Abstract
Advances in metabolic engineering and synthetic biology have facilitated the manufacturing of many valuable-added compounds and commodity chemicals using microbial cell factories in the past decade. However, due to complexity of cellular metabolism, the optimization of metabolic pathways for maximal production represents a grand challenge and an unavoidable barrier for metabolic engineering. Recently, cell-free protein synthesis system (CFPS) has been emerging as an enabling alternative to address challenges in biomanufacturing. This review summarizes the recent progresses of CFPS in rapid prototyping of biosynthetic pathways and genetic circuits (biosensors) to speed up design-build-test (DBT) cycles of metabolic engineering and synthetic biology.
Keywords: Cell-free protein synthesis, Metabolic pathway optimization, Genetic circuits, Metabolic engineering, Synthetic biology
1. Introduction
Metabolic engineering and synthetic biology are one of the most promising solutions to address sustainability and global climate change challenges through the development of efficient cell factories for producing fuels, chemicals, and pharmaceutics. Introduction of a biosynthetic pathway containing multiple heterologous genes is generally the first step to enable microbial synthesis. For practical applications, pathway gene expression levels must be carefully fine-tuned and balanced to maximize product titer, rate, and yield (TRY) [1]. However, iterative design-build-test (DBT) cycles are generally required to optimize metabolic pathways, making the development of efficient cell factories rather time-consuming. Although numerous synthetic biology tools have been developed, such as gene copy number tuning [2,3] and combinatorial transcriptional regulation [4], the build and test of the combinatorial biosynthetic pathway library remains a grand challenge for metabolic engineering. For example, it is reported that more than 100 person-years are needed to commercialize biobased 1,3-propanediol [1]. Therefore, the development of novel enabling synthetic biology tools to accelerate DBT cycles will be critical for the construction and optimization of microbial cell factories.
Cell-free protein synthesis system (CFPS) has been widely used for recombinant protein expression, particularly toxic proteins and membrane proteins that are difficult to express in vivo [[5], [6], [7], [8], [9]]. Recently, CFPS is further developed as an enabling platform for rapid prototyping of biosynthetic pathways and genetic circuits to address the challenges in metabolic engineering and synthetic biology. Compared with the conventional in vivo systems, CFPS activates biological machineries without the boundary of cell membranes and cell walls, and the open environment allows for direct monitoring and manipulation of transcription, translation and metabolism [10]. In addition, CFPS uses linear DNAs (i.e. PCR products) as templates for transcription, which bypasses the time-consuming and laborious gene-cloning and microorganism transformation steps and allows rapid prototyping in a high-throughput manner [11]. Different from the in vivo strategies by manipulating the complex transcription and translation machineries, metabolic pathway gene expression levels can be simply controlled by adjusting linear DNA concentrations supplemented to CFPS with coupled transcription-translation reactions under no resource limitation conditions [11]. Currently, CFPS has been successfully applied to the construction and optimization of metabolic pathways [[12], [13], [14], [15], [16], [17]] and genetic circuits [[18], [19], [20], [21], [22]]. It has been reported that the CFPS can reduce the time to build metabolic pathways and genetic circuits from days to hours. Notably, biosensors based on the optimized genetic circuits hold great potentials for rapidly testing the constructed metabolic pathway libraries in both in vivo and in vitro metabolic engineering systems in a high throughout manner [23,24]. The combined rapid prototyping of metabolic pathways (build) and genetic circuits (test) open unique opportunities to accelerate DBT cycles for metabolic engineering and synthetic biology (Fig. 1).
Fig. 1.
CFPS as an enabling platform to accelerate design-build-test cycles of metabolic engineering and synthetic biology.
As protein synthesis in CFPS has been reviewed in detail [[25], [26], [27]], this review covers the most recent update on the applications of CFPS in rapid prototyping for metabolic engineering and synthetic biology. More specifically, we focus on the construction and optimization of metabolic pathways and genetic circuits (biosensors) using CFPS. The methods to prepare the CFPS extracts and the corresponding energy regeneration systems will be briefly reviewed as well. Finally, the challenges and future perspectives on CFPS for metabolic engineering and synthetic biology applications will be discussed.
2. Construction of CFPS systems
CFPS systems have been developed for decades due to its versatility and broad applications [26]. Transcription-translation process can be accomplished by cell extracts from Escherichia coli and some other organisms [27]. Due to the high efficiency, flexibility, and low cost, E. coli CFPS system is the most commonly used. The traditional E. coli S30 extract preparation method have been simplified and streamlined to decrease the cost while maintain the productivity [28,29]. Notably, most of the CFPS systems rely on T7 bacteriophage mechanism (T7 promoter and T7 RNA polymerase pair), enabling high yield production of recombinant proteins [28,29]. Recently, a more versatile and flexible E. coli crude cell extract was developed to allow expression and regulation using both endogenous (i.e. sigma70-based promoters) and exogenous (i.e. T7 promoter) transcription and translation mechanisms [30]. In other words, the new system maintained high yield expression capabilities of existing CFPS systems, but also preserve endogenous regulatory mechanisms for metabolic engineering and synthetic biology applications.
