ABSTRACT
The development and activity of a cold-adapting microbial community was monitored during low-temperature anaerobic digestion (LtAD) treatment of wastewater. Two replicate hybrid anaerobic sludge bed-fixed-film reactors treated a synthetic sewage wastewater at 12°C, at organic loading rates of 0.25–1.0 kg chemical oxygen demand (COD) m−3 d−1, over 889 days. The inoculum was obtained from a full-scale anaerobic digestion reactor, which was operated at 37°C. Both LtAD reactors readily degraded the influent with COD removal efficiencies regularly exceeding 78% for both the total and soluble COD fractions. The biomass from both reactors was sampled temporally and tested for activity against hydrolytic and methanogenic substrates at 12°C and 37°C. Data indicated that significantly enhanced low-temperature hydrolytic and methanogenic activity developed in both systems. For example, the hydrolysis rate constant (k) at 12°C had increased 20–30-fold by comparison to the inoculum by day 500. Substrate affinity also increased for hydrolytic substrates at low temperature. Next generation sequencing demonstrated that a shift in a community structure occurred over the trial, involving a 1-log-fold change in 25 SEQS (OTU-free approach) from the inoculum. Microbial community structure changes and process performance were replicable in the LtAD reactors.
Keywords: anaerobic digestion; psychrophilic, hydrolysis; microbial community structure; adaptation
This paper explores low-temperature anaerobic digestion of a synthetic sewage-based wastewater with a focus on microbial community adaptation when using a mesophilic starting community.
INTRODUCTION
High-rate anaerobic digestion (AD) of domestic wastewaters is both successful and well established at full scale in tropical regions (Bowen et al.2014). Low-strength anaerobic treatment of wastewater at ambient temperatures in areas with a temperate climate, however, calls for efficient AD processes capable of operating below 20°C. Numerous successful laboratory-scale low-temperature [<20°C] anaerobic digestion (LtAD) trials have been undertaken over the past decade for a range of waste streams (e.g. Connaughton, Collins and O'Flaherty 2006; Enright et al.2009; McKeown et al.2012; Gouveia et al.2015). Yet, despite laboratory-scale success and the economical and environmental advantages of LtAD, full-scale implementation has not yet come to fruition. Moreover, many successful LtAD studies have focused on less complex wastewater and, as such, do not address the issue of solids hydrolysis (Petropoulos et al.2017). It has been reported that hydrolysis rates decrease as temperatures drop and suspended solids subsequently may accumulate in AD reactors, causing a reduction in treatment efficiencies and biomass washout (Elmitwalli et al.2001; Singh and Viraraghavan 2002; Lew et al.2003). Recent studies have, however, demonstrated efficient treatment of sewage by LtAD (Smith, Skerlos and Raskin 2013; Keating et al.2016) using reactors designed to retain biomass and particulates.
Efficient long-term treatment cannot rely solely on physical entrapment. The degradation of organic matter to methane during LtAD is dependent on the microbial community structure (Raskin et al.1994) and the activity (Lettinga et al.1999; Foresti, Zaiat and Vallero 2006; Cavicchioli 2015) of the reactor biomass, which are strongly influenced by temperature. The requirement for a psychrophilic, or psychrotolerant, inoculum for successful LtAD has been proposed as being advantageous. The use of a truly psychrophilic inoculum (from naturally cold environments) has been tested by Xing, Zhao and Zuo (2010) and Petropoulos et al. (2017) with promising results. A disadvantage of this approach is that this type of biomass is non-granular and may not have high levels of activity against some substrates. Granular seed inocula are particularly advantageous for biomass settling and retention in high-rate AD reactors (van Lier et al.2001; Sakar, Yetilmezsoy and Kocak 2009). Granular biomass also provides a more rapid start-up time (Elmitwalli et al.2001) and can prevent acidification (Neves, Oliveira and Alves 2004). Using mesophilically cultivated granular inocula for psychrophilic treatment without prior efforts to cold-adapt has been deployed in numerous studies, with varying degrees of success (Rebac et al.1995; Langenhoff and Stuckey 2000; Smith, Skerlos and Raskin 2013). In light of this information, long-term assessments into cold acclimation (how a community adapts to this change in its environment), activity (how active this community will be) and maturation (the sustainability of this adaptation and activity) of cold-adapting AD communities and how these impact treatment efficiencies warrants deeper investigation.
In practice, full-scale treatment facilities still work as a classic ‘black-box’ systems with the microbial community structure and diversity largely unknown. As of yet, there have not been sufficient advances to link what we know about the microbial communities to process optimisation and bio-monitoring of AD a larger scale. Identifying the structure of the microbial community during stable and unstable periods of operation is crucial to understanding treatment parameters, but this in itself is not straightforward. The diversity of the microbial community within an ecosystem is essential for stability, productivity and sustainability (Girvan et al.2005). This is true for AD reactors, regardless of operating temperature. Levén, Eriksson and Schnürer (2007) reported a higher diversity of species at lower temperatures during operation during mesophilic and thermophilic conditions. Authors have also described a further increase in microbial diversity from mesophilic to psychrophilic conditions (Bialek et al.2012). The reproducibility of bacterial community structure in reactor systems is debated owing to high functional redundancy and microbial population disparity between reactors and waste streams. It has been reported that changes in microbial community structure in suspended biomass systems can occur, even during stable operation (Fernández et al.1999), while other authors have reported no changes in a microbial structure, despite perturbation (Akarsubasi et al.2005). In contrast, other authors Collins, Mahony and O'Flaherty (2006) and Madden et al. (2010) found mirroring microbial communities in identical parallel granular reactor set-ups.
The objective of this study was two-fold: (i) to examine the development of microbial structure and activity of a cold-adapting community in replicated parallel LtAD reactors treating a complex, but defined, wastewater; and (ii) to investigate if mirrored microbial community development occurred in the separate LtAD reactors seeded with the same mesophilically cultivated biomass. We hypothesised that a mesophilic granular inoculum would demonstrate cold adaptation as well reproducible reactor performance and reproducible microbial community development in LtAD reactors with stable input and operational parameters.
MATERIALS AND METHODS
Reactor design, set-up and operation
This study employed two glass laboratory-scale hybrid sludge bed/fixed-film reactors (R1 and R2) [2.8 l working volume] as described by (Hughes et al.2011). Both reactors were seeded with 20 g volatile suspended solids (VSS) l−1 of anaerobic biomass. Anaerobic sludge granules were obtained from a mesophilic, full-scale, internal circulation reactor, located at the Carbery Milk Products plant in Co. Cork, Ireland. The VSS content of the granules was 119 g VSS l−1. The substrate used was synthetic sewage (SYNTHES; Aiyuk and Verstraete 2004) at 500 mg l−1 CODTot. The reactors were operated at 12°C in a trial lasting for 889 days. The trial was divided into five phases, each representative of a different hydraulic retention time (HRT) and organic loading rate (OLR; Table 1). The filter unit was replaced on Day 434.
Table 1.
Reactor operation phases and associated operational conditions.
| Phase | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Days | 0–104 | 105–259 | 260–559 | 560–665 | 666–889 |
| HRTa | 48 | 36 | 24 | 18 | 12 |
| TEMPb | 12 | 12 | 12 | 12 | 12 |
| OLRc | 0.25 | 0.33 | 0.5 | 0.63 | 1 |
| VLRd | 0.5 | 0.67 | 1 | 1.33 | 2 |
| SLRe | 0.03 | 0.03 | 0.05 | 0.06 | 0.1 |
| SLRf | 0.01 | 0.02 | 0.03 | 0.03 | 0.05 |
| UVg | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
Temperature (°C).
Hydraulic retention time (h).
Organic loading rate (kg COD m−3 d−1*.
Volumetric loading rate (m3 Wastewater m−3 Reactor d−1).
Sludge loading rate (kg COD kg[VSS]−1 d−1)*.
Sludge loading rate (m3Wastewater kg[VSS]−1 d−1).
Up-flow velocity (m h−1). *Values calculated based on influent concentration of 500 mg l−1 CODTot.
Reactor effluent analyses
Reactor effluent was sampled daily and also combined into a weekly composite sample for total, chemical oxygen demand (COD) (CODTot), soluble COD (CODSol), suspended COD (CODSus) and colloidal COD (CODCol) determinations according to Standard Methods (APHA 2005). Protein and polysaccharide concentrations in the effluent were determined by the Lowry method (Lowry et al.1951) and the DuBois method (DuBois et al.1956), respectively. The concentration of volatile fatty acids (VFAs) in the effluent was determined by chromatographic analysis in a Varian Saturn 2000 GC/MS system (Varian Inc., Walnut Creek, CA). Biogas analysis was performed by gas chromatography (Varian Inc., Walnut Creek, CA) according to Standard Methods (APHA 2005)
Biomass characterisation
Specific methanogenic activity testing
To evaluate changes in the hydrolytic and methanogenic capabilities of the seed (Day 0) and reactor biomass (sampled on days 105, 260, 666 and 889), samples were screened using the specific methanogenic activity (SMA) testing method using the pressure transducer technique, as described previously (Colleran et al.1992; Keating et al.2016).
