Abstract
The interplay between inflammation and metabolism is widely recognized, yet the underlying molecular mechanisms remain poorly characterized. Using experimental database mining and genome-wide gene expression profiling methods, we found that in contrast to other TNFAIP8 family members, TNFAIP8L2 (TIPE2) was preferentially expressed in human myeloid cell types. In addition, Tnfaip8l2 expression drastically decreased in lipopolysaccharide (LPS)- stimulated macrophages. Consequently, Tnfaip8l2 deficiency led to heightened expression of genes that were enriched for leukocyte activation and lipid biosynthesis pathways. Furthermore, mitochondrial respiration rate was increased in Tnfaip8l2-deficient macrophages, as measured by Seahorse metabolic analyzer. Taken together, these results indicate that Tnfaip8l2 serves as a “brake” for immunometabolism, which needs to be released for optimized metabolic reprogramming as well as mounting effective inflammatory responses. The unique anti-inflammatory and metabolic-modulatory function of TNFAIP8L2 renders it a novel therapeutic target for cardiovascular diseases and cancer.
Keywords: inflammation, lipid metabolism, immunometabolism, cardiovascular diseases, cancer
Introduction
There are four members in the TNFAIP8 (TNF alpha induced protein 8) family, which includes TNFAIP8, TNFAIP8L1 (TIPE1), TNFAIP8L2 (TIPE2), and TNFAIP8L3 (TIPE3). We found recently that germline deletion of TNFAIP8L2 causes fatal inflammation and hypersensitivity to Toll-like receptor stimulation[1]. Crystal structure of TNFAIP8L2 reveals a unique structural fold, which is shared by all members of the TNFAIP8 family, with no significant homology to other known proteins[2]. Although TNFAIP8L2 has emerged as a critical regulator of immunity that maintains immune homeostasis, its additional cellular function and the mechanisms of its action are poorly understood.
Recently, there has been considerable interest in the role of cellular metabolism as a central regulator of immunity. In adaptive immune cells, induction of de novo fatty acid synthesis is essential for activation-induced proliferation and differentiation of effector T cells[3, 4], while fatty acid catabolism via β-oxidation is important for the differentiation of regulatory T cells[5]. The role of fatty acid metabolism in innate immune cells, however, is controversial[6, 7]. De novo fatty acid synthesis was proposed to support fatty acid oxidation that is required during M2 macrophage differentiation. However, it was recently demonstrated that fatty acid oxidation is dispensable in early activation of M2 macrophages. Thus, the role of fatty acid metabolism in immune cells, especially in the innate immune cells such as macrophages, remains to be elucidated.
In this study, we tested a novel hypothesis that TNFAIP8L2 is involved in metabolic regulation in the immune cells. Using microarray analysis and experimental database mining method, we found that TNFAIP8L2 is exclusively expressed in myeloid cells. TNFAIP8L2 gene expression is reduced in response to proinflammatory stimuli in macrophages, which serves as an immune “brake” molecule that inhibit inflammation, lipid biosynthesis genes, and mitochondrial respiration. Thus, TNFAIP8L2 is an immune checkpoint molecule in myeloid cells and targeting TNFAIP8L2 may serve as a novel therapeutic target for inflammatory and metabolic diseases.
Results
1. TNFAIP8 family members are differentially expressed in human tissues
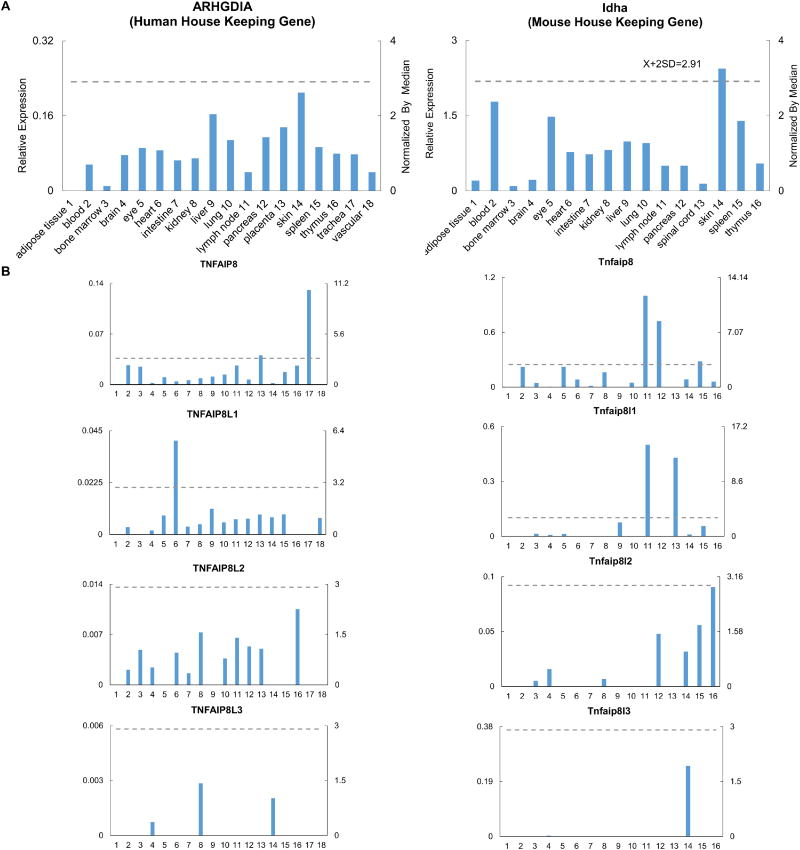
The expression profile of TNFAIP8 family members, especially in humans, is poorly characterized. We hypothesized that to fulfill overlapping yet distinct cellular functions, TNFAIP8 family members are differentially expressed in tissues. To examine this hypothesis, a database mining method that we developed, was used to examine experimentally verified expression profiles of mRNA transcripts of four TNFAIP8 family members including TNFAIP8, TNFAIP8L1, TNFAIP8L2, and TNFAIP8L3 in NCBI-UniGene database. The expression of these four genes were examined in 18 human tissues and 16 mouse tissues. The results showed that the TNFAIP8 family members are differentially expressed. TNFAIP8, TNFAIP8L1, and TNFAIP8L2 are ubiquitously expressed in human tissues, whereas TNFAIP8L3 is not constitutively expressed in most human tissues (Figure 1 and Table 1). In addition, TNFAIP8 is highly expressed in human placenta and trachea tissues and TNFAIP8L1 is highly expressed in human heart tissue (Table 1). In mouse, Tnfaip8 is also ubiquitously expressed in tissues, but Tnfaip8l1, Tnfaip8l2, and Tnfaip8l3 have a limited expression profile in mouse tissues, compared to their expression in humans (Figure 1 and Table 2). Tnfaip8 is highly expressed in the lymph node, pancreas, and spleen, and Tnfaip8l1 is highly expressed in the lymph node and spinal cord (Table 2). The differential expression pattern of TNFAIP8 family members in human and mouse suggested that they have distinct tissue functions, despite their structural similarities.
