Abstract
The ability to create cell-derived decellularized matrices in a dish gives researchers the opportunity to possess a bioactive, biocompatible material made up of fibrillar proteins and other factors that recapitulates key features of the native structure and composition of in vivo microenvironments. By using cells in a culture system to provide a natural ECM, decellularization allows for a high degree of customization through the introduction of selected proteins and soluble factors. The culture system, culture medium, cell types, and physical environments can be varied to provide specialized ECMs for wide-ranging applications to study cell–ECM signaling, cell migration, cell differentiation, and tissue engineering purposes. This chapter describes a procedure for performing a detergent and high pH-based extraction that leaves the native, cell-assembled ECM intact while removing cellular materials. We address common evaluation methods for assessing the ECM and its composition as well as potential uses for a decellularized ECM.
1 INTRODUCTION
The extracellular matrix (ECM) is an essential and complex meshwork of collagens, glycoproteins, proteoglycans, and polysaccharides. For any given tissue type, the ECM is assembled into the appropriate architectural design consisting of the correct array of components and molecular stoichiometries to enhance the formation of multiprotein complexes and the polymerization of the key fibrous networks required for the tissue-specific activities of resident cells. ECM functions rely on its unique features including large multidomain protein components and the organization of ECM protein polymers (Hynes & Yamada, 2012; Mecham, 2011). We do not know all of the proteins that constitute an ECM or how they vary in ECMs from different tissue types. Nor can we build an ECM with appropriate architecture from pure protein components or synthetic materials. However, through decellularization procedures, we can isolate the ECM from cell cultures or tissues as a fibrous network in a form that is amenable to structure/function studies (Cukierman, Pankov, Stevens, & Yamada, 2001; Mao & Schwarzbauer, 2005).
The goal of decellularization is to remove cells while maintaining the organization and composition, as much as possible, of the ECM. The insoluble, highly stable core structure of the ECM allows one to extract cellular components, leaving behind an interconnected fibrillar network of ECM protein polymers. This core is assembled by and interacts with cells and also controls the types, amounts, and distributions of many matrix-associated components and modulatory factors. Proteomic analyses of differentially extracted tissue fractions combined with bioinformatics analyses using ECM protein motifs have defined core matrisome and matrisome-associated proteins (Hynes & Naba, 2012; Naba et al., 2012). The human genome contains almost 300 genes encoding core matrisome proteins (collagens, glycoproteins, proteoglycans) and another ~800 genes for associated proteins (e.g., proteases, growth factors, mucins). Decellularization creates a material that retains the ECM architecture and key structural components of the core but can lack associated modulatory proteins or soluble factors needed to stimulate tissue-specific cell functions (Mao & Schwarzbauer, 2005). These factors can be added back to potentially restore partial tissue-specific functions to the decellularized matrix (Williams & Schwarzbauer, 2009).
In an effort to regenerate tissues for implant, whole tissues or tissue sections, such as lung, liver, or tendon, have been decellularized (Faulk, Wildemann, & Badylak, 2015; Hoganson, Bassett, & Vacanti, 2014; Jung, Bhuiyan, & Ogle, 2016; Lovati, Bottagisio, & Moretti, 2016). The resulting ECM “scaffolds” can be reseeded with stem cells or tissue-appropriate cells in attempts to regenerate a functional organ or tissue (Chen et al., 2017; Hoshiba et al., 2016; Yu, Alkhawaji, Ding, & Mei, 2016). Because of the density and volume of cells and matrix in a typical organ, removing cells requires rather harsh techniques. Various combinations of physical, chemical, and biological treatments are generally used, such as freeze–thaw cycles, protease and nuclease treatments, and detergent or acid extractions, and have been applied over periods of days to weeks (Jung et al., 2016; Kawecki et al., 2017). Ultimately, the decellularized ECM scaffold lacks immunogenic components making it suitable for implant but, in the process, it seems likely that many components have been extracted or denatured and molecular interactions have been perturbed. While these perturbations can be one disadvantage of these methods, one of the advantages is that the resulting decellularized ECM scaffold retains core components and some of the mechanical properties found in the native tissue (Chen et al., 2017).
Because of the cell nonautonomy of the ECM, using a cell culture model in which all components derive from a single cell type facilitates dissection of its biological capabilities. Decellularization of a cell culture yields a fibrillar matrix, which for mesenchymal cell types means a matrix in which fibronectin and type I collagen predominate (Singh, Bandini, Donnelly, Schwartz, & Schwarzbauer, 2014). After decellularization, the matrix retains the thickness and fibrillar organization as before cell removal (Mao & Schwarzbauer, 2005). In the case of NIH 3T3 cells, the matrix retains a thickness of approximately 10–15 μm when cultured for ~1 week. Studies have shown that compared to a planar protein-coated surface, a decellularized ECM has distinct effects on cell proliferation, migration, matrix assembly, signaling, and other cell responses (Cukierman et al., 2001; Harris et al., 2017; Hellewell, Rosini, & Adams, 2017; Mao & Schwarzbauer, 2005, 2006; Vlodavsky, 2001) leading some to refer to the matrix as “three-dimensional (3D).” Although there is disagreement about whether the ECM is truly 3D, its effects on cell functions are substantially different from the protein-coated surfaces that are widely used to study cell adhesion-dependent activities.
The methods for decellularization of cell cultures are generally milder than those mentioned earlier for tissues. As a result, critical protein interactions and native ECM architectures can be maintained. One disadvantage of the milder procedures is that certain cellular components such as actin filaments and chromatin might not be completely removed (Hoshiba et al., 2016; Lu, Hoshiba, Kawazoe, & Chen, 2012). Additional washing steps, enzymatic treatments, and postdecellularization screening can be used to determine the quality of the ECM.
ECM deposition and decellularization procedures can be applied to cells grown in a variety of formats including: on planar surfaces, inside of tubes, within or on the fibers of porous synthetic scaffolds, and on gels (biologic or synthetic). Because the cell culture-derived decellularized matrix can be quite fragile, incorporating the matrix into a synthetic scaffold can provide added stability during various manipulations (Goyal et al., 2017). Once decellularized, the ECM network provides a unique substrate for studying cell behaviors such as migration, proliferation, and cell differentiation and for molecular and structural analyses of protein composition, colocalization, and biophysical properties. Customization of the ECM can be achieved by adding back matrisome-associated proteins after decellularization or by coculturing different cell types prior to decellularization.
