Summary
A dramatic difference in global DNA methylation between male and female cells characterizes mouse embryonic stem cells (ESCs), unlike somatic cells. We analyzed DNA methylation changes during reprogramming of male and female somatic cells and in resulting induced pluripotent stem cells (iPSCs). At an intermediate reprogramming stage, somatic and pluripotency enhancers are targeted for partial methylation and demethylation. Demethylation within pluripotency enhancers often occurs at ESC binding sites of pluripotency transcription factors. Late in reprogramming, global hypomethylation is induced in a female-specific manner. Genome-wide hypomethylation in female cells affects many genomic landmarks, including enhancers and imprint control regions, and accompanies the reactivation of the inactive X chromosome. The loss of one of the two X chromosomes in propagating female iPSCs is associated with genome-wide methylation gain. Collectively, our findings highlight the dynamic regulation of DNA methylation at enhancers during reprogramming and reveal that X chromosome dosage dictates global DNA methylation levels in iPSCs.
Keywords: induced pluripotency, iPSC, DNA methylation, epigenetics, embryonic stem cells, ESC, pluripotency, reprogramming, X chromosome inactivation
Graphical Abstract
Highlights
-
•
Female iPSCs are globally hypomethylated
-
•
Hypomethylation affects multiple genomic features including imprint control regions
-
•
Loss of X chromosome restores global methylation but not at imprint control regions
-
•
Methylation changes at enhancers in male and female reprogramming intermediates
Somatic cells can be reprogrammed to iPSCs, inducing reactivation of the inactive X chromosome. Using genome-scale DNA methylation analyses, Plath, Pasque, and colleagues show that iPSCs adopt sex-specific differences in global DNA methylation that correlate with the presence of two active X chromosomes. Upon culture, female iPSCs lose one of the two X chromosomes and adopt male-like DNA methylation.
Introduction
Pluripotent stem cells (PSCs) possess the remarkable ability to self-renew indefinitely and differentiate into any cell type of the body, and therefore bear significant potential for cell therapy, disease studies, and fundamental research. A number of procedures can be used to obtain PSCs, including the derivation of embryonic stem cells (ESCs) from the mammalian pre-implantation embryo and the reprogramming of somatic cells by nuclear transfer or transcription factor overexpression (Takahashi and Yamanaka, 2006). Considerable effort is being directed at studying the molecular properties of PSCs including those obtained following reprogramming of somatic cells to induced PSCs (iPSCs). Accumulating evidence suggests that DNA methylation plays a crucial role during reprogramming of somatic cells to iPSCs (Koche et al., 2011). For example, inhibition of the major DNA methyltransferase DNMT1 can enhance the efficiency of reprogramming (Mikkelsen et al., 2008). In this context, it is notable that sex chromosome content influences the global methylation state in ESCs (Zvetkova et al., 2005). However, to date the effect of sex chromosomes on DNA methylation dynamics during reprogramming remains poorly understood.
Global DNA methylation levels differ significantly between male and female mouse ESC lines (Habibi et al., 2013, Ooi et al., 2010, Shirane et al., 2016, Zvetkova et al., 2005). Male ESCs have equivalent global levels of DNA methylation to female and male differentiated cells, such as mouse embryonic fibroblasts (MEFs), whereas female ESCs have up to 75% less DNA methylation (Habibi et al., 2013). Compared with male ESCs, demethylation in female ESCs occurs in most genomic contexts including regulatory regions such as CpG islands, promoters, enhancers, and imprint control regions (ICRs), as well as repeat regions such as major and minor satellites, long interspersed nuclear elements (LINEs), and long terminal repeats (LTRs) (Choi et al., 2017a, Choi et al., 2017b, Habibi et al., 2013, Hackett et al., 2013, Ooi et al., 2010, Yagi et al., 2017, Zvetkova et al., 2005).
The global hypomethylation of the female mouse ESC genome has been attributed to the presence of two active X chromosomes (Xa), since a gain in overall methylation takes place after the loss of one of the two X chromosomes (reaching the XO state) (Choi et al., 2017a, Zvetkova et al., 2005). The increased dosage of X-linked gene Dusp9 in XaXa female ESCs was shown to contribute to the hypomethylation occurring in female ESCs (Choi et al., 2017a). The presence of two active X chromosomes in female ESCs was also shown to delay exit from pluripotency (Schulz et al., 2014). Altogether, these data indicate that the X chromosome status is an important regulator of the DNA methylation landscape and differentiation dynamics of ESCs.
Reprogramming of female somatic cells to iPSCs induces the reactivation of the inactive X chromosome (Xi) (Maherali et al., 2007). Thus, like mouse ESCs, female mouse iPSCs have two active X chromosomes, which enables them to undergo random X chromosome inactivation upon differentiation (Maherali et al., 2007; reviewed in Pasque and Plath, 2015). Notably, the reactivation of the Xi occurs very late in the reprogramming process, specifically in those cells that already express critical pluripotency factors (Pasque et al., 2014). The influence that Xi reactivation (X chromosome reactivation, XCR) may play on global DNA methylation during the female reprogramming process remains to be investigated. A comprehensive analysis of DNA methylation during female and male cell reprogramming to iPSCs, and the correlation with the X chromosome state, are critical to clarifying this important point. Our earlier study that examined DNA methylation of microsatellites suggested that female iPSCs become hypomethylated as a result of reprogramming (Maherali et al., 2007), suggesting that female-specific methylation dynamics may be at play in reprogramming to pluripotency. Interestingly, a recent paper showed that female cells undergo a transient global hypomethylation event during the reprogramming process but reach a similarly high methylation state as male iPSCs at the end (Milagre et al., 2017), raising the question of how these changes in methylation relate to the X chromosome state.
Analyzing the dynamics of DNA methylation during the generation of iPSCs is complicated by the low efficiency and heterogeneity with which the establishment of iPSCs takes place. Early in reprogramming, when reprogramming cultures are thought to be still relatively homogeneous, few changes in DNA methylation were found while histone modifications change more dramatically (Koche et al., 2011, Polo et al., 2012). Moreover, studies that examined promoters in sorted reprogramming subpopulations or heterogeneous reprogramming cultures at various time points toward the generation of partially reprogrammed cells and iPSCs suggested that changes in DNA methylation mainly take place late in reprogramming (Lee et al., 2014, Polo et al., 2012). For promoters, a gain in DNA methylation was found to take place more rapidly during reprogramming than loss (Lee et al., 2014). Binding sites for pluripotency-associated transcription factors in ESCs show focal DNA demethylation early in reprogramming cultures, resolving into larger hypomethylated regions in the pluripotent state (Lee et al., 2014). The dynamics of DNA methylation at key regulatory regions such as cell-type-specific enhancers remains to be explored during intermediate reprogramming stages. Similarly, whether differences in DNA methylation exist between male and female cells undergoing reprogramming also remains to be determined. Currently, most published comprehensive analyses of DNA methylation dynamics do not reportedly take X chromosome dosage into account (Milagre et al., 2017).
Here, we set out to define the dynamics of DNA methylation during the reprogramming of male and female MEFs to pluripotency. To this end, we analyzed genome-scale single-base-pair resolution DNA methylation maps of MEFs, reprogramming intermediates, and iPSCs, both male and female, and, for comparison, of male and female ESCs. To define kinetics and modes of male and female DNA methylation reprogramming, we focused our analysis on specific genomic features such as somatic and pluripotency enhancers, promoters, repeat elements, and ICRs in relation to the timing of XCR and X chromosome content. This effort led us to reveal targeted changes in DNA methylation at enhancer regions in reprogramming intermediates, irrespective of sex, and a female-specific, extensive global hypomethylation during reprogramming to iPSCs that occurs concomitant with XCR and is associated with the presence of two Xas. Global hypomethylation is reversed as female iPSCs are propagated and one X chromosome is lost. Our results reveal that the transcriptional activity and number of X chromosomes are key features to consider when studying reprogramming and iPSCs.
