Abstract
C4 photosynthesis has evolved repeatedly from the ancestral C3 state to generate a carbon concentrating mechanism that increases photosynthetic efficiency. This specialized form of photosynthesis is particularly common in the PACMAD clade of grasses, and is used by many of the world’s most productive crops. The C4 cycle is accomplished through cell-type-specific accumulation of enzymes but cis-elements and transcription factors controlling C4 photosynthesis remain largely unknown. Using the NADP-Malic Enzyme (NADP-ME) gene as a model we tested whether mechanisms impacting on transcription in C4 plants evolved from ancestral components found in C3 species. Two basic Helix-Loop-Helix (bHLH) transcription factors, ZmbHLH128 and ZmbHLH129, were shown to bind the C4NADP-ME promoter from maize. These proteins form heterodimers and ZmbHLH129 impairs trans-activation by ZmbHLH128. Electrophoretic mobility shift assays indicate that a pair of cis-elements separated by a seven base pair spacer synergistically bind either ZmbHLH128 or ZmbHLH129. This pair of cis-elements is found in both C3 and C4 Panicoid grass species of the PACMAD clade. Our analysis is consistent with this cis-element pair originating from a single motif present in the ancestral C3 state. We conclude that C4 photosynthesis has co-opted an ancient C3 regulatory code built on G-box recognition by bHLH to regulate the NADP-ME gene. More broadly, our findings also contribute to the understanding of gene regulatory networks controlling C4 photosynthesis.
Keywords: basic Helix-Loop-Helix, cis-element evolution, C3 and C4 photosynthesis, NADP-Malic Enzyme, PACMAD Panicoid grasses
Introduction
C3 plants inherited a carbon fixation system developed by photosynthetic bacteria, with atmospheric carbon dioxide (CO2) being incorporated into ribulose-1, 5-bisphosphate (RuBP) by the enzyme Ribulose Bisphosphate Carboxylase/Oxygenase (RuBisCO) to form the three-carbon compound (C3) 3-phosphoglycerate (Calvin and Massini 1952). However, RuBisCO can also catalyse oxygenation of RuBP, which leads to the production of 2-phosphoglycolate, a compound that is toxic to the plant cell and needs to be detoxified through an energetically wasteful process called photorespiration (Bowes et al. 1971; Sharkey 1988; Sage 2004). The oxygenase reaction of RuBisCO becomes more common as temperature increases and so in C3 plants photorespiration can reduce photosynthetic output by up to 30% (Ehleringer and Monson 1993). In environments such as the tropics where rates of photorespiration are high, C4 photosynthesis has evolved repeatedly from the ancestral C3 state (Lloyd and Farquhar 1994; Osborne and Beerling 2006). Phylogenetic studies estimate that the first transition from C3 to C4 occurred around 30 million years ago (MYA) (Christin et al. 2008, 2011; Vicentini et al. 2008). The ability of the C4 cycle to concentrate CO2 around RuBisCO limits oxygenation and so increases photosynthetic efficiency in conditions where photorespiration is enhanced (Hatch and Slack 1966; Maier et al. 2011; Christin and Osborne 2014; Lundgren and Christin 2017).
The evolution of C4 photosynthesis involved multiple modifications to leaf anatomy and biochemistry (Hatch 1987; Sage 2004). In most C4 plants, photosynthetic reactions are partitioned between two distinct cell types known as mesophyll (M) and bundle sheath (BS) cells (Langdale 2011). M and BS cells are arranged in concentric circles around veins in the so-called Kranz anatomy (Haberlandt 1904), which enables CO2 pumping from M to BS where RuBisCO is specifically located. Atmospheric CO2 is first converted to HCO3 by carbonic anhydrase (CA) and then combined with phosphoenolpyruvate (PEP) by PEP-carboxylase (PEPC) to produce oxaloacetate in the M cells. This four-carbon acid (C4) is subsequently converted into malate and/or aspartate that transport the fixed CO2 from M to BS cells (Kagawa and Hatch 1974; Hatch 1987). Three biochemical C4 subtypes are traditionally described based on the predominant type of C4 acid decarboxylase responsible for the CO2 release around RuBisCO in the BS: NADP-dependent Malic Enzyme (NADP-ME, e.g. Zea mays), NAD-dependent Malic Enzyme (NAD-ME, e.g. Gynandropsis gynandra formerly designated Cleome gynandra) and phosphoenolpyruvate carboxykinase (PEPCK). However, recent reports suggest that only the NADP-ME and NAD-ME should be considered as distinct C4 subtypes, which in response to environmental cues may involve a supplementary PEPCK cycle (Williams et al. 2012; Wang et al. 2014; Rao and Dixon 2016).
The recruitment of multiple genes into C4 photosynthesis involved both an increase in their transcript levels (Hibberd and Covshoff 2010) and also patterns of expression being modified from relatively constitutive in C3 species (Maurino et al. 1997; Penfield et al. 2004; Taylor et al. 2010; Brown et al. 2011; Maier et al. 2011) to M- or BS-specific in C4 plants (Hibberd and Covshoff 2010). Therefore, considerable efforts have been made to identify the transcription factors (TF) and the cis-elements they recognize that are responsible for this light-dependent and cell-specific gene expression (Hibberd and Covshoff 2010). Various studies suggest that different transcriptional regulatory mechanisms have been adopted during C3 to C4 evolution. One is the acquisition of novel cis-elements in C4 gene promoters that can be recognized by TFs already present in C3 plants (Matsuoka et al. 1994; Ku et al. 1999; Nomura et al. 2000), and a second possibility is the acquisition of novel or modified TFs responsible for the recruitment of genes into the C4 pathway through cis-elements that pre-exist in C3 plants (Patel et al. 2006; Brown et al. 2011; Kajala et al. 2012).
A small number of cis-elements found in different gene regions have been shown to be sufficient for the M- or BS-specific expression of C4 genes. For example, a 41-base pair (bp) Mesophyll Expression Module 1 (MEM1) cis-element was identified from the PEPC promoter of C4Flaveria trinervia and shown to be necessary and sufficient for M cell-specific accumulation of PEPC transcripts in C4Flaveria species (Gowik et al. 2004). A MEM1-like cis-element has also been found in the C4 carbonic anhydrase (CA3) promoter of Flaveria bidentis and shown to drive M cell-specific expression (Gowik et al. 2017). A second cis-element named MEM2 and consisting of 9 bp from untranslated regions has also been shown to be capable of directing M-specificity in C4G. gynandra (Kajala et al. 2012; Williams et al. 2016). Lastly, in the case of the NAD-ME gene from C4G. gynandra a region from the coding sequence generates BS-specificity (Brown et al. 2011). In contrast to these insights into cis-elements that control cell-specific expression in the C4 leaf, no TFs recognizing these cis-elements have yet been identified.
To address this gap in our understanding, a bottom-up approach was initiated to identify TFs that regulate the maize gene ZmC4-NADP-ME (GRMZM2G085019) that encodes the Malic Enzyme responsible for releasing CO2 in the BS cells. Using Yeast One-Hybrid two maize TFs belonging to the superfamily of basic Helix-Loop-Helix (bHLH), ZmbHLH128 and ZmbHLH129, were identified and functionally characterized. We show that these TFs bind two cis-elements synergistically and analysis of the NADP-ME promoters from grass species from BEP and PACMAD (Panicoideae subfamily) indicated that this regulation is likely derived from an ancestral G-box that is present in C3 species.
Results
ZmbHLH128 and ZmbHLH129 Homeologs Bind FAR1/FHY3 Binding Site cis-Elements in the ZmC4-NADP-ME Promoter
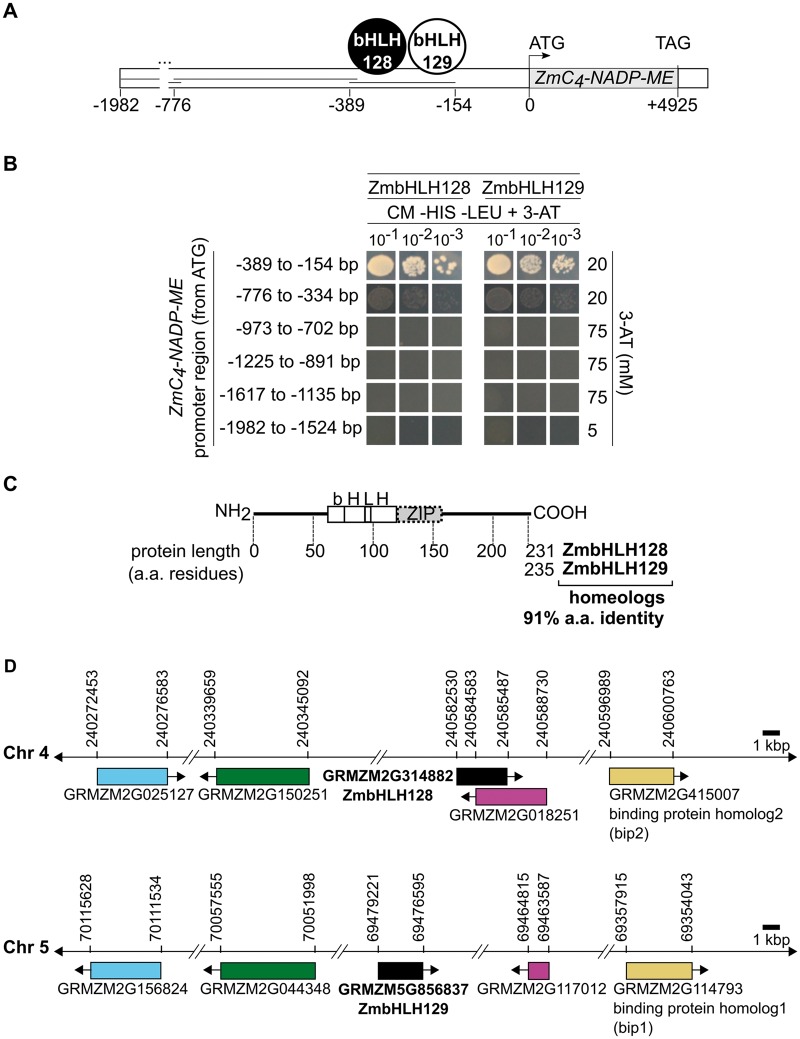
To identify TFs that interact with the ZmC4-NADP-ME gene (GRMZM2G085019), we studied the promoter region comprising 1982 bp upstream of the translational start site. This region was divided into six overlapping fragments ranging from 235 to 482 bp in length (supplementary table S1, Supplementary Material online) and used in Yeast One-Hybrid (Y1H). Each fragment was used to generate one yeast bait strain that was then used to screen a maize cDNA expression library. After screening at least 1.3 million colonies for each region of the promoter, two maize bHLH TFs known as ZmbHLH128 and ZmbHLH129 were identified. Both of these TFs bind the promoter between base pairs −389 and −154 in relation to the predicted translational start site of ZmC4-NADP-ME (fig. 1A). These interactions were confirmed by re-transforming yeast bait strains harbouring each of the six sections of the promoter with cDNAs encoding ZmbHLH128 and ZmbHLH129. Consistent with the initial findings, ZmbHLH128 and ZmbHLH129 only activated expression of the HIS3 reporter when transformed into yeast containing fragment −389 to −154 bp upstream of ZmC4-NADP-ME (fig. 1B, supplementary fig. S1, Supplementary Material online).
