Abstract
The tumor suppressor phosphatase and tensin homolog (PTEN) plays a central role in regulating phosphatidylinositol 3-kinase (PI3K) signaling, and its gene is very frequently mutated in various human cancers. Numerous studies have revealed that PTEN levels are tightly regulated by both transcriptional and posttranslational modifications, with especially ubiquitylation significantly regulating PTEN protein levels. Although several ubiquitin ligases have been reported to mediate PTEN ubiquitylation in vitro, the ubiquitin ligase that promotes PTEN degradation in vivo has not been reported. Here we took advantage of specific knockout mouse models to demonstrate that WW domain–containing E3 ubiquitin protein ligase 2 (WWP2) promotes PTEN degradation under physiological conditions, whereas another ubiquitin ligase, carboxyl terminus of Hsp70-interacting protein (CHIP), had no such effect. WWP2 knockout mice exhibited reduced body size, elevated PTEN protein levels, and reduced phosphorylation levels of the serine/threonine kinase and PTEN target AKT. In contrast, we observed no elevation of PTEN protein levels in CHIP knockout tissues and mouse embryonic fibroblasts. Furthermore, PTEN protein levels in CHIP/WWP2 double knockout mice were very similar to those in WWP2 single knockout mice and significantly higher than in WT and CHIP knockout mice. Our results demonstrate that WWP2, rather than CHIP, is an ubiquitin ligase that promotes PTEN degradation in vivo. Considering PTEN's significant role in tumor development, we propose that WWP2 may be a potential target for fine-tuning PTEN levels in anticancer therapies.
Keywords: Akt PKB, protein stability, ubiquitin ligase, gene knockout, mouse, CHIP, PTEN, WWP2
Introduction
PTEN4 is an important tumor suppressor that has lipid phosphatase activity. Cytoplasmic PTEN mainly dephosphorylates the serine/threonine kinase AKT activator phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3 (also known as PIP3)) and thus represses the phosphatidylinositol 3-kinase (PI3K)–AKT pathway and inhibits cellular process and tumor growth (1). In addition to cytoplasmic functions, nuclear PTEN also plays a critical role in suppressing tumor growth by maintaining genome stability (2, 3). Numerous lines of evidence have been provided to elucidate the importance of PTEN in tumor suppression. Germline mutations of PTEN cause Cowden disease, characterized by multiple hamartomas and predisposition to tumors of the breast, thyroid, and skin (4, 5), Bannayan–Riley–Ruvalcaba syndrome, and Lhermitte-Duclos disease (6, 7). Somatic mutations of PTEN have also been found in various human cancers (8, 9). Although homozygous deletion of Pten in mice causes early embryonic lethality, PTEN heterozygous deleted mice developed a broad spectrum of tumors, including colon adenocarcinomas, gonadostromal tumors, teratomas, and thyroid papillary adenocarcinomas (10–12). Moreover, PTEN conditional knockout mice also develop a specific type of tumors spontaneously (13). PTEN also plays important roles in metabolism and axon regeneration (14–16). PTEN transgenic mice (referred to as Super-PTEN mice) exhibit a relatively healthy metabolic state by regulating PI3K-dependent and -independent pathways (14).
Different from other haploinsufficient tumor suppressor genes, a negligible decrease in PTEN expression can sufficiently promote cancer susceptibility in mice. When PTEN levels decreased to 80%, 40% of mice developed breast cancer (17), demonstrating that the PTEN dose has important effects on tumor susceptibility and suggesting that regulation of the PTEN protein level is much more complicated than for other proteins.
Indeed, several E3 ubiquitin ligases have been demonstrated to regulate the ubiquitylation and stability of PTEN. Nedd4-1 was the first identified E3 ubiquitin ligase for PTEN. Nedd4-1 could promote both mono- and polyubiquitylation of PTEN (18, 19), which promote PTEN nuclear transport and proteasome degradation, respectively. Interestingly, despite the functional role of Nedd4-1 in promoting the monoubiquitylation of PTEN for nuclear transport and preventing PTEN from proteasome degradation, high levels of NEDD4-1 and low levels of PTEN have been found in a mouse prostate cancer model (18). The roles of Nedd4-1 were challenged later by mouse models. Both the protein level and ubiquitylation of PTEN show no difference between WT and Nedd4-1 knockout MEFs (20).
After that, XIAP, WWP2, and CHIP were identified as ubiquitin ligases for PTEN in succession (21–23). All of them show the ability to promote polyubiquitylation and degradation of PTEN, suppress PI3K–AKT signaling, and inhibit cell proliferation. In addition, XIAP and CHIP also promote monoubiquitylation of PTEN and transport PTEN from the cytoplasm to the nucleus. Clinically, immunohistochemistry analysis showed that CHIP is highly expressed in prostate cancer tissues, whereas PTEN is expressed at lower levels, implying a negative correlation between CHIP and PTEN in the tumor microenvironment.
Although several ubiquitin ligases were demonstrated to promote PTEN degradation, no study has been done to identify the physiological E3 ligase for PTEN using a mouse model. Considering the critical roles of PTEN in suppressing tumorigenesis, it is very necessary to elucidate the physiological ubiquitin ligase for PTEN. To solve this problem, here we take advantage of transgenic and knockout mouse models, demonstrating that WWP2, rather than CHIP, degrades PTEN under physiologic conditions.
Results
The phenotypes of WWP2 knockout mice resemble those of PTEN transgenic mice
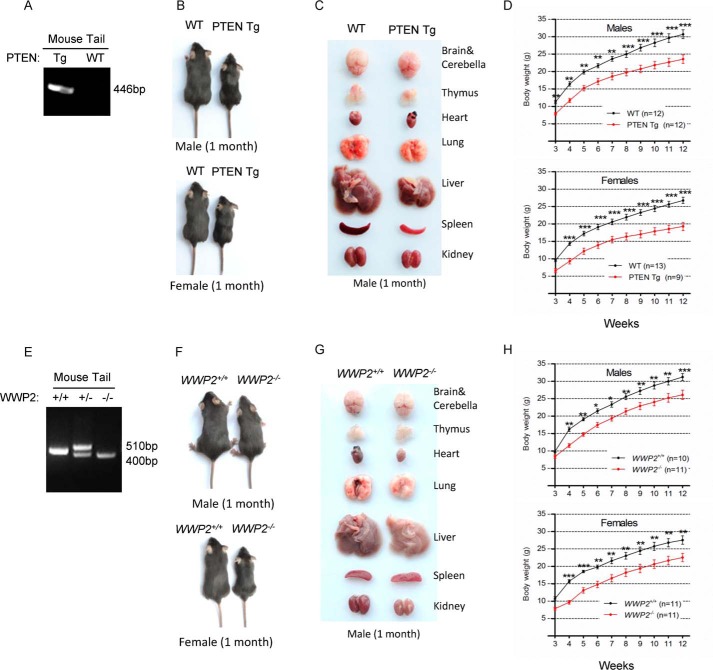
According to previous studies of the regulation between PTEN and its ubiquitin ligases, deletion of the ubiquitin ligase(s) for PTEN would lead to elevation of PTEN, meaning that the phenotypes of PTEN E3(s) knockout (KO) mice might be similar to that of Super-PTEN mice. To reciprocally validate the physical effects of PTEN up-regulation in vivo, we obtained PTEN E3 KO mice and generated PTEN transgenic mice (PTEN Tg) by adopting the same strategy with Super-PTEN mice. The genotype of PTEN Tg mice was identified through PCR (Fig. 1A). PTEN Tg mice are viable, and all organs examined from them weighed below normal, especially the liver (Fig. 1, B and C). The weight difference between WT and PTEN Tg mice, as indicated by growth curves, existed since 3 weeks of age (Fig. 1D). All of these phenotypes are consistent with the Super-PTEN data published previously (14).