Results
Genome-Scale DNA Methylation Maps during Female and Male Reprogramming
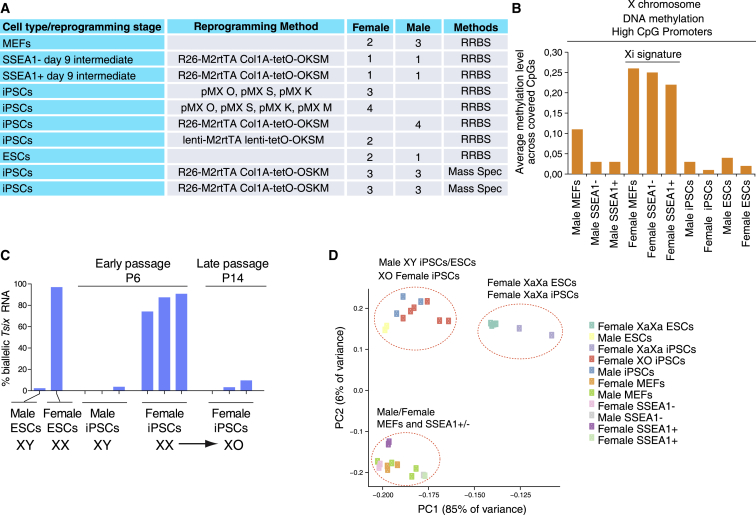
To define the global DNA methylation state and the dynamics of DNA methylation at major genomic landmarks during female cell reprogramming, we took advantage of genome-scale methylation maps generated by reduced representation bisulfite sequencing (RRBS) (Meissner et al., 2008) for specific stages of female cell reprogramming for our previous analysis of XCR kinetics (Pasque et al., 2014). In our previous study, these datasets revealed a high persistence of DNA methylation on CpG islands of the Xi in late reprogramming intermediates but absence in iPSCs, consistent with the reactivation of the Xi late in the reprogramming process after a large number of pluripotency genes are activated (Pasque et al., 2014). Specifically, methylation maps of the female reprogramming time course were available for starting MEFs, a late intermediate reprogramming state, and resulting iPSCs (Figure 1A, also summarized in Table S1). For the late intermediate, we analyzed cells at day 9 of reprogramming based on the presence of the cell surface marker stage-specific embryonic antigen 1 (SSEA1) (Polo et al., 2012), thereby circumventing the problem of heterogeneity of bulk reprogramming cultures. In agreement with previous studies, we have described that upon re-plating, the SSEA1-positive (+) cell population preferentially progresses to the iPSC state compared with the SSEA1-negative (−) cell population, and therefore is enriched for cells that can successfully progress to pluripotency (Pasque et al., 2014, Polo et al., 2012). We also had the methylation map of the SSEA1 population at day 9 of female reprogramming available (Pasque et al., 2014) (Figure 1A).
Figure 1.
Genome-Scale Methylation Dynamics during Male and Female Cell Reprogramming
(A) Detailed information of the cell types and reprogramming methods used, and the analysis method for methylation. Further details are included in Table S1.
(B) Average methylation across listed male and female cell types and reprogramming stages at high CpG-density promoters (HCPs) on the X chromosome. The Xi signature highlights the hemi-methylation of the inactive X chromosome (Xi) present specifically in female MEFs and SSEA1+/− reprogramming stages.
(C) RNA FISH analysis of Tsix biallelic expression. The counts are presented as percentage of cells with Tsix signal (monoallelic or biallelic) that show biallelic Tsix signals. The female iPSC lines analyzed at passage 6 are the same lines as those analyzed at passage 14. The same lines at passages 5, 7, and 13 were also used in Figure 2E.
(D) PCA for RRBS-derived methylation data, considering male and female MEFs, male and female SSEA1− and SSEA1+ sorted reprogramming intermediates, male iPSCs, and female XX and XO iPSCs, as well as male and female ESCs. Each color represents a cell type and each spot represents a biological replicate (different samples). PCA was performed for the mean methylation of non-repetitive autosomal 100-bp genomic tiles with at least two CpGs covered at 5× or greater.
In addition, we generated new RRBS datasets for the same stages of male cell reprogramming to enable the comparison between male and female cell reprogramming, and profiled female iPSCs at different passages to control for DNA methylation changes that take place over time in culture after these cell lines are established (Figure 1A, also summarized in Table S1). We also used different reprogramming methods and male and female iPSCs generated in different experiments and laboratories to control for method- and laboratory-dependent differences in DNA methylation in iPSCs (summarized in Table S1 and Figure 1A). To this end, iPSCs were generated by retroviral transduction of MEFs with Oct4 (O), Sox2 (S), Klf4 (K), with or without cMyc (M), or the addition of doxycycline (dox) to transgenic MEFs carrying different dox-inducible polycistronic reprogramming factor cassettes (OSKM or OKSM) (Table S1 and Figure 1A). To control for sex chromosome content and enable a comparison with the global hypomethylation of female ESCs, we also used RRBS-based methylation datasets for male and female ESC samples (Pasque et al., 2014). All pluripotent stem cells (PSCs) were maintained in serum and leukemia inhibitory factor (S/L) conditions. With these methylation datasets, we aimed to define methylation dynamics during reprogramming to pluripotency and in established iPSCs at the genome-scale level, and to determine how it is influenced by sex chromosome content and the transcriptional dose of genes on the X chromosome.
Our previous work indicated that early-passage female iPSCs possess two active X chromosomes due to XCR very late in the reprogramming process (Maherali et al., 2007, Pasque et al., 2014). Thus, female SSEA1+ reprogramming intermediates still possess an Xi but newly established iPSCs carry two active X chromosomes (Pasque et al., 2014). We also demonstrated previously that demethylation of CpG islands on the Xi occurs only late in reprogramming, after the day-9 SSEA1+ stage, consistent with the expression state of the X chromosome (Pasque et al., 2014). This dynamic is illustrated here by plotting the average DNA methylation level of promoters with high CpG content (HCPs) on the X chromosome from our RRBS datasets, revealing higher methylation in cells with an Xi including female MEFs and the female day-9 SSEA1+ and SSEA1− reprogramming subpopulations (Figure 1B). Conversely, all male cell types or reprogramming intermediates and female iPSC lines displayed much lower methylation levels of HCPs. These results are consistent with XCR taking place late during reprogramming to iPSCs, and demonstrate that the X chromosomes in all female iPSC and ESC lines are in an active expression state. These data also demonstrate the completely reprogrammed state of our iPSC lines.
Female ESCs tend to lose one of the two active X chromosomes in culture (Choi et al., 2017b, Choi et al., 2017a, Schulz et al., 2014, Yagi et al., 2017, Zvetkova et al., 2005), raising the possibility that X chromosome loss occurred in our iPSCs. To determine the number of active X chromosomes present in our female iPSC lines, we used RNA fluorescent in situ hybridization analysis (FISH). This analysis allowed us to determine the proportion of female iPSCs with two active X chromosomes as judged by biallelic expression of the X-linked gene Tsix. We found that most female iPSC lines had low biallelic Tsix count, indicative of X chromosome loss (Figure S1A and Table S1). In contrast, two female iPSC lines had >50% biallelic Tsix signals indicating the presence of two active X chromosomes in the majority of the culture (Figure S1A and Table S1). The female ESC line used in this analysis was derived from an F1 cross between two different mouse strains (Musculus/Castaneus), and was previously shown to maintain two active X chromosomes upon extended culture (Lee and Lu, 1999). To test whether iPSCs with only one Xa indeed arise from XaXa iPSCs through continued propagation, we cultured different newly derived female iPSC lines for several passages and examined X chromosome number. As previously shown for ESCs, the presence of two active X chromosomes in female iPSCs correlated with low passage number, and over time in culture the cells lose one X chromosome, becoming XO (Figure 1C) (Choi et al., 2017b, Yagi et al., 2017, Zvetkova et al., 2005). Therefore, our female iPSC lines included pluripotent cell lines with one or two active X chromosomes. We termed these XO and XaXa iPSC lines for the remaining analyses (Figure S1A and Table S1).
