Abstract
Asexual propagation and whole body regeneration are forms of nonembryonic development (NED) widespread across animal phyla and central in life history and evolutionary diversification of metazoans. Whereas it is challenging to reconstruct the gains or losses of NED at large phylogenetic scale, comparative studies could benefit from being conducted at more restricted taxonomic scale, in groups for which phylogenetic relationships are well established. The ascidian family of Styelidae encompasses strictly sexually reproducing solitary forms as well as colonial species that combine sexual reproduction with different forms of NED. To date, the phylogenetic relationships between colonial and solitary styelids remain controversial and so is the pattern of NED evolution. In this study, we built an original pipeline to combine eight genomes with 18 de novo assembled transcriptomes and constructed data sets of unambiguously orthologous genes. Using a phylogenomic super-matrix of 4,908 genes from these 26 tunicates we provided a robust phylogeny of this family of chordates, which supports two convergent acquisitions of NED. This result prompted us to further describe the budding process in the species Polyandrocarpa zorritensis, leading to the discovery of a novel mechanism of asexual development. Whereas the pipeline and the data sets produced can be used for further phylogenetic reconstructions in tunicates, the phylogeny provided here sets an evolutionary framework for future experimental studies on the emergence and disappearance of complex characters such as asexual propagation and whole body regeneration.
Keywords: asexual reproduction, Botryllinae, cross-contamination, Polyandrocarpa, regeneration, tunicate
Introduction
Most animals reproduce sexually (White 1978). Male and female gametes combine their genetic material in a zygote, develop by embryogenesis, and produce individuals that are genetically different from their parents. This reproductive strategy is regarded as evolutionarily advantageous by allowing rapid adaptation to constantly changing environments; for example, it can efficiently eliminate deleterious mutations or rapidly incorporate beneficial mutations (reviewed by West et al. 1999). However, other reproductive modes co-exist within the metazoans. Nearly half of all animal phyla contain species that are able to propagate asexually via agametic reproduction (i.e., cloning), and the number is higher if one includes parthenogenetic species (Bell 1982; Hughes and Cancino 1985; Nilsson Sköld et al. 2009; Brusca et al. 2016). Organisms that reproduce clonally mobilize somatic cells, either as undifferentiated progenitors or terminally differentiated cells, to produce new individuals genetically identical to their parent. Agametic asexual reproduction is often linked with extensive regenerative capacity, as many clonally propagating organisms can regenerate their body entirely, including the germline (Hughes 1989; Sanchez-Alvarado and Yamanaka 2014). This relationship strongly suggests that clonal propagation and regeneration are two processes of “nonembryonic development” (NED) that involve common cellular and molecular mechanisms (Martinez et al. 2005; Agata et al. 2007; Bosch 2007; Galliot and Chera 2010; Gurley et al. 2010).
Some animals undergoing NED can form colonies, where individuals remain either physically connected to each other or develop clusters of genetically identical modules termed zooids (e.g., in tunicates), ramets (e.g., in bryozoans), or polyps (e.g., in cnidarians; Boardman et al. 1973). Depending on the species, the modules can be either morphologically identical or different, so that physiological functions, like reproduction or feeding can be subdivided among all units of the colony. This type of growth and modular organization has important physiological and ecological consequences, such as allowing competitive dominance over solitary forms on space-limited hard substrata (Jackson 1977) or reducing gene flow compared with solitary species (Jackson and Coates 1986). Solitary organisms show an inverse relationship between body size and reproductive rate due to reduced metabolic rate with high body mass. However, colonial species escape this limitation and show mass-specific metabolic rates related to module size rather than colony size. Thus, colonial species can grow faster than solitary species and propagate indefinitely without loss in reproductive potential (Hughes 1989; Jackson and Coates 1986; Burgess et al. 2017). Therefore, widespread sexual reproduction by gamete production only represents one facet of species perpetuation and evolutionary diversification of metazoans.
Species employing asexual clonal reproduction are distributed across animal phyla, including nonbilaterian animals such as placozoans, cnidarians or sponges, many spiralian groups such as bryozoans, annelids or planarians, and deuterostome phyla such as hemichordates, echinoderms or tunicates, with the notable exception of vertebrates (Hughes 1989; Bely and Nyberg 2010; Brusca et al. 2016; Alié et al. 2015; Funayama et al. 2010). Regenerative abilities are also widespread and unevenly distributed across metazoans (reviewed in Tiozzo and Copley 2015). While many authors consider regeneration an ancestral trait for animals (Sánchez Alvarado 2000; Bely and Nyberg 2010), the more scattered phylogenetic distribution and diversity of agametic asexual reproductive mechanisms is suggestive of multiple independent acquisitions (Nilsson Sköld et al. 2009). For example, in annelids asexual reproduction by fission was gained at least 19 times (Zattara and Bely 2016). In anthozoan cnidarians fission has likely arisen from an aclonal ancestor at least four times (Geller et al. 2005). In scleractinian corals, on the other hand, coloniality is reported to have been lost at least six times (Barbeitos et al. 2010). It is difficult to reconstruct the cellular and molecular mechanisms underlying the evolution of NED by comparing distant phyla, first because in many cases phylogenetic relationships are unresolved, and because, the cell types involved and their behaviors are hardly comparable at such phylogenetic scales (Carlson 2007). Therefore, in order to understand how novel reproductive and regenerative strategies evolved and the directionality of these transitions (i.e., gains or losses) it is essential to compare closely related species undergoing NED or lacking such traits with well-defined phylogenetic relationships (Tiozzo and Copley 2015; Nogueira et al. 2016).
Ascidians belong to the subphylum Tunicata, the closest living relatives to vertebrates (Delsuc et al. 2006) in which it is possible to find strictly sexually reproducing species and others that combine sexual reproduction with different forms of budding and whole body regeneration (Kürn et al. 2011; Brown and Swalla 2012; Piette and Lemaire 2015). Whereas solitary ascidians reproduce only sexually, colonial species can develop both sexually and via NED. Interestingly, colonial and solitary species are scattered across the phylogenetic tree of ascidians, suggesting that NED, and therefore coloniality, has been acquired and/or lost several times independently (Brown and Swalla 2012). Ascidians are polyphyletic and classically divided in three orders—Aplousobranchia, Phlebobranchia, and Stolidobranchia—in each of which budding mechanisms are very different, further supporting convergent acquisitions of coloniality (Brown and Swalla 2012). The order Stolidobranchia comprises three traditionally recognized families (Molgulidae, Pyuridae, and Styelidae). Molgulidae and Pyuridae are solitary, whereas approximately one third of Styelidae are colonial species (WoRMS Editorial Board 2017; fig. 1A–F). Thus, Stolidobranchia has long been considered a suitable group to study the plastic evolution of NED and coloniality (Zeng et al. 2006; Pérez-Portela et al. 2009; Brown and Swalla 2012).
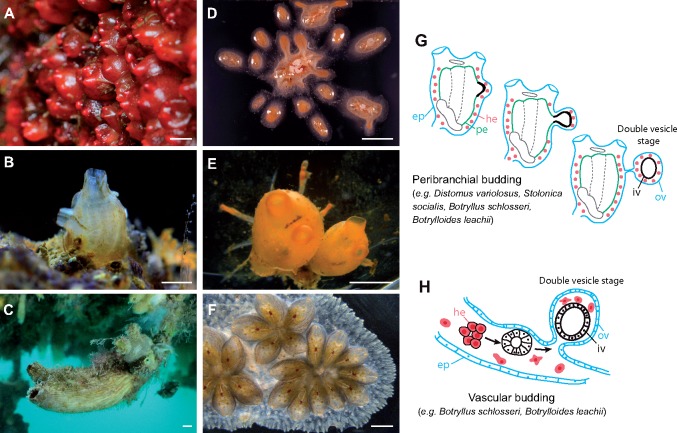
Fig. 1.
(A–C) Solitary Styelidae. (A) Dendrodoa grossularia. (B) juvenile of Styela plicata. (C) Styela clava © Thomas Wilfried Station Biologique de Roscoff. (D–F) Colonial Styelidae. (D) Polyandrocarpa misakiensis. (E) Stolonica socialis. (F) Botryllus schlosseri (courtesy of L. Ricci). (G) Schematic representation of peribranchial budding at three successive stages, from peribranchial invagination to double vesicle. (H) Schematic representation of vascular budding at three successive stages, from hemocyte clustering to double vesicle. ep: epidermis, he: hemocytes, iv: inner vesicle, ov: outer vesicle, pe: peribranchial epithelium. Scale bars: (A–C) 1 cm, (D–E) 5 mm, (F) 1 mm.
In colonial styelids, two budding modes have been described and sometimes coexist in the same species: peribranchial budding and vascular budding (fig. 1G–H). In peribranchial budding, a portion of the zooid epithelium lining the peribranchial chamber starts to bud in a stereotyped process (fig. 1G) and gives rise to new adult zooids (Selys-Longchamps 1917; Berrill 1935; Kawamura and Watanabe 1981; Fujiwara et al. 2011; Manni et al. 2014). In addition to peribranchial budding, several species also use vascular budding either for asexual propagation or in response to injury (fig. 1H). During this process circulatory cells cluster, proliferate, and develop into a new individual within the vasculature of the colony (Kawamura and Sunanaga 2010; Rinkevich et al. 2010; Ricci et al. 2016). In both processes, the surrounding epidermis envelops the bud, forming a double vesicle of monolayered tissue (fig. 1G and H) that will later undergo organogenesis. Therefore, in Styelidae the two described budding processes pass through a similar stage, the double vesicle, which has different cellular origins and is triggered by different mechanisms. Whereas peribranchial budding mainly involves epithelial morphogenesis, vascular budding is ensured by circulating putative stem cells (Laird et al. 2005).
Previous attempts to reconstruct the phylogeny of the Styelidae have not reached a consensus on the relationships between colonial versus solitary species. Kott (1985) splits the styelids into three subfamilies, solitary Styelinae, colonial Polyzoinae, and colonial Botryllinae based on morphological characters. However, molecular phylogenies based on ribosomal and mitochondrial markers have led to conflicting results, either retrieving all colonial species together in a single clade and suggesting a unique acquisition of asexual reproduction (Zeng et al. 2006; Tsagkogeorga et al. 2009; Shenkar et al 2016), or retrieving several lineages of colonial species and thus proposing multiple convergent acquisitions of asexual reproduction (Pérez-Portela et al. 2009). Whereas the former studies included only a limited number of colonial styelids, Pérez-Portela et al. (2009) included more of them but the key branches were not statistically supported.
