Abstract
Astrocytes play a critical role in regulating the interface between the cerebral vasculature and the central nervous system. Contributing to this is the astrocytic endfoot domain, a specialized structure that ensheathes the entirety of the vasculature and mediates signaling between endothelial cells, pericytes and neurons. The astrocytic endfoot has been implicated as a critical element of the glymphatic pathway and changes in protein expression profiles in this cellular domain are linked to Alzheimer’s disease pathology. Despite this, basic physiological properties of this structure remain poorly understood including the developmental timing of its formation, and the protein components that localize there to mediate its functions. Here we use human transcriptome data from male and female subjects across several developmental stages and brain regions to characterize the gene expression profile of the dystrophin associated complex (DAC), a known structural component of the astrocytic endfoot that supports perivascular localization of the astroglial water channel aquaporin-4 (AQP4). Transcriptomic profiling is also used to define genes exhibiting parallel expression profiles to DAC elements, generating a pool of candidate genes that encode gene products that may contribute to the physiological function of the perivascular astrocytic endfoot domain. We found that several genes encoding transporter proteins are transcriptionally associated with DAC genes.
Keywords: Glymphatic, AQP4, Astrocytes, Perivascular endfoot, Dystrophin-associated complex, SCR_010943, SCR_008083, SCR_003302
Graphical abstract
The astrocytic endfoot is a critical component of the neurovascular unit, yet its developmental timing and protein constituents are poorly defined. One established component is the dystrophin associated complex (DAC). Here, developing human brain transcriptome data is used to characterize temporal expression of DAC genes and identify transcriptionally linked genes.
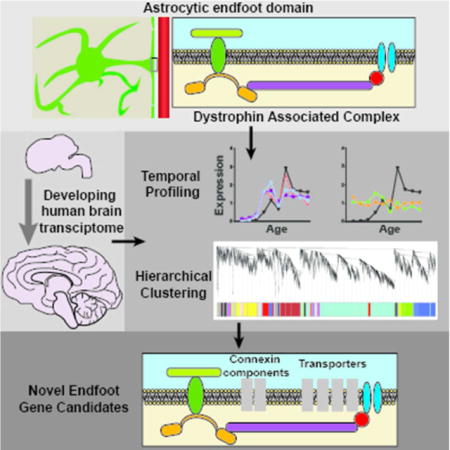
Introduction
Astrocytes contribute to several physiological functions that involve distribution or transport of water, ions and other solutes. This includes neuromodulation, waste clearance, maintenance of the blood brain barrier (BBB), and regulation of metabolic and ionic homeostasis (Broux et al. 2015; Erdo et al. 2016; Hertz et al. 2014; Hladky and Barrand 2016; Langer et al. 2016; Longden et al. 2016; Stokum et al. 2015; Tritsch and Bergles 2007). Astrocytes possess unique morphological characteristics which promote their role in these functions. This includes the non-overlapping, but gap-junction connected astrocytic syncytium which supports movement of ions, nutrients and water throughout brain tissue (Bennett et al. 2003; Nedergaard et al. 2003). Another distinct feature of astrocytes is the perivascular endfoot domain which completely ensheathes the brain microcirculation (Mathiisen et al. 2010; McCaslin et al. 2011). This structure represents a secondary barrier, restricting the movement of cells and macromolecules from the abluminal face of the BBB into the wider parenchyma (Agrawal et al. 2006; Engelhardt and Coisne 2011). Though these anatomical features are well defined, little is known regarding the molecular composition of perivascular astroglial endfeet nor the biophysical basis of their function. Here we utilize a human developmental transcriptomic database to gain insight into the developmental timing of endfoot protein expression. Furthermore, gene network analysis is used to identify novel gene products that may be involved in perivascular endfoot function.
Perivascular astrocytic endfoot processes have long been associated with dense macromolecular arrays comprised of the astroglial water channel aquaporin-4 (AQP4), which is primarily localized to these perivascular structures (Dermietzel 1973; Nielsen et al. 1997; Rash et al. 1998; Verbavatz et al. 1997). Yet the physiological relevance of these formations is still not well understood. One possible function is in the recently described “glymphatic” system, a perivascular network that supports interstitial solute clearance through a process that is dependent upon astroglial water transport (Iliff et al. 2012; Xie et al. 2013). Along this pathway, cerebrospinal fluid (CSF) and interstitial fluid (ISF) exchange across the perivascular astrocytic endfoot in an AQP4-dependent manner (Iliff et al. 2014; Iliff et al. 2012). While the dense AQP4 arrays provide a low-resistance path for water diffusion, it remains unclear what cellular processes facilitate the rapid exchange of solutes along the perivascular compartment.
A critical component of the perivascular astrocytic endfoot domain is the dystrophin associated complex (DAC). This multi-protein scaffolding complex maintains endfoot stability by intracellular interactions with astrocytic cytoskeletal components and binding extracellularly with elements of the basal lamina (Boulay et al. 2015; Lunde et al. 2015). Known protein constituents of the DAC include dystroglycan, dystrophin, dystrobrevin and α-syntrophin. The perivascular localization of AQP4 is maintained through its association with the DAC, as elimination of several DAC components including dystrophin and α-syntrophin, or their extracellular binding partners such as agrin, disrupt perivascular AQP4 localization and endfoot integrity (Bragg et al. 2006; Bragg et al. 2010; Derouiche et al. 2012; Nico et al. 2003; Noell et al. 2007; Noell et al. 2011). In the present study, we first characterize the developmental profile of DAC component expression throughout different human brain regions in order to define the temporal profile of astrocytic endfoot development. Expanding upon this analysis, an unbiased bioinformatics approach is used to identify candidate genes exhibiting similar regional and developmental expression profiles to genes of the DAC complex. These genes encode products that may contribute to perivascular endfoot function.
Materials and Methods
Data Sources
Links to access all of the data used for this study, including the Allen Brain Institute data and the Barres Brain RNA-seq data, are available in the “Data Accessibility” section.
Allen Brain Institute Developing Human Brain Database
The data used for analysis came from the Allen Brain Institute Developing Human Brain Database (2011; Miller et al. 2014). This data contained RNA-seq based transcriptome data from 42 individuals ranging in ages from 7 post-conception weeks to 40 years. Sex of the individuals was balanced approximately evenly (19 females, 23 males). RNA-seq data was collected from 16 brain regions within each individual with over 20,000 genes read from each region, with the exception of tissue collected at the earliest prenatal stage (8–9 post-conception weeks) at which larger regional dissections were made (10 total regions). Detailed description of tissue collection and processing is available in the documentation associated with the database (http://help.brain-map.org//display/devhumanbrain/Documentation). Data was downloaded on October 24th 2016. Entrez IDs (as provided by Allen Brain Institute) were used for protein identification and subsequent analysis.
Weighted Gene Correlation Network Analysis
Weighted Gene Correlation Network Analysis (WGCNA) was implemented using the WGCNA package in R (http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/, RRID: SCR_003302) (Langfelder and Horvath 2008; Langfelder and Horvath 2012). Due to the lack of full male and female datasets for multiple ages, a single network correlation network was constructed for each brain region of which there were 16 in total. In the high-threshold network construction, initial filtering was applied prior to the analysis to eliminate spurious genes that had fragments per kilobase per million (FPKM) values of less than 10 in 90% or more of total reads. This resulted in approximately 8000 genes per brain region. For the second, less-conservative WGCNA, this threshold was adjusted to FPKM values of less than 3 in 95% or more of total reads, resulting in approximately 16000 genes per region. Following standard practices, soft-thresholding was used to establish a connection weight between each gene pair across gender and age within each brain region (Zhang and Horvath 2005). Using this soft-thresholding power, a topographical overlap matrix was generated and a dendrogram relating gene expression was derived. Leveraging the developed dendrogram, gene modules were generated using hierarchical clustering.
Cluster-Based Analysis
The gene modules in which DAC genes were most highly expressed were extracted from each brain region’s WGCNA-based clusters. In the high threshold analysis, of the 16 brain regions analyzed, 2 (amygdaloid complex and cerebellar cortex) did not demonstrate clustering of DAC genes and were excluded from further consideration. In the low threshold analysis, all 16 regions exhibited clustering of at least 2 DAC genes. Genes with astrocyte-specific expression were extracted by thresholding out genes with less than 50% of total expression derived from astrocytes. Cell type expression values were derived from a separate cell-type RNA-seq transcriptome database which resulted in 50–100 astrocytic genes per cluster (Zhang et al. 2014). Drawing on the consistent joint clustering of DAC genes, Pearson’s correlation coefficient was used to identify additional genes highly co-expressed with DAC complex genes across subjects. First, a gene was only considered as co-expressing with DAC after a conservative correlation of r>0.65 (p<1e−5) was observed with at least 2 of the DAC genes. Next, genes were only selected that demonstrated this significant correlation to the DAC genes AQP4, DTNA, SNTA1 or DAG1 in at least 11 of 14 brain regions.
