Abstract
Natural products with significant biological activities continuously act as rich sources for drug discovery and development. To harness the potential of these valuable compounds, robust methods need to be developed for their rapid and sustainable production. Cell-free biosynthesis of pharmaceutical natural products by in vitro reconstruction of the entire biosynthetic pathways represents one such solution. In this review, we focus on in vitro biosynthesis of two important classes of natural products, polyketides (PKs) and nonribosomal peptides (NRPs). First, we summarize purified enzyme-based systems for the biosynthesis of PKs, NRPs, and PK/NRP hybrids. Then, we introduce the cell-free protein synthesis (CFPS)-based technology for natural product production. With that, we discuss challenges and opportunities of cell-free synthetic biology for in vitro biosynthesis of natural products.
Keywords: Cell-free synthetic biology, Purified enzymes, Cell-free protein synthesis, Polyketides, Nonribosomal peptides, In vitro biosynthesis, Natural products
1. Introduction
Nature has extraordinarily rich natural products, which are synthesized by living organisms ranging from tiny microorganisms to giant plants on this planet. Natural products are a large family of low molecular weight organic compounds with diverse chemical structures. These natural compounds have significant biological activities that act as abundant sources for drug discovery and development [1]. Over the past 30 years, more than 50% of new drugs available in the pharmaceutical market are natural products and their derivatives [2]. The important classes of natural products include, but not limited to, the well-known polyketides (PKs) and nonribosomal peptides (NRPs) that are produced by polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively, which are found in various microorganisms like the Streptomyces species [3]. PKs and NRPs possess a broad spectrum of biological activities (e.g., antibiotic, immunosuppressant, and anticancer, etc.) and are used in many clinical applications [1,4]. For example, erythromycin (PK) and daptomycin (NRP) are clinically important antibiotics; the NRP/PK hybrid compound epothilone has been developed as an antitumor agent [1]. Due to their multiple uses in human medicine, the demand for these pharmaceutical natural products is continuously growing [2].
Traditionally, pharmaceutical natural products are extracted directly from their native producers like plants. However, these native producers often suffer from the low productivity of interesting molecules, demonstrating this solution is not environmentally friendly, sustainable, and cost-effective. Another strategy utilizes chemical synthesis to produce natural products of medicinal importance, however, the structural complexity of many natural compounds makes chemical synthesis hardly feasible or practical. As a current alternative approach, metabolic engineering and synthetic biology studies have tried to utilize surrogate microbes, for instance, Escherichia coli and Saccharomyces cerevisiae, to produce intricate pharmaceutical molecules by reconstitution of their entire gene clusters in the host cells [[5], [6], [7], [8]]. Despite its success in the past, this approach still remains problematic to obtain high yields, which are mainly caused by metabolic burden inhibiting host cell growth, incorrect folding of heterologous proteins, lack of post-translational modification enzymes, and unavailability of necessary precursors in heterologous hosts. In order to tackle these in vivo problems, in vitro, cell-free, platforms have recently been developed and are emerging as powerful systems for the biomanufacturing of therapeutic proteins, low-value biocommodities, and value-added chemicals [[9], [10], [11], [12], [13]].
Generally, in vitro cell-free biomanufacturing systems separate cell growth (catalyst synthesis) from target product formation (catalyst utilization). Because of the absence of cell walls, these open cell-free systems allow for easy manipulation, monitoring, optimization, and sampling. In addition, in vitro cell-free platforms have many advantages over in vivo microbial systems, such as (i) high product yields that can be achieved by eliminating the synthesis/maintenance of cell biomass, removing undesired side pathways, and preventing the formation of by-products; (ii) fast reaction rates enabled by better mass transfer due to the lack of cell membrane; and (iii) tolerance of toxic precursors, intermediates, and products [9,10,14]. As a result, various products are produced via in vitro reconstruction of different biosynthetic pathways in a single reaction vessel. To this end, two cell-free systems are being commonly used: purified enzyme system and crude cell extract system [10,13].