Figure 1. The gene expression profiles of TNFAIP8 family members in human and murine tissues.
A. Data presentation format (The data presented in X-, Y-axis, and tissue order of ARHGDIA and Idha are applied to all the human and mouse genes examined respectively). As an example, the gene expression profiles of human housekeeping gene Rho GDP dissociation inhibitor (GDI) alpha (ARHGDIA) in the eighteen tissues are presented, with the tissue names and position numbers shown on the X-axis. The gene expression data were normalized by the β-actin (Hs. 520640) expression data from the same tissue, which are presented on the left Y-axis. The expression ratios among tissues were generated by normalizing the arbitrary units of the gene in the tissues with the median level of the arbitrary units of the gene in all the tissues which are presented on the right Y-axis. In order to define confidence intervals for statistically higher expression levels of given genes, we calculated the confidence intervals of tissue expression for three housekeeping genes [ the mean X+ 2 x standard deviations (SD) = 2.83] including ARHGDIA (Hs. 159161), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs. 544577), and ribosomal protein S27a (RPS27A, Hs. 311640). The expression variations of given genes in tissues, when they were larger than 2.83-fold, were defined as the high expression levels with statistical significance (the right Y-axis). To define confidence intervals for statistically higher expression levels of given genes in 14 mouse tissues, we calculated the confidence intervals of tissue expression [the mean X + 2 x standard deviations (SD) = 3.0] for three mouse house keeping genes including Lactate dehydrogenase A (Ldha, Mm. 29324), non-POU-domain-containing octamer binding protein (Nono, Mm. 280069), and ribosomal protein L32 (Rpl32, Mm. 104368). The expression variations of given genes in tissues, when they were larger than 3.0-fold, were defined as the high expression levels with statistical significance (the right Y-axis). B. The expression profiles of TNFAIP8 family in human (Left column, with TNFAIP8 protein with capital letters) and mouse (right column, with Tnfaip8 protein designated with lowercase letters) tissues.
Table 1.
TNFAIP8 protein family are differentially expressed in human tissues. The colored values indicate high expression levels with statistical significance among all tissues.
| Human | TNFAIP8 | TNFAIP8L1 | TNFAIP8L2 | TNFAIP8L3 |
|---|---|---|---|---|
| adipose tissue | – | – | – | – |
| blood | 2.15 | 0.45 | 0.45 | – |
| bone marrow | 1.99 | – | 1.04 | – |
| brain | 0.19 | 0.24 | 0.52 | 0.36 |
| eye | 0.80 | 1.18 | – | – |
| heart | 0.36 | 5.82 | 0.96 | – |
| intestine | 0.48 | 0.49 | 0.34 | – |
| kidney | 0.71 | 0.63 | 1.56 | 1.40 |
| liver | 0.89 | 1.59 | – | – |
| lung | 1.11 | 0.75 | 0.79 | – |
| lymph node | 2.12 | 0.94 | 1.40 | – |
| pancreas | 0.55 | 0.98 | 1.14 | – |
| placenta | 3.24 | 1.23 | 1.07 | – |
| skin | 0.16 | 1.07 | – | 1.00 |
| spleen | 1.39 | 1.25 | – | – |
| thymus | 2.11 | – | 2.26 | – |
| trachea | 10.47 | – | – | – |
| vascular | – | 1.02 | – | – |
Table 2.
TNFAIP8 protein family are differentially expressed in mouse tissues. The colored values indicate high expression levels with statistical significance among all tissues.
