ser247 in Mus musculus) that are phosphorylated upon nutrient supplementation to adjust the overall rates of protein synthesis (Ruvinsky and Meyuhas, 2006). We found that in free-living species, the phosphorylation sites are preserved from yeasts to humans, whereas in parasites they are either mutated to non-Ser residues or the whole C-terminal is fully degenerated, suggesting that parasites are unable to sense nutrient availability via the eS6-dependent mechanism (fig. 5).
Fig. 5.
Variations in protein eS6 as a possible adaptation to specific lifestyles. The figure shows a multiple sequence alignment for eukaryotic ribosomal protein eS6 (C-terminal fragment). The C-terminal eS6 segment endows ribosomes with sensitivity to nutrients: it harbors serine residues (highlighted by asterisks) that are phosphorylated in response to hormones and nutrient availability to readjust the overall rate of protein synthesis in a eukaryotic cell. The figure shows that the phosphorylation sites remain conserved in free-living species but are degenerated in parasites, suggesting the lack of a nutrient sensor within parasitic ribosomes.
Although not comprehensive, the examples mentioned earlier illustrate that even within a single domain of life species keep diversifying their ribosomes by degrading or evolving new protein features. Hence, there is possibly a whole universe of unexplored ribosome features that adapt ribosomes to unique strategies of protein synthesis, ribosome biogenesis, and possibly other activities of the ribosomal proteins in a live cell.
Discussion
Here we have classified structural variations in conserved ribosomal proteins across the three domains of life. We showed that current automated tools of comparative analysis systematically underestimate diversity of ribosomal proteins. This underestimation is primarily caused by an inability to deal with poor sequence conservation and lack of secondary structure of the nonglobular extensions of ribosomal proteins. By resolving these limitations through manually curated structural alignments, we uncovered a new class of local structural variations that appear to adapt ribosomal proteins to the specific requirements of environment, ribosome composition, and cell separation into the nucleus and the cytoplasm.
Implications to Evolution of Ribosomal Proteins
One question that arises from our work is how the structurally conserved cores of ribosomal proteins are related to the last common ancestor cores? In other words, do the structurally conserved cores represent the most ancient parts of ribosomal proteins? Although this question cannot be answered with certainty, our analysis suggests that the last common ancestors could rather resemble ribosomal proteins from modern bacteria. We believe that when archaeal and eukaryotic species emerged, ribosomes got bigger and more complex. At this time, many segments of the ribosomal proteins were locally remodeled to evolve new biological activities, such as the ability to bind rRNA expansion segments or recruit eukaryote-specific factors of ribosome biogenesis, to name a few. This local remodeling of proteins in archaeal and eukaryotic species resulted in a “mosaic” pattern of sequence conservation—a pattern in which universally conserved protein segments were intermingled with variable protein segments. If this understanding is correct, then the conserved core of the ribosome does not represent the last common ancestor core but rather a part of it that has survived extensive local remodeling of protein structures upon transition from bacteria to archaea and eukaryotes or upon diversifications of environments.
Another question raised by our work is why some structurally conserved segments of ribosomal proteins have highly diverged sequences between different domains of life—to the extent that they cannot be properly aligned in multiple sequence alignments? A good illustration to this is protein uS4 in which α-helices 1 and 2 have universally conserved tertiary structure and position in the ribosome, yet their sequence is only moderately conserved between different domains of life (supplementary fig. S6, Supplementary Material online). Comparison of ribosome structures suggests that this sequence divergence is to some extent caused by different biological roles played by the protein segments in bacterial and eukaryotic ribosomes (supplementary fig. S7, Supplementary Material online). In bacteria, α-helices 1 and 2 form a hydrophobic core of uS4 by interacting with bacteria-specific N- and C-terminal extensions of uS4 (supplementary fig. S7, Supplementary Material online). In eukaryotes, the very same α-helices serve as a protein–protein interface between protein uS4 and eukaryote-specific protein eS30 (supplementary fig. S7, Supplementary Material online). The example of uS4 illustrates that the local repurposing of ribosomal protein structures does not necessarily require remodeling of the tertiary structure but might cause marked divergence of protein sequence between the domains of life.