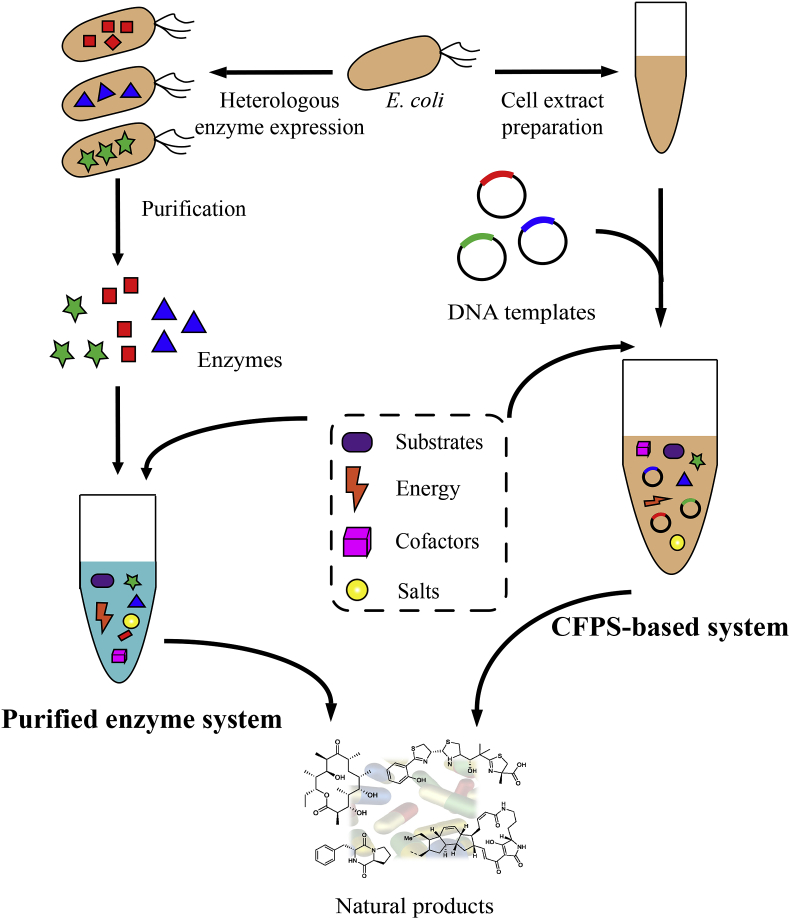
Cell-free biosynthesis of proteins, bulk chemicals, and value-added compounds, etc. has been well summarized in several outstanding reviews [9,10,13,14]. In this review, we focus on cell-free biosynthesis of pharmaceutical natural products with an emphasis on PKs and NRPs (Fig. 1). First, we summarize purified enzyme-based natural product biosynthesis. Then, we introduce crude cell extract systems for natural product production, especially, with the cell-free protein synthesis (CFPS)-based technology. Finally, we discuss challenges and opportunities of cell-free synthetic biology for in vitro biosynthesis of natural products.
Fig. 1.
In vitro biosynthesis of pharmaceutical natural products with purified enzyme-based system and cell-free protein synthesis (CFPS)-based system.
2. Purified enzyme-based biosynthesis of natural products
The biosynthesis of PKs and NRPs occurs by means of successive condensation reactions of simple monomeric building blocks. Typically, the large multimodular PKS and NRPS enzymes catalyze monomers acyl-CoA thioesters (e.g., acetyl-CoA and malonyl-CoA, etc.) and amino acids (proteinogenic and nonproteinogenic), respectively, to form the final complex products with additional modifications like methylation, epimerization, and glycosylation [3,15]. The chemical logic for the assembly of PKs and NRPs is similar [3]. The core catalytic domains of each PKS module include an acyltransferase (AT), a ketosynthase (KS), and an acyl carrier protein (ACP), which carry out three essential chemical reactions: acyl-CoA selection, C-C bond formation, and acyl transfer, respectively. Similarly, each NRPS module also consists of three core domains: an adenylation (A) domain responsible for the selection of substrate, a peptidyl carrier protein (PCP) responsible for the translocation of growing peptide chain, and a condensation (C) domain responsible for peptide bond formation. The ACP in PKSs and the PCP in NRPSs are often called the thiolation (T) domain. To functionalize the PKS and NRPS assembly lines, all the T domains have to be posttranslationally modified by transferring the 4′-phosphopantetheinyl prosthetic group from coenzyme A (CoA) to a conserved serine residue on the T domain by a phosphopantetheinyl transferase (PPTase) like the Sfp-type PPTase from Bacillus subtilis [16,17]. The enzyme Sfp was found to be the most promiscuous PPTase and often heterologously coexpressed with PKS and NRPS genes to generate active holo-enzymes.
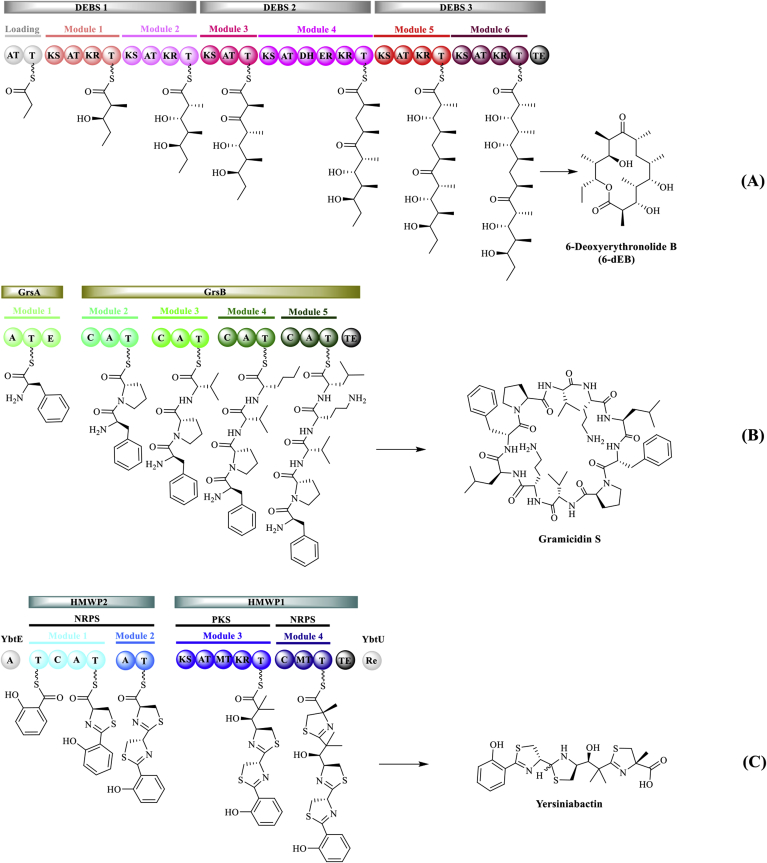
In vitro reconstruction of purified enzymes has been widely used to characterize individual enzymes, identify intermediates, and elucidate entire natural product biosynthetic pathways [18,19]. While there are many studies on individual enzymes, we primarily focus on the in vitro reconstitution of PK and NRP biosynthetic pathways with a set of purified enzymes to generate core scaffolds and final products, which include the PKs 6-deoxyerythronolide B and enterocin, the NRPs terrequinone A and pacidamycin, as well as the PK/NRP hybrids yersiniabactin and ikarugamycin. Three representative biosynthetic pathways of PK, NRP, and PK/NRP hybrid are shown in Fig. 2.
