Abstract
The dried rhizomes of Coptis chinensis have been extensively used in heat clearing, dampness drying, fire draining, and detoxification by virtue of their major bioactive components, benzylisoquinoline alkaloids (BIAs). However, C. teeta and C. chinensis are occasionally interchanged, and current understanding of the molecular basis of BIA biosynthesis in these two species is limited. Here, berberine, coptisine, jatrorrhizine, and palmatine were detected in two species, and showed the highest contents in the roots, while epiberberine were found only in C. chinensis. Comprehensive transcriptome analysis of the roots and leaves of C. teeta and C. chinensis, respectively, identified 53 and 52 unigenes encoding enzymes potentially involved in BIA biosynthesis. By integrating probable biosynthetic pathways for BIAs, the jatrorrhizine biosynthesis ill-informed previously was further characterized. Two genes encoding norcoclaurine/norlaudanosoline 6-O-methyltransferases (Cc6OMT1 and Cc6OMT2) and one gene encoding norcoclaurine-7OMT (Ct7OMT) catalyzed enzymatically O-methylate (S)-norcoclaurine at C6 that yield (S)-coclaurine, along with a smaller amount of O-methylation occurred at C7, thereby forming its isomer (isococlaurine). In addition, scoulerine 9-OMT (CtSOMT) was determined to show strict substrate specificity, targeting (S)-scoulerine to yield (S)-tetrahydrocolumbamine. Taken together, the integration of the transcriptome and enzyme activity assays further provides new insight into molecular mechanisms underlying BIA biosynthesis in plants and identifies candidate genes for the study of synthetic biology in microorganisms.
Keywords: benzylisoquinoline alkaloid, biosynthesis, Coptis, O-methyltransferases, transcriptome
Introduction
Coptis chinensis is a perennial herb that belongs to the family Ranunculaceae. Among 16 species distributed all over the world, 6 distribute in Sichuan, Guizhou, Yunnan, and Shanxi provinces of western China (Zhang Y. et al., 2015; Xue et al., 2017). For centuries, its dried rhizomes have been widely used in traditional Chinese medicine (TCM) (Kamath et al., 2009; Xiang et al., 2016) for heat clearing, dampness drying, fire draining, and detoxification (Zhang et al., 2011; Yang et al., 2017). Currently, C. chinensis is a commonly used TCM with definitive curative effects in clinic, and 108 patented drugs have been produced from C. chinensis in China (Feng et al., 2008; Wang et al., 2014). Due to overexploitation, the wild resources of this herb are now endangered. To meet the ever increasing demand of this herb, studies on the biosynthesis of the bioactive components of C. chinensis are important to ensure the reasonable utilization of resources and the use of synthetic biology through microorganism engineering.
The major active components in C. chinensis are benzylisoquinoline alkaloids (BIAs) (Kamath et al., 2009; Chen et al., 2017). Of great interest in this field are the identification of effective separation methods and assessment of their efficacy in the treatment of various diseases (Tjong et al., 2011; Zhang et al., 2011; Cui et al., 2016; Friedemann et al., 2016; Kim et al., 2016). Chemical investigations have determined that the major BIAs in C. chinensis are protoberberine alkaloids, which include berberine, palmatine, jatrorrhizine, coptisine, columbamine, magnoflorine, and epiberberine (Lv et al., 2016; Yang et al., 2017), with berberine as the predominant component (Tang et al., 2009; Ye et al., 2009; Lv et al., 2016). These compounds have been shown to be efficacious in antiviral, anti-inflammatory, and antimicrobial activity, for dispelling dampness, for removing toxicosis, and in detoxification (Tjong et al., 2011; Zhang et al., 2011; Cui et al., 2016; Friedemann et al., 2016; Kim et al., 2016). However, current understanding of the biosynthesisof BIAs in C. chinensis remains limited due to insufficient genomic resources.
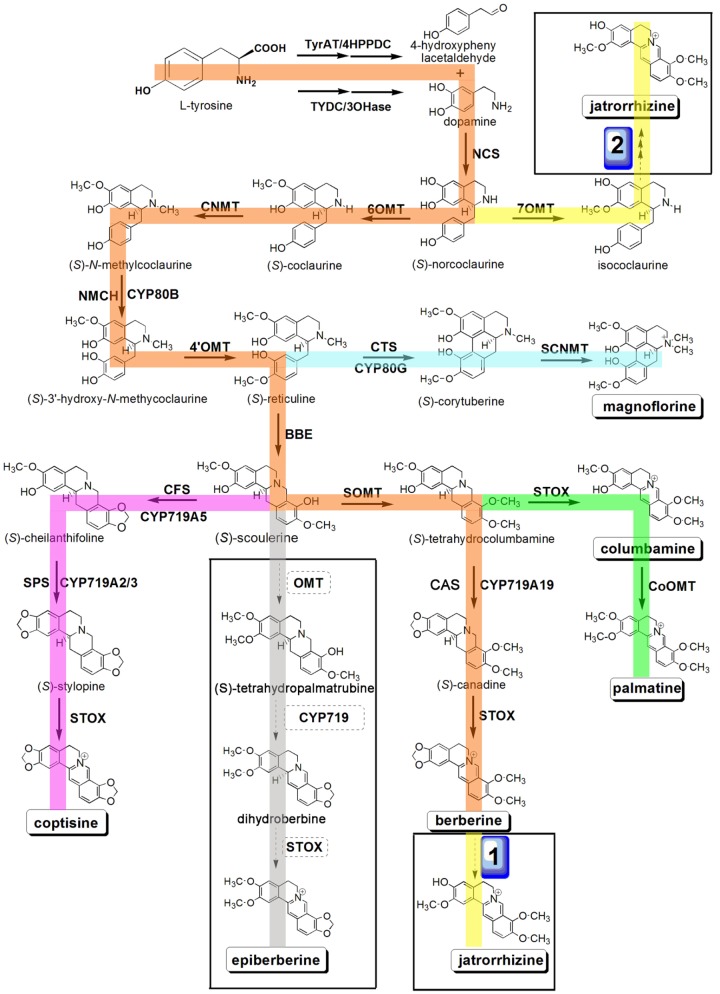
The biosynthetic pathways for several BIAs have been reported in a number of plant species, such as berberine in C. japonica, sanguinarine in Eschscholzia californica, and morphine in Papaver somniferum, and many enzymes involved in BIA biosynthesis in these plants have been characterized (Morishige et al., 2010; Hagel and Facchini, 2013; Sato, 2013). The biosynthesis of BIAs begins with the conversion of L-tyrosine to dopamine and 4-hydroxyphenylacetaldehyde, which are then condensed to (S)-norcoclaurine by (S)-norcoclaurine synthase (NCS) (Samanani and Facchini, 2002; Samanani et al., 2004; Minami et al., 2007). Three methyltransferases [(S)-norcoclaurine/norlaudanosoline 6-O-methyltransferase (6OMT), (S)-coclaurine-N-methyltransferase (CNMT), and (S)-3′-hydroxy-N-methylcoclaurine-4′-O-methyltransferase (4′OMT)] and one cytochrome P450 [P450, (S)-N-methylcoclaurine 3′-hydroxylase (NMCH)] are involved in catalyzing the conversion of (S)-norcoclaurine to (S)-reticuline, which is a central intermediate for the production of different BIAs, including protoberberine, benzophenanthridine, and morphinan alkaloids (Morishige et al., 2000; Choi et al., 2002; Samanani et al., 2006; Sato et al., 2007; Ziegler and Facchini, 2008; Hagel and Facchini, 2013; Beaudoin and Facchini, 2014; Yamada et al., 2015). BIA biosynthesis consists of several steps that involve oxidation reactions catalyzed by P450s and O-methylation processes catalyzed by members of the OMT family that participate in the synthesis of the intermediate product, (S)-reticuline, followed by multistep transformations that yield different end-products (Chapple, 1998; Mizutani and Sato, 2011; Dang and Facchini, 2012; Takemura et al., 2013; Chang et al., 2015) (Figure 1).
FIGURE 1.
Putative pathways for BIA biosynthesis in C. teeta and C. chinensis. Enzymes found in this study are boxed. Abbreviations: TyrAT, L-tyrosine aminotransferase; 4HPPDC, 4-hydroxyphenylpuruvate decarboxylase; TYDC, tyrosine decarboxylase; 3OHase, tyrosine/tyramine 3-hydroxylase; NCS, (S)-norcoclaurine synthase; 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, (S)-coclaurine N-methyltransferase; NMCH, N-methylcoclaurine 3′-hydroxylase; 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; BBE, berberine bridge enzyme; CTS, corytuberine synthase; SCNMT, (S)-corytuberine-N-methyltransferase; SOMT, (S)-scoulerine 9-O-methyltransferase; CAS, (S)-canadine synthase; STOX, (S)-tetrahydroprotoberberine oxidase; CoOMT, columbamine O-methyltransferase; CFS, (S)-cheilanthifoline synthase; SPS, (S)-stylopine synthase.
