Abstract
For the microviruses and the leviviruses, bacteriophages with small single-stranded genomes, host lysis is accomplished by expression of a single gene that encodes an inhibitor of cell wall synthesis. In contrast, phages with double-stranded DNA genomes use a more complex system involving, at minimum, an endolysin, which degrades peptidoglycan, and a holin, which permeabilizes the membrane in a temporally programmed manner. To explore the basis of this difference, a chimera was created in which lysis gene E of the microvirus ϕX174 replaced the entire lysis cassette of phage λ, which includes the holin gene S and the endolysin gene R. The chimeric phage was viable but more variability was observed both in the distribution of plaque sizes and in the burst sizes of single cells, compared to the isogenic S+ parent. Using different alleles of E, it was found the average burst size increased with the duration of the latent period, just as observed with S alleles with different lysis times. Moreover, within a set of missense E alleles, it was found that variability in lysis timing was limited and almost exclusively derived from changes in the level of E accumulation. By contrast, missense mutations in S resulted in a wide variation in lysis times that was not correlated with levels of accumulation. We suggest that the properties of greater phenotypic plasticity and lesser phenotypic variation make the function of holin proteins more genetically malleable, facilitating rapid adaptation towards a lysis time that would be optimal for changed host and environmental conditions. The inferior malleability of single-gene systems like E would restrict their occurrence to phages in which coding capacity is the overriding evolutionary constraint.
INTRODUCTION
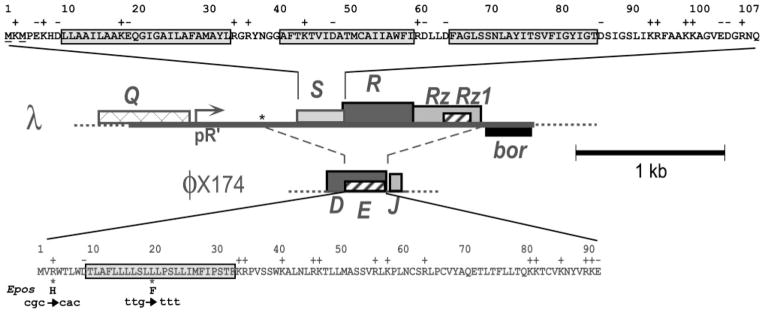
Phages use two fundamentally different host lysis strategies (Young & Wang, 2006). Double-stranded DNA (dsDNA) and double-stranded RNA (dsRNA) phages use a holin-endolysin system (Young et al., 2000). The endolysin, or lysozyme, is an enzyme that has one or more murein-degrading activities. The holin is a small membrane protein that controls the activation of the endolysin or its access to the murein (Wang et al., 2000; Xu et al., 2004, 2005) and thus constitutes the ‘clock’ of the bacteriophage infection cycle. The best-studied holin, the S product of the λ S gene (Fig. 1), accumulates in the membrane throughout the late gene expression period, without affecting membrane integrity or energization, until suddenly, at an allele-specific time, it triggers ‘hole formation’ (Gründling et al., 2001). This allows the endolysin, protein R, which accumulates fully folded and active in the cytoplasm, to escape across the bilayer and attack the cell wall. Lysis follows within seconds. In addition to the holin and endolysin, dsDNA phages often encode other lysis proteins, including antiholins that regulate holin function (Young & Wang, 2006) and, in phages of Gram-negative hosts, Rz and Rz1 proteins, which form a complex that connects the inner and outer membranes (Summer et al., 2007).
Fig. 1.
Sequences and genes of the λ S and ϕX174 E lysis proteins and the context of the λ lysis cassette. Top: the amino acid sequence of the S107/S105 dual start sequence is shown; S105, the holin, starts at the second methionine. The three transmembrane domains (TMDs) are boxed. Bottom: the amino acid sequence of E is shown, with its putative TMD boxed. The E gene is embedded out of frame within the D gene, which encodes the ϕX174 morphogenetic scaffolding protein. Below the E protein sequence, the two mutations in Epos are shown. Middle: the λ lysis cassette is depicted, transcribed from the late promoter, pR′, and flanked by the Q and bor genes; the dashed lines indicate the region of the cassette replaced by ϕX174 DNA in the E chimeras. The asterisk indicates the position (coordinate 44953) of the upstream single-nucleotide mutation in λ*E and λ*Epos.
In contrast, lytic single-strand nucleic acid (ssDNA, ssRNA) phages like the microviruses (e.g. ϕX174; 5.4 kb), the leviviruses (e.g. MS2; 3.5 kb), and the alloleviruses (e.g. Qβ; 4.2 kb) accomplish lysis of the host without encoding a muralytic enzyme. Instead, in all three cases, lysis is accomplished by expression of a single gene: gene E (Fig. 1), gene L and gene A2 in ϕX174, MS2 and Qβ, respectively. The mechanism by which lysis is effected has been elucidated for the E and A2 proteins, which have been shown to be specific inhibitors of the conserved murein biosynthesis enzymes MraY and MurA, respectively (Bernhardt et al., 2001a, b).
Although the small genome size of the ssRNA and ssDNA phages undoubtedly favoured the evolution of these single-gene systems, it is unclear why they are not employed by any known dsDNA phage. One notion was that the lytic lesion supported by the single-gene system was sufficient for the release of small phages like ϕX174 but not for the much larger, more complex dsDNA phages. This idea was based on a model for E-mediated lysis in which the E protein would oligomerize into a ‘transmembrane tunnel’ that spanned the entire envelope of the host cell (Witte et al., 1990b, 1997). The tunnel would be adequate for release of the small ϕX174 virion but not for a large, complex dsDNA phage like λ. However, the elucidation of the molecular mechanism of E, as an inhibitor of MraY, and the complete cell lysis that derives from it, leaves the transmembrane tunnel hypothesis untenable. Thus there is no fundamental functional limitation that disqualifies single-gene lysis systems for the context of dsDNA phages.