In the present work, we used Illumina-based RNA-seq comparisons to address phylogenetic relationships between colonial and solitary styelids. We used an original pipeline to combine de novo assembled transcriptomes of 18 stolidobranchs with 8 ascidian genomes to build a supermatrix of orthologous genes, which lead to a maximally supported tree topology. Our results showed that NED was acquired at least twice independently in the family Styelidae and led us to uncover a novel budding mechanism in Polyandrocarpa zorritensis. In light of these results, we discuss the evolution of asexual reproduction and coloniality emphasizing the evolutionary plasticity of asexual development in tunicates.
Results
Species Selection from 18S rRNA-COI Phylogeny
In order to a priori select species that span the diversity of colonial and solitary styelids, we first conducted a preliminary phylogenetic analysis based on a concatenated alignment of 18S rRNA and mitochondrial COI markers, combining data from Pérez-Portela et al. (2009), publicly available sequences, and sequences obtained from 12 specimens that we collected (see Materials and Methods and supplementary fig. 1, Supplementary Material online). Whereas most of the deep nodes were not statistically supported, the topology of the resulting tree was helpful to guide a posteriori selection of the colonial and solitary species to be used in the study. We used colonial ascidians from all different clades recovered in the phylogeny, as well as solitary species of Styela, Polycarpa, and Asterocarpa that were located in different phylogenetic lineages (supplementary fig. 1, Supplementary Material online).
In order to test the monophyly of styelids, we chose an outgroup that is a priori paraphyletic, including four Pyuridae, two Molgulidae, and four other distantly related species belonging to the genera Ciona and Phallusia (order Phlebobranchia). The final list of selected species is given in supplementary table 1, Supplementary Material online. Outgroup species of choice were mainly selected by the availability of the full genome sequence, which later simplified the steps of data set construction (supplementary table 1, Supplementary Material online).
A New Pipeline for the De Novo Assembly of Ascidian Transcriptomes
We generated cDNA libraries from the 16 species we collected, from most of which 30–50 million reads were sequenced using the Illumina HiSeq 4000 platform (table 1). The number of sequence reads, accession numbers, and other basic statistics to assess the quality of the transcriptomes are shown in table 1. The initial assembly of these reads using Trinity software (Grabherr et al. 2011) led to transcriptomes composed of between 76,825 and 410,479 contigs depending on the species (supplementary table 1, Supplementary Material online). These values are far higher than expected from current knowledge of ascidian genome complexity (Dehal et al. 2002; Voskoboynik et al. 2013; Velandia-Huerto et al. 2016). We therefore sought to eliminate redundant and low-quality contigs by developing a custom pipeline (detailed in fig. 2A and Materials and Methods). Briefly, we used stringent parameters to filter reads, contigs and ORFs based on sequence quality, redundancy, length, and expression levels. We additionally detected and removed cross-contaminations between samples that were sequenced in the same batch using CroCo (Simion et al. 2018). An average of 5% of all filtered contigs were detected as cross-contaminants (supplementary table 1, Supplementary Material online), and most cross-contaminations arose between samples sequenced in the same Illumina run (supplementary fig. 2 and supplementary table 2, Supplementary Material online).
Table 1.
Species from Which De Novo Transcriptomes Were Assembled and Included in Our Study.
| Species | Accession Number | Sampling Location | Tissues Used for De Novo Transcriptome | n Raw Reads | n Contigs | N50 | Average Length | Busco Index |
|---|---|---|---|---|---|---|---|---|
| Asterocarpa humilis | PRJNA422120 | Brest (France) | 1 adult (no tunic, no stomach) | 39,963,596 | 24,341 | 1,644 | 1,180.46 | 88% |
| Botrylloides leachii | SAMN04161391-98 | Otago (New Zealand) | Embryos and regenerative tissues | 315,587,750 | 42,036 | 1,365 | 903 | 96% |
| Botryllus schlosseri | SRX726446-59, 67-80 | Santa Barbara (USA) | Whole colonies | 621,300,622 | 65,586 | 1,224 | 905 | 95% |
| Dendroda grossularia | PRJNA422120 | Roscoff (France) | 1 adult + larvae (no tunic, no stomach) | 42,386,509 | 57,564 | 1,194 | 852.74 | 95% |
| Distomus variolosus | PRJNA422120 | Roscoff (France) | 1 adult (no tunic) | 39,810,612 | 44,516 | 1,458 | 1,004.97 | 92% |
| Eusynstyela tincta | PRJNA422120 | São Sebastião (Brazil) | 1 adult (no tunic) | 33,225,966 | 39,494 | 837 | 694 | 74% |
| Microcosmus sabatieri | PRJNA422120 | Banyuls (France) | Siphon | 29,425,964 | 22,951 | 1,471 | 1,022.8 | 93% |
| Polyandrocarpa misakiensis | PRJNA422120 | Misaki (Japan) | Asexual zooids and buds | 39,178,452 | 52,545 | 1,194 | 851.48 | 96% |
| Polyandrocarpa zorritensis | PRJNA422120 | La Spezia (Italy) | 2 adults and buds | 158,133,887 | 57,781 | 1,197 | 816.91 | 96% |
| Polycarpa aurata | PRJNA422120 | Cebu (Philippines) | 1 adult (no tunic, no stomach) | 34,876,779 | 29,348 | 1,443 | 982.17 | 95% |
| Polycarpa mamillaris | PRJNA422120 | Banyuls (France) | Oocytes | 12,985,467 | 25,221 | 1,599 | 1,056.42 | 96% |
| Polycarpa pomaria | PRJNA422120 | Brest (France) | 1 adult (no tunic) | 33,554,939 | 41,730 | 1,386 | 949.48 | 95% |
| Pyura dura | PRJNA422120 | Banyuls (France) | Gonads | 33,619,672 | 17,739 | 1,854 | 1,216.9 | 96% |
| Stolonica socialis | PRJNA422120 | Roscoff (France) | 1 adult | 35,870,064 | 43,474 | 1,431 | 934.34 | 94% |
| Styela canopus | PRJNA422120 | São Sebastião (Brazil) | 1 adult (mantle) | 44,826,592 | 42,420 | 1,047 | 815.45 | 87% |
| Styela clava | PRJNA422120 | Brest (France) | 1 adult (no tunic, no stomach) | 47,686,633 | 26,553 | 1,647 | 1,164.48 | 89% |
| Styela plicata | PRJNA422120 | Banyuls (France) | Embryos | 38,186,700 | 19,288 | 1,870 | 1,253.51 | 97% |
| Polycarpa sp. | PRJNA422120 | Roscoff (France) | 1 adult (no tunic) | 43,522,115 | 44,672 | 1,311 | 918.30 | 90% |
Fig. 2.
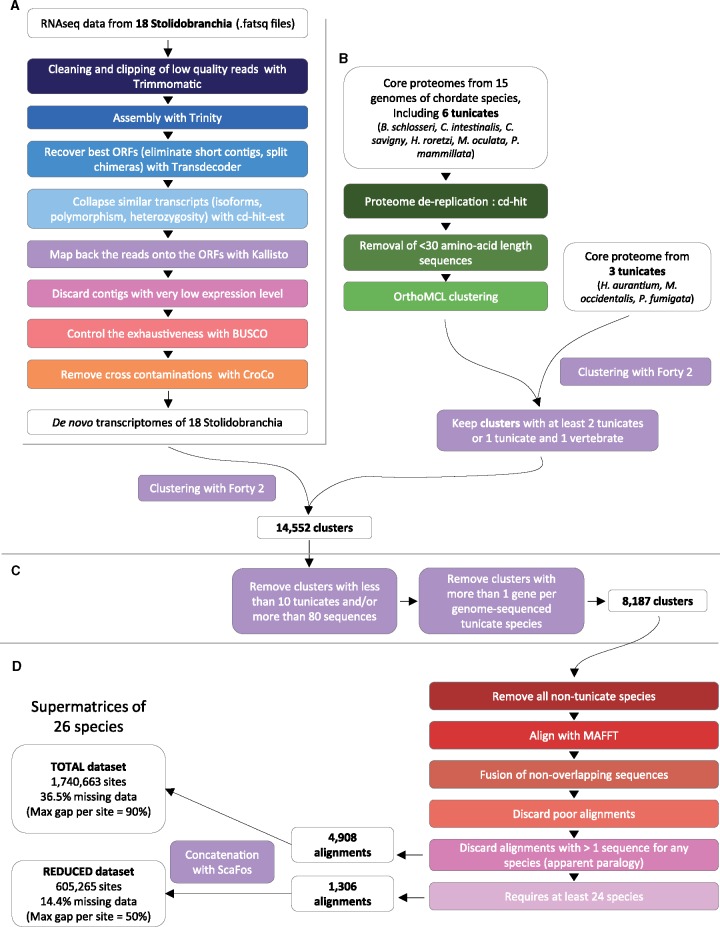
Graphical summary of the data set construction protocol used in this study (see Materials and Methods for the details of each step). (A) Part of the pipeline corresponding to the de novo transcriptome assembly. (B) Part of the clustering pipeline resulting in 14,552 clusters deduced from 9 tunicate genomes and 18 de novo transcriptomes. (C) Part of the clustering pipeline leading to 8,187 clusters with no apparent paralogy in genome-sequenced species. (D) Part of the pipeline corresponding to alignment and building of the FULL and REDUCED data sets devoid of paralogy for any species.
On average, our pipeline retained ∼20% of initial contigs (supplementary table 1, Supplementary Material online). BUSCO indices ranged from 87% and 96% (except for Eusynstyela tincta for which it was 74%), showing that our pipeline did not massively reduce gene numbers but rather eliminated undesired redundancy and/or contaminations. As an example, whereas Trinity assembly of reads from Polycarpa aurata gave 118,198 contigs with BUSCO index of 95% we ended up with 29,348 contigs. Final transcriptomes are available at https://github.com/AlexAlie/styelidae, last accessed April 2018, and raw Illumina reads can be retrieved from NCBI BioProject PRJNA422120.