Substrate (Protein) degradation assays to assess substrate depletion curve for the determination of K, Amax and Km
The maximum specific activity (Amax), the maximum initial velocity (Vmax), the apparent half-saturation constant (Km) and the first-order hydrolysis constant (k) of the seed inoculum and reactor biomass were evaluated on a protein source (solubilised skimmed milk powder). These rates were determined using substrate depletion assays, which were set up similarly to the SMA test described above and the kinetic parameters calculated as described by Bialek, Cysneiros and O'Flaherty (2013). Tests were performed, in triplicate; at 12°C and 37°C using biomass and protein concentrations of 2 g VSS l−1 and 2 g COD vial−1, respectively.
DNA/RNA co-extraction from biomass
Genomic DNA and RNA was extracted from granular biomass samples taken from R1 and R2 on Days 0 (I), Days 105 (P1), 236 (P2a), 296 (P2b), 392 (P3a), 531 (P3b), 546 (P3c), 666 (P4) and at the end of the trial (Day 889). Biomass was sampled from the fixed-film filter at two points:—mid-trial (Day 454) and at the end of the trial (Day 889). The nucleic acids were co-extracted by a modification of a phenol extraction method and processed as outlined by Keating et al. (2016).
Quantitative-polymerase chain reaction
Quantitative polymerase chain reaction (qPCR) was carried out for Archaeal and Bacterial domains using DNA and cDNA generated from granular biomass sampled from R1 and R2 and the fixed-film filter as described above. qPCR was performed using a LightCycler 480 (Roche, Manheim, Germany). The primers 1369F and 1492R and Taqman probe TM1389F (5′-CTTGTACACACCGCCCGTA-3′) were used for bacterial analysis (Suzuki, Taylor and DeLong 2000). The primers 787F and 1059R and Taqman probe 915F (5′-AGGAATTGGC-GGGGGAGCAC-3′) were used for archaeal analysis (Yu et al.2005). Standard curves were prepared using plasmids containing the full-length 16S rRNA gene sequence from a representative bacterial (Escherichia coli) and archaeal (Methanosarcina bakeri) strain. The plasmids were extracted using a Plasmid Extraction kit (BIOLINE). A PCR reaction was then carried out using the primer pairs described above. This product was cleaned using QIAQuick PCR Clean Up kit (Qiagen, Crawley, UK) according to manufacturer's instructions. To construct the RT-PCR cDNA standard curves were produced from cDNA prior in vitro transcription of the target mRNA by using the MEGAshortscript T7 kit (Ambion) as described by Smith et al. (2006). The concentration of standards was measured in duplicate using a Qubit system (Invitrogen) and converted into copy concentration. A 10-fold serial dilution series (109–101 copies ml−1) was generated for each standard solution and analysed, in duplicate, with its corresponding primer and probe set. qPCR cycling conditions can be found in Keating et al. (2016).
Illumina Miseq analysis
Terminal Restriction Fragment Length Polymorphism was used as a screening step to select samples for next generation sequencing (data not shown)-outlined in Keating et al. (2016). Subsequently, DNA and cDNA from reactor biomass sampled on Day 0 (Seed), Days 296 (P2b), 531 (P3b), Take-Down (E-Day 889) and from the filter upon take-down (FE) were sent for Miseq Illumina analysis at MR DNA (Shallowater, Texas, USA). Universal 16S rRNA primer pair targeting the V4 region were used, 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′GGACTACHVGGGTWTCT-AAT-3′)—for paired-end sequencing with the forward primer in each pair containing a barcode sequence. Amplicons were pooled and purified using calibrated Ampure XP beads (Bechman Coulter). This product was prepared using the Illumina TruSeq DNA library protocol. The DNA library was processed on a Solexa Miseq machine according to the manufacturer's instructions. Sequences were analysed using an OTU-free approach using the DADA2 algorithm (Callahan et al.2016). We used the standard workflow given at http://benjjneb.github.io/dada2/tutorial.html that learns the error model from the data first, dereplicates the reads and then runs the DADA2 algorithm separately on forward and reverse reads. Finally, merging the overlapping reads from both forward reduced sequence variants and reverse reads to give 1396 unique sequences (SEQs), which were then used to create sequence tables for the different samples. The representative SEQs were then taxonomically classified against the Silva 123 database with assign_taxonomy.py script from Qiime (Caporaso et al.2010). To find the phylogenetic distances between SEQs, we multisequence aligned the SEQs against each other using mafft v7.040 (Katoh and Standley 2013) and FastTree v2.1.7 (Price, Dehal and Arkin 2010). Finally, the make_otu_table.py from Qiime was employed to combine abundance table with taxonomy information. Raw sequences were submitted to the SRA database under bioproject submission number SUB3108010.
Statistical analysis
GraphPad Prism software (San Diego, California, USA) was used for calculating Student's t-test based on reactor effluent parameters and qPCR data. A significance level of 95% (P < 0.05) was selected. Further statistical analyses of the sequencing data were performed via the software R, version 3.4.1 (http://www.R-project.org/) using the SEQS tables and data generated as described previously and metadata. For community analysis, we used the package ‘Vegan’ (Oksanen et al.2013). The following alpha diversity measures were used: Fisher's alpha; Pielou's evenness; Richness; Shannon and Simpson. We used Vegan's aov() to calculate pair-wise ANOVA P-values and drew these on top of alpha diversity figures. To calculate Unifrac distances, we used the package ‘Phyloseq’ (McMurdie and Holmes 2013). Principal co-ordinate analysis (PCoA) plot of community data (SEQs) were made using different distance measures (Vegan's capscale() function): Bray Curtis; Unweighted Unifrac; and Weighted Unifrac. The samples were grouped for different treatments as well as the mean ordination value and spread of points (ellipses were drawn using Vegan's ordiellipse() function that represent the 95% confidence interval of the standard errors). To find SEQs that are significantly different between different conditions, we used DESeqDataSetFromMatrix() function from DESeq2 (Love, Huber and Anders 2014) package with adjusted P-value (after accounting for all comparisons) cut-off of 0.01 and minimum log fold change of 1. After performing multiple testing corrections, it reports SEQs that have log-fold changes between multiple conditions. The statistical workflows for the above can be found at http://userweb.eng.gla.ac.uk/umer.ijaz#bioinformatics.
RESULTS
Reactor performance
Both reactors treated the synthetic sewage wastewater successfully, with COD removal efficiencies in excess of 80% generally recorded, corresponding to low effluent COD concentrations of typically less than 120 mg CODTot l−1 at applied OLRs up to 0.63 kg CODTot m−3 day−1 (Table 2). The performance was sustained during the long-term trial, with minor fluctuations, until the loading rate was increased to 1.0 kg CODTot m−3 day−1 from Day 666 (Table 1), at which point the efficiency of the process decreased somewhat in R1, although CODTot removal rates of c. 60% were maintained (Table 2).
Table 2.
Average effluent CODTot, CODSus, CODCol and CODSol values during the five phases of reactor operation for R1 and R2. a; concentration in mg l−1, b; removal efficiency percentage, c; standard deviation, d; VFA:COD ratio based on average VFA concentrations and CODSol for each phase.
| Sample | Total COD | Suspended COD | Colloidal COD | Soluble COD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Conc)a | (RE)b | (SD)c | (Conc) | (RE) | (SD) | (Conc) | (RE) | (SD) | (Conc) | (RE) | (SD) | |
| R1 Phase 1 | 73 | 86 | ±11 | 22 | 71 | ±8 | 11 | 60 | ±0.1 | 41 | 84 | ±2 |
| R2 Phase 1 | 140 | 73 | ±4 | 62 | 17 | ±0.2 | 23 | 17 | ±5 | 56 | 78 | ±2 |
| R1 Phase 2 | 61 | 88 | ±4 | 18 | 75 | ±6 | 4 | 84 | ±2 | 40 | 83 | ±1 |
| R2 Phase 2 | 76 | 85 | ±6 | 31 | 58 | ±0.7 | 13 | 27 | ±0.7 | 32 | 86 | ±4 |
| R1 Phase 3 | 110 | 79 | ±3 | 41 | 46 | ±6 | 25 | 9 | ±2 | 45 | 82 | ±1 |
| R2 Phase 3 | 103 | 80 | ±19 | 39 | 47 | ±15 | 24 | 14 | ±4 | 40 | 84 | ±2 |
| R1 Phase 4 | 124 | 75 | ±25 | 46 | 36 | ±13 | 39 | 0 | ±6 | 44 | 84 | ±3 |
| R2 Phase 4 | 105 | 79 | ±8 | 27 | 63 | ±14 | 33 | 0 | ±5 | 46 | 83 | ±0.2 |
| R1 Phase 5 | 215 | 59 | ±12 | 125 | 0 | ±0.5 | 25 | 9 | ±1 | 67 | 73 | ±2 |
| R2 Phase 5 | 114 | 78 | ±5 | 61 | 19 | ±0.9 | 15 | 45 | ±1 | 37 | 85 | ±3 |
| VFA:COD (Ratio)d | Phase 1 | Phase 2 | Phase 3 | Phase 4 | Phase 5 | |||||||
| R1 | 0.07 | 0.1 | 0.14 | – | 1.47 | |||||||
| R2 | 0.04 | 0.14 | 0.48 | – | 0.56 | |||||||
Replicability of reactor performance
During Phase 1, a significant difference (P < 0.05) in performance was observed between the two reactors. Reactor 2 (R2) average COD concentrations were much higher than reactor 1 (R1) for all COD fractions (Table 2). However, this can be attributed to a start-up period of ∼56 days for R2, while no start-up period was observed for R1. Both systems performed well upon commencement of the second phase. Transient increases in the effluent concentrations of the CODTot, CODSus and CODCol fractions from both reactors were observed upon further reduction of the applied HRT during Phase 3 (Table 2). The CODSol removal, however, was not noticeably affected by this change (Table 2).
