Summary
To determine the role for mutations of MECP2 in Rett syndrome, we generated isogenic lines of human induced pluripotent stem cells, neural progenitor cells, and neurons from patient fibroblasts with and without MECP2 expression in an attempt to recapitulate disease phenotypes in vitro. Molecular profiling uncovered neuronal-specific gene expression changes, including induction of a senescence-associated secretory phenotype (SASP) program. Patient-derived neurons made without MECP2 showed signs of stress, including induction of P53, and senescence. The induction of P53 appeared to affect dendritic branching in Rett neurons, as P53 inhibition restored dendritic complexity. The induction of P53 targets was also detectable in analyses of human Rett patient brain, suggesting that this disease-in-a-dish model can provide relevant insights into the human disorder.
Keywords: MECP2, Rett syndrome, disease in a dish, senescence
Highlights
-
•
Development of a patient-specific model of human Rett syndrome
-
•
Loss of function of MECP2 leads to induction of p53
-
•
MECP2 null neurons show evidence of cellular senescence
-
•
Inhibition of p53 can restore dendritic branching in MECP2 null neurons
In this report, Lowry and colleagues found that loss of MECP2 has a more profound effect as pluripotent stem cells are terminally differentiated toward neurons. The loss of MECP2 leads to induction of P53 protein and subsequent senescence pathways including an SASP gene program, which appears to be a cause of diminished dendritic branching in Rett neurons.
Introduction
Rett syndrome is a disease associated with loss of function mutations in the gene MECP2, which was originally identified as encoding a methylated DNA binding protein (Chen et al., 2001, Meehan et al., 1992). Patient symptoms include microcephaly, intellectual disability, facial dysmorphia, and seizure activity (Bird, 2008). Studies in murine models recapitulate many of the patient phenotypes and have recently identified a role for Mecp2 particularly in inhibitory neurons (Tomassy et al., 2014). These studies demonstrated that loss of MECP2 can lead to defects in transcription (Chen et al., 2003, Lee et al., 2014), dendritic branching (Zhou et al., 2006), nuclear size (Chen et al., 2001), and AKT signaling (Li et al., 2013).
MECP2 has also been described as a transcription factor with specific targets (Chen et al., 2003, Zhou et al., 2006), and more broadly as either a transcriptional activator (Li et al., 2013) or repressor (Cross et al., 1997, Nan et al., 1997). However, despite decades of research on MECP2, it is still unclear how mutations in this protein lead to patient symptoms (Chen et al., 2001, Marchetto et al., 2010). To confirm findings made in other models and further study these in a human system, some have turned to modeling Rett syndrome in vitro by taking advantage of disease-in-a-dish approaches. This involves making human induced pluripotent stem cells (hiPSCs) from patient somatic cells, or using genome engineering to introduce mutations into wild-type (WT) human pluripotent stem cells. In the current study, we also sought to mitigate the effect of genetic background and variability of differentiation by taking advantage of several isogenic lines of hiPSCs that either express the WT allele or the mutant allele leading to cells that express or lack MECP2 (Tchieu et al., 2010). This allowed for detailed molecular analyses of hiPSCs, neural progenitor cells (NPCs), and neurons with and without MECP2 under the same genetic background. In comparing neurons from Rett patients as well as those with MECP2 silenced by small interfering RNA (siRNA), it is clear that loss of MECP2 leads to induction of P53 and senescence, potentially opening an avenue of investigation for this intellectual disability syndrome.
Results
A Human Model of Rett Syndrome In Vitro
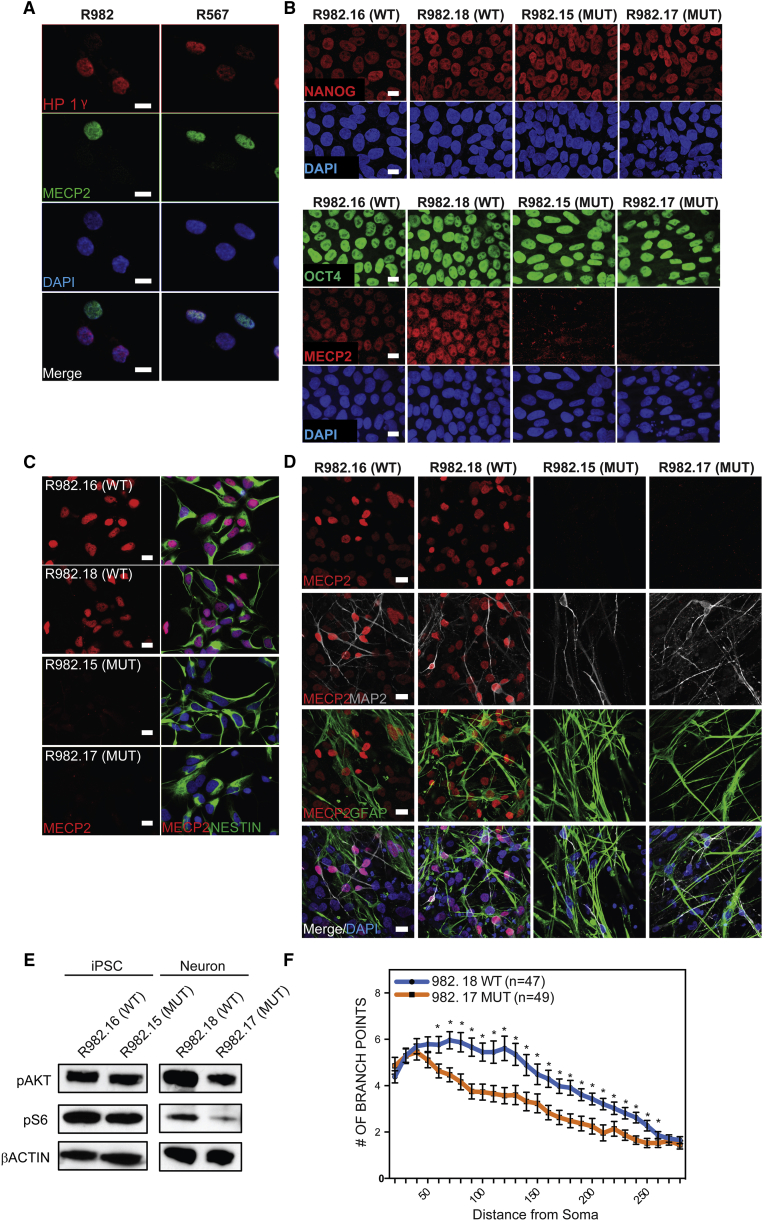