Figure 1.
WWP2 KO mice have similar phenotypes as PTEN transgenic mice. A, genotyping of the BAC-PTEN Tg mice generated. PCR analysis of genomic DNA from the mouse tails was performed. B and C, WT and PTEN Tg mice were compared for body and organ size (1 month). D, growth curves indicate that PTEN overexpression results in reduced body mass (3 weeks after birth, weighed once a week). Line graphs represent mean ± S.E. (error bars) of body weight obtained from the indicated number of littermates. The body weights were analyzed by two-way ANOVA with Bonferroni post-test (PTEN Tg versus WT; **, p < 0.01; ***, p < 0.001). E, genotyping of WWP2 WT and KO mice; genomic DNA is from the mouse tails. F and G, WWP2 WT and KO mice were compared for body and organ size (1 month). H, WT mice had a significantly higher body weight than WWP2 knockout mice (3 weeks after birth, weighed once a week). Line graphs represent mean ± S.E. (error bars) of body weight obtained from the indicated number of littermates. The body weights were analyzed by two-way ANOVA with Bonferroni post-test (WWP2−/− versus WWP2+/+; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
WWP2 is one of the E3s that specifically promotes PTEN degradation. WWP2 KO mice are alive but exhibit developmental retardation compared with WT mice; thus, we obtained WWP2 KO mice and used PCR technology to characterize them (Fig. 1E). Surprisingly, we found that the phenotypes of WWP2 KO mice resemble those of PTEN Tg mice, showing reduced body and organ size as well as body weight (Fig. 1, F–H). As shown by growth curves of WWP2 WT and KO mice, the smaller body phenotype of WWP2 KO mice occurred in the early stage of development and continued to adulthood. These results prompted us to speculate that the phenotypes of WWP2 KO mice may result from elevation of PTEN protein levels.
Deficiency of WWP2 in vivo elevates PTEN protein levels and antagonizes PI3K–AKT signaling
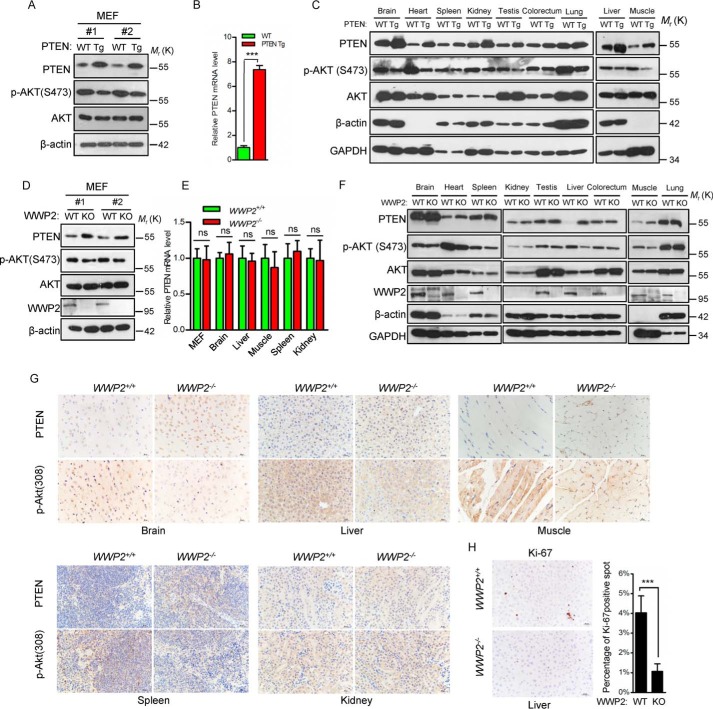
We analyzed PTEN protein levels in mouse embryonic fibroblasts (MEFs) of WWP2 KO mice by Western blotting. Consistent with PTEN Tg mice (Fig. 2, A and B), increased PTEN protein levels and decreased phosphorylated AKT (Ser-473) levels were observed in WWP2−/− MEFs (Fig. 2D), whereas the PTEN mRNA level showed no difference between WT and WWP2−/− MEFs and multiple tissues (Fig. 2E), indicating that WWP2 deletion does not regulate PTEN expression at the transcriptional level. To further confirm these results under physiological conditions, we analyzed PTEN protein levels in several major organs and tissues. As expected, PTEN levels in the brain, heart, liver, muscle, and spleen of WWP2 KO mice were increased (Fig. 2F), which is consistent with PTEN Tg mice (Fig. 2C), but the PTEN protein levels in other tissues showed no difference between WWP2 WT and KO mice (Fig. 2F). The results of immunohistochemical staining consistently showed elevated PTEN protein expression in brain, liver, muscle, and spleen tissues of WWP2 KO mice (Fig. 2G). Because PTEN inhibits cell proliferation by negatively regulating the PI3K–AKT pathway, we investigated Ki-67 staining in liver tissue from WT and WWP2 KO mice. Indeed, there was significantly less Ki-67 staining in WWP2 KO livers (Fig. 2H), demonstrating decreased cell growth, which may be a major reason for the reduced liver size.
Figure 2.