| Mouse | Tnfaip8 | Tnfaip8l1 | Tnfaip8l2 | Tnfaip8l3 |
|---|---|---|---|---|
| adipose tissue | – | – | – | – |
| blood | 2.62 | – | – | – |
| bone marrow | 0.53 | 0.40 | 0.16 | – |
| brain | 0.03 | 0.22 | 0.50 | 0.02 |
| eye | 2.62 | 0.35 | – | – |
| heart | 0.98 | – | – | – |
| intestine | 0.15 | – | – | – |
| kidney | 1.91 | – | 0.21 | – |
| liver | – | 2.15 | – | – |
| lung | 0.57 | – | – | – |
| lymph node | 11.79 | 14.26 | – | – |
| pancreas | 8.53 | – | 1.51 | – |
| spinal cord | – | 12.24 | – | – |
| skin | 1.00 | 0.29 | 1.00 | 1.97 |
| spleen | 3.32 | 1.60 | 1.77 | – |
| thymus | 0.71 | – | 2.86 | – |
2. TNFAIP8L2 is primarily expressed in the myeloid cells in human
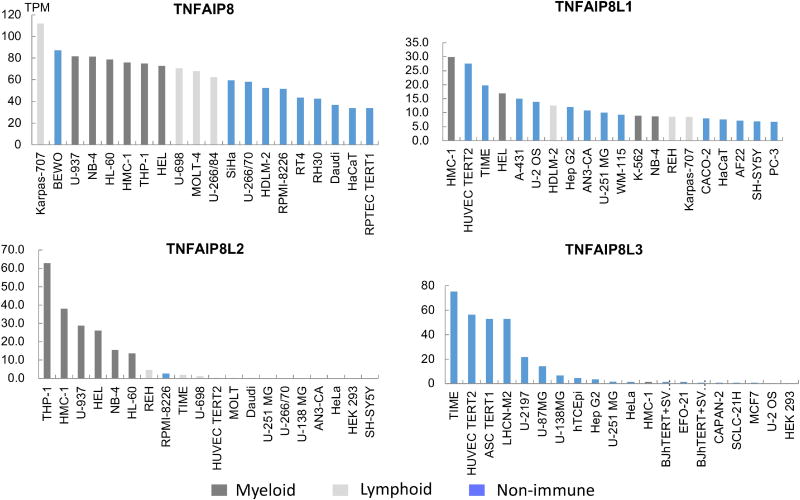
Although we did not see an enrichment of TNFAIP8L2 in both human and mouse tissues (Figure 1), we hypothesized that TNFAIP8L2 gene expression could be limited to certain cell types. For this purpose, we examined the transcript abundance of TNFAIP8 family members in a panel of 56 human cell lines, which could be categorized into different groups (myeloid, lymphoid, or non-immune cell types) based on the organs that they were obtained from (Figure 2). The results showed that TNFAIP8 and TNFAIP8L2 are highly expressed in the immune cell types. Although TNFAIP8 is highly expressed both in the myeloid and lymphoid lineage, TNFAIP8L2 transcripts are found exclusively in the myeloid lineage. By contrast, TNFAIP8L3 is highly expressed in the non-immune cell types, whereas TNFAIP8L1 is expressed both in the immune and non-immune cell types. Despite that TNFAIP8L2 is ubiquitously expressed in human tissues (Figure 1), its expression profile is limited to that of myeloid lineage, suggesting that the expression of TNFAIP8L2 in the non-immune tissues is likely due to the presence of myeloid cells.
Figure 2. The gene expression profiles of TNFAIP8 family members in human cells.
Transcription abundance of TNFAIP8 family members are examined in the Human Protein Atlas project. The analyzed cell lines are divided into 3 color-coded groups according to the organ they were obtained from. The RNA-sequencing results generated are reported as number of Transcripts per Kilobase Million (TPM). Top 20 most expressed out of 56 cell lines examined are shown.
3. Tnfaip8l2 negatively regulates inflammation and metabolism in the macrophage
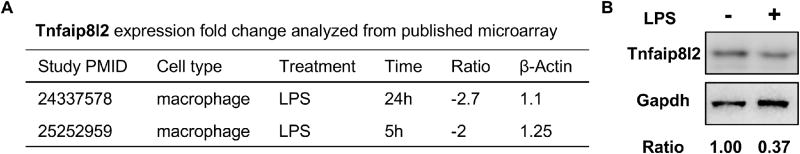
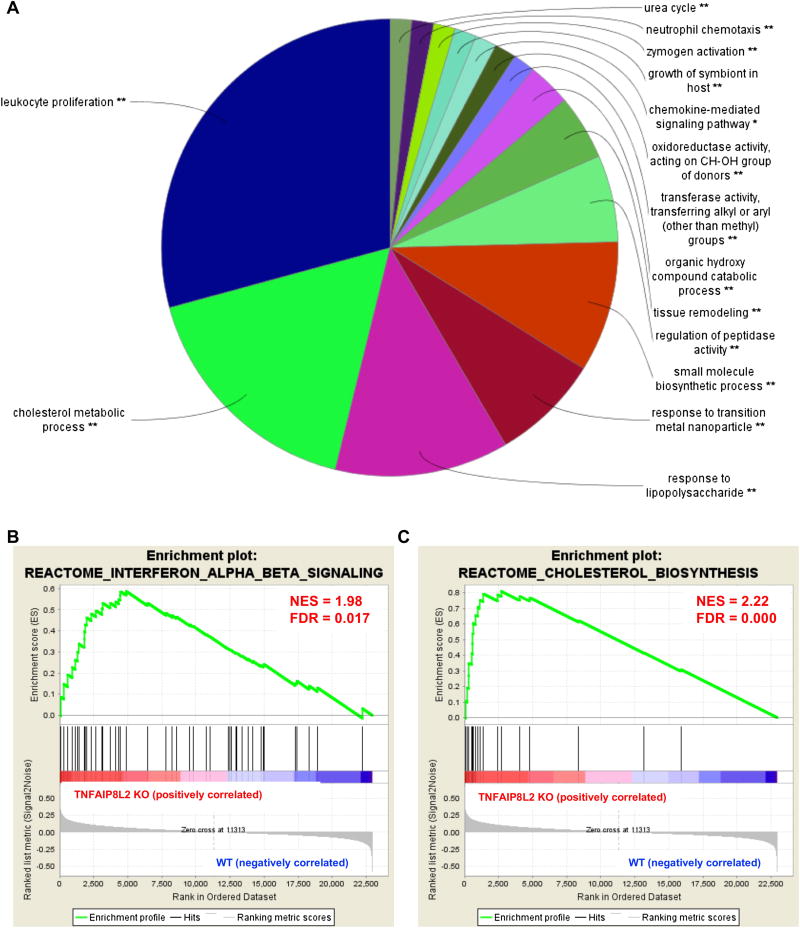
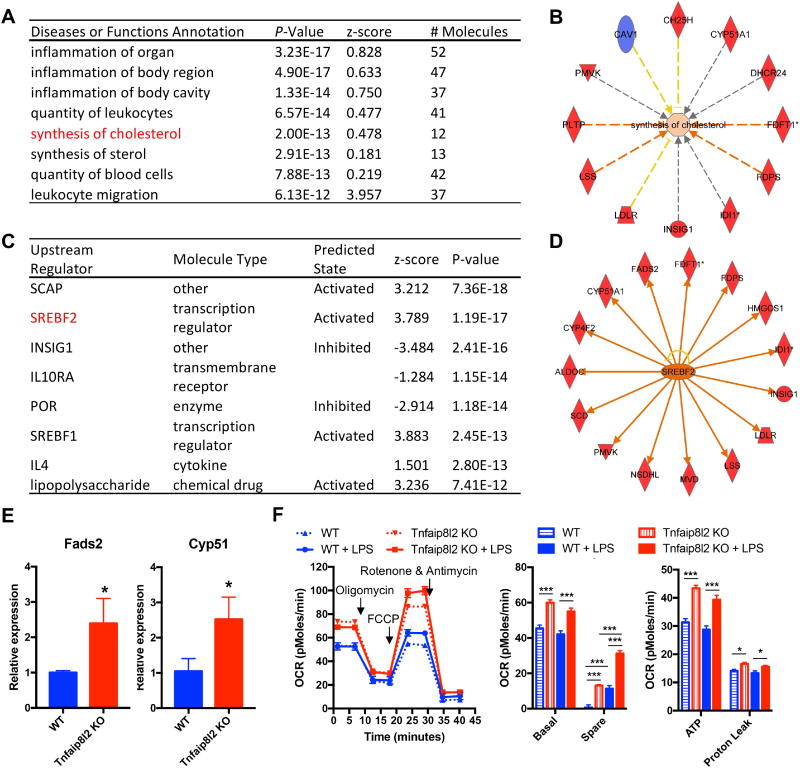
Next, we hypothesized that Tnfaip8l2 gene expression could be changed in myeloid cells in response to proinflammatory stimulation. To test this, we examined Tnfaip8l2 expression in mouse macrophages after lipopolysaccharide (LPS) stimulation and found that it is downregulated by more than two-folds in two separate microarray datasets (Figure 3A). We performed Wester Blots and confirmed that Tnfaip8l2 was downregulated more than two folds in the bone marrow-derived macrophages (BMDMs) treated with LPS for 1 hour (Figure 3B). These results correlated well with our previous findings from BMDMs [8] and macrophage cell lines [9]. To determine the specific cellular functions of Tnfaip8l2, we performed microarray analysis in the BMDMs of Tnfaip8l2 knockout (KO) mice stimulated by LPS (100 ng/mL) in comparison to those of wild type control mice. The results showed that Tnfaip8l2 was the most significantly downregulated gene in the dataset, suggesting the high quality of our microarray analysis (Table 3). We then performed pathway analysis using ClueGO software to visualize the non-redundant biological terms for the genes that were changed by Tnfaip8l2 gene deficiency. The ClueGO network was created with kappa statistics and reflected the relationships between the terms based on the similarity of their associated genes and the related terms which share similar associate genes were fused to reduce redundancy. The results showed that “Leukocyte Proliferation” and “Response to lipopolysaccharide” terms were significantly enriched comparing Tnfaip8l2 KO BMDMs with WT BMDMs (Figure 4A). In addition, several smaller clusters such as “Positive regulation of apoptotic signaling pathway” were also significantly changed in response to Tnfaip8l2 KO. Interestingly, two clusters including “cholesterol metabolic process” and “small molecule biosynthetic process” were also affected by TNFAIP8L2. To determine whether these signaling pathways were increased or decreased in Tnfaip8l2 KO BMDMs, we performed Gene Set Enrichment Analysis (GSEA), which is a computational method that determines whether a priori defined set of genes shows statistically significant, concordant differences between two biological states. We found that “interferon signaling” and “cholesterol biosynthesis” were positively correlated in Tnfaip8l2 KO BMDMs in comparison to WT BMDMs (Figure 4B and Figure 4C), suggesting that TNFAIP8L2 negatively regulated inflammation and lipid biosynthesis pathways. To determine the specific pathways that are most sensitive to TNFAIP8L2 deficiency, we utilized INGENUITY pathway analysis. Correlating with previous analyses, the results showed that genes involved in “inflammation of organ” and “synthesis of cholesterol" were significantly affected by TNFAIP8L2 (Figure 5A and 5B). In addition, when we examined the top upstream regulators of TNFAIP8L2-regulated genes, we found that “SREBP cleavage-activating protein (SCAP)” and “Sterol regulatory element-binding transcription factor 2 (SREBF2)” were predicted to be the most likely upstream regulators and were also predicted to be activated after Tnfaip8l2 KO (Figure 5C). SREBPs are encoded by the genes SREBF1 and SREBF2 and they are master transcription factors in cholesterol and fatty acid biosynthesis. When we examined the genes that were regulated by SREBF2, we found that 15 genes such as Fatty Acid Desaturase 2 (Fads2), Stearoyl-CoA Desaturase (Scd) were unidirectional upregulated in response to Tnfaip8l2 deficiency (Figure 5D). Moreover, Scd was the most upregulated genes (4.14-folds) in Tnfaip8l2 KO BMDMs in comparison to WT BMDMs (Table 4). Scd is a key enzyme in fatty acid metabolism, which is important for the production of phosphoinositides, eicosanoids, and sphingolipids. We performed q-PCR and confirmed that key fatty acid/cholesterol metabolic enzymes Fads2 and Cyp51 were significantly upregulated by more than two folds in Tnfaip8l2- deficient macrophages (Figure 5E). Moreover, mitochondrial respiration profiling by Seahorse XF analyzer revealed that both basal and spare mitochondrial respiration rate were increased in Tnfaip8l2-deficient macrophages (Figure 5F). Basal mitochondrial respiration is linked to both ATP production and proton leak. Our results showed that only ATP production, but not proton leak, was significantly induced in Tnfaip8l2 KO macrophages (Figure 5F). Importantly, mitochondrial respiration was increased in Tnfaip8l2 KO BMDMs under both basal condition and 1-hour LPS stimulation. LPS treatment induced spare respiratory capacity in wild type BMDMs, which was further increased in LPS-treated Tnfaip8l2 KO BMDMs (Figure 5F). As previously reported, macrophages do not significant alter their basal respiration at early time point (1-2h) after LPS stimulation (Figure 5F), but they undergo a strong metabolic remodeling (low respiration, high glycolysis) after long-term LPS challenge, which is a hallmark of macrophage activation[10]. For this reason, we examined mitochondrial metabolism of WT and Tnfaip8l2- deficient BMDM after 18-hour LPS stimulation. The results showed that none of the mitochondrial parameters was significantly changed in Tnfaip8l2 KO BMDMs (Supplemental Figure 3). These results indicated that Tnfaip8l2 is involved in the regulation of mitochondrial metabolism of macrophages under both basal and acute inflammatory settings, but it is not involved in the regulation of mitochondrial respiration after chronic LPS stimulation. Taken together, these results indicate for the first time that TNFAIP8L2 negatively regulates mitochondrial respiration and the genes involved in lipid biosynthesis.