The particulate proportion of the influent (CODSus) was degraded/retained in both reactors until concentrations in effluent from R1 increased from Day 329 (Phase 3) and subsequently effluent CODCol concentrations also increased. This suggested that suspended solids might have been degraded to colloidal particles. Similarly, effluent CODSus and CODCol in R2 increased during this period. These fractions of COD remained elevated in effluent from both reactors, until the filter matrix was changed on Day 434 (Phase 4). The fourth period of reactor operation was characterised by efficient and stable process performance by both systems, with removal efficiencies of CODTot and CODSol routinely >75% (Table 2). However, colloidal particles were not degraded/retained by either R1 or R2 (0% removal). The removal of the CODTot, CODSol and CODSus fractions was not significantly different (P > 0.05) between the replicate reactors during phases 2–4.
The final operational phase (Phase 5) was defined by an HRT of 12 h and an applied OLR of 1 kg COD m−3 d−1. The response to this HRT change perturbation was distinct in both reactors. A period of biomass washout, lasting two weeks, upon commencement of the phase was observed in R1, with effluent CODTot concentrations reaching 1 g l−1, composed primarily of suspended solids, before slowly decreasing over a period of 20–25 days. In contrast, R2 displayed no obvious response to the HRT change. The effluent VFA to COD ratio was highest during this phase (Table 2). A period of ∼100 days of stable operation was then recorded in both reactors before R1 effluent values began to fluctuate again, with CODSus concentrations reaching 440 mg l−1 on Day 868. R2 also displayed a period of less efficient performance from Day 819, where CODSus and CODSol increased (reaching below 130, and 150 mg l−1, respectively). An increase in effluent VFA concentrations was recorded from Day 805, to reach a range of 10–20 mg l−1 (data not shown). It was demonstrated that effluent CODTot and CODSol were significantly different (P < 0.05) between both systems during this final phase of the trial.
Protein was completely hydrolysed/degraded in both reactors throughout the trial with removal efficiencies of c. 100% (Table 3). Carbohydrate (the polysaccharide portion) was also completely degraded/retained in both reactors (Table 3) until the filter matrix was changed on Day 434. Following this, effluent carbohydrate concentrations from R1 reached 34 mg l−1 on Day 490 (data not shown). The removal of carbohydrates was not significantly different between systems during phases 1–4, but during the final phase of operation P was < 0.05.
Table 3.
Average effluent Carbohydrate and Protein values throughout the five phases of reactor operation for R1 and R2. a; concentration in mg l−1, b; removal efficiency percentage, c; standard deviation.
| Sample | Carbohydrate | Protein | ||||
|---|---|---|---|---|---|---|
| (Conc)a | (RE)b | (SD)c | (Conc) | (RE) | (SD) | |
| R1 Phase 1 | 0 | 100 | ±0 | 0.05 | 100 | ±0.04 |
| R2 Phase 1 | 0 | 100 | ±0 | 0.01 | 100 | ±0.05 |
| R1 Phase 2 | 0.3 | 100 | ±1 | 0.08 | 99 | ±0.05 |
| R2 Phase 2 | 0.1 | 100 | ±0.05 | 0.03 | 100 | ±0.1 |
| R1 Phase 3 | 7.08 | 91 | ±1 | 0.09 | 99 | ±0.04 |
| R2 Phase 3 | 4.1 | 95 | ±1 | 0.07 | 99 | ±1 |
| R1 Phase 4 | 12 | 84 | ±1 | 0 | 100 | ±0 |
| R2 Phase 4 | 24.3 | 69 | ±3 | 0.02 | 100 | ±0.02 |
| R1 Phase 5 | 9.4 | 88 | ±8 | 0.01 | 100 | ±0.02 |
| R2 Phase 5 | 2.1 | 98 | ±1 | 0 | 100 | ±0.06 |
Microbial activity and cold adaptation
The granular biomass was sampled temporally from each reactor throughout the trial and tested for its activity against hydrolytic and methanogenic substrates under mesophilic and psychrophilic conditions to; assess: (i) the activity of the microbial population; (ii) if the microbial populations were adapting to psychrophilic conditions; and (iii) if the activity and adaptation developed at the same rate in both reactor systems.
Hydrolysis
The hydrolysis rate constant (k) results demonstrated that, throughout the trial, biomass activity increased when tests were carried out under both mesophilic and psychrophilic conditions. In fact, the hydrolysis rate at 12°C during phase 4 was increased by ∼20 times in biomass from both reactors, compared to the seed inoculum. In biomass from both reactors, Amax (Table 4) increased at both temperatures tested, with psychrophilic activity at the end of the trial being comparable to (R1), or greater than (R2) the mesophilic activity. Km for mesophilic hydrolysis increased throughout the trial for both reactors, indicating a decrease in substrate affinity at the higher temperature. Km at the lower temperature decreased over time, indicating an increase in substrate affinity for both biomass sources under these conditions.
Table 4.
Hydrolysis kinetic assays of reactor biomass at 37°C and 12°C, based on a skimmed milk protein source. a; Maximum substrate utilising rate gCOD gProtein−1 d−1. b; Apparent half-saturation rate constant gProtein l−1. c; Maximum initial velocity gProtein l−1 d−1 for R1 and R2, d: Hydrolysis rate constant d−1. Values are the mean of triplicates ± standard deviation in brackets.
| Sample | Amaxa | Kmb | Vmax c | kd |
|---|---|---|---|---|
| Seed 37°C | 15 (±3) | 1.1 (±0.41) | 0.9 (±1.07) | 0.9 (±0.58) |
| Seed 12°C | 18 (±0.13) | 2.5 (±0.03) | 0.3 (±0.05) | 0.3 (±0.06) |
| R1 Phase 1 37°C | 12 (±1) | 1.8 (±0.36) | 2.2 (±1.13) | 1 (±0.42) |
| R2 Phase 1 37°C | 104 (±10) | 2.7 (±0.07) | 0.8 (±0.23) | 0.8 (±0.31) |
| R1 Phase 1 12°C | 40 (±7) | 1 (±0.34) | 1.1 (±0.4) | 1.3 (±0.06) |
| R2 Phase 1 12°C | 19 (±2.52) | 4.3 (±3) | 2.7 (±2.19) | 0.9 (±0.21) |
| R1 Phase 2 37°C | 145 (±26.5) | 3.9 (±2.12) | 8.8 (±8.39) | 5.7 (±2.18) |
| R2 Phase 2 37°C | 164 (±16) | 3.1 (±2.23) | 7.5 (±6.72) | 2.1 (±0.91) |
| R1 Phase 2 12°C | 127.5 (±45) | 1.6 (±0.11) | 0.1 (±0.04) | 0.8 (±0.16) |
| R2 Phase 2 12°C | 35 (±27) | 1.4 (±0.13) | 0.1 (±0.06) | 1.2 (±0.58) |
| R1 Phase 3 37°C | 94 (±26) | 1.6 (±0.43) | 3.3 (±3.94) | 1.6 (±0.37) |
| R2 Phase 3 37°C | 62 (±9.5) | 2.8 (±0.14) | 3.1 (±0.70) | 2.2 (±0.41) |
| R1 Phase 3 12°C | 52 (±32) | 2.6 (±0.19) | 0.3 (±0.09) | 1.1 (±0.32) |
| R2 Phase 3 12°C | 91 (±29) | 1.9 (±0.22) | 0.3 (±0.16) | 1.5 (±0.33) |
| R1 Phase 4 37°C | 257 (±32) | 1.8 (±0.27) | 1 (±1.07) | 0.9 (±0.58) |
| R2 Phase 4 37°C | 15 (±1) | 2.1 (±2.02) | 2 (±2) | 1.9 (±1.18) |
| R1 Phase 4 12°C | 40 (±18) | 0.5 (±0.04) | 0.1 (±0.01) | 3.2 (±0.89) |
| R2 Phase 4 12°C | 51 (±5) | 0.3 (±0.08) | 1.7 (±1.57) | 4.7 (±2.92) |
| R1 End 37°C | 89 (±33) | 2.8 (±1.58) | 2.3 (±3.73) | 1.4 (±0.787) |
| R2 End 37°C | 72 (±8) | 1.2 (±0.16) | 1 (±0.27) | 2.9 (±0.216) |
| R1 End 12°C | 68 (±7) | 1.8 (±0.37) | 0.4 (±0.23) | 1.8 (±0.714) |
| R2 End 12°C | 83 (±22) | 2.2 (±0.19) | 0.6 (±0.15) | 1.6 (±0.383) |
Acetogenic and direct methanogenic substrates
The initial inoculum had a high SMA at 37°C, with acetoclastic activity being six times higher than hydrogenotrophic activity (Table 5). While SMA against acetate was only slight (7±1 ml Methane (CH4) g [VSS]−1 d−1) at 12°C, hydrogenotrophic activity (39±19 ml Methane (CH4) g [VSS]−1 d−1) was comparable to that measured at 37°C (Table 5). By the end of the first phase of the trial, reactor biomass SMA at 37°C had increased, with the hydrogenotrophic activity having increased to a level 10 times and 3 times greater than the seed biomass in R1 and R2, respectively. At the lower temperature, there was little change in SMA values compared to the inoculum.