Three holin genes have been subjected to extensive mutational analysis: λ S (Gründling et al., 2000a; Johnson-Boaz et al., 1994; Raab et al., 1988), T4 t (Ramanculov & Young, 2001) and PRD1 XXXV (Rydman & Bamford, 2003). In all three cases, the timing of holin-mediated lysis was found to be extraordinarily allele-specific, with missense mutations throughout each of the holin genes conferring profound changes in the timing of lysis, either shortening or lengthening the latent period. It has been suggested that the genetic plasticity of holins confers a selectable advantage on dsDNA phages because it allows them to rapidly evolve towards optimum lysis timing for any particular environmental condition and host character (Bull et al., 2004). The existence of such an ideal lysis time, for a given host and environmental milieu, has been predicted on a theoretical basis (Wang et al., 1996). This mathematical model suggests that, to effect the best exponential increase in phage titre, the length of the infection cycle should increase when host cell concentration, host quality or the affinity of the phage for the host is reduced, and should decrease if these parameters improve. Support for this idea was obtained in competition experiments with mutants of the T4-like phage RB49 differing only in the holin allele (Abedon et al., 2003).
To test the notion that the functional plasticity of holins confers a fitness advantage to holin-endolysin systems when compared to single-gene lysis systems, we have constructed chimeric λ phages in which the lysis gene E of ϕX174 replaces the entire lysis gene cassette of λ. The chimeric phages were compared to λ in terms of the timing and extent of lysis in bulk liquid culture, and the dispersion of burst sizes within populations of cells. In addition, the mutational plasticity of lysis timing was assessed for E and S, starting with alleles of comparable lysis kinetics. The results are discussed in terms of a model for the contribution of holin-mediated timing to the evolutionary fitness of bacteriophages.
METHODS
Media, chemicals and general methods
Standard Luria–Bertani (LB) broth was used for all bacterial cultures growth and agar plates. When indicated, the medium was supplemented with ampicillin (Amp, 100 μg ml−1), chloramphenicol (Cam, 10 μg ml−1) and kanamycin (Kan, 40 μg ml−1). IPTG and arabinose were used for induction at a final concentration of 1 mM and 0.2 %, respectively. Lysis profiles were obtained by monitoring OD550 after thermal or IPTG and arabinose induction, as described previously (Tran et al., 2005). Phage plating was performed as previously described (Bläsi et al., 1999). To permit meaningful comparison of plaque sizes, phage and indicator bacteria were pre-incubated together for 30 min at 25 °C to ensure complete pre-adsorption prior to initiating infection.
Bacterial strains, bacteriophages and plasmids
The prototroph MDS12 tonA : : Tn10 (Kolisnychenko et al., 2002; Tran et al., 2005), carrying deletions of all the cryptic prophage sequences of Escherichia coli, was used as a non-complementing indicator strain for phage plating and host for all constructions and lysogenic inductions. The same strain carrying the plasmid pS105 (see below) was used for complementing lysis defects. Substitution of the various E alleles for the λ lysis cassette was done by recombination of the indicated plasmid constructs with λΔ(SR), which is λΔ(stf tfa) : : cat cI857 Δ(SR), as previously described (Gründling et al., 2000b). The isogenic phage designated λS was constructed in the same way, using the plasmid pSwt for recombination (see below), and thus has the genotype λΔ(stf tfa) : : cat cI857 (R. White, unpublished). Lysogenization with these phages was accomplished by infecting MDS12 tonA : : Tn10 at low multiplicity, plating for survivors on LB-Cam at 30 °C, and screening candidate lysogens for single-copy prophages using PCR (Powell et al., 1994).
The plasmid pQ, a low-copy plasmid carrying Q, which encodes the λ late gene activator, has been described (Gründling et al., 2001). Inductions were done with an isogenic lacIQ derivative of MDS12 tonA : : Tn10, carrying pQ to supply the Q protein for transactivation of pRW and pRE derivatives. pRW was derived from pRE (Park et al., 2006), a medium-copy plasmid which has a multiple cloning site under the control of the λ late promoter, by inserting the λ bor gene (λ nt 46421 to 46772) after the multiple cloning site. pRW was the backbone used for construction of pRWE and pRWEpos, carrying the parental and Epos4B alleles of the ϕX174 E lysis gene. In these plasmids, nt 544 to nt 840 of ϕX174, spanning the 273 bp of E, were inserted into the EcoRI and BamHI sites of pRW. The coding sequence for the c-myc epitope tag (EQKLISEEDL) was added at the end of the E or Epos sequence in pRWE or pRWEpos, using PCR with primers carrying an EcoRI site (EEcoRIFor: GAGCAGGAATTCGTCGCTGCGTTGAGG) at the 5′ end of the coding sequence and the c-myc sequence and a BamHI site (EcmycBamHIRev: GACGAGGGATCCTTACAGATCTTCTTCAGAGATCAGTTTCTGCTCCTTCCGCACGTA) at the 3′ end. The amplified products were digested with EcoRI and BamHI and ligated into the corresponding sites on the expression vectors, generating pRWEcmyc and pRWEposcmyc, respectively. The plasmid pS105 has the S105 allele of the S gene in the context of the complete λ lysis cassette and the wild-type (wt) λ late promoter region (Smith et al., 1998). The plasmid pSwt was made by site-directed mutagenesis of pS105 to restore the wt S gene dual start motif.
