Abstract
Harmonia axyridis (Coleoptera: Coccinellidae) is a polyphagous insect that is an important biological agent used to control agricultural and forestry pests. The role of functional genes in H. axyridis based on quantitative real-time PCR (qRT-PCR) is increasingly well understood to investigate biology, physiology, feeding behavior and the role of important genes in physiological processes. Quantitative real-time PCR (qRT-PCR) is a powerful and reliable technique to quantify gene expression. Using qRT-PCR, expression levels of target genes are determined based on the levels of internal reference genes; therefore, reference genes need to be stably expressed under specific experimental conditions. However, there have been no studies on the stability of reference genes used in H. axyridis. In this study, we systematically investigated expression profiles of nine candidate reference genes from H. axyridis, including β-actin (ACTIN); elongation factor 1 α (EF1A); ribosomal proteins L10, L18, L28, S13, and S15 (RPL10, RPL18, RPL28, RPS13 and RPS15); glyceraldehyde-3-phosphate dehydrogenase (GAPDH); and superoxide dismutase (SOD). Four analytical methods (geNorm, BestKeeper, NormFinder, and the ΔCt method) were used to evaluate the suitability of these genes as internal reference genes for three biotic factors (developmental stage, tissue, and sex) and two abiotic treatments (temperature and photoperiod). RefFinder, a comprehensive evaluation platform integrating the four analytical methods, was used to rank the overall stability of these reference genes. Among the nine candidate genes, different reference genes were identified as having the most stable expression across biotic and abiotic factors. Genes encoding ribosomal proteins typically had the most stable expression, though EF1A was the most stable across developmental stages and photoperiods. To validate the suitability of these reference genes, heat shock protein 90 (HSP90) was chosen as a target. Significant up-regulation in HSP90 expression level in response to both low and high temperature was observed when using the most suitable reference genes but not when using an arbitrarily selected reference gene. The reference genes identified in this study will provide the basis for future functional genomics research in H. axyridis and will also facilitate the establishment of a standardized qRT-PCR program for other related insects.
Introduction
Harmonia axyridis (Coleoptera: Coccinellidae) is a polyphagous insect that is native to northeast Asia and has been widely introduced as a biological control against pest aphids. Unfortunately, it has unintentionally spread to many other countries [1]. The role of functional genes is increasingly well understood, allowing for them to be used in species like H. axyridis to investigate biology, physiology, and feeding behavior [2–5], as well as the role of important genes in physiological processes [6]. In addition, the identification of functional genes involved in host-parasite interactions is an important next step in understanding population dynamics [7].
Quantitative real-time PCR (qRT-PCR) is a reliable and reproducible method for gene quantification [8]. Although qRT-PCR has become one of the most-used techniques in molecular biology research, analysis of gene expression is limited by the integrity and quality of RNA samples, reverse transcription, normalization, and PCR efficiency [9, 10]. The typical method for normalizing gene expression data is to simultaneously measure the expression levels of one or more reference genes (also called an endogenous control or housekeeping genes), which are involved in basic and ubiquitous cellular functions and typically exhibit stable and constitutive expression levels across various biotic and abiotic conditions [8]. There are many reference genes that have been widely used for the normalization of qRT-PCR data, such as ribosomal protein, β-actin (ACTIN), elongation factor 1 α (EF1A), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and superoxide dismutase (SOD) [11–15]. Because reference genes are necessary for survival, it is often thought that there is little fluctuation in the transcription of these genes. For example, ribosomal proteins are important components of ribosomes and play important roles in intracellular protein biosynthesis, cell differentiation, and DNA repair, among other functions [16]. EF1A plays an important role in translation by catalyzing the GTP-dependent binding of aminoacyl-tRNA to the acceptor site of the ribosome [17]. SOD, which is a metalloenzyme capable of catalyzing the dismutation of the superoxide anion to elemental molecular oxygen and hydrogen peroxide, is widely distributed in insects [18]. ACTIN is a major component of the cellular skeleton, which maintains structural integrity and gives shape to cells [17]. GAPDH plays a role in energy metabolism [19]. However, many studies have demonstrated that these widely used reference genes are differentially expressed under different experimental conditions [17, 20]. Differentially expressed genes could usually make significantly biological changes across various tissues, developmental stages, sex, and other samples from diverse experimental conditions. Thus, it is important to identify genes with stable expression so that specific changes in gene expression can be evaluated.
Some recent studies have attempted to determine the stability of reference genes in arthropods [15, 21–23]. Yang et al. [24, 25] and Pan et al. [26] identified stable reference genes in three species of ladybeetles: Coleomegilla maculate, Coccinella septempunctata, and Hippodamia convergens. However, there are no available data on the most appropriate genes to use for H. axyridis target gene normalization under different conditions and at varied developmental stages.
The objectives of this study were to identify appropriate H. axyridis reference genes for real-time qRT-PCR experiments under various conditions and to provide the basis for future functional genomics research in H. axyridis. Nine commonly used reference genes, including ACTIN, EF1A, ribosomal proteins L10, L18, L28, S13, and S15 (RPL10, RPL18, RPL28, RPS13, and RPS15), GAPDH, and SOD from H. axyridis were tested. The effectiveness of these genes for expression normalization was further validated by qRT-PCR analysis of the well-studied target heat shock protein 90 (HSP90) gene. Many studies have demonstrated that heat-shock proteins act as molecular chaperones and play an important role in cellular responses to environmental stressors, including sublethal heat and cold shocks, infections, environmental contaminants [6, 27–29]. H. axyridis is the focus of agriculture and forestry production pest control strategies worldwide. And studying the temperature adaptability of H. axyridis is very important. Thus, it is of interest to study HSPs like HSP90 that may confer thermal tolerance. Based on our analysis of HSP90 and commonly used reference genes, we recommend specific combinations of reference genes to use for qRT-PCR analysis of different biotic and abiotic experimental conditions.
Materials and methods
Insects
Harmonia axyridis (Coleoptera: Coccinellidae) was purchased from a commercial company in Beijing (Beijing Kuoye Tianyuan Biological Technology Co., Ltd., http://www.kuoye.com/). H. axyridis larvae and adults were fed with the aphid Aphis craccivora Koch (Hemiptera: Aphididae). Ladybeetles were reared in a growth chamber located at Beijing Academy of Agriculture and Forestry Sciences at a temperature of 23±1°C, with a 16L:8D photoperiod and 70% relative humidity.