A New Data Set of Orthologous Genes for Ascidian Phylogeny
Combining our refined de novo transcriptome assemblies with sets of genes derived from genomes allowed us to build a high quality data set of orthologous tunicate genes (fig. 2). The contigs of our 18 transcriptomes (fig. 2A) were first attributed to 14,552 clusters that were built using 18 chordate genomes (fig. 2B, supplementary table 3, Supplementary Material online). Among them we selected 8,187 clusters with no apparent paralogy in tunicate genomes (fig. 2C). After removing all nontunicate species, the contigs within each cluster were aligned (fig. 2D) and we performed alignment curation by joining end-to-end fragments of single transcripts that were split due to imperfect transcriptome assembly (fig. 2D, step “fusion of nonoverlapping sequences”). The combination of high quality transcriptomes, fusion of fragmented sequences, and subsequent filtering (see Materials and Methods) led to the retention of 4,908 alignments devoid of apparent paralogy in tunicates (fig. 2D, TOTAL data set of 1,740,663 amino acid sites and 36.5% missing data). A reduced version of this data set comprising 1,306 genes was selected, in which virtually all species were present (fig. 2D, REDUCED data set of 605,265 amino acid sites and 14.4% missing data). Functional annotations showed that the genes belong to different families, functional groups, and have different kinds of protein domains (supplementary table 4, Supplementary Material online). Both TOTAL and REDUCED matrices are provided at https://github.com/AlexAlie/styelidae, last accessed April 2018.
A Robust Phylogeny of Styelid Ascidians
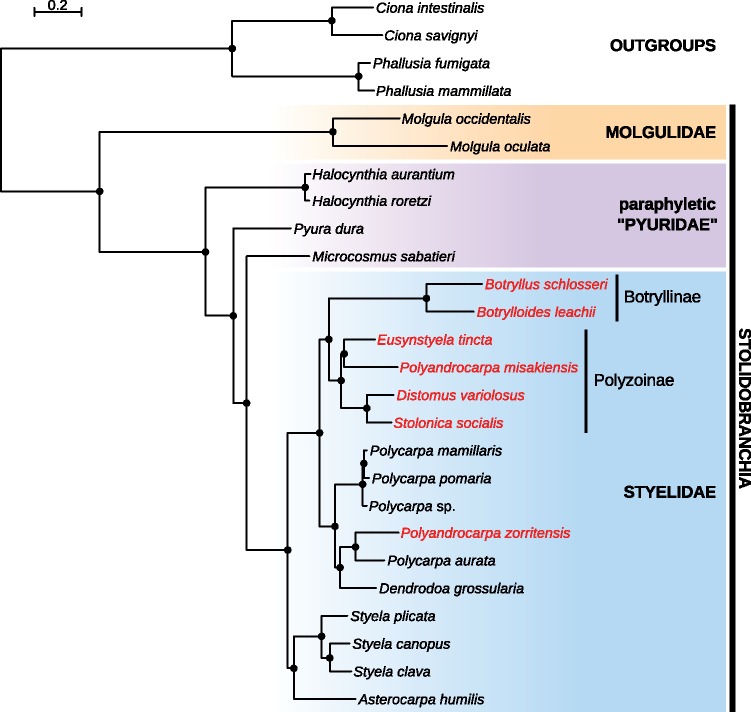
We conducted maximum likelihood analyses using models with (LG4X) and without (LG) site heterogeneity, as well as Bayesian inference under the site-heterogeneous CAT mixture model. All three strategies generated the same topology, with maximum bootstrap and maximum posterior probabilities obtained for all branches (fig. 3 and supplementary figs. 3–5, Supplementary Material online). We retrieved a monophyletic Stolidobranchia, in which a monophyletic Styelidae was embedded within a paraphyletic Pyuridae, and in which a monophyletic Molgulidae was the sister-group to all other Stolidobranchia. Within Styelidae, a clade of solitary Styela and Asterocarpa was the sister-group to all the rest (fig. 3). Colonial species, that is, species adopting NED, fell in two distinct locations in our inferred phylogeny. First, we recovered a clade containing Botryllus schlosseri, Botrylloides leachii, Polyandrocarpa misakiensis, Eusynstyela tincta, Distomus variolosus, and Stolonica socialis (fig. 3). In all six species belonging to this clade, colonial growth occurs by peribranchial budding, and therefore we named this clade the peribranchial budding clade (PB clade). In this clade, B. schlosseri and B. leachii (Botryllinae) are also able to regenerate colonies through vascular budding (fig. 3). The Botryllinae was the sister group to a clade containing four Polyzoinae: E. tincta together with P. misakiensis and S. socialis together with D. variolosus. The fifth Polyzoinae and last colonial species of our sampling, P. zorritensis grouped together with solitary Polycarpa and Dendrodoa grossularia, as sister to P. aurata (fig. 3). Therefore, the subfamily Polyzoinae appeared as polyphyletic, with P. zorritensis being more closely related to solitary species than to other colonial styelids. The Polycarpa genus was also polyphyletic, with P. aurata being separated from other Polycarpa species. The distant phylogenetic position of P. zorritensis apart from the PB clade led us to further describe budding in this species.
Fig. 3.
Phylogenetic relationships between Stolidobranchia, with a focus on Styelidae, inferred from the REDUCED supermatrix (1,306 genes, 605,265 aa, 14.4% missing data) using the site-heterogeneous CAT + F81 + Γ4 model. All phylogenetic analyses in this study on both FULL and REDUCED data sets inferred the same topology with maximum bootstrap support and maximum posterior probabilities for all branches. Colonial species (capable of asexual reproduction via NED) are depicted in red, solitary species in black. The three other species trees obtained in this study are provided as supplementary figures 3–5, Supplementary Material online.
Novel Budding in P. zorritensis
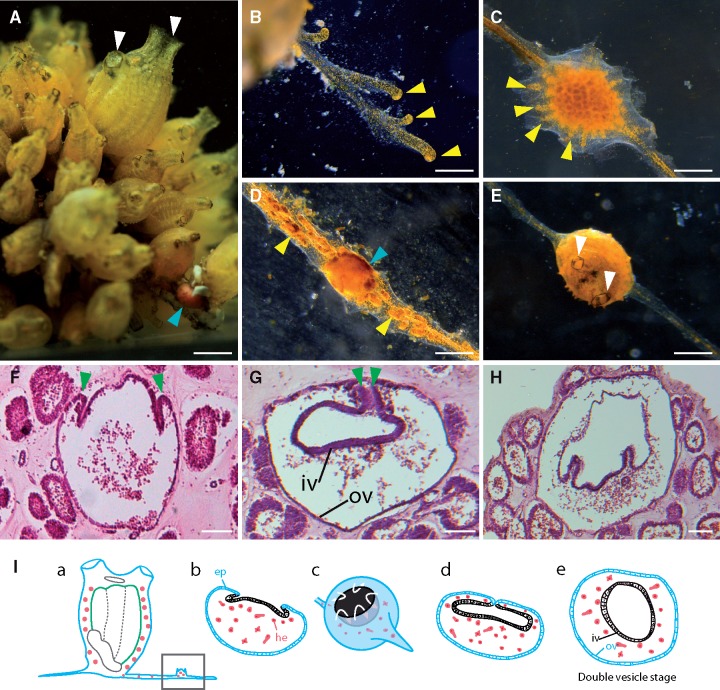
In a previous report, budding of P. zorritensis was described as vascular, involving “blood vessel wall and lymphocytes” (Brunetti and Mastrototaro 2004), but this was based on in vivo observations only. To gain more insights into the cellular processes leading to budding, we conducted histological observations of P. zorritensis budding. We observed that the budding process in this species differed from those known in other ascidians. After a few days in culture, the adult zooid emits stolons (fig. 4A, B) that consist of a single blood vessel bearing short blind ramifications, named ampullae, and surrounded by a thin layer of tunic (fig. 4B). Budding starts at several spots along each stolon, where ampullae become very numerous, short, and oriented in every direction, taking a flower-shape (fig. 4C). The growing bud appears in the center of these ramifications in form of a round vesicle (fig. 4D) and later develops into a new functional zooid (fig. 4E). Histological sections at successive stages showed that the vascular epidermis of the central stolon invaginates (fig. 4F), until invagination borders come into contact and fuse with each other (fig. 4G). This process results in the formation of an inner vesicle, separated from the outer vesicle by mesenchymal space. A few hours later, the inner vesicle shows distinct foldings (fig. 4H) and later organ primordia become visible. Figure 4I summarizes the budding process until the double vesicle stage.
Fig. 4.
Budding process in Polyandrocarpa zorritensis. (A) A colony of young adults of P. zorritensis, with a visible bud at the base of the colony (light blue arrowhead), white arrowheads: siphons. (B) A ramifying stolon, running onto the substrate (glass dish). (C) Budding site along a stolon, characterized by numerous ampullae oriented in every direction. (D) A new zooid in the process of forming (blue arrowhead), which externally appears as a round vesicle with no apparent organs. In (B)–(D) yellow arrowheads show ampullae. (E) A very young oozoid in which siphons recently opened (white arrowheads). (F–H) Histological sections of three successive budding stages. (F) Beginning of invagination. (G) Fusion of invaginating borders. Green arrowheads: invagination borders. (H) Beginning of organogenesis. (I) Diagram summarizing the process of budding: (a) A bud (squared) arises along a vascular stolon. (b) Initiation of invagination, the prospective inner vesicle is in black, whereas the stolon wall is in blue. (c) The same stage in a “3D” view, white arrows show the invagination movement. (d) Fusion of the inner invaginating borders. (e) Double-vesicle stage. ep: epidermis, he: hemocytes, iv: inner vesicle, ov: outer vesicle. Scale bars: (A) 2 mm, (B–E) 1 mm, (F–H) 25 µm.