Because transcription and translation are both energy-intensive processes, it is critical to develop an inexpensive and durable energy regeneration system to maintain efficient expression of heterologous genes and metabolic pathways. The simplest method is to add high energy source compounds directly to fuel in vitro transcription and translation, such as phosphoenolpyruvate (PEP) and creatine phosphate (CP) [26]. Unfortunately, these high energy phosphate bond containing molecules can be degraded rapidly by phosphatase in the cell extract. One strategy is to feed PEP periodically to regenerate adenosine triphosphate (ATP), which extended protein synthesis from ∼20 min to at least 80 min and increased the production of recombinant proteins accordingly [31]. However, the accumulation of inorganic phosphate in the reaction mixture inhibited protein synthesis. A new approach for ATP regeneration without the accumulation of inorganic phosphate was developed by introducing Pediococcus sp. pyruvate oxidase [32], converting pyruvate to acetyl phosphate. The released acetyl phosphate can be used for ATP regeneration by endogenous acetyl kinase. Later, glucose [33] and glycolytic intermediates (fructose-1,6-bisphosphate [34]) were determined to be cheap and efficient energy sources for ATP regeneration (PANOx system) without the accumulation of inorganic phosphate. In addition, maltose [35], maltodextrin [36], and soluble starch [37] were also found to be good energy sources for CFPS, which could not only allow recycling of inorganic phosphate, but also maintain a relatively homeostatic environment with stable pH.
3. CFPS driven metabolic engineering (CFPS-ME)
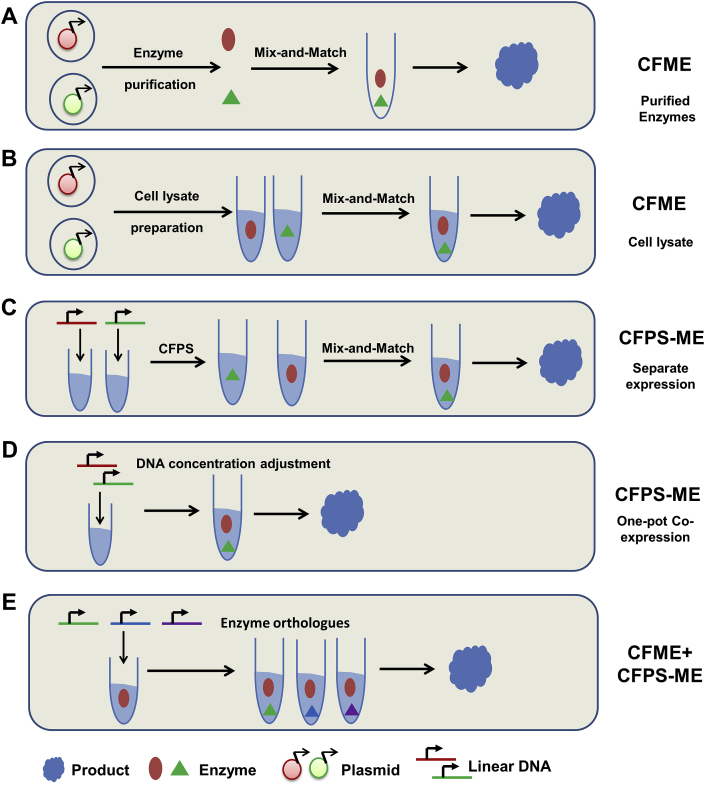
With the development of metabolic engineering and synthetic biology, cell-free metabolic engineering (CFME) has been constructed to produce biomaterials, biofuels, and drug precursors [38]. Initially, CFME was developed by assembling a metabolic pathway using purified enzymes (Fig. 2A). For example, fatty acid biosynthetic pathway was reconstituted in vitro and used to investigate and engineer fatty acid production quantitatively [39]. However, in vivo protein expression and purification of every pathway protein are laborious and time-consuming. To speed up DBT cycles for metabolic engineering, Jewett group [12] constructed a new CFME framework based on crude cell lysate for rapid building and optimizing biosynthetic pathways (Fig. 2B). In this framework, each pathway gene was overexpressed individually in E. coli, which was used as the source strain to make crude cell lysate. Then the CFME crude lysate was mixed in a combinatorial manner to initiate DBT cycles, exploring the pathway design space with the highest efficiency (product yield).
Fig. 2.