Cognizant of the fact that differentiation from hPSCs is highly variable across individual lines, culture conditions, and time, we developed an isogenic model to study Rett syndrome in vitro to remove the confound of genetic background (Tchieu et al., 2010). Because female patients with Rett syndrome are usually heterozygous for mutant alleles of MECP2, fibroblasts isolated from these patients display a mosaic pattern where roughly half the cells express either the mutant or WT allele. This is shown in Figure 1A, where fibroblasts isolated from two patients with distinct mutant alleles of MECP2 (R982 and R567) showed that roughly half the cells expressed MECP2 while the other half lacked detectable amounts of this protein. Patient descriptions are provided in Figure S1. Reprogramming to iPSCs using a small set of transcription factors has been shown to happen at the clonal level, such that individual reprogramming events in single fibroblasts generate isolated hiPSC clones (Winkler et al., 2010). Therefore, reprogramming of mosaic fibroblast cultures from two different patients generated single hiPSC clones that either expressed MECP2 protein or lacked it (Figure 1B) (method described in a previous study; Sahakyan et al., 2016). In addition, our work and that of others has shown that under standard conditions, the inactive X chromosome in human fibroblasts does not reactivate upon reprogramming to the pluripotent state (Tchieu et al., 2010), which is distinct from murine reprogramming (Maherali et al., 2007).
Figure 1.
Generation of the Isogenic Model of Rett Syndrome In Vitro
(A) Fibroblasts isolated from Rett syndrome patients (R982 and R567) heterozygous for MECP2 mutations exhibit a mosaic pattern of MECP2 expression due to random XCI.
(B) Multiple isogenic hiPSC lines were produced from patient 982 with a typical Yamanaka protocol yielding individual isogenic clones with and without MECP2 expression from the same patient, as judged by NANOG and OCT4 staining.
(C) Specification of hiPSCs derived from patient 982 toward neural progenitor cells yielded homogeneous cultures of NPCs with and without MECP2.
(D) Terminal differentiation of NPCs derived from patient 982 toward neurons and glia by growth factor withdrawal as measured by immunostaining for MAP2 and GFAP.
(E) MECP2+ and MECP2− hiPSCs and neurons were assayed by western blot with antibodies that recognize the active forms of Akt and its downstream target S6.
(F) Sholl assay of dendritic complexity was performed on WT versus MUT neurons derived from patient 982.
∗p < 0.05 according to Student's t test. Bar graphs represent means ± SEM. Scale bars on images indicate 10 μm.
Thus, we were able to create multiple lines of hiPSCs with and without MECP2 from individual patients and thereby control for differences in genetic background (shown in Figure 1B are clones made from patient 982; clones from 567 look similar). The hiPSCs generated from fibroblasts of both patients appeared to be unaffected by the lack of MECP2, expressed all appropriate markers, and successfully generated teratomas upon injection into the testes of immunocompromised mice, consistent with previous hiPSC models for loss of MECP2 (Figure S2A) (Cheung et al., 2011, Hotta et al., 2009). Lack of MECP2 in patient-derived cells and specificity of antibody was also confirmed by western blot (Figure S2B).
Importantly, we never observed reactivation of the silenced X chromosome that would have resulted in re-expression of the WT allele of MECP2 in any cultures regardless of differentiation status or passage. This is consistent with previous data showing that, despite evidence for erosion of isolated portions of the silenced X chromosome (Mekhoubad et al., 2012), many portions of the inactivated X remain silenced even through reprogramming or differentiation (Patel et al., 2017, Tchieu et al., 2010). To measure the effect of any potential XCI erosion, we performed a DNA methylation analysis on the X chromosome on the lines from patient 982. This analysis showed that while some erosion of XCI was detectable across the X chromosome, there was not a significant difference between any of the lines (Figure S3A), and methylation at the MECP2 locus specifically was unchanged between the lines (Figure S3B).
As Rett syndrome primarily afflicts the nervous system and MECP2 is most highly expressed in neurons, we first generated NPCs from all of the hiPSCs lines following standard protocols (Patterson et al., 2012). Across at least two lines per patient with and without MECP2, we measured the rate of neuralization, the morphology of NPCs, and expression of typical marker genes. We were unable to detect consistent differences in these properties between multiple clones of both WT and MECP2 null lines derived from both patients (Figures 1C and S2C). Furthermore, the growth rate of NPCs with and without MECP2 was not consistently different in NPCs made from either patient (Figure S2D). Next, the NPCs were further differentiated by a non-directed differentiation approach that yields both neurons and glia (growth factor withdrawal; Patterson et al., 2012) (Figure 1D). All NPCs from both patients produced neurons and glia at the same rate (Figures S2E and S2F).