Standard DNA manipulations, PCR and DNA sequencing
Plasmid DNA isolation, DNA amplification by PCR, DNA transformation, DNA sequencing and Quikchange (Stratagene) site-directed mutagenesis were performed as previously described (Tran et al., 2005). Primers were from Integrated DNA Technologies, and were used without further purification. All enzymes were purchased from New England Biolabs, except for Pfu polymerase, which was from Stratagene. The Laboratory for Plant Genome Technology at the Texas Agricultural Experiment Station performed the automated fluorescent sequencing.
SDS-PAGE and Western blotting
SDS-PAGE and Western blotting were performed generally as described previously (Tran et al., 2005). Quantification of the production of c-myc-tagged E proteins from induced lysogens was done by growing designated thermo-inducible cultures in LB-Cam at 30 °C to OD550 ~0.5, aerating at 42 °C for 15 min and then aerating at 37 °C for an additional 5 min. At this time, well before lysis of any of the cultures, 5 ml aliquots were withdrawn and the cells collected by centrifugation for 30 min in a clinical benchtop centrifuge at 4 °C. Pellets were resuspended in SDS-PAGE buffer [4 % SDS, 110 mM Tris/HCl (pH 6.8), 10 % glycerol, 9 % β-mercaptoethanol]; volumes were chosen to normalize final OD550 units per μl. The samples were boiled for 5 min prior to SDS-PAGE; 100 μl of each sample was resolved on a 10 % Tris-Tricine gel at 100 V for 4–5 h. Gels were blotted to nitrocellulose overnight using a semi-dry blotting apparatus. Blots were washed, incubated with antibodies (c-myc monoclonal antibody 9E10 from Covance, at 1 : 1000 dilution; goat anti-mouse-HRP, from Pierce, at 1 : 1000 dilution), and developed as described previously (Bernhardt et al., 2002a). For the analysis of c-myc-tagged E production in the collection of E mutants, MDS12 lacIQ tonA : : Tn10 carrying pQ and the indicated pRW derivative was grown in LB-Amp-Kan at 37 °C, induced with IPTG+arabinose at OD550 0.5, and aerated for 20 min before 5 ml samples were harvested by centrifugation in the cold. Processing of the samples for immunoblotting was the same as for the lysogenic inductions, except that PVDF membranes were used for E blots.
Burst size dispersion
Lysogenic cultures were grown at 30 °C to OD550 0.2 and diluted 106-fold in PBS buffer (120 mM NaCl, 10 mM NaH2PO4, pH 7.5) to give ~100 c.f.u. ml−1. Then 3 μl aliquots of this dilution were added to each well of a 96-well microtitre plate, which already contained 100 μl pre-warmed LB. The lysogens were induced by incubating the microtitre dishes at 42 °C for 20 min and the plates were further incubated at 37 °C for 2 h to allow all cells to lyse. The total p.f.u. in each well was determined by plating its entire contents on a lawn of MDS12 tonA : : Tn10.
RESULTS
Generation of λE constructs
To explore the functionality of single-gene lysis in comparison to the well-characterized holin-endolysin lysis system of phage λ, it was necessary to replace the λ lysis cassette (Fig. 1) with the lysis gene E of ϕX174. To accomplish this substitution, λΔ(SR) was induced in trans to the plasmid pRWE, which carries the E gene flanked by the sequences proximal and distal to the lysis cassette, immediately downstream of the λ late promoter. Plaque-forming recombinants were detected in the lysate, but the frequency was extremely low: approximately 104-fold lower than observed with an isogenic S+R+ plasmid (Table 1). This result suggested that, in addition to the recombination event, a mutation was required to generate the plaque-forming phenotype. Sequencing revealed a single base change, C to T, at position 44953 in the λ DNA, 49 nt upstream from E. When this mutation was incorporated into the initial pRWE clone, plaque-forming recombinants were recovered at a high frequency, confirming that this mutation (indicated by ‘*’; e.g. pRW*) is necessary for the ability of λE to form plaques on lawns of E. coli. Accordingly, the plaque-forming λE construct is designated λ*E. An identical experiment starting with the Epos allele, which carries two point mutations in the 5′ end of the E gene that are known to increase synthesis of the E protein (Bernhardt et al., 2002a), also yielded plaque-forming recombinants at high frequency, irrespective of the presence of the mutation at 44953 (Table 1). This suggests that the upstream mutation in λ*E confers plaque-forming ability on the recombinant phage by increasing the expression level of the wt E gene.
Table 1.
Frequency of plaque-forming recombinants
| Plasmid | P.f.u.† | P.f.u.‡ (complemented) | Frequency of plaque-forming recombinants |
|---|---|---|---|
| pRWE | 4 × 102 | 1.2 × 1010 | 3.3 × 10−8 |
| pRWEpos | 3.6 × 106 | 7.6 × 109 | 4.7 × 10−4 |
| pRW* E | 3.0 × 106 | 5.8 × 109 | 5.0 × 10−4 |
| pRW* Epos | 3.7 × 106 | 1.1 × 1010 | 3.4 × 10−4 |
| pSwt | 1.3 × 107 | 7.8 × 109 | 1.6 × 10−3 |
An asterisk designates plasmids with C to T mutation at coordinate 44953; see text.
Titre on non-complementing lawn, MDS12 tonA : : Tn10.
Titre on complementing lawn, MDS12 tonA : : Tn10/pS105.