Gene Ontology analysis
To explore protein functions overrepresented in the “endfoot enriched” modules, gene ontology analysis was used. Gene ontology was performed using the limma package in R (http://bioinf.wehi.edu.au/limma/, RRID: SCR_010943) (Ritchie et al. 2015). Only human pathway information was probed, with a false discovery rate cutoff of 0.05. Genes with a p-value for over-representation of p<0.001 were reported.
Subcellular localization mapping
Localization and associated functions of identified proteins was determined through literature review and the CellWhere database (http://cellwhere-myo.rhcloud.com/) (Zhu et al. 2015). The UniProt and Gene ontology functions were used to assess cell localization.
Results
DAC genes are highly expressed in astrocytes
To examine the transcriptome profile of the astrocytic endfoot, we first defined a set of “candidate endfoot genes” which encode proteins associated with the established roles of the DAC in perivascular endfoot physiology. This included 5 genes: four of which encode known DAC complex proteins: SNTA1 (alpha-syntrophin), DTNA (dystrobrevin), DMD (dystrophin), DAG1 (dystroglycan) as well as AQP4 (Figure 1A). Though histological analysis has suggested the presence of numerous proteins at the perivascular endfoot domain, genes encoding DAC proteins were chosen based on their function as a scaffolding complex and integral role for endfoot stability (Bragg et al. 2006; Bragg et al. 2010; Nico et al. 2003; Noell et al. 2007; Noell et al. 2011). AQP4 interacts with the DAC complex, contributes to neurovascular and glymphatic pathway functions of the endfoot and is thought to be a mediator of cell adhesion (Hiroaki et al. 2006; Iliff et al. 2012; Sun et al. 2016). All 5 genes have been previously validated at the endfoot domain histologically (Table 2).
Figure 1. Cell type expression profile of DAC proteins.
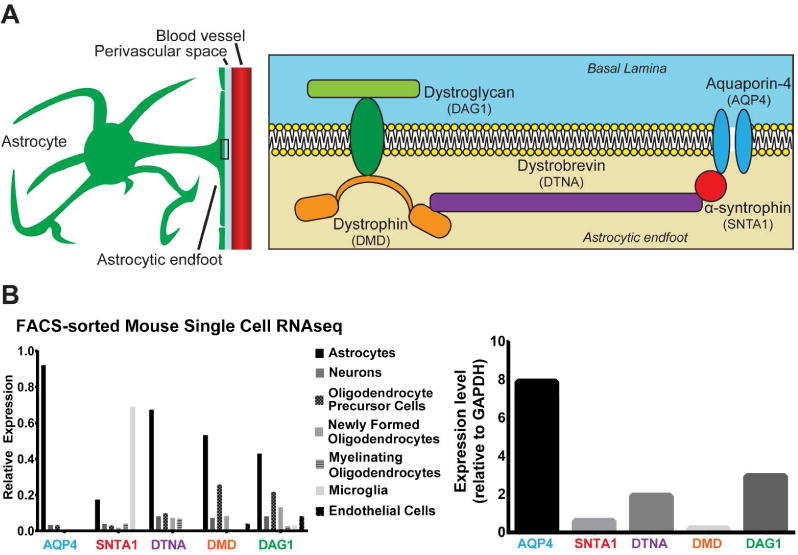
(A) Schematic of known components of the dystrophin associated complex, and a primary binding partner at the astrocytic endfoot: AQP4. (B) Percentage of total expression of DAC proteins in each cell type as measured by the Barres Brain RNA-seq Database (left), as well as a quantification of total expression relative to a control gene (Gapdh, right).
Table 2.
Genes highly correlated to expression of DAC genes.
| Category | Previously described at the astrocytic endfoot? | Gene | Protein Name | Literature References |
|---|---|---|---|---|
| “Endfoot candidate genes” | Yes | AQP4 | Aquaporin 4 | (Nielsen et al. 1997; Simard et al. 2003) |
| SNTA1 | Syntrophin, alpha 1 | (Bragg et al. 2006; Neely et al. 2001) | ||
| DTNA | Dystrobrevin, alpha | (Bragg et al. 2010; Ueda et al. 2000) | ||
| DMD | Dystrophin | (Enger et al. 2012; Frigeri et al. 2001) | ||
| DAG1 | Dystroglycan | (Noell et al. 2011; Zaccaria et al. 2001) | ||
| “Correlated to endfoot candidate genes” | Yes | S1PR1 | sphingosine-1-phosphate receptor 1 | (Nishimura et al. 2010) |
| TIMP3 | TIMP metallopeptidase inhibitor 3 | (Manousopoulou et al. 2016) | ||
| SLC4A4 | solute carrier family 4 (sodium bicarbonate cotransporter), member 4 | (Majumdar et al. 2008) | ||
| CYBRD1 | cytochrome b reductase 1 | (Loke et al. 2013) | ||
| SLC1A2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | (Langer et al. 2016; Schreiner et al. 2014) | ||
| SLC1A3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | (Langer et al. 2016; Schreiner et al. 2014) | ||
| GJA1 | gap junction protein, alpha 1, 43kDa | Boulay et al. 2015; Simard et al. 2003 | ||
| GJB6 | gap junction protein, beta 6, 30kDa | (Boulay et al. 2015; Nagy et al. 1999) | ||
| AGT | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | (Wosik et al. 2007) | ||
| ATP1B2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | (Brignone et al. 2011) | ||
| AXL | AXL receptor tyrosine kinase | (Miner et al. 2015) | ||
| NDRG2 | NDRG family member 2 | (Flugge et al. 2014) | ||
| MLC1 | Megalencephalic leukoencephalopathy with subcortical cysts 1 | (Boor et al. 2007) | ||
| No | PPAP2B | phosphatidic acid phosphatase type 2B | – | |
| PRODH | proline dehydrogenase (oxidase) 2 | – | ||
| GLUD1 | glutamate dehydrogenase 1 | – | ||
| CPE | cytoplasmic polyadenylation element binding protein 3 | – | ||
| CLDN10 | Claudin 10 | – | ||
| AK4 | adenylate kinase 4 | – | ||
| PPP1R3C | Protein phosphatase 1, regulatory (inhibitor) subunit 3C | – | ||
| DIO2 | Deiodinase, iodothyronine, type II | – | ||
| SLC39A12 | Solute carrier family 39 (zinc transporter), member 12 | – | ||
| SLC25A18 | Solute carrier family 25 (mitochondrial carrier), member 18 | – | ||
| F3 | Coagulation factor III (thromboplastin, tissue factor) | – | ||
| TST | Thiosulfate sulfurtransferase (rhodanese) | – | ||
| HTRA1 | HtrA serine peptidase 1 | – | ||
| ATP1A2 | ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide | – | ||
| TRIL | TLR4 interactor with leucine rich repeats | – | ||
| NTSR2 | Neurotensin receptor 2 | – | ||
| BAALC | Brain and acute leukemia, cytoplasmic | – | ||
| PBXIP1 | Pre-B-cell leukemia homeobox interacting protein 1 | – | ||
| LRIG1 | Leucine-rich repeats and immunoglobulin-like domains 1 | – | ||
| RANBP3L | RAN binding protein 3-like | – | ||
| ACSBG1 | Acyl-CoA synthetase bubblegum family member 1 | – | ||
| AMOT | Angiomotin | – | ||
| SFXN5 | Sideroflexin 5 | – | ||
| FXYD1 | FXYD domain containing ion transport regulator 1 (phospholemman) | – | ||
| ACSS1 | Acyl-CoA synthetase short-chain family member 1 | – | ||
| CLU | Clusterin | – | ||
| SLC9A3R1 | Solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | – | ||
| FAM69C | Family With Sequence Similarity 69 Member C, C18orf51 | – |
List of the 5 “endfoot candidate genes” used, along with literature demonstrating localization at the astrocytic endfoot domain. Below, the list of 41 genes derived from the correlation analysis of genes in the “endfoot associated” WGCNA clusters across brain regions, with any published literature regarding their known localization to the astrocytic endfoot domain. Genes in the gray section have been previously described to encode gene products found at the endfoot domain, while those in white have not been validated.