CFPS enabled rapid prototyping of metabolic pathways. Cell-free metabolic engineering (CFME) using purified enzymes (A) and crude cell lysate (B) as well as CFPS driven metabolic engineering (CFPS-ME, C and D) have demonstrated applications in the construction and optimization of biosynthetic pathways. (E) CFME and CFPS-ME can be combined for pathway enzyme discovery.
Readers of interest are directed to recent reviews on CFME based on purified enzymes or crude cell lysates [40]. This review mainly focuses on CFPS-ME, where pathway genes are directly expressed in CFPS for subsequent biocatalysis. In other words, gene cloning, bacterial transformation, in vivo protein expression, cell lysate preparation, and/or protein purification are all eliminated, further improving the speed of DBT cycles for metabolic engineering and synthetic biology. Pathway genes can be expressed individually (Fig. 2C) or in an all-in-one manner (Fig. 2D) in CFPS. In the former case, each pathway gene was expressed separately, and subsequent pathway optimization was carried out in a mix-and-match fashion. In the latter case, all pathway genes were added to CFPS to express the whole metabolic pathway in one pot, and the expression level of pathway genes was fine-tuned by controlling the amount of DNA supplemented into CFPS.
The first example to combine CFPS with metabolic engineering is the efforts to reconstitute the in vitro UDP-N-acetylglucosamine (UDP-GlcNAc) biosynthetic pathway, including a glucokinase (YqgR), an N-acetyl-glucosamine-phosphate mutase (Agm1), and an N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) [17]. By simply mixing the CFPS individually expressed pathway proteins, the 3-step biosynthetic pathway was successfully constructed to produce UDP-GlcNAc from GlcNAc. Co-express of yqgR, agm1, and glmU in CFPS was also performed to reconstitute the UDP-GlcNAc biosynthetic pathway in one pot reaction. The same group further extended the construction of a 6-step biosynthetic pathway leading to the production of UDP-N-acetylmuramyl pentapeptide, a precursor of the bacterial peptidoglycan [16]. All the metabolic pathway enzymes (murA, murB, murC, murD, murE, and murF) were expressed simultaneously in CFPS, with the final product successfully detected. Although metabolic pathway optimization by mix-and-match or adjusting DNA concentration was not attempted, the successful reconstitution of the whole biosynthetic pathway in CFPS opens new opportunities for metabolic engineering and synthetic biology applications.
Recently, Jewett group systematically evaluated the potential of CFPS for rapid prototyping of biosynthetic pathways [12]. More specifically, n-butanol biosynthesis, including 11 steps of endogenous metabolism (glycolysis, from glucose to acetyl-CoA) and 6 steps of exogenous biosynthesis (Clostridia n-butanol fermentative pathway containing AtoB, Hbd, Crt, Ter, and AdhE, from acetyl-CoA to n-butanol) was adopted as a case study for both CFME and CFPS-ME. In CFME methods, recombinant E. coli strains with each pathway gene overexpressed individually were used to make crude cell lysates, and mix-and-match was then performed to optimize n-butanol biosynthesis, which increased n-butanol production for ∼3-fold (from 0.51 g/L to 1.4 g/L). To further accelerate DBT cycles for metabolic engineering, the authors tried to integrate CFPS into metabolic engineering (CFPS-ME). Initially, no n-butanol production was detected with all 5 pathway genes expressed in CFPS. Rapid pathway prototyping and debugging identified the expression of AdhE as rate-limiting. By increasing the concentration of the AdhE plasmid, n-butanol production in CFPS was restored. However, the production in CFPS-ME (∼0.5 g/L) was still much lower than that in CFME. Later it was found that the performance of biosynthetic pathways was negatively affected by CFPS integration, mainly caused by the accumulation of inorganic phosphate during in vitro protein synthesis. The use of non-phosphorylated energy sources decreased phosphate accumulation and increased n-butanol production [15].
Wu et al. also took advantage of CFPS for rapid prototyping of 1,4-butanediol (BDO) biosynthesis [13]. CFPS was demonstrated as a platform for exploring the design space of metabolic pathways. CFPS was not only used to verify pathway gene expression and pathway functionality, but also identified the rate-limiting step of the BDO pathway (the conversion of 4-hydroxybutyrate to downstream metabolites). Combinatorial optimization of various enzyme expression levels and increasing downstream enzyme expression levels dramatically improved BDO production [13]. Similarly, CFPS was used to explore the design space of the violacein pathway [41]. The buildup of prodeoxyviolacein intermediate indicates unbalanced pathway gene expression levels. Violacein production was improved with higher vioC and vioD expression levels (higher VioC and VioD DNA concentrations). A following-up study from the same group carried out system-level studies of CFPS as an enabling platform for metabolic engineering and synthetic biology applications [14]. The exploration of the design space of the BDO pathway allowed rapid tuning of pathway enzyme expression levels and screening enzyme variants with improved catalytic activities for improved product yield. For the first time, it was demonstrated that CFPS-ME results can be directly applied to in vivo strain development.