Fig. 2.
Representative biosynthetic pathways: (A) PK: 6-deoxyerythronolide B (6-dEB), (B) NRP: gramicidin S, and (C) PK/NRP hybrid: yersiniabactin. Catalytic domains: AT, acyltransferase; KS, ketosynthase; KR, ketoreductase; DH, dehydratase; ER, enoylreductase; TE, thioesterase; A, adenylation; C, condensation; T, thiolation; E, epimerase; MT, methyltransferase; Re, reductase.
The most well studied PK assembly line is the deoxyerythronolide B synthase (DEBS) that synthesizes 6-deoxyerythronolide B (6-dEB, the aglycone core of erythromycin antibiotics) [20,21]. DEBS consists of three multienzyme polypeptides DEBS1 (370 kDa), DEBS2 (380 kDa), and DEBS3 (332 kDa), each of which contains two modules harboring several catalytic domains [22]. Although 6-dEB has been successfully produced by in vivo heterologous expression of three DEBS genes in E. coli [23], in vitro reconstitution of the full DEBS system from purified protein components for 6-dEB synthesis was only achieved recently [24]. To realize in vitro 6-dEB synthesis, the three large DEBS proteins have to be expressed, purified, and activated (phosphopantetheinylation of T domains by PPTase). For this purpose, three DEBS enzymes were heterologously expressed in an engineered strain E. coli BAP1 with genomic incorporation of the PPTase sfp gene. Although DEBS2 and DEBS3 were well expressed and purified, DEBS1 was expressed poorly in the E. coli, making the production of enough amounts of pure DEBS1 enzyme difficult. To overcome this problem, the DEBS1 gene was divided into three fragments to allow the expression of three smaller polypeptides, which are a loading didomain, module 1, and module 2. With this strategy, each dissociated protein was successfully expressed, purified, and ready for in vitro reconstitution. When all five purified components were mixed with required substrates, the expected product 6-dEB was detected by LC-MS analysis, albeit the yield was not reported in the paper [24].
Another elegant example is in vitro total biosynthesis of the polyketide antibiotic enterocin from a PKS pathway in the marine bacterium Streptomyces maritimus, which includes nine enzyme components (EncA/B, -C, -D, -N, -M, -K, -R, and FabD – an acyltransferase that is borrowed from Streptomyces glaucescens) [25]. In order to obtain active enzymes, the nine proteins were heterologously expressed in either E. coli or Streptomyces. For instance, EncC is an acyl carrier protein (ACP, T domain) that needs to be activated by PPTase to form holo-EncC. This was achieved by the coexpression of EncC and a PPTase Svp protein from Streptomyces verticillus in E. coli [26]. Overexpression of EncM (a favorskiiase flavoprotein) in E. coli only generated apoprotein, whereas by switching the host to Streptomyces lividans, holo-EncM was obtained with a brightly pigmented yellow color. Having obtained all active enzymes, total biosynthesis of enterocin was performed by the combination of nine enzymes, two precursors (benzoic acid and malonyl-CoA), and necessary cofactors (S-adenosyl-l-methionine (SAM), NADPH, ATP and Mg2+). However, the last enzymatic conversion step from 5-deoxyenterocin to enterocin by EncR (cytochrome P450 hydroxylase) was inhibited by SAM, which led to a low production of enterocin. To circumvent the inhibitory effect, eight enzymes except for EncR were first reconstituted in a single pot for the formation of 5-deoxyenterocin. After extraction of the reaction mixture, 5-deoxyenterocin was subjected to the last EncR-catalyzed reaction to yield the final product enterocin. Notably, this in vitro two-step enzymatic cascade forms ten C-C bonds, five C-O bonds, and seven chiral centers in the polyketide enterocin with an approximately 25% overall yield based on the precursor benzoic acid.