Factors
Effects of the following factors on reference gene expression were measured: development stage, tissue, sex, temperature, and photoperiod. The different developmental stages included eggs (30), first-instar larvae (15), second-instar larvae (10), third-instar larvae (2), fourth-instar larvae (2), pupae (1), and sex (one male and one female adult). The head, midgut, and carcass (body with head and viscera removed) were dissected from fourth instar larvae (10 for each replication) under a microscope (Invitrogen, Carlsbad, CA) and stored in TRIzol reagent. To determine the effect of sex on reference gene expression, one adult female and male were collected separately and placed in 1.5 ml centrifuge tubes. To examine the influence of temperature, groups of two third-instar larvae were separately exposed to 5°C, 20°C, and 35°C for 3 h in a constant-temperature incubator (16L:8D photoperiod, 70±10% RH). To test the effect of photoperiod, groups of two third-instar larvae were kept in a constant-temperature incubator (23±1°C, 70±10% RH) with a 16L:8D, 12L:12D, or 8L:16D photoperiod for 2 d. Each experiment was repeated three times. All samples were quickly frozen in liquid nitrogen after collection and stored at -80°C in 1.5 ml centrifuge tubes for subsequent total RNA extraction.
Selection of gene sequences and primer design
We selected nine housekeeping genes and one target gene from our H. axyridis transcriptome data: ACTIN, EF1A, GAPDH, RPL10, RPL18, RPL28, RPS13, RPS15, SOD, and HSP90. The mfold web server (http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form) was used to predict the secondary structure of the DNA template using the following settings: melting temperature, 60°C; DNA sequence, linear; Na+ concentration, 50 mM; and Mg++ concentration, 3 mM. The default settings were used for the remaining parameters [30]. Primers were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) with the following settings: GC content between 40–60%; melting temperature of 60°C; and PCR product size between 100–200 base pairs. Excluded regions were defined based on the results of mfold analysis, and the default settings were used for the remaining parameters. PCR primer sequences used for quantification of the ten genes are shown in Table 1.
Table 1. Sequences and amplicon characteristics of qRT-PCR primers for nine candidate reference genes and one target gene.
| Gene Symbol | Gene Name | Primer sequences (5’-3’) | Length (bp) | Efficiency (%) | R2 | Linear regression equation |
|---|---|---|---|---|---|---|
| ACTIN | B-actin | CTCTTGACCGAAGCCCCATT | 114 | 104.1 | 0.9996 | y = -3.2264x+14.485 |
| GGAGAGTACGGCTTGGATGG | ||||||
| EF1A | Elongation factor 1 alpha | TCACCGGAACATCTCAAGCC | 103 | 100.6 | 0.9976 | y = -3.3084x+14.218 |
| GCGTGTTCACGAGTTTGTCC | ||||||
| RPL10 | Ribosomal protein L10 | AAGGAACCGTAGCCCGAGTA | 124 | 100.4 | 0.99 | y = -3.3084x+14.218 |
| TTTTGACGACCGGGGAACTT | ||||||
| RPL18 | Ribosomal protein L18 | AACCCGGTCGTGAAAACCTT | 134 | 97.8 | 0.9978 | y = -3.3765x+14.343 |
| TGCTTTCACAATTCTGGCACG | ||||||
| RPL28 | Ribosomal protein L28 | CAGAACCTAGCAACCTCACCA | 122 | 101.7 | 0.9995 | y = -3.2829x+15.442 |
| AGGTCTCTGACAGACTACGGT | ||||||
| RPS13 | Ribosomal protein S13 | ACAGACGAAGTGTCCCAACA | 135 | 96.0 | 0.9988 | y = -3.4204x+13.669 |
| CCTGAGCAACTCCAAGGGAAT | ||||||
| GAPDH | Glyceraldehyde-3-phosphate | GTGAGAGGGATCCCAAAGCC | 181 | 94.7 | 0.9961 | y = -3.455x+17.471 |
| TCGAGATTGACACCGCAGAC | ||||||
| RPS15 | Ribosomal protein S15 | CGCCTAGATTCGATGTCCCA | 108 | 100.9 | 0.9993 | y = -3.3002x+14.5 |
| CCATGATGCCACCGCTAGTA | ||||||
| SOD | Superoxide dismutase | TCAGCTGGAGCACACTTCAA | 186 | 97.3 | 0.9955 | y = -3.3822x+15.218 |
| ATGTACAACCAGGGTGCGAC | ||||||
| HSP90 | Small heat shock protein | TTGGACCCGAAATGCTGACG | 145 | 106.0 | 0.9988 | y = -3.1857x+17.542 |
| AGGAACAAACAGGAGTGCCC |
RNA extraction and cDNA synthesis
Total RNA was extracted using the Trizol method. Each sample was homogenized with 1 ml TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. The quality and quantity of RNA were assessed with a Thermo Scientific NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The quality of the nucleic acid sample was considered good if the OD ratio (A260/A280) was between 1.8 and 2.05 [31]. Complementary DNA (cDNA) was synthesized from 1 μg total RNA using the PrimeScript™RT reagent Kit with gDNA Eraser (perfect Real Time) (TAKARA, Japan) according to the manufacturer’s protocol. The cDNA was diluted 10-fold for subsequent qRT-PCR studies.
qRT-PCR
Real-time qPCR was carried out in 20 μl reactions containing 2.0 μl cDNA, 10 μl SYBR Premix Ex TaqTM II (Takara, Japan), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), 0.4 μl Rox Reference Dye II (Takara, Japan) and 5.6 μl nuclease free water using an ABI PRISM 7500 Real-time PCR System (Applied Biosystems, USA). The amplification conditions for qRT-PCR were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 34 s. After all reactions, melting curve analysis (from 60 to 95°C) was done to ensure consistency and specificity of the amplified product. A 10-fold dilution series of cDNA from the whole body of adults was used for a standard curve. The corresponding qRT-PCR efficiencies (E) were calculated using the equation: E = (10[−1/slope] − 1) × 100 [32].
Constancy analysis of candidate reference genes
The constancy of candidate genes was evaluated using the ΔCT method [33], GeNorm [8], NormFinder [34], and BestKeeper [35]. We also used the online software RefFinder (http://fulxie.0fees.us/?type=reference&ckattempt=1&i=1) to further evaluate the suitability of reference genes by analyzing the results of the four algorithms.
Both geNorm and Normfinder require the conversion of raw cycle threshold (Ct) values into relative quantities. Initially, geNorm calculates a gene expression stability value (M) and then compares the pairwise variation (V) with other genes. Using microarray data as a training set for the algorithm, the pairwise variation between two sequential normalization factors, Vn/Vn+1, is calculated. The optimal number of reference genes required for accurate normalization is determined based on a cutoff Vn/Vn+1 value of 0.15. NormFinder calculates gene expression stability for all samples in any number of groups based on intra- and inter-group variation and combines these values to provide a gene rank order based on the variation in gene expression. BestKeeper uses raw data (Ct values) and PCR efficiency (E) to compute best-suited standards and combines them into an index. The comparative ΔCT method compares the relative expression of pairs of genes within each sample to select the optimal reference gene. Finally, RefFinder evaluates and screens reference genes by integrating the results of the above four major software programs.