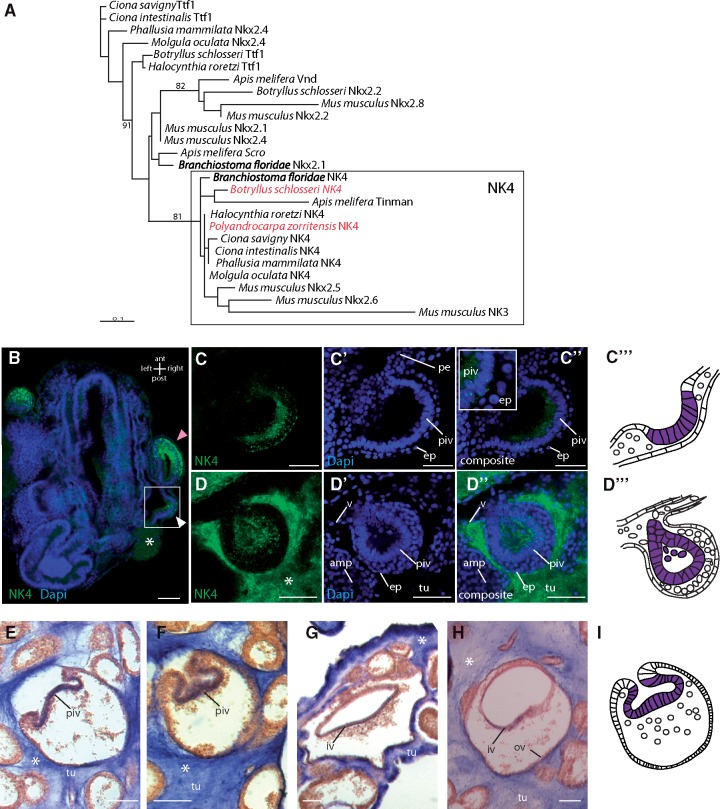
Expression of NK4 Delineates the Prospective Inner Vesicle in B. schlosseri and P. zorritensis
In order to find molecular markers of budding tissues across styelids, we studied the expression of seven candidate genes based on known expression in Botryllus (supplementary table 6, Supplementary Material online) and identified the homeobox containing NK4, having a similar expression pattern in the prospective territory of the future inner vesicle in both P. zorritensis and B. schlosseri (fig. 5). NK4 is the name given to the ascidian orthologue of NkX2.5/2.6/Tinman family of homeobox cardiac determinants (fig. 5A, Wang et al. 2013). During peribranchial budding in B. schlosseri this gene was first expressed in the thickened, evaginating peribranchial epithelium, neither beyond this region of budding nor in the surrounding epidermis (fig. 5B, C‴ and supplementary fig. 6, Supplementary Material online). When the inner vesicle was fully formed, NK4 became restricted to the anterior-right side of the budlet (fig. 5B). Similarly, during the earliest stages of vascular budding in B. schlosseri, NK4 was specifically expressed in the growing inner vesicle (fig. 5D–D‴). NK4 was not detected either in circulating cells or in the vascular epidermis of the ampullae (fig. 5D–D″), but because the tunic stains nonspecifically, we cannot rule out expression in the epidermal outer vesicle of the budlet. In P. zorritensis the earliest detected expression of NK4 began during epidermal invagination, and expanded throughout the whole invaginating region but not in the noninvaginating epithelium (fig. 5E). The expression persisted until the invagination borders fused to each other (fig. 5F–H). Thus, NK4 expression defines the onset of the budding epithelia in both B. schlosseri and P. zorritensis in three different budding modes.
Fig. 5.
Expression of the NK4 gene in Botryllus schlosseri and Polyandrocarpa zorritensis. (A) Phylogenetic analysis of Nkx2.5/2.6 family members showing the orthology between B. schlosseri and P. zorritensis NK4 genes (NK4 orthology group was defined as the smallest group including Botryllus NK4, Ciona NK4, and mouse NKx2.5 and 2.6). (B–C‴) NK4 expression in peribranchial bud of B. schlosseri. In (B) the zooid bears three buds at three different stages (white arrowhead shows the youngest one and pink arrowhead shows the oldest one). (C–C″) Close-up view of NK4 expression in the early peribranchial bud, insert in (C″) is highly magnified to show the unlabeled epidermis. (C‴) Diagram of NK4 expression in the peribranchial bud. (D–D″) Close-up view of NK4 expression in a vascular bud at the double vesicle stage. (D‴) Diagram of NK4 expression in the inner vesicle of a vascular bud. (E–I) Expression of NK4 in bud of P. zorritensis. (E) Early invagination. (F) Closure of invagination borders. (G) Right after closure. (H) Late double vesicle stage. (I) Diagram showing NK4 expression in the forming inner vesicle of P. zorritensis. Asterisks show nonspecific staining in the tunic, amp: ampullae. ep: epidermis, iv: inner vesicle, ov: outer vesicle, pe: peribranchial epithelium, piv: prospective inner vesicle, tu: tunic, v: vessel. Scale bars: (B) and (D–D″) 50 µm, (C–C″) 25 µm, (E–H) 25 µm.
Discussion
In this study, we generated 18 new transcriptomes of solitary and colonial ascidians belonging to the Styelidae family. We designed a bioinformatic pipeline that allowed us to conduct a phylogenomic analysis from heterogeneous sources of samples, including genomic and transcriptomic data. We provided a robust phylogeny of a family of chordates, which permitted us to trace the evolution of a complex character, that is, NED, and to highlight a previously undescribed mechanism of asexual development in colonial ascidians.
A Reference Set of Orthologous Genes for Tunicate Phylogeny
De novo transcriptome assembly often yields a high number of redundant contigs due to assembly artifacts, but also because of heterozygosity, isoforms, and polymorphism generated when several individuals are pooled together for RNA extraction (Cahais et al. 2012). Contaminations from other organisms, such as epibionts or micro and macroorganisms present in the ascidian pharynx, as well as poorly assembled rare transcripts, may also participate to increase the number of undesired contigs. In addition, some colonial ascidians form chimeric colonies (Kassmer et al. 2015), which adds another source of polymorphism to the transcriptome assembly. Reducing such redundancy and eliminating contaminations have proven essential for downstream analyses such as gene expression, functional enrichment, and orthology assessment (Davidson and Oshlack 2014; Philippe et al. 2017). In this paper, we designed an original procedure that was aimed at increasing transcriptome quality. The importance of good quality transcriptome assemblies only now is starting to be recognized (Cahais et al. 2012; Cabau et al. 2017) and therefore our new approach represents a straightforward solution to this problem with light computational demands. The merging step using CD-HIT-EST (fig. 2A) led to a drastic reduction of contig numbers (supplementary table 1, Supplementary Material online), showing that part of the redundancy comes from heterozygosity, assembly artifacts, or polymorphism of colonial species. Further drastic reduction of contig numbers was obtained from ORF selection, showing that many contigs assembled by Trinity had small ORF size (smaller than our threshold of 90 aa), possibly due to the inclusion of rare transcripts that split into multiple small ORFs.
Cross-contaminations in RNA-seq experiments are known to arise when RNA molecules from one sample accidentally pass to another sample, leading to a misattribution of sequencing reads to the contaminated sample (Ballenghien et al. 2017). Because of the Illumina sequencing depth, even extremely minor contamination events can be sequenced and result in unwanted assembled transcripts (Ballenghien et al. 2017; Simion et al. 2018). The level of cross-contamination was important in our samples, ranging between 0.07% and 24% of the contigs, depending on the species. Therefore removing cross-contaminants using recently developed CroCo software (Simion et al. 2018) turned out to be a crucial step. Interestingly, the level of cross-contamination was of the same order of magnitude regardless of whether RNA was extracted in the same or different laboratories, but it was much higher between samples sequenced on the same Illumina lane than between samples sequenced on different lanes performed at different dates (supplementary fig. 2, Supplementary Material online). These observations show that, even if all care is taken during tissue collection and RNA extractions, cross-contaminations mainly arise at the sequencing platform, when samples are handled for library preparation or Illumina sequencing. It is important to note that the amount of cross contaminants produced by sequencing facilities is drastically variable, as we observed an average of 0.2% of cross contaminants for samples handled by one company and an average of 7.3% of cross contaminants for samples handled by a different company (supplementary table 1, Supplementary Material online). Our study represents a typical case that highlights a recurrent problem that cannot be underestimated (Ballenghien et al. 2017; Simion et al. 2018), as it could produce artifacts not only for phylogenetic reconstruction, but also in any RNA-seq study.
Phylogenomics is believed to outperform single-gene phylogeny by adding a high number of genes that will supposedly convey sufficient phylogenetic signal (Philippe et al. 2011). However, if the genes selected for the data matrix are not conscientiously curated, a large amount of signal noise from sequences that deviate from the species tree can result in a fallacious phylogeny (Philippe et al. 2011). Careful selection of orthologous genes is therefore indispensable. The choice of the probabilistic model that best fits the data is also crucial in order to reduce systematic errors such as long-branch attraction between fast evolving taxa (Lartillot et al. 2007; Philippe et al. 2011). We built 4,908 alignments devoid of apparent paralogy by selecting single-copy genes thanks to a customized pipeline that was facilitated by genomic data available for eight ascidian species as well as by the elimination of spurious redundancy in the de novo assembled transcriptomes. All phylogenetic analyses conducted under site-homogeneous (LG) and site-heterogeneous (LG4X and CAT) models led to the same topology with maximum statistical support for all nodes. Therefore, the data set produced here turned out to be particularly appropriate to robustly recover phylogenetic relationships of Styelidae and relatives at the family level. It can thus be used as a reference set of orthologous genes in future tunicate phylogenetic studies. The REDUCED data set of 1,306 alignments shared by at least 24 species provided the same resolution and can be used as an alternative when the computational power is limited, for instance when using elaborated evolutionary models.
Evolutionary and Developmental Implications for Asexual Reproduction and Coloniality
A diverse range of budding modes has been reported across the whole subphylum of tunicates. In planktonic thaliaceans, buds arise from strobilation of stolons that contain tissues from all the three germ layers (Godeaux et al. 1998; Piette and Lemaire 2015). In phlebobranch ascidians such as Perophora viridis, buds arise from a stolonial vessel composed of an epidermis and containing a transverse mesenchymal septum (Freeman 1964; Kawamura et al. 2007). In aplousobranch ascidians, budding involves different kinds of tissues and mechanisms, from epithelial epicardium in Aplidium during abdominal strobilation, to mesenchymal septum during stolonial budding in Clavelina (Berrill 1950; Nakauchi 1982). NED mechanisms in different orders are so diverse that unifying principles are difficult to identify and study. Moreover, a full understanding of the evolution of asexual reproduction and regeneration in tunicates is limited by the current state of the tunicate phylogeny, in which the relationships among orders have only recently started to be resolved (Turon and López-Legentil 2004; Tsagkogeorga et al. 2009; Tatián et al. 2011; Shenkar et al. 2016; Kocot et al. 2018, Delsuc et al. 2018). However, focusing on one particular order or family allows one to decipher phylogenetic relationships between closely related solitary and colonial species and thus to reconstruct the evolution of NED at the intrafamily level (Shenkar et al. 2016).