Fig. 1.
ZmbHLH128 and ZmbHLH129 homeologs bind the ZmC4-NADP-ME promoter. (A) Schematic representation of the ZmC4-NADP-ME promoter, divided into fragments used as baits in Y1H screenings, and the ZmbHLH TFs identified. ATG and TAG are the translational start codon and the stop codon of the ZmC4-NADP-ME ORF, respectively. ZmbHLH position on the scheme indicates that they bind between the base pairs −389 and −154 in relation to the ATG. (B) Analysis of ZmbHLH-pZmC4-NADP-ME binding specificity. Each of the six yeast bait strains was transformed with both ZmbHLHs (pAD-GAL4-2.1::TF vectors) and positive interactions selected on CM -HIS -LEU + 3-AT [yeast Complete Minimal medium lacking histidine and leucine amino acids, and supplemented with 3-amino-1, 2, 4-triazole (3-AT), a competitive inhibitor of the HIS3 gene product]. (C) Schematic representation of bHLH and ZIP protein domains, and respective position in protein sequences. (D) Schematic representation of ZmbHLH128 and ZmbHLH129 (black) and four additional maize homeolog gene pairs located in syntenic regions of chromosomes 4 and 5. Homeolog genes are indicated by colour. Arrows indicate direction of transcription of each gene. Genomic coordinates provided from the B73 RefGen_v3 assembly version.
ZmbHLH128 and ZmbHLH129 possess a bHLH domain followed by a contiguous leucine zipper (ZIP) motif (fig. 1C). This bHLH domain is highly conserved between both ZmbHLHs and consists of 61 amino acids that can be separated into two functionally distinct regions. The first is a basic region located at the N-terminal end of the bHLH domain and is involved in DNA binding, and the second is a Helix-Loop-Helix region mediating dimerization towards the carboxy-terminus (fig. 1C) (Murre et al. 1989; Toledo-Ortiz et al. 2003). ZmbHLH128 and ZmbHLH129 share 91% amino acid identity (fig. 1C) and they are encoded by homeolog genes located in syntenic regions of maize chromosomes 4 and 5 (fig. 1D, supplementary table S2, Supplementary Material online).
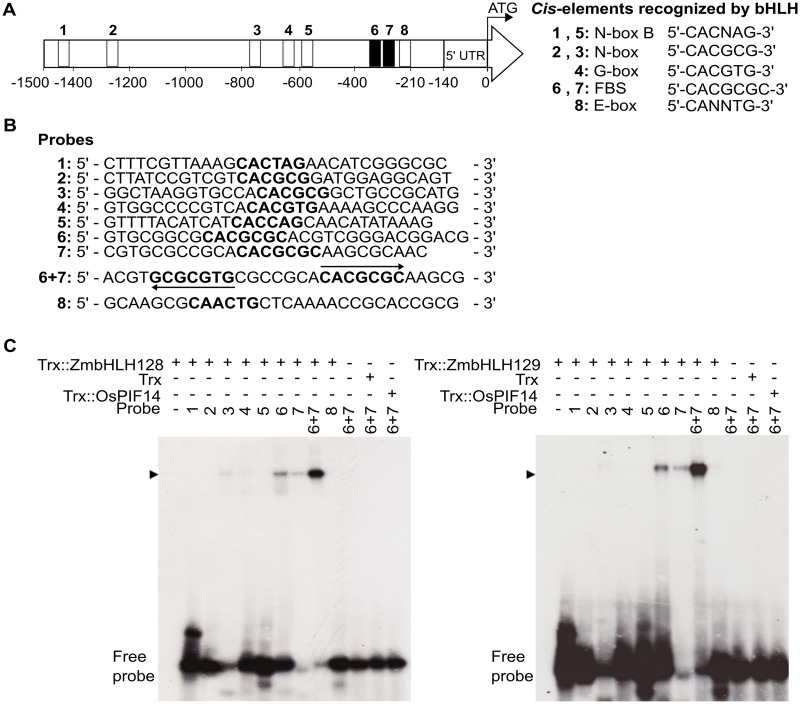
Although ZmbHLH128 and ZmbHLH129 both possess three amino acids involved in G-box binding (K9, E13, and R17) (Massari and Murre 2000; Li et al. 2006), this family of TFs has also been shown to bind to N-box (5′-CACGCG-3′), N-box B (5′-CACNAG-3′) and FBS (FAR1/FHY3 Binding Site, 5′-CACGCGC-3′) motifs (Sasai et al. 1992; Ohsako et al. 1994; Fisher and Caudy 1998; Kim et al. 2016). Therefore, the ZmC4-NADP-ME promoter was assessed for additional cis-elements to which ZmbHLH128 and ZmbHLH129 might bind. A total of eight such cis-elements were found, consisting of two N-boxes B, two N-boxes, one G-box, two FBSs, and one E-box (fig. 2A). Electrophoretic Mobility Shift Assays (EMSA) were used to test whether ZmbHLH128 and ZmbHLH129 were able to interact with each of these cis-elements in vitro (fig. 2B and C). Consistent with the Y1H findings, EMSA showed that recombinant Trx::ZmbHLH128 and Trx::ZmbHLH129 proteins caused an uplift of radiolabeled probes containing FBS cis-elements (probes 6, 7, and 6 + 7) (fig. 2C), positioned between nucleotides −389 and −154 in relation to the predicted translational start site (see fig. 1A). ZmbHLH128 also showed weak binding to probe 3 that contained a N-box cis-element that was not bound by ZmbHLH128 or ZmbHLH129 in Y1H (see fig. 1B), and signal intensity was similar to that observed from probe 7 (fig. 2C). We cannot exclude however that relatively weak binding to probe 7 is due to it being three nucleotides-shorter than the other probes (fig. 2B). Trx alone and OsPIF14 (a bHLH known to bind the N-box motif; Cordeiro et al. 2016) were used as negative controls (fig. 2C). The two FBS motifs, in probe 6 + 7, are separated by a short 7 bp spacer sequence and are found in opposite orientations (fig. 2B). The increase in band intensities detected when both cis-elements were combined (fig. 2C) suggests that they function synergistically. Overall, these data indicate that ZmbHLH128 and ZmbHLH129 target 21 bp of DNA sequence (7 bp FBS, 7 bp spacer, and 7 bp FBS).
Fig. 2.
ZmbHLH128 and ZmbHLH129 bind two FBS cis-elements present in ZmC4-NADP-ME promoter. (A) Schematic representation of position and nucleotide sequence of eight cis-elements recognized by bHLH that were identified in the ZmC4-NADP-ME promoter. FBS stands for FHY3/FAR1 Binding Site and it is a N-box-containing motif. (B) EMSA probe sequences used to test in vitro binding affinity of ZmbHLH128 and ZmbHLH129 to cis-elements (highlighted in bold). Arrows indicate that the FBS cis-elements are present in opposite orientations. (C) EMSAs showing in vitro binding affinity of Trx::ZmbHLH128 (gel on the left) and Trx::ZmbHLH129 (gel on the right) to the radiolabeled probes described in (B). Arrowheads indicate uplifted ZmbHLH–DNA probe complexes. Free probe indicates unbound DNA probes.
ZmbHLH128 and ZmbHLH129 Form Both Homo- and Heterodimers and ZmbHLH129 Impairs trans-Activation by ZmbHLH128
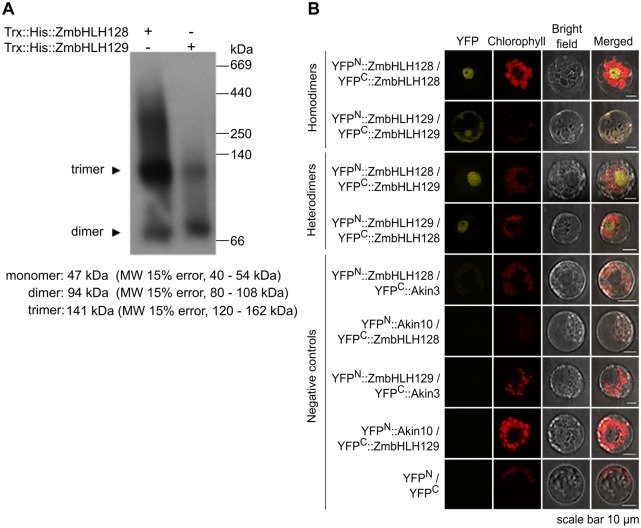
Because ZmbHLH128 and ZmbHLH129 bind the FBS cis-elements in close proximity but also possess domains mediating protein dimerization, we next investigated whether these proteins form homo- and/or heterodimers. In vitro, the recombinant Trx::ZmbHLH128 and Trx::ZmbHLH129 proteins formed homodimers (fig. 3A). To confirm this interaction in vivo, as well as to test for heterodimerization, Bimolecular Fluorescence Complementation Assays (BiFC) in maize protoplasts were performed. While negative controls produced no YFP fluorescence, ZmbHLH128 and ZmbHLH129 formed both homo- and heterodimers (fig. 3B). With the exception of ZmbHLH129 homodimers whose location extended to the cytoplasm and plasma membrane, in each case YFP signal was specifically localized to the nucleus (fig. 3B). Nuclear localization of these ZmbHLH proteins supports their roles as transcriptional regulators.
Fig. 3.