The antitumor NRP product terrequinone A, identified in the fungi Aspergillus species, is biosynthesized by a five-enzyme (TdiA-E) pathway [27]. TdiA is a single-module NRPS that lacks a traditional condensation (C) domain. Instead, TdiA consists of three individual domains, which are an adenylation (A) domain, a thiolation (T) domain, and a thioesterase (TE) domain. Each of the three domains is unique. The A domain, unlike a typical NRPS A domain that selects amino acids as its substrates [3], is special for recognition of the substrate α-keto acid (phenylpyruvate) rather than the α-amino acid (l-tryptophan). Such keto acid activating A domains are unusual but have been demonstrated in the NRPS modules of cereulide [28] and valinomycin [29]. The TE domain, located at the C-terminus of the final NRPS module, traditionally catalyzes the release of final product by either hydrolysis or cyclization [3,18]. However, the TdiA TE domain was found to perform a novel chemistry function for catalyzing C-C bond formation. The T domain of TdiA was activated by the purified PPTase Sfp in an in vitro reaction. Briefly, the other four enzymes are TdiB, an indole prenyltransferase; TdiC, an oxidoreductase; TdiD, a pyridoxal-5′-phosphate-dependent aminotransferase that generates TdiA's substrate phenylpyruvate from l-tryptophan; and TdiE, an unknown protein but with weak homology to SAM-dependent methyltransferase. All five proteins were overproduced and purified from E. coli, followed by reconstitution of the full terrequinone A biosynthetic pathway in vitro. With the five enzymes, the antitumor fungal secondary metabolite terrequinone A was biosynthesized from the precursor l-tryptophan via the sequential reactions of oxidation (TdiD), dimerization (TdiA), and bisprenylation (TdiB/C/E). More importantly, this work reveals the mechanisms of the unique TE-catalyzed carbon-carbon bond formation of TdiA and the two distinct prenylations of TdiB. However, further mechanistic work needs to be carried out to uncover more details about these enzymes like the still-uncharacterized role of TdiE.
While typical NRP assembly lines with large multimodular NRPSs are encoded within a single protein, a highly dissociated NRPS with several freestanding domains and modules was reported for the biosynthesis of uridyl peptide antibiotic pacidamycins [30]. Pacidamycins are produced by Streptomyces coeruleorubidus, which contain two nonproteinogenic amino acid building blocks meta-tyrosine (m-Tyr) and 2,3-diaminobutyric acid (DABA) and three other proteinogenic amino acids (l-alanine, l-phenylalanine, and l-tryptophan). These building blocks are assembled by nine proteins (PacD, -H, -I, -J, -L, -N, -O, -P, -U) that constitute a highly dissociated NRPS assembly line [30,31]. Among them, several proteins are standalone enzymes with interesting functions. For instance, PacH, a freestanding T domain, acts as a key carrier component on which the tetrapeptide framework is assembled. Another freestanding PacI (C domain) protein, with a novel catalyzing ability to condense peptide and nucleoside substrates, catalyzes the release of the tetrapeptidyl intermediate from PacH by a nucleoside moiety. In addition, one exceptional feature of this assembly is that it does not start at the N-terminal residue amino acid 1 and proceed to the C-terminal residue amino acid 5. Instead, the assembly of the pacidamycin scaffold is initiated by the activation and methylation of the core residue 3 (i.e., DABA), which is tethered via its carboxyl as a thioester to PacH, and then the chain is built from the middle outward in both directions [32]. All nine Pac proteins were overexpressed in either E. coli BL21 or BAP1 cells if the T domains need to be activated by the PPTase Sfp. Thereafter, complete in vitro reconstitution of all purified enzymes for the biosynthesis of pacidamycins was accomplished, showing another elegant nonribosomal biosynthetic paradigm for synthesizing complex peptidyl nucleoside antibiotics from simple nucleoside and amino acid building blocks.
Because multimodular PKS and NRPS have similar chemical logic and protein machinery, hybridization of them naturally occurs generating mixed PKS/NRPS assembly lines that can biosynthesize PK/NRP hybrids [3]. One example is the PK/NRP hybrid yersiniabactin, an iron-chelating siderophore natively produced by the plague bacterium Yersinia pestis [33]. The hybrid PKS/NRPS assembly line yersiniabactin synthetase comprises three NRPS modules and one PKS module that are divided into two large proteins, HMWP1 (350 kDa) and HMWP2 (230 kDa) [34]. In addition, two small standalone proteins are involved in the biosynthetic pathway, a salicylate adenylation (A) domain YbtE (60 kDa) and a reductase domain YbtU (41 kDa). These four proteins work together to select, activate, and incorporate one salicylate, three cysteines, and one malonyl moiety into the final product yersiniabactin. Overexpression of all proteins were performed in the E. coli BL21(DE3) strain, followed by purification with nickel chelate chromatography. All T domains of the PKS/NRPS hybrid were activated by the purified PPTase Sfp in an in vitro reaction. With purified enzymes (YbtE, HMWP1, HMWP2, and YbtU), yersiniabactin was fully synthesized in vitro by 22 chemical operations with a net throughput of 1.4 products per minute. This study represents the first case for complete in vitro reconstitution of a PKS/NRPS hybrid system, enabling the biosynthesis of complex hybrid natural products and the elucidation of the specificity of the catalytic domains at the PKS/NRPS interface.