The identification and characterization of related genes involved in the synthesis of bioactive components is essential for understanding the molecular mechanism of biosynthetic pathways. With the rapid development of RNA sequencing technology, it is easy to capture the candidate genes at a genome-wide scale. In a recent study, Chen et al. (2017) analyzed major components of C. chinensis from three biotopes; namely, Zhenping, Zunyi, and Shizhu, and found that berberine was highest in concentration, followed sequentially by coptisine, palmatine, and epiberberine in all three accessions. Among these accessions, the drug quality of the accession from Shizhu may be the highest (Fan et al., 2012). Further transcriptomic investigation and correlation analysis of chemical components revealed four candidate genes encoding aspartate aminotransferase, polyphenol oxidase, primary-amine oxidase, and tyrosine decarboxylase (TYDC). However, functional characterization of these candidate genes has not been performed to date.
Of the six species of C. chinensis distributed in China, C. chinensis Franch., C. deltoid C.Y. Chen et Hsiao, and C. teeta Wall. are respectively called “Weilian,” “Yalian,” and “Yunlian” in the Chinese Pharmacopeia (version 2010) (Yang et al., 2012; Li et al., 2013). Among these, C. chinensis is the most important raw material because it is widely distributed and has a high yield, whereas C. teeta is an endangered species (Zhang Y. et al., 2015). Previously, people thought that the three species could be used interchangeably because of their similar phytochemical contents (mainly alkaloids) (Lv et al., 2016). However, accumulating evidence shows that they differ in their pharmacological activities. For example, Feng et al. (2011) determined that the rhizomes of C. chinensis have higher antibacterial activity than C. deltoidea and C. teeta. Therefore, analysis of their chemical compositions is essential for promoting their appropriate pharmacological use.
In the present study, we carried out a comprehensive transcriptome study of C. chinensis and C. teeta by means of next-generation RNA-seq technology. Combining our present results with previous reports (Rüffer et al., 1983; Ikezawa et al., 2008; Ziegler et al., 2009; Hagel et al., 2015; He et al., 2017), we propose the biosynthetic pathways for berberine, palmatine, jatrorrhizine, coptisine, columbamine, magnoflorine, and epiberberine, and identify candidate genes that are involved in the biosynthesis of BIAs. Moreover, we characterized 6OMT and norcoclaurine-7OMT (7OMT), which catalyze O-methylation at C6 on (S)-norcoclaurine to yield (S)-coclaurine, along with a smaller amount of O-methylation at C7, thereby forming its isomer, isococlaurine. SOMT 9-O-methylates (S)-scoulerine specifically to yield (S)-tetrahydrocolumbamine. Taken together, our study further promotes the molecular and biochemical understanding of BIA production in Coptis and provides substantial genetic resources for further research.
Materials and Methods
Plant Materials
C. teeta was collected from Nujiang, Yunnan Province (26°54′N, 98°97′E and altitude: 2771 m), while C. chinensis was collected from an area managed by the Guangxi Botanical Garden of Medicinal Plants in Nanning, China. The roots and leaves were harvested separately, immediately frozen in liquid nitrogen, and stored at -80°C until use.
cDNA Library Construction, Sequencing and de Novo Assembly
Total RNA of roots and leaves from two Coptis species were extracted using RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The quantity and quality of total RNA were detected by the NanoDrop system (Thermo Scientific, United States) and gel electrophoresis, respectively. Then, at least 4 μg of total RNA samples were used to construct mRNA libraries and deep sequencing by NEBNext® UltraTM RNA Library Prep Kit and Illumina sequencing on a Hiseq 2000 platform, respectively. After RNA-seq, raw reads were firstly transformed into clean reads by removing adaptors, low-quality and unknown nucleotides. Then, the clean reads were de novo assembled using the Trinity program (k-mer = 25, group pairs distance = 300) with default parameters (Liscombe et al., 2005).
Functional Annotation, CDS Prediction and Phylogenetic Analysis
For functional annotations, the assembled unigenes were blasted against public databases (E-value < 1 × 10-5), including the NCBI non-redundant protein (NR) database1, the SwissProt database2, the euKaryotic Ortholog Groups (KOG) database3, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database4, and the protein family (Pfam) database (version 26.0) (Finn et al., 2014). Based on NR database annotation, GO unigene annotations were obtained by Blast2GO program (Conesa et al., 2005). Then, GO functional classification was performed by WEGO software (Ye et al., 2006). Finally, CDSs of unigenes were predicted using BLSATX and ESTscan (Iseli et al., 1999). In addition, a neighbor-joining tree was built by Clustal X 2.0 and MEGA 5.0 with deduced amino acid of OMTs from two Coptis and other plants.
High-Performance Liquid Chromatography Analysis
0.2 g dried powder of two Coptis leaves, roots and rhizomes was respectively extracted with 50 mL of hydrochloric acid-methanol mixed liquor (1:100, v/v) for 30 min, and sonicated for 30 min. For determining the main bioactive components of two Coptis species, an Agilent 1260 High-performance liquid chromatography (HPLC) system (Agilent Technologies, Santa Clara, CA, United States) was used. Chromatographic separation was performed on the chromatographic column Agilent Zorbax SB-C 18 (250 mm × 4.6 mm, 5 μm, Agilent Technologies) at a column temperature of 30°C, the flow rate was fixed at 1 mL/min. The mobile phase consisted of acetonitrile-0.05mol/L potassium dihydrogen phosphate solution (50:50, v/v) containing 0.1% sodium dodecyl sulfate, and separation was achieved by an isocratic elution. Detection was performed at 345 nm (Zhang G.H. et al., 2015; He et al., 2017). The content of berberine, coptisine, jatrorrhizine, palmatine, and epiberberine were calculated from standard curves. Authentic berberine, palmatine, jatrorrhizine, coptisine and epiberberine were purchased from JK chemical (Beijing, China).
Recombinant Protein Expression and Purification
Full-length cDNAs of OMTs including Cc6OMT1, Cc6OMT2, Ct7OMT, and CtSOMT were obtained by PCR amplification using primers (Supplementary Table S5), and cloned into the pCzn1 vector (Zoonbio Biotechnology, China). The vector was introduced into the Escherichia coli line Arctic-Express (Zoonbio Biotechnology, China) for protein expression. The expression of the recombinant protein was induced by 0.5 mM of IPTG at 37°C for 4 h. The cells were harvested by centrifugation and resuspended in binding buffer, and the suspension was subsequently homogenized by 1 h of 200Wsonication (Vibra Cell VC 505 Sonicator; Sonics & Materials, Newtown, CT, United States). Cell debris was subsequently removed with 10-min centrifugation at 12,000 rpm. The purified inclusion body fist were denatured with 2 and 8 M urea, then renatured using 0.15 M NaCl. The protein was purified by Ni-IDA-Sepharose CL-6B (Spectrum Chemical Manufacturing, United States) under the manufacturer’s instructions. The purity of the His-tagged protein was determined by SDS–PAGE followed by Coomassie Brilliant Blue staining.
Determination of Enzyme Activity
The standard enzyme assay for OMTs activity was performed using a reaction mixture in 50 μl of 25 mM Tris–HCl (pH 8.0), 25 mM sodium ascorbate, 0.1 mM S-adenosyl-L-methionine, 100 μM potential alkaloid substrate, and 50 μg of purified recombinant enzyme. Assays were carried out at 30°C for 1 h and terminated by adding 200 μL of ethanol. Controls were performed with denatured purified His-tagged proteins prepared by boiling in water for 20 min (Robin et al., 2016). Products were centrifuged at 14,000 rpm for 10 min, and 2 μL supernatants were subjected to UPLC-QTOF-MS/MS system (Waters Corporation, Milford, MA, United States), which consisted of a UPLC I-Class instrument (Waters Corporation, Singapore). UPLC was carried out using a Waters Acquity UPLC HSS T3 C18 column (2.1 mm × 100 mm, 1.8 μm) at a flow rate of 0.5 mL⋅min-1, and the temperature was set at 35°C. The mobile phases were 0.1% formic acid in H2O (A) and acetonitrile (B), with the following gradient: 2% B (0–1.5 min), 2–30% B (1.5–4 min), 30–60% B (4–6 min), 60–80% B (6–7.5 min), 80% B (7.5–8 min), and 2% B (8–10 min). Mass spectrometry was performed on a definition accurate mass quadrupole time-of-flight (Q-TOF) Xevo G2-S mass spectrometer (Waters MS Technologies, Manchester, United Kingdom) equipped with electrospray ionization (ESI) source. Eluate was applied to the mass analyzer using an electrospray ionization interface operating in positive mode with the following conditions: capillary voltage of 3.0 kV, cone voltage of 40 V, source temperature of 100°C, desolvation temperature of 400°C, cone gas flow of 50 L/h, and desolvation gas flow of 700 L/h. The energy for collision-induced dissociation (CID) was set to 6 V for the precursor ion, and the MS/MS fragment information was obtained using a collision energy ramp from 35 to 50 eV in MSE mode. To ensure the mass accuracy and reproducibility of the optimized MS condition, leucine encephalin (m/z 556.2771 in positive mode) was used as the reference lock mass (Yang et al., 2015). Compounds were identified based on retention times and ESI [+]-CID spectra compared with authentic standards. Authentic (S)-norcoclaurine, (S)-coclaurine, (S)-scoulerine and (S)-tetrahydrocolumbamine were purchased from JK chemical (Beijing, China).