Differences in DNA Methylation in iPSCs Correlate with the Number of Active X Chromosomes
To assess the methylation state in the reprogramming stages and different cell types, we first performed principal component analysis (PCA), focusing specifically on differentially methylated autosomal CpGs (based on Fisher's exact test with 5% false discovery rate) for all possible pairwise cell type comparisons to avoid any influence from the differential sex chromosome content and epigenetic state. The PCA analysis revealed that male and female MEFs clustered together with the male and female SSEA1− and SSEA1+ reprogramming subpopulations, and a strong separation of these from all PSC types (Figure 1D). These results are in agreement with the absence of major changes in DNA methylation taking place between MEFs and the day-9 SSEA1 intermediates and a dramatic reorganization of DNA methylation between the SSEA1 state and iPSCs (Knaupp et al., 2017, Lee et al., 2014, Milagre et al., 2017, Polo et al., 2012).
Notably, in all of our female iPSC lines profiled with RRBS, the number of active X chromosomes correlated with differences in DNA methylation. Specifically, iPSCs and ESCs clustered into two main groups (Figure 1D). The first group contained male iPSCs and ESCs as well as the XO female iPSC lines. The second group contained the female XaXa iPSCs and XaXa ESCs (Figure 1D). Therefore, differences in DNA methylation correlate with the X chromosome status of iPSCs. These data suggested that the first principal component, which correlated with the number of active X chromosomes and captured the separation of XO/XY and XX ESCs/iPSCs as well as most variation in methylation (Figure 1D), corresponded to the previously described global difference in methylation between established male and female PSCs or XO and XX female PSCs (Choi et al., 2017b, Choi et al., 2017a, Habibi et al., 2013, Ooi et al., 2010, Yagi et al., 2017, Zvetkova et al., 2005). Conversely, the second principal component corresponded to methylation differences associated with reprogramming progression. Thus, sex chromosome content influences DNA methylation dynamics during reprogramming.
iPSCs with Two Active X Chromosomes Are Hypomethylated and Gain Male-like Global Methylation upon X Chromosome Loss
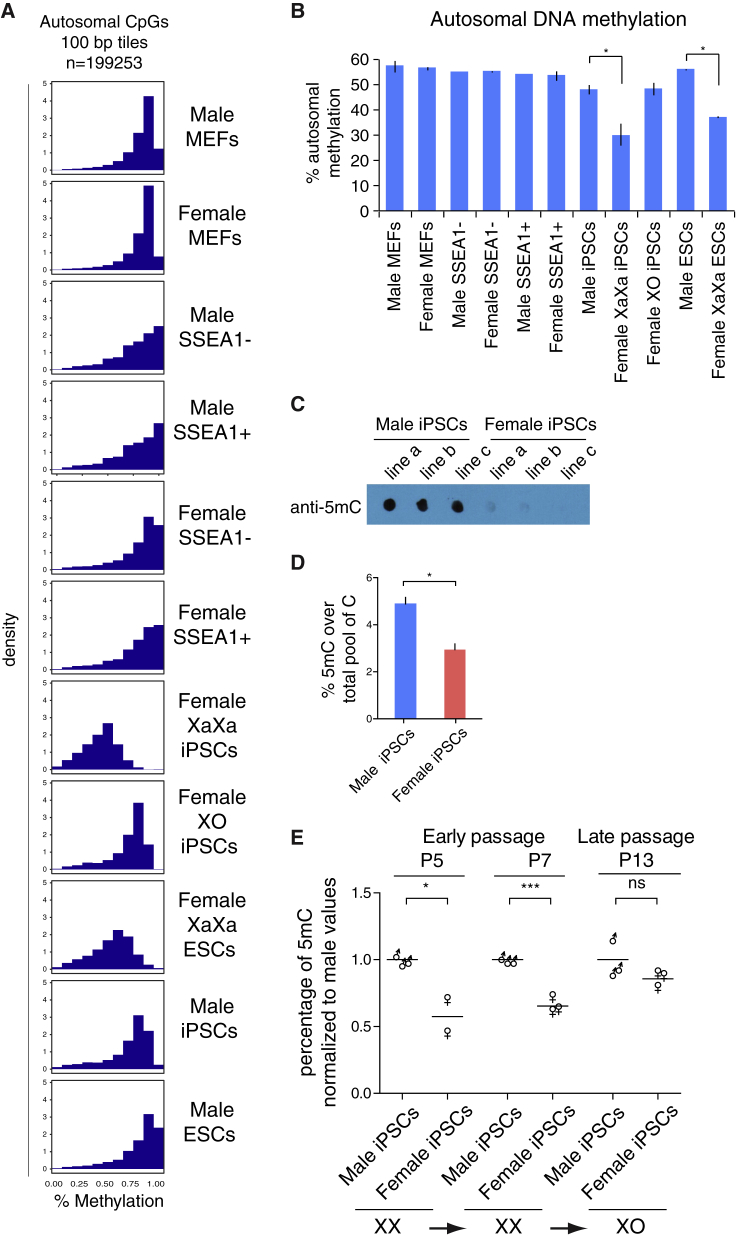
In mouse ESCs the number of active X chromosomes correlates with genome hypomethylation, with XaXa ESCs displaying global demethylation compared with XY or XO ESCs, while DNA hypomethylation is typically lost over time in culture due to the loss of one of the two X chromosomes (Choi et al., 2017b, Choi et al., 2017a, Habibi et al., 2013, Hackett et al., 2013, Ooi et al., 2010, Schulz et al., 2014, Yagi et al., 2017, Zvetkova et al., 2005). To assess the global dynamics of methylation during female cell reprogramming, we next analyzed the methylation levels across the genome (Figure 2A). As before, we excluded sex chromosomes and considered only autosomal CpGs to not be influenced by any potential methylation changes on the X chromosome due to XCR. The distribution of autosomal methylation values was similar across all cell types, displaying a pattern skewed toward hypermethylation. However, compared with the other cell types, both female XaXa ESCs and iPSCs had a pronounced reduction in highly methylated genomic regions (those carrying 90%–100% methylation). A clear reduction in overall methylation in XaXa female ESCs and iPSCs compared with their male counterparts was also obvious when plotting the average level of autosomal methylation (Figure 2B). These data showed that XaXa iPSCs are globally hypomethylated compared with the starting female MEFs, SSEA1− and SSEA1+ reprogramming intermediates, as well as male iPSCs and female XO iPSCs, with a pattern similar to XaXa ESCs.
Figure 2.
Global Gain in Genome Methylation upon Loss of One of the Two X Chromosomes in iPSCs
(A) Histograms showing the distribution of methylation levels of 100-bp tiles captured by RRBS genome wide for the indicated male and female cell types and reprogramming intermediates. Only 100-bp tiles on autosomes, but not on sex chromosomes, with at least two CpGs at coverage 5× or greater were considered for this analysis. The methylation level ranges from 0 (no methylation) to 1 (100% methylation).
(B) Mean autosomal CpG methylation level over covered CpGs (1-kb tiles) across autosomes in indicated male and female cell types and reprogramming stages. Two-tailed unpaired t test, ∗p < 0.01. Error bars are SEM. The test was performed on male iPSCs and XaXa iPSCs only.
(C) Dot blot analysis of 5mC in three female and three male iPSC lines.