The developmental transcriptome data used to quantify expression of these genes was collected by regional microdissection of tissue, with no cell type specificity. To ensure that information derived from these data is directly applicable to astrocytic endfeet and not influenced by expression by multiple cell types, cell type expression of the selected DAC genes was characterized. FACs sorting-derived, single cell transcriptome data from the mouse brain obtained from the Barres Brain RNA-seq Database was used (Zhang et al. 2014). The results demonstrate abundant expression of all 5 genes in astrocytes, with specific astroglial expression relative to other major brain cell types including neurons, microglia, endothelial cells and oligodendrocytes noted for all gene products with the exception of Snta1 which is present in both astrocytes and microglia (Figure 1B). We also sought to evaluate the relative abundance of each DAC-associated gene. Aqp4, Dag1 and Dtna demonstrate significantly elevated FPKM values relative to a constitutively stable gene (Gapdh) while Dmd and Snta1 expression is lower. Together, these results suggest that the selected candidate genes are significantly expressed in astrocytes.
DAC genes demonstrate two temporally distinct expression profiles
To characterize temporal expression patterns through development, mean transcript expression of endfoot gene products was quantified throughout the total brain and across prenatal and postnatal time points through adulthood. To assure that changes seen do not purely reflect changes in overall astroglial expression, DAC gene expression was also compared to the general astrocytic gene S100β. Quantification of mean gene expression level at each developmental time point reveal that the endfoot gene products stratify into two groups (Figure 2 A–E). Expression of AQP4, DTNA and SNTA1 demonstrate substantial increases in expression beginning at the “Late Prenatal” stage, while DAG1 and DMD maintain a consistent expression level throughout development (Figure 2D–E). Interestingly, when compared to S100β, both DAC profiles were independent (Figure 2F). Though the AQP4, DTNA and SNTA1 share a similar expression profile in later developmental stages, expression of these DAC genes increases significantly during late prenatal stages while S100β does not increase until “Early Infancy” (Figure 2G). Results of statistical analysis of these data are available in Supplemental Table 1. Although included for in Figure 2 for visual reference, data from “Late Infancy” could not be subjected to statistical analysis because of insufficient sample size. These data suggest that the genes comprising the DAC are not entirely transcriptionally linked and instead reflect two distinct transcriptional profiles.
Figure 2. Temporal expression profile of DAC proteins during human development.
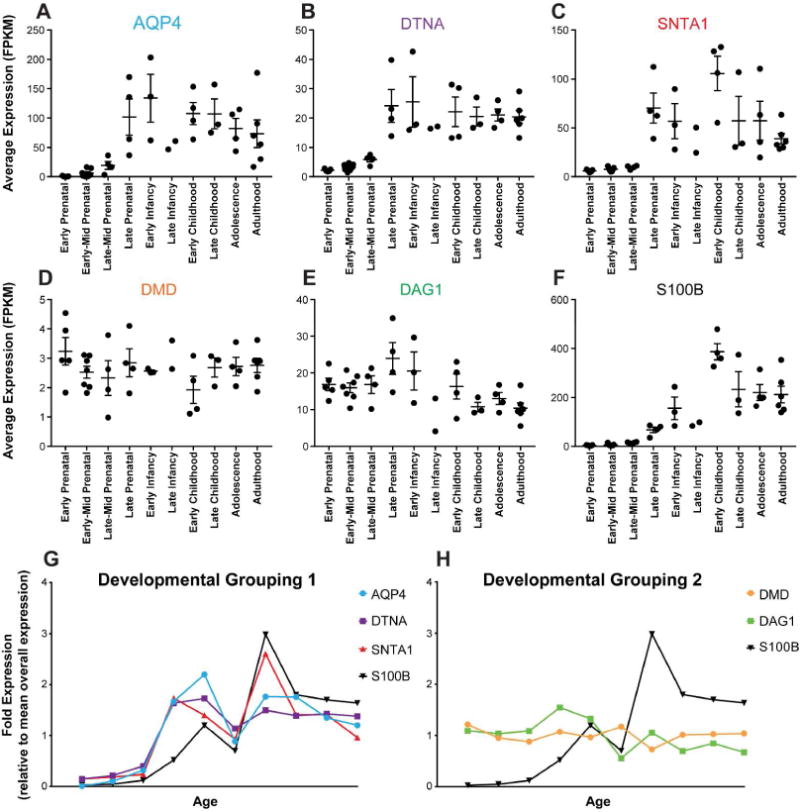
(A–F) Mean normalized RNA-seq FPKM values with standard error for DAC genes at ten developmental stages. Statistical analyses available in Supplemental Table 1, One-way ANOVA, Multiple comparisons. (G) Mean expression of genes at each developmental stage relative to total sample mean values. DAC candidate genes have been grouped based on similar developmental timing profiles. This clusters AQP4, DTNA, and SNTA1 together (left), and DMD and DAG1 together (right). Both are compared to the astrocytic gene S100β.
Weighted Gene Correlation Network Analysis reveals clustered expression of DAC genes
Prompted by the two temporally distinct DAC gene groupings, we next sought to probe the transcriptome database for novel candidate genes that might be transcriptionally linked to the DAC proteins. To cluster DAC expression patterns into broader gene expression groups within each brain region, WGCNA was performed to generate an unbiased, biologically motivated hierarchical clustering of genes across developmental age (Figure 3, Supplemental Data Set 1). These clusters were probed for expression of the candidate endfoot proteins. For all brain regions examined, only two did not result in clustering of at least two of the candidate DAC proteins within a single cluster: amgydaloid complex and cerebellar cortex (Table 1). An “endfoot enriched” cluster was defined as the cluster that included at least two of the 5 probe DAC genes. In the 14 of 16 regions that demonstrated endfoot gene clustering, AQP4 expression was present in all of the “endfoot enriched” clusters. DTNA and SNTA1 were each in 11 of the 14 “endfoot enriched” clusters. These results are indicative of highly correlated expression of three of the candidate DAC genes, AQP4, DTNA and SNTA1, across most brain regions throughout development.
Figure 3. Example WGCNA results for primary auditory cortex.
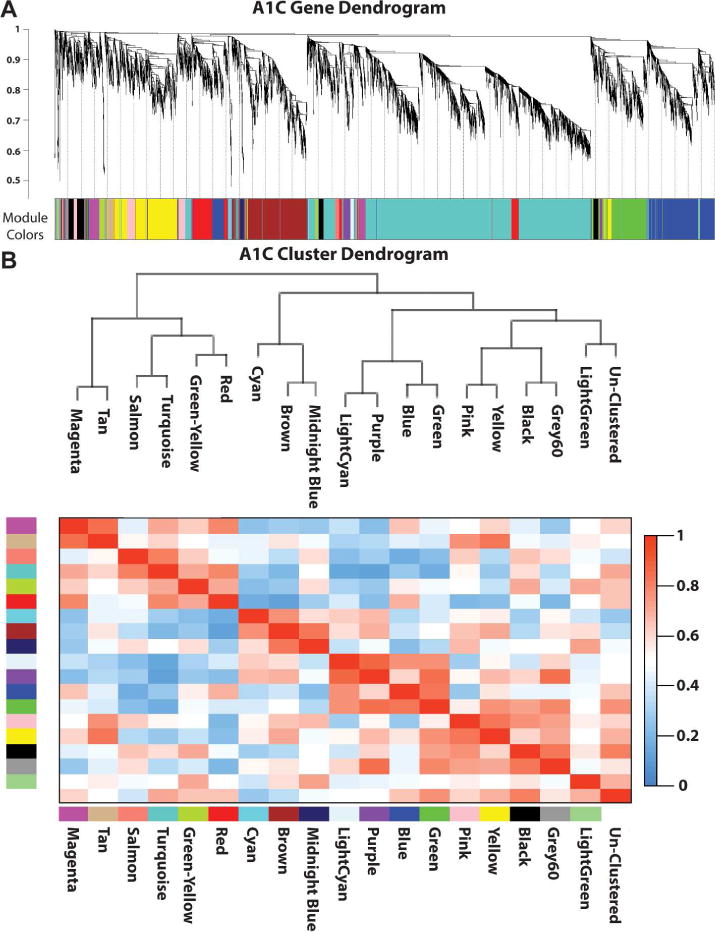
(A) Relationship between each gene in the A1C RNA-seq transcriptome. Hierarchical branching of genes is described by the dendrogram (top) while the colors correspond to the cluster each gene was assigned (bottom). (B) Relatedness between clusters generated by WGCNA analysis. Dendrogram demonstrating the relatedness between the clustered modules (top), while the correlation between any two dendrograms is represented by the heat map (bottom).
Table 1.