Reverse Transcription Quantitative Real-Time PCR Analysis
Total RNA isolated from leaves, roots and rhizomes of the two Coptis species was used for reverse transcription quantitative real-time PCR (qRT-PCR) analysis. cDNA was transcribed from 2 μg purified RNA using HiScript QRT SuperMix (Vazyme, Nanjing, China). 2 μL (100 ng/μL) of cDNA in 20 μL solution system composed of 2 × SYBR Green Master mix (TaKaRa) was used for gene expression analysis in a Roche LightCycler 2.0 system (Roche Applied Science, Branford, CT, United States). The primers used in this study are listed in Supplementary Table S6. PCR amplification conditions were: 95°C for 30 s; 40 cycles of 95°C for 20 s, 55°C for 20 s and 72°C for 30 s. The actin gene was chosen as a reference gene to control for normalization. The relative changes in gene expression levels were calculated using the 2-ΔΔCt method. For each target gene, the experiment was carried out with three biological replicates.
Statistical Analysis
Data shown are the mean ± standard deviation of three biological replicates. Mean differences were compared using the statistical software data processing system (SPSS 17.0). The significant differences between samples were statistically evaluated by the Students’s t-test for HPLC and Tukey test for qRT-PCR.
Results and Discussion
Morphology and Quantitative Analysis of Major BIAs in Two Coptis Species
While the distribution of C. teeta is restricted to high-altitude regions, including Yunnan Province in China, C. chinensis is widely distributed in China. To differentiate C. teeta from C. chinensis, we randomly collected twenty plants of C. teeta and C. chinensis, respectively and compared their morphology. The aboveground of C. teeta is upright and compact with long petiole and more shoots, while that of C. chinensis is loose with short petiole and less shoots (Figure 2). In contrast, the belowground of C. teeta has small rhizomes and less roots but contains foraging branches, while C. chinensis has large rhizomes and more roots but has no foraging branches (Figure 2). Foraging branches (vegetative propagules) are mainly used for asexual reproduction, which is an adaptation to high altitudes as the environment of these regions is unfavorable for sexual reproduction (Yang et al., 2013). Therefore, the morphological evolution of C. teeta coincides with its geographical distribution.
FIGURE 2.
Morphology comparison of roots and shoots between C. teeta and C. chinensis. Three-year-old C. teeta and C. chinensis were collected from Yunnan and Guangxi, respectively.
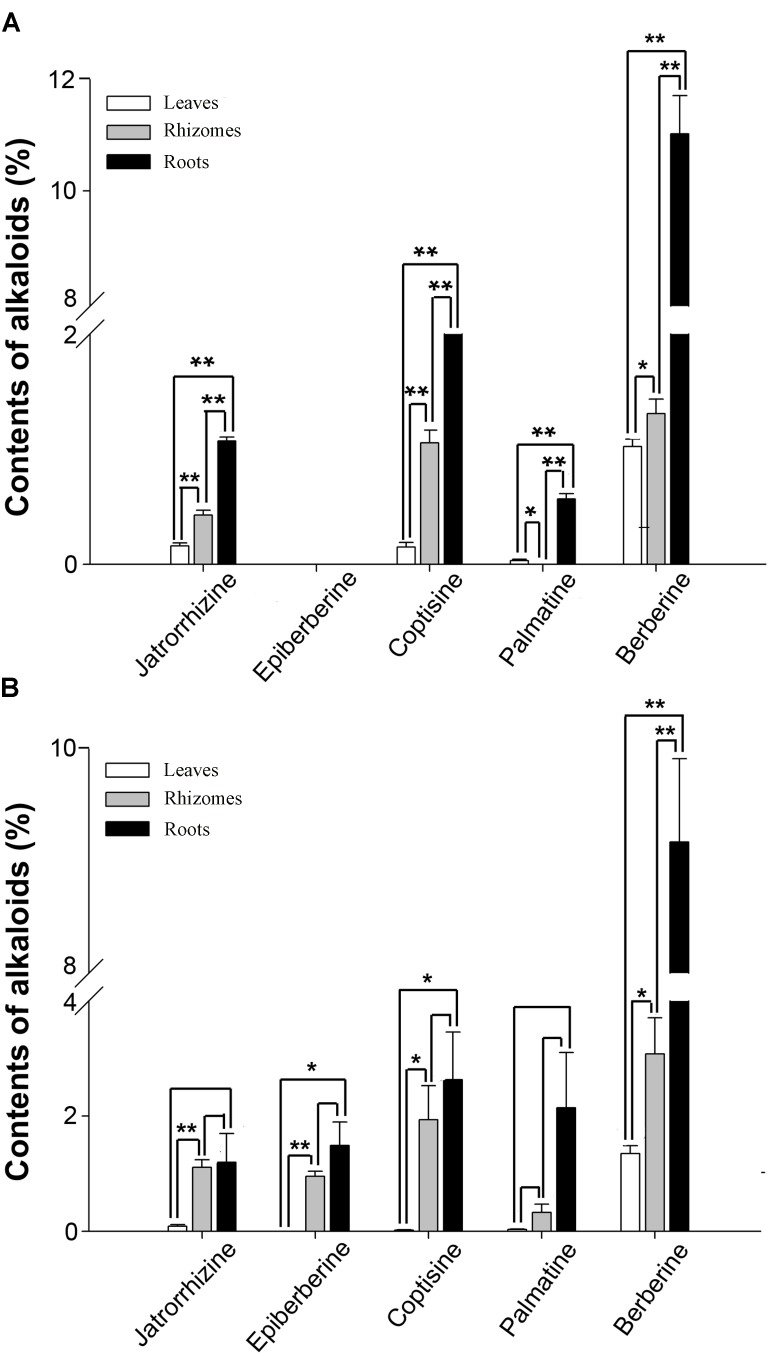
According to previous reports, berberine, coptisine, jatrorrhizine, and palmatine constitute the main BIAs of C. teeta and C. chinensis (Kamath et al., 2009; Chen et al., 2017; Yang et al., 2017). HPLC analysis of leaves, roots, and rhizomes of C. teeta and C. chinensis indicated the presence of berberine, coptisine, jatrorrhizine, and palmatine in both species (Figure 3). The predominant BIA was berberine, followed by coptisine, and jatrorrhizine and palmatine was the lowest among them. Notably, epiberberine was only detected in C. chinensis. In terms of different tissues, the four major BIAs were most abundant in the roots, followed by rhizomes and leaves, which is consistent with the use of roots in TCM. In leaves, there were no significant differences between berberine and palmatine in two Coptis species, while coptisine and jatrorrhizine were 8 and 2 times higher in C. teeta than that in C. chinensis, respectively. In rhizomes, berberine, coptisine, and jatrorrhizine were 2 times higher in C. chinensis than that in C. teeta. Morever, palmatine was detected only in C. chinensis. In roots, there were no significant differences among berberine, coptisine and jatrorrhizine in two Coptis species, whereas palmatine was 4 times higher in C. chinensis than that in C. teeta (Figure 3). The results of the present study thus further illustrate the similarities and differences in BIA contents and types between C. teeta and C. chinensis. Such differences may explain the reported variations in pharmacological activities between the two species (Feng et al., 2011). Further studies on their pharmacological activity will also be helpful in providing a scientific basis for using a particular Coptis species for treating a specific condition.
FIGURE 3.
The contents of five main BIAs in three organs of C. teeta (A) and C. chinensis (B). ∗P < 0.05, ∗∗P < 0.01.
Illumina Sequencing, de Novo Assembly, and Functional Annotation
To obtain a comprehensive C. teeta and C. chinensis transcriptome, 12 cDNA libraries, CtLF| CtRT and CcLF| CcRT, were constructed from total RNA extracted from the roots and leaves of two Coptis species, respectively. Three biological replications for each tissue were sequenced using the Illumina HiSeq 2000 sequencing platform. After removal of the adaptor sequences, ambiguous and low-quality reads (Q20 < 20), a total of 282,639,316 clean reads (total length: 39,748,135,308 bp; 39.7 Gb) and 312,917,026 clean reads (total length: 43,953,381,053; 44.0 Gb) were respectively generated. The Q20 (sequencing error rate < 1%) and GC percentages of CtLF| CtRT were 98.3 and 43.82%, respectively, and those of CcLF| CcRT were 98.33 and 43.66%, respectively (Supplementary Table S1). With the aid of the short-reads assembly software Trinity (Grabherr et al., 2011), the clean reads of the CtLF| CtRT cDNA libraries were de novo assembled into 81,823 unigenes, with an N50 length of 1,536 bp and a mean length of 810 bp (Supplementary Table S1, Supplementary Figure S1, and Supplementary Data S1). The other CcLF| CcRT cDNA libraries generated a total of 78,499 unigenes, with an average length of 784 bp and an N50 length of 1,491 bp (Supplementary Table S1, Supplementary Figure S1, and Supplementary Data S2). The high-quality reads obtained in this study were deposited in the NCBI SRA database (accession number SRA588794).