The high efficiency of CFME and rapid DBT cycle of CFPS-ME can be combined for enzyme discovery (Fig. 2E). Taking the n-butanol biosynthetic pathway as an example, four enzymes in the pathway has been heterologously overexpressed in the crude cell lysate source strains, DNAs (either plasmids or linear PCR products) encoding orthologs of the lacked enzyme were supplemented to CFPS to initiate DBT cycles. In a few hours, the authors studied 4 Ter and 3 AdhE orthologs for n-butanol production by integrating CFME and CFPS-ME [12].
4. CFPS for the construction and optimization of biosensors and genetic circuits
Although advances in synthetic biology and metabolic engineering have made it possible to create large populations of metabolic pathway variants and strain mutants (build), a bigger challenge for metabolic engineers is to find the best producing strain from millions of possibilities (test). Biosensors that can detect intracellular and extracellular metabolites and signals have found broad applications and drawn tremendous attention in metabolic engineering [23,24]. Because natural biosensors are adapted to microorganism evolution rather than overproduction of any targeted metabolite, engineering of biosensing parameters are generally required for practical metabolic engineering applications, such as increasing the detection limits and dynamic ranges, altering detection specificities, and improving signal/noise ratios [23]. Considering the open nature and rapid prototyping capabilities, CFPS serves an important platform to explore a larger design space for the construction and optimization of biosensors and genetic circuits. While biosensors with diverse mechanisms and metabolic engineering applications have been reviewed before [23,24], we focus on the application of CFPS in the construction and optimization of biosensors based on transcription factors (TF) and RNAs.
4.1. CFPS for rapid prototyping of TF based biosensors and genetic circuits
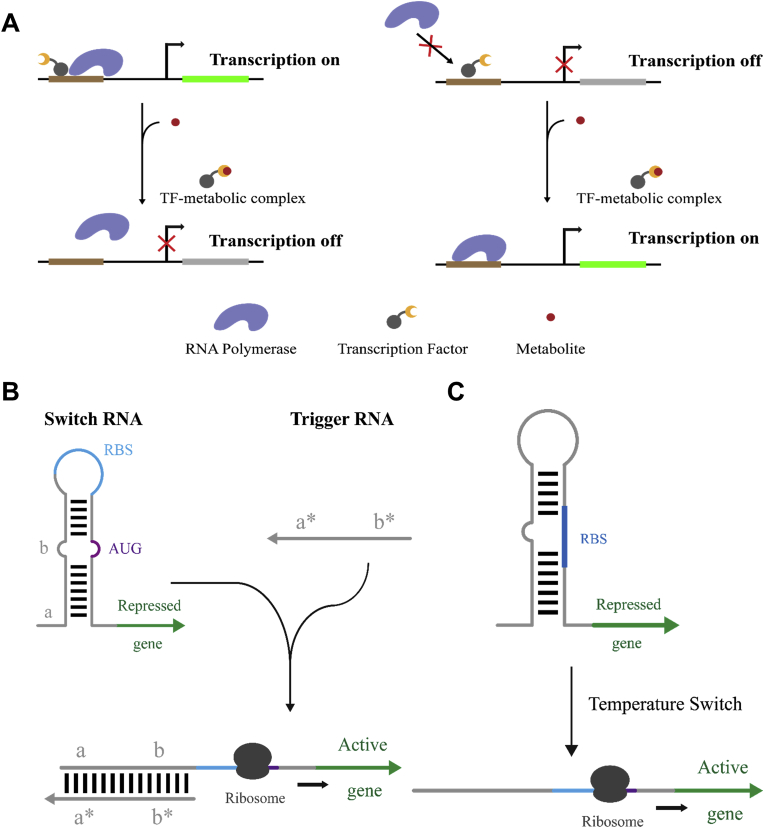
Transcription factors are proteins that regulate gene expression at the transcription level by binding to specific DNA sequences. Thus, several naturally existed metabolite responsive transcription factors have been adopted to construct biosensors. For example, metabolite binding mediated transcription repression (Fig. 3A, left) and transcription activation (Fig. 3A, right) has been widely applied for metabolic engineering [23].
Fig. 3.
CFPS enabled rapid prototyping of biosensors and genetic circuits based on transcriptional factor (A) and RNAs (B and C). (A) Metabolite responsive transcriptional factor based biosensors. The biosensor output can be repressed (left) or activated (right) by metabolite binding (adapted from Ref. [24]). (B) Toehold switches containing a switch RNA and a trigger RNA. (The figure adapted from Ref. [48]). (C) A temperature biosensor based on RNA.