ZmbHLH128 and ZmbHLH129 form both homo- and heterodimers. (A) Western blot of BN-PAGE for the recombinant proteins Trx::His::ZmbHLH128 and Trx::His::ZmbHLH129. Gel was loaded with equivalent amount of protein. Recombinant proteins were immunodetected using α-His antibody. MW indicates molecular-weight size marker. (B) Protein interactions between ZmbHLH128 and ZmbHLH129 were tested by BiFC in maize mesophyll protoplasts co-transformed with constructs expressing ZmbHLH128 and ZmbHLH129 fused to N- and C-terminal YFP domains. YFPN and YFPC indicate split N- and C-terminal YFP domains, respectively.
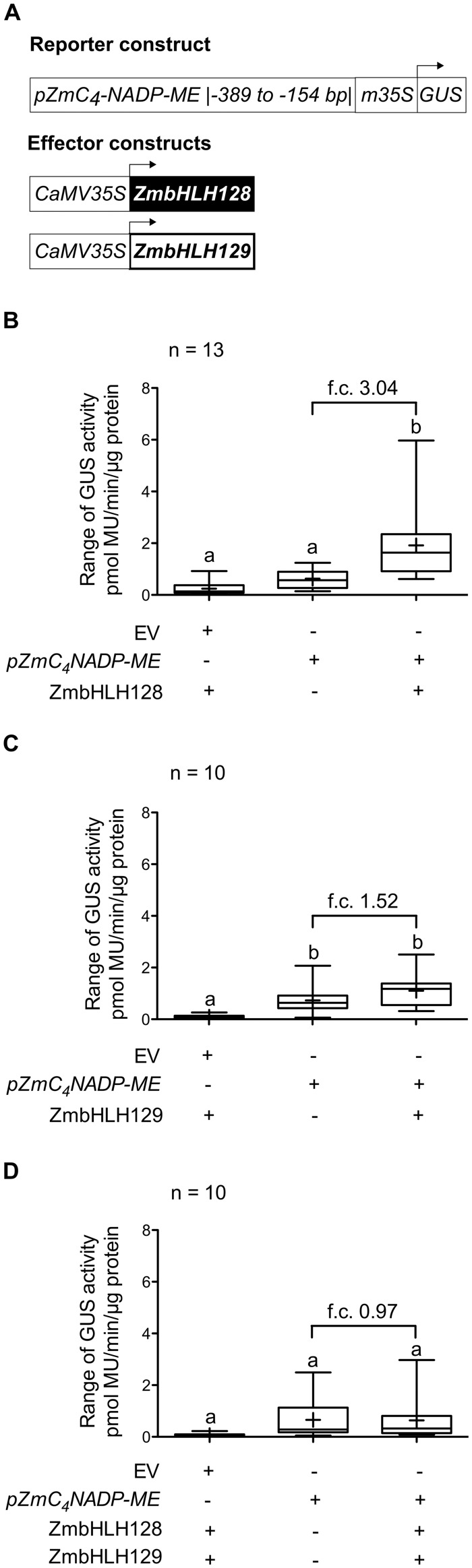
To test the capacity of ZmbHLH128 and ZmbHLH129 to regulate transcription, transient expression assays were performed in leaves of Nicotiana benthamiana. The GUS reporter gene driven by the fragment of pZmC4-NADP-ME to which ZmbHLH128 and ZmbHLH129 bind was used as reporter, while the full-length ZmbHLH128 and ZmbHLH129 CDS sequences driven by the constitutive CaMV35S promoter were used as effectors (fig. 4A). Co-infiltration of this reporter with the ZmbHLH128 effector resulted in an increase in GUS activity, indicating that ZmbHLH128 can act as a transcriptional activator (fig. 4B). In contrast, ZmbHLH129 showed no intrinsic trans-activation activity (fig. 4C). In order to test whether the ZmbHLH128-ZmbHLH129 heterodimers had a different trans-activation activity from ZmbHLH128 or ZmbHLH129 homodimers, leaves were co-infiltrated with the reporter and both effectors simultaneously. Interestingly, the trans-activation activity observed for the ZmbHLH128 alone (fig. 4B) was lost when this TF was co-expressed with its homeolog ZmbHLH129 (fig. 4D).
Fig. 4.
ZmbHLH129 impairs trans-activation of the ZmC4-NADP-ME promoter by ZmbHLH128. (A) Schematic representation of reporter and effector constructs used in transient expression assays in leaves of N. benthamiana. Reporter construct contains GUS gene driven by the minimal CaMV35S promoter (m35S) fused to pZmC4-NADP-ME (−389 to −154 bp). Effector constructs contain the ZmbHLH128 or ZmbHLH129 CDS driven by the full CaMV35S promoter. (B–D) Box plots (2.5–97.5 percentiles) showing GUS activity, expressed in picomoles of the reaction product 4-methylumbelliferone (MU) generated per minute per microgram of protein, in leaves agro-infiltrated with reporter and the following effector constructs: (B) ZmbHLH128, (C) ZmbHLH129, and (D) ZmbHLH128 and ZmbHLH129. Different letters denote differences in experimental data that are statistically significant (One-way ANOVA, Tukey test, P ≤ 0.05, n = 10-13). EV indicates pGWB3i empty vector (no promoter fragment cloned). Cross inside box plots indicates mean. f.c. indicates fold-change.
The G-Box-Based cis-Element Pair Recognized by ZmbHLH128 and ZmbHLH129 in NADP-ME Promoters Operates Synergistically
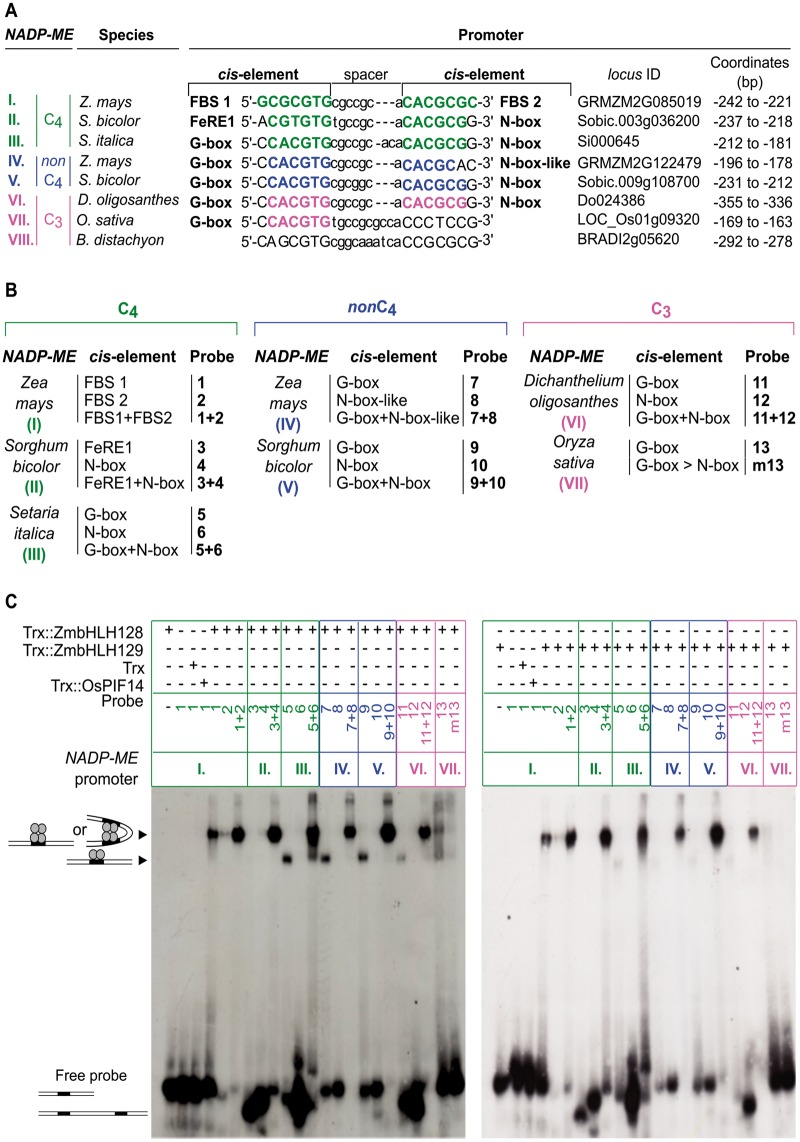
To understand whether the two FBS cis-elements identified in the promoter of ZmC4-NADP-ME (see fig. 2) are associated with the evolution of C4 photosynthesis, we investigated whether they are conserved in promoters of other NADP-MEs from C3 and C4 grass species. Three C3 species (Dichanthelium oligosanthes, Oryza sativa, and Brachypodium distachyon) and three C4 species (Z. mays, Sorghum bicolor, and Setaria italica) were assessed (fig. 5A). Within the C4 species, Z. mays and S. bicolor possess two plastidic NADP-ME isoforms: one that is used in C4 photosynthesis (C4-NADP-ME, GRMZM2G085019, and Sobic.003g036200) and a second one not involved in the C4 cycle (nonC4-NADP-ME, GRMZM2G122479, and Sobic.009g108700) (Alvarez et al. 2013; Emms et al. 2016). In contrast, S. italica possesses only one plastidic NADP-ME isoform that is used in the C4 cycle (C4-NADP-ME, Si000645) (Alvarez et al. 2013; Emms et al. 2016).
Fig. 5.
The G-box-based cis-element pair recognized by ZmbHLH128 and ZmbHLH129 in NADP-ME promoters operates synergistically. (A) Sequence alignment of the two FBS cis-elements present in ZmC4-NADP-ME promoter against homologous cis-elements present in other promoters of genes encoding plastidic NADP-ME. C4 grasses: Z. mays, S. bicolor and S. italica; C3 grasses: D. oligosanthes, O. sativa, and B. distachyon. Plastidic NADP-MEs are color-coded: green for C4, blue for nonC4 and magenta for C3. Cis-elements are highlighted in bold and colored according to the NADP-ME they belong to. FBS stands for FHY3/FAR1 Binding Site and FeRE1 for Iron Responsive Element 1. (B) EMSA probes used to test in vitro binding affinity of ZmbHLH128 and ZmbHLH129 to each cis-element described in (A). Probe sequences are listed in supplementary table S3, Supplementary Material online. (C) EMSA assays showing in vitro binding affinity of Trx::ZmbHLH128 (gel on the left) and Trx::ZmbHLH129 (gel on the right) proteins to the probes described in (B). Arrowheads indicate uplifted ZmbHLH-DNA probe complexes. Free probe indicates unbound DNA probes.