Implications to Biology of Ribosomal Proteins
By highlighting unique structural features of ribosomal proteins in different domains of life, our work will help to build more accurate homology models for experimental and computational studies of ribosomes from different species. This work will also contribute to three major directions of ribosome biology.
First, ribosomal proteins are essential players of protein synthesis, and their diversity may reflect unknown species-specific strategies of protein synthesis and ribosome biogenesis. Showing that most of the variable segments represent nonglobular protein extensions, our work could help uncover an underexplored and complex biology of these hallmark features of ribosomal proteins. Nonglobular extensions are present in 23 of 33 conserved ribosomal proteins in bacteria and in 27 conserved proteins in eukaryotes (Ban et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001; Ben-Shem et al. 2011; Klinge et al. 2011; Rabl et al. 2011). They are thought to govern rRNA folding during ribosome biogenesis (Klein et al. 2004; Timsit et al. 2009). Some of the extensions, like those of uL3, uL22, uS7, uS12, form the inner walls of ribosomal active sites, and their mutations and natural sequence variants confer resistance to numerous drugs, such as linezolid, tiamulin, or valnemulin and anisomycin due to mutations in uL3 (Klitgaard et al. 2015); oxazolidinones, macrolides, and chloramphenicol due to deletions in uL4 (Wolter et al. 2005); erythromycin due to deletions in uL22 (Wilcox et al. 2001; Zaman et al. 2007); or emetine due to mutations in uS11 (Madjar et al. 1983). Some other extensions control the rate and accuracy of protein synthesis by directly contacting ribosomal ligands, such as tRNAs (uS13, uS19 and uL5), mRNA (uS2, uS3, bacterial uS4, uS7 and uS11), and nascent peptides (uL4 and uL22) (Kramer et al. 2009; Rozov et al. 2015). In eukaryotes, the extensions also accommodate nuclear localization signals and binding sites for eukaryote-specific chaperones to ensure safe delivery of highly positively charged ribosomal proteins to the nucleolus of eukaryotic cells (Preissler and Deuerling, 2012; Melnikov et al. 2015; Pillet et al. 2017). Finally, several protein extensions carry extraribosomal activities, such as extensions of uL5 and uL18 that inhibit oncoprotein Mdm2 to control programmed cell death (Deisenroth and Zhang, 2011), or the extension of uL13 that controls translation of inflammatory mRNAs during immune response (Mazumder et al. 2003). Given this broad range of biological activities, exploring structural variations in ribosomal proteins will help us to better understand species-specific strategies of protein synthesis and ribosome biogenesis, which may eventually enable species-specific targeting of protein synthesis by therapeutics.
Second, ribosomal proteins comprise a rare group of ubiquitous cell components that are used as universally abundant markers of biodiversity and evolution. Together with 17 aminoacyl-tRNA synthetases, 6 translation factors, 2 enzymes involved in modifying RNA and protein, and 5 core subunits of RNA and DNA polymerases, ribosomal proteins comprise a small group of as few as ∼60 proteins that are present in every organism with known genome sequence (Koonin, 2000). It is therefore not surprising that when species face new challenges—such as new environment, metabolism, or cell architecture—ribosomal proteins should inevitably adapt to these changes, since they cannot be functionally replaced or reinvented. Hence, it is possible, even probable, that variations in sequence and structure of ribosomal proteins represent complex records of adaptive changes in these essential proteins. Our finding that high temperatures or parasitic lifestyles appear to induce change in protein structures is empowering because it suggests that—through genome sequencing and homology modeling—we may eventually predict optimal growth conditions, lifestyles, cellular architecture, and possibly many other properties of poorly studied, uncultured, or extinct species.