WGCNA-based Clustering of DAC proteins across 16 brain regions.
| Region | Region Abbreviation | Included Genes | Number of Modules | “Endfoot” Module | Genes in “Endfoot” module | “Candidate endfoot gene” in module |
|---|---|---|---|---|---|---|
| Primary auditory cortex | A1C | 8321 | 19 | Brown | 902 | AQP4, DTNA, SNTA1 |
| Amygdaloid complex | AMY | 8537 | 20 | – | – | – |
| Cerebellar cortex | CBC | 8514 | 26 | – | – | – |
| Dorsolateral prefrontal Cortex | DFC | 8689 | 23 | Green | 544 | AQP4, DTNA, SNTA1 |
| Hippocampus | HIP | 8639 | 19 | Cyan | 119 | AQP4, DTNA |
| Posteroventral (inferior) parietal cortex | IPC | 8300 | 21 | Yellow | 697 | AQP4, DTNA, SNTA1 |
| Inferolateral temporal cortex | ITC | 8437 | 23 | Green | 465 | AQP4, DTNA |
| Primary motor cortex | M1C | 8551 | 16 | Yellow | 982 | AQP4, DTNA, SNTA1, DAG1 |
| Mediodorsal nucleus of thalamus | MD | 7968 | 19 | Green | 700 | AQP4, SNTA1, DAG1 |
| Anterior (rostral) cingulate cortex | MFC | 8447 | 19 | Red | 542 | AQP4, DTNA, SNTA1 |
| Orbital frontal cortex | OFC | 8402 | 18 | Black | 416 | AQP4, DTNA, SNTA1 |
| Primary somatosensory cortex | S1C | 8568 | 17 | Yellow | 921 | AQP4, SNTA1 |
| Posterior (caudal) superior temporal cortex | STC | 8580 | 21 | Green | 613 | AQP4, SNTA1 |
| Striatum | STR | 8620 | 27 | Pink | 388 | AQP4, DTNA |
| Primary visual cortex | V1C | 8472 | 19 | Yellow | 841 | AQP4, DTNA, SNTA1 |
| Ventrolateral prefrontal cortex | VFC | 8621 | 24 | Yellow | 588 | AQP4, DTNA, SNTA1 |
Summary of the clustering resulting from WGCNA of gene expression in each brain region. The cluster which contained the greatest number of candidate DAC genes is listed along with the genes that were associated with it.
Although they did not consistently cluster with the other DAC candidate genes, we next sought to determine if DMD and DAG1 display an independent co-clustering as suggested by the similarity of their temporal expression profiles (Figure 2). Thresholding used in the initial WGCNA eliminated DMD from analysis due to a low expression level across samples (Figure 1B). To address this, a second WGCNA with a lower inclusion threshold was performed (Supplemental Data Set 2). In this analysis, DMD did not demonstrate co-clustering with other candidate endfoot genes in any of the 16 region. DAG1 clustered with the other candidate genes in 4 of the 16 regions. With this reduced threshold all three remaining candidate genes (AQP4, DTNA and SNTA1) clustered together in 9 of the 16 regions, and at least 2 of the 3 clustered together in 14 of the 16 regions investigated. These data support the co-clustering of the 3 genes demonstrated in the original WGCNA, and suggest that DMD and DAG1 despite similarities in temporal expression, are not linked transcriptionally.
Identification of candidate genes transcriptionally associated with DAC-encoding genes
Utilizing the gene pools generated by the WGCNA, we next aimed to identify candidate genes that may be co-expressed with DAC genes. An “endfoot enriched” cluster gene list was collected for all 14 brain regions that exhibited co-clustering of candidate genes (Supplemental Table 2). In total, 1623 genes co-clustered with at least 1 DAC candidate gene in at least 1 region. The genes were then “ranked” based on a series of inclusion criteria to identify genes highly associated with the DAC candidate genes. The cluster gene list was generated based on the high threshold WGCNA results and thus associations with DMD were not assessed. Only genes that had a significant Pearson’s correlation coefficient with at least 2 of the 4 remaining DAC candidate genes in 11 or more of the 14 brain regions were analyzed further. Remaining genes were then assessed for astrocytic specificity based on the Barres Brain RNA-seq Database. This highly conservative criteria was utilized to offset the heterogeneity in the available data set. A list of 41 genes was generated (Table 2). A literature review suggests that 13 of these gene products (Table 2, Gray) have previously been implicated histologically at the astrocytic endfoot compartment or brain perivascular space, while the remaining 28 genes encode novel endfoot candidate proteins. These data support the role of the previously defined proteins at the perivascular endfoot. The full list represents a transcriptionally linked unit that may be functionally connected at the endfoot domain.
Characterization of gene product functions and developmental profiles for identified candidates reveals enrichment for transporter encoding genes
We next asked whether these transcriptional associations might reveal functional or structural units that are co-regulated at the gene expression level. To gain insight into what role the DAC-associated candidate genes play in endfoot biology, the known functions and properties of the proteins they encode were analyzed by gene ontology (GO) annotations for molecular function (mf), cellular components (cc) and biological function (bf) in the genes identified as highly co-expressing with DAC genes (Supplemental Data Set 3). GO analysis was also run for the enriched cluster within each brain region. Each region individually demonstrated enrichment for transporters, consistent with the list of highly associated genes. Assessment of functions enriched in the GO analysis reveals a significant overrepresentation of genes encoding membrane transporters (Table 3, p <.00001). 11 of the 42 genes encode proteins associated with transporter functions. A literature review reveals that 6 of these 11 transporters have been previously reported at the endfoot domain, while the other 5 are novel candidates.
Table 3.
Molecular functions associated with WGCNA-derived candidate genes
| Molecular Function (MF) | Gene Ontology (GO) identifier | P-Value | Number of Genes | Genes |
|---|---|---|---|---|
| sodium ion transmembrane transporter activity | GO:0015081 | 8.03E-06 | 5 | ATP1A2, ATP1B2, SLC1A2, SLC1A3, SLC4A4 |
| transmembrane transporter activity | GO:0022857 | 2.59E-05 | 10 | ATP1A2, ATP1B2, GJA1, FXYD1, SLC1A2, SLC1A3, SLC4A4, SLC25A18, SFXN5, SLC39A12 |
| transporter activity | GO:0005215 | 3.9606E-05 | 11 | ATP1A2, ATP1B2, GJA1, FXYD1, SLC1A2, SLC1A3, SLC4A4, SLC25A18, SFXN5, SLC39A12, MLC1 |
| ion transmembrane transporter activity | GO:0015075 | 4.25941E-05 | 9 | ATP1A2, ATP1B2, GJA1, FXYD1, SLC1A2, SLC1A3, SLC4A4, SFXN5, SLC39A12 |
| substrate-specific transporter activity | GO:0022892 | 5.49191E-05 | 10 | ATP1A2, ATP1B2, GJA1, FXYD1, SLC1A2, SLC1A3, SLC4A4, SFXN5, SLC39A12, MLC1 |
| substrate-specific transmembrane transporter activity | GO:0022891 | 8.06906E-05 | 9 | ATP1A2, ATP1B2, GJA1, FXYD1, SLC1A2, SLC1A3, SLC4A4, SFXN5, SLC39A12 |
| active transmembrane transporter activity | GO:0022804 | 8.27759E-05 | 6 | ATP1A2, ATP1B2, SLC1A2, SLC1A3, SLC4A4, SLC25A18 |
Molecular functions identified by GO analysis as enriched within the list of genes highly correlated to the DAC candidate genes (P<.00001). All functions are related to transporter functions, and produced a list of 11 total genes.
We next sought to characterize the developmental profile of the identified transporter genes relative to the DAC candidate genes. With the exception of ATP1B2, all of the transporter genes undergo an increase in expression beginning at late prenatal stages, concurrent with AQP4, DTNA and SNTA1 expression profiles (Figure 4). Conversely, ATP1B2 shows a relatively stable level of expression across all developmental stages, similar to the profile of DMD and DAG1. Statistical characterization of the developmental profiles of these genes is available in Supplemental Table 3.
Figure 4. Developmental profile of WGCNA-derived candidate transporter genes.