Figure 3. Tnfaip8l2 is downregulated in macrophages after lipopolysaccharide (LPS) stimulation.
A. Tnfaip8l2 expression was examined in two published microarrays that examined the response of macrophage to LPS challenge. B. Bone marrow-derived macrophages from mice were challenged with LPS (100 ng/mL) for 1 hour and Western blotting was performed. PMID: PubMed ID.
Table 3.
Top ten most downregulated genes in TNFAIP8L2 knockout mice.
| Gene Symbol | Full Name | Location | Fold Change |
|---|---|---|---|
| Tnfaip8l2 | TNF alpha induced protein 8 like 2 | Cytoplasm | −2.69 |
| Snora75 | small nucleolar RNA, H/ACA box 75 | Other | −2.27 |
| Colec12 | collectin subfamily member 12 | Plasma Membrane | −2.19 |
| Mir221 | microRNA 221 | Cytoplasm | −2.17 |
| n-R5s136 | nuclear encoded rRNA 5S 136 | Other | −2.17 |
| Slc6a4 | solute carrier family 6 member 4 | Plasma Membrane | −2.15 |
| Olfml3 | olfactomedin like 3 | Extracellular Space | −2.07 |
| Igf2bp3 | insulin like growth factor 2 mRNA binding protein 3 | Cytoplasm | −2.06 |
| Abhd3 | abhydrolase domain containing 3 | Plasma Membrane | −2.04 |
| Mir181b-1 | microRNA 181a-1 | Cytoplasm | −1.97 |
Figure 4. TNFAIP8L2 negatively regulates gene signatures of inflammation and lipid biosynthesis.
Bone marrow-derived macrophages from either wild type mice or Tnfaip8l2 gene knockout (KO) mice (pooled sample of 5 in each group) were challenged with LPS (100 ng/mL) for 1 hour and microarray was performed. A. ClueGO pathway enrichment analysis of the genes that are changed by more than 1.5-folds in Tnfaip8l2 gene KO mice. B. Enrichment plots from Gene Set Enrichment Analysis (GSEA) of “interferon signaling” and “cholesterol biosynthesis”.
Figure 5. TNFAIP8L2 negatively regulates genes related to transcription factor Sterol regulatory element-binding protein 2 (SREBP-2) and mitochondrial respiration.
Bone marrow-derived macrophages from either wild type mice or Tnfaip8l2 gene knockout (KO) mice were challenged with LPS (100 ng/mL) for 1 hour and microarray with Ingenuity Pathway Analysis (A to D), qPCR (E), and mitochondrial respiration measurement by Seahorse XF96 analyzer (F) were performed. A. Top enriched diseases or functions annotation of the genes that are changed by more than 1.5-folds in Tnfaip8l2 gene KO mice. B. Tnfaip8l2 KO macrophages are positively enriched with the “synthesis of cholesterol” gene signature. C. Top upstream regulator analysis of the genes that are changed by more than 1.5-folds in Tnfaip8l2 gene KO mice. D. The 15 SREBP2-dependent genes that are induced by more than 1.5-folds in Tnfaip8l2 gene KO mice. Blue color indicates decreased gene expression, while red color indicates increased gene expression in Tnfaip8l2 KO macrophages. E. Confirmation of Fads2 and Cyp51 gene expression by qPCR (n=3 in each group). F. BMDMs from WT or Tnfaip8l2 KO mice were treated with vehicle control or LPS (100ng/mL) for 1 hour and oxygen consumption rate (OCR) was measured using Seahorse XF96 analyzer. Afterwards, XF Mito Stress Test was performed by sequential adding of Oligomycin (2 μM), FCCP (2 μM), and Rotenone (1 μM). Experiment plot (left) and quantification of four mitochondrial parameters (right) were shown (n=7 to 8 in each group). For all panels, data are expressed as mean ± SEM.*, p<0.05, **, p<0.01, ***, p<0.001, NS, not significant.
Table 4.