In the phylogeny presented here, the solitary Styela and Asterocarpa form an early-diverging clade together with the families Pyuridae and Molgulidae, the latter being exclusively composed of solitary species. Therefore, the most parsimonious interpretation is that the last common ancestor of Styelidae was a solitary animal, and as a consequence NED and coloniality probably arose at least twice independently in Styelidae in two branches: 1) the monophyletic PB clade, that is, Polyzoinae + Botryllinae; and 2) P. zorritensis. An alternative interpretation of NED evolution, considering its acquisition in deeper nodes, can only be explained with multiple independent reversions to solitary lifestyle (see examples of different scenarios in supplementary fig. 7A, Supplementary Material online). In general, our topology of major Stolidobranch clades is in accordance with previously reported phylogenies that retrieved paraphyletic pyurids (Zeng et al. 2006; Tsagkogeorga et al. 2009; Shenkar et al. 2016). Within styelids, our result contrasts with previous phylogenies based on ribosomal and mitochondrial genes showing either a single acquisition (Zeng et al. 2006; Tsagkogeorga et al. 2009; Shenkar et al. 2016) or at least five independent acquisitions of coloniality (Pérez-Portela et al. 2009). However, these studies either included a limited number of colonial styelids, notably lacking P. zorritensis, or their topologies received poor statistical support.
In the phylogenomic tree resulting from this study, only members of Botryllinae are known to perform vascular budding within the PB clade (for colony growth or regeneration), while in Polyzoinea only peribranchial budding has been reported (Selys-Longchamps 1917; Berrill 1950; Abbott 1953; Watanabe and Tokioka 1972; Kawamura and Watanabe 1981). Thus the species tree implies that vascular budding is a Botryllinae innovation, and secondarily acquired in an ancestor that already possessed the peribranchial budding. Our understanding of budding evolution could be further improved in the future by including more colonial styelids to our phylogenomic data set. For instance, adding species of the genera Polyzoa (peribranchial budding), Metandrocarpa (peribranchial budding), and Symplegma (vascular/peribranchial budding).
The fact that budding tissues are of a different nature between P. zorritensis and the PB clade species reinforces the hypothesis that NED has been acquired convergently. Brunetti and Mastrototaro (2004) provided a general description of P. zorritensis and defined NED in this species as a form of vascular budding. However, a detailed morphological description was not provided. In the Stolidobranchia, only peribranchial budding and vascular budding have been extensively studied. Both these NEDs pass through a double vesicle stage (fig. 1G, H), where the outer vesicle in peribranchial budding derives from the parental epidermis and gives rise to the epidermis of the new zooid, and an inner vesicle, which will give rise to most of the internal organs, derives either from a portion of peribranchial epithelia (fig. 1G) or from population(s) of circulating cells (fig. 1H), in peribranchial and vascular budding respectively. The histological analyses of P. zorritensis budding show that the new zooids originate from the blood vessel epithelia but follow completely different ontogenetic trajectories, in which both the inner and the outer vesicles derive from the same tissue, namely the vascular epidermis (fig. 4). To our knowledge, this mode of budding has never been described in any other species.
Each of these three events of NED acquisition involved nonhomologous tissues for the formation of the inner vesicle (vascular epidermis, peribranchial epithelium and hemocytes, respectively) but it remains unknown whether budding is triggered by orthologous genes. The example of NK4 highlights the importance of having a robust phylogenetic framework to select relevant species for comparative studies but also to interpret gene expression patterns in an evolutionary perspective. NK4 is a highly conserved transcription factor involved in heart specification during ascidian embryogenesis (Wang et al. 2013, Prünster et al. unpublished). It also shows localized expression patterns in the anterior endoderm of larve and juveniles in Ciona intestinalis, and in the ventral ectoderm of larvae in both C. intestinalis and Molgula occidentalis (Christiaen et al. 2010; Stolfi et al. 2014). In B. schlosseri and P. zorritensis NK4 shows an additional domain of expression restricted to the tissues of the emerging bud, thus suggesting a putative new role in NED. By exploring the function of NK4 and by adding the description of other bud-specific molecular markers it will be possible to establish whether homologous programs are directing budding in different context and species, and to explore the plastic and modular nature of gene regulatory networks that are responsible for nonembryonic developmental processes (Ricci et al. 2016).
New Insights on Styelidae Systematics and Future Directions
Relationships of major Styelidae clades have been debated for more than a century. Berrill (1948) considered that all colonial species should be unified as a natural group because he assumed that they all perform peribranchial budding. In contrast Kott (1985), in her revision of Australian ascidians, suspected that budding modes may be more diverse than expected and proposed that “many species [of Polyandrocarpa] have a closer relationship with species of Polycarpa than they do with one another […]” and that “Accurate resolution of their taxonomy must await information on the process of vegetative reproduction, which may indicate their true phylogeny.” Our phylogenetic tree and the histological investigations on budding mode reconcile the views of Berrill and Kott. As Berrill suspected all peribranchial budding species form a monophyletic group, and as Kott suspected, the genus Polyandrocarpa is polyphyletic and P. zorritensis has an original budding mode.
The phylogenetic position of P. zorritensis is further supported by its gonad morphology, which is of the polycarpid type, that is, a large gonad containing numerous male and female lobules all together embedded in a common sac, also found in Polycarpa and Dendrodoa (Monniot and Monniot 1972; Monniot 2016). In contrast, other colonial styelids have either sex-separated gonads or small hermaphroditic gonads with no more than two testes (Monniot and Monniot 1972). It would be informative to incorporate into the existing phylogenomic matrix other Polyandrocarpa species having polycarpid gonad, e.g. Polyandrocarpa arianae (Monniot 2016), and to compare their budding mechanisms. Interestingly, our phylogenetic tree is also in agreement with the view that the acquisition of brooding predates the evolution of coloniality (Brown and Swalla 2012), and that internal brooding was secondarily lost in the oviparous P. misakiensis (see supplementary fig. 7B, Supplementary Material online for comparison between alternative scenarios). However, broader sampling of PB clade species would be necessary to better trace the evolution of this character.
Conclusions
Experimental approaches to study macroevolutionary transitions requires a priori definitions of homologous characters, but also the identification of the most suitable species in which to trace the evolution of such characters, as well as the knowledge of the phylogenetic relationships among these species. The phylogeny provided in this paper sets an initial framework to characterize the emergence and disappearance of other complex sets of characters, such as agametic asexual propagation and whole body regeneration. In this regard, the tools we generated already draw our attention to the plasticity of NED in an ascidian family and led to the discovery of an undescribed form of asexual development. Comparing the cellular and molecular mechanisms behind each form of NED can also lead to the discovery of conserved developmental modules that regulate regeneration and asexual development. The phylogenetic framework presented here needs further taxonomic sampling to fully understand the evolutionary transitions to NED, but we can already use these findings to trace and study other characters with known plastic evolution, such as larval photoreceptors (Sorrentino et al. 2001), adultation (Jeffery 2007) or brooding strategies.
Materials and Methods
Animal Collection and Identification
Sampling locations are provided in table 1. Brazilian collections were authorized by license No. 44082-1 of the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio). Before relaxation and fixation in menthol and 10% formalin, animals were kept for ∼12 h in running, grossly filtrated sea water, shortly after which they were fixed. Solitary species are not externally recognizable and need to be dissected, formaldehyde-fixed, and carefully examined. For this reason, a single individual cannot be used for both morphology-based identification and RNA extraction, since fresh tissues have to be frozen very rapidly to get good quality RNA. Therefore, we used the following strategy: when two individuals looked similar, one was dedicated to morphology-based identification and fixed in 4% formaldehyde, the other was used for RNA extraction and flash-frozen in liquid nitrogen. From both, a small piece of tissue was first removed and transferred in 80% ethanol in order to extract DNA and proceed to mitochondrial cytochrome oxidase subunit I (COI) and nuclear 18S rRNA barcoding. By this method, both individuals could be compared at the DNA level, allowing confirmation that the morphology-based identified specimen was of the same species as the frozen one, from which the transcriptome was to be sequenced. When COI and/or 18S rRNA sequences had been deposited in databases by others, morphology-based species determination was confirmed by sequence identity. Before flash-freezing specimens in liquid nitrogen, the tunic and digestive tract were removed in order to decrease the level of contaminating RNA. Concerning colonial species, three zooids or pieces of the same colony were fixed in ethanol, formaldehyde, and liquid nitrogen respectively. In some colonial species, the zooids were so small and so deeply embedded in the tunic that digestive tract and tunic could not be removed before fixation. In these cases, the animals were starved for 24 h and the tunic was cleaned by brush and forceps, as much as possible.
DNA Barcoding and Phylogenetic Analyses Based on COI and 18S rRNA
COI was amplified using primers modified from Folmer et al. (1994), Meyer (2003), and Schwendinger and Giribet (2005). Ribosomal 18S rRNA gene was amplified using 18S-TF and 18S-TR primers from Carreras-Carbonell et al. (2005). A list of primers used for each species is provided in supplementary table 5, Supplementary Material online. For Brazilian samples, DNA was extracted with DNeasy Blood & Tissue kit (Qiagen) following manufacturer’s instructions, and then quantified on a NanoDrop 2000 Spectrophotometer (Thermo Scientific). The PCR amplifications were carried out in 20 µl reactions with 0.15 µl GoTaq polymerase in supplied buffer (Promega), 10 mM dNTPs, and 10 mM of each primer. Cycling parameters were as follows: a 94 °C denaturation for 5 min, followed by 35 cycles of: 94 °C denaturation for 30 s, 45 °C annealing for COI or 66 °C for 18S for 50 s, 72 °C extension for 1 min, and a final extension at 72 °C for 5 min. The PCR products were labeled using BigDye Terminator ver. 3.1 Cycle Sequencing kit chemistry (Applied Biosystems), and sequenced on an Applied Biosystems 3730 DNA sequencer. For French samples, DNA was extracted as follows: tissues was first digested by 200 µg/ml Proteinase K at 37 °C for 30 min then mashed with a sterile tip; nucleic acids were purified by phenol/chloroform/isoamyl alcohol (25:24:1), precipitated in sodium chloride 0.2 M and finally quantified on a NanoDrop 2000 Spectrophotometer (Thermo Scientific). Cycling parameters were the same as in Brazil and France. Successful amplifications were sequenced on an ABI 3730 at Genewiz, Inc. (South Plainfield, NJ, USA). New COI and 18S rRNA sequences have were in GenBank (supplementary table 5, Supplementary Material online for accession numbers).