Here, we describe the method that we use for decellularization (Engler, Chan, Boettiger, & Schwarzbauer, 2009; Mao & Schwarzbauer, 2005). It is a mild treatment of cell cultures with a hypotonic solution, nonionic detergent, and multiple washes based on the original procedure developed by Chen, Murray, Segal, Bushnell, and Walsh (1978). Other procedures have been described and are discussed in Section 2.5.3.
2 DECELLULARIZATION OF CELL-DERIVED ECM
2.1 CELL SOURCE FOR ECM PRODUCTION
Most adherent cell types assemble some amount of ECM. The ability to maintain ECM organization during decellularization depends on the density of the ECM network as densely connected fibrils will stabilize the ECM structure during the extraction. A dense matrix is usually accomplished by growing cells past confluence for extended periods with regular medium changes to stimulate continued matrix assembly. Thus in addition to ECM density, cells that are able to survive past confluence are advantageous for this procedure. For cells that do not assemble significant matrix on their own, these can be cocultured with matrix-producing cells to generate a hybrid matrix from both cell components. For example, a coculture of matrix-producing NIH 3T3 fibroblasts with primary rat neonatal Schwann cells that do not assemble a matrix by themselves generates an ECM containing fibronectin produced by fibroblasts and Schwann cells (Harris et al., 2017).
Immunofluorescence staining is the most direct way to screen cells for ECM production. First, cells should be visualized daily by phase microscopy from the time they reach confluence until 7–8 days postconfluence. This allows one to assess cell survival in very dense cultures. Second, cells growing on glass coverslips are fixed and stained with anti-ECM protein antibodies and fluorescent secondary antibodies (Wierzbicka-Patynowski, Mao, & Schwarzbauer, 2004). Microscopic visualization is used to determine the density of the ECM proteins. Fibronectin is abundantly produced by most mesenchymal cells and is a useful marker for ECM. Type I collagen can also be monitored in cultures grown in medium containing ascorbate. Levels of specific ECM components can also be examined by immunoblotting of cell lysates (Wierzbicka-Patynowski et al., 2004).
2.2 MATERIALS AND REAGENTS FOR DECELLULARIZATION
2.2.1 Solutions
Sterile PBS (Ca2+-Mg2+-free)
Deionized (DI) water
Wash buffer 1: 100mM Na2HPO4, 2mM MgCl2, 2mM EGTA (pH 9.6)
Wash buffer 2: 10mM Na2HPO4, 300mM KCl (pH 7.5)
Lysis buffer: 8mM Na2HPO4, 1% NP-40 (pH 9.6)
Wash buffer 1, wash buffer 2, and lysis buffer should be stored at 4°C. For long-term storage, aliquots can be frozen at −20°C
2.2.2 Materials
The standard protocol described in the next section is based on cells growing in a 24-well plate, with or without a glass coverslip. For other culture plate sizes, cell seeding densities and volumes of solutions should be adjusted based on surface areas. Surfaces of plates or coverslips can be coated with 0.1% gelatin, which may help to maintain ECM attachment to the surface during decellularization.
2.3 DECELLULARIZING CELL-DERIVED MATRIX
2.3.1 Cell culture
A suitable substrate on which to culture the cells must be selected with glass-bottom dishes, polystyrene culture dishes, and coverslips among the options. Examples of applications for each include: glass-bottom dishes utilized for live cell imaging assays, polystyrene culture dishes for solubilization assays or observation with an inverted microscope, and coverslips utilized for immunofluorescence experiments mounted on microscope slides. Sterilize coverslips by autoclaving in a glass Petri dish or other heat-stable container and store at room temperature.
Seed cells at a subconfluent density such that the cells become confluent in 2 days. For NIH 3T3 cells, seed approximately 2 × 105 cells/well in 1mL of medium (10% BCS, DMEM). The seeding density should be determined empirically on a cell type-by-cell type basis.
At confluence, replace the medium with fresh medium. To increase collagen production and matrix incorporation, add 50μg/mL ascorbic acid (Sigma) to the medium. Culture the cells for another 3 days, 5 days from cell seeding. Exchange the medium on day 4 if necessary.
2.3.2 Decellularization procedure
Warm PBS, wash buffers, and lysis buffer to 37°C.
Remove culture medium and rinse cells twice with 1mL PBS. In this and all subsequent steps, slowly add the solution along the wall of the well and aspirate gently to remove the solution.
Wash culture with 1mL wash buffer 1, three times.
Add 1mL lysis buffer and incubate for 15min at 37°C. From this step onward, take special care not to dislodge the ECM in subsequent steps through forceful pipetting or aspiration. During lysis steps, check cells by phase microscopy to monitor the extent of cell lysis and loss of nuclear integrity.
Remove lysis buffer and add 1mL fresh lysis buffer to cells. Incubate for 40–60min at 37°C. The time can be extended or lysis buffer exchanged again if cell lysis appears incomplete by microscopic examination. The matrix should also be visible by phase microscopy.
Remove lysis buffer and rinse the matrix with wash buffer 2, 1mL per wash, three to five times. Then wash four times with 1mL DI water. After the final wash, replace with PBS and store at 4°C. Samples can be stored for at least several months with minimal degradation or structural breakdown of the ECM.
2.4 PREPARING STERILE DECELLULARIZED ECM
When using the decellularized ECM for recellularization or tissue engineering applications, it is necessary to take the proper steps to ensure the sterility of the matrix. This can be accomplished in two ways: carry out the decellularization procedure under sterile conditions or sterilize the matrix after decellularization. For the first approach, we sterile filter all solutions with 0.22-μm filters in a laminar flow hood. All steps of the procedure are performed in the hood with sterile materials and solutions to maintain sterility throughout the decellularization. In the case of nonsterile decellularization, the ECM can be treated after the procedure, but before storing, using irradiation or ethylene oxide (Dai, Ronholm, Tian, Sethi, & Cao, 2016). However, these sterilization techniques might damage or degrade ECM proteins or fibril structure.