Table 5.
Maximum specific methanogenic activity (SMA) of reactor biomass at 37°C and 12°C presented as ml Methane (CH4) g [VSS]−1 d−1 for R1 and R2. Values are the mean of triplicates ± standard deviation in brackets.
| Sample | Propionate | Butyrate | Ethanol | Acetate | H2/CO2 |
|---|---|---|---|---|---|
| Seed 37°C | 84 (±9) | 523 (±30) | 561 (±140) | 300 (±33) | 50 (±5) |
| Seed 12°C | 5 (±2) | 3 (±2) | 7 (±4) | 7 (±1) | 39 (±19) |
| R1 Phase 1 37°C | 100 (±26) | 155 (±19) | 470 (±39) | 345 (±20) | 570 (±115) |
| R2 Phase 1 37°C | 30 (±3) | 55 (±18) | 166 (±45) | 179 (±32) | 134 (±6) |
| R1 Phase 1 12°C | 8 (±3) | 2 (±15) | 33 (±13) | 14 (±11) | 15 (±2) |
| R2 Phase 1 12°C | 5 (±3) | 6 (±1) | 51 (±18) | 14 (±7) | 31 (±3) |
| R1 Phase 2 37°C | 136 (±20) | 147 (±35) | 230 (±97) | 79 (±47) | 65 (±41) |
| R2 Phase 2 37°C | 87 (±4) | 187 (±10) | 307 (±46) | 11 (±6) | 193 (±41) |
| R1 Phase 2 12°C | 17 (±8) | 27 (±21) | 39 (±2) | 0 | 11 (±3) |
| R2 Phase 2 12°C | 22 (±2) | 2 (±2) | 12 (±3) | 0 | 24 (±1) |
| R1 Phase 4 37°C | 23 (±2) | 65 (±9) | 257 (±92) | 201 (±18) | 170 (±15) |
| R2 Phase 4 37°C | 40 (±22) | 94 (±7) | 21 (±9) | 353 (±50) | 308 (±21) |
| R1 Phase 4 12°C | 1 (±1) | 3 (±1) | 31 (±6) | 51 (±22) | 19 (±9) |
| R2 Phase 4 12°C | 3 (±1) | 12 (±8) | 46 (±12) | 42 (±6) | 98 (±23) |
| R1 End 37°C | 23 (±13) | 91 (±13) | 153 (±33) | 329 (±66) | 231 (±29) |
| R2 End 37°C | 34 (±2) | 73 (±14) | 171 (±16) | 181 (±29) | 110 (±6) |
| R1 End 12°C | 2 (±1) | 9 (±1) | 21 (±6) | 26 (±7) | 38 (±3) |
| R2 End 12°C | 4 (±2) | 8 (±3) | 29 (±4) | 28 (±3) | 35 (±4) |
R2 biomass sampled during the second phase had an increased SMA against all substrates tested at 37°C, with the exception of acetate. In contrast, R1 biomass displayed decreased activity for all substrates tested, except for propionate. Biomass from both systems tested at the psychrophilic condition demonstrated similar activity ranges with increased activity noted against propionate (Table 5). Interestingly, no acetoclastic activity was detected in either reactor biomass at this point.
At the end of the fourth phase, the SMA of biomass from both systems was within a similar range. Under psychrophilic conditions, the SMA profile had increased for both R1 and R2, with comparable levels of activity in both biomass sources (Table 5). At the end of trial, the SMA at 37°C was comparable for R1 and R2 biomass samples, with the exception of the direct methanogenic substrates, for which SMA was significantly lower in R2 biomass. The SMA at 12°C against all substrates was in a similar range for both R1 and R2 and compared to the initial inoculum activity on ethanol and acetate had increased by ∼3 times (Table 5).
Microbial community structure
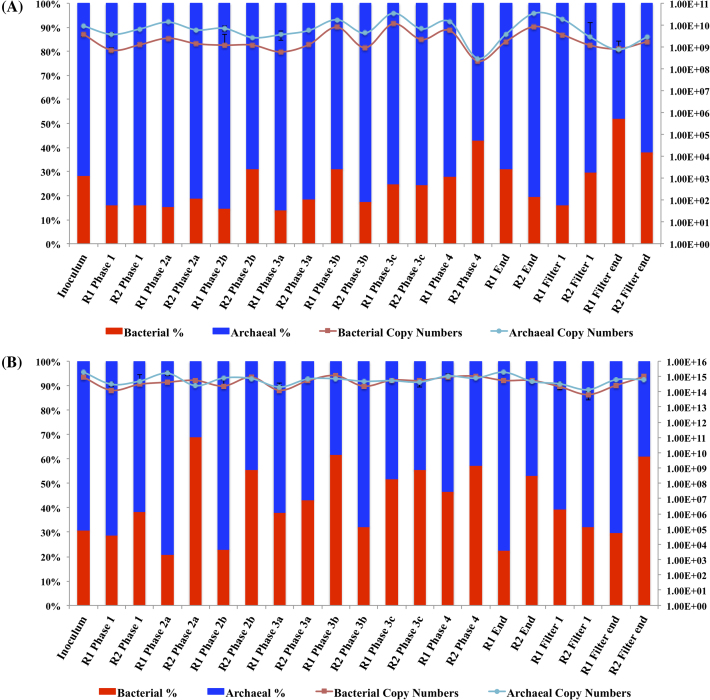
Bacterial and archaeal numbers were quantified throughout the reactor trial. The bacterial and archaeal profiles were generally reproducible in both systems with copy numbers in the range of 3 × 108–3.5 × 1010 (copies g-1) and 2.4 × 108–1.2 × 1010 (copies g-1), respectively (Fig. 1A). The ratio of bacteria to archaea in R1 and R2 biomass was broadly similar also, but deviations were noted, for example, in R2 on Day 296 (Phase 2b) and Day 666 (Phase 4-start of Phase 5), whereby the bacterial population increased to 31% and 43% of the total population, respectively (1.2 × 109 and 2.3 × 108 copies g-1) and was greater than in R1 biomass during these times. However, no significant difference (P > 0.05) was found throughout the trial. A 1-log reduction of the total bacterial and archaeal gene copy numbers was also observed in R2 biomass on Day 666 (Phase 4-start of Phase 5; Fig. 1A). The filter communities were distinct from the other sampling points with a greater proportion of bacterial to archaeal cells. This was due to a reduction in the numbers of archaeal cells relative to the granular biomass (Fig. 1A).
Figure 1.
(A) qPCR data of Bacterial and Archaeal 16S copy numbers (per g biomass) on the right y-axis from biomass samples (x-axis) throughout the trial corresponding to the ratio of bacteria to archaea (expressed as a percentage) on the left y-axis. (B) qPCR data of Bacterial and Archaeal 16S rRNA transcripts copy numbers (per g biomass) on the right y-axis from biomass samples (x-axis) throughout the trial corresponding to the ratio of bacteria to archaea (expressed as a percentage) on the left y-axis.