Although in C3B. distachyon no homologous cis-elements to the FBSs in the ZmC4-NADP-ME promoter were detected, in O. sativa one G-box was found in the same position as FBS 1 from Z. mays. Moreover, in the other promoters, cis-elements that can bind bHLH proteins were present in pairs (fig. 5A). In both the C3 and C4 grasses these cis-element pairs flank a spacer that is highly conserved in sequence and length (7–9 bp) (fig. 5A). The C4-NADP-ME promoters from Z. mays and S. bicolor share a common mutation in the third nucleotide position of the alignment (A→G) (fig. 5A). Two additional mutations are specific to Z. mays (the first and last nucleotides of FBS 1 and FBS 2, respectively), while one is S. bicolor-specific (C→T at the fourth position) (fig. 5A). It is possible that mutations unique to Z. mays or S. bicolor are neutral and the main impact on C4-NADP-ME gene expression is due to mutation in the third nucleotide in the common ancestor of Z. mays and S. bicolor. Alternatively, it is also possible that both this mutation in the last common ancestor and species-specific modifications impacted on gene expression of C4-NADP-ME.
To test if ZmbHLH128 and ZmbHLH129 bind the cis-elements identified from these additional species EMSA was performed on each cis-element separately as well as the cis-element pairs found in each NADP-ME promoter (fig. 5B and C,supplementary table S3, Supplementary Material online). ZmbHLH128 and ZmbHLH129 showed low binding affinity for the single G-box identified in the O. sativa promoter (probe 13) and binding affinity was not increased by mutating the G-box to a canonical N-box (probe m13) (fig. 5B and C). This low binding affinity behaviour for single G-box cis-elements was consistent for all the NADP-ME promoters containing G-boxes (probes 5, 7, 9, and 11) (fig. 5B and C). Although both ZmbHLHs did not show binding affinity for the additional N-boxes or N-box-like alone (probes 6, 8, 10, and 12) (fig. 5B and C), when these additional motifs were acquired and formed a pair with the ancestral G-box, binding affinity was increased (probes 5 + 6, 7 + 8, 9 + 10, and 11 + 12) and led to an increased uplift compared with the G-boxes alone (probes 5, 7, 9, and 11) (fig. 5B and C). Given the similar length of probes 1, 2, 1 + 2, 5, 7, 9, and 11 (24–30 bp) (supplementary table S3, Supplementary Material online), it is possible that this difference in migration of ZmbHLH–probe complexes results from the binding of bHLH to G-boxes in a lower oligomeric state (supplementary fig. S2, Supplementary Material online), which based on the literature must be dimers (De Masi et al. 2011). Strong binding of cis-element pairs was also observed when the ancestral G-box evolved into either FBS or FeRE1 elements found in C4Z. mays and S. bicolor (probes 1 + 2 and 3 + 4) (fig. 5B and C). In the C4Z. mays promoter, both ZmbHLHs showed binding affinity for single FBS cis-elements (probes 1 and 2) in the highest oligomeric state (fig. 5B and C, supplementary fig. S2, Supplementary Material online).
Since ZmbHLH128 and ZmbHLH129 showed weak binding to single cis-elements, we tested their binding by mutating these cis-elements in probes with the pairs (supplementary fig. S3, Supplementary Material online). For each pair, three mutant probes were designed: two in which the two cis-elements were mutated individually (keeping one cis-element wild-type) and one in which both cis-elements were mutated simultaneously (supplementary table S3, Supplementary Material online). Competition experiments were performed using radiolabeled wild-type probes (with cis-element pairs) and 200- to 400-fold excess of unlabeled wild-type and mutant probes (supplementary fig. S3, Supplementary Material online). Binding of both ZmbHLHs to the labeled wild-type probes could be efficiently out-competed by unlabeled wild-type and mutant probes in which the following cis-elements were not mutated: FBS 1 (in Z. mays C4-NADP-ME, probe 1 + m2-A, supplementary fig. S3A, Supplementary Material online); FBS 2 (in Z. mays C4-NADP-ME, probe m1 + 2-B, supplementary fig. S3A, Supplementary Material online); N-box (in S. bicolor C4-NADP-ME, probe m3 + 4-E, supplementary fig. S3B, Supplementary Material online); and G-box (in S. italica C4-NADP-ME, probe 5 + m6-G, supplementary fig. S3C, Supplementary Material online; Z. mays nonC4-NADP-ME, probe 7 + m8-J, supplementary fig. S3D, Supplementary Material online; S. bicolor nonC4-NADP-ME, probe 9 + m10-M, supplementary fig. S3E, Supplementary Material online; and D. oligosanthes C3-NADP-ME, probe 11 + m12-P, supplementary fig. S3F, Supplementary Material online). These EMSA competition experiments thus confirmed binding of ZmbHLH128 and ZmbHLH129 to the cis-elements described above. Taken together, the results indicate that a second cis-element recognized by bHLH TFs is acquired in the promoters of genes encoding plastidic NADP-ME and that each cis-element pair operates synergistically to allow interaction with either ZmbHLH128 or ZmbHLH129 in C3 and C4 grasses (fig. 5, supplementary figs. S2 and S3, Supplementary Material online).
Given the binding affinity in vitro of ZmbHLH128 and ZmbHLH129 to the G-box in the ZmnonC4-NADP-ME promoter (probes 7 and 7 + 8, fig. 5C), we tested their binding ability in planta. Transient expression assays were performed in leaves of N. benthamiana co-infiltrated with GUS reporter gene driven by a ZmnonC4-NADP-ME promoter fragment containing the cis-element pair G- and N-box-like (−368 to −143 bp) and the effector constructs ZmbHLH128 and ZmbHLH129 (supplementary fig. S4A, Supplementary Material online). Compared with the reporter alone, co-infiltration of ZmnonC4-NADP-ME reporter and the ZmbHLH128 and ZmbHLH129 effectors did not impact on GUS activity in tobacco system (supplementary fig. S4B–D, Supplementary Material online). These results suggest that although ZmbHLH128 on its own binds both the ZmC4-NADP-ME and ZmnonC4-NADP-ME promoters in vitro (probes 1, 2, 1 + 2, 7, and 7 + 8, fig. 5B and C), this might not be the case in planta (supplementary fig. S4, Supplementary Material online).
Acquisition of N-Box-Derived cis-Elements in NADP-ME Promoters Facilitates ZmbHLH128 and ZmbHLH129 Binding in PACMAD Panicoid Grasses
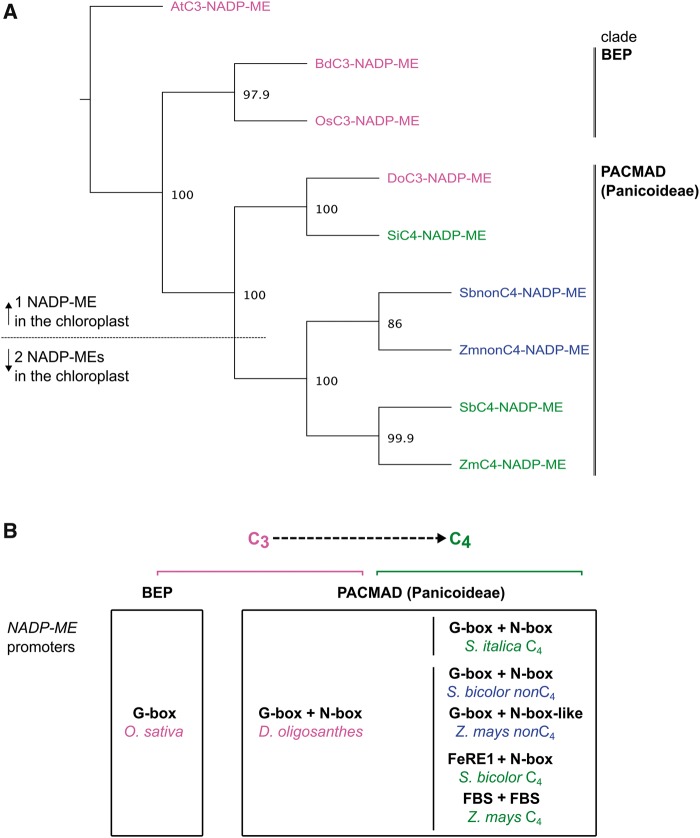
Phylogenetic analysis of the genes encoding C3 and C4 plastidic NADP-MEs reflects previously reported grass species phylogeny (fig. 6A) (Grass Phylogeny Working Group II 2012). It inferred two main clades: one formed by C3 BEP species (B. distachyon and O. sativa) and a second formed by C3 (D. oligosanthes) and C4 Panicoid species of the PACMAD clade (S. italica, S. bicolor, and Z. mays) (fig. 6A).
Fig. 6.
Acquisition of N-box-derived cis-elements in NADP-ME promoters facilitates ZmbHLH128 and ZmbHLH129 binding in PACMAD Panicoid grasses. (A) Phylogenetic tree of genes encoding plastidic NADP-ME from C3 and C4 grass species. C3: B. distachyon (Bd), O. sativa (Os), and D. oligosanthes (Do); C4: S. italica (Si), S. bicolor (Sb) and Z. mays (Zm). NADP-MEs are color-coded: magenta for C3, blue for nonC4 and green for C4. NADP-ME genomic sequences were aligned using MUSCLE, and the phylogenetic tree inferred by NJ method (1000 bootstrap pseudoreplicates, node numbers indicate bootstrap values). Gene encoding C3 plastidic NADP-ME from A. thaliana (AtC3-NADP-ME) was used as outgroup. (B) Diagram representing C3 to C4 molecular evolution of homologous bHLH binding cis-elements identified in promoters of genes encoding plastidic NADP-ME. Dashed arrow indicates intermediate evolutionary steps from C3 to C4. Vertical lines indicate two independent C4 origins of S. italica and S. bicolor/Z. mays (Paniceae and Andropogoneae tribes, respectively).