Previous studies have also shown that loss of MECP2 in neurons can lead to a decrease in AKT signaling (Li et al., 2013). A similar pattern was observed here in mutant neurons generated from Rett patient hiPSCs as measured by phosphorylation of AKT and S6, while hiPSCs themselves did not seem to be affected by loss of MECP2 (Figure 1E). Dendritic complexity has been shown extensively to be reliant on MECP2 expression in various models of Rett syndrome, and we found a statistically significant decrease in complexity in neurons made in the absence of MECP2 by Sholl assay (Figure 1F). In addition, we observed qualitative differences in basic neuronal morphology between WT and mutant neurons, where the neurons lacking MECP2 had shorter, thicker processes, and their soma was not as well defined.
Loss of MECP2 Affects the Transcriptome of Neurons
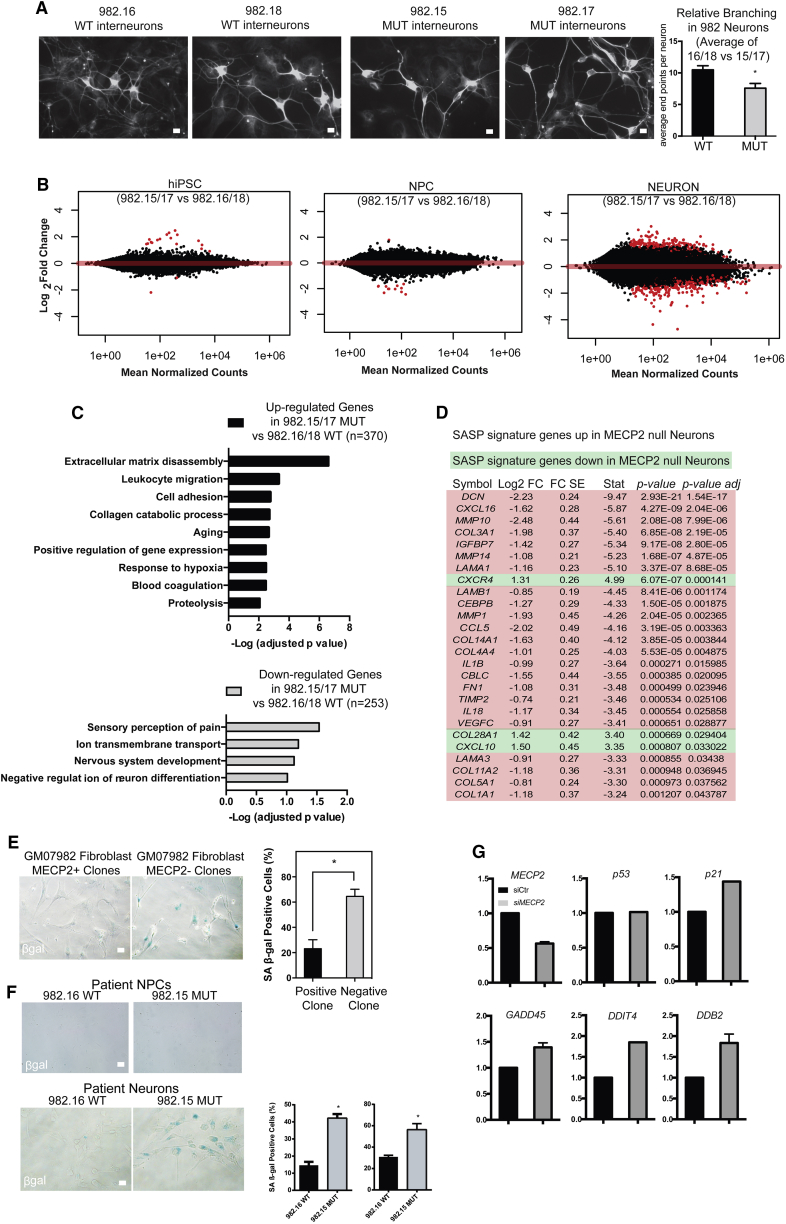
It has been suggested that loss of MECP2 only affects gene expression in neurons as opposed to the hPSCs and NPCs from which they were derived (Li et al., 2013). We sought to determine whether gene expression was affected in hiPSCs, NPCs, or neurons in this patient-derived in vitro model. To optimize the search for molecular effects of loss of MECP2 in neurons, we generated defined neuronal cultures by following the newly established 3i (three inhibitor) method to create interneurons (Figure 2A) (Maroof et al., 2013). Interneurons are particularly relevant in the study of Rett syndrome as interneuron-specific deletion of Mecp2 in mice recapitulates many of the disease symptoms (Ito-Ishida et al., 2015, Tomassy et al., 2014). We validated the quality of differentiation at each step by immunostaining for markers typical of particular cell types (SOX2, SOX1, and NESTIN as well as FOXG1 and NKX2.1 for NPCs; and Tuj1, MAP2, and GABA for interneurons) in both WT and MUT cultures followed by quantification (Figures S4A and S4B). While methods for derivation from pluripotent stem cells are effective at making interneurons, these cultures are not pure. As such, we first ensured that the proportion of neurons present in the cultures for comparison were not consistently different (Figure S4C). We then assessed whether interneurons lacking MECP2 also showed diminished dendritic branching. In fact, in patient-derived interneurons made by 3i, defects in dendritic branching as measured by the number of endpoints were clearly observed (Figure 2A).