To annotate the assembled unigenes, the sequences were searched against four public protein databases: NR, Swiss-Prot, KOG, and KEGG. A total of 38,398 unigenes (46.93%) in C. teeta and 36,745 unigenes (46.81%) in C. chinensis were annotated in the public databases. Additionally, in C. teeta, 34,888 (42.64%), 29,615 (36.19%), 17,444 (21.32%), and 26,109 (31.91%) unigenes were annotated in the NR, Swiss-Prot, KEGG, and KOG databases, respectively. However, in C. chinensis, 34,273 (43.66%), 28,040 (35.72%), 15,992 (20.37%), and 24,285 (30.94%) unigenes were annotated in the NR, Swiss-Prot, KEGG, and KOG databases, respectively (Supplementary Table S2).
In the GO analyses, 23,066 unigenes from C. teeta were classified into three classes, including cellular components (9,155 unigenes), biological processes (9,371 unigenes), and molecular functions (4,540 unigenes). In C. chinensis, 26,005 unigenes were classified into three categories; namely, cellular components (11,046 unigenes), biological processes (10,108 unigenes), and molecular functions (4,851 unigenes) (Supplementary Figure S2). Approximately 40,493 and 37,121 unigenes from C. teeta and C. chinensis were respectively annotated and grouped into 24 KOG categories. In both C. teeta and C. chinensis, the largest cluster was general function prediction, which is related to only basic physiological and metabolic functions and accounted for 8,921 unigenes (22.03%) and 8,373 unigenes (22.56%), respectively. However, the smallest category was cell motility, which represented 0.03% of the annotated unigenes (Supplementary Figure S3). KEGG pathway analysis is helpful for understanding the biological functions and interactions of genes. A total of 5,894 unigenes from C. teeta were annotated in the KEGG database and assigned to 124 biological pathways. The largest pathway was the metabolic pathway, which consisted of 4,917 unigenes, included biosynthesis of other secondary metabolites (333 unigenes), which included BIA biosynthesis. In contrast, a total of 7,285 unigenes were annotated in the KEGG database and were assigned to 124 biological pathways in C. chinensis. Similar to C. teeta, the metabolic pathway that contained 6,066 unigenes was the largest one, included the biosynthesis of other secondary metabolites (406 unigenes) (Supplementary Figure S4).
Transcripts Encoding Enzymes Involved in BIA Biosynthesis
Benzylisoquinoline alkaloid synthesis starts from tyrosine, which forms the precursor (S)-reticuline via a multi-step enzymatic reaction. (S)-reticuline is then transformed into different proberberines, including berberine, coptisine, palmatine, columbamine, and magnoflorine. To date, biosynthetic pathways for berberine, palmatine, coptisine, columbamine, and magnoflorine have been characterized in other plant species (Ikezawa et al., 2008; Ziegler et al., 2009; Beaudoin and Facchini, 2014; Hagel et al., 2015). However, pathways for jatrorrhizine and epiberberine have not been determined to date. On the basis of previous reports and present transcriptome data, we hereby propose the biosynthetic pathways for berberine, palmatine, jatrorrhizine, coptisine, columbamine, magnoflorine, and epiberberine (Beecher and Kelleher, 1983; Rüffer et al., 1983) (Figure 1). According to the KEGG pathway, we discovered the transcripts encoding almost all the known enzymes for (S)-reticuline biosynthesis in our Illumina dataset, which included L-tyrosine aminotransferase (TyrAT), tyrosine decarboxylase (TYDC), tyrosine/tyramine 3-hydroxylase (3OHase), NCS, 6OMT, CNMT, NMCH, and 4′OMT (Table 1 and Supplementary Tables S3, S4). Berberine is widely distributed in plants and has been documented intensively (Sato and Yamada, 1984; Ikuta and Itokawa, 1988; Hagel and Facchini, 2013; Sato, 2013). Thus, several genes encoding proteins associated with berberine biosynthesis have been identified and characterized. These include the berberine bridge enzyme (BBE; Bird et al., 2003; Minami et al., 2008), which converts (S)-reticuline to berberine, SOMT (Takeshita et al., 1995), (S)-canadine synthase (CAS; Ikezawa et al., 2003), and (S)-tetrahydroprotoberberine oxidase (STOX; Matsushima et al., 2012; Sato, 2013). However, the pathway of epiberberine, which is closely related to berberine in structure, has not been determined. We propose that scoulerine is subsequently O-methylated at C2, which forms methylenedioxy bridges and undergoes oxidation to yield epiberberine, which is catalyzed by members of the OMT, CYP719, and OX families that consist of 18, 4, and 2 candidate genes, respectively. The coptisine, palmatine, columbamine, and magnoflorine biosynthesis pathways have previously been described as follows. For all of these pathways, (S)-scoulerine is first formed from (S)-reticuline by BBE (Facchini et al., 1996; Winkler et al., 2006). The conversion of (S)-scoulerine to coptisine begins with the formation of two methylenedioxy bridges by cheilanthifoline synthase (CFS) and stylopine synthase (SPS), which are members of the CYP719 family, forming (S)-stylopine. CFS and SPS have been isolated from E. californica (Ikezawa et al., 2009; Mizutani and Sato, 2011) and Argemone mexicana (Chavez et al., 2011) and characterized. Subsequent oxidation of (S)-stylopine by STOX yields coptisine (Matsushima et al., 2012). (S)-scoulerine is sequentially converted to (S)-tetrahydrocolumbamine by SOMT, to columbamine by STOX, and to palmatine by columbamine O-methyltransferase (CoOMT) (Pienkny et al., 2009; Ziegler et al., 2009). In magnoflorine biosynthesis, (S)-reticuline is first oxidized to (S)-corytuberine by (S)-corytuberine synthase (CTS), and subsequently, (S)-corytuberine-N-methyltransferase (SCNMT) converts (S)-corytuberine to magnoflorine (Ikezawa et al., 2008) (Table 1, Figure 1, and Supplementary Tables S3, S4).
Table 1.
Unigenes involved in benzylisoquinoline alkaloid biosynthesis in C. teeta and C. chinensis.
| Gene name (EC number) | C. teeta | C. chinensis |
|---|---|---|
| TYDC, tyrosine decarboxylase (4.1.1.25) | 5 | 4 |
| 3OHase, tyrosine/tyramine 3-hydroxylase (1.14.16.2) | 6 | 7 |
| TyrAT, L-tyrosine aminotransferase (2.6.1.5) | 2 | 3 |
| 4HPPDC, 4-hydroxyphenylpuruvate decarboxylase (4.1.1.80) | 0 | 0 |
| NCS, (S)-norcoclaurine synthase (4.2.1.78) | 14 | 14 |
| 6OMT, (S)-norcoclaurine 6-O-methyltransferase (2.1.1.128) | 2 | 3 |
| CNMT, (S)-coclaurine N-methyltransferase (2.1.1.140) | 5 | 4 |
| NMCH, N-methylcoclaurine 3′-monooxygenase (1.14.13.71) | 2 | 1 |
| 4′OMT, 3′-hydroxy-N-methyl-(S)-coclaurine 4′-O-methyltransferase (2.1.1.116) | 2 | 1 |
| BBE, berberine bridge enzyme (1.21.3.3) | 8 | 8 |
| SOMT, (S)-scoulerine 9-O-methyltransferase (2.1.1.117) | 1 | 1 |
| CAS, (S)-canadine synthase (1.14.21.5) | 2 | 2 |
| STOX, (S)-tetrahydroprotoberberine oxidase (1.3.3.8) | 1 | 1 |
| CTS, (S)-corytuberine synthase (1.14.21.-) | 1 | 1 |
| CoOMT, columbamine O-methyltransferase (2.1.1.118) | 2 | 2 |
| CFS, (S)-cheilanthifoline synthase (1.14.21.2) | 0 | 0 |
| SPS, (S)-stylopine synthase (1.14.21.1) | 0 | 0 |
| All number of unigenes | 53 | 52 |
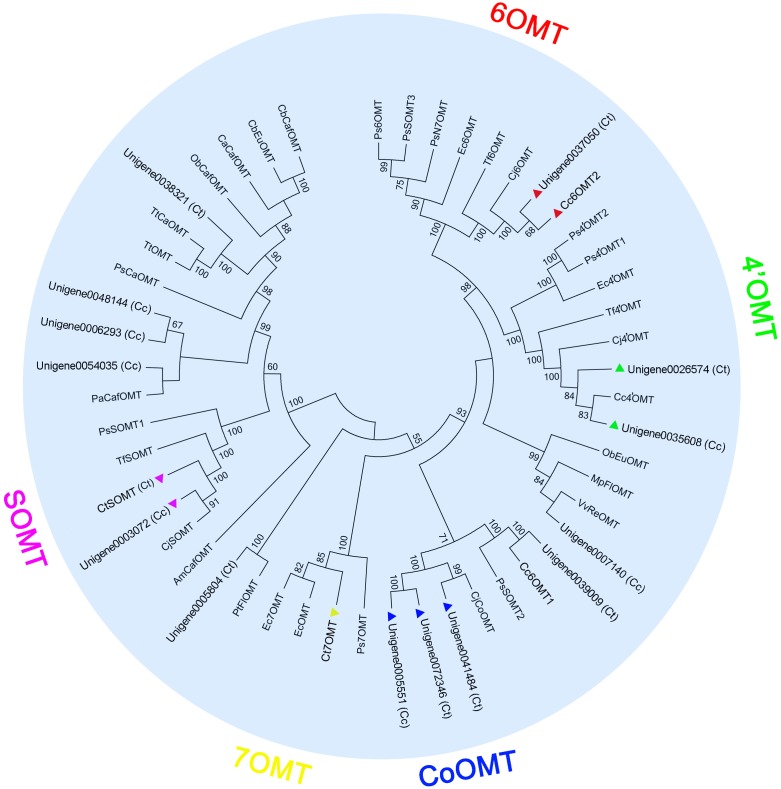
Structurally, jatrorrhizine is a special protoberberine because it contains an unusual 7-O-methylation pattern. Previous studies have suggested that jatrorrhizine is synthesized via ring-opening of berberine (Beecher and Kelleher, 1983; Rüffer et al., 1983) (Figure 1, pathway 1). However, the enzyme catalyzing this ring-opening reaction has not been investigated to date. Six-O-methyltransferase catalyzes the formation of 6-O-methylnorlaudanosoline accompanied by a smaller amount of 7-O-methylnorlaudanosoline, which initiates its biosynthesis using norlaudanosoline instead of reticuline; however, testing for the relevant enzyme activity has not been conducted (Beecher and Kelleher, 1983; Rüffer et al., 1983). In the present study, we obtained 18 unigenes encoding OMT from our transcriptome data. Phylogenetic tree analysis revealed that unigenes 0037050 (Ct) and 0033238 (Cc) (designated as Cc6OMT2, MH165876) had high sequence identity with Cj6OMT, whereas unigenes 0026574 (Ct) and 0035608 (Cc) were clustered closely with 4′OMTs from C. coptis and other plants. Unigenes 0042287 (Ct) (designated as CtSOMT, MH165874) and 0003072 (Cc) had high sequence homology with CjSOMT, whereas unigenes 0041484 (Ct), 0072346 (Ct), and 0005551 (Cc) had high sequence homology with CjCoOMT. Unigene 0046714 (Ct) (designated as Ct7OMT, MH165877) clustered closely with 7OMT, and eight other OMTs [unigenes 0042981 (Cc) (designated as Cc6OMT1, MH165875), 0039009 (Ct), 0007140 (Cc), 0005804 (Ct), 0054035 (Cc), 0006293 (Cc), 0048144 (Cc), and 0038321 (Ct)] were clustered together with different known OMTs (Figure 4).