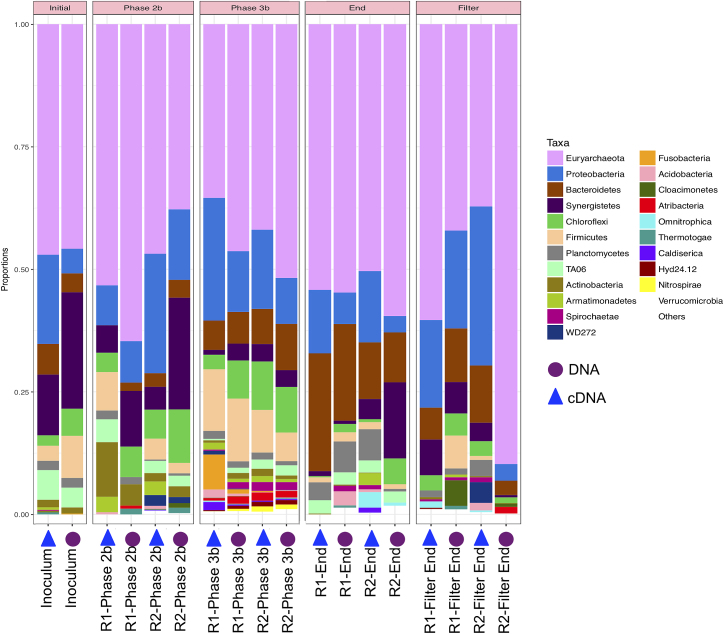
The 16S rRNA transcripts (Fig. 1B) varied from 6.5 × 1013 to 1.15 × 1015 copies g−1 (21%–69%) for bacterial cells and 1.4 × 1014 to 2 × 1015 copies g−1 (31%–79%) for archaeal cells, these numbers were greater than those observed through DNA-based analysis. Deviations were again noted in the proportion of bacteria to archaea between the systems in biomass sampled from Day 236 (Phase 2a), Day 296 (Phase 2b), Day 531 (Phase 3b), Day 666 (Phase 4-start of Phase 5), End and Filter End (Day 889). The greatest deviation was noted in the biomass sampled on Day 531 (Phase 3b) in which R1 had the highest bacterial transcripts recorded (1.2 × 1015 copies g−1), comprising 62% of the total sample pool (Fig. 1B). This contrasted with the same time point in R2, when bacterial copy numbers were 2.3 × 1014 (copies g−1), comprising just 32% of the total sample pool (Fig. 1A). Despite these deviations, the bacterial and archaeal transcript numbers were reproducible between the systems and no significant difference was found between the systems (P > 0.05). Nextgeneration sequencing was carried out to identify the bacterial and archaeal populations. The major bacterial populations identified included representatives of the Proteobacteria (8%–52%), mainly Deltaproteobacteria based on a DNA-based analysis and Gammaproteobacteria, based on the cDNA-derived sequences (Fig. 2). The Synergistetes (1.5%–44%) and the Bacteroidetes, mainly Flavobacteria, Sphingobacteria and Bacteroidea (0%–52%) were also present in the reactors throughout the trial. Chloroflexi (0%–19%), Firmicutes comprised mainly Clostridia and Bacilli (0%–24%). Less abundant, or occasionally present, bacterial groups included the Fusobacteria (0%–11%), the Actinobacteria (Actinobacteria and Coriobacteria; 0%–24%), the Planctomycetes (Phycisphaerae and Planctomycetales; 0%–14%), Acidobacteria (Halophagae; 0%–6.5%), and <5% abundance; Caldiserica, Chlorobi, Gemmatimonadetes, Hyd24–12, Omnitrophica, Spirochaetae, Thermotogae, TM6, WD272, Verrucomicrobia and rare phyla; Candidate division SR1, Cyanobacteria, Deferribacteres, Dictyoglomi, Elusimicrobia, Gracilibacteria, Fibrobacteres, Hydrogenedentes, Lentisphaerae, Nitrospirae, Parcubacteria, SHA-109.
Figure 2.
Taxa-plot of the percentage abundance of bacterial and archaeal phyla identified per sample. Samples are grouped according to phase ‘Initial’, ‘Phase 2’, ‘Phase 3’, ‘End’ and ‘Filter’.
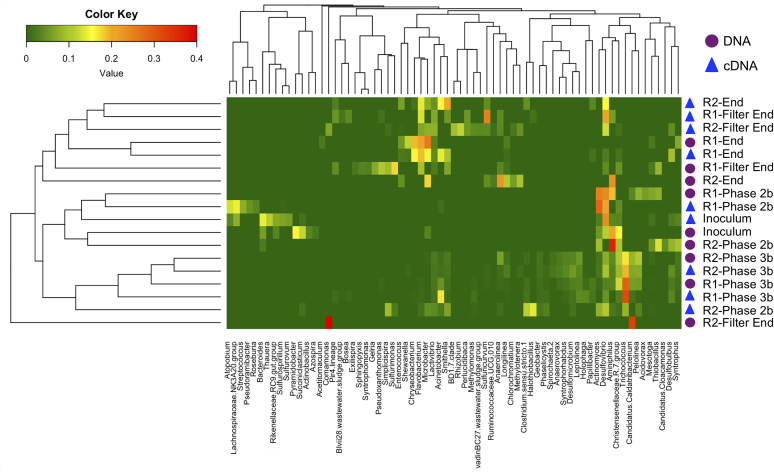
Heatmap analysis was employed to visualise temporal variations in the bacterial populations in both reactors and similarity matrices were used in tandem (Fig. 3). In the heatmap of the bacterial genera, the sequences clustered together based on time, and DNA or cDNA origin. An exception to this was the R2 DNA biomass sample from the pumice filter unit, which formed a separate branch. This was due to the apparent increased abundance presence of Commamonas and Candidatus Caldatribacterium and this sample also demonstrated decreased species richness.
Figure 3.
Heatmap analysis for the bacterial throughout the trial showing the dominant genera (>2%) and Bray–Curtis similarity between samples and between the dominant genera.
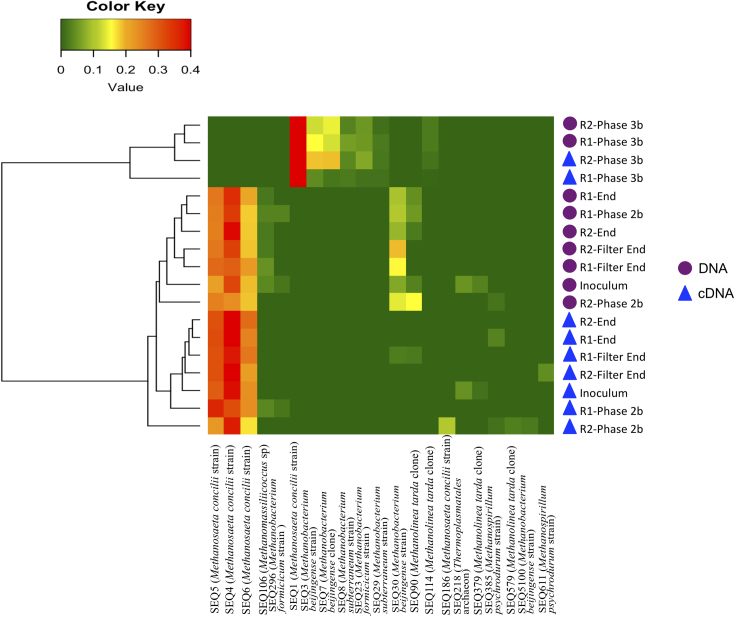
The archaeal portion of the community was dominated by sequences identified as Methanosaeta concilii strain X16932 throughout the trial (Fig. 4). Methanobacterium, Methanolinea and Methanospirillum sequences were also present. Biomass from ‘Phase 3b’ branched separately due to apparent decreases in hydrogenotrophic methanogens.
Figure 4.
Heatmap analysis for the archaeal fraction throughout the trial showing the dominant sequences (>2%) and Bray–Curtis similarity between samples.
Microbial community development over time
Community comparisons
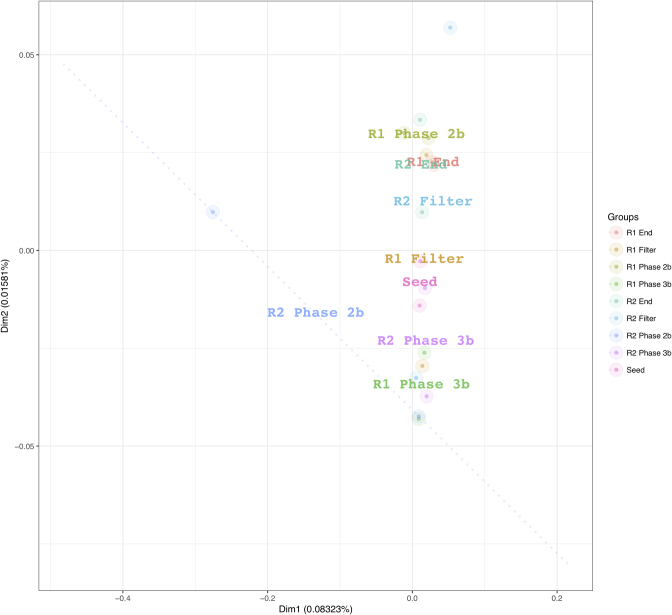
To follow the bacterial and archaeal community over time and to compare the development of the mesophilic ‘seed’ within each reactor, alpha-diversity matrices (Richness, Shannon, Simpson, Alpha and Evenness) were compared at the SEQ level. Samples from the ‘Seed’, ‘R1’ and ‘R2’ demonstrated similar observed values (Figure S1, Supporting Information). No significant difference was found between the ‘Seed’, ‘R1’ and ‘R2’; however, a large variation could be observed within the values per group. Subsequently, PCoA was carried out at SEQ level using unweighted Unifrac (ß-diversity metric) on the phylogenetic distance of sequences to visualise the similarities and dissimilarities in the microbial communities. Figure 5 demonstrates that the sequences from the replicated reactors group together based on time—‘Seed’, ‘Phase 2b’, ‘Phase 3b’, ‘Filter’ and ‘End’ rather than reactor origin. However, it must be noted that while the sequences grouped together based on sampling period they were not found to be significantly different from each other.