Evaluation of target gene expression
The target gene H. axyridis HSP90 was used to evaluate the performance of nine candidate reference genes. We estimated up- or down-regulation of the HSP90 gene in H. axyridis across different temperatures. Relative expression of HSP90 was calculated using the formula (2−ΔΔCT) [36].
Results
Evaluation of primer specificity and amplification efficiency
The primer specificity of nine candidate reference genes and one target gene (HSP90) was evaluated by PCR. Visualization of PCR products by 2% agarose gel electrophoresis revealed a single amplicon of expected size for each primer pair (S1 Fig). Furthermore, gene-specific amplification was confirmed by a single peak in real-time melting curve analysis (S2 Fig). A 5-point standard with known RNA standard concentrations was used to estimate the amplification efficiencies, which ranged from 94.7% to 106%. The coefficients of all 10 genes based on the linear regression were >0.990 (Table 1).
Expression profiling of reference genes
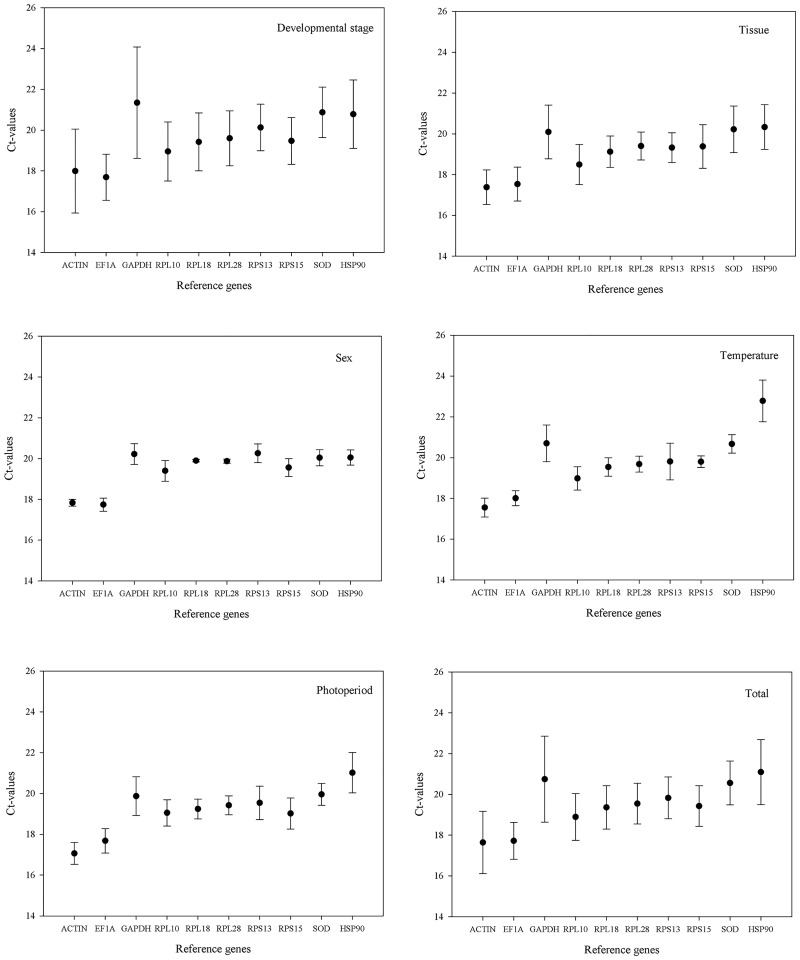
Mean Ct values of ten candidate genes of developmental stage, tissue, sex, temperature, photoperiod and total ranged from 17.692 (EF1A) to 21.355 (GAPDH); 17.384 (ACTIN) to 20.325 (HSP90); 17.744 (EF1A) to 20.258 (RPS13); 17.554 (ACTIN) to 22.783 (HSP90); 17.056 (ACTIN) to 21.017 (HSP90); 17.640 (ACTIN) to 21.094 (HSP90). The Ct values of all the tested samples were between 17.056 to 22.783. Among the candidate genes, ACTIN and EF1A had the highest accumulation of the transcript among different conditions, while GAPDH and HSP90 had the lowest accumulation of transcript levels, and the remaining genes had intermediate transcript expression levels (Fig 1).
Fig 1. Expression profiles of the nine housekeeping genes and the target gene HSP90 in H. axyridis under different factors.
Expression level is indicated by cycle threshold (Ct) value. Samples included different development stages, sexes, tissues, photoperiods, and temperatures. Values are means±SD.
Stability of reference genes across biotic conditions
Developmental stage
The most stable genes based on developmental stage were RPL18, RPL28, and RPL10 according to geNorm. EF1A, RPS13, and RPS15 were the most stable according to Normfinder, BestKeeper, and the △Ct method (Table 2).
Table 2. Stability of reference gene expression across biotic conditions.