Lysis phenotypes of the chimeric λE phages
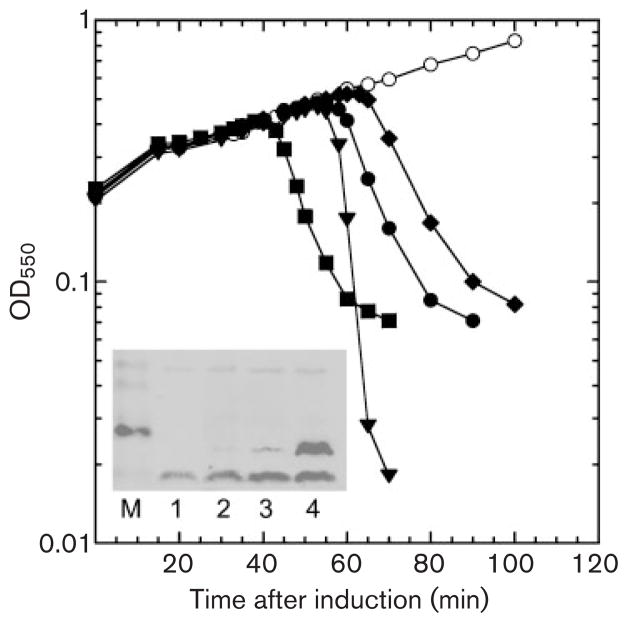
To assess the overall lysis function of the E single-gene system in the λ context, several phenotypes associated with lysis were examined for the three recombinant phages, λ*E, λEpos, and λ*Epos, in comparison with the parental phage, which will be designated λS for clarity. This set of four phages was first compared in terms of lysis in bulk liquid culture, using thermal induction of single-prophage lysogens to permit synchronous induction of the infection cycle throughout the subject cultures (Fig. 2). As expected, all the phages effected lysis of their hosts. Moreover, the presence of the pos and the upstream mutations accelerated lysis synergistically in the E constructs, with λ*Epos lysing before λS, and the two other E constructs, λEpos, and λ*E, lysing later. Western blot analysis confirmed that the shorter latent periods correlated with increased E production (Fig. 2, inset).
Fig. 2.
Induced lysis of λS and the λE chimeras. MDS12 tonA : : Tn10 cells with the indicated prophages were thermally induced by shifting the aerating cultures from 30 °C to 42 °C at time zero for 15 min and then to 37 °C for the duration of the experiment. ○, λΔ(SR); ◆, λ*E; ●, λEpos; ■, λ*Epos; ▼, λS. Inset: E protein production in inductions of chimeric prophages. Inductions of chimeric phages carrying cmyc-tagged versions of the three E alleles were analysed for production of the E protein by immunoblotting with anti-c-myc antibody. Lanes: M, mass standards; 1, λΔ(SR); 2, λ*E-cmyc; 3, λEpos-cmyc; 4, λ*Epos-cmyc.
Unlike with the λS culture, cultures lysed by any of the chimeric phages retained a significant fraction (~20 %) of the pre-lysis turbidity. This was due to the presence of large numbers of non-refractile ‘ghosts’ in the latter cultures; no ghosts or any large cellular remnants were visible in the λS culture. This is consistent with the previous observation that rod-shaped, empty cell ghosts are produced in ϕX174 infections and with inductions of the cloned E gene (Witte et al., 1990a), reflecting the mechanism of E-mediated lysis, which involves the inhibition of peptidoglycan synthesis rather than its degradation (Bernhardt et al., 2000, 2001a, 2002b). Despite the production of ghosts, no more than 11 % of the total virions produced were trapped inside sedimentable debris in the λE constructs (Table 2). Moreover, λ virion production was inversely related to the length of the latent period for the three lytic E constructs. This finding, coupled with the calculated average burst sizes, also shows that there is nothing about the E-mediated lysis pathway that impairs the efficiency of λ morphogenesis (Table 2).
Table 2.
Characteristics of phages with S and E lysis functions
| Phage | Burst† (p.f.u. per cell) | Trapped‡ (%) | EOP§ | Plaque size|| |
|---|---|---|---|---|
| λS | 125 | – | 1.19 (±0.16) | 1.00 (±13 %) |
| λ*E | 143 | 7 | 0.70 (±0.13) | 0.32 (±38 %) |
| λEpos | 112 | 7 | 0.67 (±0.08) | 0.33 (±33 %) |
| λ*Epos | 44 | 11 | 1.06 (±0.23) | 0.45 (±24 %) |
Calculated average burst size per induced lysogenic cell; >1000 plaques were counted in each case.
Fraction of p.f.u. retained in sedimentable cells and released by mechanical cell disruption.
Relative efficiency of plating, as calculated from titre on non-complementing lawn divided by titre on complementing lawn.
Average plaque size, relative to λS (=2.25 mm), and standard deviation, based on measuring 100, 198, 220 and 64 plaques for λS, λ*E, λEpos, and λ*Epos, respectively, as a percentage of the average diameter.