Third, because of their ancient origin, ribosomal proteins serve as markers of the early evolutionary events that resulted in the origin of life on our planet (Lecompte et al. 2002; Klein et al. 2004; Smith et al. 2008). Ribosomes are thought to be the most ancient enzymes that have survived more than 3.5 billion years of evolution (Wittmann, 1982; Bokov and Steinberg, 2009; Fox, 2010; Harish and Caetano-Anolles, 2012; Petrov et al. 2014), and the extensions of ribosomal proteins are thought to represent perhaps the first primitive proteins to be produced by the ancient protein synthesis machinery (Hsiao et al. 2009). Because the extensions interact with the most conserved segments of rRNA, it is tempting to think that these protein features have very ancient origin. However, our data show that the extensions are rapidly evolving: Not only do many of them have totally dissimilar structures in different domains of life, but their structures may also vary within a single domain of life in an apparent adaptation to specific environments and lifestyles. Furthermore, many innovative activities of the protein extensions—such as those of the nuclear localization signals or rRNA expansion-binding sites—have likely coemerged with the first eukaryotic cells (∼1.5 billion years ago), which is at least ∼2 billion years later than the origin of pioneering bacterial species. Therefore, rather than being rudimentary molecular fossils buried in the ribosome interior, protein extensions represent highly innovative protein segments whose structures reflect ribosome adaptation to specific environments and lifestyles.
Conclusions
Our work summarizes an unexplored diversity of conserved ribosomal proteins and provides a resource to help elucidate its physiological roles in different species. Like Darwin who observed how animal bodies change their shapes to adapt to new environments (Darwin, 1859), we may now see how similar changes are occurring at a scale ∼100,000,000 times smaller, in the individual molecules that inhabit every living cell on our planet. Knowledge of structural variations in these ubiquitous molecules will help better understand species-specific adaptations of cellular molecules and will bring us closer to resolving the enigma of the origin and early evolution of life on Earth.
Note Added in Proof
Shortly after this paper appeared online, we were contacted by Temple Smith (Boston University), who pointed out that we did not compare our findings to the previously proposed model of ribosomal protein sequence block structure (Vishwanath et al. 2004). That study claimed that, in the large ribosomal subunit, the majority of bacterial and archaeal ribosomal proteins contained taxon-specific segments intermingled with conserved protein segments. Our findings fully agree with the block structure model as shown in (Figs. 2, supplementary table S1, Supplementary Material online). However, we demonstrate that the taxon-specific segments in ribosomal proteins are not strictly immutable within each taxon; some vary slightly in structure from species to species, especially in organisms with unusual lifestyles or living in extreme habitats (Figs. 4, 5).
Materials and Methods
Comparison of Ribosome Structures
The ribosome structures along with the corresponding electron density maps were retrieved from the protein databank (https://www.rcsb.org; last accessed April 6, 2018) (Berman et al. 2000) or the electron microscopy databank (www.emdatabank.org; last accessed April 6, 2018) (Lawson et al. 2016) and were visualized and manually inspected by using Coot (Emsley et al. 2010) and USCF Chimera (Pettersen et al. 2004) (supplementary table S1, Supplementary Material online). The disordered protein segments were excluded from the analysis based on elevated B-factor values (typically above 200) and poor electron densities to avoid potential artifacts of structural refinements. The structural alignments of homologous proteins were initially done using FatCat (Ye and Godzik, 2004). Then, the structures were morphed where necessary to correct structural differences caused by ribosome conformational changes, such as movements of the central protuberance in the large ribosomal subunit, intersubunit bridges, and head-to-body junction in the small ribosomal subunit. The aligned homologous proteins were then manually compared in the complete ribosome structure. To define structurally conserved and variable protein segments, variations in protein sequence were dismissed. Instead, protein segments were assigned as structurally conserved if their secondary structure is conserved between the three domains of life and variable if their secondary structure is different in different domains of life. For protein segments that lack secondary structure in all the three domains of life (e.g., nonglobular protein extensions), each protein residue was assigned as structurally conserved if its Cα-atoms would fall within 2.0 Å of the corresponding atom in the homologous protein structure, and there was no other Cα atom nearer. Thus, in our approach, we disregarded protein sequence and instead described conservation of the three-dimensional folds of the polypeptide chains of ribosomal proteins. The structural alignments with annotation of variable and conserved protein segments were attached as the (supplementary fig. S1, Supplementary Material online). They can be viewed by using the PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC.). We also provided a text summary of the variable protein segments in ribosomal proteins (supplementary table S3, Supplementary Material online).