Recently, Greunke et al. reported in vitro total biosynthesis of ikarugamycin with an iterative PKS/NRPS machinery IkaA (340 kDa) and two reductases IkaB (70 kDa) and IkaC (41 kDa) [35]. During the biosynthesis process, IkaA catalyzes the formation of an unstable intermediate tetramic acid from the precursors of acetyl-CoA, malonyl-CoA, and l-ornithine. Afterwards, the intermediate tetramic acid is cyclized to ikarugamycin by the enzymes IkaB and IkaC. For the production of three required enzymes, the genes (ikaA, ikaB, and ikaC) were amplified from the native producer Streptomyces sp. Tü6239, followed by heterologous expression in E. coli BL21(DE3). The large PKS/NRPS system IkaA and the reductase IkaB were solubly expressed, which are ready for the following purification and in vitro reconstitution. However, IkaC was only expressed in insoluble inclusion bodies. To circumvent this problem, a homologous ikaC gene from Salinispora arenicola was cloned and expressed in E. coli, giving rise to soluble IkaC. Two apo-T domains of the PKS/NRPS hybrid were activated by the PPTase Sfp as well to generate holo-IkaA. Eventually, in vitro total synthesis of ikarugamycin in a one-pot reaction was achieved with the three recombinant enzymes IkaA/B/C, which, taken together, install 17 individual bonds (15 C-C and 2 C-N bonds) and set 8 chiral centers in the final PK/NRP hybrid product.
3. CFPS-based biosynthesis of natural products
While purified enzyme-based cell-free systems, as mentioned above, have been used to produce complex natural products, i.e., PKs, NRPs, and PK/NRP hybrids, by in vitro reconstruction of their biosynthetic pathways, this strategy often suffers from time-consuming cloning steps, laborious purification process, insoluble expressed enzymes, enzyme instability and so on. Alternatively, cell-free protein synthesis (CFPS), in recent years, has emerged as a powerful platform technology to recombinant protein biosynthesis for applications in biotechnology and synthetic biology [9,36,37]. So far, crude extract based CFPS systems have been developed from various prokaryotic and eukaryotic organisms, each with their own distinct advantages and opportunities [38]. Therefore, different CFPS systems have been broadly utilized for producing a wide variety of active protein products such as therapeutic vaccines [39], antibodies [40], virus-like particles [41], membrane proteins [42], metalloproteins [43], and proteins harboring non-standard amino acids [44]. In addition, CFPS has been applied for the rapid prototyping of biosynthetic pathways [45,46] and for the advancement of paper-based diagnostics [47,48].
Despite multiple types of proteins have been synthesized by the CFPS technology, expression of enzymes like PKS and NRPS from natural product biosynthetic pathways is very rare. In vivo heterologous expression of PKS and NRPS is possible, but still remains challenging in part due to their complex multidomain structures and large size with molecular weights normally ranging from one hundred to several hundreds of kilodaltons (kDa). In this context, CFPS offers an alternative protein production platform with potential advantages for the expression of natural product biosynthetic enzymes. For example, the cell-free environment allows for design-build-test cycles to be performed without the need to reengineer organisms, DNA for pathway enzymes is directly input, and substrates and cofactors needed for protein expression can be controlled and maintained at defined concentrations. To investigate the potential of CFPS for studying and engineering natural product pathways, recently, two NRPS proteins involved in the first steps of gramicidin S biosynthesis were expressed by using the CFPS platform technology [49].
Gramicidin S is a nonribosomal cyclodecapeptide antibiotic, produced by the bacterium Brevibacillus brevis (formerly known as Bacillus brevis) via two NRPS enzymes GrsA (126 kDa, one module) and GrsB (510 kDa with four modules) [50,51]. However, a natural shunt product of the gramicidin S pathway is the D-Phe-L-Pro diketopiperazine (DKP) formed by spontaneous intramolecular cyclization of the substrates of the first two NRPS modules in the pathway, namely, GrsA and GrsB1 (121 kDa, the first module of GrsB) [52,53]. GrsA, as a single starter module, contains an A domain, a T domain, and an epimerization (E) domain that converts the substrate l-phenylalanine to d-phenylalanine. In the second module GrsB1, the A domain activates the substrate l-proline to install on the T domain, followed by peptide bond formation between D-Phe and L-Pro with the condensation (C) domain, ultimately resulting in the production of D-Phe-L-Pro DKP. In order to establish an in vitro platform for D-Phe-L-Pro DKP biosynthesis, NRPS proteins GrsA and GrsB1 were expressed using an E. coli-based CFPS system with DNA as a direct input [49]. Individual plasmids containing the genes grsA and grsB1 were used as DNA templates to express the two enzymes in CFPS reactions. With cell extracts prepared from the strain E. coli BL21 Star(DE3), both proteins GrsA and GrsB1 were expressed in full-length with soluble yields of 106 μg/mL and 77 μg/mL, respectively. Importantly, cell-free expressed apo-NRPSs GrsA and GrsB1 could be converted to their functional (holo) form by adding the purified PPTase Sfp to the post-CFPS reaction mixture. Afterwards, GrsA and GrsB1 were coexpressed in a single-pot mixture allowing in vitro reconstitution of the partial NRPS assembly line for product formation. Finally, the target product D-Phe-L-Pro DKP was successfully detected by the LC-MS/MS analysis with correct D-L stereochemistry. Even without optimization of the cell-free production system, the final DKP yield reached 12 mg/L, which is higher than the previous reported yield of 9 mg/L from cell-based recombinant protein expression in E. coli [54]. This work, for the first time, notably demonstrates that the CFPS platform is robust to produce “difficult-to-express” proteins like the large multimodular NRPS enzymes, and more importantly, complex natural products that could be biosynthesized via the in situ CFPS expressed enzymes.