Firstly, Chappell et al. developed and validated an entire in vitro approach for rapid prototyping of DNA regulatory elements [42], which laid solid foundation for biosensor and genetic circuits optimization in CFPS. Importantly, the authors demonstrated that the most frequently used DNA regulatory elements behaved consistently in vivo and in vitro. Based on this platform, Wen et al. further optimized a modular DNA-encoded biosensor (LasR transcription factor and LasR responsive promoters) in CFPS to measure a bacterial biomarker (3-oxo-C12-HSL) of Pseudomonas aeruginosa infection from human sputum samples [43]. Recently, Murray group developed a framework to generate new biosensors that can respond to different small molecules of interest by combining computational protein design (CPD) and rapid prototyping in CFPS [44]. The development of a vanillin biosensor based on a tetR-family repressor (qacR) was demonstrated as a case study. CPD was firstly used to obtain vanillin-responding qacR mutants, which were rapidly prototyped in CFPS by placing green fluorescent protein (GFP) downstream of the qacA promoter sequence. In addition, two engineered qacR mutants were further tested in vivo, and the vanillin biosensors were demonstrated their capability of vanillin sensing in a more complex system.
Transcription factors have been designed and tested in CFPS to detect bacterial quorum sensing signals [20,42,45]. Kawaguchi et al. developed a cell-free assay rapid screening of quorum-sensing signal N-acyl homoserine lactones (AHL). As the time-consuming steps were eliminated, biosensor assay time was dramatically reduced from more than 24 h to less than 3 h [45]. To explore the quorum sensing systems for building complex microbial consortia, Halleran et al. [20] characterized three Lux-type quorum sensing systems, including transcription factors (LuxR, RpaR, LasR), their cognate promoters as well as cognate AHLs for crosstalk in CFPS. By comparing the results tested in vivo and in vitro, the authors verified that CFPS could predict the quorum sensing behaviors (i.e. crosstalk) observed in vivo.
In order to build complex synthetic gene circuits consisted by multi-functional modules, mathematical models and CFPS were combined to design and verify novel synthetic biocircuits [21]. The authors chose the AraC-arabinose activation system as a feedforward loop, a transcription factor TetR as the repressor, deGFP with the corresponding promoter as output signals. The reporter deGFP could be activated by AraC and repressed by TetR. First, the circuit design was optimized by placing all circuit components into CFPS and regulating the parameters in the circuit model. Then the genetic circuits were characterized in CFPS by simply mixing cell-free extract, buffer, linear DNAs (AraC, TetR and deGFP), and the inducer arabinose. Finally, the genetic circuit components were assembled into a plasmid and tested in E. coli cells. As a result, the dynamics of the circuit were consistent in vivo and in vitro. CFPS was further explored for the construction and optimization of biological networks [18]. Specifically, the authors characterized and compared novel three, four, and five genetic ring oscillators in vivo and in vitro. CFPS was firstly employed to rapidly characterize circuit components as well as analyze the complete networks. When implemented in cells, the authors found that oscillation periods in cells matched CFPS results for all networks tested.
4.2. CFPS for rapid prototyping of RNA based biosensors and genetic circuits
While TF-based biosensors control transcription initiation, RNA-based biosensors are more versatile by regulating transcription termination, translation initiation, and RNA stability [23]. In addition, computational tools to rationally design RNAs have been developed and could provide relative predictable control over gene expression [46]. Therefore, RNA-based biosensors will have broad applications in metabolic engineering and synthetic biology.
Paper-based synthetic gene networks based on riboswitches were reported. Pardee et al. utilized toehold switches, without sequence constraint of trigger RNA by moving the RBS to a loop region of the hairpin, to demonstrate that complicated synthetic gene networks with colorimetric outputs could be implemented in the freeze-dried, paper-based cell free reactions (Fig. 3B) [47,48]. Furthermore, this technology was verified that it can rapidly prototype complex gene circuits and be programmed for in vitro diagnostics.
RNA-based biosensors have been designed to sense temperature, light, and osmolarity. Murray group [22] designed a series of RNA-based temperature sensors using thermodynamic computations and cell-free breadboards (CFPS). RNA thermometers undergo conformation changes at different temperatures (Fig. 3C). The authors changed an existing thermometer sequence one base at a time and constructed a library of temperature sensors. Rapid characterization in CFPS allowed the authors to create a toolbox of RNA-based circuit elements responsive to a wide range of temperatures. Sadler et al. designed thermometers by introducing hairpins between the Shine-Dalgarno sequence and complementary sequences within the gene of interest [49]. The thermometers regulated high-yield protein expression in CFPS with corresponding temperatures in the range of 30–37 °C. Furthermore, protein expression efficiency could be tuned by small variations of the coding sequence. Moreover, CFPS has also been explored for rapidly prototyping RNA networks (RNA genetic circuitry). For example, Takahashi et al. used CFPS to test the response time of an RNA transcription cascade, which was approximately 5 min per step of the cascade [19]. At the same time, temperature and regulator threshold tuning could affect this response time. In addition, the authors also prototyped a new RNA network with an RNA single input module and compared the performance both in vivo and in vitro.