WWP2 depletion elevated PTEN protein levels and inhibited cell proliferation. A, MEFs of WT and PTEN Tg mice were used to detect the levels of PTEN and AKT phosphorylation. B, analysis of mRNA levels in PTEN WT and PTEN Tg MEFs. The bar graph shows the mean ± S.D. (error bars) obtained from n = 3 littermate MEFs. ***, p < 0.001, Student's t test. C, immunoblotting of PTEN and p-AKT (Ser-473) in tissues from WT and PTEN Tg littermates (1 month). D, Western blot analysis of the PTEN and p-AKT (Ser-473) levels in WWP2+/+ and WWP2−/− MEFs. E, quantification of PTEN mRNA levels in MEFs and tissues obtained from WWP2+/+ and WWP2−/− littermates (1 month). The bar graph shows mean ± S.D. (error bars) obtained from n = 3 littermates. ns, not significant, Student's t test. F, immunoblotting of PTEN and p-AKT (Ser-473) in tissues from adult WWP2+/+ and WWP2−/− littermates (1 month). G, immunohistochemistry analysis of protein levels of PTEN and p-AKT (Thr-308) in WWP2 WT and knockout tissues (1 month). Magnification, ×40; scale bars = 20 μm. H, representative Ki-67 immunohistochemical staining in the liver of WWP2 WT and KO mice (1 month). The bar graph shows mean ± S.D. (error bars) of n = 3 independent experiments. ***, p < 0.001, Student's t test.
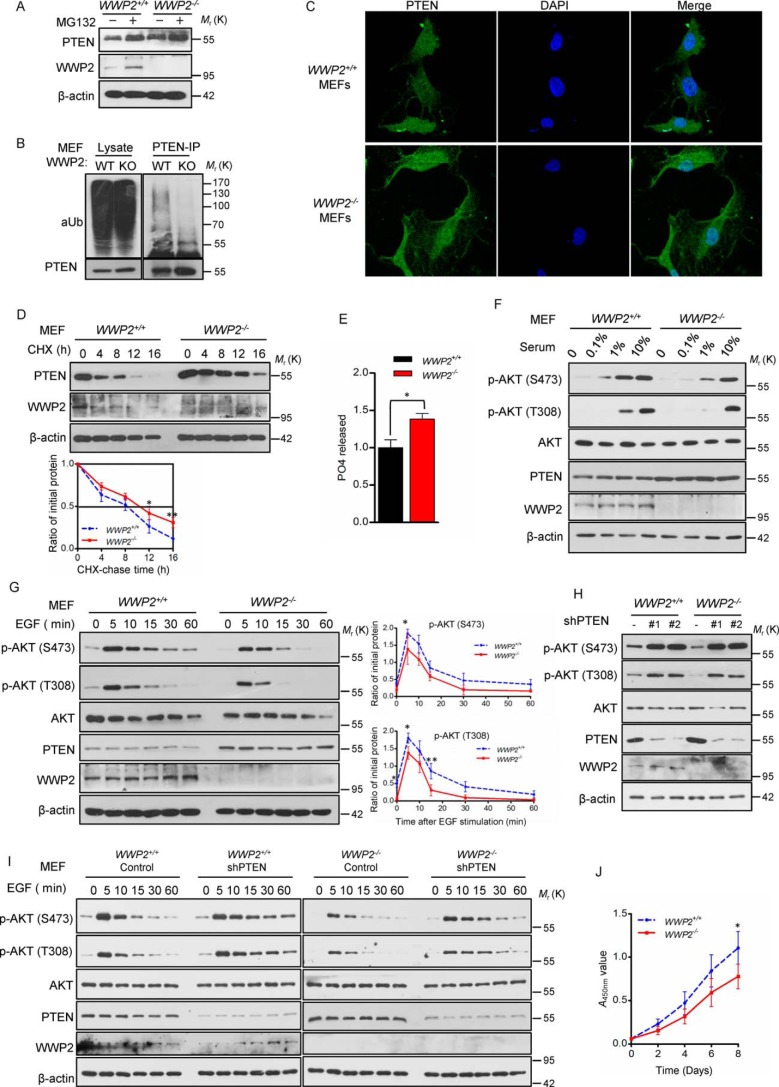
As reported previously, PTEN ubiquitylation and protein stability could be regulated by WWP2 upon overexpression or knockdown of WWP2 in cultured cells (22). We found that the proteasome inhibitor MG132 effectively blocks the ubiquitin proteasome pathway in MEFs and leads to PTEN accumulation (Fig. 3A). To determine the overall level of PTEN ubiquitylation, WWP2−/− and WWP2+/+ MEFs were pretreated with MG132 for 6 h and lysed for an ubiquitylation assay. The results showed that WWP2 deletion decreases the level of PTEN ubiquitylation in MEFs (Fig. 3B). As described previously, siRNA-mediated knockdown of WWP2 hardly influenced the subcellular localization of PTEN (22), and the same effects were observed in WWP2−/− MEFs, but the fluorescence intensity of PTEN was indeed more than that of WWP2+/+ MEFs (Fig. 3C). To determine PTEN stability in WWP2+/+ and WWP2−/− MEFs, MEFs were treated with the protein synthesis inhibitor cycloheximide, and the stability of PTEN was enhanced in WWP2−/− MEFs (Fig. 3D). Because cytoplasmic PTEN inhibits the PI3K–AKT pathway by dephosphorylating PtdIns(3,4,5)P3 to PtdIns(4,5)P2 and thus mediates its tumor-suppressor function (24), a PtdIns(3,4,5)P3 phosphatase assay was applied to evaluate the ability of PTEN to restrain AKT activation in WWP2 WT and KO MEFs. Notably, WWP2 KO resulted in an increase in the PTEN capacity to hydrolyze PtdIns(3,4,5)P3 in MEFs compared with the WT control (Fig. 3E). Furthermore, depletion of WWP2 led to later activation in a gradient serum concentration assay (Fig. 3F). In contrast to AKT phosphorylation activated at 0.1% serum concentration in WWP2+/+ MEFs, AKT phosphorylation in WWP2−/− MEFs was retarded to 1% serum concentration. Furthermore, an EGF stimulation assay also confirmed a shorter duration of AKT signaling activation in WWP2−/− MEFs (Fig. 3G). Importantly, knockdown of PTEN in WWP2 KO MEFs rescued AKT phosphorylation (Fig. 3, H and I). We also observed that deletion of WWP2 in vivo significantly attenuates MEF proliferation (Fig. 3J). Hence, we conclude that depletion of WWP2 in mice leads to elevation of PTEN protein levels and inhibition of AKT signaling.
Figure 3.