Comparison of Protein Sequences
The sequences that were used in the study were retrieved from NCBI Protein databank by using BLAST and custom scripts to remove duplicates and sequences of ribosome-like proteins. The retrieved sequences were manually inspected for completeness, and partial sequences were replaced by complete isoforms from the same or closely related species. For species containing numerous copies of ribosomal proteins’ genes, such as S. cerevisiae and Arabidopsisthaliana, we used isoforms A and 1, respectively. For bacterial species in which several ribosomal proteins are encoded by two genes (corresponding to Zn-coordinating and Zn-free isoforms of ribosomal proteins), we used the genes coding for Zn-coordinating isoforms of ribosomal proteins. The retrieved sequences were then used to create multiple sequence alignments by Muscle (Edgar, 2004). The aligned sequences were attached to the manuscript as the (supplementary Data S2). Sequence similarities were calculated by using Sequence Manipulation Suite (http://www.bioinformatics.org/sms2/ident_sim.html; last accessed April 6, 2018) with default definitions of similar amino acids.
Homology Modeling of Ribosomal Proteins
The homology models of ribosomal protein uS17 from thermophilic bacteria were created by using Robetta (Kim et al. 2004) and then docked in the structure of 23S rRNA from T. thermophilus (pdb id 4y4o) (Polikanov et al. 2015) for illustration purposes. The figures were made by using the PyMOL Molecular Graphics System (Version 2.0 Schrödinger, LLC.), JalView (Waterhouse et al. 2009), and Adobe Illustrator.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to members of Dieter Söll’s (Yale University, New Haven, CT), Tom Steitz’s (Yale University, New Haven, CT), and Marat Yusupov’s (IGBMC, France) laboratories for valuable discussions, critical reading, and commenting on the manuscript and to Catherine Dunlavey (St. Andrews University, United Kingdom), Kyle Hoffman and Hui-Si Kwok (both at Yale University, New Haven, CT ) for serving as generalist reviewers and for their editorial support during preparation of the manuscript, and Victor de Jager for his advice on bioinformatics analysis of ribosomal proteins. This work was supported by the National Institutes of Health (R35 GM122560 to D.S.).
References
- Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli LL (2011). Structure and function of organellar ribosomes as revealed by cryo-EM In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes: structure, function, and dynamics. Springer Link. [Google Scholar]
- Ahmed T, Shi J, Bhushan S.. 2017. Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res. 4514:8581–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V.. 2015. The structure of the human mitochondrial ribosome. Science 3486230:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R.. 2013. Structures of the human and Drosophila 80S ribosome. Nature 4977447:80–85. [DOI] [PubMed] [Google Scholar]
- Armache JP, Anger AM, Marquez V, Franckenberg S, Frohlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R.. 2013. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Res. 412:1284–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N.. 2015. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol. 223:269–271. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA.. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 2895481:905–920. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, Lafontaine DLJ, Lindahl L, Liljas A, Lipton JM et al. , . 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 24:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M.. 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 3346062:1524–1529. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE.. 2000. The protein data bank. Nucleic Acids Res. 281:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N.. 2017. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 364:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV.. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 4577232:977–980. [DOI] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray Press. [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y.. 2011. The Ribosomal Protein-Mdm2-p53 Pathway and Energy Metabolism: bridging the Gap between Feast and Famine. Genes Cancer 24:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai N, Brown A, Amunts A, Ramakrishnan V.. 2017. The structure of the yeast mitochondrial ribosome. Science 3556324:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K.. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66(Pt 4):486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Z, Matzov D, Krupkin M, Wekselman I, Paukner S, Zimmerman E, Rozenberg H, Bashan A, Yonath A.. 2015. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci U S A. 11243:E5805–E5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V.. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 1574:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 29:a003483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogala M, Becker T, Beatrix B, Armache JP, Barrio-Garcia C, Berninghausen O, Beckmann R.. 2014. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 5067486:107–110. [DOI] [PubMed] [Google Scholar]