4. Conclusions and perspectives
Cell-free synthetic biology is emerging as a powerful and robust platform for the next-generation biomanufacturing without using intact cells. In this review, we focus on cell-free biosynthesis of pharmaceutical natural products, in particular, with an emphasis on polyketides (PKs) and nonribosomal peptides (NRPs). For this purpose, two cell-free systems commonly used are (i) purified enzyme system and (ii) crude cell extract system. Full reconstitution of natural product biosynthetic pathways with purified enzymes in both PKs and NRPs contexts has allowed the elaboration of remarkable complexity from simple building blocks as aforementioned cases. This strategy not only significantly contributes to decipher novel chemical logic and enzymatic machinery of PKS and NRPS assembly lines, but also provides efficient platforms for in vitro total biosynthesis of natural products [18,19]. In addition, due to the flexibility of open cell-free systems, precursor analogues could be easily added to the reaction, allowing the formation of natural product analogues with potential novel bioactivities as shown in an example of the NRP beauvericin [55]. While purified enzyme systems succeeded in the past, some challenges are still remaining. These include high costs of protein purification, protein degradation, cofactor regeneration, longevity of reactions, and ability to scale, etc., which limit mass production of desired natural products of interest. To address these issues, crude cell extract systems, alternatively, may provide more advantages and opportunities for natural product biosynthesis.
During the past two decades, crude extract based cell-free protein synthesis (CFPS) systems have been greatly developed, which result in the dramatic increase of protein yields, active reaction durations, and reaction volumes [9]. The variety of different CFPS technology platforms has further enabled the in vitro production of proteins with diverse complexity and species origin. In this context, two large multimodular NRPS enzymes from Brevibacillus brevis were coexpressed by an E. coli-based CFPS system, which led to cell-free biosynthesis of the antibiotic D-Phe-L-Pro diketopiperazine (DKP) via the in situ CFPS expressed enzymes [49]. This work represents a key proof-of-principle for how one could apply CFPS to synthesize natural products.
In addition, with the rapid development of next-generation DNA sequencing and genome mining technologies, more and more potential natural product gene clusters have been identified from the genome data of Streptomyces microorganisms (Gram-positive bacteria with high GC-content genomes (>70% GC)) [56,57]. However, identifying novel natural products from these data derived clusters is a significant challenge because overexpression of gene clusters necessary to synthesize natural products remains difficult. Utilization of the native Streptomyces producers or heterologous hosts (e.g., E. coli, yeast, etc.) for the production of natural products is often hampered by low productivity [[5], [6], [7], [8]]. Therefore, it is necessary to establish a robust and high-throughput method for the rapid expression of increasing biosynthetic pathways. Although E. coli-based CFPS systems have been developed for more than twenty years to express a variety of proteins with enhanced yields (>1000 μg/mL) [9], these systems may not be able to efficiently express GC-rich genes that are originated from Streptomyces due to, for instance, codon usage bias, solubility issues, and post-translational modifications. In order to solve this problem, high-yielding Streptomyces-based CFPS systems were recently developed with the goal to express high GC-content genes that are involved in the biosynthesis of PKs and NRPs [[58], [59], [60]]. The Streptomyces-based CFPS system showed a significant increase of the solubility of high GC genes-encoded proteins as compared to an E. coli-based CFPS system, demonstrating the Streptomyces CFPS system is beneficial for the expression of GC-rich genes [58]. However, current protein yields with the Streptomyces-based CFPS system (<200 μg/mL) remain lower than the E. coli CFPS system (>1000 μg/mL) [60]. In order to enhance the productivity of the Streptomyces CFPS system, further efforts need to be taken to improve the overall protein yields through physicochemical optimization, bioprocess engineering, and strain engineering strategies. Taken together, we envision that CFPS systems, especially, the newly established Streptomyces CFPS platforms, will contribute significantly to express complex natural product gene clusters (e.g., PKS and NRPS) from various Streptomyces species, enabling the synthesis and discovery of novel natural products in vitro.