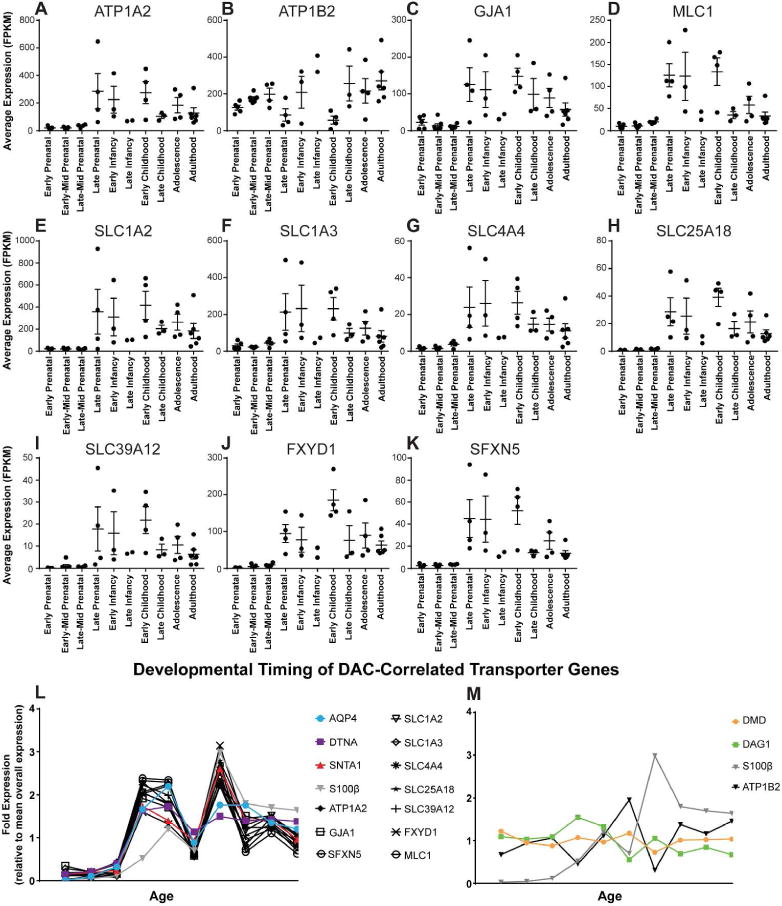
(A–K) Temporal expression profile of the candidate transporter genes identified by WGCNA. Mean FPKM values with standard error from each individual are represented for each time point. Statistical analyses available in Supplemental Table 1, One-way ANOVA, Multiple comparisons. (L–M) Mean expression of WGCNA-derived candidate genes at each developmental stage relative to total sample mean values. Expression of similarly grouped probe genes AQP4, DTNA, and SNTA1, as well as the generic astrocyte gene S100β are overlaid to demonstrate temporal similarities.
Discussion
In the present study, we investigated the human developmental expression profile of genes which encode proteins that contribute to perivascular astroglial endfoot function. We first defined the temporal expression profile of genes associated with DAC components and the water channel AQP4, which are known to localize to astrocytic endfeet and contribute to its function. Using the unbiased gene clustering technique WGCNA, we observed that DAC components, including most prominently AQP4, DTNA and SNTA1 clustered in terms of their expression profiles across brain regions and throughout development. Based on these clusters, we defined genes associated with DAC elements and AQP4 throughout the course of human brain development. The genes identified by this analysis include both proteins which have been previously reported at the endfoot domain, as well as entirely novel candidates. Our results suggest that components of the astrocytic endfoot complex may undergo concurrent upregulation during development. Furthermore, in addition to the water channel AQP4, components of the DAC may interact with, and be transcriptionally linked to a number of ion and solute transporters.
Temporal analysis of DAC gene expression revealed two distinct profiles within the complex. While DMD and DAG1 demonstrate a consistent level of expression across development and aging, expression of the other three components (AQP4, DTNA and SNTA1) undergo significant age-linked fluctuation. These data suggest that the DAC complex is not transcriptionally linked as a whole, but rather that it is regulated through two or more transcriptional modules. DMD and DAG1 have been suggested to directly interact with cytoskeletal and extracellular matrix components respectively (Gesemann et al. 1998; Michalak and Opas 1997; Szabo et al. 2004). These proteins may represent the “core” of the DAC complex that facilitate its membrane localization properties. Complementary to this, SNTA1 and DTNA express multiple protein binding domains (Constantin 2014) and may serve as interchangeable scaffolding components of the DAC that link the core to functional units such as signaling and transporter proteins. Further investigation is necessary to determine whether the apparent transcriptional co-regulation of DAC proteins facilitates functional subdomains of the complex.
AQP4, DTNA and SNTA1 demonstrate a significant elevation in expression beginning at late prenatal stages (25–39 post-conception weeks) and a slow decline in expression after early childhood (19 months-5 years of age). This rise and fall in expression exhibits key differences from expression of the general astrocytic gene S100β. It is worth noting that recent studies suggest astrocytes are a highly heterogeneous cell type, and it is possible that the expression profile of DAC genes may be consistent with an astrocytic subtype not enriched for S100β (John Lin et al. 2017). The greatest differences from S100β expression are seen both in late prenatal development and in the aging brain. Elevated expression of these genes at the earliest stages of astrogliogenesis may reflect a role of astrocytes in distributing secreted factors critical to developmental processes such as blood brain barrier maintenance and neuronal maturation (Blanchette and Daneman 2015; Chau et al. 2015; Gato et al. 2014; Lehtinen et al. 2011). In the aging brain, the decline in expression of these proteins may help explain age dependent changes seen in the localization of AQP4 to the endfoot domain (Zeppenfeld et al. 2017).
With its unique location at the neurovascular interface, the endfoot is also critical to generation and maintenance of fluid and ionic homeostasis in the brain. Previous research in rats has shown that total water content in the brain decreases postnatally and similarly the total brain amount of key ions Na+, K+, and Cl− all decline during early postnatal weeks (Erecinska et al. 2005; Vernadakis and Woodbury 1962). Consistent with this, regulation of the extracellular space of the brain is greatest during this period. Measurement of the extracellular volume fraction in developing rats demonstrates the most robust changes occur during early postnatal stages (Lehmenkuhler et al. 1993; Sykova 2005; Vorisek and Sykova 1997). Accordingly, several DAC proteins found at the astrocytic endfoot, including AQP4, have been shown to markedly increase expression during the first two weeks of postnatal development in rodents, suggesting a role for these proteins regulating developmental chances in extracellular volume and ion homeostasis (Lunde et al. 2015; Wen et al. 1999). This notion is supported by the observation of increased total brain water with Aqp4 gene deletion (Nagelhus and Ottersen 2013). The decline in expression of these proteins with aging also has implications for our understanding of fluid and solute movement systems in the pathophysiology of the aging brain.
Since the initial description of the glymphatic system, the cellular basis of solute movement through the brain and the associated driving forces have remained poorly understood. While the initial characterization of glymphatic function suggested that arterial pulsation drives convective movement of CSF through the brain interstitial space (Iliff et al. 2012; Iliff et al. 2013), more recent computational modeling studies have suggested that these forces may be insufficient to drive bulk flow through these spaces under physiological conditions (Jin et al. 2016; Smith et al. 2015). Most recently, intracellular flow and dispersion models have been elaborated as potential alternative mechanisms to explain the rapid exchange of solutes between the CSF and the ISF along perivascular pathways (Asgari et al. 2015; Asgari et al. 2016). Together these illustrate the need for a more thorough understanding of the constituents of the astrocytic endfoot that may contribute to fluid and solute movement across this domain. The identification of several transporter genes transcriptionally linked to expression of DAC genes has major implications may provide valuable insight into the mechanisms that facilitate fluid movement as described in the glymphatic pathway.
Among the identified transporters are components of the connexin complex GJA1 (encoding connexin 43) and GJB6 (encoding connexin 30). These gap junction proteins are astrocyte specific and play a role in maintenance of the interconnectivity of the astrocytic syncytium (Giaume et al. 2010). Interestingly both GJA1 and GJB6 have been observed at the astrocytic endfoot (Boulay et al. 2015; Nagy et al. 1999; Simard et al. 2003), and GJA1 has been suggested to functionally couple endfeet domains. Fenestrations resulting from inter-endfoot process gaps along with gap junction coupling between endfeet may contribute to macromolecular transport in this space. Development of an understanding of how homeostasis is maintained at the neurovascular unit has important implications for a wide range of pathophysiological conditions including cerebral edema and cerebral amyloid angiopathy (Thrane et al. 2014; Thrane et al. 2015; Wilcock et al. 2009). Our data support the notion that connexins may be a critical component for movement of large solutes and macromolecules in the context of glymphatic function. Several other molecular and ionic transporters were also highly represented within the identified candidate genes. In addition to the connexin genes, 10 other genes with previously defined roles in ion or molecule transport were identified. Furthermore, these genes show largely congruent age-dependent decline in expression along with AQP4. These genes may provide important new insight into mechanisms that underlie the age dependent impairment in the function of the glymphatic pathway (Kress et al. 2014). Five of the identified ionic transporters (ATP1A2, ATP1B2, SLC1A2, SLC1A3 and SLC4A4) are known sodium membrane transporters. Expressional association of these sodium transporters with AQP4 is of particular interest as it may help explain how the osmotic gradients necessary for abundant movement of water across the astrocytic endfoot are mediated. Many of the transporters were also associated with transport of molecules. These are also of interest as they may provide insight into the sort of solutes that are commonly transported though this domain. If co-localization of these transporter genes with DAC components at the astrocytic endfoot is validated histologically, these genes would provide valuable insight into both the ionic and molecular basis of solute movement across this cellular domain (Figure 5).
Figure 5. Proposed schematic of DAC interactions with transporters at the endfoot domain.