Figure 2.
Loss of MECP2 Is Associated with Differential Gene Expression in Neurons
(A) Immunostaining neurons generated from patient 982 for TuJ1, a neuronal-specific marker. Right: quantification of dendritic complexity by counting endpoints.
(B) Volcano plots of differentially expressed genes (DEGs) in hiPSCs, NPCs, and neurons.
(C) Gene ontological analysis of DEGs increased versus decreased in MECP2 null neurons.
(D) An examination of SASP genes in neurons.
(E) Patient skin-derived clones of fibroblasts lacking MECP2 showed strong β-gal activity, while those of WT fibroblasts did not.
(F) Top: the senescence assay applied to neural progenitors derived from Rett patients did not show significant senescence activity. Bottom: patient-derived neuronal cultures showed a strong increase in the absence of MECP2 (quantification across independent lines shown on the right).
(G) RT-PCR for P53 targets after siRNA treatment of WT neurons.
Bar graphs represent means ± SEM. ∗p < 0.05. Scale bars on images indicate 10 μm.
We therefore proceeded with deep RNA sequencing (RNA-seq; >120 million reads per sample) of hiPSC, NPC, and interneuron cultures. With such sequencing depth, it was possible to analyze the RNA-seq reads for the known mutations present in the patients from which these lines were made (Figure S4D). This analysis demonstrated that each line studied expressed strictly either the WT or mutant allele of MECP2, and that XCI status was unchanged even after extensive differentiation to neurons. We quantified the expression level of MECP2 in WT cells across these three stages of development and found that the average reads per kilobase of transcript per million mapped reads RPKM was 3.1 for hiPSCs, 4.3 for NPCs, and 7.75 for interneuron-enriched cultures. This is consistent with consensus that MECP2 is enriched in neuronal cells, but also demonstrates that it could potentially be relevant to hiPSC and NPC physiology as well. However, high stringency analyses (false discovery rate <0.05) of the RNA-seq data yielded very few gene expression changes due to loss of MECP2 in hiPSCs or NPCs derived from Rett patients (Figure 2B), consistent with Li et al. (2013). On the other hand, interneuron cultures made from patient 982 showed many gene expression changes when comparing two individual WT and MUT clones (Figure 2B). Gene ontology analysis uncovered many neuronal physiology-related pathways that were downregulated due to loss of MECP2 in neurons, while genes associated with extracellular remodeling and cell migration appeared to be induced (Figure 2C).
Probing the RNA-seq data, we also found that MECP2 null interneuron cultures showed a strong increase in a group of genes that are known to be induced by senescent cells, known as the senescence-associated secretory program (SASP). The vast majority of SASP genes that were changed in MECP2 null neurons were upregulated as opposed to downregulated, suggesting a robust pattern of SASP induction (Figure 2D). The only previous report linking MECP2 loss to senescence was performed by partial silencing of this protein in mesenchymal stem cells, but the results were consistent with those shown here for patient-derived MECP2 null fibroblasts (Squillaro et al., 2010). The induction of SASP was intriguing in light of the fact that, while attempting to make clones of fibroblasts from patients with Rett syndrome, we repeatedly found that clones lacking MECP2 did not expand well after passage (14 MECP2 null clones were created, none expanded), while clones expressing the WT allele expanded without a problem (42 MECP2+ clones were created, we attempted to expand four of them, and all four expanded).
To determine whether MECP2 null fibroblasts encounter senescence, we performed an assay to detect endogenous beta-galactosidase activity, which is known to be a hallmark of this process (Wang et al., 2013). Indeed, MECP2 null fibroblasts showed strong activity in this senescence assay (Figure 2E). We did not encounter such difficulties with clonal expansion once hiPSCs or hiPSC-derived NPCs were made from patients, presumably because during reprogramming, telomerase is strongly induced to restore telomere length at least beyond the critical threshold (Marion et al., 2009, Suhr et al., 2009). In fact, our RNA-seq data showed that hiPSCs made from patients had very high expression of TERT, and NPCs still expressed moderate levels, while neurons did not express appreciable levels (average RPKM for TERT: hiPSC, 8.8; NPC, 1.6; neuron, 0.006). Importantly, the same endogenous galactosidase activity assay on interneurons showed a dramatic increase in senescence activity in neurons lacking MECP2 (Figure 2F). These data indicate that loss of MECP2 leads to not only induction of SASP but also a bona fide senescence program in neurons.
Induction of P53 in the Absence of MECP2
Cellular senescence programs are known to be regulated by P53, which can then activate various response pathways downstream, such as DNA repair and apoptosis (Vaziri and Benchimol, 1996). Interestingly, P53 induction due to telomere shortening was previously shown to cause defects in dendritic branching (Ferron et al., 2009), which is also the dominant phenotype in Rett syndrome. To begin to look for hallmarks of P53 induction in the absence of MECP2, we performed RT-PCR for P53-related targets in cells with silencing of MECP2 by siRNA (Figure S4E). This assay suggested that decreased MECP2 levels led to induction of P53-related target genes such as P21, GADD45, DDIT4, and DDB2 (Figure 2G).