Figure 5.
PCoA plot based on unweighted Unifrac of DNA and cDNA sequences from R1 and R2 biomass samples. For each group, the legends are drawn at the mean value of the samples of that group.
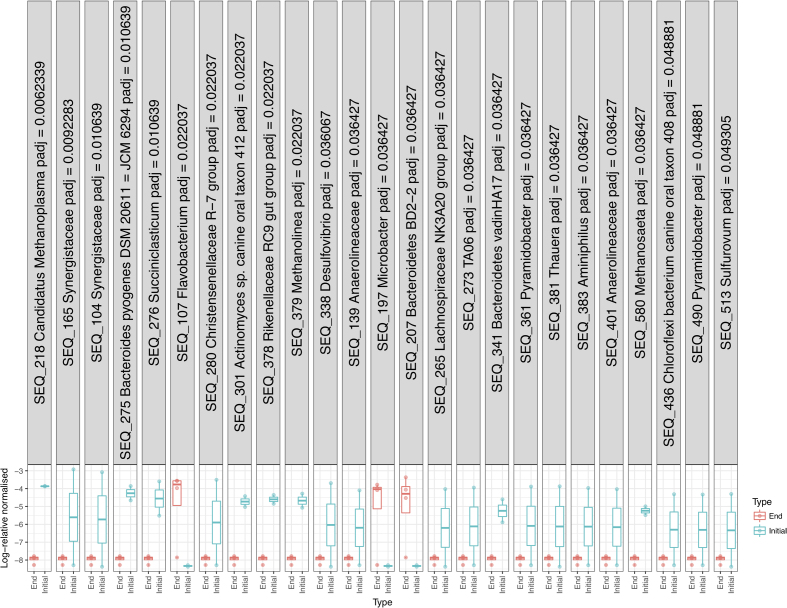
As the PCoA data indicated that the samples clustered based on time period, sampled analysis of significant species contributing to beta-diversity was carried out to identify what species were responsible for differences in these groupings. Analysis of the significant species contributing to beta-diversity was carried out at genus level at a 2-log and 1-log fold change for ‘Phase 2b’, ‘Phase 3b’, ‘Filter’ and ‘End’ whereby direct comparisons could be made between R1 and R2. The results demonstrated that there were no significantly different species between the replicate reactors at each of these phases (data not shown). This analysis was then repeated at SEQ level for the reactor phases (Figure S2a–c, Supporting Information). In the case of ‘Phase 2b’ SEQ4, SEQ5, SEQ6 and SEQ139 were greater in R1 (S2a, Supporting Information). SEQ4, 5 and 6 were found to be a Methanosaeta concilii strain X16932 and SEQ139 were found to be an uncultured Anaerolinaceae bacterium clone (Table S3, Supporting Information). In ‘Phase 3b’ a total of 24 SEQs were significantly different between R1 and R2 (S2b, Supporting Information). Of these 19 were greater in R1 (SEQS 36, 46, 184, 159, 17, 45, 50, 122, 210, 27, 111, 281, 19, 320, 53, 67, 103 and 92) and 5 (SEQs 69, 414, 512, 546, and 430) were greater in the R2 samples. There was no significant difference between the communities in R1 and R2 filter unit communities. There were only two sequences that were significantly different between R1 and R2 biomasses at the end of the trial (Figure S2c, Supporting Information). These were SEQ44 that was greater in R1 and SEQ235 that was greater in R2. SEQ44 was identified as an uncultured Synergistetes bacterium and SEQ235 was identified as Chryseobacterium species strain SE19. Significant species was also used to assess the maturation of the granular biofilm and the species contributing at a 1-log fold difference between the seed inoculum and the R1 and R2 end biomass (Fig. 6). From this 25 SEQS were identified as significantly different. SEQs 218, 165, 104, 275, 280, 301, 378, 379, 338, 139, 265, 273, 341, 361, 381, 383, 401, 580, 436, 490 and 513 were more abundant in the seed inoculum. While SEQs 107, 197 and 207 were more abundant in the biomass upon take down of the reactors (Fig. 6; S3, Supporting Information). Interestingly, SEQ107 was identified as a psychrotolerant species—Flavobacterium sinopsychrotolerans (Xu et al.2011). The identities of all significant SEQs are described in Table S3 (Supporting Information).
Figure 6.
Significant SEQs contributing to beta-diversity at an SEQ level at a 1-log fold change was assessed between the seed community (Initial) and the biomass taken from the end of the trial for both R1 and R2 (End).
DISCUSSION
Though we have not tested real sewage, we have demonstrated a sufficient capacity of the microbial community for sustained low-temperature degradation of a complex wastewater. Indeed, the removal efficiencies of these systems exceeded those reported in previous low-temperature trials carried out in a traditional UASB [upflow anaerobic sludge bed reactor] (Bandara et al.2012). This study demonstrated that a mesophilic inoculum rapidly acclimated to psychrophilic conditions to allow efficient COD removal to occur in both reactors. There were indications of a capacity for enhanced bacterial activity at 12°C, as evidenced by the protein hydrolysis assays. Km values throughout the trial increased at 12°C and decreased at 37°C, indicating an increase in substrate affinity at lower temperatures. The literature that substrate affinity will decrease at lower temperatures for psychrophiles, mesophiles and thermophiles (Nedwell 1999), but this often reflects only short-term studies. Our results point towards the emergence of psychrophilic proteolytic activity that was mirrored in both systems. While psychrophilic microorganisms may not be crucial for successful remediation of waste streams from a process steering aspect, the possibility to develop truly psychrophilic consortia could open important new opportunities for AD technology (Sekiguchi et al.2001). With respect to the archaeal populations, SMA data revealed that the microbial consortia became psychrotolerant for methanogenic substrates, rather than truly psychrophilic, a finding commonly reported in the literature (Lettinga et al.1999; O'Flaherty, Collins and Mahony 2006). Our study demonstrates that a psychrophilic or cold-adapted ‘seed’ was not necessary as a starting inoculum for successful stable anaerobic digestion at low temperatures. Bowen et al. (2014) reported that a mesophilic inoculum from an anaerobic suspended biomass sewage sludge reactor was not successful for LtAD, but this biomass had much lower SMA than the high-rate granular sludges used as inocula here, and in previous successful LtAD trials (e.g. Collins, Mahony and O'Flaherty 2006; Madden et al.2013; Keating et al.2016). It is likely that the retention of the anaerobic biomass in hybrid sludge bed fixed-film reactors supported the development of the reactor microbial community to function efficiently at lower temperatures. Moreover, the trial lasted 889 days, which may have provided sufficient time for the maturation of cold-adapted populations to allow for increased loading rates to be applied. This strategy for low-temperature sewage treatment offers a significant advantage over suspended biomass systems. In suspended biomass systems biomass washout would occur and the microbial population may be more sensitive to immigration and selective pressures of the influent (Vanwonterghem et al.2014).
We have demonstrated that stable, long-term, high-rate anaerobic digestion of a relatively complex wastewater, in the form of synthetic sewage, was possible and even efficient, at low-operating temperature. Reactor performance data indicated that the systems were functionally robust and stable, via the efficient effluent degradation with COD removal efficiencies for CODTot and CODSol of >73% (Table 2), despite incremental increases in the OLR applied over the course of the trial. Perhaps surprisingly, we have also shown that under these conditions hydrolysis was not rate-limiting at 12°C with evidence suggesting that CODSus in the synthetic wastewater were readily degraded to CODCol, despite the absence of wastewater-borne lipases associated with non-synthetic wastewaters (Petropoulos et al. 2017). SYNTHES was used so we could strictly define the influent and remove the variability associated with using real sewage—to be sure the microbial community development was not impacted by external variables. While SYNTHES carries a similar proportion of particulate COD (31%) to real sewage (30%) [Aiyuk and Verstraete 2004] a disadvantage is however, that starch comprises the complex carbohydrate portion, which may be easier to degrade than complex cellulosic materials that would be present in real sewage. No accumulation of solids was observed in the granular sludge bed in agreement with previous work (Keating et al.2016). The physical entrapment of solids within the pumice matrix of the hybrid reactor may have facilitated subsequent degradation.