(D) Mass spectrometry analysis of 5mC in iPSCs. 5mC content is expressed as the percentage of 5mC in the total pool of cytosine for six male and five female iPSC lines. Data are presented as mean + SEM. Two-tailed unpaired t test, n = 2 experiments, ∗p < 0.0005.
(E) Mass spectrometry analysis of 5mC in iPSCs at different passages. 5mC content is expressed as the percentage of 5mC in the total pool of cytosine, normalized to the mean 5mC level in male samples in each time point analyzed. The average for 3 male and 2–3 female lines are shown for different passages. Two-tailed unpaired t test, n = 2 experiments, ∗p < 0.05, ∗∗∗p < 0.001; ns, not significant. The XaXa and XO state of the cell lines used was quantified for the same lines in Figure 1C.
To confirm the hypomethylated state of XaXa iPSCs, we performed two additional assays. First, we measured 5-methylcytosine (5mC) levels by dot blot analysis for three early-passage female iPSC lines and three male iPSC lines derived independently, in a different lab, and using a different reprogramming system (Figure 2C and Table S1). Second, we analyzed global DNA methylation levels in these same lines as well as another three independent XaXa iPSC lines and three independent male iPSCs lines by mass spectrometry (Figure 2D and Table S1). In both cases, female iPSCs showed reduced global levels of DNA methylation, validating the RRBS approach. Therefore, hypomethylation in female XaXa iPSCs does not depend on the reprogramming method used to generate iPSCs, the lab of origin, or the assay used to measure DNA methylation. These findings show that during reprogramming female cells transition toward a globally hypomethylated genome and that global hypomethylation occurs very late in reprogramming, after the SSEA1+ stage. Thus, sex chromosome content influences DNA methylation dynamics and global DNA methylation levels at the end of the reprogramming process. Consequently, the sex of the starting cells should be considered when delineating DNA methylation dynamics during reprogramming and in iPSCs.
The observation that female XO iPSCs show DNA methylation levels comparable with those of male iPSCs suggests that global female-specific hypomethylation is reversed as a result of X chromosome loss upon passage. To test this idea, we measured global 5mC and 5-hydroxymethylcytosine (5hmC) levels over time by mass spectrometry in cells propagated for several passages during which X chromosome loss takes place as determined by Tsix RNA FISH (Figure 1C). Loss of one of the X chromosomes correlated with gain of DNA methylation for both 5mC and 5hmC (Figures 2E and S2A). Thus, iPSCs display the same strong relationship between the presence of two active X chromosomes and global hypomethylation as female mouse ESCs (Habibi et al., 2013, Ooi et al., 2010, Schulz et al., 2014, Zvetkova et al., 2005). We conclude that global DNA hypomethylation correlates with the presence of two active X chromosomes during reprogramming to iPSCs.
In Addition to the Non-repetitive Genome, Hypomethylation in Female iPSCs Affects the Repetitive Genome as well as ICRs
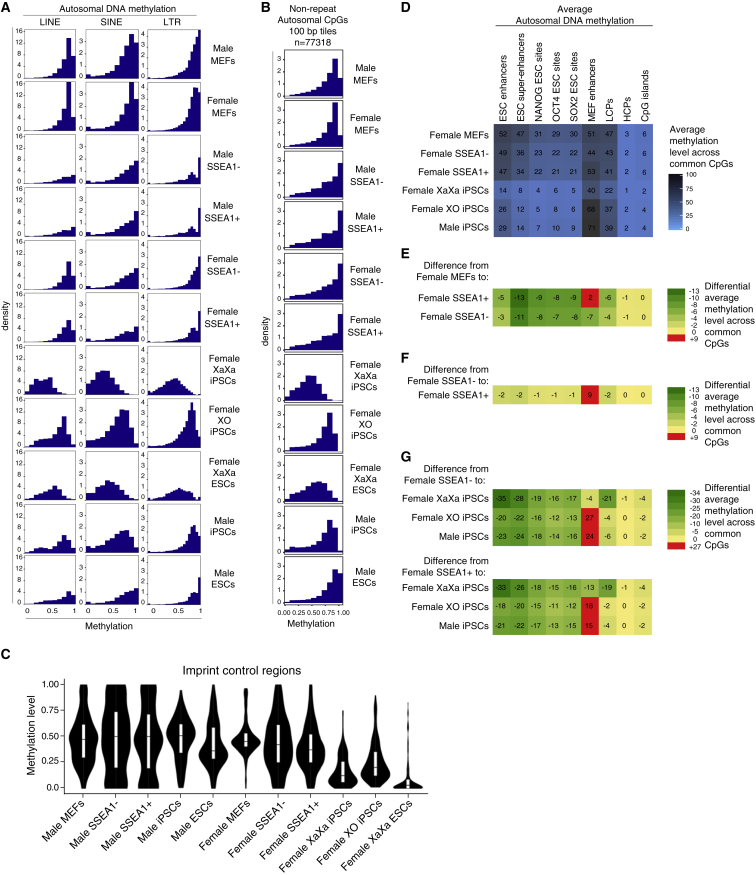
One characteristic of the female XaXa ESC genome is hypomethylation of repetitive elements (Ooi et al., 2010, Zvetkova et al., 2005). We therefore examined the distribution of methylation across repetitive elements including LINEs, short interspersed nuclear elements (SINEs), and LTRs. We found that LINEs, SINEs, and LTRs were hypermethylated in male and female MEFs, male and female SSEA1− and SSEA1+ reprogramming stages, as well as in male iPSCs and ESCs and female XO iPSCs (Figure 3A). These elements exhibited lower methylation in female XaXa iPSCs and ESCs, consistent with their XX- and pluripotency-specific hypomethylation (Figure 3A).
Figure 3.
Changes in Methylation at Repeat Elements, ICRs, and cis-Regulatory Sites during Reprogramming
(A) As in Figure 1C but for LINEs, SINEs, and LTRs.
(B) Histograms showing the distribution of methylation levels of non-repeat CpGs captured by RRBS genome wide for the indicated male and female cell types and reprogramming intermediates. Only 100-bp tiles on autosomes, but not on sex chromosomes, with at least two CpGs with coverage >5× across all cell types/reprogramming stages were filtered with repeat masker to exclude repeat regions. The methylation level ranges from 0 (no methylation) to 1 (100% methylation).
(C) Distributions of CpG methylation levels for ICRs, shown as violin plots.
(D) Average autosomal CpG methylation values for various cis-regulatory sites in female reprogramming and female XX and XO iPSCs as well as male iPSCs for comparison, as indicated. Coverage-weighted mean methylation was computed for each feature using CpGs with coverage 5× or greater. LCPs and HCPs refer to low and high CpG-density promoters, respectively.
(E) Heatmap of changes in average methylation for features shown in (D) for the transition from female MEFs to female SSEA1− and SSEA1+ sorted reprogramming subpopulations. The scale indicates the extent of loss or gain in average methylation. Numbers represent percent methylation.
(F) As in (E), except for the transition from female SSEA1− to the female SSEA1+ reprogramming stages.
(G) As in (E), except for the transition from female SSEA1− and SSEA1+ stages, respectively, to female XaXa iPSCs, female XO iPSCs, and male iPSCs, respectively. Note that the color scale is different from (E) and (F) but the numbers can be compared directly across (D) to (G).