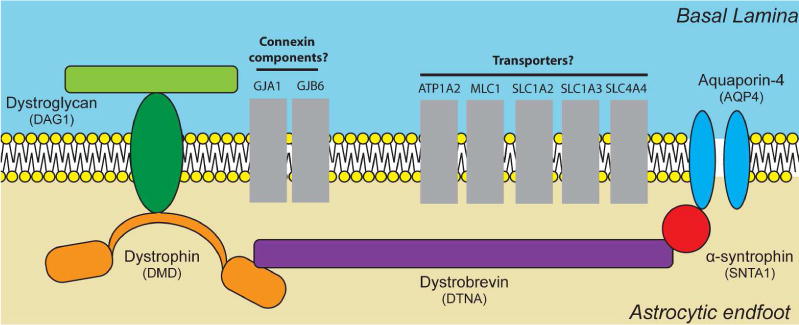
Diagram representing the possible interaction between gene products encoded by a subset of the candidate genes identified by WGCNA. All gene products illustrated have been previously reported in the literature to have a role at the astrocytic endfoot domain. Genes are clustered by established functional roles. Proteins are represented in grey as further biochemical and histological validation is necessary to confirm endfoot localization and physical interactions.
There are several limitations to our study. Primary among these is the data set upon which our analysis was performed as many of the time points had small sample sizes. This is demonstrated by the “Late Infancy” developmental stage, from which only two samples were available. Furthermore, many of these did not have adequate coverage between genders. As previously noted, an additional drawback for our characterization is the RNA-seq data utilized was not astrocyte specific and instead encompasses genes expressed in all brain cell types. To diminish these effects and generate a conservative candidate gene list, strict inclusion criterion were utilized, however the thresholds utilized were arbitrarily defined. Finally, it is important to note that analysis was limited to assessment of transcriptional expression, with no experimental validation of protein localization. Associated gene expression does not necessitate co-localization at the astrocytic endfoot. Despite this, 13 of the 41 endfoot correlated genes identified encode proteins previously described at the endfoot domain. Though experimental validation is necessary, these findings support the concept of a transcriptionally linked endfoot unit.
In this study, our data has defined a transcriptionally linked DAC-associated unit, along with new candidate genes for this region. If confirmed by biochemical studies, these proteins represent novel targets for understanding and therapeutically treating impaired waste clearance in the aging brain.
Supplementary Material
Significance Statement.
The astrocytic endfoot plays a critical role in several brain homeostatic functions yet the expression profile at this domain is poorly defined in both the developing and aging brain. Here, an unbiased transcriptomic-based approach is used to identify the gene expression profile of a key structural component of the endfoot domain, and identify novel candidate genes that may encode protein products transcriptionally linked to known endfoot components. These data provide a pool of novel targets for defining basic endfoot physiology and therapeutic treatment targets for diseases thought to involve dysfunction at the astrocytic endfoot such as Alzheimer’s disease.
Acknowledgments
Support/Grant information: This study was funded by research grant support from the American Heart Association (12SDG11820014, JJI), NINDS (NS089709, JJI) and the Paul G. Allen Family Foundation (JJI).
Footnotes
Associate Editor: Jerome Badaut, PhD
Conflicts of Interest
Dr. Jeffrey Iliff reports serving as a consultant for Shire Pharmaceuticals and GlaxoSmithKline. Dr. Iliff’s research is also funded in part through a research collaboration with GlaxoSmithKline.
Author Contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: M.S. and J.I. Acquisition of data: M.S. Analysis and interpretation of data: M.S., C.M. and J.I. Drafting of the manuscript: M.S. and J.I. Critical revision of the manuscript for important intellectual content: C.M. and J.I. Statistical analysis: M.S. and C.M. Obtained funding: J.I. Study supervision: J.I.
Data Accessibility
The data used to determine cell-type expression of DAC proteins (Fig. 1B) was obtained from the Barres Mouse Brain RNA-Seq Database (2014, available at: http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) (Zhang et al. 2014).
The data used for the assessment of DAC gene transcriptional profiles in the developing human brain (Figures 2–4) was obtained from the BrainSpan Atlas of the Developing Human Brain (2010, available at: http://www.brainspan.org, RRID: SCR_008083) (Miller et al. 2014). Detailed description of sample demographic information is available “Documentation” tab of the website. Briefly, at all developmental time points, samples were taken approximately equally from both sexes, based on availability (19 females, 23 males). Abnormal samples were excluded based on a variety on selection criteria including chromosomal abnormalities, exposure to drug or alcohol abuse, malformation, etc.
References
- 2011. BrainSpan: Atlas of the Developing Human Brain.
- Agrawal S, Anderson P, Durbeej M, van Rooijen N, Ivars F, Opdenakker G, Sorokin LM. Dystroglycan is selectively cleaved at the parenchymal basement membrane at sites of leukocyte extravasation in experimental autoimmune encephalomyelitis. The Journal of experimental medicine. 2006;203(4):1007–1019. doi: 10.1084/jem.20051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari M, de Zelicourt D, Kurtcuoglu V. How astrocyte networks may contribute to cerebral metabolite clearance. Sci Rep. 2015;5:15024. doi: 10.1038/srep15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari M, de Zelicourt D, Kurtcuoglu V. Glymphatic solute transport does not require bulk flow. Scientific reports. 2016;6:38635. doi: 10.1038/srep38635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MV, Contreras JE, Bukauskas FF, Saez JC. New roles for astrocytes: gap junction hemichannels have something to communicate. Trends in neurosciences. 2003;26(11):610–617. doi: 10.1016/j.tins.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Daneman R. Formation and maintenance of the BBB. Mechanisms of development. 2015;138(Pt 1):8–16. doi: 10.1016/j.mod.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Boor I, Nagtegaal M, Kamphorst W, van der Valk P, Pronk JC, van Horssen J, Dinopoulos A, Bove KE, Pascual-Castroviejo I, Muntoni F, Estevez R, Scheper GC, van der Knaap MS. MLC1 is associated with the dystrophin-glycoprotein complex at astrocytic endfeet. Acta neuropathologica. 2007;114(4):403–410. doi: 10.1007/s00401-007-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay AC, Saubamea B, Cisternino S, Mignon V, Mazeraud A, Jourdren L, Blugeon C, Cohen-Salmon M. The Sarcoglycan complex is expressed in the cerebrovascular system and is specifically regulated by astroglial Cx30 channels. Frontiers in cellular neuroscience. 2015;9:9. doi: 10.3389/fncel.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg AD, Amiry-Moghaddam M, Ottersen OP, Adams ME, Froehner SC. Assembly of a perivascular astrocyte protein scaffold at the mammalian blood-brain barrier is dependent on alpha-syntrophin. Glia. 2006;53(8):879–890. doi: 10.1002/glia.20347. [DOI] [PubMed] [Google Scholar]
- Bragg AD, Das SS, Froehner SC. Dystrophin-associated protein scaffolding in brain requires alpha-dystrobrevin. Neuroreport. 2010;21(10):695–699. doi: 10.1097/WNR.0b013e32833b0a3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignone MS, Lanciotti A, Macioce P, Macchia G, Gaetani M, Aloisi F, Petrucci TC, Ambrosini E. The beta1 subunit of the Na,K-ATPase pump interacts with megalencephalic leucoencephalopathy with sub cortical cysts protein 1 (MLC1) in brain astrocytes: new insights into MLC pathogenesis. Human molecular genetics. 2011;20(1):90–103. doi: 10.1093/hmg/ddq435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broux B, Gowing E, Prat A. Glial regulation of the blood-brain barrier in health and disease. Seminars in immunopathology. 2015;37(6):577–590. doi: 10.1007/s00281-015-0516-2. [DOI] [PubMed] [Google Scholar]
- Chau KF, Springel MW, Broadbelt KG, Park HY, Topal S, Lun MP, Mullan H, Maynard T, Steen H, LaMantia AS, Lehtinen MK. Progressive Differentiation and Instructive Capacities of Amniotic Fluid and Cerebrospinal Fluid Proteomes following Neural Tube Closure. Developmental cell. 2015;35(6):789–802. doi: 10.1016/j.devcel.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin B. Dystrophin complex functions as a scaffold for signalling proteins. Biochimica et biophysica acta. 2014;1838(2):635–642. doi: 10.1016/j.bbamem.2013.08.023. [DOI] [PubMed] [Google Scholar]
- Dermietzel R. Visualization by freeze-fracturing of regular structures in glial cell membranes. Die Naturwissenschaften. 1973;60(4):208. doi: 10.1007/BF00599446. [DOI] [PubMed] [Google Scholar]
- Derouiche A, Pannicke T, Haseleu J, Blaess S, Grosche J, Reichenbach A. Beyond polarity: functional membrane domains in astrocytes and Muller cells. Neurochemical research. 2012;37(11):2513–2523. doi: 10.1007/s11064-012-0824-z. [DOI] [PubMed] [Google Scholar]