2.5 MODIFICATIONS TO THE DECELLULARIZATION PROTOCOL
2.5.1 Matrix density
The density of the matrix can be increased by extending the postconfluence cell culture time. For some applications, we have grown NIH 3T3 cell cultures for 8 days after reaching confluence (10 days total) (Harris et al., 2017; Singh et al., 2014). Lysing cells and removing debris are more difficult with a denser matrix. We modify the protocol by extending the second lysis buffer incubation to 1h and 15min. Additional time or a third incubation with lysis buffer can be added as necessary to thoroughly lyse cells as visualized by phase microscopy. A wash with 0.5% deoxycholate in wash buffer 2 can be added if nuclear material remains after the lysis buffer steps (Chen et al., 1978).
2.5.2 Scaling up cell culture
To prepare decellularized matrices on surfaces larger than a 24-well plate, scale up the cell seeding density proportionally, that is, seed the cells at the same number per cm2. Wash and lysis volumes will also increase: use 2–3mL for a 35-mm dish, 3–4mL for a 60-mm dish, and 5–6mL for a 100-mm dish. Test wash and lysis volumes to assure reproducible decellularization.
2.5.3 Other decellularization procedures
Our decellularization protocol was adapted from a method originally developed to study the organization of fibronectin matrix fibrils using scanning and transmission electron microscopy approaches (Chen et al., 1978). Other methods have been described many of which are variations of an ECM denuding procedure first applied to study endothelial cell matrices (Robinson & Gospodarowicz, 1984). In this protocol, cells are removed by extraction with ammonium hydroxide and Triton X-100. Detailed protocols as well as a video demonstration of the approach are available (Cukierman et al., 2001; Franco-Barraza, Beacham, Amatangelo, & Cukierman, 2016; Hellewell et al., 2017; Vlodavsky, 2001).
2.6 POTENTIAL PITFALLS AND TROUBLESHOOTING
A major variable that can affect results is the fragility of a cell-derived decellularized ECM. It is very important to slowly add solutions and to be very gentle when aspirating liquid off of the cell-assembled ECM to reduce the chances of dislodging it from the surface. Coating the initial surface with gelatin prior to cell seeding may help to bind the ECM to the surface. Manual aspiration with a Pasteur pipette and rubber bulb is usually gentler than vacuum aspiration. To limit the number of sites at which the ECM is disturbed, liquid can be added and removed at the same location on the side of the well or dish each time.
If cellular proteins or nucleic acids are not completely removed, make sure that the pH of wash buffer 1 and the lysis buffer is at 9.6, as the high pH is important for cell lysis. Further solubilization of DNA will occur upon storage so after several days background DAPI staining should be reduced. If you are using a hydrogel or other nonstandard surface as a culture platform, extra washes or multiple changes of lysis buffer may be required to completely exchange the buffer for the water in the gel and to maintain the high pH.
3 ASSESSMENT OF DECELLULARIZED MATRIX
3.1 EVALUATION OF NUCLEAR CONTENT
Decellularized matrices are visible by phase microscopy immediately after the decellularization protocol (Fig. 1A). The distribution of fibrils and the presence of certain ECM proteins in the decellularized matrix can be determined by indirect immunofluorescence. Two samples, a culture before decellularization and the decellularized ECM, are fixed and stained with relevant ECM protein antibodies and imaging is used to compare the similarity of matrix before and after the procedure. After incubation in PBS for a week, ECM can be stained for fibronectin and nuclei (4′,6-diamidino-2-phenylindole (DAPI)) to confirm a three-dimensional fibrillar ECM as well as the absence of nuclear material. As shown in Fig. 1B, NIH 3T3 fibroblasts assembled a dense fibronectin-rich matrix and a subsequent decellularization protocol left them devoid of fibroblasts while retaining fibronectin fibrils. Soluble and insoluble fibronectin matrix fractions can be quantified using a deoxycholate lysis assay (Wierzbicka-Patynowski et al., 2004). The main criterion for successful decellularization is the lack of residual nuclear material visualized through DAPI staining. Other more stringent procedures call for <200bp DNA fragment length to confirm successful decellularization, as foreign DNA is directly correlated to immune reactions (Nagata, Hanayama, & Kawane, 2010). This condition can be analyzed through either commercially available kits (PicoGreen dsDNA assay (ThermoFisher) for example) or by gel electrophoresis.
FIG. 1.
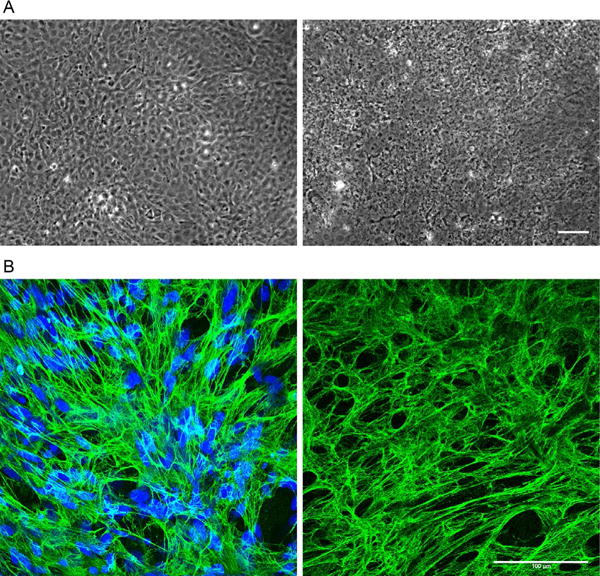
Cultures and ECM before and after decellularization. Phase contrast and immunofluorescently stained images showing NIH 3T3 cells before (left) and after (right) decellularization. (A) Phase contrast images captured at 6 days of culture. (Left) Prior to undergoing decellularization, the monolayer consists of highly confluent cells. (Right) One week after decellularization, a fibrillar network with some cellular debris but lacking intact cell bodies is visible. (B) Immunofluorescence images of NIH 3T3 cells stained with antiserum against fibronectin (green) and with DAPI (blue). The fibrillar networks of fibronectin look very similar before (left) and 1 week after decellularization (right). No DNA was detected after decellularization (right). Scale bars are 100μm.