Plaque morphology variance and burst size dispersion
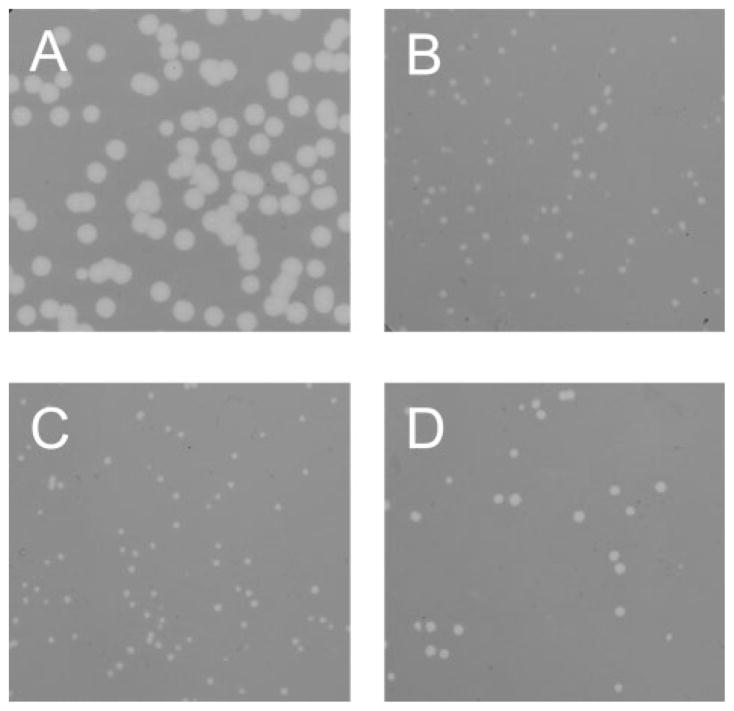
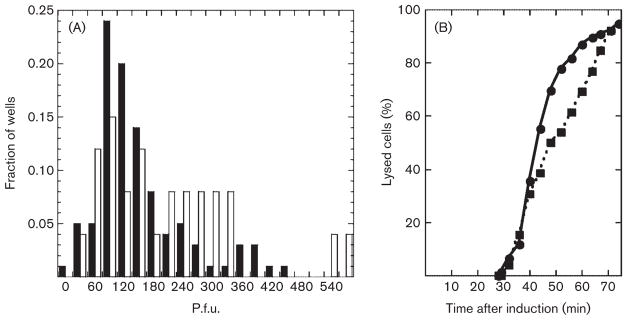
While the efficiency of plating of the λE constructs was not significantly different from that of λS (Table 2), there was a dramatic difference in plaque morphology. Under conditions where virions were quantitatively preadsorbed to the indicator cells, λS formed large plaques of nearly uniform size (Fig. 3, Table 2). In contrast, all three lytic λE constructs formed plaques of smaller and more variable size (Table 2). Moreover, when replated, phages isolated from both small and large plaques generated the same variable size distribution (not shown). This suggested to us that, at the level of an individual cell, there was significant variability in the latent period with any of the λE constructs. It is not practical to test this notion by direct observation of infected cells. However, since progeny virions accumulate linearly shortly after the beginning of late gene expression (Reader & Siminovitch, 1971; Wang, 2006), the burst size should reflect the length of the latent period. Thus, measurement of the dispersion in burst size for individual cells should allow us to detect a signficant variation in time of their lysis. Cells lysogenic for λS or λEpos, which have approximately the same lysis time, were distributed to wells of a microtitre plate at an average of 0.3 cells per well, induced, and the total progeny in each well measured by plating. At this average number of cells per well, only 3.7 % of the wells should contain two or more cells, so the titres in the non-zero wells should mostly reflect the bursts from individual lytic events. As shown in Fig. 4, the distribution of burst sizes from the λEpos lysogen was significantly more disperse that from λS, consistent with a greater cell-to-cell variation in latent period or time of lysis. If the same data are replotted with the assumption that the burst size per cell varies linearly with the lysis time (Fig. 4B), it is clear that the lysis of the induced λS culture is significantly more saltatory than that of λEpos.
Fig. 3.
Plaque morphology: lawns of cells of MDS12 tonA : : Tn10 plated with (A) λS, (B) λ*E, (C) λEpos and (D) λ*Epos.
Fig. 4.
Burst size dispersion of λS and λEpos. (A) Values on the x-axis are bins with the total number of p.f.u. in a well; except for the ‘0’ bin, which represents 0–15 p.f.u., each bin spans 30 p.f.u. and is labelled with the midpoint of the bin. For example, ‘60’ represents 45–75 p.f.u.. For clarity, only alternate bins are labelled. Filled bars, λS; open bars, λEpos. (B) Data from (A) are converted into lysis times, with the assumptions that phage accumulate intracellularly at 7.7 p.f.u. min−1, beginning at 28 min after induction and that samples with less than 360 p.f.u. contained a single lysogenic cell at the time of induction. ●, λS; ■, λEpos.
Lysis time plasticity of S and E
In a previous study aimed at identifying transmembrane domains, the S holin was subjected to cysteine scanning, in which multiple locations throughout the protein were substituted with a Cys residue. Remarkably, nearly every substitution resulted in altered lysis timing, with mutant timing phenotypes both advanced and retarded from the parental lysis time (Gründling et al., 2000a). To see if this functional plasticity extended to E, 14 missense changes in E were chosen by a randomizing computer algorithm and constructed in the vector pRW*. In parallel, 10 missense changes in S were chosen in the same way and constructed in the vector pRE. The complete sets of E and S mutant plasmids were characterized for their lysis timing phenotype by transformation into a host carrying pQ and induction with IPTG and arabinose (see Methods). For this experiment, the S105 allele, which encodes only the holin product S105 (Fig. 1), was used as the parental S allele to avoid potential complications stemming from effects on the S107 antiholin.