Top ten most upregulated genes in TNFAIP8L2 knockout mice
| Gene Symbol | Full Name | Location | Fold Change |
|---|---|---|---|
| Scd1 | stearoyl-CoA desaturase | Cytoplasm | 4.14 |
| Rps3a3 | ribosomal protein S3A3 | Other | 2.94 |
| Mnd1 | meiotic nuclear divisions 1 | Nucleus | 2.78 |
| Fcrls | Fc receptor-like S, scavenger receptor | Plasma Membrane | 2.71 |
| 2010005H15Rik | cystatin A | Cytoplasm | 2.42 |
| Ltb4r1 | leukotriene B4 receptor | Plasma Membrane | 2.42 |
| Fads2 | fatty acid desaturase 2 | Plasma Membrane | 2.31 |
| LOC102634459 | uncharacterized LOC102634459 | Other | 2.17 |
| Gm3571 | farnesyl diphosphate synthetase pseudogene | Other | 2.15 |
| Stfa1 | stefin A1 | Cytoplasm | 2.12 |
Next, we compared the tissue/cell expression profile of TNFAIP8L2 with BCL3 and SQSTM1, which we and the others previously identified as macrophage “brake” molecules[11, 12]. The results showed that both BCL3 and SQSTM1 were ubiquitously expressed in mouse and human tissues (Supplemental Figure 1 and Supplemental Table 1). BCL3 was highly expressed in mouse but not human spleen, while Sqstm1 was highly expressed in human skin and mouse liver. Moreover, unlike TNFAIP8L2, BCL3 and SQSTM1 were not preferentially expressed in human myeloid cells, but ubiquitously expressed in the immune and non-immune cells (Supplemental Figure 2). Furthermore, neither Bcl3 nor Sqstm1 was downregulated, but rather increased, in LPS-treated macrophages (Supplemental Table 2), while Tnfaip8l2 was downregulated in the same microarray studies (Figure 3A). Taken together, these results highlighted the unique role of myeloid-restricted TNFAIP8L2.
4. TNFAIP8L2 is induced in response to hyperlipidemia conditions
The findings that TNFAIP8L2 negatively regulates lipid biosynthesis pathway and the fact that TNFAIP8 family members including TNFAIP8L2 are lipid transfer proteins prompted us to examine the hypothesis that the gene expressions of TNFAIP8 family members might be reciprocally regulated by hyperlipidemia conditions. To examine this hypothesis, we firstly examined the expression profile of Tnfaip8, Tnfaip8l1, and Tnfaip8l2, and Tnfaip8l3 in three independent studies which studied the effects of high fat diet on adipose tissues. We found that Tnfaip8l2 gene expression were significantly induced (Fold change between 2.75 folds to 3.59 folds) after high fat feeding in the three datasets (Table 5). In addition, Tnfaip8 was also significantly induced in these datasets after high fat feeding, but to a lesser degree (fold change between 1.27 folds to 1.88). We also examined the gene expression of TNFAIP8 family members in different hyperlipidemia mouse models and found that Tnfaip8l2 gene expressions were also significantly induced in the aortas of Apolipoprotein E (ApoE) KO mice (mouse model for atherosclerosis), in the livers of Scd1 KO (mouse models for severe hypercholesterolemia) and in the adipose tissue of Gpr120 KO mice (mouse model for obesity) (Table 6). Similarly, Tnfaip8 was also significantly induced in these studies, while Tnfaip8l1 level did not change significantly and Tnfaip8l3 gene expression was not available in these studies. Collectively, these results indicate Tnfaip8 and Tnfaip8l2 gene expressions are induced in different tissues in response to high fat feeding or hyperlipidemia conditions. It is not clear, however, whether these effects were due to gene expression changes in the adipocyte or due to increased immune cell infiltration in these tissues.
Table 5.
High fat diet induces TNFAIP8L2 in white adipose tissue.
| GEO ID | Comparison | Tnfaip8 | Tnfaip8l1 | Tnfaip8l2 | Tnfaip8l3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| FC | P val. | FC | P val. | FC | P val. | FC | P val. | ||
| GSE32095 | chow vs high fat diet | 1.88 | 0.00576 | 2.93 | 0.00254 | 2.75 | 0.00472 | – | – |
| GSE36033 | chow vs high fat diet | 1.27 | 0.00525 | 1.40 | 0.0599 | 2.77 | 0.000217 | 0.91 | 0.136 |
| GSE63198 | chow vs high fat diet | 1.52 | 0.00002 | – | – | 3.59 | 0.000000 | – | – |
FC, fold change.
Table 6.
Hyperlipidemia condition induces TNFAIP8L2 in various hyperlipidemia mouse models.
| GEO ID | Comparison | Tnfaip8 | Tnfaip8l1 | Tnfaip8l2 | Tnfaip8l3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| FC | P val. | FC | P val. | FC | P val. | FC | P val. | ||
| GSE2372 | WT vs ApoE KO | 1.35 | 0.0249 | 0.86 | 0.468 | 2.61 | 0.00023 | – | – |
| GSE19286 | WT vs ApoE KO | 1.42 | 0.0312 | 1.06 | 0.509 | 1.34 | 0.0246 | – | – |
| GSE3889 | WT vs Scd1 KO | 2.55 | 7.87E–07 | 0.61 | 0.533 | 1.60 | 0.000191 | – | – |
| GSE32095 | WT vs Gpr120 KO | 1.69 | 0.0432 | 0.77 | 0.241 | 3.06 | 0.00364 | – | – |
FC, fold change. ApoE, apolipoprotein E. WT, wild-type, KO, knockout.
Discussion
The results reported here argue that TNFAIP8L2 protein can function as an immune checkpoint “brake” in the macrophages. In order to achieve efficient immune response, immune checkpoint molecule TNFAIP8L2 needs to be suppressed to achieve full-blown inflammation. The specific mechanisms underlying LPS-induced TNFAIP8L2 gene downregulation remain to be identified. On one hand, it was suggested that in macrophages activated by LPS, Tnfaip8l2 was among the top 25 most highly repressed super enhancers. On the other hand, previous mouse-human experimental epigenetic analysis revealed that TNFAIP8L2 was significantly hypomethylated in high-fat-diet mice and obese humans, and was significantly induced in the corresponding subjects[13]. These results indicate that TNFAIP8L2 might be downregulated/induced by an alteration of epigenetic status in response to proinflammatory stimulation or hyperlipidemia conditions.
The relationship between increased mitochondrial respiration and heightened inflammation in Tnfaip8l2 KO BMDMs remains to be examined. Several factors, including fatty acid oxidation, glycolysis, and mitochondrial modifications, are involved in the regulation of mitochondrial respiration. Future experiments are warranted to determine whether inhibitors of each of these factors could revert the inflammatory phenotype in Tnfaip8l2 KO BMDMs.