To identify the major protein components in the matrix, SDS-PAGE and either Coomassie or silver staining can be used to detect the total protein profile in the matrix (Mao & Schwarzbauer, 2005). Levels of specific components, including intracellular contaminants, can be determined by quantitative immunoblotting. Immunofluorescence staining and immunoblotting are also useful quality control measures of the ECM after storage.
3.2 QUANTIFICATION OF MATRIX FIBRIL ALIGNMENT
3.2.1 Fast Fourier transform analysis to assess fibrillar alignment
ECM patterning is critical in determining tissue formation and function. The orientation of ECM fibrils acts as a guide to direct cell spreading, migration, and phenotype. It is thus useful to have tools to objectively determine whether an ECM is aligned or how the alignment of different matrices compare. Alignment of matrix fibrils can be quantified using a fast Fourier transform (FFT) analysis. This approach allows for morphological quantification of alignment by transforming the spatial pixel information of immunofluorescence images to the Fourier space (Ayres et al., 2008; Harris et al., 2017; Singh et al., 2014). A protocol for quantifying fiber alignment with FFT theory and detailed instructions has been documented previously (Taylor, Cao, Talauliker, & Lifshitz, 2013). As an example, we demonstrate this approach using immunofluorescence images of decellularized matrices assembled by NIH 3T3 cells cultured for 6 days on a chemical interface patterned to produce an “aligned” matrix (Fig. 2A) or an unpatterned surface to give an “unaligned” ECM (Fig. 2B). The images were imported into NIH ImageJ software where analysis produces the FFT images shown on the right in Fig. 2A and B.
FIG. 2.
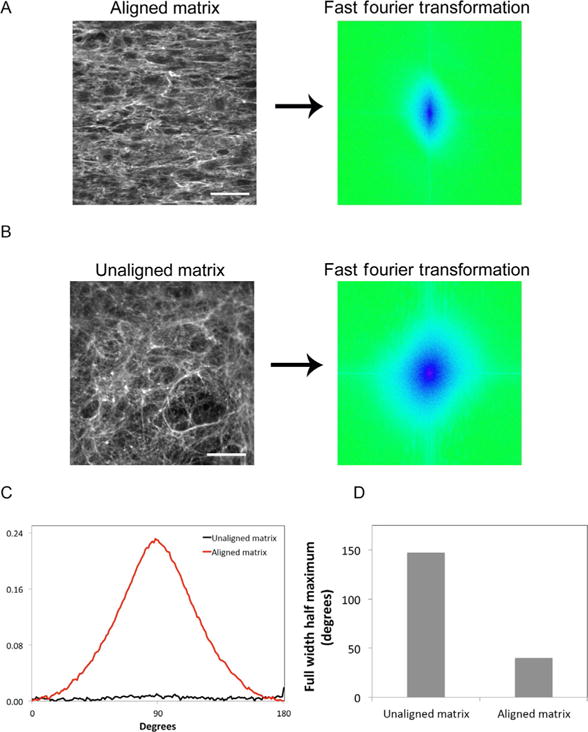
Fast Fourier transform analysis of matrix fibril alignment. Decellularized matrices from NIH 3T3 cells were stained with anti-FN antiserum. Immunofluorescence images show (A, left) aligned matrix fibrils from cells cultured on a striped chemical pattern and (B, left) unaligned matrix from cells grown on an unpatterned substrate. Scale bar is 100μm. (A and B, right) 2D FFT analysis transforms spatial information from the immunofluorescence staining pattern to the frequency domain. These FFT images represent pixel alignment of those patterns. The FFT images are artificially colored to show frequency domain information in blue. A line through the FFT image origin would indicate perfect alignment. (B, right) The circular pattern shows no alignment of pixels; (A, right) alignment gives an oval-shaped pattern. (C) FFT pixels were radially summed around a 360 point perimeter. A circular pattern with no alignment gives an equal summation around the perimeter with no peaks, while the aspect ratio of the oval pattern generates peaks at 90 and 270 degrees. Radial summation from aligned and unaligned matrices is graphed from 0 to 180 degrees after averaging and normalization. (D) The full width-half maximum values were calculated for each plot in (C). A larger full width half maximum signifies less alignment.
3.2.2 Quantification of FFT results
Further quantification of data in the FFT image can be achieved using the “oval profile” plugin downloaded into ImageJ. FFT pixels are summed for 360 radii around the center of the FFT image and used to produce a line graph showing pixel intensity values. Alignment in the image will generate peaks separated by 180 degrees, at 90 and 270 degrees (Fig. 2C). A flat line indicates no alignment in the original image. Values can be normalized and plotted for comparison of alignment of ECM fibrils across different matrices (Fig. 2C). For a direct comparison of line graphs, a “full width-half maximum” value can be calculated from the width of the peak at half of the maximum value of the peak; larger widths equal less alignment and smaller widths show higher alignment of fibrils (Fig. 2D). This approach allows quantification and direct comparison of the alignment of fibrous materials and can be applied to natural matrices as well as engineered tissue-mimetic substrates.
4 EXPLOITING A DECELLULARIZED MATRIX
4.1 STUDYING THE ECM AND CELL–ECM INTERACTIONS
A decellularized matrix is a unique material for studying cell–ECM interactions and cell responses to matrices with different fibril organizations, compositions, or biophysical properties. The ability to use any number of cell types to generate the matrix as well as the ability to add purified proteins during assembly or after decellularization makes this a manipulable system that can be tailored to address a variety of questions and experimental protocols. The simplest experiment, cell adhesion and spreading, involves carefully adding a suspension of cells to a well containing a decellularized matrix substrate. For example, Schwann cells seeded at low density spread on the decellularized fibroblast ECM and adopted a polygonal morphology. Certain cell projections (visualized by staining actin filaments) are coaligned with fibronectin fibrils in the matrix (Fig. 3). The distributions of fibrils and cell adhesion sites can be followed over time to determine how cells restructure the ECM fibrils and how matrix organization directs cell morphology and signaling (Hakkinen, Harunaga, Doyle, & Yamada, 2011).
FIG. 3.