Successful high-rate AD is contingent on well-functioning microbial communities. Stable community structures are maintained through syntrophic interactions between the bacterial and archaeal communities (Schnürer, Zellner and Svensson 1999). Low temperatures had been thought to limit these syntrophic interactions (Kotsyurbenko 2005). However, the communities represented in our systems were well balanced from the commencement of the trial, as indicated by negligible VFA accumulation in the reactor effluents and the diverse bacterial and archaeal populations found in the active fraction (cDNA-based analysis) throughout the trial. Interestingly, members of the Synergistetes were dominant members of the AD community in this trial. These are generally only found in frequencies of 1% or less in most AD systems (Godon et al.2005), but in this study their abundance increased up to 44% of bacterial sequences (Fig. 2). Isolated members of the Synergistetes partner syntrophic relationships with the methanogens in the degradation of amino acids with the production of VFAs (Vartoukian, Palmer and Wade 2007). Thus, the Synergistetes may be important for LtAD reactor function and may play a role in the low-temperature metabolism of proteins observed in reactor biomass. Perhaps, the nature of SYNTHES selected for a protein/amino acid degrading community or their high prevalence in the seed inoculum (coming from a dairy treatment facility) allowed for their development in this trial.
Adaptation involved a temporal shift in the microbial community structure over the course of the study. However, the replicate reactors maintained a remarkably similar microbial community profile to each other and this development was, in fact, reproducible down to genus level with no significant difference found between the reactors at each phase. Indeed, in the take-down biomass only two sequences were significantly different between the reactors (S3, Supporting Information). This was mirrored in the reactor performance data, whereby the reactors exhibited significant long-term reproducibility (889 days) during treatment of the synthetic sewage substrate (Table 2). Fluctuations in COD removal rates generally occurred at similar points in both reactors, indicating that degradation was occurring through biological activity, rather than by physical entrapment of the COD fractions. Two divergences in behaviour were, however, identified between the two reactors, based on process performance despite there being no significant differences between the communities at genus level. Firstly, an initial variation was observed upon start-up of the replicated systems. An immediate start-up was observed in R1 whereby all COD fractions were degraded, while the start-up of R2 took considerably longer (∼56 days). While this variation was found to be significant, no definitive cause could be identified, as molecular sampling was not carried out during the cold-adapting period so as not to disturb initial community development. Considering that CODSol removal was similar and highly efficient in both reactors during phase 1 (pointing to efficient microbial activity), the cause may have reflected a greater potential for leaching of CODSus particles or the loss of flocculent biomass from R2. Secondly, the commencement of Phase 5 led to a period of ∼5 weeks perturbation in R1, which was not mirrored in R2. qPCR data from the start of this phase showed a 1-log reduction of the total bacterial and archaeal gene copy numbers were observed in R2 (Fig. 1A). Changes in the microbial community structure were missed at this time point; however, sequencing results prior to this (from Phase 3) indicated that samples from this time point clustered together and no significant difference was found at genus level.
As stated previously there were no significant changes (1 or 2-log) in the microbial populations present between reactors at each time period at genus level as demonstrated by significant species contributing to beta-diversity analyses. This statistical measurement indicated that time was the driver of microbial community structure rather than reactor identity. Comparisons were then made at a sequence level in order to elucidate further the species that were different between the systems and the species diverging from the ‘seed’ inoculum. From the sequences outlined in Table S3 (Supporting Information), it is worth noting sequences associated with granule formation and granule integrity (Methanosaeta concilii species and Anaerolinea species). Anaerolinea species dominated the Chloroflexi phyla in the reactor systems. The Chloroflexi metabolise primary substrates in wastewater such as carbohydrates and cellular matter (Yamada et al.2005). Anaerolinea species belong to Subphylum 1 an elusive phylum comprising environmental clones (Hugenholtz, Goebel and Pace 1998). They form web-like structures on the outside of granules in mesophilic and thermophilic systems and thus are thought to be important for granule structure (Sekiguchi et al.1998). Given their stable dominance in these reactors further characterisation of their role in low-temperature systems would be valuable. Methanosaeta dominated the archaeal communities in both systems as demonstrated by sequencing analysis (Figs 2 and 4; Table S3, Supporting Information). Methanosaeta concilii is a key organism in granulation in these anaerobic systems (Hulshoff Pol et al.2004). The distinctive solely acetate utilising acetoclastic Methanosaeta are known to dominate in steady state reactors in which acetate concentrations are low (McMahon et al.2001). VFA analyses indicated that in-reactor acetate values were negligible throughout the trial. Moreover, acetoclastic methanogens have been seen to be dominant at low temperatures (Chin, Lukow and Conrad 1999). However, this is in contrast to reports by several authors that suggest that acetoclastic activity is impacted by lower temperatures and that under these conditions hydrogenotrophic methanogens dominate and facilitate efficient VFA degradation (Nozhevnikova et al.2000; Collins et al.2005; Connaughton, Collins and O'Flaherty 2006). In Phase 3, Anaerolinea-like species were reduced in R1 in comparison to R2 (following this R1 demonstrated biomass washout upon commencement of Phase 5). Biomass sampled from Phase 2 demonstrated that Methanosaeta like species were reduced in R2 in comparison to R1 (prior to this phase R2 demonstrated reduced performance). While this data are not conclusive, the close monitoring of such species is crucial to granule integrity may provide an opportunity to link granule health with process performance and to develop means to promote their growth in poorly performing systems.
CONCLUSIONS
Overall this study revealed that a cold-adapted or psychrophilic ‘seed’ inoculum was not necessary for efficient LtAD of wastewater. Our work demonstrated reproducible process performance and mirrored microbial community development between replicated systems. The nature of the reactor system allowed for the retention of biomass allowing sufficient cold-adapted communities to develop and mature, to the point where increased activity at low temperature developed within the hydrolytic and methanogenic populations. Next generation sequencing identified a number of possible cold-adapted species and increased abundance of the Synergistetes and Anaerolinea phyla that warrant further targeted investigations to determine their possible future biotechnological relevance.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Aoife Duff and Dr. Eoin Gunnigle for the provision of culture strains.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work was supported by Science Foundation Ireland, through a Charles Parsons Award (06_CP_E006) and an Investigator Programme Grant (14/IA/2371); and the Irish Environmental Protection Agency (2014-W-LS-7). CJS is supported by Science Foundation Ireland Starting Investigator-COFUND fellowship (11/SIRG/B2159). UZI is funded by a NERC fellowship NE/L011956/1.
Conflict of interest. None declared.