WWP2 deletion in mice enhances PTEN stability and inhibits AKT signaling. A, Western blotting showing PTEN levels in WWP2 WT and KO MEFs after MG132 treatment. B, WWP2+/+ and WWP2−/− MEFs were treated with MG132, and the lysates were immunoprecipitated (IP) with anti-PTEN antibody, followed by immunoblotting with anti-Ub antibody. C, subcellular localization of PTEN was compared between primary MEFs derived WWP2+/+ and WWP2−/− mice using immunofluorescence. PTEN antibody was used (green); nuclei were counterstained with DAPI (blue). Magnification, ×1,000. D, half-life analysis of PTEN in WWP2+/+ and WWP2−/− MEFs. MEFs were treated with cycloheximide (CHX, 20 μg ml−1) and collected at the indicated times for Western blotting. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to β-actin). The levels of PTEN protein were analyzed by two-way ANOVA with Bonferroni post-test (WWP2−/− versus WWP2+/+; *, p < 0.05; **, p < 0.01). E, catalytic activity of immunoprecipitated PTEN from the indicated MEFs was tested in phosphatase assays using PtdIns(3,4,5)P3 as substrate. PO4 released is normalized over levels of WT MEFs. The bar graph shows mean ± S.D. (error bars) obtained from n = 3 littermate MEFs. *, p < 0.05; Student's t test. F, immunoblot analysis of cell lysates derived from WWP2+/+ and WWP2−/− MEFs treated with different concentrations of serum. G, WWP2+/+ and WWP2−/− MEFs were serum-starved and treated with EGF for various times; cells were collected for immunoblot analysis. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to AKT). The levels of p-AKT (Ser-473) and p-AKT (Thr-308) were analyzed by two-way ANOVA with Bonferroni post-test (WWP2−/− versus WWP2+/+; *, p < 0.05; **, p < 0.01). H, Akt signaling was analyzed in WWP2 WT and KO MEFs before and after PTEN knockdown. I, immunoblot analysis of EGF stimulation for various times in WWP2+/+ and WWP2−/− MEFs before and after PTEN knockdown. J, growth curves of WWP2−/− and WWP2+/+ MEFs. Line graphs represent mean ± S.D. (error bars) obtained from n = 3 littermate MEFs. The A450 nm values of MEFs were analyzed by two-way ANOVA with Bonferroni post-test (WWP2−/− versus WWP2+/+; *, p < 0.05).
Physiological depletion of CHIP fails to cause changes in PTEN protein levels
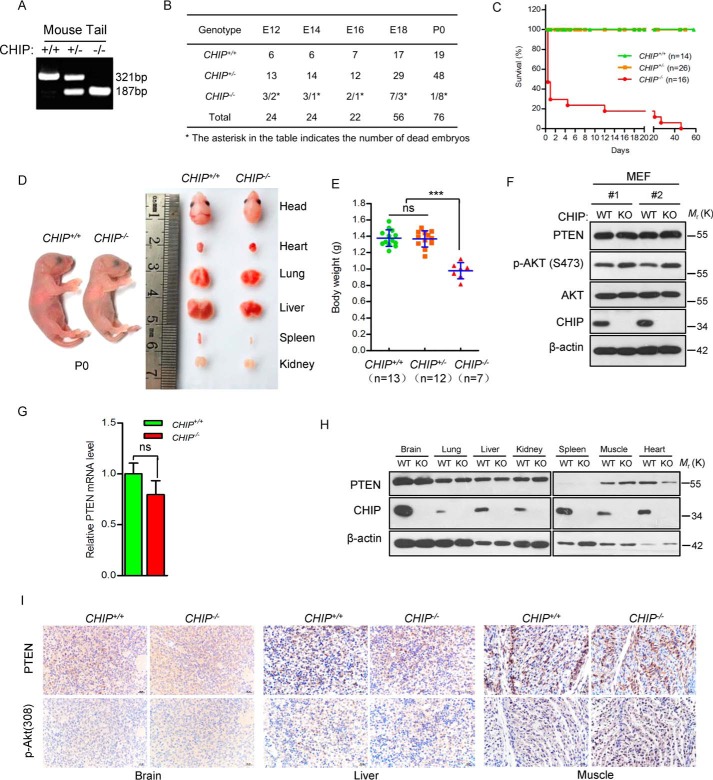
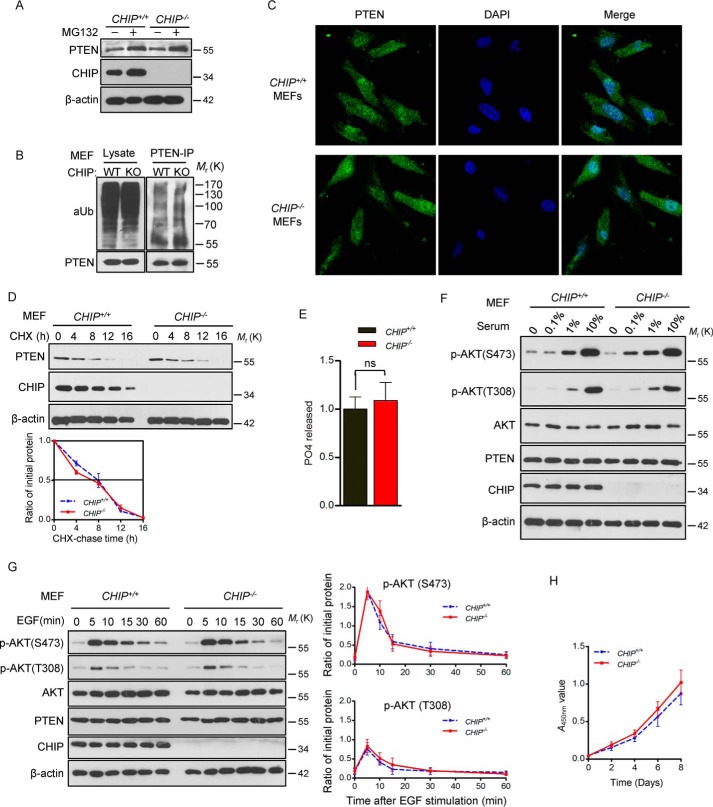
To systematically describe the regulation of PTEN protein level by E3 ubiquitin ligases under physiological conditions, CHIP KO mice were raised and analyzed. We found that CHIP KO mice showed a very low birth rate (11.84%, 9 of 76), suggesting that homozygous depletion of CHIP leads to embryonic lethality. Therefore, we attempted to dissect pregnant mice and trace the livability from the embryonic day 12 (E12) to the birth day (P0). The results showed that 31.82% (7 of 22) CHIP KO mice died at the embryonic stage (Fig. 4B), and the other CHIP KO mice died 2 months after birth. Further investigation showed that CHIP KO mice are smaller than WT mice in weight and tissue size (Fig. 4, D and E). Although previous studies have shown that CHIP acts as an important E3 of PTEN and regulates the stability of PTEN, no change was detected in PTEN protein levels in CHIP−/− MEFs as well as mRNA levels compared with CHIP+/+ MEFs (Fig. 4, F and G). In addition, similar PTEN protein levels were observed in all tissues examined between CHIP WT and KO mice (Fig. 4, H and I). However, there is the remarkable fact that the p-AKT (Ser-473) level shows a small increase in CHIP−/− MEFs, although PTEN remains at normal levels.