All sequences were aligned and trimmed using Geneious R7 (Biomatters LTD). We aligned the sequences with those from Pérez-Portela et al. (2009) and some available on Genbank (NCBI; see supplementary table 5, Supplementary Material online for accession numbers) using CLUSTALW (Higgins et al. 1996) and trimmed and confirmed by eye. The 18S rRNA alignment was 801 bp and the COI alignment was 679 bp. As in Pérez-Portela et al. (2009), we concatenated the COI and 18S rRNA sequences. Two partitions were specified, one for each gene. We conducted a Maximum Likelihood analysis using the program RAxML v.8 (Stamatakis 2014) with nucleotide substitution model GTR + G + I, and run 100 bootstrap replicates on our alignment of 100 sequences.
RNA Isolation and cDNA Sequencing
Information on tissue used for RNA extraction is provided in table 1. For the species collected in Roscoff, total RNA was extracted in the laboratory of S.Tiozzo (France), using and NucleoSpin RNA II kit (Macherey-Nagel), including a DNAase treatment, and following manufacturer’s instructions. For P. misakiensis, RNA was extracted in the laboratory of K. Kawamura (Japan), using NucleoSpin RNA kit (Takara-Clontech). For species collected in Brazil, RNA was extracted in the laboratory of F. Brown (Brazil) using a NucleoSpin RNA Plus kit (Macherey-Nagel). Total RNA were sent to Beijing Genome Institute (BGI, China) where cDNA libraries were prepared before sequencing. For species collected in Banyuls, RNA was extracted in the laboratory of S. Darras (France) using Trizol (Life Technologies), followed by purification using the RNeasy kit (Qiagen) with a DNase treatment. cDNA preparation and sequencing were performed by Sistemas Genomicos (Spain). RNA-seq data were obtained using Illumina HiSeq 4000 technology. The exact number of reads per species is given in table 1.
De Novo Transcriptome Assembly
For all the species for which de novo transcriptomes were assembled, we applied the following eight-step pipeline: 1) Illumina reads were clipped and trimmed to eliminate low quality regions using Trimmomatic v. 0.35 (Bolger et al. 2014) with the following parameters: PE reads_1.fq reads_2.fq ILLUMINACLIP: adapters/TruSeq3-PE.fa: 2: 30: 10 LEADING: 36 TRAILING: 36 SLIDINGWINDOW: 4: 34 MINLEN: 100 and then sequence quality was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/; last accessed April 2018); 2) reads were assembled with Trinity v. 2.4.0 (Grabherr et al. 2011) using default parameters; 3) open reading frames (ORF’s) of a minimum of 90 amino acids were kept using Transdecoder v. 3.0.1 (https://github.com/TransDecoder/TransDecoder/wiki; last accessed April 2018); 4) very similar ORF’s (e.g., putative isoforms or alleles) were collapsed using CD-HIT-EST v4.6 (Fu et al. 2012) with default parameters; 5) reads were then mapped back onto the remaining contigs using Kallisto v. 0.42.4 (Bray et al. 2016) with default parameters; 6) density plots were drawn with R, using hist() and density() functions. On the basis of these plots, contigs with low expression levels were eliminated. The threshold above which contigs were kept corresponds to the lower peak of the density plot; 7) exhaustiveness was assessed by running BUSCO (Simão et al. 2015) against metazoan database; 8) cross-contaminations were removed using CroCo (Simion et al. 2018). Number of contigs at each step for each species is given in supplementary table 1, Supplementary Material online.
Data Availability
The 18 de novo assembled transcriptomes can be downloaded from https://github.com/AlexAlie/styelidae; last accessed April 2018. Raw Illumina reads obtained in this study can be retrieved from NCBI BioProject PRJNA422120. Accession numbers or Illumina reads from B. schlosseri (Rodriguez et al. 2014) and B. leachii (Zondag et al. 2016) are provided in table 1.
H. roretzi and H. aurantium gene prediction are accessible via ANISEED (https://www.aniseed.cnrs.fr/aniseed/download/download_data; last accessed April 2018). Molgula oculata and M. occidentalis transcripts were retrieved from ANISEED. C. robusta data can be downloaded from http://ghost.zool.kyoto-u.ac.jp/download_kh.html, and C. savignyi data from Ensembl (ftp://ftp.ensembl.org/pub/release-91/fasta/ciona_savignyi/cdna/).
Construction of a New Set of Orthologous Alignments
We selected 15 chordate species for which genomic data were available (i.e., six tunicates, six vertebrates, one cephalochordate, one echinoderm, and one hemichordate, see supplementary table 3, Supplementary Material online), and we retained only the longest transcript for each gene of their corresponding proteomes. These proteomes were further dereplicated using CD-HIT (-c 1.0; Fu et al. 2012) and sequences <30 amino acids were removed. These 15 proteomes were clustered using OrthoMCL (-I 1.3, Li et al. 2003). We later added three new proteomes from tunicate species for which genomic data became available (supplementary table 3, Supplementary Material online) into the clusters using Forty 2 (https://bitbucket.org/dbaurain/42/). The few sequences that were simultaneously added to different clusters were discarded, except in the cluster in which taxonomic diversity was the highest. We discarded all resulting clusters that did not possess at least: 1) two tunicate sequences or 2) one tunicate and one vertebrate sequence, retaining 14,552 clusters.
Gene Augmentation, Selection, and Concatenation
Transcriptomic data from 18 stolidobranchs including 16 styelids (table 1) were added to the 14,552 clusters of orthologous sequences using Forty 2. Clusters that contained apparent paralogy for at least one tunicate species among those that correspond to the genomic data set (i.e., species belonging to the Ciona, Phallusia, Molgula, and Halocynthia genera) or which contained <10 tunicate species or which contained >80 sequences were discarded. All nontunicate sequences were removed from the remaining 8,187 clusters in order to improve the accuracy of the subsequent aligning step, a. All clusters were aligned using MAFFT (–maxiterate 1000 –localpair, Katoh and Standley 2013). For every alignment, nonoverlapping sequences were fused as they most likely correspond to sequence fragments from the same transcript. We used an extended definition of a “nonoverlap” between two sequences: if actually overlapping, the total overlap is smaller than 200 amino acids and the two sequences differ by less than five amino acid positions. This allowed us to concatenate 64,340 fragmented transcripts (out of the initial 219,099 transcripts) into 28,188 longer ones. We then removed 4,631 low-quality transcripts (i.e., that had >70% missing data and were shorter than 100 amino acids). Alignments were then checked again for apparent paralogy, this time for all species, and we only kept the 4,908 alignments that showed no paralogy. We also selected alignments containing at least 24 species (out of the 26 total species) to create a smaller and more complete phylogenomic data set of 1,306 alignments. These alignments were concatenated into supermatrices using ScaFos (Roure et al. 2007), available on https://github.com/AlexAlie/styelidae; last accessed April 2018.
Functional annotation was done through the DAVID online service (Huang et al. 2009).
Phylogenetic Analyses
The assembled supermatrices as explained above were analyzed using several models of sequence evolution to assess the relative reliability of the different species to infer the phylogenies we might infer. The FULL supermatrix was analyzed under the Maximum Likelihood framework using the site-homogeneous model of sequence evolution LG + Γ4 + F implemented in IQ-TREE (Nguyen et al. 2015) and node support was estimated with 100 bootstrap replicates. The REDUCED supermatrix was analyzed with the exact same approach, as well as using the site-heterogeneous LG4X + R4 + F sequence evolution model (also in IQ-TREE). Lastly, we also analyzed the REDUCED supermatrix under a Bayesian framework using the site-heterogeneous CAT + F81+Γ4 model implemented in PhyloBayes (Lartillot et al. 2013). We ran two independent MCMC chains until the two topologies obtained perfectly converged (maxdiff = 0 with a 50% burnin).
In Situ Hybridization
In situ hybridization on B. schlosseri was performed as in Ricci et al. (2016). In situ hybridization on P. zorritensis was performed on paraffin sections as follows. Samples at the desired stage were fixed in paraformaldehyde (PFA) 4%, DMSO 1%, DEPC 0.2%, RNase inhibitor (Sigma, R7397-30ML) 1/500 (=thereafter 1XPR), in Phosphate Buffer Saline (PBS 1×), then washed twice in PBS 1× and then dehydrated by graded methanol bath of 15 min (from 25% to 100% in PBS + 1XPR), then samples were immediately processed or stored at -20 °C. For paraffin embedding, samples were washed 2 × 10 min in butanol 1, then 1 h in butanol 1/paraffin at 60 °C, then overnight in paraffin at 60 °C. Paraffin blocks were allowed to solidify 8 h at room temperature and then were stored at 4 °C no more than two days. Paraffin sections were performed on Leica Microtome, at a thickness of 15 µm and attached on SuperFrost glass slides and allowed to dry overnight at 37 °C. All tools were previously washed with RNAse decontamination solution. Then in situ hybridizations were performed as follows: Slides were dried at 55 °C for 30 min, followed by two baths of xylene of 10 min each, then rehydrated by graded ethanol baths and then washed 2 × 10 min in a solution PBST (PBS 1× + 0.1% Tween 20) + 1XPR, for 5 min. Slides were then treated with Proteinase K (2.5 ng/µl in PBST + RNAse inhibitor) for 10 min at 37 °C, and immediately rinsed in PBST + 1XPR, and bath for 2 min in PBST. Then they were postfixed in PFA 4% + glutaraldehyde 0.2% in PBST + 1XPR for 30 min at room temperature, and rinsed in in PBST + 1XPR 3 × 5 min. Slides were pre hybridized in buffer (Formamide deionized 65%; 5× SSC; 1× denhardt’s solution; 0.1% tween 20; 5 mg/ml torula yeast RNA; 50 µg/ml heparine; 1% Dextran sulfate) for 1 h, after what probes at a final dilution of 1.5–1.7 ng/µl in 300 µl of hybridization buffer, for 44 or 68 h at 58 °C. Chambers of hybridization (FRAME-SEAL BIORAD SLF 3001) were applied to the slides who accepted 300 µl of probe dilution. The slides were rinsed in 2× SSC pH 4.5, 50% Formamide, 0.1% Tween 20 + 1XPR, 3 × 30 min at 58 °C, then rinsed in PBST + PR 2 × 5 min and in MABT (MAB 1× + 0.1% Tween 20) + RNase inhibitor 1× 2 × 5 min (each bath). Then the slides were blocked in MABT, 2% Blocking solution, 10% goat serum for 1 h at RT. 500 µl of antidig-AP (1/2,000 in MABT, 2% blocking solution, 10% goat serum) was deposited directly on the slides overnight at 4 °C. Slides were rinsed in MABT 2 × 5 min, then rinsed TMNT filtered (tris–HCl ph 9.5 100 mM, NaCl 100 mM, MgCl2 50 mM, Levamisole 10 mM, 0.1% Tween 20) 2 × 5 min. Then NBT/BCIP in TMNT was directly applied on the slides until the coloration developed. The reaction was stopped by 2 × 5 min rinse in PBST. Then slides were dehydrated in successive baths of ethanol (50%, 70%, 90%, 100%), 3 min each, and finally in xylene for 15 min and mounted in Permount medium.