| Biotic conditions | Reference genes | geNorm | Normfinder | BestKeeper | △Ct | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Developmental Stage | ACTIN | 1.309 | 7 | 2.340 | 8 | 1.339 | 7 | 2.577 | 8 |
| EF1A | 0.710 | 5 | 0.099 | 1 | 0.910 | 1 | 1.209 | 1 | |
| GAPDH | 1.619 | 8 | 2.506 | 9 | 1.981 | 8 | 2.705 | 9 | |
| RPL10 | 0.503 | 2 | 0.806 | 6 | 1.160 | 6 | 1.333 | 6 | |
| RPL18 | 0.390 | 1 | 0.791 | 5 | 1.160 | 6 | 1.287 | 4 | |
| RPL28 | 0.390 | 1 | 0.784 | 4 | 1.083 | 5 | 1.291 | 5 | |
| RPS13 | 0.681 | 4 | 0.571 | 2 | 0.953 | 2 | 1.268 | 2 | |
| RPS15 | 0.634 | 3 | 0.622 | 3 | 0.999 | 3 | 1.275 | 3 | |
| SOD | 0.918 | 6 | 1.010 | 7 | 1.020 | 4 | 1.632 | 7 | |
| Tissue | ACTIN | 0.624 | 6 | 0.587 | 7 | 0.745 | 6 | 0.858 | 7 |
| EF1A | 0.513 | 3 | 0.499 | 4 | 0.666 | 4 | 0.778 | 5 | |
| GAPDH | 0.851 | 8 | 1.223 | 9 | 1.109 | 9 | 1.325 | 9 | |
| RPL10 | 0.533 | 4 | 0.507 | 5 | 0.730 | 5 | 0.768 | 4 | |
| RPL18 | 0.343 | 1 | 0.415 | 3 | 0.558 | 1 | 0.713 | 3 | |
| RPL28 | 0.471 | 2 | 0.316 | 1 | 0.582 | 2 | 0.711 | 2 | |
| RPS13 | 0.343 | 1 | 0.404 | 2 | 0.620 | 3 | 0.704 | 1 | |
| RPS15 | 0.557 | 5 | 0.547 | 6 | 0.808 | 7 | 0.798 | 6 | |
| SOD | 0.715 | 7 | 0.790 | 8 | 0.848 | 8 | 1.001 | 8 | |
| Sex | ACTIN | 0.159 | 2 | 0.221 | 3 | 0.127 | 3 | 0.399 | 3 |
| EF1A | 0.251 | 3 | 0.250 | 4 | 0.246 | 4 | 0.427 | 4 | |
| GAPDH | 0.429 | 7 | 0.484 | 8 | 0.455 | 8 | 0.568 | 7 | |
| RPL10 | 0.395 | 6 | 0.428 | 7 | 0.493 | 9 | 0.532 | 6 | |
| RPL18 | 0.103 | 1 | 0.117 | 1 | 0.043 | 1 | 0.357 | 1 | |
| RPL28 | 0.103 | 1 | 0.181 | 2 | 0.071 | 2 | 0.377 | 2 | |
| RPS13 | 0.467 | 8 | 0.526 | 9 | 0.420 | 7 | 0.600 | 8 | |
| RPS15 | 0.320 | 4 | 0.334 | 6 | 0.402 | 6 | 0.471 | 5 | |
| SOD | 0.362 | 5 | 0.331 | 5 | 0.279 | 5 | 0.471 | 5 | |
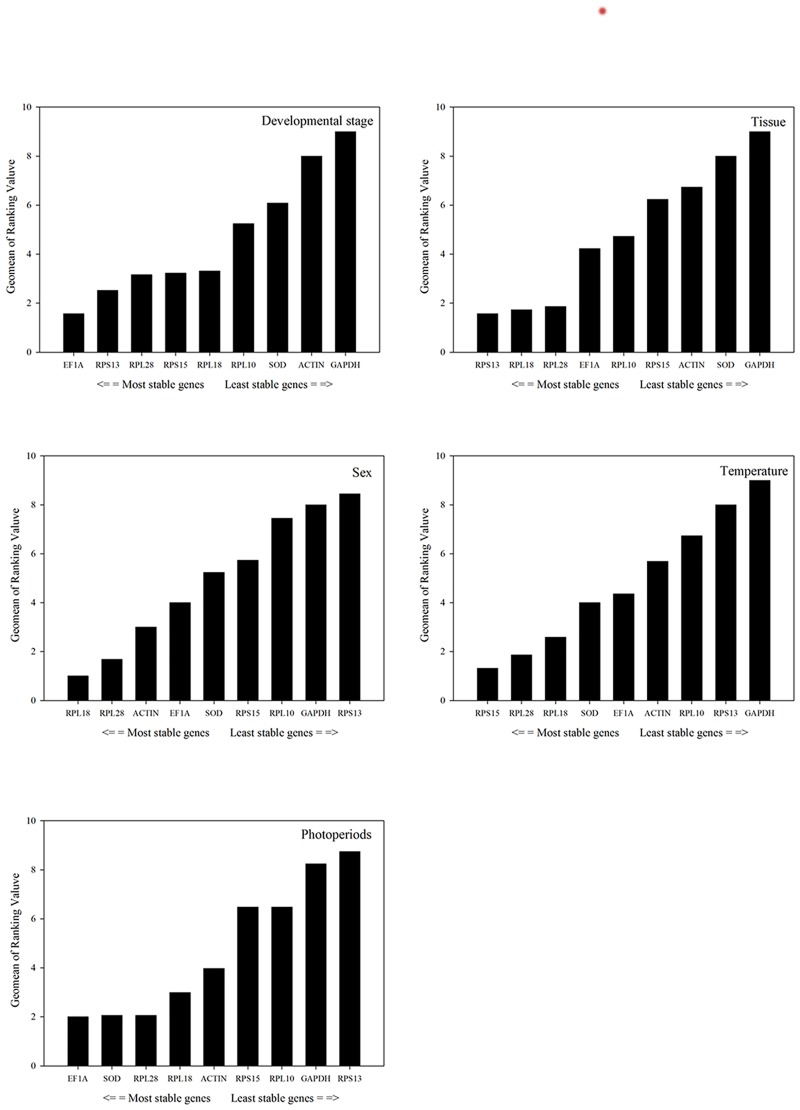
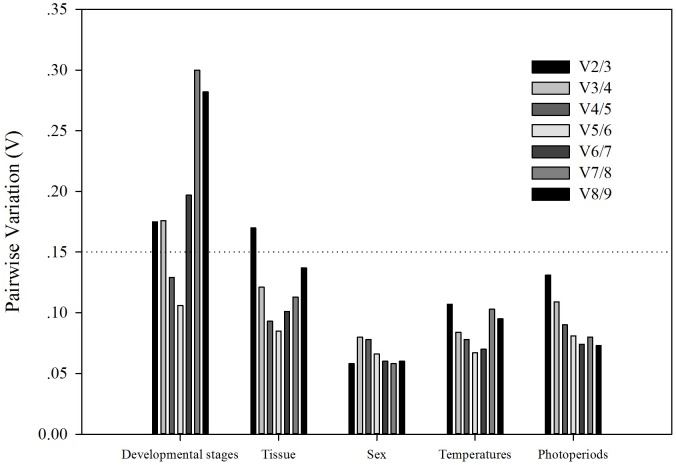
Based on RefFinder, the ranking of reference genes from the most to the least stable was: EF1A, RPS13, RPL28, RPS15, RPL18, RPL10, SOD, ACTIN, and GAPDH (Fig 2). From GeNorm analysis, the pair-wise values of V2/3 and V3/4 were both above the cut-off value of 0.15 while the pair-wise value of V4/5 was <0.15 (Fig 3). A value <0.15 indicates that supplemental reference genes are unnecessary. Based on RefFinder and convenient operation, EF1A, RPS13, and RPL28 were determined to be the best reference genes across different developmental stages (Table 3).
Fig 2. The stability of the expression levels of nine housekeeping genes across different treatments based on RefFinder stability value.
A lower Geomean value indicates more stable expression.
Fig 3. Pairwise variation (Vn/Vn+1) analysis of the candidate reference genes.