Autosomal CpGs in non-repeat regions display a similar behavior across reprogramming stages and iPSCs as LINEs, SINEs, and LTRs (Figure 3B), demonstrating the widespread extent of the global demethylation wave very late in female reprogramming and the gain of methylation upon the loss of one of the two X chromosomes in propagating female iPSCs. To understand whether hypomethylation in XX iPSCs and ESCs affects different CpG density outside of repeats, we selected all non-repetitive DNA segments that possess a ratio of observed to expected CpG dinucleotides (i.e., CpG density) of higher than 0.55 and grouped them into bins of increasing CpG density. We found that CpG-rich regions of a CpG density between 0.55 and 0.80 were generally more methylated than those with a CpG density >0.8 regardless of cell identity (Figures S2B and S2C). Moreover, female XX iPSCs and ESCs carried less methylation regardless of CpG density (Figures S2B and S2C). Together these data indicate that the pluripotent female XaXa state alters DNA methylation levels not only of repeats but also genome wide and across different CpG densities.
We next investigated whether methylation in differentially methylated regions (DMRs) associated with ICRs was affected in female XX iPSCs or during female cell reprogramming, especially given that they are subject to hypomethylation specifically in female ESCs (Choi et al., 2017b, Hackett et al., 2013, Yagi et al., 2017). We found that for DMRs of ICRs in ESCs (Meredith et al., 2015), methylation was high across all male cell types, starting female MEFs, and the female SSEA1 reprogramming populations (Figures 3C and S2D; Table S2). ICR methylation was drastically depleted in female XX iPSCs toward levels observed in female ESCs (Figures 3C and S2D; Table S2). Thus, ICRs are largely resistant to DNA demethylation throughout reprogramming and become erased as XaXa iPSCs are established. These results obtained for iPSCs grown in S/L are reminiscent of imprint erasure upon dual ERK (mitogen-activated protein kinase/extracellular signal regulated kinases) and GSK3b (glycogen synthase kinase 3b) inhibition (2i) in XY and XX ESCs, or in S/L XX ESCs (Choi et al., 2017b, Hackett et al., 2013, Yagi et al., 2017). Furthermore, X chromosome loss in our female iPSCs did not completely restore imprint methylation, despite reacquisition of global DNA methylation (Figures 3C and S2D). Poor developmental potential has recently been attributed to imprint erasure in XX ESCs (Choi et al., 2017b, Yagi et al., 2017) and correlate with loss of Zrsr1 imprint in iPSCs (Chang et al., 2014). Here, we found evidence of imprint erasure for the imprint control region of Zrsr1 in female XX and XO iPSCs (Table S2). Together, these results indicate that imprints are not only erased in XX iPSCs but also are not re-established upon global genome remethylation in XO iPSCs, which suggests that female iPSCs, regardless of the number of Xas, may have a similarly poor developmental potential as female ESCs (Choi et al., 2017b, Yagi et al., 2017).
Methylation Changes during Female Reprogramming at cis-Regulatory Elements
Next, we focused our analysis of DNA methylation on cis-regulatory sites during female reprogramming. To this end, we determined average methylation levels (Figure 3D) in ESC-specific enhancers (Koche et al., 2011), ESC super-enhancers (Hnisz et al., 2013), binding sites for the transcription factors NANOG (N), OCT4 (O), and SOX2 (S) in ESCs (OSN binding sites) (Chronis et al., 2017), MEF-specific enhancers (Koche et al., 2011), low CpG-content promoters (LCPs), HCPs, and CpG islands. Interestingly, we found that within several of these cis-regulatory regions, changes in average methylation levels were already apparent in both SSEA1 subpopulations, and more pronounced in XaXa iPSCs (Figures 3D–3G).
To identify genomic features particularly prone to methylation changes in the SSEA1+ and SSEA1− reprogramming populations, we analyzed changes in average methylation from female MEFs to the SSEA1− and SSEA1+ reprogramming subpopulations, respectively (Figure 3E). We also determined changes in average methylation between the SSEA1− and SSEA1+ reprogramming stages and from the SSEA1− and SSEA1+ intermediates to the iPSC types (Figures 3F and 3G). Several conclusions arose from these analyses: first, we noticed a preferential loss of methylation within ESC-regulatory regions particularly for ESC super-enhancers and OSN ESC binding sites in both SSEA1+ and SSEA1− reprogramming intermediates (Figure 3E). The smaller prototypical ESC enhancers and LCPs showed fewer substantial changes in their average methylation level in the SSEA1 intermediates. Because super-enhancers play key roles for the maintenance of cell identity and contain a high number of pluripotency transcription factor binding sites (Hnisz et al., 2013), methylation changes in these regulatory regions may be of particular interest for the mechanistic understanding of somatic cell reprogramming to iPSCs. The average methylation level of ESC super-enhancers and OSN binding sites differed dramatically between MEFs and SSEA1− but only relatively little between SSEA1− and SSEA1+ cells (Figures 3D and 3E). Thus, at these sites loss of methylation takes place to a surprisingly high extent in the female SSEA1− intermediate. Together these data suggest that ESC super-enhancers and OSN ESC binding sites initiate their demethylation relatively early in female reprogramming, i.e., before SSEA1 expression is induced. This finding is consistent with the binding of the reprogramming factors within ESC super-enhancers already early in reprogramming (Chronis et al., 2017). Despite methylation changes taking place at the female SSEA1+/− stages, additional changes in average DNA methylation in ESC-specific cis-regulatory sites occurred after the SSEA1+ stage as cells move on toward the XaXa iPSC state (Figures 3D, 3F, and 3G).
Second, we found that MEF enhancers became less methylated in the SSEA1− sorted intermediate stage, whereas they slightly gained methylation in SSEA1+ intermediate relative to the starting MEFs (Figures 3D and 3E). Thus, cells that have a lowered ability to progress toward the iPSC state displayed lower methylation levels at MEF enhancers than those with an increased propensity to reprogram. These data indicate that different cis-regulatory sites are differentially affected in the SSEA1+ and SSEA1− female reprogramming stages. In male iPSCs, MEF enhancers strongly gained methylation compared with prior reprogramming stages (Figures 3D and 3G). Interestingly, in XaXa iPSCs the methylation level of MEF enhancers was reduced compared with the SSEA1+ reprogramming intermediate, consistent with the globally hypomethylated state of these cells (Figures 3F and 3G). However, these sites maintained a higher methylation level in XaXa iPSCs than in any other elements analyzed (Figure 3D).
Third, for the average methylation levels of HCPs and CpG islands, little change was apparent throughout reprogramming, consistent with constitutive hypomethylation of CpG-rich regions (Figures 3D–3G, S2B, and S2C: CpG density at 0.8 and higher).
Fourth, we observed that ESC-specific cis-regulatory sites, MEF enhancers, and LCPs were more strongly demethylated in female XaXa iPSCs compared with female XO iPSCs and male iPSCs (Figures 3D and 3G). The loss of the X chromosome in propagating female iPSCs (XO iPSCs) increased methylation within these genomic regions to an average level similar to that in male iPSCs (Figures 3D, 3F, and 3G). MEF enhancers displayed the most pronounced increase in methylation level in male iPSCs and XO iPSCs compared with XaXa iPSCs (Figures 3D and 3G), consistent with their silent state in pluripotent cells. Conversely, even in male iPSCs, ESC-specific cis-regulatory sites displayed a methylation level lower than in SSEA1 subpopulations, suggestive of greater binding by transcription factors at these regions in the pluripotent state (Figures 3D, 3F, and 3G). These data therefore indicate that global female demethylation also affects cis-regulatory elements in female iPSCs as a result of the presence of two active X chromosomes and is subsequently reversed upon X chromosome loss, but the extent of this reversal differs among cis-regulatory sites.
In summary, we conclude that methylation changes at cis-regulatory regions already begin before the induction of the pluripotent state in female reprogramming, and that methylation dynamics during reprogramming and in propagated female iPSCs differ according to the type of genomic elements considered and the number of active X chromosomes. In intermediate female reprogramming stages, changes are enriched at cis-regulatory regions that are critical to maintain the somatic and pluripotency transcriptional programs. Our results indicate that somatic and pluripotency enhancers already adopt some of the characteristics of their pluripotency state early in female reprogramming, before the global and female-specific hypomethylation wave occurs in the process.