FIGURE 4.
Phylogenetic tree of OMTs. Phylogenetic tree was constructed based on the deduced amino acid sequences from C. teeta and C. chinensis OMTs (triangles of different colors) and other plant OMTs. Abbreviations and GenBank accession numbers for the sequences used are as follows: EcOMT, putative E. californica OMT (ACO90220.1); CjCoOMT, C. japonica columbamine OMT (Q8H9A8.1); TtOMT, Thalictrum tuberosum catechol OMT (AAD29845.1); TtCaOMT, T. tuberosum catechol OMT (AAD29843.1); PsCaOMT, P. somniferum catechol OMT (AAQ01670.1); PsN7OMT, P. somniferum norreticuline 7OMT (ACN88562.1); Ps6OMT, P. somniferum norcoclaurine 6OMT (AAP45315.1); Ps4′OMT2, P. somniferum 3′-hydroxy-N-methylcoclaurine 4′OMT2 (AAP45314.1); Ps4′OMT1, P. somniferum 3′-hydroxy-N-methylcoclaurine 4′OMT1 (AAP45314.1); Cc4′OMT, C. chinensis 3′-hydroxy-N-methylcoclaurine 4′OMT (ABY75613.1); Cj4′OMT, C. japonica 3′-hydroxy-N-methylcoclaurine 4′OMT (Q9LEL5.1); Tf4′OMT, T. flavum 3′-hydroxy-N-methylcoclaurine 4′OMT (AAU20768.1); Cj6OMT, C. japonica norcoclaurine 6OMT (Q9LEL6.1); Tf6OMT, T. flavum norcoclaurine 6OMT (AAU20765.1); VvReOMT, Vitis vinifera resveratrol OMT (CAQ76879.1); PtFlOMT, Populus trichocarpa flavonoid OMT predicted protein (XP_002312933.1); CjSOMT, C. japonica scoulerine 9OMT (Q39522.1); TfSOMT, T. flavum scoulerine 9OMT(AAU20770.1); PaCafOMT, Picea abies caffeate OMT (CAI30878.1); CaCafOMT, Capsicum annuum caffeate OMT (AAG43822.1); PsOMT1, P. somniferum SOMT1 (JN185323); PsOMT2, P. somniferum SOMT2 (JN185324); PsOMT3, P. somniferum SOMT3 (JN185325); ObEuOMT, Ocimum basilicum eugenol OMT (AAL30424.1); MpFlOMT, Mentha X piperita flavonoid 8OMT (AAR09600.1); ObCafOMT, O. basilicum caffeate OMT (AAD38189.1); CbEuOMT, Clarkia breweri (iso)eugenol OMT (AAC01533.1); CbCafOMT, C. breweri caffeate OMT (O23760.1); AmCafOMT, Ammi majus caffeate OMT (AAR24095.1); Ec7OMT, E. californica reticuline 7OMT (BAE79723.1); Ps7OMT, P. somniferum reticuline 7OMT (AAQ01668.1); Ec4′OMT, E. californica 3′-hydroxy-N-methylcoclaurine 4′OMT (BAM37633.1); and Ec6OMT, E. californica OMT (BAM37634.1).
Functional Characterization of OMT Candidates
Jatrorrhizine is structurally similar to berberine, functions in detoxification and imparts bactericidal properties and hypoglycemic effects (Arens et al., 1985; Moody et al., 1995; Volleková et al., 2003); however, its biosynthetic pathway remains unclear. Molecular structural analysis indicates that jatrorrhizine contains an unusual 7-O-methylation pattern that makes it difficult to deduce its biosynthesis from reticuline, the common precursor of BIAs. Previous studies involving 14C isotopic tracers have shown that 6OMT catalyzes the formation of 6-O-methylnorlaudanosoline along with a smaller amount of 7-O-methylnorlaudanosoline; thus, it is probable that the 7-O-methylation pattern of jatrorrhizine is already established at the norlaudanosoline step (Beecher and Kelleher, 1983; Rüffer et al., 1983). In the present study, five 6OMT unigenes; namely, 0039009 (Ct), 0037050 (Ct), 0071836 (Cc), Cc6OMT1, Cc6OMT2, and one 7OMT unigene, Ct7OMT, were selected to study their function by in vitro enzyme activity analysis. Figure 5A shows that incubation of Cc6OMT1, Cc6OMT2 and Ct7OMT with (S)-norcoclaurine (mass-to-charge ratio [m/z] 272.13) yielded two peaks with m/z 286.14 at 3.55 min and 3.83 min, respectively. A molecular formula (C17H20NO3) was obtained by their accurate molecular weight and element composition analysis. One product with m/z 286.14 at 3.55 min was identified as (S)-coclaurine by comparison of retention time and mass spectra with authentic standard. Based on accurate molecular weight and molecular formula, another product with m/z 286.14 at 3.83 min was proposed as isococlaurine which contains a 7-O-methylation pattern. This means that Cc6OMT1 and Cc6OMT2 are capable of O-methylating (S)-norcoclaurine at C6, yielding (S)-coclaurine along with a smaller amount of O-methylation at C7, thereby forming its isomer, isococlaurine, and Ct7OMT also O-methylates (S)-norcoclaurine at C6, yielding (S)-coclaurine, along with O-methylation of (S)-norcoclaurine at C7, yielding isococlaurine (Figure 5A and Supplementary Figure S5). Subsequently, isococlaurine can be further transformed into jatrorrhizine through a series of enzymatic reactions (Figure 1, pathway 2). This result validates the previous results of feeding experiments with distant single or doubly labeled precursors (Rüffer et al., 1983), and shows that jatrorrhizine can be formed by (S)-norcoclaurine C7 O-methylation, thereby providing a molecular basis for jatrorrhizine synthesis via (S)-norcoclaurine. Despite their names suggesting strict functions, characterized OMTs of BIAs often display diversify functions in different species. For example, P. somniferum 6OMT specifically 6-O-methylated (R,S)-norcoclaurine, while 6OMT also 7-O-methylated (R,S)-isoorientaline. 6OMT from C. japonica efficiently 6-O-methylated (R,S)-norcoclaurine, (R,S)-norlaudanosoline, and (R,S)-laudanosoline, but weakly O-methylated (R,S)-6-O-methylnorlaudanosoline, (S)-scoulerine, and (R,S)-coclaurine, clearly not at the already O-methylated 6-position in each case (Sato et al., 1993; Morishige et al., 2000; Chang et al., 2015). Therefore, it is acceptable that Cc6OMT from C. chinensis was able to 6-O-methylate (S)-norcoclaurine, yielding (S)-coclaurine along with a smaller amount of O-methylation at C7. For 7OMT, P. somniferum 7OMT 7-O-methylated (R,S)-reticuline, (R,S)-orientaline, and (R,S)-protosinomenine, (R,S)-isoorientaline. Conversely, our present results showed that Ct7OMT from C. teeta mainly O-methylates (S)-norcoclaurine at C6, yielding (S)-coclaurine, along with O-methylation of (S)-norcoclaurine at C7, yielding isococlaurine that has been confirmed for the first time in all species. Therefore, we suggest that Ct7OMT should be more properly referred to as 6-/7- O-methyltransferase (Ct6/7OMT), which may be related to the particularity of C. teeta. Overall, depending on different species, both 6OMT and 7OMT often display a range of substrate, which contribute overlapped and divergent functions.