- Engelhardt B, Coisne C. Fluids and barriers of the CNS esta blish immune privilege by confining immune surveillance to a two-walled castle moat surrounding the CNS castle. Fluids and barriers of the CNS. 2011;8(1):4. doi: 10.1186/2045-8118-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enger R, Gundersen GA, Haj-Yasein NN, Eilert-Olsen M, Thoren AE, Vindedal GF, Petersen PH, Skare O, Nedergaard M, Ottersen OP, Nagelhus EA. Molecular scaffolds underpinning macroglial polarization: an analysis of retinal Muller cells and brain astrocytes in mouse. Glia. 2012;60(12):2018–2026. doi: 10.1002/glia.22416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdo F, Denes L, de Lange E. Age-associated physiological and pathological changes at the blood-brain barrier: A review. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2016 doi: 10.1177/0271678X16679420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erecinska M, Cherian S, IAS Brain development and susceptibility to damage; ion levels and movements. Current topics in developmental biology. 2005;69:139–186. doi: 10.1016/S0070-2153(05)69006-0. [DOI] [PubMed] [Google Scholar]
- Flugge G, Araya-Callis C, Garea-Rodriguez E, Stadelmann-Nessler C, Fuchs E. NDRG2 as a marker protein for brain astrocytes. Cell and tissue research. 2014;357(1):31–41. doi: 10.1007/s00441-014-1837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigeri A, Nicchia GP, Nico B, Quondamatteo F, Herken R, Roncali L, Svelto M. Aquaporin-4 deficiency in skeletal muscle and brain of dystrophic mdx mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15(1):90–98. doi: 10.1096/fj.00-0260com. [DOI] [PubMed] [Google Scholar]
- Gato A, Alonso MI, Martin C, Carnicero E, Moro JA, De la Mano A, Fernandez JM, Lamus F, Desmond ME. Embryonic cerebrospinal fluid in brain development: neural progenitor control. Croatian medical journal. 2014;55(4):299–305. doi: 10.3325/cmj.2014.55.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesemann M, Brancaccio A, Schumacher B, Ruegg MA. Agrin is a high-affinity binding protein of dystroglycan in non-muscle tissue. The Journal of biological chemistry. 1998;273(1):600–605. doi: 10.1074/jbc.273.1.600. [DOI] [PubMed] [Google Scholar]
- Giaume C, Koulakoff A, Roux L, Holcman D, Rouach N. Astroglial networks: a step further in neuroglial and gliovascular interactions. Nature reviews Neuroscience. 2010;11(2):87–99. doi: 10.1038/nrn2757. [DOI] [PubMed] [Google Scholar]
- Hertz L, Gibbs ME, Dienel GA. Fluxes of lactate into, from, and among gap junction-coupled astrocytes and their interaction with noradrenaline. Frontiers in neuroscience. 2014;8:261. doi: 10.3389/fnins.2014.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, Mizoguchi A, Fujiyoshi Y. Implications of the aquaporin-4 structure on array formation and cell adhesion. Journal of molecular biology. 2006;355(4):628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- Hladky SB, Barrand MA. Fluid and ion transfer across the blood-brain and blood-cerebrospinal fluid barriers; a comparative account of mechanisms and roles. Fluids and barriers of the CNS. 2016;13(1):19. doi: 10.1186/s12987-016-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff JJ, Chen MJ, Plog BA, Zeppenfeld DM, Soltero M, Yang L, Singh I, Deane R, Nedergaard M. Impairment of glymphatic pathway function promotes tau pathology after traumatic brain injury. J Neurosci. 2014;34(49):16180–16193. doi: 10.1523/JNEUROSCI.3020-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff JJ, Wang M, Liao Y, Plogg BA, Peng W, Gundersen GA, Benveniste H, Vates GE, Deane R, Goldman SA, Nagelhus EA, Nedergaard M. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med. 2012;4(147):147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff JJ, Wang M, Zeppenfeld DM, Venkataraman A, Plog BA, Liao Y, Deane R, Nedergaard M. Cerebral arterial pulsation drives paravascular CSF-interstitial fluid exchange in the murine brain. J Neurosci. 2013;33(46):18190–18199. doi: 10.1523/JNEUROSCI.1592-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin BJ, Smith AJ, Verkman AS. Spatial model of convective solute transport in brain extracellular space does not support a “glymphatic” mechanism. The Journal of general physiology. 2016;148(6):489–501. doi: 10.1085/jgp.201611684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin CC, Yu K, Hatcher A, Huang TW, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, Deneen B. Identification of diverse astrocyte populations and their malignant analogs. Nature neuroscience. 2017;20(3):396–405. doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress BT, Iliff JJ, Xia M, Wang M, Wei HS, Zeppenfeld D, Xie L, Kang H, Xu Q, Liew JA, Plog BA, Ding F, Deane R, Nedergaard M. Impairment of paravascular clearance pathways in the aging brain. Ann Neurol. 2014;76(6):845–861. doi: 10.1002/ana.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer J, Gerkau NJ, Derouiche A, Kleinhans C, Moshrefi-Ravasdjani B, Fredrich M, Kafitz KW, Seifert G, Steinhäuser C, Rose CR. Rapid sodium signaling couples glutamate uptake to breakdown of ATP in perivascular astrocyte endfeet. Glia. 2016 doi: 10.1002/glia.23092. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R Functions for Ro bust Correlations and Hierarchical Clustering. Journal of statistical software. 2012;46(11) [PMC free article] [PubMed] [Google Scholar]
- Lehmenkuhler A, Sykova E, Svoboda J, Zilles K, Nicholson C. Extracellular space parameters in the rat neocortex and subcortical white matter during postnatal development determined by diffusion analysis. Neuroscience. 1993;55(2):339–351. doi: 10.1016/0306-4522(93)90503-8. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, D’Ercole AJ, Wong ET, LaMantia AS, Walsh CA. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69(5):893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke SY, Siddiqi NJ, Alhomida AS, Kim HC, Ong WY. Expression and localization of duodenal cytochrome b in the rat hippocampus after kainate-induced excitotoxicity. Neuroscience. 2013;245:179–190. doi: 10.1016/j.neuroscience.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Longden TA, Hill-Eubanks DC, Nelson MT. Ion channel networks in the control of cerebral blood flow. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2016;36(3):492–512. doi: 10.1177/0271678X15616138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde LK, Camassa LM, Hoddevik EH, Khan FH, Ottersen OP, Boldt HB, Amiry-Moghaddam M. Postnatal development of the molecular complex underlying astrocyte polarization. Brain structure & function. 2015;220(4):2087–2101. doi: 10.1007/s00429-014-0775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar D, Maunsbach AB, Shacka JJ, Williams JB, Berger UV, Schultz KP, Harkins LE, Boron WF, Roth KA, Bevensee MO. Localization of electrogenic Na/bicarbonate cotransporter NBCe1 variants in rat brain. Neuroscience. 2008;155(3):818–832. doi: 10.1016/j.neuroscience.2008.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousopoulou A, Gatherer M, Smith C, Nicoll JA, Woelk CH, Johnson M, Kalaria R, Attems J, Garbis SD, Carare RO. Systems proteomic analysis reveals that clusterin and tissue inhibitor of metalloproteinases 3 increase in leptomeningeal arteries affected by cerebral amyloid angiopathy. Neuropathology and applied neuro biology. 2016 doi: 10.1111/nan.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiisen TM, Lehre KP, Danbolt NC, Ottersen OP. The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia. 2010;58(9):1094–1103. doi: 10.1002/glia.20990. [DOI] [PubMed] [Google Scholar]