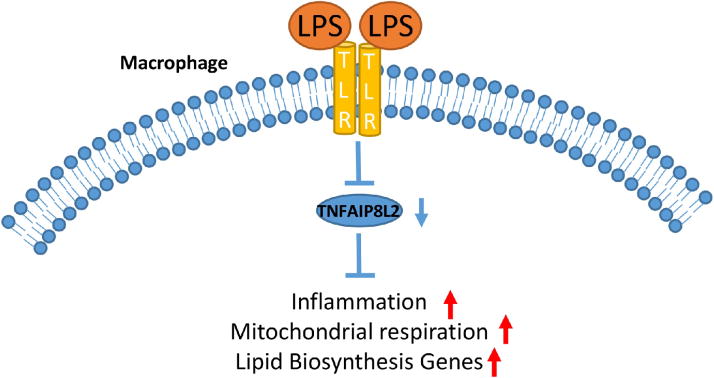
We propose a new working model to summarize our findings (Figure 6). During inflammation, TNFAIP8L2 gene expression is downregulated, potentially due to alteration of its epigenetic status. The downregulated TNFAIP8L2 results in upregulated mitochondrial respiration, lipid biosynthesis gene expression, and inflammation. Despite current therapies such as lipid-lowering statins, cardiovascular diseases, including myocardial infarction, stroke, and peripheral arterial disease, remain the number one cause of death in the United States. The development of cardiovascular diseases is fueled by aberrant lipid metabolism and malfunction of the immune system. The unique dual anti-inflammatory and lipid-inhibitory function of TNFAIP8L2 reported here renders TNFAIP8L2 an ideal novel therapeutic target for cardiovascular diseases.
Figure 6. A working model of TNFAIP8L2.
In macrophages stimulated with danger signals such as Toll-like receptor (TLR) ligand lipopolysaccharide (LPS), TNFAIP8L2 gene expression is downregulated, potentially due to alteration of its epigenetic status. The decreased TNFAIP8L2 leads to increased mitochondrial respiration, lipid biosynthesis, and inflammatory gene expression.
Materials and Methods
Chemicals and Antibodies
All chemicals were from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Lipopolysaccharides from Escherichia coli O55:B5 was purchased from Sigma-Aldrich (#L2880, Lot number 113M4068V; Alabaster, Alabama). The primers (5′-3′) used for the real-time PCR are: mouse Fads2: GCTCATCCCTATGTACTTCCAG and CTCCCAAGATGCCGTAGAAAG; and mouse Cyp51: CAAAGACTGAAAGACTCCTGGG and TCTCCAACACAACGATGACG. Housekeeping 18s ribosomal RNA primers were purchased from QuantiTect Primer Assay (Qiagen, CA). Anti-Tnfaip8l2 antibody (#15940-1-AP) was purchased from proteintech (Chicago, IL). Anti-GAPDH antibody (#2118) was purchased from Cell Signaling (Danvers, MA).
Animals Samples
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) Guidelines and Authorization for the use of Laboratory Animals and were approved by the IACUC of University of Pennsylvania School of Medicine, which confirmed to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. All mice used were on a C57BL/6 background.
Tissue expression profiles of genes encoding TNFAIP8 family members in human and mouse
An experimental data mining strategy, as we reported, was used to analyze the expression profiles of mRNA transcripts of genes in immune system and other tissues in humans and mice by mining experimentally verified mRNA expressions in the expressed sequence tag (EST) databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) UniGene (http://www.ncbi.nlm.nih.gov/ sites/entrez?db=unigene). Transcripts per million of genes of interest were normalized with that of house-keeping β-actin in each given tissue to calculate the arbitrary units of gene expression. A confidence interval of the expression variation of house-keeping genes was generated by calculating the mean plus two times that of the standard deviation of the arbitrary units of three randomly selected house- keeping genes (ARHGDIA, GADPH, and RPS27A in human; Ldha, Nono, and Rpl32 in mouse) normalized by β-actin in the given tissues (Fig. 2A). If the expression variation of a given gene in the tissues was larger than the upper limit of the confidence interval (the mean plus two times the standard deviation) of the house-keeping genes, the high expression levels of genes in the tissues were considered statistically significant. Gene transcripts lower than one per million were technically presented as no expression.
Transcript level profiling in human cell lines
The transcript levels of TNFAIP8 family members in human cell lines were examined in the Human Protein Atalas. Briefly, the cell lines in the human protein atlas have been analyzed by RNA-Seq to estimate the transcript abundance of each protein-coding gene.
Transcriptomic database mining
Published microarrays were analyzed using GEO2R from NCBI-GEO datasets. Briefly, GEO2R performs comparisons of microarray data tables using the GEOquery and limma (Linear Models for Microarray Analysis) R packages from the Bioconductor project. The GEOquery R package imports GEO data into R data structures, while the limma R package is one of the post widely used statistical tests for identifying differentially expressed genes.
Microarray
Bone marrow-derived macrophages (BMDMs) were collected from wild type and Tnfaip8l2 knockout mice (pooled sample of 5 in each group) and stimulated with lipopolysaccharide (LPS) for 1 hour. RNA was collected and microarray was performed. The results were analyzed using the Transcriptome Analysis Console (TAC) software from Affymetrix. Pathway analysis was performed using both ClueGo, Ingenuity Pathway Analysis (IPA), and Gene Set Enrichment Analysis (GSEA). Briefly, ClueGo is a Cytoscape plug-in that visualizes the non-redundant biological terms for large clusters of genes in a functionally grouped network. The ClueGO network is created with kappa statistics and reflects the relationships between the terms based on the similarity of their associated genes. IPA is a web-based software application that goes beyond pathway analysis by identifying key upstream regulators to explain expression patterns and predicting downstream effects on biological and disease processes. GSEA is a computational method that determines whether a priori defined set of genes shows statistically significant, concordant differences between two biological states. GSEA does not focus on only significantly/highly changed genes, but examines all the genes that belongs to a certain biological process instead.