FIGURE 5.
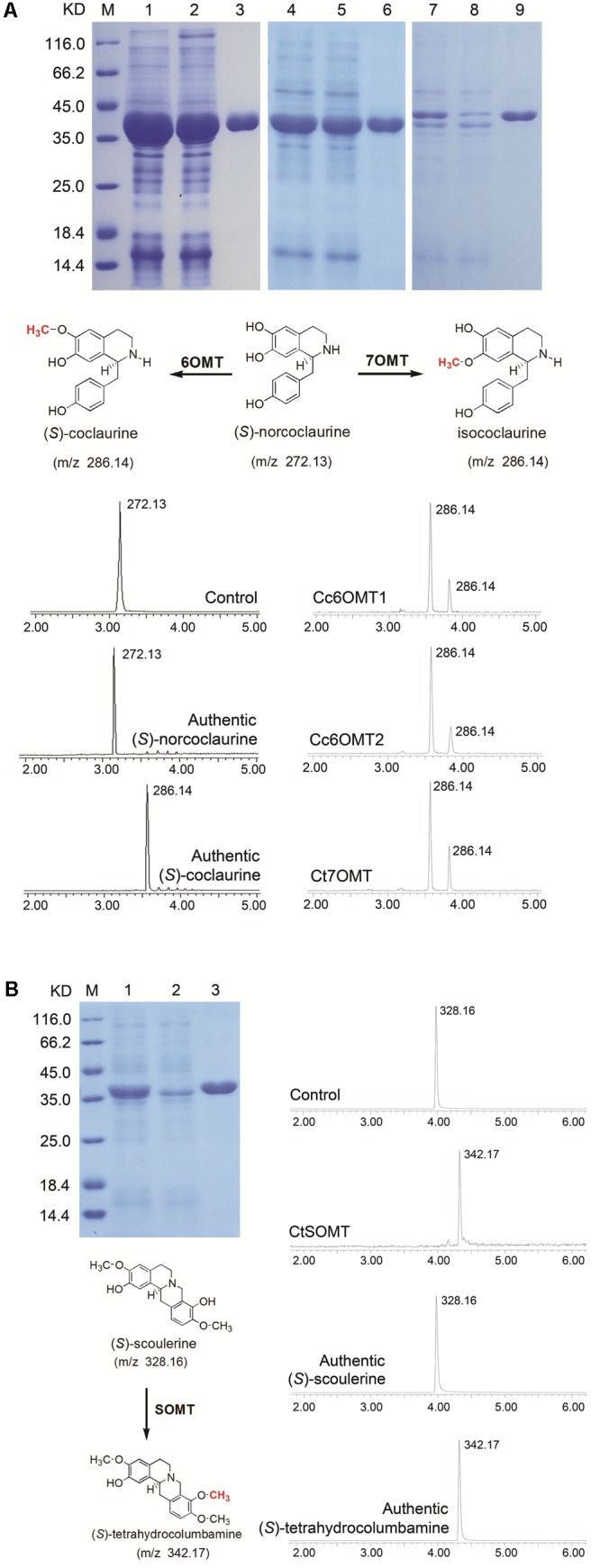
Extract ion chromatograms showing the O-methylation activity of recombinant OMTs on various substrates. (A) His-tag purified recombinant OMTs on 12% SDS–PAGE gel, Lane M: Protein Marker, Lane 1: Unpurified Cc6OMT1, Lane 2: Flow through Cc6OMT1, Lane 3: Elution Cc6OMT1; Lane 4: Unpurified Cc6OMT2, Lane 5: Flow through Cc6OMT2, Lane 6: Elution Cc6OMT2; Lane 7: Unpurified Ct7OMT, Lane 8: Flow through Ct7OMT, Lane 9: Elution Ct7OMT. Verified reaction equation, the top (control) substrate with denatured purified OMTs proteins, in vitro assay products of Cc6OMT1, Cc6OMT2, Ct7OMT with (S)-norcoclaurine, respectively. (B) His-tag purified recombinant OMT on 12% SDS–PAGE gel, Lane M: Protein Marker, Lane 1: Unpurified CtSOMT, Lane 2: Flow through CtSOMT, Lane 3: Elution CtSOMT; verified reaction equation; the top (control) substrate with denatured purified OMT protein, in vitro assay product of CtSOMT with (S)-scoulerine. Products were identified based on retention times and ESI[+]-CID spectra using authentic standards, 1. (S)-norcoclaurine, 2. (S)-coclaurine, 3. (S)-scoulerine, 4. (S)-tetrahydrocolumbamine.
Furthermore, previous studies have shown that SOMT1 sequentially 9- and 2-O-methylates (S)-scoulerine, yielding (S)-tetrahydrocolumbamine and (S)-tetrahydropalmatine, respectively (Dang and Facchini, 2012). By comparing their molecular structures, we propose that epiberberine is synthesized through O-methylation of (S)-scoulerine at C2 (Figure 1). Thus, two unigenes, CtSOMT and 0003072 (Cc), were selected to study their enzyme activity. The results showed incubation of CtSOMT with (S)-scoulerine (m/z 328.16) yielded only one reaction product identified as tetrahydrocolumbamine (m/z 342.17) based on authentic standards. It means that CtSOMT has stringent substrate specificity because 9OMT targets (S)-scoulerine, but it could not O-methylate (S)-scoulerine at C2 (Figure 5B and Supplementary Figure S6). This may be attributable to the fact that C. chinensis has no (S)-tetrahydropalmatine, thereby leading to the inability of SOMT to 2-O-methylate (S)-tetrahydrocolumbamine.
Expression Patterns of Putative Genes Involved in BIA Biosynthesis
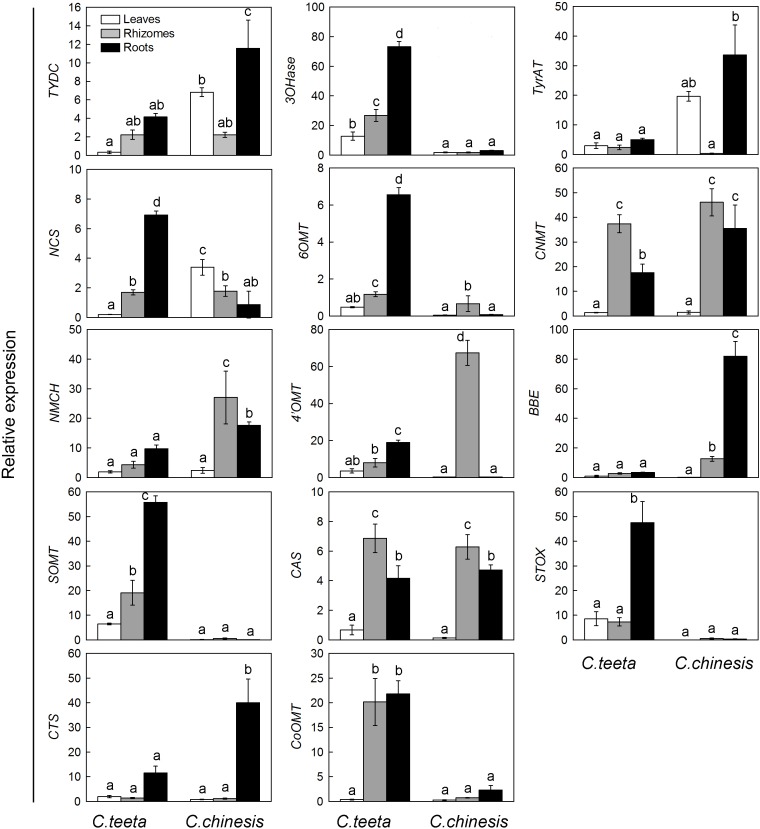
To investigate the expression patterns of candidate genes involved in BIA biosynthesis in two Coptis species, the expression levels of 14 unigenes related to protoberberine biosynthesis in the roots, rhizomes, and leaves of C. teeta and C. chinensis were performed by qRT-PCR. Figure 6 indicates that most genes were upregulated in the roots rather than in the leaves of the two Coptis species. In C. teeta, the highest expression was observed in the roots and the lowest in the leaves, whereas some genes were more highly expressed in the rhizomes than in the roots in C. chinensis, such as NCS, 6OMT, 4′OMT, and SOMT. The enzymes encoded by the genes NCS, 6OMT, and 4′OMT are located upstream of the BIA synthetic pathway from a key branch-point intermediate (S)-reticuline, whereas SOMT is involved in the synthesis of berberine and palmatine. The differential gene expression patterns of two Coptis species may have resulted from their growth environment, climate, topography, other environmental factors, and cultivation conditions (Chen et al., 2017). Moreover, these results coincided with the higher contents of major BIAs in the roots of the two Coptis species (Figure 3). BIA synthesis and accumulation in Thalictrum (Ranunculaceae) were in the pith and cortex of the roots and rhizomes (Samanani et al., 2005; Lee et al., 2013), suggesting that the main sites of BIA synthesis and accumulation in C. chinensis were probably in the roots.