P. zorritensis Husbandry and Histology
Colonies of P. zorritensis were collected in Taranto and La Spezia (Italy), fixed on glass slides and cultured in a circulating sea water system at 24 °C. The developing buds were fixed in 4% PFA in phosphate-buffered saline (PBS, pH 7.4) over night at 4 °C, dehydrated in a graded series of ethanol and butanol and then included in paraffin wax. Transversal section (5–6 µm think) were cut using a hand operated microtome and stained with hematoxylin and eosin (H&E) following this protocol: stain for 3 min in Mayer’s hematoxylin (Hematoxylin Solution, Mayer’s; Sigma–Aldrich), wash for 10 min in running tap water, stain for 15 s in Eosin Y solution (1 g Eosin Y powder in 20 ml Mq water and 80 ml 95% EtOH), rinse in 95% EtOH, dehydrated and mounted.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We would like to thank Philippe Dru and Faisal Bekkouche for help with bioinformatics resources, Laurent Gilletta, Loann Gissat, Regis Lasbleiz, Sophie Collet, and Camille Lejeune for setting up the aqua-culture systems. Françoise Monniot for advice on taxonomy and sample identification. Sophie Booker, Elodie Bourrigaud, Laurent Lévêque, Ronan Garnier, Francesco Mastrototaro, and the Service Mer of Banyuls-sur-mer, EMBRC-France for sample collection. We thank Martine Fioroni for providing literature and old manuscripts that were difficult to obtain. This work was supported by FAPESP Jovem Pesquisador grant (JP 2015/50164-5) to F.B., ANR grant (ANR-14-CE02-0019-01) to S.T., FAPESP postdoctoral fellowship (grant number 2015/14052-8) to L.H., ANR grant (ANR-13-BSV2-0011-01) to P.L. and C.D.; ISEM 2018-057-SUD to P.D. and E.J.P.D., ANR grant (ANR-11-JSV2-007-01) to S.D.
Author Contributions
A.A., L.H., F.B., and S.T. conceived and designed the study, collected and identified biological material and wrote the manuscript. A.A., L.H., and P.S. designed the assembly and clustering pipeline and performed sequence analyses. P.S. conducted orthology and phylogenomic analyses. A.A. and M.S. established P. zorritensis culture and conducted histological analyses. M.M.P. conducted NK4 in situ hybridization on B. schlosseri. A.A. and L.H. conducted DNA barcoding and phylogenetic analysis. S.L. performed NK4 in situ hybridization on P. zorritensis. E.D. and F.D. participated in the pipeline design and supported part of the transcriptome sequencing. P.L. and C.D. sequenced genomes and provided gene models. S.D. and K.K. provided RNA for several species and participated to species identification. All authors read and approved the final version of the manuscript.
References
- Abbott D. 1953. Asexual reproduction in the colonial ascidian Metandrocarpa taylori. Univ. Calif. publ. zool. 61(1):1–78. [Google Scholar]
- Agata K, Saito Y, Nakajima E.. 2007. Unifying principles of regeneration I: epimorphosis versus morphallaxis. Dev Growth Differ. 492:73–78. [DOI] [PubMed] [Google Scholar]
- Alié A, Hayashi T, Sugimura I, Manuel M, Sugano W, Mano A, Satoh N, Agata K, Funayama N.. 2015. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci U S A. 11251:E7093–E7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballenghien M, Faivre N, Galtier N.. 2017. Patterns of cross-contamination in a multispecies population genomic project: detection, quantification, impact, and solutions. BMC Biol. 151:25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeitos MS, Romano SL, Lasker HR.. 2010. Repeated loss of coloniality and symbiosis in scleractinian corals. Proc Natl Acad Sci U S A. 10726:11877–11882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality. Berkeley: University of California Press. [Google Scholar]
- Bely AE, Nyberg KG.. 2010. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol. 253:161–170. [DOI] [PubMed] [Google Scholar]
- Berrill NJ. 1935. Studies in Tunicate development Part IV – Asexual reproduction. Proc Philos Trans Roy Soc B. 225526:327–379. [Google Scholar]
- Berrill NJ. 1948. The gonads, larvae, and budding of the polystyelid ascidians Stolonica and Distomus. J Mar Biol Assoc UK. 2703:633–650. [Google Scholar]
- Berrill NJ. 1950. The Tunicata, with an account of the British species. Vol. 133 London: The Ray Society; p. 354. [Google Scholar]
- Boardman RS, Cheetham AH, Oliver WA.. 1973. Animal colonies. Development and function through time. Stroudsburg (PA: ): Dowden, Hutchinson & Ross. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 3015:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG. 2007. Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol. 3032:421–433. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 345:525–527. [DOI] [PubMed] [Google Scholar]
- Brown FD, Swalla BJ.. 2012. Evolution and development of budding by stem cells: ascidian coloniality as a case study. Dev Biol. 3692:151–162. [DOI] [PubMed] [Google Scholar]
- Brunetti R, Mastrototaro F.. 2004. The non-indigenous stolidobranch ascidian Polyandrocarpa zorritensis in the Mediterranean: description, larval morphology and pattern of vascular budding. Zootaxa 5281:1–8. [Google Scholar]
- Brusca RC, Moore W, Shuster SM.. 2016. Invertebrates. Sunderland (MA: ): Sinauer Associates, Inc., Publishers. [Google Scholar]
- Burgess SC, Ryan WH, Blackstone NW, Edmunds PJ, Hoogenboom MO, Levitan DR, Wulff JL.. 2017. Metabolic scaling in modular animals. Invertebr Biol. 1364:456–472. [Google Scholar]
- Cabau C, Escudié F, Djari A, Guiguen Y, Bobe J, Klopp C.. 2017. Compacting and correcting Trinity and Oases RNA-Seq de novo assemblies. PeerJ 5:e2988.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahais V, Gayral P, Tsagkogeorga G, Melo-Ferreira J, Ballenghien M, Weinert L, Chiari Y, Belkhir K, Ranwez V, Galtier N.. 2012. Reference-free transcriptome assembly in non-model animals from next-generation sequencing data. Mol Ecol Resour. 125:834–845. [DOI] [PubMed] [Google Scholar]
- Carlson BM. 2007. Principles of regenerative biology. Burlington (MA: ): Academic Press. [Google Scholar]
- Carreras-Carbonell J, Macpherson E, Pascual M.. 2005. Rapid radiation and cryptic speciation in Mediterranean triplefin blennies (Pisces: tripterygiidae) combining multiple genes. Mol Phylogenet Evol. 373:751–761. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Levine M.. 2010. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev Biol. 3402:179–187. [DOI] [PubMed] [Google Scholar]
- Davidson NM, Oshlack A.. 2014. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 157:410.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM.. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 2985601:2157–2167. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H.. 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 4397079:965–968. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Philippe H, Tsagkogeorga G, Simion P, Tilak MK, Turon X et al. , . 2018. A phylogenomic framework and timescale for comparative studies of tunicates. BMC Biol. 161:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R.. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 35:294–299. [PubMed] [Google Scholar]
- Freeman G. 1964. The role of blood cells in the process of asexual reproduction in the tunicate Perophora viridis. J Exp Zool. 156:157–184. [DOI] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. 2012. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics 2823:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Isozaki T, Mori K, Kawamura K.. 2011. Expression and function of myc during asexual reproduction of the budding ascidian Polyandrocarpa misakiensis. Dev Growth Differ. 539:1004–1014. [DOI] [PubMed] [Google Scholar]
- Funayama N, Nakatsukasa M, Mohri K, Masuda Y, Agata K.. 2010. Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol Dev. 123:275–287. [DOI] [PubMed] [Google Scholar]
- Galliot B, Chera S.. 2010. The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 209:514–523. [DOI] [PubMed] [Google Scholar]
- Geller JB, Fitzgerald LJ, King CE.. 2005. Fission in sea anemones: integrative studies of life cycle evolution. Integr Comp Biol. 454:615–622. [DOI] [PubMed] [Google Scholar]
- Godeaux J, Bone Q, Braconnot JC.. 1998. Anatomy of Thaliaceans In: Bone Q, editor. The biology of pelagic tunicates. Oxford, New York, Tokyo (Japan: ): Oxford University Press; p. 1–24. [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al. , . 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 297:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A.. 2010. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol. 3471:24–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ.. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383–402. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 41:44–57. [DOI] [PubMed] [Google Scholar]
- Hughes RN. 1989. A functional biology of clonal animals. London, New York: Chapman and Hall; p. 331 [Google Scholar]
- Hughes RN, Cancino JM.. 1985. An ecological overview of cloning in Metazoa In: Jackson JBC, Buss LW, Cook RE, editors. Population biology and evolution of clonal organisms. New Haven (CT: ): Yale University Press; p. 153–186. [Google Scholar]
- Jackson JBC, Coates AG.. 1986. Life cycles and evolution of clonal (modular) animals. Philos Trans Roy Soc B. 3131159:7–22. [Google Scholar]
- Jackson JBC. 1977. Competition on marine hard substrata: the adaptive significance of solitary and colonial strategies. Am Nat. 111980:743–767. [Google Scholar]
- Jeffery WR. 2007. Chordate ancestry of the neural crest: new insights from ascidians. Semin Cell Dev Biol. 184:481–491. [DOI] [PubMed] [Google Scholar]