The pairwise variation was analyzed by GeNorm software. A value <0.15 indicates that normalization would not be dramatically changed by the inclusion of an additional reference gene.
Table 3. Stability of reference gene expression across abiotic conditions.
| Abiotic conditions | Reference genes | geNorm | Normfinder | BestKeeper | △Ct | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | ||
| Temperature | ACTIN | 0.392 | 4 | 0.416 | 6 | 0.418 | 7 | 0.605 | 5 |
| EF1A | 0.42 | 5 | 0.415 | 5 | 0.265 | 2 | 0.616 | 6 | |
| GAPDH | 0.646 | 8 | 0.840 | 9 | 0.737 | 9 | 0.934 | 9 | |
| RPL10 | 0.460 | 6 | 0.439 | 7 | 0.412 | 6 | 0.638 | 7 | |
| RPL18 | 0.312 | 1 | 0.278 | 3 | 0.407 | 5 | 0.533 | 3 | |
| RPL28 | 0.312 | 1 | 0.247 | 2 | 0.318 | 3 | 0.519 | 2 | |
| RPS13 | 0.563 | 7 | 0.799 | 8 | 0.606 | 8 | 0.903 | 8 | |
| RPS15 | 0.340 | 2 | 0.177 | 1 | 0.180 | 1 | 0.506 | 1 | |
| SOD | 0.361 | 3 | 0.302 | 4 | 0.362 | 4 | 0.557 | 4 | |
| Photoperiod | ACTIN | 0.495 | 4 | 0.415 | 5 | 0.422 | 2 | 0.609 | 5 |
| EF1A | 0.472 | 3 | 0.258 | 1 | 0.429 | 3 | 0.535 | 1 | |
| GAPDH | 0.595 | 7 | 0.629 | 8 | 0.725 | 8 | 0.755 | 8 | |
| RPL10 | 0.523 | 5 | 0.533 | 7 | 0.503 | 5 | 0.679 | 7 | |
| RPL18 | 0.426 | 1 | 0.361 | 4 | 0.450 | 4 | 0.580 | 4 | |
| RPL28 | 0.426 | 1 | 0.354 | 3 | 0.429 | 3 | 0.572 | 2 | |
| RPS13 | 0.631 | 8 | 0.636 | 9 | 0.627 | 7 | 0.759 | 9 | |
| RPS15 | 0.549 | 6 | 0.420 | 6 | 0.577 | 6 | 0.619 | 6 | |
| SOD | 0.439 | 2 | 0347 | 2 | 0.404 | 1 | 0.574 | 3 | |
Tissue
Across tissue samples, RPL18, RPS13, and RPL28 were the most stable reference genes based on geNorm, Normfinder, BestKeeper, and the △Ct method (Table 2), and the ranking of reference gene stability across different tissues based on RefFinder was: RPS13, RPL18, RPL28, EF1A, RPL10, RPS15, ACTIN, SOD, and GAPDH (Fig 2). From geNorm analysis, the pair-wise value of V3/4 was <0.15 (Fig 3). Thus, RPS13, RPL18, and RPL28 were considered the most suitable reference genes for comparisons across different tissues (Table 4).
Table 4. Recommended H. axyridis reference genes for various experimental conditions.
| Experimental Conditions | Reference Genes | ||
|---|---|---|---|
| Developmental stage | EF1A | RPS13 | RPL28 |
| Tissue | RPS13 | RPL18 | RPL28 |
| Sex | RPL18 | RPL28 | |
| Photoperiod | EF1A | SOD | |
| Temperature | RPS15 | RPL28 | |
Sex
According to geNorm, RPL18, RPL28, and EF1A were the most stable reference genes based on sex. Normfinder, BestKeeper and the △Ct method all identified RPL18, RPL28, and ACTIN as the most stable genes across sexes (Table 2).
According to RefFinder, the ranking of reference gene stability based on sex was: RPL18, RPL28, ACTIN, EF1A, SOD, RPS15, RPL10, GAPDH, and RPS13 (Fig 2). The pair-wise value of V2/3 was <0.15 based on geNorm data (Fig 3). Thus, RPL18 and RPL28 were considered the most stable reference genes across sexes (Table 4)
Stability of reference genes under abiotic conditions
Temperature
Based on geNorm, Normfinder, and the △Ct method, RPL18, RPL28, and RPS15 were the most stable reference genes across temperature treatments. According to BestKeeper, RPS15, EF1A, and RPL28 were the most stable (Table 3).
According to RefFinder, the ranking of reference genes from the most to least stable was: RPS15, RPL28, RPL18, SOD, EF1A, ACTIN, RPL10, RPS13, and GAPDH (Fig 2). The value of V2/3 from geNorm analysis was <0.15 (Fig 3). Therefore, RPS15 and RPL28 were considered the most stable reference genes across temperature treatments (Table 4).
Photoperiod
EF1A, SOD, and RPL28 were the most stable reference genes across photoperiod treatments according to Normfinder and the △Ct method; RPL18, RPL28, and SOD were the most stable according to geNorm; and SOD, ACTIN, EF1A, and RPL28 were the most stable according to BestKeeper (Table 3).
According to RefFinder, the ranking of reference genes across photoperiod treatments from the most to least stable was: EF1A, SOD, RPL28, RPL18, ACTIN, RPS15, RPL10, GAPDH, and RPS13 (Fig 2). The value of V2/3 from geNorm analysis was <0.15 (Fig 3). Therefore, EF1A and SOD were considered the most stable reference genes across photoperiod treatments (Table 4)
Target gene expression
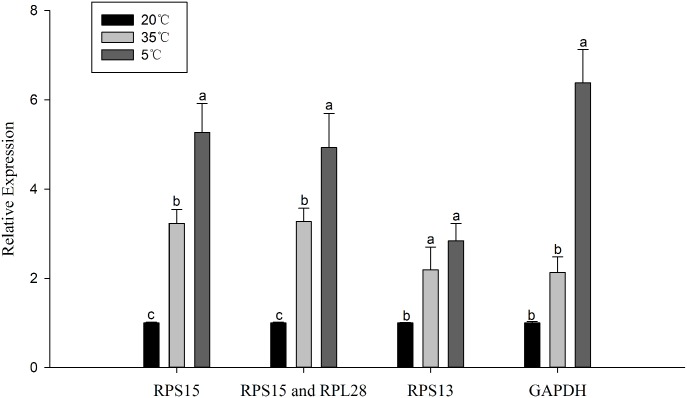
To test the effect of reference genes on the calculation of target gene expression, the relative expression level of the target gene HSP90 under different temperature treatments (5, 20, 35°C) was normalized to the most and least stable reference genes. The relative expression level of HSP90 significantly differed between temperature treatments. When the most stable reference gene, RPS15, was used, HSP90 was significantly up-regulated under low temperature (5°C) and high temperature (35°C), and the relative expression at 5°C was significantly higher than at 35°C (Fig 4). Similar results were obtained using RPS13 and RPL28. However, the expression level of HSP90 was not significantly different between the 5°C and 35°C samples when using an unstable reference gene (such as RPS13). Similarly, the expression of HSP90 was not significantly different between 35°C and 20°C samples when the least stable reference gene, GADPH, was used (Fig 4).