FIGURE 6.
Expression patterns of candidate unigenes involved in BIA biosynthesis in C. teeta and C. chinensis. The transcripts were analyzed by qRT-PCR, with actin as an internal standard.
Conclusion
We conducted transcriptomic analysis of C. teeta and C. chinensis and obtained a total of 81,823 and 78,499 unigenes, respectively, which provide valuable genetic resources for these invaluable Chinese herb medicines. We propose the integrated biosynthetic pathways of berberine, palmatine, jatrorrhizine, coptisine, columbamine, magnoflorine, and epiberberine, and identified 53 and 52 unigenes involved in the biosynthesis of the protoberberine alkaloids in the two Coptis species C. teeta and C. chinensis, respectively. Further enzyme activity testing in vitro demonstrated that two 6OMTs and one 7OMT were able to 6-O-methylate (S)-norcoclaurine, yielding (S)-coclaurine along with a smaller amount of O-methylation at C7, whereas SOMT specifically catalyzed O-methylation at C2 on (S)-scoulerine, yielding (S)-tetrahydrocolumbamine. These results provide opportunities for the de novo production of active ingredients by microorganism engineering.
Author’s Note
This paper is dedicated to Mrs. Li, Guanghui Zhang’s wife, who has been suffering from ataxia for many years. We are grateful to those who are willing to help her. Please contact zgh73107310@163.com.
Author Contributions
WF and S-CY conceived the study. S-MH, Y-LL, KC, XW, YD, GC, J-JZ, Q-MZ, Z-LQ, J-LY, and XZ performed the experiments and carried out the analysis. S-MH, WF, S-CY, and G-HZ designed the experiments and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Funding. This work was supported financially by the National Natural Science Foundation of China (81460583), the Pilot Project for Establishing Social Service System through Agricultural Science and Education in Yunnan Province, the Medical Plant Unit (2014NG003), and the Project of Young and Middle-Aged Talent of Yunnan Province (2014HB011).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00731/full#supplementary-material
References
- Arens H., Fischer H., Leyck S., Römer A., Ulbrich B. (1985). Antiinflammatory compounds from Plagiorhegma dubium cell culture1. Planta Med. 51 52–56. 10.1055/s-2007-969392 [DOI] [PubMed] [Google Scholar]
- Beaudoin G. A., Facchini P. J. (2014). Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta 240 19–32. 10.1007/s00425-014-2056-8 [DOI] [PubMed] [Google Scholar]
- Beecher C. W. W., Kelleher W. J. (1983). The incorporation of berberine into jatrorrhizine. Tetrahedron Lett. 24 469–472. 10.1016/S0040-4039(00)81439-2 [DOI] [Google Scholar]
- Bird D. A., Franceschi V. R., Facchini P. J. (2003). A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15 2626–2635. 10.1105/tpc.015396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Hagel J. M., Facchini P. J. (2015). Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol. 169 1127–1140. 10.1104/pp.15.01240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C. (1998). Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 311–343. 10.1146/annurev.arplant.49.1.311 [DOI] [PubMed] [Google Scholar]
- Chavez M. L., Rolf M., Gesell A., Kutchan T. M. (2011). Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 507 186–193. 10.1016/j.abb.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Chen H., Deng C., Nie H., Fan G., He Y. (2017). Transcriptome analyses provide insights into the difference of alkaloids biosynthesis in the Chinese goldthread (Coptis chinensis Franch.) from different biotopes. PeerJ 5:e3303. 10.7717/peerj.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. B., Morishige T., Shitan N., Yazaki K., Sato F. (2002). Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277 830–835. 10.1074/jbc.M106405200 [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Cui L., Liu M., Chang X., Sun K. (2016). The inhibiting effect of the Coptis chinensis polysaccharide on the type II diabetic mice. Biomed. Pharmacother. 81 111–119. 10.1016/j.biopha.2016.03.038 [DOI] [PubMed] [Google Scholar]
- Dang T. T., Facchini P. J. (2012). Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159 618–631. 10.1104/pp.112.194886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini P. J., Penzes C., Johnson A. G., Bull D. (1996). Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 112 1669–1677. 10.1104/pp.112.4.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G., Zhang M. Y., Zhou X. D., Lai X. R., Yue Q. H., Tang C., et al. (2012). Quality evaluation and species differentiation of Rhizoma coptidis by using proton nuclear magnetic resonance spectroscopy. Anal. Chim. Acta 747 76–83. 10.1016/j.aca.2012.08.038 [DOI] [PubMed] [Google Scholar]
- Feng X., Yan D., Zhao K. J., Luo J. Y., Ren Y. S., Kong W. J., et al. (2011). Applications of microcalorimetry in the antibacterial activity evaluation of various Rhizoma coptidis. Pharm. Biol. 49 348–353. 10.3109/13880209.2010.523428 [DOI] [PubMed] [Google Scholar]
- Feng Y., Luo W. Q., Zhu S. Q. (2008). Explore new clinical application of Huanglian and corresponding compound prescriptions from their traditional use. China J. Chin. Mater. Med. 33 1221–1225. [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42 222–230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedemann T., Ying Y., Wang W., Kramer E. R., Schumacher U., Fei J., et al. (2016). Neuroprotective effect of Coptis chinensis in MPP[Formula: see text] and MPTP-Induced Parkinson’s disease models. Am. J. Chin. Med. 44 907–925. 10.1142/S0192415X16500506 [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel J. M., Facchini P. J. (2013). Benzylisoquinoline alkaloid metabolism–a century of discovery and a brave new world. Plant Cell Physiol. 54 647–672. 10.1093/pcp/pct020 [DOI] [PubMed] [Google Scholar]
- Hagel J. M., Morris J. S., Lee E. J., Desgagné-Penix I., Bross C. D., Chang L., et al. (2015). Metabolome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 15:220. 10.1186/s12870-015-0594-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. M., Song W. L., Cong K., Wang X., Dong Y., Cai J., et al. (2017). Identification of candidate genes involved in isoquinoline alkaloids biosynthesis in Dactylicapnos scandens by transcriptome analysis. Sci. Rep. 7:9119. 10.1038/s41598-017-08672-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. (2008). Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283 8810–8821. 10.1074/jbc.M705082200 [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. (2009). CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 28 123–133. 10.1007/s00299-008-0624-8 [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inouye K., et al. (2003). Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 278 38557–38565. 10.1074/jbc.M302470200 [DOI] [PubMed] [Google Scholar]
- Ikuta A., Itokawa H. (1988). “Berberine; production through plant (Thalictrum spp.) cell culture,” in Medicinal and Aromatic Plants I, ed. Bajaj Y. P. S. (Berlin: Springer; ), 282–298. 10.1007/978-3-642-73026-9_15 [DOI] [Google Scholar]
- Iseli C., Jongeneel C. V., Bucher P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 99 138–148. [PubMed] [Google Scholar]
- Kamath S., Skeels M., Pai A. (2009). Significant differences in alkaloid content of Coptis chinensis (Huanglian), from its related American species. Chin. Med. 4 1–17. 10.1186/1749-8546-4-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Ahn S., Rhee H. I., Lee D. C. (2016). Coptis chinensis Franch. extract up-regulate type I helper T-cell cytokine through MAPK activation in MOLT-4 T cell. J. Ethnopharmacol. 189 126–131. 10.1016/j.jep.2016.05.046 [DOI] [PubMed] [Google Scholar]
- Lee E. J., Hagel J. M., Facchini P. J. (2013). Role of the phloem in the biochemistry and ecophysiology of benzylisoquinoline alkaloid metabolism. Front. Plant Sci. 4:182. 10.3389/fpls.2013.00182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yan D., Ma L., Xiong Y., Yan C., Li B., et al. (2013). A quality evaluation strategy for Rhizoma coptidis from a variety of different sources using chromatographic fingerprinting combined with biological fingerprinting. Chin. Sci. Bull. 58 4092–4100. 10.1007/s11434-013-5861-8 [DOI] [Google Scholar]
- Liscombe D. K., MacLeod B. P., Loukanina N., Nandi O. I., Facchini P. J. (2005). Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66 1374–1393. 10.1016/j.phytochem.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Lv X., Li Y., Tang C., Zhang Y., Zhang J., Fan G. (2016). Integration of HPLC-based fingerprint and quantitative analyses for differentiating botanical species and geographical growing origins of Rhizoma coptidis. Pharm. Biol. 54 3264–3271. 10.1080/13880209.2016.1223699 [DOI] [PubMed] [Google Scholar]
- Matsushima Y., Minami H., Hori K., Sato F. (2012). Pathway engineering of benzylisoquinoline alkaloid biosynthesis in transgenic California poppy cells with ectopic expression of tetrahydroberberine oxidase from Coptis japonica. Plant Biotechnol. 29 473–481. 10.5511/plantbiotechnology.12.1101a [DOI] [Google Scholar]