5. Summary and perspective
Thanks to the open environment and linear DNAs templated transcription and translation capabilities, CFPS offers a flexible and powerful platform for metabolic engineering and synthetic biology. CFPS enabled rapid prototyping of metabolic pathways and genetic circuits (biosensors) significantly accelerates the DBT cycles for pathway optimization and strain development.
Besides the construction and optimization metabolic pathways and genetic circuits, the open and rapid phenotyping advantages of CFPS have found novel synthetic biology applications. For example, Marshall et al. adopted CFPS for rapid and scalable characterization of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technologies [50]. CFPS was demonstrated the applications in measuring dynamics of DNA cleavage and gene repression, predicting CRISPR interference efficiency, determining the specificities of anti-CRISPR proteins, and characterizing protospacer-adjacent motifs, all in a fast and scalable manner.
The rapid prototyping capabilities of CFPS can be further enhanced by integrating high throughput technologies, such as the robotic systems [15,51] and microfluidics [52]. The open nature and ease of operation of CFPS, various components can be mixed using a robotic system in a high throughput manner to explore the design space for optimal performance. Karim et al. used liquid handling robotics to optimize physiochemical parameters for n-butanol production in CFPS [15]. Most recently, Hori et al. combined CFPS and droplet microfluidics to rapidly test genetic circuits. In this system, millions of parameter combinations were assayed per hour [52].
Although relatively simple, the transcription and translation of CFPS still involves non-linear regulations and large-dimensional impact factors. Therefore, computational tools should be incorporated for a better design of metabolic pathways and genetic circuits. Guo and Murray constructed a novel functional biological network motif from scratch by integrating mathematical modeling and CFPS rapid prototyping [21]. Caschera et al. combined liquid handling robotics and machine learning for optimizing protein production in CFPS [51]. Specifically, the authors created a library of ∼106 combinations in a 16-dimensional space (i.e. promoter, ribosome binding site, and terminator variants). By using a liquid handling robotic workstation, CFPS reactions were implemented in an iterative and high-throughput manner. The machine learning algorithm learned from the CFPS reaction results and offered a better design of CFPS reactions. The robotic workstation performed the next generation CFPS reactions to validate and polish the computational models. After 8 iterations of the DBT cycles (performed in 8 days), the optimal experimental design increased CFPS protein yield ∼3.5-fold by testing only 0.014% of the total possible combinations. Although the machine learning algorithm was demonstrated for improving protein yield in CFPS, the machine learning strategies could be further developed to optimize metabolic pathways and genetic circuits.
In summary, CFPS has been emerging as an enabling platform for rapid prototyping of biosynthetic pathways and genetic circuits and attracts increasing attention in metabolic engineering and synthetic biology. Nevertheless, the potential of CFPS in speeding up DBT cycles will be not fully demonstrated until the integration of advanced high throughout technologies (i.e. automation and microfluidics) and artificial intelligence (i.e. machine learning).
Acknowledgement
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21606205, 21576232 & 21506185), the Fundamental Research Funds for the Central Universities, and the Startup Fund from Zhejiang University.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Jiazhang Lian, Email: jzlian@zju.edu.cn.
Zhinan Xu, Email: znxu@zju.edu.cn.