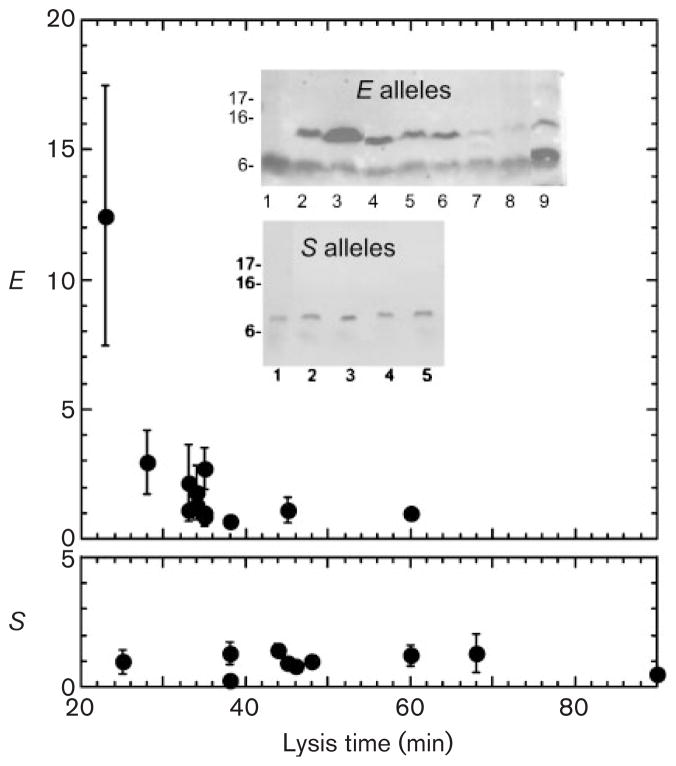
As shown in Table 3, 8 of the 14 E alleles were indistinguishable from the wild-type and three others conferred an absolute lysis defect. One change, R3D, advanced lysis by 7 min, and two others, P65R and T72P, retarded lysis by 10 min and 25 min, respectively. When these alleles were analysed by Western blotting in samples taken at 20 min after induction (before the earliest lysis time), there was clearly increased accumulation of E protein for the R3D allele (Fig. 5), as previously reported for another early lysing allele, ER3H, which was isolated in the context of the ϕX174 phage (Bernhardt et al., 2000). Also, much less E protein accumulated in the case of the alleles with retarded lysis. Thus lysis time was largely invariant, and what variation could be achieved was correlated with the net production of E protein. In contrast, 9 of the 10 randomly chosen missense changes in S affected lysis timing, four alleles advancing lysis between 5 and 23 min, four alleles retarding lysis between 10 and 42 min, and one allele losing the ability to support saltatory lysis. Moreover, Western blot analysis of samples taken at 20 min after induction revealed that the wt and mutant S proteins accumulated to similar levels (Fig. 5). Thus not only were timing mutants much more frequent in the panel of random S missense alleles than in the E panel; the timing differences for the S mutants could not be correlated with altered expression levels.
Table 3.
Lysis time of random missense S and E alleles
Cells carrying plasmid-borne clones of the indicated E or S105 alleles and plasmid pQ were induced and monitored for OD550. Change in lysis time is relative to the parental allele in each case: pRW*E for E and pS105 for S. –, Allele gave a lysis time within 3 min of the wt. An asterisk (*) indicates mutant E alleles previously isolated by selecting for plaque-formation by ϕX174 on a slyD lawn (Bernhardt et al., 2002a) and included here for comparative purposes. The G83I allele of S105 exhibited a very delayed triggering phenotype, with a gradual decline in OD550 beginning approximately 50 min after wt lysis.
| E allele | Change in lysis time (min) |
|---|---|
| wt | wt |
| R3H L19F (Epos4b)* | −12 |
| R3H (Epos6)* | −12 |
| L19F (Epos5)* | – |
| R3D | −7 |
| T5S | – |
| L16A | – |
| S17I | – |
| C61S | – |
| N43R | – |
| S53R | – |
| K57A | – |
| K90F | – |
| P21S | Non-lytic |
| L24P | Non-lytic |
| L42P | Non-lytic |
| P65R | +10 |
| T72P | +25 |
| S allele | |
| wt | wt |
| A12T | – |
| L25G | +42 |
| Y31I | −10 |
| M50G | −5 |
| I53Y | +20 |
| G66E | −23 |
| V77T | −10 |
| G83I | +50 |
| I87Y | +12 |
| V101T | – |
Fig. 5.
Expression of E and S mutants with different lysis times. Inset: representative immunoblots of induced cultures of MDS12 tonA : : Tn10 pQ hosts carrying the pRW* clones of mutants described in Table 3. Upper gel, E alleles, blotted with anti-c-myc antibody; lanes: 1, pRW vector; 2, pRW*E; 3, pRWEpos; 4, R3D; 5, T5S; 6, S17I; 7, P21S; 8, L24P; 9, P65R. Lower gel, S alleles, blotted with anti-S; lanes: 1, S wt; 2, M50G; 3, I53Y; 4, G66E; 5, V77T. The graphs show the relative amount of E or S expression, compared to the parental allele (*E or Swt), versus time of lysis. Only alleles that supported clear lysis are plotted.
DISCUSSION
The experiments reported here were aimed at determining if single-gene lysis systems had features that placed them at a competitive disadvantage when compared to the holin-endolysin systems universally found in dsDNA phage. λ was chosen as a test-bed because synchronous culture-wide lysogenic induction allows precise discrimination of lysis kinetics, unlike infection, which is inherently asynchronous. To compare the functionality of S and E, the ϕX174 E gene was substituted for the entire lysis cassette of λ. We found that an additional mutation in the λ DNA upstream of the E gene was required for plaque formation by the λE construct. This mutation dramatically increases the amount of E protein produced in infected cells or induced lysogens, allowing host lysis. In any case, this upstream mutation found in λ*E or the previously identified pos mutation, known to increase the expression level of E, or a combination of both, resulted in λE chimeras that effect host lysis with kinetics that are within the range of known λS alleles (Gründling et al., 2000a; Johnson-Boaz et al., 1994; Raab et al., 1988) and generate plaques with comparable efficiency. Presumably this reflects the fact that in ϕX174 transcription of the E gene, which occurs constitutively from all the known phage promoters throughout the infection cycle, is significantly higher than in the λ chimera, where late genes are expressed from the single late promoter. In addition, in the λ infection cycle, but not in ϕX174 infections, the dsDNA templates are steadily packaged into proheads, thus depleting the transcribable pool during late gene expression. This set of E-dependent dsDNA phages were examined for features that might explain why dsDNA phages with single-gene lysis systems are not found in nature.