Based on the observed nucleotide modifications in cis-elements recognized by bHLH TFs, we propose a model relating to the recruitment of NADP-ME into C4 photosynthesis in grasses (fig. 6B). This proposes that an ancestral G-box found in the NADP-ME promoter of the common ancestor of C3 BEP O. sativa and C4 Panicoid grasses has been conserved during the evolution of C4 photosynthesis. However, in the Panicoideae subfamily of the PACMAD clade a second cis-element recognized by bHLH is present such that the NADP-ME gene from the C3 species D. oligosanthes and genes encoding plastidic nonC4-NADP-ME from C4S. bicolor and Z. mays all contain a G- and N-box/N-box-like pair. In C4S. italica this cis-code has been retained in the C4-NADP-ME, but in S. bicolor and Z. mays the original G-box has evolved to become either a FeRE1 or a FBS element, respectively (fig. 6B). No G-box motifs are, however, present in the promoter of genes encoding cytosolic NADP-ME from S. bicolor and Z. mays. Overall, these results suggest that the acquisition of N-box-derived cis-elements have facilitated ZmbHLH128 and ZmbHLH129 binding to promoters of genes encoding plastidic NADP-ME in the PACMAD (Panicoideae subfamily).
Discussion
ZmbHLH128 and ZmbHLH129 Homeologs Interact with Maize C4- and nonC4-NADP-ME Promoters in vitro Showing Different trans-Activation Activity in planta
In this study, we showed that ZmbHLH128 and ZmbHLH129 form a maize homeolog pair resulting from the recent maize whole genome duplication (WGD) event that occurred 5–12 million years ago. This WGD occurred 5–16 million years after C4 photosynthesis evolved in the Andropogoneae tribe of the PACMAD clade (17–21 MYA) (Christin et al. 2008, 2009). As the length of exons 1 and 2 and the total number of amino acids in the mature protein of ZmbHLH128 are more similar to sorghum ortholog SbbHLH66 (supplementary fig. S5, Supplementary Material online), we propose that ZmbHLH129 has diverged more from the ancestral gene. Both of these TFs bind two FBS cis-elements that are in close proximity in the maize C4-NADP-ME (GRMZM2G085019) promoter. Although ZmbHLH128 has been predicted in silico to regulate C4 photosynthesis (Wang et al. 2014), as far as we are aware, this is the first report of its functional characterization. ZmbHLH128 alone activates ZmC4-NADP-ME gene expression, while ZmbHLH129 alone shows no trans-activation activity on this promoter. As the duplication event that generated ZmbHLH129 took place after the evolution of C4 photosynthesis, it seems possible that this gene is not required for C4 photosynthesis. ZmbHLH128 and ZmbHLH129 form heterodimers and despite ZmbHLH128 activating the expression of ZmC4-NADP-ME its regulatory activity is impaired by its homeolog ZmbHLH129. To explain this impairment, we hypothesize different scenarios that may occur in vivo: either ZmbHLH128 and ZmbHLH129 act as heterodimers and ZmbHLH128 loses its DNA binding activity when combined with ZmbHLH129 or they act as homodimers and compete directly for the same FBSs, toward which ZmbHLH129 has a higher binding affinity. The former scenario has been described for bZIP TFs from Arabidopsis, where bZIP63 has negative effects on the formation of bZIP1–DNA complexes probably due to conformational differences between bZIP1 homodimer and bZIP1-bZIP63 heterodimers (Kang et al. 2010). The latter scenario has been reported for the maize Dof1 and Dof2 TFs. Dof1 is a transcriptional activator of light-regulated genes in leaves, however, in stems and roots, this TF is not able to regulate those genes since the repressor Dof2 is expressed there and blocks Dof-specific cis-elements (Yanagisawa and Sheen 1998).
In addition to the capacity of ZmbHLH128 and ZmbHLH129 to interact with FBSs found in the maize C4-NADP-ME promoter, both ZmbHLHs were shown to bind in vitro to the promoter of maize nonC4-NADP-ME (GRMZM2G122479) that possesses the cis-element pair G- and N-box-like. In planta, however, ZmbHLH128 and ZmbHLH129 showed no trans-activation activity on this promoter. It is well known that primary DNA sequence and its structural properties are determinants of DNA binding specificity in vivo (Rohs et al. 2009) and so it is possible that both ZmbHLHs display increased in vivo binding specificity for the FBS pair in the ZmC4-NADP-ME promoter than for the G- and N-box-like pair in the ZmnonC4-NADP-ME promoter. Therefore, ZmbHLH128 seems to affect the level of expression of NADP-ME as it activates the ZmC4-NADP-ME promoter through the pair formed by two FBSs but the same trend was not observed for the ZmnonC4-NADP-ME promoter with the G- and N-box pair. In addition, we hypothesize that these modifications of promoter sequences may also affect light/circadian regulation of the ZmC4-NADP-ME gene as FBS cis-elements have been described in promoters of circadian-clock-regulated and light-responsive genes (Lin et al. 2007, 2011; Kim et al. 2016). The mutation of two close FBSs in the promoter of the circadian-clock gene EARLY FLOWERING 4 (ELF4) proved to be sufficient to abolish its rhythmic expression (Li et al. 2011). More broadly, our findings also contribute to the understanding of gene regulatory networks controlling C4 photosynthesis.
The G-Box-Based cis-Element Pair Present in NADP-ME Promoters Synergistically Bind Either ZmbHLH128 or ZmbHLH129
We identified a cis-element pair recognized by bHLH that occupy homologous positions in NADP-ME promoters from C3 and C4 grasses. These cis-elements flank a short spacer and operate synergistically to facilitate interaction with ZmbHLH128 and ZmbHLH129. We suggest a mechanism by which these TFs may be recruited to the cis-elements associated with C4 photosynthesis. We propose that one cis-element is sufficient to recruit a bHLH homodimer (G-box) or tetramer (N-box or FBS in promoters where the ancestral G-box is no longer present); however, the presence of a second cis-element in the vicinity increases bHLH binding affinity (supplementary fig. S2, Supplementary Material online). It is possible that both cis-elements are brought together through the interaction with a bHLH tetramer formed by two dimers, which may involve DNA bending (supplementary fig. S2, Supplementary Material online). Therefore, this cis-element pair could operate synergistically to confer stabilization of bHLH binding. This mechanism of TF-DNA assembly has previously been proposed for MADS-domain TFs that can bind two nearby CArG boxes through DNA looping and formation of tetrameric complexes (Theissen 2001; Theissen and Saedler 2001; Melzer et al. 2009; Smaczniak et al. 2012; Smaczniak et al. 2017). In this case, and consistent with our results, MADS-domain TFs were found to bind single CArG boxes either as dimers or tetramers, however, when their target gene promoters contain CArG box pairs they bind as tetramers (Smaczniak et al. 2012). It has been proposed that the probability of DNA loop formation increases with shorter distances between cis-elements due to the low elastic bending energy required to bring the protein dimers together (Agrawal et al. 2008). Interestingly, in all NADP-ME promoters assessed in this study except rice and Brachypodium the two cis-elements were found to be in close proximity, which may encourage DNA looping. In addition to the spacer length, its sequence appears highly conserved. This is consistent with evidence suggesting that nucleotides outside core cis-elements affect TF binding specificity by providing genomic context and influencing three-dimensional structure (Atchley et al. 1999; Martínez-Garcia et al. 2000; Grove et al. 2009; Gordân et al. 2013). For example, Cbf1 and Tye7 are yeast bHLHs that show preference for a subset of G-boxes present throughout the yeast genome (Gordân et al. 2013). These differences in binding preferences were observed not just in vivo but also in vitro and so DNA sequences flanking core G-boxes were found to explain this differential bHLH-G-box binding (Gordân et al. 2013).
The mechanism proposed here for how bHLH TFs interact with their target cis-elements suggests that these DNA sequences are not randomly arranged in gene promoters and may affect how cis-element specificity is achieved. Indeed, in some promoters bound by bHLH TFs two or more cis-elements were found to be clustered. For example, two overlapping FBSs were reported in the 400 bp upstream of the translational start site of the gene encoding ELF4 (Li et al. 2011). Also, pairs of G- and N-boxes were found to be highly enriched in promoters targeted by the bHLH PIF1 (Kim et al. 2016). It is possible that multiple cis-elements serve to recruit additional TFs for in vivo cooperative binding.
C4 Photosynthesis Co-Opted an Ancient C3Cis-Regulatory Code Built on G-Box Recognition by bHLH Transcription Factors
Finally, from this study we propose a model that summarizes how molecular evolution of cis-elements recognized by bHLHs may relate to the recruitment of NADP-ME into C4 photosynthesis. C4 photosynthesis is an excellent example of convergent evolution (Sage et al. 2011; Christin et al. 2013) as it has evolved independently over 60 times in angiosperms (Sage et al. 2011; Sage 2016) and at least 22 times in grasses (Grass Phylogeny Working Group II 2012). How this repeated evolution has come about is not fully understood. Our model contributes to our understanding of C4 evolution and is based on the following findings: first, in rice, which belongs to the BEP clade that contains no C4 species, only one copy of a G-box was present in the NADP-ME promoter. In contrast, cis-element pairs recognized by ZmbHLH128 and ZmbHLH129 in NADP-ME promoters seem to be common in the Panicoideae subfamily of the PACMAD clade that contains independent C4 lineages. For example, in the PACMAD Panicoid grasses a G- and N-box pair was identified in C3D. oligosanthes (Do024386) and appears to be reasonably conserved in C4 species. However, in the case of the C4-NADP-MEs from S. bicolor and Z. mays (Sobic.003g036200 and GRMZM2G085019) these elements have diversified. Both of these grass species belong to the C4 tribe Andropogoneae in which the plastidic NADP-ME isoform that is used in C4 photosynthesis (C4-NADP-ME) evolved by duplication from an ancestral plastidic NADP-ME that still exists and is not involved in the C4 cycle (nonC4-NADP-ME, Sobic.009g108700 and GRMZM2G122479) (Tausta et al. 2002; Maier et al. 2011; Alvarez et al. 2013). In contrast, C4S. italica together with C3D. oligosanthes belong to the grass tribe Paniceae in which only one plastidic NADP-ME isoform is known to exist (Si000645 and Do024386) (Alvarez et al. 2013; Emms et al. 2016). Surprisingly, the cis-element pair identified in the C4-NADP-ME promoter from S. italica (G- and N-box) was found to be closer to those occurring in the C3 and nonC4-NADP-ME promoters from D. oligosanthes, S. bicolor, and Z. mays (G- and N-box/N-box-like) than to those occurring in the C4-NADP-ME promoters from S. bicolor and Z. mays (FeRE1 and N-box or FBS and FBS, respectively). A similar trend has previously been observed (Alvarez et al. 2013) and may be explained by the independent evolutionary origin of C4 photosynthesis in grass tribes formed by S. italica (Paniceae) or S. bicolor/Z. mays (Andropogoneae).