Western Blot Analysis
Protein extracts were collected from BMDMs. Protein concentrations were determined by the bicinchoninic acid (BCA) assay with BSA standards. Proteins were separated on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were blocked with 5% BSA in TBS containing 0.1% Tween 20. Membranes were incubated with primary antibodies overnight at 4°C. Membranes were then washed extensively with TBS and incubated with the appropriate horseradish peroxidase-labeled secondary antibodies for 1 hour at room temperature. Afterward, membranes were incubated with enhanced chemiluminescence (ECL) substrate for horseradish peroxidase (Pierce/Thermo, Rockford, IL) and the ECL intensity was detected by Odyssey Fc (LI-COR, NE). The expression levels of proteins as indicated by the ECL intensity were measured with Image Studio software (LI-COR).
Real-time PCR
Total RNA was isolated from BMDMs with RNeasy Mini Kit (Qiagen) following the manufacturer’s suggestion. 2μg of total RNA were reverse transcribed to generate complementary DNA (cDNA) using SuperScript II RT (Invitrogen, CA). The mRNA expression levels of genes were determined by quantitative real-time PCR (qRT-PCR) with the SYBR-green dye (Invitrogen) on the 7500 Fast PCR system (Applied Biosystems, CA).
Seahorse XF96 Analyzer
Seahorse XF96 analyzer was used to assess four mitochondrial parameters, including basal respiration, proton leak, ATP production, and spare respiratory capacity. Briefly, BMDMs were seeded at 50K per well in 96-well plates and cultured overnight. Culturing media was changed to modified DMEM media (1mM sodium pyruvate, 10mM glucose, 2mM glutamine) and placed into a 37°C non-CO2 incubator for 1 hour. XF Cell Mito Stress Test Assay (2μM Oligomycin, 2μM FCCP, 1μM Antimycin&Rotenone) was performed afterwards to determine the four different mitochondria parameters.
Statistical Analysis
Data were expressed as the mean ± standard error of the mean (SEM) throughout the manuscript. For comparisons between two groups, two-tailed Student t test was used for evaluation of statistical significance or, when the data were not normally distributed, a nonparametric Mann-Whitney U test was used. For comparisons across multiple groups, one-way ANOVA with Bonferroni post-test adjustment was used or, when the data were not normally distributed, the data were analyzed using one-way ANOVA with the Kruskal-Wallis test, followed by pairwise comparison using the Dunn test. Data shown are representative of two to three independent experiments. NS, not significant; *, p<0.05; **, p<0.01; ***, p<0.001.
Supplementary Material
Highlights.
TNFAIP8L2 was preferentially expressed in human myeloid cell types.
Tnfaip8l2 expression drastically decreased after lipopolysaccharide treatment in macrophages.
Tnfaip8l2 deficiency led to heightened gene expression enriched for leukocyte activation and lipid biosynthesis pathways.
Tnfaip8l2 negatively regulated mitochondrial respiration in response to lipopolysaccharide.
Hyperlipidemia condition upregulated Tnfaip8l2 gene expression in the adipose tissue.
Acknowledgments
This work was partially supported by the National Institutes of Health (NIH) Grants to YHC (R01- AI-099216 and R01-AI-136945) and NIH hematopoiesis training grant to XL (T32-DK007780).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- 1.Sun H, Gong S, Carmody RJ, Hilliard A, Li L, Sun J, Kong L, Xu L, Hilliard B, Hu S, Shen H, Yang X, Chen YH. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell. 2008;133:415–426. doi: 10.1016/j.cell.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Wang J, Fan C, Li H, Sun H, Gong S, Chen YH, Shi Y. Crystal structure of TIPE2 provides insights into immune homeostasis. Nat Struct Mol Biol. 2009;16:89–90. doi: 10.1038/nsmb.1522. [DOI] [PubMed] [Google Scholar]
- 3.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, Tschirner SK, Gorinski N, Gohmert M, Mayer CT, Huehn J, Ponimaskin E, Abraham WR, Muller R, Lochner M, Sparwasser T. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 5.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, Yan C, Du H, Abumrad NA, Urban JF, Jr, Artyomov MN, Pearce EL, Pearce EJ. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nomura M, Liu J, Rovira Gonzalez-Hurtado E, II, Lee J, Wolfgang MJ, Finkel T. Fatty acid oxidation in macrophage polarization. Nat Immunol. 2016;17:216–217. doi: 10.1038/ni.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Zhuang G, Chai L, Wang Z, Johnson D, Ma Y, Chen YH. TIPE2 controls innate immunity to RNA by targeting the phosphatidylinositol 3-kinase-Rac pathway. J Immunol. 2012;189:2768–2773. doi: 10.4049/jimmunol.1103477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gus-Brautbar Y, Johnson D, Zhang L, Sun H, Wang P, Zhang S, Zhang L, Chen YH. The anti-inflammatory TIPE2 is an inhibitor of the oncogenic Ras. Mol Cell. 2012;45:610–618. doi: 10.1016/j.molcel.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garaude J, Acin-Perez R, Martinez-Cano S, Enamorado M, Ugolini M, Nistal-Villan E, Hervas-Stubbs S, Pelegrin P, Sander LE, Enriquez JA, Sancho D. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat Immunol. 2016;17:1037–1045. doi: 10.1038/ni.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 12.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, McGeough MD, Ellisman MH, Seki E, Gustafsson AB, Hoffman HM, Diaz-Meco MT, Moscat J, Karin M. NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, Li Y, Rodriguez V, Drong A, Hussain M, Lindgren C, McCarthy M, Naslund E, Zierath JR, Wong GW, Feinberg AP. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell metabolism. 2015;21:138–149. doi: 10.1016/j.cmet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.