References
- 1.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164(6):1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Ajikumar P.K., Xiao W.H., Tyo K.E., Wang Y., Simeon F., Leonard E. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lian J., Jin R., Zhao H. Construction of plasmids with tunable copy numbers in Saccharomyces cerevisiae and their applications in pathway optimization and multiplex genome integration. Biotechnol Bioeng. 2016;113(11):2462–2473. doi: 10.1002/bit.26004. [DOI] [PubMed] [Google Scholar]
- 4.Du J., Yuan Y., Si T., Lian J., Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012;40(18):e142. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lian J., Ma Y., Cai J., Wu M., Wang J., Wang X. High-level expression of soluble subunit b of F1F0 ATP synthase in Escherichia coli cell-free system. Appl Microbiol Biotechnol. 2009;85(2):303–311. doi: 10.1007/s00253-009-2055-z. [DOI] [PubMed] [Google Scholar]
- 6.Kai L., Kaldenhoff R., Lian J., Zhu X., Dotsch V., Bernhard F. Preparative scale production of functional mouse aquaporin 4 using different cell-free expression modes. PLoS One. 2010;5(9):e12972. doi: 10.1371/journal.pone.0012972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Lian J., Kai L., Huang L., Cen P., Xu Z. Enhanced functional expression of aquaporin Z via fusion of in situ cleavable leader peptides in Escherichia coli cell-free system. Enzym Microb Technol. 2014;55:26–30. doi: 10.1016/j.enzmictec.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Junge F., Haberstock S., Roos C., Stefer S., Proverbio D., Dotsch V. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. N Biotechnol. 2011;28(3):262–271. doi: 10.1016/j.nbt.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Klammt C., Schwarz D., Lohr F., Schneider B., Dotsch V., Bernhard F. Cell-free expression as an emerging technique for the large scale production of integral membrane protein. FEBS J. 2006;273(18):4141–4153. doi: 10.1111/j.1742-4658.2006.05432.x. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y. Cell-free synthetic biology: engineering in an open world. Synth Syst Biotechnol. 2017;2(1):23–27. doi: 10.1016/j.synbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z.Z., Yeung E., Hayes C.A., Noireaux V., Murray R.M. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth Biol. 2014;3(6):387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 12.Karim A.S., Jewett M.C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab Eng. 2016;36:116–126. doi: 10.1016/j.ymben.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Wu Y.Y., Culler S.J., Khandurina J., Van Dien S., Murray R.M. Prototyping 1,4-butanediol (BDO) biosynthesis pathway in a cell-free transcription-translation (TX-TL) system. BioRxiv. 2017 [Google Scholar]
- 14.Wu Y.Y., Sato H., Huang H., Culler S.J., Khandurina J., Nagarajan H. System-level studies of a cell-free transcription-translation platform for metabolic engineering. BioRxiv. 2017 [Google Scholar]
- 15.Karim A.S., Heggestad J.T., Crowe S.A., Jewett M.C. Controlling cell-free metabolism through physiochemical perturbations. Metab Eng. 2017;45:86–94. doi: 10.1016/j.ymben.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Sheng J., Huang L., Zhu X., Cai J., Z X Reconstitution of the peptidoglycan cytoplasmic precursor biosynthetic pathway in cell-free system and rapid screening of antisense oligonucleotides for Mur enzymes. Appl Microbiol Biotechnol. 2014;98(4):1785–1794. doi: 10.1007/s00253-013-5467-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J., Huang L., Lian J., Sheng J., Cai J. Z X. Reconstruction of the UDP-N-acetylglucosamine biosynthetic pathway in cell-free system. Biotechnol Lett. 2010;32(10):1481–1486. doi: 10.1007/s10529-010-0315-8. [DOI] [PubMed] [Google Scholar]
- 18.Niederholtmeyer H., Sun Z.Z., Hori Y., Yeung E., Verpoorte A., Murray R.M. Rapid cell-free forward engineering of novel genetic ring oscillators. Elife. 2015;4:e09771. doi: 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription translation (TX-TL) systems. ACS Synth Biol. 2015;4(5):503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halleran A.D., Murray R.M. Cell-free and in vivo characterization of Lux, Las, and Rpa quorum activation systems in E. coli. ACS Synth Biol. 2018;7(2):752–755. doi: 10.1021/acssynbio.7b00376. [DOI] [PubMed] [Google Scholar]
- 21.Guo S., Murray R.M. Prototyping and implementation of a novel feedforward loop in a cell-free transcription-translation system and cells. BioRxiv. 2017 [Google Scholar]
- 22.Sen S., Apurva D., Satija R., Siegal D., Murray R.M. Design of a toolbox of RNA thermometers. ACS Synth Biol. 2017;6(8):1461–1470. doi: 10.1021/acssynbio.6b00301. [DOI] [PubMed] [Google Scholar]
- 23.Liu D., Evans T., Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Rogers J.K., Taylor N.D., Church G.M. Biosensor-based engineering of biosynthetic pathways. Curr Opin Biotechnol. 2016;42:84–91. doi: 10.1016/j.copbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 2012;30(5):1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swartz J. Developing cell-free biology for industrial applications. J Ind Microbiol Biotechnol. 2006;33(7):476–485. doi: 10.1007/s10295-006-0127-y. [DOI] [PubMed] [Google Scholar]