Figure 4.
Depletion of CHIP results in death of mice but did not affect PTEN protein levels. A, DNA was extracted from mouse tails, and the first lane shows CHIP KO. B, the mortality rate of three CHIP+/+, CHIP+/−, and CHIP−/− genotypes mice at the indicated stages. C, Kaplan–Meier survival analysis of mice with the indicated genotypes; CHIP KO mice have short survival after birth (female and male). D, comparison of body and organ size of CHIP WT and KO mice on P0 (male). E, comparison of body weights of CHIP+/+ (n = 13), CHIP+/− (n = 12), and CHIP−/− (n = 7) newborn mice (P0, female and male). The scatterplot shows the mean ± S.D. (error bars) obtained from the indicated number of littermates. ***, p < 0.001; ns, not significant; Student's t test. F, analysis of the PTEN and p-AKT(Ser-473) protein levels of CHIP WT and KO MEFs. G, quantification of PTEN mRNA levels in MEFs obtained from CHIP+/+ and CHIP−/− littermates. The bar graph shows the mean ± S.D. (error bars) obtained from n = 3 littermate MEFs; Student's t test. H, immunoblot of PTEN in tissues from newborn CHIP+/+ and CHIP−/− littermates (P0, male). I, PTEN and p-AKT(Thr-308) expression by immunohistochemistry in tissues from WT and CHIP KO mice (P0, male). Magnification, ×40; scale bars = 20 μm.
CHIP is dispensable for the regulation of PTEN stability
Monoubiquitylation of PTEN promotes its translocation into the nucleus, which results in activation of AKT, whereas PTEN protein levels are not affected. CHIP had been demonstrated to prompt PTEN nuclear shuttling (23). We supposed that activation of AKT in CHIP−/− MEFs may be due to the nuclear translocation of PTEN. However, immunofluorescence assays showed no difference in subcellular location of PTEN between CHIP+/+ and CHIP−/− MEFs (Fig. 5C). To further explore the regulation of PTEN stability by CHIP in MEFs, we treated cells with MG132 and found that the protein level of PTEN was similar in CHIP WT and KO MEFs before and after treatment. The ubiquitylation assay also showed no difference in PTEN ubiquitylation between WT and CHIP−/− MEFs (Fig. 5B). We tested the half-life of PTEN, and the results showed, compared with WT MEFs, that PTEN stability was unchanged in CHIP−/− MEFs (Fig. 5D). Regarding phosphatase activity, CHIP+/+ and CHIP−/− MEFs were similar (Fig. 5E). To investigate the influence of CHIP KO on AKT signaling, we treated WT and CHIP−/− MEFs with different concentrations of serum and found that CHIP−/− MEFs are more sensitive to low concentrations of serum (1%) than WT MEFs (Fig. 5F), and an EGF stimulation assay also showed that absence of CHIP leads to slight activation of AKT signaling in MEFs (Fig. 5G) but has no effect on cell proliferation (Fig. 5H). Collectively, these results indicate that CHIP depletion in mice has no effect on PTEN protein levels but slightly promotes AKT phosphorylation, indicating that there is a mechanism that CHIP physiologically regulates AKT signaling independent of PTEN.
Figure 5.
CHIP knockout does not affect PTEN stability but slightly activates AKT signaling. A, Western blot showing PTEN levels in CHIP WT and KO MEFs after MG132 treatment. B, CHIP+/+ and CHIP−/− MEFs were treated with MG132, and the lysates were immunoprecipitated (IP) with anti-PTEN antibody followed by immunoblotting with anti-Ub antibody (aUb). C, subcellular localization of PTEN was compared between primary MEFs derived from CHIP+/+ and CHIP−/− mice using immunofluorescence. PTEN antibody was used (green); nuclei were counterstained with DAPI (blue). Magnification, ×1000. D, half-life analysis of PTEN in CHIP+/+ and CHIP−/− MEFs. MEFs were treated with cycloheximide (CHX, 20 μg ml−1) and collected at the indicated times for Western blotting. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to β-actin). The levels of PTEN protein were analyzed by two-way ANOVA (CHIP−/− versus CHIP+/+; p > 0.05, not significant). E, phosphatase catalytic activity of immunoprecipitated PTEN from the indicated MEFs was tested. PO4 released is normalized over levels of WT MEFs. The bar graph shows mean ± S.D. (error bars) obtained from n = 3 littermate MEFs. ns, not significant; Student's t test. F, MEFs treated with different serum concentrations and then immunoblotted with the indicated antibodies. G, CHIP+/+ and CHIP−/− MEFs were serum-starved and treated with EGF for the indicated duration; cells were collected for immunoblot analysis. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to AKT). The levels of p-AKT(Ser-473) and p-AKT(Thr-308) were analyzed by two-way ANOVA (CHIP−/− versus CHIP+/+; p > 0.05, not significant). H, growth curves of CHIP−/− and CHIP+/+ MEFs. Line graphs represent mean ± S.D. (error bars) obtained from n = 3 littermate MEFs. The A450 nm values of MEFs were analyzed by two-way ANOVA (WWP2−/− versus WWP2+/+; p > 0.05, not significant).