- Kassmer SH, Rodriguez D, Langenbacher AD, Bui C, De Tomaso AW.. 2015. Migration of germline progenitor cells is directed by sphingosine-1-phosphate signalling in a basal chordate. Nat Commun. 61:8565.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 304:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Sugino Y, Sunanaga T, Fujiwara S.. 2007. Multipotent epithelial cells in the process of regeneration and asexual reproduction in colonial tunicates. Dev Growth Differ. 501:1–11. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Sunanaga T.. 2010. Hemoblasts in colonial tunicates: are they stem cells or tissue-restricted progenitor cells? Dev Growth Differ. 521:69–76. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Watanabe H.. 1981. Studies of Japanese compound styelid ascidians. III. A new, possibly asexual Polyandrocarpa from Shimoda Bay. Publ Seto Mar Biol Lab. XXVI(4/6):425–436. [Google Scholar]
- Kocot KM, Tassia MG, Halanych KM, Swalla BJ.. 2018. Phylogenomics offers resolution of major tunicate relationships. Mol Phylogenet Evol. 121:166–173. [DOI] [PubMed] [Google Scholar]
- Kott P. 1985. The Australian Ascidiacea. Part 1, Phlebobranchia and Stolidobranchia. Mem Qd Mus. 21:1–440. [Google Scholar]
- Kürn U, Rendulic S, Tiozzo S, Lauzon RJ.. 2011. Asexual propagation and regeneration in colonial ascidians. Biol Bull. 2211:43–61. [DOI] [PubMed] [Google Scholar]
- Laird DJ, De Tomaso AW, Weissman IL.. 2005. Stem cells are units of natural selection in a colonial ascidian. Cell 1237:1351.. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H.. 2007. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 7(Suppl 1):S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J.. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 624:611–615. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 139:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni L, Gasparini F, Hotta K, Ishizuka KJ, Ricci L, Tiozzo S, Voskoboynik A, Dauga D.. 2014. Ontology for the asexual development and anatomy of the colonial chordate Botryllus schlosseri. PLoS ONE. 95:e96434.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VG, Menger GJIII, Zoran MJ.. 2005. Regeneration and asexual reproduction share common molecular changes: upregulation of a neural glycoepitope during morphallaxis in Lumbriculus. Mech Dev. 1225:721–732. [DOI] [PubMed] [Google Scholar]
- Meyer CP. 2003. Molecular systematics of cowries (Gastropoda: cypraeidae) and diversification patterns in the tropics. Biol J Linnean Soc. 793:401–459. [Google Scholar]
- Monniot F. 2016. A new species of Polyandrocarpa (Ascidiacea, Styelidae) in the Mediterranean Sea. Zootaxa 41321:87–096. [DOI] [PubMed] [Google Scholar]
- Monniot C, Monniot F.. 1972. Clé mondiale des genres d’Ascidies. Key to Ascidian Genera of the world. Ach Zool Exp Gén. 113:311–367. [Google Scholar]
- Nakauchi M. 1982. Asexual development of ascidians: its biological significance, diversity, and morphogenesis. Am Zool. 224:753–763. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 321:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson Sköld H, Obst M, Sköld M, Åkesson B.. 2009. Stem cells in asexual reproduction of marine invertebrates In: Rinkevich B, Matranga V. editors. Stem cells in marine organisms. Dordrecht (the Netherlands: ): Springer; p. 105. [Google Scholar]
- Nogueira AF, Costa CM, Lorena J, Moreira RN, Frota-Lima GN, Furtado C, Robinson M, Amemiya CT, Darnet S, Schneider I.. 2016. Tetrapod limb and sarcopterygian fin regeneration share a core genetic programme. Nat Commun. 7:13364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Portela R, Bishop J, Davis A, Turon X.. 2009. Phylogeny of the families Pyuridae and Styelidae (Stolidobranchiata, Ascidiacea) inferred from mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol. 503:560–570. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DT, Manuel M, Wörheide G, Baurain D.. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 93:e1000602.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Vienne de DM, Ranwez V, Roure B, Baurain D, Delsuc F.. 2017. Pitfalls in supermatrix phylogenomics. Eur J Tax. 283:1–25. [Google Scholar]
- Piette J, Lemaire P.. 2015. Thaliaceans, the neglected pelagic relatives of ascidians: a developmental and evolutionary enigma. Q Rev Biol. 902:117–145. [DOI] [PubMed] [Google Scholar]
- Ricci L, Cabrera F, Lotito S, Tiozzo S.. 2016. Redeployment of germ layers related TFs shows regionalized expression during two non-embryonic developments. Dev Biol. 4161:235–248. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Rosner A, Rabinowitz C, Lapidot Z, Moiseeva E, Rinkevich B.. 2010. Piwi positive cells that line the vasculature epithelium, underlie whole body regeneration in a basal chordate. Dev Biol. 3451:94–104. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Sanders EN, Farell K, Langenbacher AD, Taketa DA, Hopper MR, Kennedy M, Gracey A, De Tomaso AW.. 2014. Analysis of the basal chordate Botryllus schlosseri reveals a set of genes associated with fertility. BMC Genomics. 15:1183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H.. 2007. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol Biol. 7(Suppl 1):S2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado AS. 2000. Regeneration in the metazoans: why does it happen? Bioessays 226:578.. [DOI] [PubMed] [Google Scholar]
- Sanchez-Alvarado A, Yamanaka S.. 2014. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 1571:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendinger P, Giribet G.. 2005. The systematics of south—east Asian genus Fangensis Rambla (Opiliones: cyphophthalmi: stylocellidae). Invertebr Syst. 194:297–323. [Google Scholar]
- Selys-Longchamps MD. 1917. Sur le bourgeonnement des Polystiélinés Stolonica et Heterocarpa avec quelques notes sur l’anatomie de ces deux genres. Bull Sci France Belg, L(3): 170–276. [Google Scholar]
- Shenkar N, Koplovitz G, Dray L, Gissi C, Huchon D.. 2016. Back to solitude: solving the phylogenetic position of the Diazonidae using molecular and developmental characters. Mol Phylogenet Evol. 100:51–56. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 3119:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simion P, Belkhir K, François C, Veyssier J, Rink JC, Manuel M, Philippe H, Telford MJ.. 2018. A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 161:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino M, Manni L, Lane NJ, Burighel P.. 2001. Evolution of cerebral vesicles and their sensory organs in an ascidian larva. Acta Zool. 813:243–258. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolfi A, Lowe EK, Racioppi C, Ristoratore F, Brown CT, Swalla BJ, Christiaen L.. 2014. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. Elife 3:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatián M, Lagger C, Demarchi M, Mattoni C.. 2011. Molecular phylogeny endorses the relationship between carnivorous and filter-feeding tunicates (Tunicata, Ascidiacea). Zool Scr. 406:603–612. [Google Scholar]
- Tiozzo S, Copley R.. 2015. Reconsidering regeneration in metazoans: an evo-devo approach. Front Ecol Evol. 3. doi:10.3389/fevo.2015.00067. [Google Scholar]
- Tsagkogeorga G, Turon X, Hopcroft RR, Tilak MK, Feldstein T, Shenkar N, Loya Y, Huchon D, Douzery EJP, Delsuc F.. 2009. An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol Biol. 9:187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turon X, López-Legentil S.. 2004. Ascidian molecular phylogeny inferred from mtDNA data with emphasis on the Aplousobranchiata. Mol Phylogenet Evol. 332:309–320. [DOI] [PubMed] [Google Scholar]
- Velandia-Huerto CA, Gittenberger AA, Brown FD, Stadler PF, Bermúdez-Santana CI.. 2016. Automated detection of ncRNAs in the draft genome sequence of a colonial tunicate: the carpet sea squirt Didemnum vexillum. BMC Genomics. 17:691.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, Neff NF, Sahoo D, Newman AM, Pushkarev D, Koh W, Passarelli B, Fan HC, Mantalas GL, Palmeri KJ et al. , . 2013. The genome sequence of the colonial chordate, Botryllus schlosseri. Elife. 2:00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Razy-Krajka F, Siu E, Ketcham A, Christiaen L.. 2013. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 1112:e1001725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Tokioka T.. 1972. Two new species and one possibly new race of social styelids from Sagami Bay with remarks on their life history especially the mode of budding. Publ Seto Mar Biol Lab. 195:327–345. [Google Scholar]
- West SA, Lively CM, Read AF.. 1999. A pluralist approach to sex and recombination. J Evol Biol. 126:1003–1012. [Google Scholar]
- White MJD. 1978. Modes of speciation. San Francisco (CA: ): W.H. Freeman. [Google Scholar]
- WoRMS Editorial Board. 2017. World Register of Marine Species [accessed 2017 December 07]. Available from: http://www.marinespecies.org at VLIZ. doi:10.14284/170.
- Zattara EE, Bely AE.. 2016. Phylogenetic distribution of regeneration and asexual reproduction in Annelida: regeneration is ancestral and fission evolves in regenerative clades. Invertebr Biol. 1354:400–415. [Google Scholar]
- Zeng L, Jacobs MW, Swalla BJ.. 2006. Coloniality has evolved once in Stolidobranch Ascidians. Integr Comp Biol. 463:255–268. [DOI] [PubMed] [Google Scholar]
- Zondag LE, Rutherford K, Gemmell NJ, Wilson MJ.. 2016. Uncovering the pathways underlying whole body regeneration in a chordate model, Botrylloides leachi using de novo transcriptome analysis. BMC Genomics. 171:114.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 18 de novo assembled transcriptomes can be downloaded from https://github.com/AlexAlie/styelidae; last accessed April 2018. Raw Illumina reads obtained in this study can be retrieved from NCBI BioProject PRJNA422120. Accession numbers or Illumina reads from B. schlosseri (Rodriguez et al. 2014) and B. leachii (Zondag et al. 2016) are provided in table 1.
H. roretzi and H. aurantium gene prediction are accessible via ANISEED (https://www.aniseed.cnrs.fr/aniseed/download/download_data; last accessed April 2018). Molgula oculata and M. occidentalis transcripts were retrieved from ANISEED. C. robusta data can be downloaded from http://ghost.zool.kyoto-u.ac.jp/download_kh.html, and C. savignyi data from Ensembl (ftp://ftp.ensembl.org/pub/release-91/fasta/ciona_savignyi/cdna/).