Single-CpG-Level Methylation Changes during Reprogramming
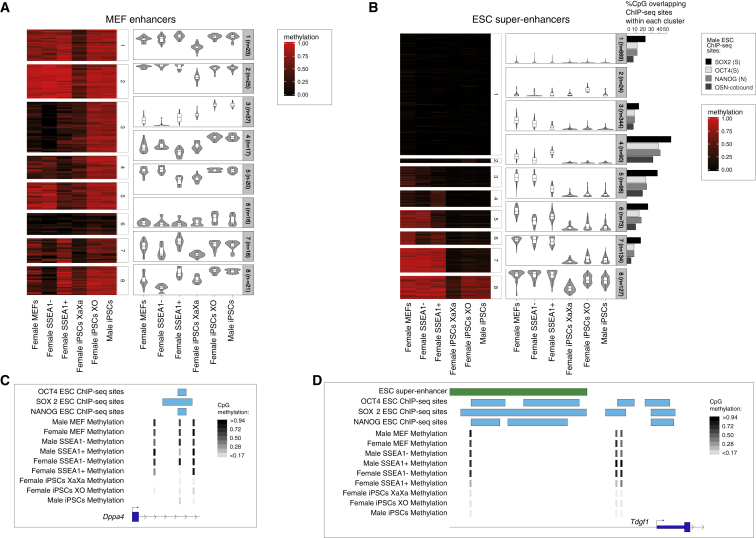
To further analyze the methylation changes within cis-regulatory sites, we next turned to the analysis of single-CpG-resolution methylation measurements provided by RRBS to define methylation changes during reprogramming. This analysis enabled us to assess changes in DNA methylation at single CpG resolution, which could be masked in averages. To identify single-CpG-level methylation variation, we performed k-means clustering of all differentially methylated CpGs within selected genomic features, including MEF enhancers, ESC super-enhancers, ESC enhancers, and NANOG binding sites in ESCs, to group single CpGs with similar changes during female reprogramming and in various iPSCs (Figures 4A, 4B, S3A, and S3B). This analysis further defined the dynamic regulation of DNA methylation with single-CpG resolution within enhancers during female reprogramming and revealed expected as well as unexpected single-CpG-level DNA methylation changes.
Figure 4.
Single-CpG-Based Analysis of Methylation during Reprogramming
(A) Heatmap showing k-means clustering of single-CpG methylation values located within MEF enhancers across all chromosomes in the indicated cell types and reprogramming stages (left). Violin plots (right) show the distribution of CpG methylation levels for each cluster across all samples. The number of CpGs in each cluster is also given.
(B) As in (A), but for ESC super-enhancers. In addition, the proportion of CpGs overlapping O, S, N ESC binding sites, or those sites co-bound by OSN (OSN) in ESCs, within each cluster, is indicated.
(C) RRBS data depicting a CpG overlapping with an OSN ESC binding site at the Dppa4 locus. The data show a focal loss of DNA methylation in the SSEA1+ intermediate but not in starting MEFs or SSEA1− sorted cells. The demethylated region is even further expanded in PSCs.
(D) As in (C), except for the Tdgf1 locus where CpG demethylation in specific to the female SSEA1+ intermediates.
Considering MEF enhancers (Figure 4A), we found that single CpGs in several clusters gained methylation toward the pluripotent state, as expected from averages measurements. However, we also resolved different clusters of single CpGs with different methylation kinetics during female reprogramming, becoming demethylated in the SSEA1− cell population and hypermethylated in the SSEA1+ subpopulation (cluster 8), or more methylated only in XO and XY PSCs (clusters 3, 4, and 6). Other CpGs were ectopically demethylated in female XaXa iPSCs (clusters 1 and 2) or in XaXa iPSCs and SSEA1− and SSEA1+ intermediates, respectively (clusters 5 and 7), which may contribute to the reprogramming state of these cells. We conclude that CpG-specific and reprogramming stage-specific changes in DNA methylation take place throughout female reprogramming. This highlights the importance of examining single-CpG-level DNA methylation during the reprogramming process.
Within ESC super-enhancers during female cell reprogramming (Figure 4B), a large proportion of CpGs was constitutively hypomethylated (clusters 1 and 2). The remaining CpGs covered in our RRBS analysis in all samples started with higher methylation in MEFs, which decreased toward XaXa iPSCs (clusters 3–8), as expected from average measurements (Figure 3C). For a large fraction of these CpGs, methylation was detected in XO and male iPSCs, and their methylation level was lower in XaXa iPSCs compared with XO or male iPSCs (clusters 5–8), indicating that they were subject to female-specific hypomethylation in XaXa iPSCs, which is reset to the male iPSC level upon loss of the second X chromosome. A decrease in methylation was already apparent in the female SSEA1− or SSEA1+ subpopulations, respectively, or both (clusters 3–6), indicative of early loss of methylation of a portion of CpG sites within ESC super-enhancers (Figure 4B). We also found a CpG associated with the pluripotency gene Dppa4 that lost methylation in the SSEA1+ intermediates, but not in SSEA1− cells or in neighboring CpGs (Figure 4C). An additional example in SSEA1+ intermediates was a cis-regulatory site of the Tdgf1 gene located within an ESC super-enhancer, which was demethylated in female SSEA1+ reprogramming intermediates but not in SSEA1− cells (Figure 4D). Finally, cluster 4 had, unexpectedly, increased methylation specifically in female SSEA1+ cells (Figure 4A). While the relevance of this observation is not clear, many of these sites were associated with known pluripotency genes such as Dppa3, Esrrb, Nanog, Otx2, Sox2, Tbx3, Utf1, and Nr5a2 (not shown). We made similar observations for ESC enhancers (Figures S3A and S3B).
To detect male DNA methylation dynamics, we also performed k-means clustering with male reprogramming samples only (Figures S3C–S3E). We were not able to directly compare the same CpG sites in male and female reprogramming intermediates due to the limited overlap in RRBS datasets. Nevertheless, we could detect single-CpG methylation changes in male reprogramming with a similar pattern as for female reprogramming with the exception of the global, female-specific DNA hypomethylation (Figures 4C and S3C–S3E). We conclude that CpG-specific and reprogramming stage-specific changes in enhancer DNA methylation take place during male and female reprogramming to iPSCs.
Dynamic Methylation in ESC Super-Enhancers and ESC Enhancers Occurs at Pluripotency Transcription Factor Binding Sites in Reprogramming Intermediates Irrespective of Sex
To define whether changes in DNA methylation during reprogramming correlate with pluripotency transcription factor binding sites, we assessed the overlap of ESC binding sites of OCT4, SOX2, and NANOG, with our female and male CpG methylation ESC super-enhancer and ESC enhancer clusters (Figures 4B, S3A, S3D, and S3E). Interestingly, ESC super-enhancer CpGs in clusters 1 and 3–7, which displayed different trajectories of methylation changes from MEFs to pluripotency, were located in regions bound by OCT4, SOX2, and NANOG in ESCs (Figure 4B). Of note, CpGs that become demethylated in SSEA1− cells and/or SSEA1+ cells (clusters 4–6) are most strongly enriched for OSN binding in ESCs. OSN binding sites in ESC super-enhancers and ESC enhancers also showed single-CpG DNA methylation changes in male reprogramming intermediates (Figure S3D). Surprisingly, methylation gains in SSEA1+ intermediates correlated both in male and female reprogramming with strong pluripotency factor transcription factor binding in iPSCs (cluster 4 in female reprogramming, Figure 4B; and cluster 6 in male reprogramming, Figure S3D; also see cluster 6, Figure S3D; cluster 1, Figure S3E). Given the CpG methylation changes at OSN binding sites within ESC super enhancers, methylation dynamics similar to those of CpGs in ESC super-enhancers were seen for CpGs within ESC binding sites of the pluripotency factor NANOG (Figure S3B). Thus, at ESC super-enhancers and ESC enhancers, several ESC binding sites of OSN change methylation in different directions early in reprogramming.