In summary, we believe cell-free synthetic biology will play more and more important roles in the next-generation biomanufacturing. Given the flexibility of cell-free systems, biosynthesis beyond the cell holds tremendous potential to create affordable and efficient in vitro artificial biosynthetic factories for the rapid synthesis, study, and discovery of pharmaceutical natural products.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Katz L., Baltz R.H. Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 2.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;9:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach M.A., Walsh C.T. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Felnagle E.A., Jackson E.E., Chan Y.A., Podevels A.M., Berti A.D., McMahon M.D., Thomas M.G. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H., Boghigian B.A., Armando J., Pfeifer B.A. Methods and options for the heterologous production of complex natural products. Nat Prod Rep. 2011;28:125–151. doi: 10.1039/c0np00037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ongley S.E., Bian X., Neilan B.A., Müller R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep. 2013;30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- 7.Li J., Neubauer P. Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol. 2014;31:579–585. doi: 10.1016/j.nbt.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Luo Y., Li B.Z., Liu D., Zhang L., Chen Y., Jia B., Zeng B.X., Zhao H., Yuan Y.J. Engineered biosynthesis of natural products in heterologous hosts. Chem Soc Rev. 2015;44:5265–5290. doi: 10.1039/c5cs00025d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley Q.M., Karim A.S., Jewett M.C. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol J. 2015;10:69–82. doi: 10.1002/biot.201400330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y.-H.P. Production of biocommodities and bioelectricity by cell-free synthetic enzymatic pathway biotransformations: challenges and opportunities. Biotechnol Bioeng. 2010;105:663–677. doi: 10.1002/bit.22630. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y. Cell-free synthetic biology: engineering in an open world. Synth Syst Biotechnol. 2017;2:23–27. doi: 10.1016/j.synbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi H., Okano K., Honda K. Modules for in vitro metabolic engineering: pathway assembly for bio-based production of value-added chemicals. Synth Syst Biotechnol. 2017;2:65–74. doi: 10.1016/j.synbio.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rollin J.A., Tam T.K., Zhang Y.-H.P. New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green Chem. 2013;15:1708–1719. [Google Scholar]
- 15.Walsh C.T. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 16.Lambalot R.H., Gehring A.M., Flugel R.S., Zuber P., LaCelle M., Marahiel M.A., Reid R., Khosla C., Walsh C.T. A new enzyme superfamily – the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 17.Quadri L.E.N., Weinreb P.H., Lei M., Nakano M.M., Zuber P., Walsh C.T. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl Carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 18.Sattely E.S., Fischbach M.A., Walsh C.T. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat Prod Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 19.Lowry B., Walsh C.T., Khosla C. In vitro reconstitution of metabolic pathways: insights into nature's chemical logic. Synlett. 2015;26:1008–1025. doi: 10.1055/s-0034-1380264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes J., Haydock S.F., Roberts G.A., Bevitt D.J., Leadlay P.F. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348:176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 21.Donadio S., Staver M.J., McAlpine J.B., Swanson S.J., Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 22.Khosla C., Tang Y., Chen A.Y., Schnarr N.A., Cane D.E. Structure and mechanism of the 6-deoxyerythronolide B synthase. Annu Rev Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 23.Pfeifer B.A., Admiraal S.J., Gramajo H., Cane D.E., Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 24.Lowry B., Robbins T., Weng C.H., O'Brien R.V., Cane D.E., Khosla C. In vitro reconstitution and analysis of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2013;135:16809–16812. doi: 10.1021/ja409048k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Q., Xiang L., Izumikawa M., Meluzzi D., Moore B.S. Enzymatic total synthesis of enterocin polyketides. Nat Chem Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 26.Izumikawa M., Cheng Q., Moore B.S. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J Am Chem Soc. 2006;128:1428–1429. doi: 10.1021/ja0559707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balibar C.J., Howard-Jones A.R., Walsh C.T. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- 28.Magarvey N.A., Ehling-Schulz M., Walsh C.T. Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J Am Chem Soc. 2006;128:10698–10699. doi: 10.1021/ja0640187. [DOI] [PubMed] [Google Scholar]
- 29.Jaitzig J., Li J., Süssmuth R.D., Neubauer P. Reconstituted biosynthesis of the nonribosomal macrolactone antibiotic valinomycin in Escherichia coli. ACS Synth Biol. 2014;3:432–438. doi: 10.1021/sb400082j. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W., Ostash B., Walsh C.T. Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics. Proc Natl Acad Sci USA. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W., Ntai I., Bolla M.L., Malcolmson S.J., Kahne D., Kelleher N.L., Walsh C.T. Nine enzymes are required for assembly of the pacidamycin group of peptidyl nucleoside antibiotics. J Am Chem Soc. 2011;133:5240–5243. doi: 10.1021/ja2011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh C.T., Zhang W. Chemical logic and enzymatic machinery for biological assembly of peptidyl nucleoside antibiotics. ACS Chem Biol. 2011;6:1000–1007. doi: 10.1021/cb200284p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry R.D., Balbo P.B., Jones H.A., Fetherston J.D., DeMoll E. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology. 1999;145:1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 34.Miller D.A., Luo L., Hillson N., Keating T.A., Walsh C.T. Yersiniabactin synthetase: a four-protein assembly line producing the nonribosomal peptide/polyketide hybrid siderophore of Yersinia pestis. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 35.Greunke C., Glöckle A., Antosch J., Gulder T.A. Biocatalytic total synthesis of ikarugamycin. Angew Chem Int Ed. 2017;56:4351–4355. doi: 10.1002/anie.201611063. [DOI] [PubMed] [Google Scholar]