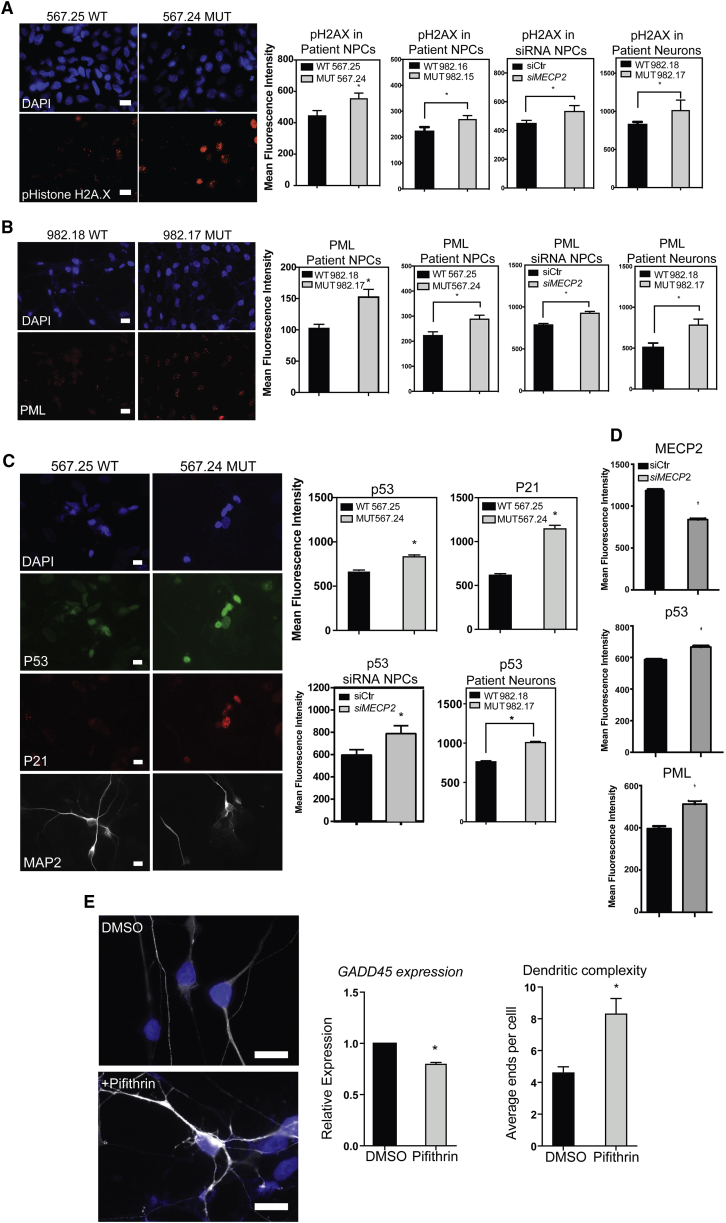
To determine the effect of loss of MECP2 in relation to cell-stress pathways at the protein level, we performed immunostaining for H2AX, PML, P53, and P21 in neurons with and without MECP2. Staining for each of these markers showed strong increases in expression/levels of these markers of cell stress in patient-derived NPCs, neurons, and also after silencing of MECP2 in both NPCs and neurons (Figures 3A–3D). WT NPCs with silencing of MECP2 by siRNA and neurons lacking MECP2 also showed clear induction of these marks.
Figure 3.
Loss of MECP2 Leads to Induction of DNA Damage and P53
(A) Immunostaining of patient NPCs, NPCs with siRNA against MECP2, and patient neurons showed a strong increase in H2aX in the absence of MECP2.
(B) Immunostaining of patient NPCs, NPCs with siRNA against MECP2, and patient neurons.
(C) Immunostaining for P53 and p21, a target of P53.
(D) Immunostaining after siRNA silencing of MECP2 in WT neuronal cultures.
(E) Treatment of MECP2 null neurons with DMSO or Pifithrin, followed by immunostaining with antibody for TuJ1 shows a change in dendritic branching. Bottom left: RT-PCR for GADD45, a P53 target gene, showed that Pifithrin reduced P53 activity. Bottom right: quantification of branching phenotype across three independent experiments.
∗p < 0.05 according to Student's t test. Bar graphs represent means ± SEM. Scale bars on images indicate 10 μm.
Blocking Induction of P53 Can Rescue Dendritic Branching Defects Due to Loss of MECP2
Previous evidence from a murine model of telomere shortening as a result of loss of telomerase complex (TERT) led to defects in dendritic branching, and this effect was strictly dependent on induction of P53 (Ferron et al., 2009). A more recent study also showed that experimentally aging the neural lineage with telomerase inhibition led to neurons with signs of aging, including reduced dendritic branching (Vera et al., 2016). Therefore, we posited that inhibition of P53 in MECP2 null neurons could potentially restore appropriate dendritic branching. To determine whether blocking the action of P53 could improve dendritic branching in MECP2 null interneurons, we took advantage of Pifithrin-α, a potent inhibitor of P53 target gene activation (Bassi et al., 2002). Treatment of MECP2 null interneurons with Pifithrin-α showed evidence of P53 inhibition as measured by RT-PCR for GADD45 (Vaziri and Benchimol, 1996), a target gene important for DNA repair (Figure 3E). After 24–48 hr of P53 inhibition by Pifithrin-α, MECP2 null interneurons appeared to adopt an improved neuronal morphology typified by increased physical distinction between the soma and neurites, longer, thinner neurites, as well as increased dendritic branching as shown and quantified in Figure 3E. These data provide evidence that neurons without MECP2 induce P53 activity, which then inhibits the formation of complex neuronal processes.