Taken together, our findings suggest that an ancestral G-box in combination with N-box-derived cis-elements form the basis of the synergistic binding of either ZmbHLH128 or ZmbHLH129 to NADP-ME promoters from PACMAD Panicoid grasses. Nucleotide diversity in cis-elements recognized by bHLH TFs has been suggested as one of the mechanisms by which these TFs are involved in complex and diverse transcriptional activity (Toledo-Ortiz et al. 2003). We, therefore, cannot exclude the possibility that the gene encoding the plastidic NADP-ME from C3 BEP B. distachyon (BRADI2g05620) can also be bound by ZmbHLH128 or ZmbHLH129 despite none of the typical cis-elements recognized by bHLH being identified in the promoter. Given recent evidence indicating that the bHLH TF family is often recruited into C4 photosynthesis regulation (Huang and Brutnell 2016), we suggest that the observed nucleotide modifications in the cis-element pair present in C4-NADP-ME promoters from S. bicolor and Z. mays may underlie changes in bHLH binding specificity in vivo and, therefore, contribute to the NADP-ME recruitment into C4 photosynthesis in the Andropogoneae tribe from the PACMAD clade. The presence of a bHLH duplicate (ZmbHLH129) that seems not to be required for C4 photosynthesis and has evolved to repress the activity of its homeolog (ZmbHLH128) is unique to maize as this homeolog gene pair resulted from the maize WGD. Therefore, we hypothesize that the single orthologous bHLH in all the other Panicoid species of the PACMAD clade activates C4-NADP-ME gene expression. This agrees with the hypothesis that C4 photosynthesis has on multiple occasions made use of cis-regulators found in C3 species and, therefore, that the recruitment of C4 genes was made through minor rewiring of pre-existing regulatory networks (Reyna-Llorens and Hibberd 2017). We conclude that regulation of C4 genes can be based on an ancient code founded on a G-box present in the BEP clade as well as the Panicoideae of the PACMAD clade. Acquisition of a second cis-element recognized by bHLH in Panicoid grasses appears to have facilitated synergistic binding by either ZmbHLH128 or ZmbHLH129. Although this G-box-based cis-code has remained similar in S. italica, it has diverged in maize and sorghum. Thus, different C4 grass lineages may employ slightly different molecular circuits to regulate orthologous C4 photosynthesis genes.
Materials and Methods
Plant Growth Conditions and Collection of Leaf Samples
To construct the cDNA expression library, maize plants (Z. mays L. var. B73) were grown at 16 h photoperiod with a light intensity of 340–350 μmol m−2 s−1, at day/night temperature of 28/26°C, and 70% relative humidity. Two light regimes were used: (1) nine days in 16 h photoperiod; and (2) nine days in 16 h photoperiod followed by a 72 h dark treatment. In both experiments, sample collection was performed under 16 h photoperiod. Third leaves grown in the former and latter light regimes were harvested respectively at time points covering the Zeitgeber times (ZT) −0.5, 0.5, 2 h, and ZT 1, 2, 4, 8, 12, 15.5 h. For isolation of maize mesophyll protoplasts, maize plants were grown for 10 days at 25°C, 16 h photoperiod (60 μmol m−2 s−1), and 70% relative humidity. For transient expression assays in planta, N. benthamiana (tobacco) plants were grown for 5 weeks at 22°C, 16 h photoperiod (350 μmol m−2s−1), and 65% relative humidity. After agro-infiltration of tobacco leaves, plants were left to grow into the same growth conditions and leaf discs (2.5 cm in diameter) collected 96 h post-infection.
Generation of Yeast Bait Strains
Yeast bait strains were generated as previously described (Ouwerkerk and Meijer 2001; Serra et al. 2013). Yeast strain Y187 (Clontech) was used to generate six bait strains carrying overlapping fragments of the ZmC4-NADP-ME (GRMZM2G085019) promoter cloned into the yeast integrative vector pINT1-HIS3 (Ouwerkerk and Meijer 2001) as NotI-SpeI or XbaI-SpeI fragments (supplementary table S1, Supplementary Material onlline). The ZmC4-NADP-ME promoter region was defined as the 1,982 bp upstream of the predicted translational start site (ATG). To assess self-activation/HIS3 leaky expression, yeast bait strains were titrated in complete minimal medium (CM) lacking histidine, with increasing concentrations of 3-amino-1, 2, 4-triazole (3-AT, up to 75 mM).
Construction of cDNA Expression Library
Total RNA was extracted from third leaves of maize seedlings using TRIzol reagent (Invitrogen), following the manufacturer’s instructions. RNA samples from nine time points (described in ‘plant growth conditions and collection of leaf samples’) were pooled in equal amounts for mRNA purification using the PolyATract mRNA Isolation System IV (Promega). A unidirectional cDNA expression library was prepared using the HybriZAP-2.1 XR cDNA Synthesis Kit and the HybriZAP-2.1 XR Library Construction Kit (Stratagene), following the manufacturer’s instructions. Four micrograms of mRNA were used for first strand cDNA synthesis. After in vivo excision and amplification of the pAD-GAL4-2.1 phagemid vector, this maize cDNA expression library was used to transform yeast bait strains.
Yeast One-Hybrid (Y1H) Screening and Validation
Yeast bait strains were transformed with 1 μg of maize cDNA expression library according to Ouwerkerk and Meijer (2001) and Serra et al. (2013). At least, 1.3 million yeast colonies of each yeast bait strain transformed with the maize cDNA expression library were screened in CM -HIS -LEU supplemented with 3-AT: 5 mM (−1982 to −1524 bp), 20 mM (−389 to −154 bp, −776 to −334 bp) or 75 mM (−973 to −702 bp, −1225 to −891 bp, −1617 to −1135 bp). Plasmids from yeast clones that actively grew on selective medium were extracted. To know whether the isolated clones encoded transcription factors (TFs), the cDNA insert was sequenced and the results analyzed using BLAST programes. To validate DNA-TF interactions in yeast, isolated plasmids encoding TFs were re-transformed into the yeast bait strain in which they were found to bind. To assess TF binding specificity, plasmids encoding TFs were also transformed into the yeast bait strains to which they do not bind.
Yeast Cell Spotting
Yeast bait strains transformed with plasmids encoding TFs were grown overnight until log or mid-log phase at 30°C in liquid yeast CM medium supplemented with Histidine (CM +HIS -LEU). Cultures were normalized to an OD600 of 0.4, spotted onto solid medium CM +HIS -LEU or CM -HIS -LEU + 3-AT, and grown for 3 days at 30°C.
Isolation and Transformation of Maize Mesophyll Protoplasts
Maize mesophyll protoplasts were isolated from 10-day-old maize greening plants and transformed according to Lourenço et al. (2013) with minor modifications. Mid-section of newly matured second leaves was digested in a cell wall digestive medium containing 1.5% (w/v) cellulase R-10 (Duchefa), 0.3% (w/v) macerozyme R-10 (Duchefa), 10 mM MES (pH 5.7), 0.4 M mannitol, 1 mM CaCl2, 0.1% (w/v) BSA and 5 mM β-mercaptoethanol. Several leaf blades were stacked and cut perpendicularly to the long axis into 0.5–1 mm slices and quickly transferred to digestive medium (25 ml digestive medium for each set of 10 leaf blades). Purity and integrity of isolated protoplasts were examined under light microscopy. Mesophyll protoplasts were quantified and its abundance adjusted to 2 × 106 protoplasts ml−1. Transformed protoplasts were resuspended in 1.25 ml of incubation solution [0.6 M mannitol, 4 mM MES (pH 5.7) and 4 mM KCl] and incubated in 24-well plates for 18 h at room temperature under dark.
Bimolecular Fluorescence Complementation (BiFC) Assay
To generate BiFC constructs, full-length coding sequences (CDS) of ZmbHLH128 (GRMZM2G314882) and ZmbHLH129 (GRMZM5G856837) were PCR-amplified using respectively the following pairs of attB-containing primers: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTNNATGATGAACTGCGCCGGA-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTNCTAAGCATTAGGCGGCCAG-3′, and 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTNNATGATGGACTGCGCTGGA-3′/5′-GGGGACCACTTTGTACAAGAAAGCTGGGTNCTAAGCATTTGGGGGCCAG-3′ (underlined sequences indicate attB Gateway adaptors). ZmbHLH128 and ZmbHLH129 CDS were recombined into pDONR221 (Invitrogen) to obtain Entry clones through BP-Gateway reaction (Invitrogen), following the manufacturer’s instructions. CDS were then recombined into vectors YFPN43 and YFPC43 through LR-Gateway reaction (Invitrogen) to raise a translational fusion with N- and C-terminal domains of yellow fluorescent protein (YFP), respectively. Final BiFC constructs were denominated as YFPN::ZmbHLH128, YFPN::ZmbHLH129, YFPC::ZmbHLH128, and YFPC::ZmbHLH129. Maize mesophyll protoplasts were transformed with 6 μg of each of the BiFC constructs. Protoplasts transformed with YFPN::Akin10 (Arabidopsis SNF1 Kinase Homolog 10), YFPC::Akin3 (Arabidopsis SNF1 Kinase Homolog 3) and YFPN43 and YFPC43 empty vectors were used as negative controls. Transformations were performed in triplicate. YFP fluorescence and chlorophyll autofluorescence signals were observed under a confocal microscope (Leica SP5).
Transient Expression Assays in planta
For the transient expression assays in tobacco leaves, reporter and effector constructs were generated in the Gateway binary vectors pGWB3i [pGWB3 containing an intron-tagged β-glucuronidase (GUS) open reading frame (Berger et al. 2007)] and pGWB2 (Tanaka et al. 2012), respectively.