We conclude that changes in single-CpG methylation occur at cis-regulatory elements such as ESC enhancers at the ESC binding sites of pluripotency transcription factors in a reprogramming stage-specific manner, irrespective of sex. Thus, changes in methylation early in reprogramming appear to be initiated at focal sites of high putative regulatory function, in both male and female cells, suggesting important consequences for our mechanistic understanding of reprogramming of cell identity.
Discussion
Our results indicate that the analysis of DNA methylation in reprogramming needs to consider the sex chromosome genotype. Reprogramming to iPSCs leads to the global hypomethylation of the female genome, which is not seen in male iPSCs. Since two active X chromosomes are acquired late during female cell reprogramming (Maherali et al., 2007), and hypomethylation is seen in female XX iPSCs but not in late female SSEA1+ intermediates, we propose that the reactivation of the inactive X chromosome triggers female-specific global hypomethylation during female cell reprogramming. Importantly, global hypomethylation of XX iPSCs does not depend on the reprogramming method used. For female mouse ESCs, it was shown that loss of one of the two active X chromosomes triggers a rapid remethylation of the genome to the male ESC level (Zvetkova et al., 2005). Our results are in line with similar events happening in female iPSCs during extended passaging, but not during reprogramming. Mechanistically, hypomethylation is likely due to the double dose of one or more X-linked factors responsible for inhibiting DNA methyltransferases and inhibiting pro-differentiation signals including the ERK and GSK3b pathways (Schulz et al., 2014). One such candidate is Dusp9, recently shown to contribute to the control of genome hypomethylation in female XX ESCs (Choi et al., 2017a). We envision a model in which XCR leads to a double dose of Dusp9, thereby inhibiting DNA methyltransferases (Choi et al., 2017a), followed by decreased Dusp9 dosage due to X chromosome loss, resulting in genome remethylation. In summary, XCR followed by X chromosome loss in female iPSCs explains the sex-specific DNA methylation events seen in our study. These observations might explain some of the changes described in a recent study of DNA methylation during reprogramming that did not examine the state of the X chromosome (Milagre et al., 2017). We have not excluded the possibility that X chromosome loss takes place as a result of global DNA hypomethylation. Collectively, our results highlight the sex chromosome content of a somatic cell as a critical factor to consider for studying DNA methylation changes during reprogramming. Importantly, it is not just sex that matters, but the number of active X chromosomes relative to autosomes (X chromosome dosage).
Several studies have been carried out to assess the methylation status of ICRs in iPSCs (Chang et al., 2014, Kim et al., 2013, Stadtfeld et al., 2010, Stadtfeld et al., 2012, Takikawa et al., 2013). Most agree that imprints can become deregulated in mouse iPSCs, but no consensus exists on the extent and variability of imprint erasure in iPSCs (Takikawa et al., 2013). Our results suggest that some of the reported differences in ICR methylation could be explained by differences in X chromosome status and sex and that imprints in female iPSCs are prone to deregulation. Notably, we found evidence of imprint erasure in female XX iPSCs, which was not fully re-established after global methylation gain due to X chromosome loss. Our results are consistent with recent reports in ESCs in which imprint erasure driven by MAPK inhibition is irreversible (Choi et al., 2017b, Hackett et al., 2013, Yagi et al., 2017). Imprint erasure in female iPSCs could therefore have important functional implications for the differentiation and function of female iPSCs and the specialized cell types derived from them. One prediction is that female iPSCs, in early or late passage, will generate chimeras with decreased efficiency over male iPSCs, which remains to be tested.
Our discovery of changes in DNA methylation at intermediate reprogramming stages is an important step toward understanding erasure of epigenetic memory during cell fate reprogramming. Such changes appear to be, at least in part, independent of sex, in agreement with a recent report (Milagre et al., 2017). Partial DNA demethylation in SSEA1+ intermediates was also confirmed using traditional bisulfite sequencing analysis for Dppa3 (Figure S4A). Because our male and female SSEA1 datasets were generated independently, a direct comparison is difficult, warranting a careful comparison of male and female epigenomes throughout reprogramming and in iPSCs. Nevertheless we found instances in which a single CpG within an ESC super-enhancer became hypomethylated in both female SSEA1− and SSEA1+ intermediates, as well as in female iPSCs, but not in female MEFs, for example bordering the miR290–295 cluster (Figure S4B).
Our study reveals that key regulatory regions associated with the control of cell identity genes are prone to changes in DNA methylation in reprogramming intermediates in both male and female cells. Previous reports using microarrays or bisulfite sequencing showed that promoter methylation is reset at a very late stage of reprogramming (Knaupp et al., 2017, Polo et al., 2012), consistent with analysis of methylation during human reprogramming (Cacchiarelli et al., 2015). Another study revealed global indiscriminate remethylation of somatic enhancers in human iPSCs regardless of somatic origin (Nissenbaum et al., 2013), which will be interesting to also test in the mouse. Nevertheless, we found that many CpGs, particularly those in enhancer elements, which are thought to play key roles for the maintenance of cell identity (Hnisz et al., 2013), change their methylation status earlier in reprogramming. This observation is in agreement with the finding that changes in methylation at intermediate reprogramming stages take place for several promoters and ESC binding sites of pluripotency transcription factors (Lee et al., 2014). We speculate that these methylation changes coincide with progressive changes in chromatin state and transcription factor access, allowing, for instance, the engagement of the reprogramming factors at their ESC target sites at intermediate stages of reprogramming. This idea is supported by early engagement of a subset of sites by OCT4, SOX2, and C-MYC at pluripotency super-enhancers at 48 hr of reprogramming (Chronis et al., 2017).
As reprogramming to iPSCs proceeds through multiple stages (Knaupp et al., 2017, Pasque et al., 2014), we anticipate that the isolation of reprogramming intermediates in these distinct stages of reprogramming combined with genome-wide analysis of methylation at the population or single-cell level will reveal additional dynamic methylation events during male and female somatic cell reprogramming to pluripotency. The importance of purifying cells at specific stages of reprogramming is underscored by our finding that the average loss of methylation at OSN transcription factor binding sites in SSEA1− as opposed to SSEA1+ intermediates is similar on average yet occurs at different genomic locations. Moreover, it is also possible that several cycles of DNA methylation/demethylation take place at a given site during reprogramming. A detailed analysis of DNA methylation dynamics in comparison with changes in histone modifications and reprogramming factor binding at various male and female reprogramming stages should also reveal how and if DNA methylation affects reprogramming events.
Our findings also may have important implications for the understanding of human naive pluripotency in relation to regenerative medicine applications and disease modeling, given that naive-like human ESCs show global hypomethylation and imprint erasure (Theunissen et al., 2016). The link between the status of the X chromosomes and human pluripotency is also of considerable current interest. Human PSCs grown in primed conditions have an inactive X chromosome, which is unstable due to erosion of X inactivation (Mekhoubad et al., 2012). Growing human ESCs in naive-like conditions induces XCR and the cells undergo non-random X chromosome inactivation when induced to differentiate (Theunissen et al., 2016). Whether X chromosome dosage influences the transcriptional and epigenomic landscape of human PSCs in the naive state is not clear. There have also been reports of sex-specific differences in gene expression and programmed cell death in human PSCs cultured in the primed state (Bruck et al., 2013, Ronen and Benvenisty, 2014). Understanding the interplay between the activity of sex chromosomes and pluripotency provides an exciting avenue for future studies.