Phenotypic comparison of single-gene and holin-endolysin lysis
In terms of gross lysis, no compelling defect is evident for the λE constructs, compared to λS. Lysis is complete in both cases (Fig. 2), and ~90 % or more of the virions produced are released into the medium. This suggests that any fitness advantage conferred by holin-dependent systems is not due to the inability of the E protein to allow release of phage particles that are much larger than ϕX174. Moreover, the efficiency of plating for the λE constructs was identical to that for λS, indicating that reliance upon the E protein for host lysis did not result in a significant number of unproductive infections. The most striking phenotypic differences between λS and λE were the highly variable plaque size and the greater burst size dispersion found for the E chimeras (Figs 3 and 4).
Changing the lysis time
Cysteine-scanning mutagenesis used to determine the topology of S also suggested that a large fraction of missense mutations in S would alter the triggering time of the holin protein rather than being without effect or resulting in its inactivation. Here, we have provided additional evidence in support of this conclusion by measuring the triggering time of 10 randomly chosen S missense alleles. Triggering times were altered in eight of these mutants, ranging from an advance of 25 min to a delay of 50 min (Table 3). The ease at which the triggering time for S can be altered by single mutations presumably reflects the fact that most of the residues in this protein are involved in intra- and intermolecular interactions necessary for its oligomerization into a ‘hole’.
Unlike the holins, the lysis proteins encoded by ssRNA and ssDNA phages do not disrupt the cytoplasmic membrane of the host. The E protein brings about host lysis due to its ability to inhibit MraY, an enzyme required for peptidoglycan synthesis. Thus, the timing of E-mediated lysis can only be adjusted by altering the time at which the concentration of lipid I, the product of MraY, is reduced below the level necessary for the viability of the dividing host cell. In principle, this could be achieved by mutations that change either the concentration of E in the infected cell or the affinity of E for its target, MraY. In λE, the scope for mutational changes affecting the level of E is limited to those that affect the total translatability of the mRNA or decrease the stability of the E protein. Transcriptional effects are not likely since varying the activity of pR′, which as the single λ late promoter serves all of the late genes, would also cause profound changes in the levels of the many proteins involved in morphogenesis. The previously described Epos mutants (Bernhardt et al., 2002a) are examples of alleles which cause earlier lysis due to increased levels of the E protein (Fig. 2, inset).
The frequency at which missense changes in the E protein altering its interaction with MraY result in physiologically relevant changes in lysis timing is unknown. In this study, we found that when a collection of randomly chosen mutations in E was constructed, more than half of the missense changes had no effect on lysis time, and 20 % were lysis-defective. Only one advanced lysis timing, and this mutation, R3D, was in the same codon as Epos (R3H) and, like Epos, resulted in an increase in E accumulation. Two mutations retarded lysis: P65R, by ~10 min, and T72P, by ~25 min. In both cases, the E protein was found to accumulate to lower levels, as is the case with the previously described slow-lysing allele EP29A. Thus, mis-sense mutations in E that result in an altered time of lysis appear to be rare. It is worth noting that the greater latent period dispersion for the λE constructs would be predicted to compound the defect associated with less plasticity, relative to the holin-mediated system. For example, if a population of λ*E phages were growing under media and host conditions where the parental lysis time was ideally suited for the current environment, and that environment was then changed, such that the average host density was dramatically lowered, it is predicted that a new, delayed lysis time would confer optimum fitness (Wang et al., 1996; Wang, 2006). However, even if a mutation did occur that caused a change in the level of E translation without ablating lysis completely, the intrinsic dispersion of the latent period in a population of infected cells would retard the selection of this mutant. In contrast, mutations that retard S triggering are relatively easy to obtain, and, with the tighter latent period dispersion (Fig. 4), they would be more rapidly selected from the parent population. Thus, compared to single-gene lysis, holin-mediated lysis timing shows greater phenotypic plasticity but much less phenotypic variance, and the fitness contributions of these features would be expected to be synergistic. Based on these observations, we propose that apparently universal reliance of dsDNA phages on holin-endolysin systems derives from the malleability of holins with respect to their triggering time. The lack of such malleability in single-gene lysis systems led to their disappearance from phages with sufficient coding capacity to accommodate holin-endolysin systems. Of course, the difference in malleability reflects the fundamental difference in the mechanism of lysis. E causes lysis by inhibiting MraY, thus stopping the flow of murein precursors. Most frequently this results in lysis when septation is attempted, since entirely new murein is required to complete cell division. Thus another reason for the predominance of the holin-endolysin systems is that the lytic function is independent of the host cell cycle.
Recently, it was shown that a large class of phage endolysins have intrinsic export signals, designated SAR (signal-anchor-release) domains, which confer the capacity to effect host lysis without holin function (Xu et al., 2004, 2005). Phages carrying a SAR endolysin gene make large, uniform plaques, but inactivation of the holin gene results in a heterogeneous plaque-size phenotype (Park et al., 2007). In effect, SAR endolysins represent another mode of single-gene lysis. In this case, holin-mediated control has been apparently been superimposed on the lysis system, presumably conferring a fitness advantage analogous to that which has prevented a λE-equivalent from evolving.
Prospects
The experiments reported here suggest that the apparently universal dependence of dsDNA phages on holin-endolysin lysis systems, and, conversely, the absence of simple single-gene lysis systems in these complex phages, is at least in part due to the intrinsic ability of the holin systems to evolve rapidly to a wide range different lysis times. The λE constructs, in which a single-gene lysis system has replaced the holin-endolysin lysis cassette, should allow direct testing of this and related notions in competition and long-term selection experiments in batch and chemostat under varied conditions of host density and quality.
Acknowledgments
We thank the members of the Young laboratory, past and present, for their helpful criticisms and suggestions. This work was supported by PHS grant GM27099 to R. Y., the Robert A. Welch Foundation, and the Program for Membrane Structure and Function, a Program of Excellence grant from the Office of the Vice President for Research at Texas A&M University.