To construct the reporter plasmids, promoter fragments of ZmC4-NADP-ME (GRMZM2G085019, from −389 to −154 bp) and ZmnonC4-NADP-ME (GRMZM2G122479, from −368 to −143 bp) were fused to a 136 bp minimal CaMV35S promoter (m35S) in a 3-step PCR reaction: (1) promoter sequences were amplified with long chimeric primers to introduce overlapping ends (reverse primer of pZmC4-NADP-ME/pZmnonC4-NADP-ME was designed to be complementary to the forward primer of the m35S) (supplementary table S4, Supplementary Material online); (2) promoter sequences amplified by PCR in (1) were mixed according to the fusion products of interest in a ratio of 1:1 [ZmC4-NADP-ME (−389 to −154 bp)::m35S and ZmnonC4-NADP-ME (−368 to −143 bp)::m35S] and 10 PCR cycles were run without primers (denaturation at 98°C for 10 s, 55°C for 30 s, and 72°C for 1 min); and (3) fusion products of interest were amplified with attB-containing primers (supplementary table S4, Supplementary Material online). To obtain Entry clones, promoter fragments fused to m35S were cloned into pDONR221 (Invitrogen) through BP-Gateway reaction (Invitrogen), following the manufacturer’s instructions. Promoter sequences were then recombined into the binary vector pGWB3i through LR-Gateway reaction (Invitrogen) to obtain the final reporter constructs for promoter::GUS analysis (pZmC4-NADP-ME and pZmnonC4-NADP-ME). For the effector constructs (TF driven by the CaMV35S promoter), ZmbHLH128 and ZmbHLH129 Entry clones previously generated (see BiFC assay) were directly recombined into the binary vector pGWB2 through LR-Gateway reaction (Invitrogen).
Reporter and effector constructs together with a construct harboring the silencing suppressor P1b (Valli et al. 2006) were transformed into the Agrobacterium tumefaciens strain GV301. Overnight cultures of Agrobacterium harboring reporter, effector and P1b constructs were sedimented (5000 × g for 15 min, at 4°C) and resuspended in infiltration medium (10 mM MgCl2, 10 mM MES (pH 5.6), 200 μM acetosyringone) to an OD600 of 0.3, 1, and 0.5, respectively, and mixed in a ratio of 1:1:1. Mixed Agrobacterium cultures were incubated for 2 h at 28°C and used to spot-infiltrate the abaxial side of 5-week-old tobacco leaves. As controls, tobacco leaves were agro-infiltrated with mixed cultures carrying the reporter construct alone or the empty vector pGWB3i and effector constructs. Infected leaves were analyzed at 96 h post-infiltration. Leaf discs of 2.5 cm in diameter were collected from the infiltrated spots and used for the quantification of GUS activity. GUS activity was quantified by measuring the rate of 4-methylumbelliferyl-β-d-glucuronide (MUG) conversion to 4-methylumbelliferone (MU) as described in Jefferson et al. (1987) and Williams et al. (2016). In brief, soluble protein was extracted from agro-infiltrated tobacco leaf discs by freezing in liquid nitrogen and maceration, followed by addition of protein extraction buffer. Diluted protein extracts (1:2) were incubated with 1 mM MUG for 30, 60, 90, and 120 min at 37°C in a 96-well plate. GUS activity was terminated at the end of each time point by the addition of 200 mM Na2CO3 and MU fluorescence measured by exciting at 365 nm and measuring emission at 455 nm. The concentration of MU/unit fluorescence in each sample was interpolated using a concentration gradient of MU from 1.5 to 800 μM MU.
Production of Recombinant ZmbHLH128 and ZmbHLH129
ZmbHLH128 and ZmbHLH129 full-length CDS were PCR-amplified using, respectively, the following pairs of gene specific primers 5′-GAATTCATGATGAACTGCGCCGGA-3′/5′-CTCGAGCTAAGCATTAGGCGGCCAG-3′ and 5′-GAATTCATGATGGACTGCGCTGGA-3′/5′-CTCGAGCTAAGCATTTGGGGGCCAG-3′ (underlined sequences indicate adaptors with restriction enzyme sites). ZmbHLH128 and ZmbHLH129 were cloned as EcoRI-XhoI fragments into the expression vector pET32a (Novagen), generating N-terminal Trx-tagged fusions. pET32a-Trx::ZmbHLH128 and pET32a-Trx::ZmbHLH129 constructs were confirmed by sequencing and transformed into Rosetta (DE3)pLysS competent cells (Invitrogen) for protein expression. Cells transformed with pET32a-Trx::ZmbHLH128 and pET32a-Trx::ZmbHLH129 constructs were, respectively, grown in Terrific Broth (TB) and Luria-Bertani (LB) medium to an OD600 of 0.5. Protein expression was induced with 4 mM isopropyl-d-1-thiogalactopyranoside (IPTG) and allowed to occur for 3 h (ZmbHLH128) or 5 h (ZmbHLH129) at 30°C. Protein purification was performed as described in Cordeiro et al. (2016).
Blue Native-Polyacrylamide Gel Electrophoresis (BN-PAGE) and Western Blotting
Molecular mass of oligomers co-existing in purified ZmbHLH128 and ZmbHLH129 recombinant proteins was determined by blue native polyacrylamide gel electrophoresis (BN-PAGE). Two micrograms of the recombinant proteins (Trx::His::ZmbHLH128 or Trx::His::ZmbHLH129) were resolved on a 3–12% Novex Bis–Tris NativePAGE mini gel (Life Technologies), following the manufacturer’s instructions. HMW Native Marker Kit (66–669 kDa, GE Healthcare) was used to estimate molecular mass. Resolved proteins were transferred to a polyvinylidene difluoride membrane (GE Healthcare). The membrane was destained with a 50% (v/v) methanol and 10% (v/v) acid acetic solution followed by pure methanol. For immunodetection of Trx::His::ZmbHLH128 and Trx::His::ZmbHLH129, the membrane was incubated with α-His antibody (GE Healthcare) followed by α-mouse horseradish peroxidase-conjugated antibody (abcam) for 1 h each at room temperature.
Electrophoretic Mobility Shift Assay (EMSA)
DNA probes were generated by annealing oligonucleotide pairs in a thermocycler followed by radiolabeling as described in Serra et al. (2013). DNA probe sequences and respective annealing temperatures are listed in supplementary table S3, Supplementary Material online. EMSAs were performed using 400 ng of the recombinant proteins Trx::ZmbHLH128 or Trx::ZmbHLH129, and 50 fmol of radiolabeled probes. Competition assays were performed adding 200- to 400-fold molar excess of the unlabeled probe. Trx::OsPIF14 (LOC_Os07g05010) and Trx protein, both purified by Cordeiro et al. (2016), were used as negative controls. Each protein was mixed with probes in a 10 μl reaction containing 10 mM HEPES (pH 7.9), 40 mM KCl, 1 mM EDTA (pH 8), 1 mM DTT, 50 ng herring sperm DNA, 15 μg BSA and 10% (v/v) glycerol. Binding reactions were incubated for 1 h on ice and the bound complexes resolved on a native 5% polyacrylamide gel (37.5:1). Gel electrophoresis and detection of radioactive signal were performed as described in Serra et al. (2013).
Synteny Analysis
SynFind (Tang et al. 2015) was used to identify maize syntenic chromosomal regions for ZmbHLH128 (GRMZM2G314882) and ZmbHLH129 (GRMZM5G856837) genes against Z. mays B73 RefGen_v3 genome. A table containing maize syntelog gene pairs was retrieved using SynFind tool (supplementary table S2, Supplementary Material online).
Phylogenetic Analyses
ZmbHLH128 and ZmbHLH129 were used as references to identify closely related bHLH genes of Z. mays, S. bicolor, Setaria viridis, S. italica, O. sativa, and B. distachyon, through Phytozome database (Goodstein et al. 2012). Predicted CDS were aligned using MUSCLE. The resulting alignment was used to infer a maximum likelihood phylogenetic tree, using GTR + G+I nucleotide substitution model (1,000 bootstrap pseudoreplicates) in MEGA 7 software (Kumar et al. 2016). Phylogenetic analysis of genes encoding C3 and C4 plastidic NADP-ME isoforms from B. distachyon (BRADI2g05620), O. sativa (LOC_Os01g09320), D. oligosanthes (Do024386), S. italica (Si000645), S. bicolor (Sobic.003g036200, Sobic.009G108700), and Z. mays (GRMZM2G085019, GRMZM2G122479) was performed using Geneious Pro 5.3.6 software (Kearse et al. 2012). Full-length genomic sequences were aligned using MUSCLE. Phylogenetic tree was inferred using the Neighbor Joining (1,000 bootstrap pseudoreplicates) and rooted using the gene encoding C3 plastidic NADP-ME (At1g79750) from Arabidopsis thaliana, a dicot angiosperm.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Lisete Fernandes (Escola Superior de Tecnologia da Saúde de Lisboa, Portugal) for discussions and advice concerning EMSA experiments, Cecília Arraiano Lab (ITQB-NOVA, Oeiras, Portugal) for material used in EMSA experiments, Myriam Goudet and Samuel Brockington (Department of Plant Sciences, University of Cambridge, UK) for assistance with phylogenetic analyses. This study was supported by European Union project 3to4 (Grant agreement no: 289582) and Fundação para a Ciência e Tecnologia (FCT) through research unit GREEN-it ‘Bioresources for Sustainability’ (UID/Multi/04551/2013). ARB (SFRH/BD/105739/2014), AG (SFRH/BD/89743/2012), AMC (SFRH/BD/74946/2010), PMB (SFRH/BPD/86742/2012), IAA (IF/00960/2013—POPH-QREN), and NJMS (IF/01126/2012—POPH-QREN) were funded by FCT, TSS and PG by European Union project 3to4 (Grant agreement no: 289582), and IR-L by BBSRC grant (BB/L014130).