Experimental Procedures
Cell Culture and Mice
ESCs were grown in ESC culture medium (Supplemental Experimental Procedures) and MEFs were cultured in DMEM with the same components as for ESC medium except for leukemia inhibitory factor and with 10% fetal bovine serum. All animal work carried out in this study is covered by a project license approved by the KU Leuven Animal Ethics Committee.
Reprogramming Stages
Reprogramming and the isolation of reprogramming intermediates was previously described (Pasque et al., 2014). iPSCs were picked from day-21 reprogramming cultures and grown in ESC medium on feeders. Table S1 lists all the cell lines, replicates, reprogramming method, and passage number of the primary and established cell lines used in this study. Additional information is included in Supplemental Experimental Procedures.
RRBS and Data Analysis
The generation of RRBS data used in this study was described previously (Pasque et al., 2014). At least two RRBS biological replicate libraries were constructed for each cell type analyzed, and up to four biological replicates for female MEFs and female ESCs were utilized (Table S1). Some of these datasets (GEO: GSE58109) were used previously to determine the methylation state of CpG islands on the X chromosome in different reprogramming stages (Pasque et al., 2014).
Author Contributions
Conception and Design, V.P. and K.P.; Experiments, V.P., C.C., R.K., P.P., J.S., L.V., and A.S.D.; Formal Analysis, R.K., V.P., J.L., and K.P.; Data Curation, R.K., C.C., G.B., and J.S.; Writing, V.P. and K.P., with input from all authors; Resources, K.P., V.P., and A.M.; Supervision, K.P., V.P., and A.M.
Acknowledgments
We thank members of the Plath and Pasque labs for feedback on the manuscript, F. Codrea and J. Scholes at the UCLA Broad Stem Cell Center FACS Core, B. Ghesquière at the KU Leuven Metabolomics Core, K. Koh and J. Vande Velde for help with dot blot analysis, and N. De Geest for help with mice. C.C. was supported by CIRM (TG2-01169) and Leukemia and Lymphoma Research (#10040) fellowships, G.B. by Whitcome, Dissertation Year, and QCB fellowships from UCLA, and J.L. by the UCLA Tumor Cell Biology Training Program (USHHS Ruth L. Kirschstein Institutional National Research Service Award #T32 CA009056). V.P. was supported by the CIRM training grant TG2-01169. Research in the Pasque lab is supported by The Research Foundation – Flanders (FWO) (Odysseus Return grant G0F7716N) and the KU Leuven Research Fund (BOFZAP starting grant StG/15/021BF, C1 grant C14/16/077, and project financing PF/10/019). A.M. is a New York Stem Cell Foundation Robertson Investigator. A.M. was supported by the New York Stem Cell Foundation and NIH grant P01GM099117. K.P. was supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, the Jonsson Comprehensive Cancer Center at UCLA, and the NIH (P01 GM099134, R01 GM115233) and is a Faculty Scholar of the Howard Hughes Medical Institute.
Published: April 19, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.03.019.
Contributor Information
Vincent Pasque, Email: vincent.pasque@kuleuven.be.
Kathrin Plath, Email: kplath@mednet.ucla.edu.
Accession Numbers
The GEO accession numbers for the RRBS data used in this study are GEO: GSE58109 and GSE111042.
Supplemental Information
References
- Bruck T., Yanuka O., Benvenisty N. Human pluripotent stem cells with distinct X inactivation status show molecular and cellular differences controlled by the X-Linked ELK-1 gene. Cell Rep. 2013;4:262–270. doi: 10.1016/j.celrep.2013.06.026. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Trapnell C., Ziller M.J., Soumillon M., Cesana M., Karnik R., Donaghey J., Smith Z.D., Ratanasirintrawoot S., Zhang X. Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell. 2015;162:412–424. doi: 10.1016/j.cell.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G., Gao S., Hou X., Xu Z., Liu Y., Kang L., Tao Y., Liu W., Huang B., Kou X. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Clement K., Huebner A.J., Webster J., Rose C.M., Brumbaugh J., Walsh R.M., Lee S., Savol A., Etchegaray J.-P. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell. 2017;20:706–719.e7. doi: 10.1016/j.stem.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Huebner A.J., Clement K., Walsh R.M., Savol A., Lin K., Gu H., Di Stefano B., Brumbaugh J., Kim S.Y. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H.D., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Dietmann S., Murakami K., Down T.A., Leitch H.G., Surani M.A. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.J., Choi H.-W., Jang H.J., Chung H.M., Araúzo-Bravo M.J., Scholer H.R., Do J.T. Conversion of genomic imprinting by reprogramming and redifferentiation. J. Cell Sci. 2013;126:2516–2524. doi: 10.1242/jcs.122754. [DOI] [PubMed] [Google Scholar]
- Knaupp A.S., Buckberry S., Pflueger J., Lim S.M., Ford E., Larcombe M.R., Rossello F.J., de Mendoza A., Alaei S., Firas J. Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell. 2017;21:834–845.e6. doi: 10.1016/j.stem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Koche R.P., Smith Z.D., Adli M., Gu H., Ku M., Gnirke A., Bernstein B.E., Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-S., Shin J.-Y., Tonge P.D., Puri M.C., Lee S., Park H., Lee W.-C., Hussein S.M.I., Bleazard T., Yun J.-Y. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith G.D., D'Ippolito A., Dudas M., Zeidner L.C., Hostetter L., Faulds K., Arnold T.H., Popkie A.P., Doble B.W., Marnellos G. Glycogen synthase kinase-3 (Gsk-3) plays a fundamental role in maintaining DNA methylation at imprinted loci in mouse embryonic stem cells. Mol. Biol. Cell. 2015;26:2139–2150. doi: 10.1091/mbc.E15-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagre I., Stubbs T.M., King M.R., Spindel J., Santos F., Krueger F., Bachman M., Segonds-Pichon A., Balasubramanian S., Andrews S.R. Gender differences in global but not targeted demethylation in iPSC reprogramming. Cell Rep. 2017;18:1079–1089. doi: 10.1016/j.celrep.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissenbaum J., Bar-Nur O., Ben-David E., Benvenisty N. Global indiscriminate methylation in cell-specific gene promoters following reprogramming into human induced pluripotent stem cells. Stem Cell Reports. 2013;1:509–517. doi: 10.1016/j.stemcr.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.K., Wolf D., Hartung O., Agarwal S., Daley G.Q., Goff S.P., Bestor T.H. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Plath K. X chromosome reactivation in reprogramming and in development. Curr. Opin. Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasque V., Tchieu J., Karnik R., Uyeda M., Sadhu Dimashkie A., Case D., Papp B., Bonora G., Patel S., Ho R. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen D., Benvenisty N. Sex-dependent gene expression in human pluripotent stem cells. Cell Rep. 2014;8:923–932. doi: 10.1016/j.celrep.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Schulz E.G., Meisig J., Nakamura T., Okamoto I., Sieber A., Picard C., Borensztein M., Saitou M., Blüthgen N., Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Shirane K., Kurimoto K., Yabuta Y., Yamaji M., Satoh J., Ito S., Watanabe A., Hayashi K., Saitou M., Sasaki H. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell. 2016;39:87–103. doi: 10.1016/j.devcel.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S.K., Kim S.Y., Bestor T.H., Shioda T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takikawa S., Ray C., Wang X., Shamis Y., Wu T.-Y., Li X. Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell Res. 2013;11:861–873. doi: 10.1016/j.scr.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T.W., Friedli M., He Y., Planet E., O'Neil R.C., Markoulaki S., Pontis J., Wang H., Iouranova A., Imbeault M. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi M., Kishigami S., Tanaka A., Semi K., Mizutani E., Wakayama S., Wakayama T., Yamamoto T., Yamada Y. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- Zvetkova I., Apedaile A., Ramsahoye B., Mermoud J.E., Crompton L.A., John R., Feil R., Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.