CHIP/WWP2 double KO (DKO) mice show a similar elevation of PTEN levels as WWP2 KO mice
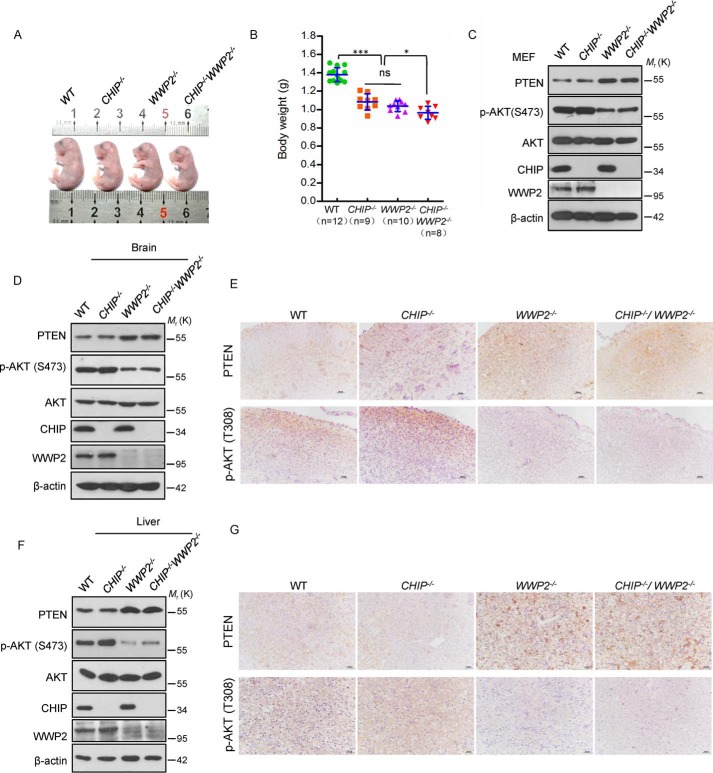
Because both CHIP and WWP2 are E3 ubiquitin ligases for PTEN, we speculated that CHIP and WWP2 have a synergistic effect on the degradation of PTEN. To verify this hypothesis, a CHIP/WWP2 DKO mouse model was generated. To avoid the influence of transient nutritional deficiency after birth on the AKT pathway, four phenotypes of mice were observed at E18. Compared with CHIP KO or WWP2 KO mice, CHIP/WWP2 DKO mice exhibited a smaller body size and lighter weight compared with CHIP or WWP2 KO alone (Fig. 6, A and B), indicating a superimposed effect of CHIP and WWP2 DKO on embryo development.
Figure 6.
Combined depletion of WWP2 and CHIP has no synergistic effect on PTEN degradation under physiological conditions. A, the size of WT, CHIP−/−, WWP2−/−, and CHIP−/− WWP2−/− mice at E18 (male). B, comparison of body weights from WT (n = 12), CHIP−/− (n = 9), WWP2−/− (n = 10), and CHIP−/−WWP2−/− (n = 8) mice (P0, female and male). The scatterplot shows the mean ± S.D. (error bars) obtained from the indicated number of littermates. *, p < 0.05; ***, p < 0.001; ns, not significant; Student's t test. C, MEF lysates from different genotypes of mice, immunoblotted with antibodies to p-AKT(Ser-473), AKT, PTEN, CHIP, and β-actin. D and F, protein levels of PTEN and p-AKT (Ser-473) in the brains and livers from four types of mice (E18, male). E and G, immunohistochemical analysis for PTEN and p-AKT (Thr-308) in the brains and livers from four types of mice (E18, male). Magnification, ×40; scale bars = 20 μm.
We next attempted to probe the level of PTEN and AKT phosphorylation in WT, CHIP−/−, WWP2−/−, and CHIP−/−WWP2−/− MEFs. Both single depletion of WWP2 and combined depletion of CHIP/WWP2 resulted in a pronounced increase in PTEN level and decrease of AKT phosphorylation compared with WT and CHIP−/− MEFs (Fig. 6C), suggesting that CHIP and WWP2 have no synergistic effect on PTEN regulation in vivo. Because deletion of CHIP in vivo causes a slight activation of AKT signaling, combined deficiency of WWP2 and CHIP also results in a little elevation of p-AKT (Ser-473) level compared with that in WWP2−/− MEFs (Fig. 6C). Similar to the results in MEFs, we also observed that the PTEN level was elevated and that the level of p-AKT (Ser-473) was decreased in the brain and liver tissues of WWP2 KO mice and CHIP/WWP2 DKO mice (Fig. 6, D–G). Therefore, these results suggest that CHIP and WWP2 do not have a synergistic effect on PTEN degradation but that WWP2 and CHIP might have a certain functional redundancy in development that still needs to be elucidated. As observed previously, CHIP KO in vivo led to elevation of p-AKT. The levels of p-AKT in the brain and liver of CHIP/WWP2 DKO mice are higher than in WWP2 KO mice (Fig. 6, D–G).
WWP2 plays a critical role in the degradation of PTEN in vivo
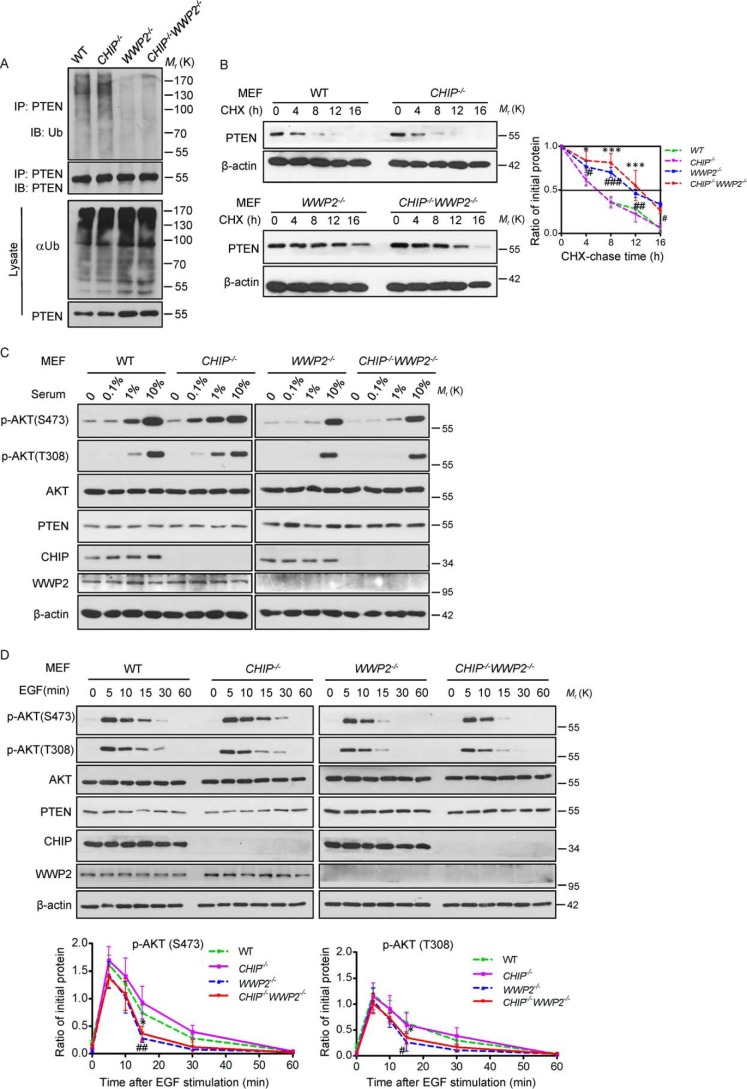
To further confirm that WWP2, but not CHIP, intrinsically ubiquitylates PTEN and controls its activity in vivo, we examined the ubiquitylation level of PTEN in four types of MEFs and found a remarkable decrease of PTEN ubiquitylation in WWP2−/− MEFs and CHIP−/−WWP2−/− MEFs compared with WT and CHIP−/− MEFs (Fig. 7A). The half-life assay was performed to investigate the stability of PTEN in these MEFs, and we found that the stability of PTEN was enhanced in WWP2−/− MEFs and CHIP−/−WWP2−/− MEFs (Fig. 7B). These results quite clearly highlight the capacity of WWP2, rather than CHIP, to ubiquitylate PTEN in vivo, and there is almost no synergy in affecting PTEN stability under physiological conditions. We next tried to detect how the AKT pathway responds to different concentrations of serum in these four kinds of MEFs. We found that, although the levels of p-AKT (Ser-473) and p-AKT (Thr-308) are low in four types of MEFs without serum stimulation, there is a delayed activation effect of AKT signaling in WWP2−/− and CHIP−/−WWP2−/− MEFs during a gradual increase in serum concentration (Fig. 7C). This result was also confirmed by the EGF stimulation assay. The duration of AKT activation was shortened in WWP2−/− MEFs and CHIP−/−WWP2−/− MEFs (Fig. 7D). All of these results highlight the critical role of WWP2 in the control of PTEN-mediated biological functions under physiological conditions.