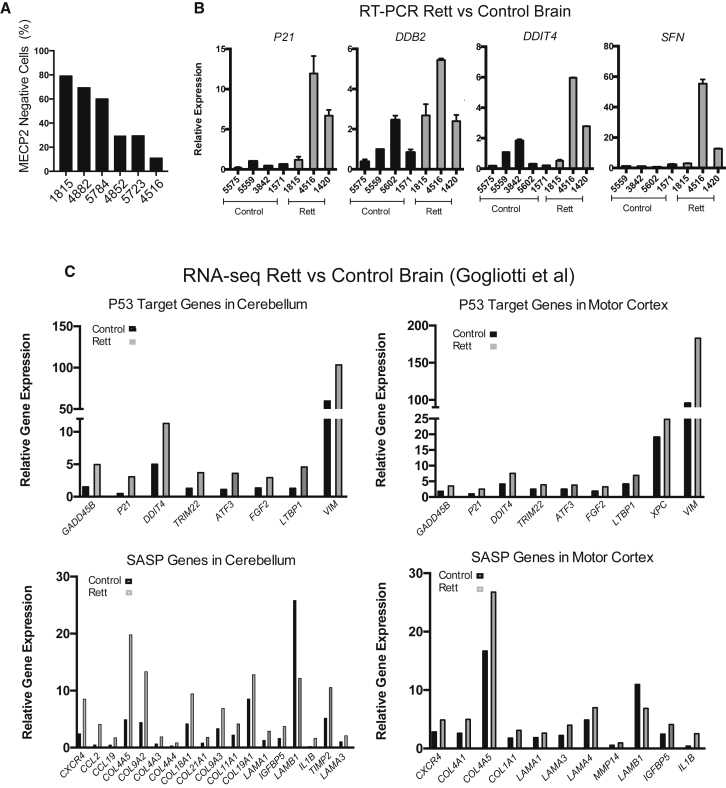
To determine whether any of the phenotypes discovered in this in vitro model of Rett syndrome have relevance to patients afflicted with the disease, we acquired tissue specimens from Rett patients and age-matched controls. We first quantified the degree of chimerism of female Rett patient neurons due to skewing of X chromosome inactivation to determine the relative ratio of neurons that express MECP2 versus those that did not. Some of the Rett patient brains showed roughly 75% of neurons lacked MECP2, while others appeared to have less than 25% MECP2 null neurons (Figure 4A).
Figure 4.
Evidence for P53 Induction in Rett Patient Neurons in Human Brain
(A) Each Rett brain sample was assessed for the percentage of neurons with and without expression of MECP2 by immunostaining.
(B) RT-PCR on RNA isolated from Rett brain versus age-matched control brains for P53 targets.
(C) A re-analysis of data published by Gogliotti et al. (2018). Shown are a sample of P53 targets and SASP genes found on lists of genes upregulated in Rett brain. All of these differentially expressed genes were derived using a corrected p value (false discovery rate) <0.05 from at least n = 5 samples from control and Rett brains.
Bar graphs represent means ± SEM.
We performed RT-PCR on samples from some of these brains to determine whether they showed signs of increased P53 activity. To ensure accurate RNA representation, we first assessed the quality of the RNA from these frozen tissues, and only proceeded with RT-PCR in samples that showed an RIN (RNA integrity number) value above 5. All the Rett patient brains we processed for RT-PCR showed induction of canonical P53 target genes (identified in Wei et al., 2006), consistent with what was observed in the patient-derived neurons in vitro (Figure 4B). Recently, a new study was published that isolated brain tissue from Rett patients and control subjects to perform an RNA-seq profile to characterize changes specific to Rett motor cortex or cerebellum. Using data provided in that study, we found that many direct P53 targets (as defined in Wei et al., 2006) and SASP signature genes were upregulated in the Rett brains in both the motor cortex and the cerebellum (Figure 4C).
Discussion
Taken together, these data demonstrate that loss of MECP2 leads to clear signs of stress such as H2AX deposition, P53/P21 induction, and initiation of a senescence program, all of which suggest that neurons in Rett syndrome could be in suboptimal health, leading to neurophysiological defects such as dendritic arborization (Zhou et al., 2006). While one paper suggested that RNAi-mediated silencing of MECP2 could promote senescence in mesenchymal cells (Squillaro et al., 2010), decades of work on Rett syndrome have not uncovered a role for MECP2 in relation to senescence in a wide variety of models such as various transgenic mouse lines, human patient postmortem analyses, and in vitro human models.
These results also raise the question of whether senescence could be common to the etiologies of other intellectual disability syndromes. The phenotypes described here show a striking similarity to those observed in hiPSCs and neural derivatives made from patients with immunodeficiency, centromeric region instability, and facial anomalies syndrome (ICF) syndrome (Yehezkel et al., 2013). Two independent studies showed that ICF patient-derived hiPSCs displayed telomere shortening that was coupled to senescence of somatic derivatives such as fibroblasts. ICF syndrome only partially overlaps with Rett syndrome in terms of patient phenotypes, but is caused by mutations in DNMT3B, a de novo DNA methyltransferase (Linhart et al., 2007). These findings together are highly relevant as DNMT3B is a key de novo methyl transferase to create methylated DNA (5mC), which is the substrate for Tet oxygenase’s to create 5-hydroxmethylated DNA (5hmC), which is known to be strongly bound by MECP2 (Mellen et al., 2012). Recently, another study showed that deletion of Tet enzymes, which are critical to generate the 5hmC mark, led to shortened telomeres (Yang et al., 2016), which is known to lead to P53 activation.