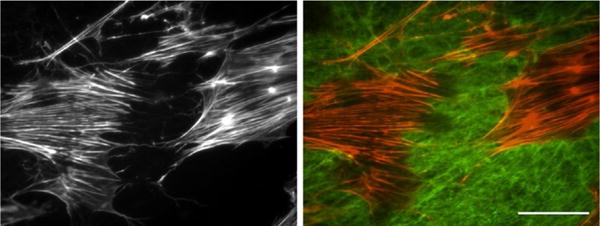
Schwann cells spreading on a decellularized matrix. Schwann cells were seeded onto a NIH 3T3 decellularized matrix, allowed to spread, and then stained with fluorescent phalloidin to visualize F-actin (red). Indirect immunofluorescence with antifibronectin antibodies was used to detect the matrix (green). The right image shows merged fibronectin and actin signals. The left image shows the actin staining only. Scale bar is 100μm.
Decellularized ECM has been targeted by researchers looking to gain a deeper understanding of how ECM architecture affects cell migration (Burns et al., 2011; Castello-Cros, Khan, Simons, Valianou, & Cukierman, 2009; Doyle et al., 2012; Mao & Schwarzbauer, 2006; Pankov et al., 2005; Petrie, Gavara, Chadwick, & Yamada, 2012; Petrie, Koo, & Yamada, 2014). Recent work nicely illustrates the novel features of cell migration on a natural decellularized matrix. A two-dimensional (2D) surface or a 3D collagen gel promotes a lamellipodia-based migration mechanism with polarized Rac1 GTPase and PIP3 signaling at the leading edge (Petrie et al., 2012). Interestingly, the natural 3D decellularized matrix promoted “lobopodia-based migration” that showed nonpolarized Rac1 and PIP3 instead using a nuclear piston mechanism for leading-edge protrusion (Petrie et al., 2014).
New insights into the effects of cell–ECM interactions on cell behaviors have been derived from studies using decellularized matrix. FN matrix assembly is stimulated when cells interact with a 3D decellularized matrix compared to 2D substrates (Green, Berrier, Pankov, & Yamada, 2009; Hakkinen et al., 2011; Mao & Schwarzbauer, 2006). Analyses of the composition or properties of decellularized matrices have been used to map the distributions of specific ECM components such as thrombospondin-1 (Hellewell, Gong, Scharich, Christofidou, & Adams, 2015), the effects of metabolic conditions such as high glucose levels on ECM protein modification by nonenzymatic glycation (Pastino, Greco, Mathias, Cristea, & Schwarzbauer, 2017), the role of matrix mechanical properties such as stiffness in controlling various cell functions (Engler et al., 2009; Soucy, Werbin, Heinz, Hoh, & Romer, 2011), and other functional effects of variations in the ECM.
4.2 EXTRACELLULAR MATRIX IN THE STEM CELL NICHE
One hurdle for controlling stem cell pluripotency in vitro is identifying a culture microenvironment that mimics the stem cell niche. Without the appropriate environment, stem cells change their self-renewal capacity, differentiate along unexpected pathways, or undergo senescence. Within the stem cell niche, the ECM provides sites for cell adhesion, growth factor storage, and relevant mechanical cues, all of which have been linked to stem cell maintenance (Brizzi, Tarone, & Defilippi, 2012). Recent work is illustrating the potential for recapitulating a native cellular environment by incorporating a cell-derived decellularized ECM into stem cell culture systems (Agmon & Christman, 2016; Cha et al., 2013; Chen et al., 1978; Lai et al., 2010; Shakouri-Motlagh, O’Connor, Brennecke, Kalionis, & Heath, 2017; Sun et al., 2011). For example, using ECMs prepared from bone marrow, Prewitz and colleagues were able to maintain pluripotent mesenchymal stem cells (MSCs) and hematopoietic stem cells in culture (Prewitz et al., 2013). Two different ECM preparations were compared and both improved the expansion of MSC populations in comparison to fibronectin-coated tissue culture plastic. They also had elasticities resembling bone marrow (0.1–0.3kPa) and were composed of similar ECM proteins (collagens, decorin, laminins, nidogens, tenascin, thrombospondins, vitronectin, and fibronectin). However, one type of ECM, derived from MSCs cultured with ascorbic acid, was more effective at promoting the expansion of hematopoietic stem cells. This report also makes an interesting technical point in showing that growing cells on a glass surface covalently coupled with fibronectin anchors the ECM and stabilizes it during decellularization.
A cell-derived ECM not only affects proliferative signaling, but can also modulate signaling through differentiation or stress response pathways. In one example, MSCs growing on a cell-derived ECM had lower intracellular levels of reactive oxygen species in comparison to tissue culture plastic (Lai et al., 2010). The osteogenic differentiation capacity of these MSCs was retained for multiple passages on the ECM. In addition to MSC differentiation, a decellularized ECM can also direct the differentiation capacity of embryonic stem cells. The composition of the ECM can have a significant effect as shown for definitive endoderm differentiation, which was dependent on the presence of laminin-111 in the matrix (Taylor-Weiner, Schwarzbauer, & Engler, 2013). Taken together, work from a number of groups, too numerous to mention in this chapter, shows the advantages of including decellularized ECM in stem cell cultures and provides useful methodologies for development of other ECM-based culture systems to mimic a particular in vivo microenvironment.
4.3 TISSUE REGENERATION
Whole-organ decellularization followed by recellularization with tissue-specific cell types in a bioreactor enables researchers to produce organ prototypes with similarities to a native tissue (Badylak, Taylor, & Uygun, 2011; Gilbert, Sellaro, & Badylak, 2006; Schenke-Layland & Nerem, 2011). Yet, a limited number of whole tissue sources are available and the decellularization procedures can affect ECM properties as described earlier in this chapter. Therefore, methods for supplementing materials with cell-derived matrices are gaining popularity in regenerative medicine applications.