Figure 7.
WWP2, rather than CHIP, regulates PTEN stability and inhibits PI3K–AKT signaling in vivo. A, WT, CHIP−/−, WWP2−/−, and CHIP−/−WWP2−/− MEFs were treated with MG132 for 6 h before harvest and then immunoprecipitated (IP) with PTEN antibody and immunoblotted (IB) with anti-Ub antibody. B, half-life analysis of PTEN in WT, CHIP−/−, WWP2−/−, and CHIP−/−WWP2−/− MEFs. MEFs were treated with cycloheximide (CHX, 20 μg ml−1) and collected at the indicated times for Western blotting. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to β-actin). The levels of PTEN protein were analyzed by two-way ANOVA with Bonferroni post-test (CHIP−/−WWP2−/− versus WT: *, p < 0.05; **, p < 0.01; ***, p < 0.001; WWP2−/− versus WT: #, p < 0.05; ##, p < 0.01; ###, p < 0.001; CHIP−/−WWP2−/− versus WWP2−/−: p > 0.05, not significant). C, four types of MEFs were treated with different serum concentrations and then immunoblotted with the indicated antibodies. D, WT, CHIP−/−, WWP2−/−, and CHIP−/−WWP2−/− MEFs were serum-starved and treated with EGF for the indicated duration; cells were collected for immunoblot analysis. Line graphs represent mean ± S.D. (error bars) of band intensity obtained through densitometric quantitation from n = 3 independent experiments (normalized to AKT). The levels of p-AKT(Ser-473) and p-AKT(Thr-308) were analyzed by two-way ANOVA (CHIP−/−WWP2−/− versus WT: *, p < 0.05; WWP2−/− versus WT: #, p < 0.05; ##, p < 0.01; CHIP−/−WWP2−/− versus WWP2−/−: p > 0.05, not significant).
Discussion
In contrast to a comprehensive understanding of PTEN regulation in cell lines, relatively little is known about modes of PTEN regulation in vivo. Although several ubiquitin ligases, including Nedd4-1, XIAP, WWP2 and CHIP, have been demonstrated to interact with and degrade PTEN in a variety of cell types in vitro, these results lack further validation in animal models, and the specific ubiquitin ligase(s) promoting PTEN ubiquitylation and degradation in vivo remain unclear. Here we revealed that WWP2 is an ubiquitin ligase and degrades the tumor suppressor PTEN under physiological conditions. Depletion of WWP2 in mice leads to elevation of protein levels and enhanced protein stability of PTEN and a similar phenotype as in PTEN Tg mice, including small body size and growth retardation. An ubiquitylation assay demonstrated that PTEN ubiquitylation is reduced in WWP2−/− MEFs. Furthermore, functional studies indicated that WWP2 plays an important role in regulating AKT signaling and cell proliferation in vivo. Based on these findings, we speculated that WWP2 is an oncogene in vivo, and thus tumorigenesis should be suppressed in WWP2 KO mice. Given the fact that depletion of WWP2 leads to elevation of PTEN in mice, generation of a tumor-induced model should be further investigated.
Similar to PTEN transgenic mice exhibiting retarded growth, we found that WWP2 KO mice also show growth retardation and increased PTEN protein levels, and there could be some correlation. Studies have shown that the smaller body size of PTEN transgenic mice is mainly due to elevations in energy expenditure, which is regulated by repressing PI3K–Akt signaling and restricting glucose uptake (14), and WWP2 has been reported to play critical roles in craniofacial development and cell self-renewal (25, 26). Collectively, in addition to increases in PTEN levels, we cannot rule out other factors that, caused by depletion of WWP2 in vivo, would lead to growth retardation of WWP2−/− mice. More work needs to be done to reveal the reason why WWP2 knockout mice are smaller than their WT littermates.
Besides WWP2, we also used CHIP KO mice to analyze the status of PTEN. Upon testing PTEN levels in MEFs and tissues from CHIP WT and KO mice, we found that CHIP did not affect PTEN stability and ubiquitination in vivo. However, we found that the phosphorylation of AKT is enhanced in CHIP−/− MEFs compared with WT MEFs, although the PTEN level is unchanged. Recently, Tawo et al. (27) reported that CHIP deficiency leads to elevated levels of the insulin receptor, which may be the reason why the PTEN protein level remains unchanged whereas AKT signaling is activated in CHIP−/− MEFs and tissues. Using CHIP/WWP2 DKO mice, we also found that CHIP does not promote PTEN degradation synergistically with WWP2 in vivo, which further verified that WWP2 is an E3 ubiquitin ligase for PTEN in vivo. In addition, here we concluded that WWP2 and CHIP have no redundancy in regulating the level of PTEN protein in mice, but they may possess some functional redundancies in the regulation of mouse embryo development because CHIP/WWP2 DKO embryos were smaller than CHIP KO and WWP2 KO embryos (Fig. 6B).
Except for Nedd4-1, which is a controversial physiological regulator of PTEN, the other identified ubiquitin ligases, including XIAP, WWP2, and CHIP, would be candidates for degrading PTEN in vivo. A previous study supported that XIAP could interact with PTEN and promotes mono- and polyubiquitylation of PTEN in MEFs, and the immunofluorescence assay indeed showed weaker nuclear immunostaining of PTEN in XIAP-deficient MEFs. We regarded XIAP as a nucleocytoplasmic shuttling regulator for PTEN in vivo; however, to find out whether XIAP affects the protein stability of PTEN and downstream signaling pathways in mice, more experimentation, even XIAP knockout mice, need to be analyzed.