Another possible interpretation of these data is that instead of a failure to mature, Rett syndrome neurons instead show aspects of premature aging. The fact that MECP2 null neurons show induction of aging-related genes, including P53 targets, and induce senescence pathways are consistent with this idea (Tan et al., 2014). On the other hand, while Rett patients suffer from a post-natal cognitive decline, and long-term survivors show phenotypes associated with Parkinson disease (Zoghbi, 2016), the typical phenotypes presented in young female patients are not consistent with premature aging. Whether the physiological response to loss of MECP2 is truly akin to premature aging or whether patients suffer from effects that are unrelated to aging is worthy of continued investigation.
Experimental Procedures
Generation of Isogenic Rett Syndrome iPSCs
Reprogramming was performed as described (Lowry et al., 2008).
Generation of Teratomas
Generation of teratomas was previously described (Lindgren et al., 2011).
Differentiation In Vitro and Analysis
Neural specification with neural rosette derivation, neuroprogenitor (NPC) purification, and further differentiation to neurons and glia were performed as described previously (Patterson et al., 2012, Patterson et al., 2014).
Immunofluorescence and Image Quantification
Immunofluorescence was performed as described previously (Patterson et al., 2014) and is described in detail in the Supplemental Information.
RT-qPCR
RT-PCR with real-time PCR measurement was carried out on a Roche 480 as described (Patterson et al., 2012, Patterson et al., 2014). The primer sequences are available in the Supplemental Information.
siRNA Gene Silencing
All knockdown experiments were performed as described previously (Patterson et al., 2014).
β-Galactosidase Senescence Assay
β-Galactosidase senescence assay was performed using the Senescence β-Galactosidase Staining Kit from Cell Signaling. The number of blue cells and number of total cells were quantified using the Cell Counter plugin in ImageJ.
Quantification of Dendritic Arborization
The stained cells were then imaged at 20×, and dendritic arbors of individual cells were traced using the Simple Neurite Tracer plugin for ImageJ. The number of process ends per cell were counted using the Cell Counter plugin for ImageJ. The number of process ends per cell are presented as mean ends per cell ± SEM.
RNA Expression Profiling
RNA-seq was performed as described previously (Gu et al., 2016). These data are available from NIH dataset GEO: GSE107399.
Author Contributions
M.O., E.K., D.A., P.L., K.F., B.S.V., J.C., C.S., J.C.P., I.G., J.L., C.C., E.K., and S.T. provided data through experimentation. M.O., E.K., and W.E.L. contributed to writing the manuscript. X.X., M.P., K.P., and W.E.L. provided financial support for this work.
Acknowledgments
We would like to acknowledge the helpful discussions and insight on this manuscript with Gail Mandel (OHSU). This work was funded by training grants to M.O. (NIH-Virology and Gene Therapy, UCLA), P.L. (CIRM, UCLA), C.S. (CIRM-Bridges, Cal-State-Northridge), and D.A. (HHURP, UCLA). W.E.L. was supported by a Rose Hills Scholar award through the Eli and Edythe Broad Center for Regenerative Medicine. W.E.L. and K.P. were supported by NIH (P015P01GM099134). This research was also supported by the Allen Distinguished Investigator Program and the Paul G. Allen Frontiers Group, and a March of Dimes Scholar Award (6-FY17-406).
Published: May 8, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.04.001.
Contributor Information
Kathrin Plath, Email: kplath@mednet.ucla.edu.
William E. Lowry, Email: blowry@mcdb.ucla.edu.