Methods to combine decellularized matrix with synthetic materials are being developed to convert synthetic polymers into bioactive or biomimetic materials with appropriate functions for tissue engineering (Datta et al., 2006; Goyal et al., 2017; Rutledge, Cheng, Pryzhkova, Harris, & Jabbarzadeh, 2014; Thibault, Scott Baggett, Mikos, & Kasper, 2010). Many procedures use static cell cultures for decellularization. However, dynamic culture conditions may have advantages especially when optimizing cell growth conditions for subsequent decellularization as was done with chondrocytes cultured in an electrospun poly(e-caprolactone) microfiber scaffold in a flow perfusion bioreactor (Liao, Guo, Grande-Allen, Kasper, & Mikos, 2010). Fibroblasts, MSCs, and other cells routinely used for decellularization assemble a dense network in which the ECM fibrils have no particular orientation. The ability to control the orientation of the fibrils provides another mechanism to control cell behaviors after reseeding. We have developed a patterned chemical interface that when deposited in micron-sized stripes onto polymeric materials controls cell orientations in alignment with the stripes and cells in turn assemble aligned matrix fibrils (Donnelly et al., 2013; Singh et al., 2014). Aligned decellularized ECM has the unique ability to direct the orientations of neurites extended by primary neurons and neural explants (Harris et al., 2017). This ECM–polymer biomaterial has potential as a nerve repair therapy for directing glial cell and neuron extension across lesions.
5 CONCLUSIONS
The decellularization procedure described here maintains the 3D architecture and core composition of the ECM, thus providing a natural substrate for cell growth that resembles the microenvironment within tissues. Many different cell types and culture conditions can be used to generate matrices for decellularization, allowing one to create in vitro models for normal tissues and for various disease states. The matrix can be directly tuned through cell cocultures, supplementation with proteins or growth factors, and chemical or enzymatic modulation of mechanical properties. Incorporation of the matrix into various synthetic constructs such as within polymeric scaffolds or on the surface of hydrogels is emerging as a new direction in biomaterials design. Thus decellularized matrices are showing significant potential as in vitro platforms for research as well as in biofriendly materials for tissue repair and regeneration in vivo.
Acknowledgments
The authors are grateful to Dr. Gary Laevsky, director of the Molecular Biology Confocal Microscopy Core Facility, a Nikon Center of Excellence, for scientific and technical assistance. We also thank the National Institutes of Health (NIH) (R01 CA160611 and P41 EB001046), the New Jersey Commission on Spinal Cord Research (CSCR15IRG002), and the Princeton Dean for Research (Innovation award). I.R. was supported by a Princeton Dept. of Molecular Biology NIH predoctoral training grant (T32 GM007388). G.M.H. was supported by a NJ Center for Biomaterials NIH postdoctoral training grant (T32EB005583).
References
- Agmon G, Christman KL. Controlling stem cell behavior with decellularized extracellular matrix scaffolds. Current Opinion in Solid State & Materials Science. 2016;20(4):193–201. doi: 10.1016/j.cossms.2016.02.001. https://doi.org/10.1016/j.cossms.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres CE, Jha BS, Meredith H, Bowman JR, Bowlin GL, Henderson SC, et al. Measuring fiber alignment in electrospun scaffolds: A user’s guide to the 2D fast Fourier transform approach. Journal of Biomaterials Science Polymer Edition. 2008;19(5):603–621. doi: 10.1163/156856208784089643. https://doi.org/10.1163/156856208784089643. [DOI] [PubMed] [Google Scholar]
- Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: Decellularization and recellularization of three-dimensional matrix scaffolds. Annual Review of Biomedical Engineering. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. https://doi.org/10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzi MF, Tarone G, Defilippi P. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Current Opinion in Cell Biology. 2012;24(5):645–651. doi: 10.1016/j.ceb.2012.07.001. https://doi.org/10.1016/j.ceb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Burns JS, Kristiansen M, Kristensen LP, Larsen KH, Nielsen MO, Christiansen H, et al. Decellularized matrix from tumorigenic human mesenchymal stem cells promotes neovascularization with galectin-1 dependent endothelial interaction. PLoS One. 2011;6(7):e21888. doi: 10.1371/journal.pone.0021888. https://doi.org/10.1371/journal.pone.0021888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello-Cros R, Khan DR, Simons J, Valianou M, Cukierman E. Staged stromal extracellular 3D matrices differentially regulate breast cancer cell responses through PI3K and beta1-integrins. BMC Cancer. 2009;9:94. doi: 10.1186/1471-2407-9-94. https://doi.org/10.1186/1471-2407-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha MH, Do SH, Park GR, Du P, Han KC, Han DK, et al. Induction of re-differentiation of passaged rat chondrocytes using a naturally obtained extracellular matrix microenvironment. Tissue Engineering Part A. 2013;19(7–8):978–988. doi: 10.1089/ten.tea.2012.0358. https://doi.org/10.1089/ten.TEA.2012.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen J, Zhang Z, Lou K, Zhang Q, Wang S, et al. Current advances in the development of natural meniscus scaffolds: Innovative approaches to decellularization and recellularization. Cell and Tissue Research. 2017;370:41–52. doi: 10.1007/s00441-017-2605-0. https://doi.org/10.1007/s00441-017-2605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LB, Murray A, Segal RA, Bushnell A, Walsh ML. Studies on inter-cellular LETS glycoprotein matrices. Cell. 1978;14(2):377–391. doi: 10.1016/0092-8674(78)90123-x. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Dai Z, Ronholm J, Tian Y, Sethi B, Cao X. Sterilization techniques for biodegradable scaffolds in tissue engineering applications. Journal of Tissue Engineering. 2016;7 doi: 10.1177/2041731416648810. 