- 36.Hodgman C.E., Jewett M.C. Cell-free synthetic biology: thinking outside the cell. Metab Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez J.G., Stark J.C., Jewett M.C. Cell-free synthetic biology: engineering beyond the cell. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zemella A., Thoring L., Hoffmeister C., Kubick S. Cell-free protein synthesis: pros and cons of prokaryotic and eukaryotic systems. ChemBioChem. 2015;16:2420–2431. doi: 10.1002/cbic.201500340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Kanter G., Voloshin A., Michel-Reydellet N., Velkeen H., Levy R., Swartz J.R. Rapid expression of vaccine proteins for B-cell lymphoma in a cell-free system. Biotechnol Bioeng. 2005;89:503–511. doi: 10.1002/bit.20283. [DOI] [PubMed] [Google Scholar]
- 40.Martin R.W., Majewska N.I., Chen C.X., Albanetti T.E., Jimenez R.B.C., Schmelzer A.E., Jewett M.C., Roy V. Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies. ACS Synth Biol. 2017;6:1370–1379. doi: 10.1021/acssynbio.7b00001. [DOI] [PubMed] [Google Scholar]
- 41.Bundy B.C., Franciszkowicz M.J., Swartz J.R. Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnol Bioeng. 2008;100:28–37. doi: 10.1002/bit.21716. [DOI] [PubMed] [Google Scholar]
- 42.Schoborg J.A., Hershewe J.M., Stark J.C., Kightlinger W., Kath J.E., Jaroentomeechai T., Natarajan A., DeLisa M.P., Jewett M.C. A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol Bioeng. 2018;115:739–750. doi: 10.1002/bit.26502. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Lawton T.J., Kostecki J.S., Nisthal A., Fang J., Mayo S.L., Rosenzweig A.C., Jewett M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol J. 2016;11:212–218. doi: 10.1002/biot.201500030. [DOI] [PubMed] [Google Scholar]
- 44.Hong S.H., Ntai I., Haimovich A.D., Kelleher N.L., Isaacs F.J., Jewett M.C. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth Biol. 2014;3:398–409. doi: 10.1021/sb400140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karim A.S., Jewett M.C. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab Eng. 2016;36:116–126. doi: 10.1016/j.ymben.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Karim A.S., Heggestad J.T., Crowe S.A., Jewett M.C. Controlling cell-free metabolism through physiochemical perturbations. Metab Eng. 2018;45:86–94. doi: 10.1016/j.ymben.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M., Bosch I., Dudley D.M., O'Connor D.H., Gehrke L., Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 48.Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D., Ferrante T., McSorley F.R., Furuta Y., Vernet A., Lewandowski M., Boddy C.N., Joshi N.S., Collins J.J. Portable, on-demand biomolecular manufacturing. Cell. 2016;167:248–259. doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 49.Goering A.W., Li J., McClure R.A., Thomson R.J., Jewett M.C., Kelleher N.L. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth Biol. 2017;6:39–44. doi: 10.1021/acssynbio.6b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krätzschmar J., Krause M., Marahiel M.A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marahiel M.A. Protein templates for the biosynthesis of peptide antibiotics. Chem Biol. 1997;4:561–567. doi: 10.1016/s1074-5521(97)90242-8. [DOI] [PubMed] [Google Scholar]
- 52.Kurotsu T., Hori K., Kanda M., Saito Y. Characterization and location of the L-proline activating fragment from the multifunctional gramicidin S synthetase 2. J Biochem. 1991;109:763–769. doi: 10.1093/oxfordjournals.jbchem.a123454. [DOI] [PubMed] [Google Scholar]
- 53.Stachelhaus T., Mootz H.D., Bergendahl V., Marahiel M.A. Peptide bond formation in nonribosomal peptide biosynthesis. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 54.Gruenewald S., Mootz H.D., Stehmeier P., Stachelhaus T. In vivo production of artificial nonribosomal peptide products in the heterologous host Escherichia coli. Appl Environ Microbiol. 2004;70:3282–3291. doi: 10.1128/AEM.70.6.3282-3291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthes D., Richter L., Müller J., Denisiuk A., Feifel S.C., Xu Y., Espinosa-Artiles P., Süssmuth R.D., Molnár I. In vitro chemoenzymatic and in vivo biocatalytic syntheses of new beauvericin analogues. Chem Commun. 2012;48:5674–5676. doi: 10.1039/c2cc31669b. [DOI] [PubMed] [Google Scholar]
- 56.Gomez-Escribano J.P., Bibb M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aigle B., Lautru S., Spiteller D., Dickschat J.S., Challis G.L., Leblond P., Pernodet J.L. Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol. 2014;41:251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- 58.Li J., Wang H., Kwon Y.C., Jewett M.C. Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol Bioeng. 2017;114:1343–1353. doi: 10.1002/bit.26253. [DOI] [PubMed] [Google Scholar]
- 59.Moore S.J., Lai H.E., Needham H., Polizzi K.M., Freemont P.S. Streptomyces venezuelae TX-TL – a next generation cell-free synthetic biology tool. Biotechnol J. 2017;12 doi: 10.1002/biot.201600678. [DOI] [PubMed] [Google Scholar]
- 60.Li J., Wang H., Jewett M.C. Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields. Biochem Eng J. 2018;130:29–33. [Google Scholar]