- Minami H., Dubouzet E., Iwasa K., Sato F. (2007). Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 282 6274–6282. 10.1074/jbc.M608933200 [DOI] [PubMed] [Google Scholar]
- Minami H., Kim J. S., Ikezawa N., Takemura T., Katayama T., Kumagai H., et al. (2008). Microbial production of plant benzylisoquinoline alkaloids. Proc. Natl. Acad. Sci. U.S.A. 105 7393–7398. 10.1073/pnas.0802981105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Sato F. (2011). Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 507 194–203. 10.1016/j.abb.2010.09.026 [DOI] [PubMed] [Google Scholar]
- Moody J. O., Bloomfield S. F., Hylands P. J. (1995). In-vitro evaluation of the antimicrobial activities of Enantia chlorantha Oliv. Extractives. Afr. J. Med. Med. Sci. 24 269–273. [PubMed] [Google Scholar]
- Morishige T., Tamakoshi M., Takemura T., Sato F. (2010). Molecular characterization of O-methyltransferases involved in isoquinoline alkaloid biosynthesis in Coptis japonica. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86 757–768. 10.2183/pjab.86.757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige T., Tsujita T., Yamada Y., Sato F. (2000). Molecular characterization of the S-adenosyl-L-methionine: 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 275 23398–23405. 10.1074/jbc.M002439200 [DOI] [PubMed] [Google Scholar]
- Pienkny S., Brandt W., Schmidt J., Kramel R., Ziegler J. (2009). Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J. 60 56–67. 10.1111/j.1365-313X.2009.03937.x [DOI] [PubMed] [Google Scholar]
- Robin A. Y., Giustini C., Graindorge M., Matringe M., Dumas R. (2016). Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 87 641–653. 10.1111/tpj.13225 [DOI] [PubMed] [Google Scholar]
- Rüffer M., Ekundayo O., Nagakura N., Zenk M. H. (1983). Biosynthesis of the protoberberine alkaloid jatrorrhizine. Tetrahedron Lett. 24 2643–2644. 10.1016/S0040-4039(00)87966-6 [DOI] [Google Scholar]
- Samanani N., Alcantara J., Bourgault R., Zulak K. G., Facchini P. J. (2006). The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J. 47 547–563. 10.1111/j.1365-313X.2006.02801.x [DOI] [PubMed] [Google Scholar]
- Samanani N., Facchini P. J. (2002). Purification and characterization of norcoclaurine synthase - The first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. Biol. Chem. 277 33878–33883. 10.1074/jbc.M203051200 [DOI] [PubMed] [Google Scholar]
- Samanani N., Liscombe D. K., Facchini P. J. (2004). Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 40 302–313. 10.1111/j.1365-313X.2004.02210.x [DOI] [PubMed] [Google Scholar]
- Samanani N., Park S. U., Facchini P. J. (2005). Cell-type specific localization of transcripts encoding nine consecutive enzymes involved in protoberberine alkaloid biosynthesis. Plant Cell 17 915–926. 10.1105/tpc.104.028654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F. (2013). Characterization of plant functions using cultured plant cells, and biotechnological applications. Biosci. Biotechnol. Biochem. 77 1–9. 10.1271/bbb.120759 [DOI] [PubMed] [Google Scholar]
- Sato F., Inui T., Takemura T. (2007). Metabolic engineering in isoquinoline alkaloid biosynthesis. Curr. Pharm. Biotechnol. 8 211–218. 10.2174/138920107781387438 [DOI] [PubMed] [Google Scholar]
- Sato F., Takeshita N., Fitchen J. H., Fujiwara H., Yamada Y. (1993). S-adenosyl-L-methionine: scoulerine-9-O-methyltransferase from cultured Coptis japonica cells. Phytochemistry 32 659–664. 10.1016/S0031-9422(00)95151-3 [DOI] [PubMed] [Google Scholar]
- Sato F., Yamada Y. (1984). High berberine-producing cultures of Coptis japonica cells. Phytochemistry 23 281–285. 10.1016/S0031-9422(00)80318-0 2463630 [DOI] [Google Scholar]
- Takemura T., Ikezawa N., Iwasa K., Sato F. (2013). Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry 91 100–108. 10.1016/j.phytochem.2012.02.013 [DOI] [PubMed] [Google Scholar]
- Takeshita N., Fujiwara H., Mimura H., Fitchen J. H., Yamada Y., Sato F. (1995). Molecular cloning and characterization of S-adenosyl-Lmethionine:scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 36 29–36. [PubMed] [Google Scholar]
- Tang J., Feng Y., Tsao S., Wang N., Curtain R., Wang Y. (2009). Berberine and Coptidis Rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. J. Ethnopharmacol. 126 5–17. 10.1016/j.jep.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Tjong Y., Ip S., Lao L., Fong H. H., Sung J. J., Berman B., et al. (2011). Analgesic effect of Coptis chinensis rhizomes (Coptidis Rhizoma) extract on rat model of irritable bowel syndrome. J. Ethnopharmacol. 135 754–761. 10.1016/j.jep.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volleková A., Kost’álová D., Kettmann V., Tóth J. (2003). Antifungal activity of Mahonia aquifolium extract and its major protoberberine alkaloids. Phytother. Res. 17 834–837. 10.1002/ptr.1256 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu W. J., Cui Y. (2014). Modern research progress of Huanglian. China J. Chin. Med. 29 1642–1645. [Google Scholar]
- Winkler A., Hartner F., Kutchan T. M., Glieder A., Macheroux P. (2006). Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 281 21276–21285. 10.1074/jbc.M603267200 [DOI] [PubMed] [Google Scholar]
- Xiang K. L., Wu S. D., Yu S. X., Liu Y., Jabbour F., Erst S., et al. (2016). The first comprehensive phylogeny of Coptis (Ranunculaceae) and its implications for character evolution and classification. PLoS One 11:e0153127. 10.1371/journal.pone.0153127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Liu Y., Ye L., Li C., Yang Q., Wang W., et al. (2017). Rapid and simultaneous analysis of five alkaloids in four parts of Coptidis Rhizoma by near-infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 188 611–618. 10.1016/j.saa.2017.07.053 [DOI] [PubMed] [Google Scholar]
- Yamada Y., Motomura Y., Sato F. (2015). CjbHLH1 homologs regulate sanguinarine biosynthesis in Eschscholzia californica cells. Plant Cell Physiol. 56 1019–1030. 10.1093/pcp/pcv027 [DOI] [PubMed] [Google Scholar]
- Yang H., Lee D. Y., Kang K. B., Kim J. Y., Kim S. O., Yoo Y. H., et al. (2015). Identification of ginsenoside markers from dry purified extract of Panax ginseng by a dereplication approach and UPLC–QTOF/MS analysis. J. Pharm. Biomed. Anal. 109 91–104. 10.1016/j.jpba.2015.02.034 [DOI] [PubMed] [Google Scholar]
- Yang J., Liu Z., Liu B., Zhu Q. (2012). Determination of Coptis chinensis’ quality by FT-NIR spectroscopy. Health 4 196–202. 10.4236/health.2012.44029 [DOI] [Google Scholar]
- Yang W. Z., Jin H., Li W. Y., Zhang Z. H., Zhao Z. L., Zhang J. Y. (2013). A study on phenotypic diversity in different populations of endangered Coptis teeta wall of Yunnan. J. Yunnan Univ. 35 719–726. [Google Scholar]
- Yang Y., Peng J., Li F., Liu X., Deng M., Wu H. (2017). Determination of alkaloid contents in various tissues of Coptis chinensis Franch. by reversed phase-high performance liquid chromatography and ultraviolet spectrophotometry. J. Chromatogr. Sci. 55 556–563. 10.1093/chromsci/bmx009 [DOI] [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., et al. (2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34 293–297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Feng Y., Tong Y., Ng K. M., Tsao S., Lau G. K., et al. (2009). Hepatoprotective effects of Coptidis rhizoma aqueous extract on carbon tetrachloride-induced acute liver hepatotoxicity in rats. J. Ethnopharmacol. 124 130–136. 10.1016/j.jep.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Zhang G. H., Ma C. H., Zhang J. J., Chen J. W., Tang Q. Y., He M. H., et al. (2015). Transcriptome analysis of Panax vietnamensis var. fuscidicus discovers putative ocotillol-type ginsenosides biosynthesis genes and genetic markers. BMC Genomics 16:159. 10.1186/s12864-015-1332-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Piao X. L., Piao X. S., Lu T., Wang D., Kim S. W. (2011). Preventive effect of Coptis chinensis and berberine on intestinal injury in rats challenged with lipopolysaccharides. Food Chem. Toxicol. 49 61–69. 10.1016/j.fct.2010.09.032 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Geng Y. F., Zhang L. L., Wang L., He L. J., Wang C., et al. (2015). Finding new sources from “using different plants as the same herb”: a case study of Huang-lian in Northwest Yunnan, China. J. Ethnopharmacol. 169 413–425. 10.1016/j.jep.2015.04.051 [DOI] [PubMed] [Google Scholar]
- Ziegler J., Facchini P. J. (2008). Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59 735–769. 10.1146/annurev.arplant.59.032607.092730 [DOI] [PubMed] [Google Scholar]
- Ziegler J., Facchini P. J., Geißler R., Schmidt J., Ammer C., Kramell R., et al. (2009). Evolution of morphine biosynthesis in opium poppy. Phytochemistry 70 1696–1707. 10.1016/j.phytochem.2009.07.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.