- Graf M, Arenz S, Huter P, Donhofer A, Novacek J, Wilson DN.. 2017. Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 455:2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N.. 2015. The complete structure of the 55S mammalian mitochondrial ribosome. Science 3486232:303–308. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Godinic-Mikulcic V, Crnkovic A, Ibba M, Weygand-Durasevic I, Ban N.. 2012. Cryo-EM structure of the archaeal 50S ribosomal subunit in complex with initiation factor 6 and implications for ribosome evolution. J Mol Biol. 418(3-4):145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar K, Kamoun S, Hiller NL, Bhattacharje S, van Ooij C. 2006. Common infection strategies of pathogenic eukaryotes. Nat Rev Microbiol. 4(12):922–931. [DOI] [PubMed] [Google Scholar]
- Harish A, Caetano-Anolles G.. 2012. Ribosomal history reveals origins of modern protein synthesis. PLoS One 73:e32776.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A.. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 1075:679–688. [DOI] [PubMed] [Google Scholar]
- Hartman H, Favaretto P, Smith TF.. 2006. The archaeal origins of the eukaryotic translational system. Archaea 21:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Fu J, Buss SN, Jossinet F, Jobe A, Zhang Q, Liao HY, Grassucci RA, Bajaj C et al. , . 2013. High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome. Nature 4947437:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel J, Burnside C, Mignot I, Leibundgut M, Boehringer D, Ban N.. 2017. The complete structure of the Mycobacterium smegmatis 70S ribosome. Cell Rep. 201:149–160. [DOI] [PubMed] [Google Scholar]
- Hsiao C, Mohan S, Kalahar BK, Williams LD.. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 2611:2415–2425. [DOI] [PubMed] [Google Scholar]
- Illergard K, Ardell DH, Elofsson A.. 2009. Structure is three to ten times more conserved than sequence–a study of structural response in protein cores. Proteins 773:499–508. [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P et al. , . 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 4146862:450–453. [DOI] [PubMed] [Google Scholar]
- Kaushal PS, Sharma MR, Booth TM, Haque EM, Tung CS, Sanbonmatsu KY, Spremulli LL, Agrawal RK.. 2014. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci U S A. 11120:7284–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusainov I, Vicens Q, Bochler A, Grosse F, Myasnikov A, Menetret JF, Chicher J, Marzi S, Romby P, Yusupova G et al. , . 2016. Structure of the 70S ribosome from human pathogen Staphylococcus aureus. Nucleic Acids Res. 4421:10491–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, Baker D.. 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32(Web Server issue):W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA.. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 3401:141–177. [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N.. 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 3346058:941–948. [DOI] [PubMed] [Google Scholar]
- Klitgaard RN, Ntokou E, Nørgaard K, Biltoft D, Hansen LH, Trædholm NM, Kongsted J, Vester B.. 2015. Mutations in the bacterial ribosomal protein l3 and their association with antibiotic resistance. Antimicrob Agents Chemother. 596:3518–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 1:99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B.. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 166:589–597. [DOI] [PubMed] [Google Scholar]
- Lawson CL, Patwardhan A, Baker ML, Hryc C, Garcia ES, Hudson BP, Lagerstedt I, Ludtke SJ, Pintilie G, Sala R et al. , . 2016. EMDataBank unified data resource for 3DEM. Nucleic Acids Res. 44(D1):D396–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O.. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 3024:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Guo Q, Zheng L, Ji Y, Xie YT, Lai DH, Lun ZR, Suo X, Gao N.. 2017. Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii. Cell Res. 2710:1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford JL Jr et al. , . 2016. Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit. Proc Natl Acad Sci U S A. 113:12174–12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjar JJ, Frahm M, McGill S, Roufa DJ.. 1983. Ribosomal protein S14 is altered by two-step emetine resistance mutations in Chinese hamster cells. Mol Cell Biol. 32:190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL.. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 1152:187–198. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M.. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 196:560–567. [DOI] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Yusupova G, Yusupov M.. 2015. Insights into the origin of the nuclear localization signals in conserved ribosomal proteins. Nat Commun. 