Supplemental Information
References
- Bassi L., Carloni M., Fonti E., Palma de la Pena N., Meschini R., Palitti F. Pifithrin-alpha, an inhibitor of p53, enhances the genetic instability induced by etoposide (VP16) in human lymphoblastoid cells treated in vitro. Mutat. Res. 2002;499:163–176. doi: 10.1016/s0027-5107(01)00273-1. [DOI] [PubMed] [Google Scholar]
- Bird A. The methyl-CpG-binding protein MeCP2 and neurological disease. Biochem. Soc. Trans. 2008;36:575–583. doi: 10.1042/BST0360575. [DOI] [PubMed] [Google Scholar]
- Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Horvath L.M., Grafodatskaya D., Pasceri P., Weksberg R., Hotta A., Carrel L., Ellis J. Isolation of MECP2-null Rett syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross S.H., Meehan R.R., Nan X., Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- Ferron S.R., Marques-Torrejon M.A., Mira H., Flores I., Taylor K., Blasco M.A., Farinas I. Telomere shortening in neural stem cells disrupts neuronal differentiation and neuritogenesis. J. Neurosci. 2009;29:14394–14407. doi: 10.1523/JNEUROSCI.3836-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti R., Fisher N., Stansley B., Jones C., Lindsley C., Conn J., Niswender C. Total RNA-sequencing of Rett syndrome autopsy samples identifies the M4 muscarinic receptor as a novel therapeutic target. J. Pharmacol. Exp. Ther. 2018;365:291–300. doi: 10.1124/jpet.117.246991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Gaeta X., Sahakyan A., Chan A.B., Hong C.S., Kim R., Braas D., Plath K., Lowry W.E., Christofk H.R. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 2016;19:476–490. doi: 10.1016/j.stem.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- Ito-Ishida A., Ure K., Chen H., Swann J.W., Zoghbi H.Y. Loss of MeCP2 in parvalbumin-and somatostatin-expressing neurons in mice leads to distinct Rett syndrome-like phenotypes. Neuron. 2015;88:651–658. doi: 10.1016/j.neuron.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Yun J.M., Woods R., Dunaway K., Yasui D.H., Lasalle J.M., Gong Q. MeCP2 regulates activity-dependent transcriptional responses in olfactory sensory neurons. Hum. Mol. Genet. 2014;23:6366–6374. doi: 10.1093/hmg/ddu358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang H., Muffat J., Cheng A.W., Orlando D.A., Loven J., Kwok S.M., Feldman D.A., Bateup H.S., Gao Q. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren A.G., Natsuhara K., Tian E., Vincent J.J., Li X., Jiao J., Wu H., Banerjee U., Clark A.T. Loss of Pten causes tumor initiation following differentiation of murine pluripotent stem cells due to failed repression of Nanog. PLoS One. 2011;6:e16478. doi: 10.1371/journal.pone.0016478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart H.G., Lin H., Yamada Y., Moran E., Steine E.J., Gokhale S., Lo G., Cantu E., Ehrich M., He T. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M.A. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R.R., Lewis J.D., Bird A.P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M., Ayata P., Dewell S., Kriaucionis S., Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Patel S., Bonora G., Sahakyan A., Kim R., Chronis C., Langerman J., Fitz-Gibbon S., Rubbi L., Skelton R.J.P., Ardehali R. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 2017;18:54–67. doi: 10.1016/j.celrep.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M., Chan D.N., Ha I., Case D., Cui Y., Handel B.V., Mikkola H.K., Lowry W.E. Defining the nature of human pluripotent stem cell progeny. Cell Res. 2012;22:178–193. doi: 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M., Gaeta X., Loo K., Edwards M., Smale S., Cinkornpumin J., Xie Y., Listgarten J., Azghadi S., Douglass S.M. let-7 miRNAs can act through notch to regulate human gliogenesis. Stem Cell Reports. 2014;3:758–773. doi: 10.1016/j.stemcr.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2016;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squillaro T., Alessio N., Cipollaro M., Renieri A., Giordano A., Galderisi U. Partial silencing of methyl cytosine protein binding 2 (MECP2) in mesenchymal stem cells induces senescence with an increase in damaged DNA. FASEB J. 2010;24:1593–1603. doi: 10.1096/fj.09-143057. [DOI] [PubMed] [Google Scholar]
- Suhr S.T., Chang E.A., Rodriguez R.M., Wang K., Ross P.J., Beyhan Z., Murthy S., Cibelli J.B. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS One. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F.C., Hutchison E.R., Eitan E., Mattson M.P. Are there roles for brain cell senescence in aging and neurodegenerative disorders? Biogerontology. 2014;15:643–660. doi: 10.1007/s10522-014-9532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sherman S., Aimiuwu O., Lingren A., Zack J.A., Clark A. Female human iPS cells retain an inactive X-chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassy G.S., Morello N., Calcagno E., Giustetto M. Developmental abnormalities of cortical interneurons precede symptoms onset in a mouse model of Rett syndrome. J. Neurochem. 2014;131:115–127. doi: 10.1111/jnc.12803. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Benchimol S. From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomere loss/DNA damage model of cell aging. Exp. Gerontol. 1996;31:295–301. doi: 10.1016/0531-5565(95)02025-x. [DOI] [PubMed] [Google Scholar]
- Vera E., Bosco N., Studer L. Generating late-onset human iPSC-based disease models by inducing neuronal age-related phenotypes through telomerase manipulation. Cell Rep. 2016;17:1184–1192. doi: 10.1016/j.celrep.2016.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wei D., Xiao H. Methods of cellular senescence induction using oxidative stress. Methods Mol. Biol. 2013;1048:135–144. doi: 10.1007/978-1-62703-556-9_11. [DOI] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Winkler T., Cantilena A., Metais J.Y., Xu X., Nguyen A.D., Borate B., Antosiewicz-Bourget J.E., Wolfsberg T.G., Thomson J.A., Dunbar C.E. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells. 2010;28:687–694. doi: 10.1002/stem.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Guo R., Wang H., Ye X., Zhou Z., Dan J., Wang H., Gong P., Deng W., Yin Y. Tet enzymes regulate telomere maintenance and chromosomal stability of mouse ESCs. Cell Rep. 2016;15:1809–1821. doi: 10.1016/j.celrep.2016.04.058. [DOI] [PubMed] [Google Scholar]
- Yehezkel S., Shaked R., Sagie S., Berkovitz R., Shachar-Bener H., Segev Y., Selig S. Characterization and rescue of telomeric abnormalities in ICF syndrome type I fibroblasts. Front. Oncol. 2013;3:35. doi: 10.3389/fonc.2013.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hong E.J., Cohen S., Zhao W.N., Ho H.Y., Schmidt L., Chen W.G., Lin Y., Savner E., Griffith E.C. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi H.Y. Rett syndrome and the ongoing legacy of close clinical observation. Cell. 2016;167:293–297. doi: 10.1016/j.cell.2016.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.