References
- Agrawal NJ, Radhakrishnan R, Purohit PK.. 2008. Geometry of mediating protein affects the probability of loop formation in DNA. Biophys J. 948:3150–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez CE, Saigo M, Margarit E, Andreo CS, Drincovich MF.. 2013. Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species. Photosynth Res. 1151:65–80. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A.. 1999. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol. 485:501–5016. [DOI] [PubMed] [Google Scholar]
- Berger B, Stracke R, Yatusevich R, Weisshaar B, Flügge UI, Gigolashvili T.. 2007. A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 505:911–916. [DOI] [PubMed] [Google Scholar]
- Bowes G, Ogren WL, Hageman RH.. 1971. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem Biophys Res Commun. 453:716–722. [DOI] [PubMed] [Google Scholar]
- Brown NJ, Newell CA, Stanley S, Chen JE, Perrin AJ, Kajala K, Hibberd JM.. 2011. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science 3316023:1436–1439. [DOI] [PubMed] [Google Scholar]
- Calvin BM, Massini P.. 1952. The path of carbon in photosynthesis. Experientia 812:445–457. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Besnard G, Samaritani E, Duvall MR, Hodkinson TR, Savolainen V, Salamin N.. 2008. Report oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr Biol. 181:37–43. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP.. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biol Evol. 511:2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Osborne CP.. 2014. The evolutionary ecology of C4 plants. New Phytol. 2044:765–781. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Osborne CP, Sage RF, Arakaki M, Edwards EJ.. 2011. C4 eudicots are not younger than C4 monocots. J Exp Bot. 629:3171–3181. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Kellogg EA, Vicentini A, Besnard G.. 2009. Integrating phylogeny into studies of C4 variation in the grasses. New Phytol. 1491:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro AM, Figueiredo DD, Tepperman J, Borba AR, Lourenço T, Abreu IA, Ouwerkerk PBF, Quail PH, Margarida Oliveira M, Saibo NJM.. 2016. Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim Biophys Acta 18592:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibés A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJM.. 2011. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 3911:4553–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehleringer JR, Monson RK.. 1993. Evolutionary and ecological aspects of photosynthetic pathway variation. Annu Rev Ecol Evol Syst. 241:411–439. [Google Scholar]
- Emms DM, Covshoff S, Hibberd JM, Kelly S.. 2016. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Mol Biol Evol. 337:1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A, Caudy M.. 1998. The function of hairy-related bHLH repressor proteins in cell fate decisions. BioEssays 204:298–306. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS.. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(D1):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML.. 2013. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 34:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P.. 2004. Cis-regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 165:1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Schulze S, Saladié M, Rolland V, Tanz SK, Westhoff P, Ludwig M.. 2017. A MEM1-like motif directs mesophyll cell-specific expression of the gene encoding the C4 carbonic anhydrase in Flaveria. J Exp Bot. 682:311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group II 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 1932:304–312. [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM.. 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 1382:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberlandt GFJ. 1904. Physiologische Pflanzenanatomie. Leipzig: Leipzig W. Engelmann. [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 8952:81–106. [Google Scholar]
- Hatch MD, Slack CR.. 1966. Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem J. 1011:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S.. 2010. The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol. 61:181–207. [DOI] [PubMed] [Google Scholar]
- Huang P, Brutnell TP.. 2016. A synthesis of transcriptomic surveys to dissect the genetic basis of C4 photosynthesis. Curr Opin Plant Biol. 31:91–99. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW.. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 613:3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Hatch MD.. 1974. C4-acids as the source of carbon dioxide for Calvin cycle photosynthesis by bundle sheath cells of the C4-pathway species Atriplex spongiosa. Biochem Biophys Res Commun. 594:1326–1332. [DOI] [PubMed] [Google Scholar]
- Kajala K, Brown NJ, Williams BP, Borrill P, Taylor LE, Hibberd JM.. 2012. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 691:47–56. [DOI] [PubMed] [Google Scholar]
- Kang SG, Price J, Lin PC, Hong JC, Jang JC.. 2010. The Arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant. 32:361–373. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C et al. , . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2812:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Kang H, Park J, Kim W, Yoo J, Lee N, Kim JJ, Yoon T, Choi G.. 2016. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell 286:1388–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku MSB, Agarie S, Nomura M, Fukayama H, Tsuchida H, Ono K, Hirose S, Toki S, Miyao M, Matsuoka M.. 1999. High-level expression of maize phosphoenol pyruvate carboxylase in transgenic rice plants. Nat Biotechnol. 171:76–80. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 337:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. 2011. C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 2311:3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan X-y, Li J, Lau O-S, Ouyang X, Dai M, Wan J et al. , . 2011. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 135:616–622. [DOI] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J et al. , . 2006. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 1414:1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H.. 2007. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 3185854:1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J, Farquhar GD.. 1994. 13C discrimination during CO2 assimilation by the terrestrial biosphere. Oecologia 99(3–4):201–215. [DOI] [PubMed] [Google Scholar]
- Lourenço T, Sapeta H, Figueiredo DD, Rodrigues M, Cordeiro A, Abreu IA, Saibo NJM, Oliveira MM.. 2013. Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol. Biol 83(4-5): 351–363. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Christin P-A.. 2017. Despite phylogenetic effects, C3–C4 lineages bridge the ecological gap to C4 photosynthesis. J Exp Bot. 682:241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Zell MB, Maurino VG.. 2011. Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C4 and C3 photosynthesis. J Exp Bot. 629:3061–3069. [DOI] [PubMed] [Google Scholar]
- Martínez-Garcia JF, Huq E, Quail PH.. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 2885467:859–863. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C.. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 202:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M, Kyozuka J, Shimamoto K, Kano-Murakami Y.. 1994. The promoters of two carboxylases in a C4 plant (maize) direct cell-specific, light-regulated expression in a C3 plant (rice). Plant J. 63:311–319. [DOI] [PubMed] [Google Scholar]
- Maurino VG, Drincovich MF, Casati P, Andreo CS, Edwards GE, Ku MSB, Gupta SK, Franceschi V.. 1997. NADP-malic enzyme: immunolocalization in different tissues of the C4 plant maize and the C3 plant wheat. J Exp Bot. 483:799–811. [Google Scholar]
- Melzer R, Verelst W, Theißen G.. 2009. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in “floral quartet”-like complexes in vitro. Nucleic Acids Res. 371:144–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D.. 1989. A new DNA binding and dimerization motif in immunoglubin enhancer binding, dauhterless, MyoD, and myc proteins. Cell 565:777–783. [DOI] [PubMed] [Google Scholar]
- Nomura M, Sentoku N, Nishimura A, Lin JH, Honda C, Taniguchi M, Ishida Y, Ohta S, Komari T, Miyao-Tokutomi M et al. , . 2000. The evolution of C4 plants: acquisition of cis-regulatory sequences in the promoter of C4-type pyruvate, orthophosphate dikinase gene. Plant J. 223:211–221. [DOI] [PubMed] [Google Scholar]
- Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M.. 1994. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 822:2743–2755. [DOI] [PubMed] [Google Scholar]
- Osborne CP, Beerling DJ.. 2006. Nature’s green revolution: the remarkable evolutionary rise of C4 plants. Philos Trans R Soc B 3611465:173–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouwerkerk PB, Meijer AH.. 2001. Yeast one-hybrid screening for DNA-protein interactions In: DNA–Protein Interactions. Current Protocols in Molecular Biology. Leiden: Leiden University Netherlands, p. 12.12.1–12.12.22. [DOI] [PubMed] [Google Scholar]
- Patel M, Siegel AJ, Berry JO.. 2006. Untranslated regions of FbRbcS1 mRNA mediate bundle sheath cell-specific gene expression in leaves of a C4 plant. J Biol Chem. 28135:25485–25491. [DOI] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA.. 2004. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 1610:2705–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X, Dixon RA.. 2016. The differences between NAD-ME and NADP-ME subtypes of C4 photosynthesis: more than decarboxylating enzymes. Front Plant Sci. 7:1525.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna-Llorens I, Hibberd JM.. 2017. Recruitment of pre-existing networks during the evolution of C4 photosynthesis. Philos Trans R Soc B 372:20160386.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B.. 2009. The role of DNA shape in protein-DNA recognition. Nature 4617268:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. New Phytol. 1612:341–370. [DOI] [PubMed] [Google Scholar]
- Sage RF. 2016. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and hall of fame. J Exp Bot. 6714:4039–4056. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin P-A, Edwards EJ.. 2011. The C4 plant lineages of planet Earth. J Exp Bot. 629:3155–3169. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S.. 1992. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 6(12B):2620–2634. [DOI] [PubMed] [Google Scholar]
- Serra TS, Figueiredo DD, Cordeiro AM, Almeida DM, Lourenço T, Abreu IA, Sebastián A, Fernandes L, Contreras-Moreira B, Oliveira MM et al. , . 2013. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol. 82(4–5):439–455. [DOI] [PubMed] [Google Scholar]
- Sharkey TD. 1988. Estimating the rate of photorespiration in leaves. Physiol Plant 731:147–152. [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QDP, Liu S, Westphal AH, Boeren S et al. , . 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U. S. A. 1095:1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Muiño JM, Chen D, Angenent GC.. 2017. Differences in DNA-binding specificity of floral homeotic protein complexes predict organ-specific target genes. Plant Cell. 298:1822–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Kimura T, Hikino K, Goto S, Nishimura M, Mano S, Nakagawa T.. 2012. Gateway vectors for plant genetic engineering: overview of plant vectors, application for bimolecular fluorescence complementation (BiFC) and multigene construction In: Barrera-Saldaña HA, editor. Genetic engineering—basics, new applications and responsabilities. InTech, p. 35–58. [Google Scholar]
- Tang H, Bomhoff MD, Briones E, Zhang L, Schnable JC, Lyons E.. 2015. SynFind: compiling syntenic regions across any set of genomes on demand. Genome Biol Evol. 712:3286–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta SL, Coyle HM, Rothermel B, Stiefel V, Nelson T.. 2002. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol Biol. 50(4–5):635–652. [DOI] [PubMed] [Google Scholar]
- Taylor L, Nunes-Nesi A, Parsley K, Leiss A, Leach G, Coates S, Wingler A, Fernie AR, Hibberd JM.. 2010. Cytosolic pyruvate, orthophosphate dikinase functions in nitrogen remobilization during leaf senescence and limits individual seed growth and nitrogen content. Plant J. 624:641–652. [DOI] [PubMed] [Google Scholar]
- Theissen G. 2001. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol. 41:75–85. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H.. 2001. Plant biology. Floral quartets. Nature 4096819:469–471. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH.. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 158:1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli A, Martin-Hernandez AM, Lopez-Moya JJ, Garcia JA.. 2006. RNA silencing suppression by a second copy of the P1 serine protease of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine protease HCPro. J Virol. 8020:10055–10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellog EA.. 2008. The age of the grasses and clusters of origins of C4 photosynthesis. Glob Chang Biol. 1412:2963–2977. [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, Si Y, Tohge T, Nunes-Nesi A, Arrivault S, Dedow LK, Bryant DW, Zhou W et al. , . 2014. Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat. Biotechnol. 3211:1158–1165. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bräutigam A, Weber APM, Zhu X-G.. 2014. Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J Exp Bot. 6513:3567–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Aubry S, Hibberd JM.. 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant. Sci. 174:213–220. [DOI] [PubMed] [Google Scholar]
- Williams BP, Burgess SJ, Reyna-Llorens I, Knerova J, Aubry S, Stanley S, Hibberd JM.. 2016. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. Plant Cell. 282:454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J.. 1998. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell. 101:75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.