REFERENCES
- Aiyuk S, Verstraete W. Sedimentological evolution in an UASB treating SYNTHES, a new representative synthetic sewage, at low loading rates. Bioresour Technol. 2004;93:269–78. [DOI] [PubMed] [Google Scholar]
- Akarsubasi AT, Ince O, Kirdar B et al. Effect of wastewater composition on archaeal population diversity. Water Res. 2005;39:1576–84. [DOI] [PubMed] [Google Scholar]
- APHA. In: Clesceri LS, Greenberg AE, Eaton AD (eds). Standard Methods for the Examination of Water and Wastewater, 20th ed American Public Health Association, Washington, DC, 2005. [Google Scholar]
- Bandara WMKRTW, Kindaichi T, Satoh H et al. Anaerobic treatment of municipal wastewater at ambient temperature: analysis of archaeal community structure and recovery of dissolved methane. Water Res. 2012;46:5756–64. [DOI] [PubMed] [Google Scholar]
- Bialek K, Cysneiros D, O'Flaherty V. Low-Temperature (10°C) anaerobic digestion of dilute dairy wastewater in an EGSB bioreactor: microbial community structure, population dynamics, and kinetics of methanogenic populations. Archaea. 2013;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek K, Kumar A, Mahony T et al. Microbial community structure and dynamics in anaerobic fluidized-bed and granular sludge-bed reactors: influence of operational temperature and reactor configuration. Microb Biotechnol. 2012;5:738–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen EJ, Dolfing J, Davenport RJ et al. Low-temperature limitation of bioreactor sludge in anaerobic treatment of domestic wastewater. Water Sci Technol. 2014;69:1004–13. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R. Microbial ecology of Antarctic aquatic systems. Nat Rev Microbiol. 2015;13:691–706. [DOI] [PubMed] [Google Scholar]
- Chin KJ, Lukow T, Conrad R. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl Environ Microbiol. 1999;65:2341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleran E, Concannon F, Golden T et al. Use of methanogenic activity tests to characterise anaerobic sludges, screen for anaerobic biodegradability and determine toxicity thresholds against individual anaerobic trophic groups and species. Water Sci Technol. 1992;25:31–40. [Google Scholar]
- Collins G, Foy C, McHugh S et al. Anaerobic biological treatment of phenolic wastewater at 15–18°C. Water Res. 2005;39:1614–20. [DOI] [PubMed] [Google Scholar]
- Collins G, Mahony T, O'Flaherty V. Stability and reproducibility of low-temperature anaerobic biological wastewater treatment. FEMS Microbiol Ecol. 2006;55:449–58. [DOI] [PubMed] [Google Scholar]
- Connaughton S, Collins G, O'Flaherty V. Development of microbial community structure and activity in a high-rate anaerobic bioreactor at 18°C. Water Res. 2006;40:1009–17. [DOI] [PubMed] [Google Scholar]
- DuBois M, Gilles KA, Hamilton JK et al. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–6. [Google Scholar]
- Elmitwalli TA, Soellner J, De Keizer A et al. Biodegradability and change of physical characteristics of particles during anaerobic digestion of domestic sewage. Water Res. 2001;35:1311–7. [DOI] [PubMed] [Google Scholar]
- Enright A-M, McGrath V, Gill D et al. Effect of seed sludge and operation conditions on performance and archaeal community structure of low-temperature anaerobic solvent-degrading bioreactors. Syst Appl Microbiol. 2009;32:65–79. [DOI] [PubMed] [Google Scholar]
- Fernández A, Huang S, Seston S et al. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti E, Zaiat M, Vallero M. Anaerobic processes as the core technology for sustainable domestic wastewater treatment: consolidated applications, new trends, perspectives, and challenges. Rev Environ Sci Biotechnol. 2006;5:3–19. [Google Scholar]
- Girvan MS, Campbell CD, Killham K et al. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ Microbiol. 2005;7:301–13. [DOI] [PubMed] [Google Scholar]
- Godon J-J, Moriniere J, Moletta M et al. Rarity associated with specific ecological niches in the bacterial world: the ‘Synergistes’ example. Environ Microbiol. 2005;7:213–24. [DOI] [PubMed] [Google Scholar]
- Gouveia J, Plaza F, Garralon G et al. Long-term operation of a pilot scale anaerobic membrane bioreactor (AnMBR) for the treatment of municipal wastewater under psychrophilic conditions. Bioresour Technol, 2015;185:225–33. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diverity. J Bacteriol. 1998;180:4765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Enright A-M, Mahony T et al. Novel Anaerobic Sewage Treatment and Bioenergy Production: High-Rate Anaerobic Digestion as a Core Technology for Sustainable Treatment of Municipal and Low-Strength Industrial Wastewaters. EPA STRIVE Report Series No. 64. Johnstown Castle: Environmental Protection Agency, 2011. [Google Scholar]
- Hulshoff Pol LW, de Castro Lopes SI, Lettinga G et al. Anaerobic sludge granulation. Water Res. 2004;38:1376–89. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biol Evoluti. 2013;30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating C, Chin JP, Hughes D et al. Biological phosphorus removal during high-rate, low-temperature, anaerobic digestion of wastewater. Front Microbiol. 2016;7:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyurbenko OR. Trophic interactions in the methanogenic microbial community of low-temperature terrestrial ecosystems. FEMS Microbiol Ecol. 2005;53:3–13. [DOI] [PubMed] [Google Scholar]
- Langenhoff AAM, Stuckey DC. Treatment of dilute wastewater using an anaerobic baffled reactor: effect of low temperature. Water Res. 2000;34:3867–75. [Google Scholar]
- Lettinga G, Rebac S, Parshina S et al. High-rate anaerobic treatment of wastewater at low temperatures. Appl Environ Microbiol. 1999;65:1696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levén L, Eriksson ARB, Schnürer A. Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol Ecol. 2007;59:683–93. [DOI] [PubMed] [Google Scholar]
- Lew B, , Beliavski M, Admon S et al. Temperature effect on UASB reactor operation for domestic wastewater treatment in temperate climate regions. Water Science and Technology. 2003;48:25–30. [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL et al. Protein measurements with the folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- Madden P, Al-Raei AM, Enright A-M et al. Effect of sulfate on low-temperature anaerobic digestion. Front Microbiol. 2013;5:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden PD, Chinalia FA, Enright AM et al. Perturbation-independent community development in low-temperature anaerobic biological wastewater treatment bioreactors. Biotechnol Bioeng. 2010;105:79–87. [DOI] [PubMed] [Google Scholar]
- McKeown RM, Hughes D, Collins G et al. Low-temperature anaerobic digestion for wastewater treatment. Curr Opin Biotechnol. 2012;23:444–51. [DOI] [PubMed] [Google Scholar]
- McMahon KD, Stroot PG, Mackie RI et al. Anaerobic codigestion of municipal solid waste and biosolids under various mixing conditions e II: microbial population dynamics. Water Res. 2001;35:1817–27. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedwell D, Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiology Ecology. 1999;30:101–11. [DOI] [PubMed] [Google Scholar]
- Neves L, Oliveira R, Alves MM. Influence of inoculum activity on the bio-methanization of a kitchen waste under different waste/inoculum ratios. Process Biochem. 2004;39:2019–24. [Google Scholar]
- Nozhevnikova AN, Rebak S, Kotsyurbenko OR et al. Anaerobic production and degradation of volatile fatty acids in low temperature environments. Water Sci Technol. 2000;41:39–46. [Google Scholar]
- O'Flaherty V, Collins G, Mahony T. The microbiology and biochemistry of anaerobic bioreactors with relevance to domestic sewage treatment. Rev Environ Sci Biotechnol. 2006;5:39–55. [Google Scholar]
- Oksanen JF, Blanchet G, Kindt R et al. Vegan: Community Ecology Package, version 2013 Dec 12;2 (9) R package version 3.0 2013.
- Petropoulos E, Dolfing J, Davenport RJ et al. Developing cold-adapted biomass for the anaerobic treatment of domestic wastewater at low temperatures (4, 8 and 15°C) with inocula from cold environments. Water Res. 2017;112:100–9. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin L, Poulsen LK, Noguera DR et al. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebac S, Ruskova J, Gerbens S et al. High-rate anaerobic treatment of wastewater under psychrophilic conditions. J Ferment Bioeng. 1995;80:499–506. [Google Scholar]
- Sakar S, Yetilmezsoy K, Kocak E. Anaerobic digestion technology in poultry and livestock waste treatment—a literature review. Waste Manage Res. 2009;27:3–18. [DOI] [PubMed] [Google Scholar]
- Schnürer A, Zellner G, Svensson BH. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol Ecol. 1999;29:249–61. [Google Scholar]
- Sekiguchi Y, Kamagata Y, Harada H, Recent advances in methane fermentation technology. Current Opinion in Biotechnology. 2001;12:277–282. [DOI] [PubMed] [Google Scholar]
- Sekiguchi Y, Kamagata Y, Syutsubo K et al. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Annu Rev Microbiol. 1998;144:2655–65. [DOI] [PubMed] [Google Scholar]
- Singh KS, Viraraghavan T. Impact of temperature on performance, microbiological and hydrodynamic aspects of UASB reactors treating municipal wastewater. In: Proceedings of Seventh Latin American Workshop and Symposium on Anaerobic Digestion. Merida, Mexico, 2002, pp. 613–20. [PubMed] [Google Scholar]
- Smith AL, Skerlos SJ, Raskin L. Psychrophilic anaerobic membrane bioreactor treatment of domestic wastewater. Water Res. 2013;47:1655–65. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Nedwell DB, Dong LF et al. Evalution of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol. 2006;8:804–15. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Taylor LT, DeLong EF. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lier JB, Tilche A, Ahring BK et al. New perspectives in anaerobic digestion. Water Sci Technol. 2001;43:1–18. [PubMed] [Google Scholar]
- Vanwonterghem I, Jensen PD, Ho DP et al. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol. 2014;27:55–64. [DOI] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. The division ‘Synergistes’. Anaerobe. 2007;13:99–106. [DOI] [PubMed] [Google Scholar]
- Xing W, Zhao Y, Zuo JE. Microbial activity and community structure in a lake sediment used for psychrophilic anaerobic wastewater treatment. J Appl Microbiol. 2010;109:1829–37. [DOI] [PubMed] [Google Scholar]
- Xu M, Xin Y, Tian J et al. Flavobacterium sinopsychrotolerans sp. nov., isolated from a glacier. Int J Syst Evol Microbiol. 2011;61:20–4. [DOI] [PubMed] [Google Scholar]
- Yamada T, Sekiguchi Y, Imachi H et al. Diversity, localization, and physiological properties of filamentous microbes belonging to chloroflexi subphylum i in mesophilic and thermophilic methanogenic sludge granules. Appl Environ Microbiol. 2005;71:7493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Lee C, Kim J et al. Group-specific primer and probe sets to detect methanogenic communities using qpcr. Biotechnol Bioeng. 2005;89:670–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.