Fig 4. The use of different reference genes to normalize the expression level of the target gene HSP90 under three temperature treatments was investigated.
The expression level was normalized by different candidate reference genes: RPS15; RPS15 and RPL28; RPS13; and GAPDH. The reference genes were selected based on expression stability across three temperature treatments. Data represent the means±SEM of three biological replicates. The comparisons were analyzed using one-way ANOVA, and different letters (a,b) denote significant differences between normalization strategies determined by Tukey test (P < 0.05).
Discussion
In the current study, the stabilities of nine reference genes were evaluated across different biotic and abiotic conditions. We found that the best reference genes varied among conditions. The ribosomal proteins typically exhibited the highest stability across different experimental conditions. Across tissues, temperatures, and sexes, RPS13, RPS15, and RPL18, respectively, were the most stable reference genes.
Consistent with our findings, ribosomal proteins have been previously reported to have the highest expression stability in several insects. For instance, in Bradysia odoriphaga, expression stability of RPS15 across temperature treatments was highest [31], and in Lucilia sericata, RPS3 and RPLP0 showed high stability across specific larval tissues [37]. in Tetranychus cinnabarinus, the highest expression stability across tissues was observed for RPS18 [38]. But expression stability was highest for RPS11 in Nilaparvata lugens [39], and for RPS13 in Sesamia inferens [40]. Our results and the results of previous studies indicate that in general ribosomal proteins are good reference genes for gene expression studies. However, an exception in our study was RPS13, which showed the least stable expression across sex and photoperiod treatment. RPS13 was also found to be the least stable reference gene across photoperiod treatments in B. odoriphaga [31].
In our study, among the nine reference genes, the expression level of EF1A was the most stable across developmental stages and photoperiod treatments. This result is consistent with previous studies concerning developmental stage in Frankliniella occidentalis [41] and developmental stage and photoperiod treatment in Danaus plexippus (L.) [42].
In our study, SOD ranked among the top three reference genes for photoperiod treatments (Table 4). In Spodoptera exigua, the expression stability of SOD was high across developmental stages and between sexest but was low across certain tissues [15].
GAPDH and ACTIN have been commonly used as internal controls in many gene expression studies [11, 43, 44]. In our study, however, ACTIN ranked low for most factors except for sex, where it was the third most stable gene. Shi et al. (2016) [31] also found that ACTIN was a good reference gene in B. odoriphaga for diet treatments. In Helicoverpa armigera, however, ACTIN was the least stable reference gene across temperature and photoperiod treatments [45], and in Tribolium castaneum exposed to Beauveria bassiana, ACTIN was not stably expressed [46]. Based on our results, GAPDH is also not an ideal reference gene in H. axyridis. Several studies in other insects have also demonstrated that the expression stability of GAPDH is low in some circumstances, such as across developmental stages in Tetranychus cinnabarinus [19]; between the labial gland and fat body in Bombus terrestris and Bombus lucorum [47]; and across different body parts in Sogatella furcifera [48]. Taken together, these results suggest that the stability of reference genes may be dependent on the insect species and other characteristics such as instar stage or tissue type.
The ranking of reference genes is not only affected by different experimental conditions or factors, but also by the tools used for ranking. In this study, for example, the most stable reference genes under different temperature treatments were RPS15, RPL28, and RPL18 using NormFinder and the △Ct method. BestKeeper also ranked RPS15 and RPS28 as the most stable, but EF1A was ranked the second most stable. The differences in ranking may result from differences in statistical algorithms. NormFinder and the △Ct method mainly analyze the pairwise variation between reference genes, and then confirm the stability of one gene in each pair. BestKeeper individually analyzes the stability among reference genes [33, 49]. RefFinder, a comprehensive evaluation platform that integrates the four algorithms we tested, was used to estimate the stability rankings of the nine reference genes. We also used GeNorm, which calculates the pairwise variation (Vn/Vn + 1) between the continuous standardization factors or NF (NFn and NFn + 1), to determine the optimal number of reference genes [8]. Based on GeNorm analysis, two reference genes were found to be sufficient for normalizing target gene expression values for sex, photoperiod, and temperature, but three reference genes were needed to normalize across developmental stages and different tissues [Fig 3]. These results suggest that it is necessary to use different combinations of reference genes to study changes in gene expression in H. axyridis in response to different factors.
In recent years, several reference genes have been used as internal controls for studying gene expression in H. axyridis under diverse experimental conditions. In this study, we found that nine candidate reference genes have different strengths and weaknesses in various conditions. For example, we recommend RPS15 and RPL28 to study gene expression in response to temperature in H. axyridis to temperature. Previously, Wang et al. [50] selected RP49 as a reference gene to study the expression of six small heat shock proteins mediating cold-hardiness in H. axyridis. Moreover, RP49 was also used as internal control to study the expression of genes in the H. axyridis trehalase and glycogen metabolic pathways [51, 52]. Vilcinskas et al. [53] also used RPS3, another member of ribosomal protein family, as a reference gene in a study of genes encoding antimicrobial peptides and proteins, while study of immunity-related genes was exclusive of our setting conditions. In addition, Tang et al. [54] selected another 18s RNA to verify the expression of cold-resistance response genes, such as E3 ubiquitin-protein ligase, transketolase, trehalase, serine/arginine repetitive matrix protein 2, glycerol kinase and sugar transporter SWEET1-like. One of the most frequently used reference genes, ACTIN, was used as the internal control to study development-related genes in the ovaries of adult H. axyridis [55], and our study showed that this gene may not be as good as other reference genes for some factors such as developmental stage, tissue, temperature, and photoperiods.
Taken together our results indicate that commonly used reference genes are often not well-suited for normalization in all qRT-PCR experiments, and the simultaneous measurement of a panel of candidate reference genes is critical for the accuracy of qRT-PCR quantification. The results of our study represent an important step for establishing a standardized gene analysis framework for H. axyridis.