6:7382.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, Yoshihama M, Kenmochi N.. 2004. RPG: the ribosomal protein gene database. Nucleic Acids Res. 32(Database issue):D168–D170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JH.. 2015. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 224:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Menetret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW.. 2014. Structure of the SecY channel during initiation of protein translocation. Nature 5067486:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM et al. , . 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci U S A. 11128:10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2513:1605–1612. [DOI] [PubMed] [Google Scholar]
- Pillet B, Mitterer V, Kressler D, Pertschy B.. 2017. Hold on to your friends: dedicated chaperones of ribosomal proteins: dedicated chaperones mediate the safe transfer of ribosomal proteins to their site of pre-ribosome incorporation. Bioessays 391:1–12. [DOI] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov SV, Soll D, Steitz TA.. 2015. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol. 224:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Deuerling E.. 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci. 377:274–283. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N.. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 3316018:730–736. [DOI] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G.. 2015. Structural insights into the translational infidelity mechanism. Nat Commun. 6:7251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O.. 2006. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 316:342–348. [DOI] [PubMed] [Google Scholar]
- Shalev-Benami M, Zhang Y, Matzov D, Halfon Y, Zackay A, Rozenberg H, Zimmerman E, Bashan A, Jaffe CL, Yonath A et al. , . 2016. 2.8-A Cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania. Cell Rep. 162:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Lee JC, Gutell RR, Hartman H.. 2008. The origin and evolution of the ribosome. Biol Direct. 3:16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohmen D, Chiba S, Shimokawa-Chiba N, Innis CA, Berninghausen O, Beckmann R, Ito K, Wilson DN.. 2015. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun. 6:6941.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Gloeckner FO.. 2006. RibAlign: a software tool and database for eubacterial phylogeny based on concatenated ribosomal protein subunits. BMC Bioinformatics 7:66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit Y, Acosta Z, Allemand F, Chiaruttini C, Springer M.. 2009. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int J Mol Sci. 103:817–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MR, Fried HM.. 1990. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 91:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanath P, Favaretto P, Hartman H, Mohr SC, Smith TF. 2004. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol Phylogenet Evol. 33:615–625. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH, Hegde RS.. 2014. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell 1577:1632–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Hegde RS.. 2015. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 259:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox SK, Cavey GS, Pearson JD.. 2001. Single ribosomal protein mutations in antibiotic-resistant bacteria analyzed by mass spectrometry. Antimicrob Agents Chemother. 4511:3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V.. 2000. Structure of the 30S ribosomal subunit. Nature 407:327–339. [DOI] [PubMed] [Google Scholar]
- Wittmann HG. 1982. Structure and evolution of ribosomes. Proc R Soc Lond B Biol Sci. 2161203:117–135. [DOI] [PubMed] [Google Scholar]
- Wolter N, Smith AM, Farrell DJ, Schaffner W, Moore M, Whitney CG, Jorgensen JH, Klugman KP.. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob Agents Chemother. 498:3554–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Sleebs BE, Triglia T, Brown A, Thompson JK, Jackson KE, Hanssen E, Marapana DS, Fernandez IS et al. , . 2017. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol. 2:17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool IG, Chan YL, Gluck A.. 1995. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 73(11-12):933–947. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A.. 2004. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32(Web Server issue):W582–W585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF.. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 2925518:883–896. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF.. 2001. The path of messenger RNA through the ribosome. Cell 1062:233–241. [DOI] [PubMed] [Google Scholar]
- Zaman S, Fitzpatrick M, Lindahl L, Zengel J.. 2007. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol Microbiol. 664:1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.