2041731416648810. https://doi.org/10.1177/2041731416648810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, Pham QP, Sharma U, Sikavitsas VI, Jansen JA, Mikos AG. In vitro generated extracellular matrix and fluid shear stress synergistically enhance 3D osteoblastic differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2488–2493. doi: 10.1073/pnas.0505661103. https://doi.org/10.1073/pnas.0505661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PE, Jones CM, Bandini SB, Singh S, Schwartz J, Schwarzbauer JE. A simple nanoscale interface directs alignment of a confluent cell layer on oxide and polymer surfaces. Journal of Materials Chemistry B, Materials for Biology and Medicine. 2013;1(29):3553–3561. doi: 10.1039/C3TB20565G. https://doi.org/10.1039/C3TB20565G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration—Myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. Journal of Cell Science. 2012;125(Pt 9):2244–2256. doi: 10.1242/jcs.098806. https://doi.org/10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Chan M, Boettiger D, Schwarzbauer JE. A novel mode of cell detachment from fibrillar fibronectin matrix under shear. Journal of Cell Science. 2009;122:1647–1653. doi: 10.1242/jcs.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk DM, Wildemann JD, Badylak SF. Decellularization and cell seeding of whole liver biologic scaffolds composed of extracellular matrix. Journal of Clinical and Experimental Hepatology. 2015;5(1):69–80. doi: 10.1016/j.jceh.2014.03.043. https://doi.org/10.1016/j.jceh.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Barraza J, Beacham DA, Amatangelo MD, Cukierman E. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Current Protocols in Cell Biology. 2016;71:10 19 11–10 19 34. doi: 10.1002/cpcb.2. https://doi.org/10.1002/cpcb.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27(19):3675–3683. doi: 10.1016/j.biomaterials.2006.02.014. https://doi.org/10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Goyal R, Vega ME, Pastino AK, Singh S, Guvendiren M, Kohn J, et al. Development of hybrid scaffolds with natural extracellular matrix deposited within synthetic polymeric fibers. Journal of Biomedical Materials Research Part A. 2017;105:2162–2170. doi: 10.1002/jbm.a.36078. https://doi.org/10.1002/jbm.a.36078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JA, Berrier AL, Pankov R, Yamada KM. beta1 integrin cytoplasmic domain residues selectively modulate fibronectin matrix assembly and cell spreading through talin and Akt-1. The Journal of Biological Chemistry. 2009;284(12):8148–8159. doi: 10.1074/jbc.M805934200. https://doi.org/10.1074/jbc.M805934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Engineering Part A. 2011;17(5–6):713–724. doi: 10.1089/ten.tea.2010.0273. https://doi.org/10.1089/ten.TEA.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris GM, Madigan NN, Lancaster KZ, Enquist LW, Windebank AJ, Schwartz J, et al. Nerve guidance by a decellularized fibroblast extracellular matrix. Matrix Biology. 2017:60–61. 176–189. doi: 10.1016/j.matbio.2016.08.011. https://doi.org/10.1016/j.matbio.2016.08.011. [DOI] [PMC free article] [PubMed]
- Hellewell AL, Gong XY, Scharich K, Christofidou ED, Adams JC. Modulation of the extracellular matrix patterning of thrombospondins by actin dynamics and thrombospondin oligomer state. Bioscience Reports. 2015;35:e00218. doi: 10.1042/BSR20140168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellewell AL, Rosini S, Adams JC. A rapid, scalable method for the isolation, functional study, and analysis of cell-derived extracellular matrix. Journal of Visualized Experiments. 2017;(119):e55051. doi: 10.3791/55051. https://doi.org/10.3791/55051. [DOI] [PMC free article] [PubMed]
- Hoganson DM, Bassett EK, Vacanti JP. Lung tissue engineering. Frontiers in Bioscience (Landmark Edition) 2014;19:1227–1239. doi: 10.2741/4278. [DOI] [PubMed] [Google Scholar]
- Hoshiba T, Chen G, Endo C, Maruyama H, Wakui M, Nemoto E, et al. Decellularized extracellular matrix as an in vitro model to study the comprehensive roles of the ECM in stem cell differentiation. Stem Cells International. 2016;2016:6397820. doi: 10.1155/2016/6397820. https://doi.org/10.1155/2016/6397820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Naba A. Overview of the matrisome—An inventory of extracellular matrix constituents and functions. Cold Spring Harbor Perspectives in Biology. 2012;4(1):a004903. doi: 10.1101/cshperspect.a004903. https://doi.org/10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Yamada KM, editors. Extracellular matrix biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- Jung JP, Bhuiyan DB, Ogle BM. Solid organ fabrication: Comparison of decellularization to 3D bioprinting. Biomaterials Research. 2016;20(1):27. doi: 10.1186/s40824-016-0074-2. https://doi.org/10.1186/s40824-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki M, Labus W, Klama-Baryla A, Kitala D, Kraut M, Glik J, et al. A review of decellurization methods caused by an urgent need for quality control of cell-free extracellular matrix’ scaffolds and their role in regenerative medicine. Journal of Biomedical Materials Research Part B, Applied Biomaterials. 2017 doi: 10.1002/jbm.b.33865. https://doi.org/10.1002/jbm.b.33865. [DOI] [PubMed]
- Lai Y, Sun Y, Skinner CM, Son EL, Lu Z, Tuan RS, et al. Reconstitution of marrow-derived extracellular matrix ex vivo: A robust culture system for expanding large-scale highly functional human mesenchymal stem cells. Stem Cells and Development. 2010;19(7):1095–1107. doi: 10.1089/scd.2009.0217. https://doi.org/10.1089/scd.2009.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JH, Guo XA, Grande-Allen KJ, Kasper FK, Mikos AG. Bioactive polymer/extracellular matrix scaffolds fabricated with a flow perfusion bioreactor for cartilage tissue engineering. Biomaterials. 2010;31(34):8911–8920. doi: 10.1016/j.biomaterials.2010.07.110. https://doi.org/10.1016/j. biomaterials.2010.07.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovati AB, Bottagisio M, Moretti M. Decellularized and engineered tendons as biological substitutes: A critical review. Stem Cells International. 2016;2016:7276150. doi: 10.1155/2016/7276150. https://doi.org/10.1155/2016/7276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Hoshiba T, Kawazoe N, Chen G. Comparison of decellularization techniques for preparation of extracellular matrix scaffolds derived from three-dimensional cell culture. Journal of Biomedical Materials Research Part A. 2012;100(9):2507–2516. doi: 10.1002/jbm.a.34150. https://doi.org/10.1002/jbm.a.34150. [DOI] [PubMed] [Google Scholar]