- McCaslin AF, Chen BR, Radosevich AJ, Cauli B, Hillman EM. In vivo 3D morphology of astrocyte-vasculature interactions in the somatosensory cortex: implications for neurovascular coupling. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31(3):795–806. doi: 10.1038/jcbfm.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Opas M. Functions of dystrophin and dystrophin associated proteins. Current opinion in neurology. 1997;10(5):436–442. doi: 10.1097/00019052-199710000-00014. [DOI] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, Bennet C, Bertagnolli D, Brouner K, Butler S, Caldejon S, Carey A, Cuhaciyan C, Dalley RA, Dee N, Dolbeare TA, Facer BA, Feng D, Fliss TP, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Howard RE, Jochim JM, Kuan CL, Lau C, Lee CK, Lee F, Lemon TA, Lesnar P, McMurray B, Mastan N, Mosqueda N, Naluai-Cecchini T, Ngo NK, Nyhus J, Oldre A, Olson E, Parente J, Parker PD, Parry SE, Stevens A, Pletikos M, Reding M, Roll K, Sandman D, Sarreal M, Shapouri S, Shapovalova NV, Shen EH, Sjoquist N, Slaughterbeck CR, Smith M, Sodt AJ, Williams D, Zollei L, Fischl B, Gerstein MB, Geschwind DH, Glass IA, Hawrylycz MJ, Hevner RF, Huang H, Jones AR, Knowles JA, Levitt P, Phillips JW, Sestan N, Wohnoutka P, Dang C, Bernard A, Hohmann JG, Lein ES. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JJ, Daniels BP, Shrestha B, Proenca-Modena JL, Lew ED, Lazear HM, Gorman MJ, Lemke G, Klein RS, Diamond MS. The TAM receptor Mertk protects against neuroinvasive viral infection by maintaining blood-brain barrier integrity. Nature medicine. 2015;21(12):1464–1472. doi: 10.1038/nm.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelhus EA, Ottersen OP. Physiological roles of aquaporin-4 in brain. Physiological reviews. 2013;93(4):1543–1562. doi: 10.1152/physrev.00011.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Jl, Patel D, Ochalski PA, Stelmack GL. Connexin30 in rodent, cat and human brain: selective expression in gray matter astrocytes, co-localization with connexin43 at gap junctions and late developmental appearance. Neuroscience. 1999;88(2):447–468. doi: 10.1016/s0306-4522(98)00191-2. [DOI] [PubMed] [Google Scholar]
- Nedergaard M, Ransom B, Goldman SA. New roles for astrocytes: redefining the functional architecture of the brain. Trends in neurosciences. 2003;26(10):523–530. doi: 10.1016/j.tins.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Neely JD, Amiry-Moghaddam M, Ottersen OP, Froehner SC, Agre P, Adams ME. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):14108–14113. doi: 10.1073/pnas.241508198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nico B, Frigeri A, Nicchia GP, Corsi P, Ribatti D, Quondamatteo F, Herken R, Girolamo F, Marzullo A, Svelto M, Roncali L. Severe alterations of endothelial and glial cells in the blood-brain barrier of dystrophic mdx mice. Glia. 2003;42(3):235–251. doi: 10.1002/glia.10216. [DOI] [PubMed] [Google Scholar]
- Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17(1):171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H, Akiyama T, Irei L, Hamazaki S, Sadahira Y. Cellular localization of sphingosine-1-phosphate receptor 1 expression in the human central nervous system. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2010;58(9):847–856. doi: 10.1369/jhc.2010.956409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noell S, Fallier-Becker P, Beyer C, Kroger S, Mack AF, Wolburg H. Effects of agrin on the expression and distribution of the water channel protein aquaporin-4 and volume regulation in cultured astrocytes. The European journal of neuroscience. 2007;26(8):2109–2118. doi: 10.1111/j.1460-9568.2007.05850.x. [DOI] [PubMed] [Google Scholar]
- Noell S, Wolburg-Buchholz K, Mack AF, Beedle AM, Satz JS, Campbell KP, Wolburg H, Fallier-Becker P. Evidence for a role of dystroglycan regulating the membrane architecture of astroglial endfeet. The European journal of neuroscience. 2011;33(12):2179–2186. doi: 10.1111/j.1460-9568.2011.07688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Hudson CS, Agre P, Nielsen S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(20):11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner AE, Durry S, Aida T, Stock MC, Ruther U, Tanaka K, Rose CR, Kafitz KW. Laminar and subcellular heterogeneity of GLAST and GLT-1 immunoreactivity in the developing postnatal mouse hippocampus. The Journal of comparative neurology. 2014;522(1):204–224. doi: 10.1002/cne.23450. [DOI] [PubMed] [Google Scholar]
- Simard M, Arcuino G, Takano T, Liu QS, Nedergaard M. Signaling at the gliovascular interface. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23(27):9254–9262. doi: 10.1523/JNEUROSCI.23-27-09254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Jin BJ, Verkman AS. Muddying the water in brain edema? Trends in neurosciences. 2015;38(6):331–332. doi: 10.1016/j.tins.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokum JA, Kurland DB, Gerzanich V, Simard JM. Mechanisms of astrocyte-mediated cerebral edema. Neurochemical research. 2015;40(2):317–328. doi: 10.1007/s11064-014-1374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Liang R, Yang B, Zhou Y, Liu M, Fang F, Ding J, Fan Y, Hu G. Aquaporin-4 mediates communication between astrocyte and microglia: Implications of neuroinflammation in experimental Parkinson’s disease. Neuroscience. 2016;317:65–75. doi: 10.1016/j.neuroscience.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Sykova E. Glia and volume transmission during physiological and pathological states. Journal of neural transmission (Vienna, Austria : 1996) 2005;112(1):137–147. doi: 10.1007/s00702-004-0120-4. [DOI] [PubMed] [Google Scholar]
- Szabo A, Jancsik V, Mornet D, Kalman M. Immunofluorescence mapping of dystrophin in the rat brain: astrocytes contain the splice variant Dp71f, but this is confined to subpopulations. Anatomy and embryology. 2004;208(6):463–477. doi: 10.1007/s00429-004-0411-4. [DOI] [PubMed] [Google Scholar]
- Thrane AS, Rangroo Thrane V, Nedergaard M. Drowning stars: reassessing the role of astrocytes in brain edema. Trends in neurosciences. 2014;37(11):620–628. doi: 10.1016/j.tins.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane AS, Rangroo Thrane V, Plog BA, Nedergaard M. Filtering the muddied waters of brain edema. Trends Neurosci. 2015;38(6):333–335. doi: 10.1016/j.tins.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Tritsch NX, Bergles DE. Defining the role of astrocytes in neuromodulation. Neuron. 2007;54(4):497–500. doi: 10.1016/j.neuron.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Ueda H, Baba T, Terada N, Kato Y, Fujii Y, Takayama I, Mei X, Ohno S. Immunolocalization of dystrobrevin in the astrocytic endfeet and endothelial cells in the rat cerebellum. Neuroscience letters. 2000;283(2):121–124. doi: 10.1016/s0304-3940(00)00925-3. [DOI] [PubMed] [Google Scholar]
- Verbavatz JM, Ma T, Gobin R, Verkman AS. Absence of orthogonal arrays in kidney, brain and muscle from transgenic knockout mice lacking water channel aquaporin-4. J Cell Sci. 1997;110(Pt 22):2855–2860. doi: 10.1242/jcs.110.22.2855. [DOI] [PubMed] [Google Scholar]
- Vernadakis A, Woodbury DM. Electrolyte and amino acid changes in rat brain during maturation. The American journal of physiology. 1962;203:748–752. doi: 10.1152/ajplegacy.1962.203.4.748. [DOI] [PubMed] [Google Scholar]
- Vorisek I, Sykova E. Evolution of anisotropic diffusion in the developing rat corpus callosum. Journal of neurophysiology. 1997;78(2):912–919. doi: 10.1152/jn.1997.78.2.912. [DOI] [PubMed] [Google Scholar]
- Wen H, Nagelhus EA, Amiry-Moghaddam M, Agre P, Ottersen OP, Nielsen S. Ontogeny of water transport in rat brain: postnatal expression of the aquaporin-4 water channel. The European journal of neuroscience. 1999;11(3):935–945. doi: 10.1046/j.1460-9568.1999.00502.x. [DOI] [PubMed] [Google Scholar]
- Wilcock DM, Vitek MP, Colton CA. Vascular amyloid alters astrocytic water and potassium channels in mouse models and humans with Alzheimer’s disease. Neuroscience. 2009;159(3):1055–1069. doi: 10.1016/j.neuroscience.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosik K, Cayrol R, Dodelet-Devillers A, Berthelet F, Bernard M, Moumdjian R, Bouthillier A, Reudelhuber TL, Prat A. Angiotensin II controls occludin function and is required for blood brain barrier maintenance: relevance to multiple sclerosis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27(34):9032–9042. doi: 10.1523/JNEUROSCI.2088-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156):373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeppenfeld DM, Simon M, Haswell JD, D’Abreo D, Murchison C, Quinn JF, Grafe MR, Woltjer RL, Kaye J, Iliff JJ. Association of Perivascular Localization of Aquaporin-4 With Cognition and Alzheimer Disease in Aging Brains. JAMA Neurol. 2017;74(1):91–99. doi: 10.1001/jamaneurol.2016.4370. [DOI] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing data base of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Malatras A, Thorley M, Aghoghogbe I, Mer A, Duguez S, Butler-Browne G, Voit T, Duddy W. CellWhere: graphical display of interaction networks organized on subcellular localizations. Nucleic acids research. 2015;43(W1):W571–575. doi: 10.1093/nar/gkv354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