- 27.Katzen F., Chang G., Kudlicki W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005;23(3):150–156. doi: 10.1016/j.tibtech.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Liu D.V., Zawada J.F., Swartz J.R. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol Prog. 2005;21(2):460–465. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 29.Kim T.W., Keum J.W., Oh I.S., Choi C.Y., Park C.G., Kim D.M. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J Biotechnol. 2006;126(4):554–561. doi: 10.1016/j.jbiotec.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Sun Z.Z., Hayes C.A., Shin J., Caschera F., Murray R.M., Noireaux V. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp. 2013;79:e50762. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D.M., Swartz J.R. Prolonging cell-free protein synthesis by selective reagent additions. Biotechnol Prog. 2000;16(3):385–390. doi: 10.1021/bp000031y. [DOI] [PubMed] [Google Scholar]
- 32.Kim D.M., Swartz J.R. Prolonging cell-free protein synthesis with a novel ATP regeneration system. Biotechnol Bioeng. 1999;66(3):180–188. [PubMed] [Google Scholar]
- 33.Calhoun K.A., Swartz J.R. Energizing cell-free protein synthesis with glucose metabolism. Biotechnol Bioeng. 2005;90(5):606–613. doi: 10.1002/bit.20449. [DOI] [PubMed] [Google Scholar]
- 34.Kim T.W., Keum J.W., Oh I.S., Choi C.Y., Kim H.C., Kim D.M. An economical and highly productive cell-free protein synthesis system utilizing fructose-1,6-bisphosphate as an energy source. J Biotechnol. 2007;130(4):389–393. doi: 10.1016/j.jbiotec.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Caschera F., Noireaux V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie. 2014;99:162–168. doi: 10.1016/j.biochi.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Zhang Y.H. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC Biotechnol. 2009;9:58. doi: 10.1186/1472-6750-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.-C., Kim T.-W., Kim D.-M. Prolonged production of proteins in a cell-free protein synthesis system using polymeric carbohydrates as an energy source. Process Biochem. 2011;46(6):1366–1369. [Google Scholar]
- 38.Zhang Y.H.P. Production of biocommodities and bioelectricity by cell-free synthetic enzymatic pathway biotransformations: challenges and opportunities. Biotechnol Bioeng. 2010;105(4):663–677. doi: 10.1002/bit.22630. [DOI] [PubMed] [Google Scholar]
- 39.Liu T.G., Vora H., Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Eng. 2010;12(4):378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 40.Dudley Q.M., Karim A.S., Jewett M.C. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol J. 2015;10(1):69–82. doi: 10.1002/biot.201400330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen P.H.B., Wu Y., Guo S., Murray R.M. Design space exploration of the violacein pathway in Escherichia coli based transcription translation cell-free system (TX-TL) BioRxiv. 2017 [Google Scholar]
- 42.Chappell J., Jensen K., Freemont P.S. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013;41(5):3471–3481. doi: 10.1093/nar/gkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen K.Y., Cameron L., Chappell J., Jensen K., Bell D.J., Kelwick R. A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples. ACS Synth Biol. 2017;6(12):2293–2301. doi: 10.1021/acssynbio.7b00219. [DOI] [PubMed] [Google Scholar]
- 44.de los Santos E.L., Meyerowitz J.T., Mayo S.L., Murray R.M. Engineering transcriptional regulator effector specificity using computational design and in vitro rapid prototyping: developing a vanillin sensor. ACS Synth Biol. 2016;5(4):287–295. doi: 10.1021/acssynbio.5b00090. [DOI] [PubMed] [Google Scholar]
- 45.Kawaguchi T., Chen Y.P., Norman R.S., Decho A.W. Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl Environ Microbiol. 2008;74(12):3667–3671. doi: 10.1128/AEM.02869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Espah Borujeni A., Mishler D.M., Wang J., Huso W., Salis H.M. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016;44(1):1–13. doi: 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pardee K., Green A.A., Ferrante T., Cameron D.E., DaleyKeyser A., Yin P. Paper-based synthetic gene networks. Cell. 2014;159(4):940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green A.A., Silver P.A., Collins J.J., Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159(4):925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sadler F.W., Dodevski I., Sarkar C.A. RNA thermometers for the PURExpress system. ACS Synth Biol. 2018;7(1):292–296. doi: 10.1021/acssynbio.7b00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall R., Maxwell C.S., Collins S.P., Jacobsen T., Luo M.L., Begemann M.B. Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol Cell. 2018;69(1) doi: 10.1016/j.molcel.2017.12.007. 146–157 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caschera F., Bedau M.A., Buchanan A., Cawse J., de Lucrezia D., Gazzola G. Coping with complexity: machine learning optimization of cell-free protein synthesis. Biotechnol Bioeng. 2011;108(9):2218–2228. doi: 10.1002/bit.23178. [DOI] [PubMed] [Google Scholar]
- 52.Hori Y., Kantak C., Murray R.M., Abate A.R. Cell-free extract based optimization of biomolecular circuits with droplet microfluidics. Lab a Chip. 2017;17(18):3037–3042. doi: 10.1039/c7lc00552k. [DOI] [PubMed] [Google Scholar]