- Mao Y, Schwarzbauer JE. Stimulatory effects of a three-dimensional micro-environment on cell-mediated fibronectin fibrillogenesis. Journal of Cell Science. 2005;118:4427–4436. doi: 10.1242/jcs.02566. [DOI] [PubMed] [Google Scholar]
- Mao Y, Schwarzbauer JE. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of a5b1 versus avb3 integrin receptors. Cell Communication & Adhesion. 2006;13:267–277. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- Mecham RP, editor. The extracellular matrix: An overview. 1st. Berlin, Heidelberg: Springer-Verlag; 2011. [Google Scholar]
- Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Molecular & Cellular Proteomics. 2012;11(4):M111.014647. doi: 10.1074/mcp.M111.014647. https://doi.org/10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140(5):619–630. doi: 10.1016/j.cell.2010.02.014. https://doi.org/10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, et al. A Rac switch regulates random versus directionally persistent cell migration. The Journal of Cell Biology. 2005;170(5):793–802. doi: 10.1083/jcb.200503152. https://doi.org/10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastino AK, Greco TM, Mathias RA, Cristea IM, Schwarzbauer JE. Stimulatory effects of advanced glycation endproducts (AGEs) on fibronectin matrix assembly. Matrix Biology. 2017;59:39–53. doi: 10.1016/j.matbio.2016.07.003. https://doi.org/10.1016/j.matbio.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. The Journal of Cell Biology. 2012;197(3):439–455. doi: 10.1083/jcb.201201124. https://doi.org/10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345(6200):1062–1065. doi: 10.1126/science.1256965. https://doi.org/10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prewitz MC, Seib FP, von Bonin M, Friedrichs J, Stissel A, Niehage C, et al. Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments. Nature Methods. 2013;10(8):788–794. doi: 10.1038/nmeth.2523. https://doi.org/10.1038/nmeth.2523. [DOI] [PubMed] [Google Scholar]
- Robinson J, Gospodarowicz D. Effect of p-nitrophenyl-beta-D-xyloside on proteoglycan synthesis and extracellular matrix formation by bovine corneal endothelial cell cultures. The Journal of Biological Chemistry. 1984;259(6):3818–3824. [PubMed] [Google Scholar]
- Rutledge K, Cheng Q, Pryzhkova M, Harris GM, Jabbarzadeh E. Enhanced differentiation of human embryonic stem cells on extracellular matrix-containing osteomimetic scaffolds for bone tissue engineering. Tissue Engineering Part C, Methods. 2014;20(11):865–874. doi: 10.1089/ten.tec.2013.0411. https://doi.org/10.1089/ten.TEC.2013.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke-Layland K, Nerem RM. In vitro human tissue models—Moving towards personalized regenerative medicine. Advanced Drug Delivery Reviews. 2011;63(4–5):195–196. doi: 10.1016/j.addr.2011.05.001. https://doi.org/10.1016/j.addr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Shakouri-Motlagh A, O’Connor AJ, Brennecke SP, Kalionis B, Heath DE. Native and solubilized decellularized extracellular matrix: A critical assessment of their potential for improving the expansion of mesenchymal stem cells. Acta Biomaterialia. 2017;55:1–12. doi: 10.1016/j.actbio.2017.04.014. https://doi.org/10.1016/j.actbio.2017.04.014. [DOI] [PubMed] [Google Scholar]
- Singh S, Bandini SB, Donnelly PE, Schwartz J, Schwarzbauer JE. A cell-assembled, spatially aligned extracellular matrix to promote directed tissue development. Journal of Materials Chemistry B, Materials for Biology and Medicine. 2014;2(11):1449–1453. doi: 10.1039/C3TB21864C. https://doi.org/10.1039/C3TB21864C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy PA, Werbin J, Heinz W, Hoh JH, Romer LH. Microelastic properties of lung cell-derived extracellular matrix. Acta Biomaterialia. 2011;7(1):96–105. doi: 10.1016/j.actbio.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li W, Lu Z, Chen R, Ling J, Ran Q, et al. Rescuing replication and osteogenesis of aged mesenchymal stem cells by exposure to a young extracellular matrix. The FASEB Journal. 2011;25(5):1474–1485. doi: 10.1096/fj.10-161497. https://doi.org/10.1096/fj.10-161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SE, Cao T, Talauliker PM, Lifshitz J. Objective morphological quantification of microscopic images using a fast Fourier transform (FFT) analysis. Current Protocols Essential Laboratory Techniques. 2013;95(Suppl 7):9 5 1–9 5 12. doi: 10.1002/9780470089941.et0905s07. https://doi.org/10.1002/9780470089941.et0905s07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Weiner H, Schwarzbauer JE, Engler AJ. Defined extracellular matrix components are necessary for definitive endoderm induction. Stem Cells. 2013;31(10):2084–2094. doi: 10.1002/stem.1453. https://doi.org/10.1002/stem.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault RA, Scott Baggett L, Mikos AG, Kasper FK. Osteogenic differentiation of mesenchymal stem cells on pregenerated extracellular matrix scaffolds in the absence of osteogenic cell culture supplements. Tissue Engineering Part A. 2010;16(2):431–440. doi: 10.1089/ten.tea.2009.0583. https://doi.org/10.1089/ten.TEA.2009.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I. Preparation of extracellular matrices produced by cultured corneal endothelial and PF-HR9 endodermal cells. Current Protocols in Cell Biology, Chapter 10. 2001 doi: 10.1002/0471143030.cb1004s01. unit 10.14. https://doi.org/10.1002/0471143030.cb1004s01. [DOI] [PubMed]
- Wierzbicka-Patynowski I, Mao Y, Schwarzbauer JE. Analysis of fibronectin matrix assembly. Current Protocols in Cell Biology, Chapter 10. 2004 doi: 10.1002/0471143030.cb1012s25. Unit 10.12. [DOI] [PubMed] [Google Scholar]
- Williams SA, Schwarzbauer JE. A shared mechanism of adhesion modulation for tenascin-C and fibulin-1. Molecular Biology of the Cell. 2009;20:1141–1149. doi: 10.1091/mbc.E08-06-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Alkhawaji A, Ding Y, Mei J. Decellularized scaffolds in regenerative medicine. Oncotarget. 2016;7(36):58671–58683. doi: 10.18632/oncotarget.10945. https://doi.org/10.18632/oncotarget.10945. [DOI] [PMC free article] [PubMed] [Google Scholar]