References
- Abedon ST, Hyman P, Thomas C. Experimental examination of bacteriophage latent-period evolution as a response to bacterial availability. Appl Environ Microbiol. 2003;69:7499–7506. doi: 10.1128/AEM.69.12.7499-7506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. Genetic evidence that the bacteriophage ϕX174 lysis protein inhibits cell wall synthesis. Proc Natl Acad Sci U S A. 2000;97:4297–4302. doi: 10.1073/pnas.97.8.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, Struck DK, Young R. The lysis protein E of ϕX174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J Biol Chem. 2001a;276:6093–6097. doi: 10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Wang IN, Struck DK, Young R. A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science. 2001b;292:2326–2329. doi: 10.1126/science.1058289. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Roof WD, Young R. The Escherichia coli FKBP-type PPIase SlyD is required for the stabilization of the E lysis protein of bacteriophage ϕX174. Mol Microbiol. 2002a;45:99–108. doi: 10.1046/j.1365-2958.2002.02984.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, Wang IN, Struck DK, Young R. Breaking free: “protein antibiotics” and phage lysis. Res Microbiol. 2002b;153:493–501. doi: 10.1016/s0923-2508(02)01330-x. [DOI] [PubMed] [Google Scholar]
- Bläsi U, Fraisl P, Chang CY, Zhang N, Young R. The C-terminal sequence of the lambda holin constitutes a cytoplasmic regulatory domain. J Bacteriol. 1999;181:2922–2929. doi: 10.1128/jb.181.9.2922-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Pfennig DW, Wang IN. Genetic details, optimization and phage life histories. Trends Ecol Evol. 2004;19:76–82. doi: 10.1016/j.tree.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Gründling A, Bläsi U, Young R. Biochemical and genetic evidence for three transmembrane domains in the class I holin, λ S. J Biol Chem. 2000a;275:769–776. doi: 10.1074/jbc.275.2.769. [DOI] [PubMed] [Google Scholar]
- Gründling A, Smith DL, Bläsi U, Young R. Dimerization between the holin and holin inhibitor of phage lambda. J Bacteriol. 2000b;182:6075–6081. doi: 10.1128/jb.182.21.6075-6081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling A, Manson MD, Young R. Holins kill without warning. Proc Natl Acad Sci U S A. 2001;98:9348–9352. doi: 10.1073/pnas.151247598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Boaz R, Chang CY, Young R. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol Microbiol. 1994;13:495–504. doi: 10.1111/j.1365-2958.1994.tb00444.x. [DOI] [PubMed] [Google Scholar]
- Kolisnychenko V, Plunkett G, III, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G. Engineering a reduced Escherichia coli genome. Genome Res. 2002;12:640–647. doi: 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Struck DK, Deaton JF, Young R. Topological dynamics of holins in programmed bacterial lysis. Proc Natl Acad Sci U S A. 2006;103:19713–19718. doi: 10.1073/pnas.0600943103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J Bacteriol. 2007;189:9135–9139. doi: 10.1128/JB.00847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- Ramanculov ER, Young R. Genetic analysis of the T4 holin: timing and topology. Gene. 2001;265:25–36. doi: 10.1016/s0378-1119(01)00365-1. [DOI] [PubMed] [Google Scholar]
- Reader RW, Siminovitch L. Lysis defective mutants of bacteriophage lambda: genetics and physiology of S cistron mutants. Virology. 1971;43:607–622. doi: 10.1016/0042-6822(71)90286-8. [DOI] [PubMed] [Google Scholar]
- Rydman PS, Bamford DH. Identification and mutational analysis of bacteriophage PRD1 holin protein P35. J Bacteriol. 2003;185:3795–3803. doi: 10.1128/JB.185.13.3795-3803.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Struck DK, Scholtz JM, Young R. Purification and biochemical characterization of the lambda holin. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summer EJ, Berry J, Tran TA, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in Gram-negative bacteria. J Mol Biol. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- Tran TAT, Struck DK, Young R. The role of holin and antiholin periplasmic domains in T4 lysis inhibition. J Bacteriol. 2005;187:6631–6640. doi: 10.1128/JB.187.19.6631-6640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN. Lysis timing and bacteriophage fitness. Genetics. 2006;172:17–26. doi: 10.1534/genetics.105.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Dykhuizen DE, Slobodkin LB. The evolution of phage lysis timing. Evol Ecol. 1996;10:545–558. [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Witte A, Bläsi U, Halfmann G, Szostak M, Wanner G, Lubitz W. ϕX174 protein E-mediated lysis of Escherichia coli. Biochimie. 1990a;72:191–200. doi: 10.1016/0300-9084(90)90145-7. [DOI] [PubMed] [Google Scholar]
- Witte A, Wanner G, Bläsi U, Halfmann G, Szostak M, Lubitz W. Endogenous transmembrane tunnel formation mediated by ϕX174 lysis protein E. J Bacteriol. 1990b;172:4109–4114. doi: 10.1128/jb.172.7.4109-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte A, Schrot G, Schon P, Lubitz W. Proline 21, a residue within the alpha helical domain of ϕX174 lysis protein E, is required for its function in Escherichia coli. Mol Microbiol. 1997;26:337–346. doi: 10.1046/j.1365-2958.1997.5781941.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Struck DK, Deaton J, Wang IN, Young R. The signal arrest-release (SAR) sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci U S A. 2004;101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Arulandu A, Struck DK, Swanson S, Sacchettini JC, Young R. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science. 2005;307:113–117. doi: 10.1126/science.1105143. [DOI] [PubMed] [Google Scholar]
- Young R, Wang IN. Phage lysis. In: Calendar R, editor. The Bacteriophages. 2. Oxford: Oxford University Press; 2006. pp. 104–126. [Google Scholar]
- Young R, Wang IN, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]