Supporting information
(TIF)
(TIF)
Acknowledgments
We greatly appreciate Dr. Xuguo Zhou (Associate professor, Department of Entomology, University of Kentucky, Lexington, USA) for the language editing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Key Research and Developing Plan (2017YFD0200400) and the China Agriculture Research System (CARS-24-C-03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Roy HE, Brown PMJ, Adriaens T, Berkvens N, Borges I, Clusella-Trullas S, et al. The harlequin ladybird, Harmonia axyridis: global perspectives on invasion history and ecology. Biological Invasions. 2016;18(4):997–1044. doi: 10.1007/s10530-016-1077-6 [Google Scholar]
- 2.Wang S, Tan XL, Michaud JP, Shi ZK, Zhang F. Sexual selection drives the evolution of limb regeneration in Harmonia axyridis (Coleoptera: Coccinellidae). Bulletin of Entomological Research. 2015;105(2):245–252. doi: 10.1017/S0007485315000036 [DOI] [PubMed] [Google Scholar]
- 3.Qin QJ, Liu S, Li S, Zhang W, He YZ. Role of photoperiod in the development and reproduction of Harmonia axyridis (Coleoptera, Coccinellidae). Biocontrol Science and Technology. 2016;26(1):116–124. doi: 10.1080/09583157.2015.1078873 [Google Scholar]
- 4.Durieux D, Fassotte B, Deneubourg JL, Brostaux Y, Vandereycken A, Joie E, et al. Aggregation behavior of Harmonia axyridis under non-wintering conditions. Insect Science. 2015;22(5):670–678. doi: 10.1111/1744-7917.12144 [DOI] [PubMed] [Google Scholar]
- 5.Reznik SY, Dolgovskaya MY, Ovchinnikov AN, Belyakova NA. Weak photoperiodic response facilitates the biological invasion of the harlequin ladybird Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae). Journal of Applied Entomology. 2015;139(4):241–249. doi: 10.1111/jen.12158 [Google Scholar]
- 6.Shen QD, Zhao LN, Xie GQ, Wei P, Yang MM, Wang SG, et al. Cloning Three Harmonia axyridis (Coleoptera: Coccinellidae) Heat Shock Protein 70 Family Genes: Regulatory Function Related to Heat and Starvation Stress. Journal of Entomological Science. 2015;50(3):168–185. [Google Scholar]
- 7.Haelewaters D, Zhao SY, Clusella-Trullas S, Cottrell TE, De Kesel A, Fiedler L, et al. Parasites of Harmonia axyridis: current research and perspectives. Biocontrol. 2017;62(3):355–371. doi: 10.1007/s10526-016-9766-8 [Google Scholar]
- 8.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7). doi: 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strube C, Buschbaum S, Wolken S, Schnieder T. Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide isomerase transcription pattern in the bovine lungworm Dictyocaulus viviparus. Gene. 2008;425(1–2):36–43. doi: 10.1016/j.gene.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 10.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR—a perspective. Journal of Molecular Endocrinology. 2005;34(3):597–601. doi: 10.1677/jme.1.01755 [DOI] [PubMed] [Google Scholar]
- 11.Van Hiel MB, Van Wielendaele P, Temmerman L, Van Soest S, Vuerinckx K, Huybrechts R, et al. Identification and validation of housekeeping genes in brains of the desert locust Schistocerca gregaria under different developmental conditions. Bmc Molecular Biology. 2009;10 doi: 10.1186/1471-2199-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of Aphid Genes by dsRNA Feeding from Plants. Plos One. 2011;6(10). doi: 10.1371/journal.pone.0025709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva AX, Jander G, Samaniego H, Ramsey JS, Figueroa CC. Insecticide Resistance Mechanisms in the Green Peach Aphid Myzus persicae (Hemiptera: Aphididae) I: A Transcriptomic Survey. Plos One. 2012;7(6). doi: 10.1371/journal.pone.0036366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu W, Xie W, Zhang Z, Wang SL, Wu QJ, Liu Y, et al. Exploring Valid Reference Genes for Quantitative Real-time PCR Analysis in Plutella xylostella (Lepidoptera: Plutellidae). International Journal of Biological Sciences. 2013;9(8):792–802. doi: 10.7150/ijbs.5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Yuan M, Shakeel M, Zhang YJ, Wang SL, Wang X, et al. Selection and Evaluation of Reference Genes for Expression Analysis Using qRT-PCR in the Beet Armyworm Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae). Plos One. 2014;9(1). doi: 10.1371/journal.pone.0084730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladror DT, Frey BL, Scalf M, Levenstein ME, Artymiuk JM, Smith LM. Methylation of yeast ribosomal protein S2 is elevated during stationary phase growth conditions. Biochemical and Biophysical Research Communications. 2014;445(3):535–541. doi: 10.1016/j.bbrc.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponton F, Chapuis M-P, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. Journal of Insect Physiology. 2011;57(6):840–850. doi: 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 18.Parker JD, Parker KM, Keller L. Molecular phylogenetic evidence for an extracellular CuZn superoxide dismutase gene in insects. Insect Molecular Biology. 2004;13(6):587–594. doi: 10.1111/j.0962-1075.2004.00515.x [DOI] [PubMed] [Google Scholar]
- 19.Sun W, Jin Y, He L, Lu WC, Li M. Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. Journal of Insect Science. 2010;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee PD, Sladek R, Greenwood CMT, Hudson TJ. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Research. 2002;12(2):292–297. doi: 10.1101/gr.217802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang CX, Pan HP, Liu Y, Zhou XG. Temperature and Development Impacts on Housekeeping Gene Expression in Cowpea Aphid, Aphis craccivora (Hemiptera: Aphidiae). Plos One. 2015;10(6). doi: 10.1371/journal.pone.0130593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang CX, Pan HP, Liu Y, Zhou XG. Stably Expressed Housekeeping Genes across Developmental Stages in the Two-Spotted Spider Mite, Tetranychus urticae. Plos One. 2015;10(3). doi: 10.1371/journal.pone.0120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu YH, Yuan M, Gao X, Kang TH, Zhan S, Wan H, et al. Identification and Validation of Reference Genes for Gene Expression Analysis Using Quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). Plos One. 2013;8(7). doi: 10.1371/journal.pone.0068059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang CX, Preisser EL, Zhang HJ, Liu Y, Dai LY, Pan HP, et al. Selection of Reference Genes for RT-qPCR Analysis in Coccinella septempunctata to Assess Un-intended Effects of RNAi Transgenic Plants (vol 7, 1672, 2016). Frontiers in Plant Science. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CX, Pan HP, Noland JE, Zhang DY, Zhang ZH, Liu Y, et al. Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Scientific Reports. 2015;5 doi: 10.1038/srep18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan HP, Yang XW, Siegfried BD, Zhou XG. A Comprehensive Selection of Reference Genes for RT-qPCR Analysis in a Predatory Lady Beetle, Hippodamia convergens (Coleoptera: Coccinellidae). Plos One. 2015;10(4). doi: 10.1371/journal.pone.0125868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13(2):127–142. doi: 10.1007/s12192-008-0023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colinet H, Lee SF, Hoffmann A. Temporal expression of heat shock genes during cold stress and recovery from chill coma in adult Drosophila melanogaster. Febs Journal. 2010;277(1):174–185. doi: 10.1111/j.1742-4658.2009.07470.x [DOI] [PubMed] [Google Scholar]
- 29.Meijering RAM, Zhang D, Hoogstra-Berends F, Henning RH, Brundel BJJM. Loss of proteostatic control as a substrate for atrial fibrillation: a novel target for upstream therapy by heat shock proteins. Frontiers in Physiology. 2012;3 doi: 10.3389/fphys.2012.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Research. 2005;33:W577–W581. doi: 10.1093/nar/gki591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi CH, Yang FS, Zhu X, Du EX, Yang YT, Wang SL, et al. Evaluation of Housekeeping Genes for Quantitative Real-Time PCR Analysis of Bradysia odoriphaga (Diptera: Sciaridae). International Journal of Molecular Sciences. 2016;17(7). doi: 10.3390/ijms17071034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2004;313(4):856–862. doi: 10.1016/j.bbrc.2003.11.177 [DOI] [PubMed] [Google Scholar]
- 33.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. Bmc Molecular Biology. 2006;7 doi: 10.1186/1471-2199-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- 35.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnology Letters. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 36.Zhu X, Li YL, Chen DX, Wu P, Yi T, Chen T, et al. Selection of Reference Genes for MicroRNA Quantitative Expression Analysis in Chinese Perch, Siniperca chuatsi. International Journal of Molecular Sciences. 2015;16(4):8310–8323. doi: 10.3390/ijms16048310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann A, Lehmann R, Beckert A, Vilcinskas A, Franta Z. Selection and Evaluation of Tissue Specific Reference Genes in Lucilia sericata during an Immune Challenge. Plos One. 2015;10(8). doi: 10.1371/journal.pone.0135093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun W, Jin Y, He L, Lu WC, Li M. Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. Journal of Insect Science. 2010;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan M, Lu YH, Zhu X, Wan H, Shakeel M, Zhan S, et al. Selection and Evaluation of Potential Reference Genes for Gene Expression Analysis in the Brown Planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) Using Reverse-Transcription Quantitative PCR. Plos One. 2014;9(1). doi: 10.1371/journal.pone.0086503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun M, Lu MX, Tang XT, Du YZ. Exploring Valid Reference Genes for Quantitative Real-Time PCR Analysis in Sesamia inferens (Lepidoptera: Noctuidae). Plos One. 2015;10(1). doi: 10.1371/journal.pone.0115979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng YT, Li HB, Lu MX, Du YZ. Evaluation and Validation of Reference Genes for qRT-PCR Normalization in Frankliniella occidentalis (Thysanoptera: Thripidae). Plos One. 2014;9(10). doi: 10.1371/journal.pone.0111369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan HP, Yang XW, Bidne K, Hellmich RL, Siegfried BD, Zhou XG. Selection of Reference Genes for RT-qPCR Analysis in the Monarch Butterfly, Danaus plexippus (L.), a Migrating Bio-Indicator. Plos One. 2015;10(6). doi: 10.1371/journal.pone.0129482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frost P, Nilsen F. Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Veterinary Parasitology. 2003;118(1–2):169–74. doi: 10.1016/j.vetpar.2003.09.020 [DOI] [PubMed] [Google Scholar]
- 44.de Boer ME, de Boer TE, Marien J, Timmermans MJTN, Nota B, van Straalen NM, et al. Reference genes for QRT-PCR tested under various stress conditions in Folsomia candida and Orchesella cincta (Insecta, Collembola). Bmc Molecular Biology. 2009;10 doi: 10.1186/1471-2199-10-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shakeel M, Zhu X, Kang TH, Wan H, Li JH. Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). Journal of Asia-Pacific Entomology. 2015;18(2):123–130. doi: 10.1016/j.aspen.2015.01.001 [Google Scholar]
- 46.Lord JC. Beauveria bassiana infection of eggs of stored-product beetles. Entomological Research. 2009;39(2):155–157. [Google Scholar]
- 47.Hornakova D, Matouskova P, Kindl J, Valterova I, Pichova I. Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Analytical Biochemistry. 2010;397(1):118–120. doi: 10.1016/j.ab.2009.09.019 [DOI] [PubMed] [Google Scholar]
- 48.An XK, Hou ML, Liu YD. Reference Gene Selection and Evaluation for Gene Expression Studies Using qRT-PCR in the White-Backed Planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Journal of Economic Entomology. 2016;109(2):879–886. doi: 10.1093/jee/tov333 [DOI] [PubMed] [Google Scholar]
- 49.Teng XL, Zhang Z, He GL, Yang LW, Li F. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in four lepidopteran insects. Journal of Insect Science. 2012;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang HJ, Shi ZK, Shen QD, Xu CD, Wang B, Meng ZJ, et al. Molecular Cloning and Induced Expression of Six Small Heat Shock Proteins Mediating Cold-Hardiness in Harmonia axyridis (Coleoptera: Coccinellidae). Frontiers in Physiology. 2017;8 doi: 10.3389/fphys.2017.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi ZK, Wang S, Wang SG, Zhang L, Xu YX, Guo XJ, et al. Effects of starvation on the carbohydrate metabolism in Harmonia axyridis (Pallas). Biology Open. 2017;6(7):1096–103. doi: 10.1242/bio.025189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi ZK, Liu XJ, Xu QY, Qin Z, Wang S, Zhang F, et al. Two novel soluble trehalase genes cloned from Harmonia axyridis and regulation of the enzyme in a rapid changing temperature. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 2016;198:10–8. doi: 10.1016/j.cbpb.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 53.Vilcinskas A, Mukherjee K, Vogel H. Expansion of the antimicrobial peptide repertoire in the invasive ladybird Harmonia axyridis. Proceedings of the Royal Society B-Biological Sciences. 2013;280(1750). doi: 10.1098/rspb.2012.2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang B, Liu XJ, Shi ZK, Shen QD, Xu YX, Wang S, et al. Transcriptome analysis and identification of induced genes in the response of Harmonia axyridis to cold hardiness. Comparative Biochemistry and Physiology D-Genomics & Proteomics. 2017;22:78–89. doi: 10.1016/j.cbd.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 55.Du WX, Zeng FR. Identification of Development-Related Genes in the Ovaries of Adult Harmonia axyridis (Pallas) Lady Beetles Using a Time-Series Analysis by RNA-seq. Scientific Reports. 2016;6 doi: 10.1038/srep39109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.