Similar to another important tumor suppressor, p53, which is degraded by multiple E3 ligases, the protein stability of PTEN have been demonstrated to be regulated by Nedd4-1, XIAP, WWP2, and CHIP, suggesting that the regulatory mechanism of PTEN stability is complicated. Here we identified that WWP2 is a physiological ubiquitin ligase for PTEN and that it regulates its protein level. As for the other nonphysiological E3 ligases, they may regulate PTEN stability in the context of some specific tumor and cellular environments. Considering the fact that PTEN protein level is closely related to the occurrence and development of tumors (17), our findings imply that WWP2 acts as a target for tumor therapy by fine-tuning the PTEN protein level.
Experimental procedures
Antibodies
Anti-STUB1 (ab134064, 1:2,000 dilution) and anti-WWP2 (ab103527, 1:500 dilution) were purchased from Abcam. Anti-ubiquitin (sc-8017, 1:500) was purchased from Santa Cruz Biotechnology. Anti-β-actin (Ac026, 1:40,000) was purchased from ABclonal Technology. Anti-PTEN (9188, 1:2,000), anti-AKT (9272, 1:2,000), anti-pThr-308–AKT (9275, 1:2,000), and anti-pSer-473–AKT (4060, D9E, 1:2,000) were purchased from Cell Signaling Technology.
Mice
CHIP knockout mice and WWP2 knockout mice were kindly proved by Prof. Zhijie Chang and Weiguo Zou, and the knockout strategies and identification processes for these mice were described previously. The PTEN transgenic mice were generated by Cyagen Biosciences, and we adopted the same strategy for Super-PTEN mice. For transgenesis, a large genomic insert (218.50 kb) containing the entire PTEN locus and cloned into the BAC vector pBACe3.6 was isolated from a mouse BAC genomic library (BAC RPCI-23-215F15 clone, RPCI library, C57BL/6J). After digestion with EcoRI, linearized BAC DNA was used for microinjection into the pronuclei of fertilized oocytes derived from intercrosses between (C57BL/6 × CBA) F1 mice. Transgenic mice were identified by PCR of tail-tip genomic DNA. The sense and antisense primers were 5′-GGCCGCTAATACGACTCAC-3′ and 5′-ATAAAATGACTTGCCATTGGGT-3′, respectively. Amplification of the transgene resulted in a 446-bp PCR product. Mice were handled in strict accordance with the Guide for the Care and Use of Laboratory Animals and the Principles for the Utilization and Care of Vertebrate Animals, and all animal work was approved by the Institutional Animal Care and Use Committee at the Beijing Institute of Radiation Medicine.
Assays with MEFs
MEFs were isolated from E12.5 embryos and cultured in α-MEM supplemented with 10% fetal bovine serum and penicillin/streptomycin using standard techniques.
Protein half-life assay
For the PTEN half-life assay, MEFs were placed in 2-cm plates, reached about 70% confluence, and then were treated with the protein synthesis inhibitor cycloheximide (Sigma, 10 μg ml−1) for the indicated duration before collection.
In vivo PTEN ubiquitylation assay
For the in vivo PTEN ubiquitylation assay, MEFs were treated with 20 μm MG132 (a proteasome inhibitor, Selleck) for 8 h before being collected. The MEFs were washed with PBS, pelleted, and lysed in HEPES buffer (20 mm HEPES (pH 7.2), 50 mm NaCl, 1 mm NaF, and 0.5% Triton X-100) plus 0.1% SDS, 20 μm MG132, and protease-inhibitor mixture. The lysates were centrifuged to obtain cytosolic proteins and incubated with anti-PTEN antibody for 3 h and protein A/G–agarose beads (Santa Cruz Biotechnology) for a further 8 h at 4 °C. Then the beads were washed three times with HEPES buffer. The proteins were released from the beads by boiling in SDS-PAGE sample buffer and analyzed by immunoblotting using an anti-ubiquitin antibody (Santa Cruz Biotechnology).
Lentivirus infection
Lentiviruses carrying shRNA targeting PTEN lentiviral vectors (GV112) were from GeneChem. The viruses were used to infect cells in the presence of Polybrene. Forty-eight hours later, MEFs were cultured in medium containing puromycin for the selection of stable clones. Clones stably knocking down PTEN were identified and verified by Western blotting. The shRNA sequences were as follows: PTEN 1, 5′-GCGCTATGTGTATTATTAT-3′; PTEN 2, 5′-TGCAGATAATGACAAGGAA-3′; nontargeting control, 5′-TTCTCCGAACGTGTCACGT-3′.
Serum and EGF stimulation
For serum dose responses, MEFs were starved overnight and then cultured with α-MEM containing 0%, 0.1%, 1%, or 10% serum for 10 min. For EGF stimulation, MEFs were treated with serum-free α-MEM for 12 h before stimulation and, the next day, incubated with serum-free Dulbecco's modified Eagle's medium supplemented with EGF (50 ng/ml) for the indicated duration. MEFs were lysed, and protein extracts were prepared for immunoblotting.
Immunoblotting
Total cellular proteins were subjected to SDS-PAGE and electrotransferred to nitrocellulose membranes (Bio-Rad). Membranes were probed with primary antibody overnight at 4 °C, followed by horseradish peroxidase–conjugated anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories) for 1 h at room temperature. Detection was performed using SuperSignal West FemtoTM substrate (Thermo Scientific).
Immunohistochemistry
The tissues were washed with PBS and fixed for 72 h with 4% paraformaldehyde/PBS at 4 °C. The samples were dehydrated, embedded in paraffin, and sectioned into 3.5-μm-thick transverse sections. The sections were deparaffinized, treated with 3% H2O2 for 15 min, microwaved in 10 mm citric sodium (pH 6.0) for 15 min to unmask antigens, rinsed in PBS, and then incubated with rabbit polyclonal anti-p-AKT (Thr-308, Cell Signaling Technology, 1:500), anti-PTEN (Cell Signaling Technology, 1:200), and anti-Ki-67 (Abcam, 1:100) overnight at 4 °C. The secondary antibody was incubated for 30 min at 37 °C. Signal amplification and detection were performed using the diaminobenzidine system according to the manufacturer's instructions.
Fluorescence microscopy
To detect the subcellular localization and content of PTEN, after fixation with 4% paraformaldehyde and permeabilization in 0.2% Triton X-100 (PBS), cells were incubated with the indicated PTEN and PTEN antibodies (dilution 1:100, Cell Signaling Technology) for 8 h at 4 °C, followed by incubation with TRITC-conjugated or FITC-conjugated secondary antibody (dilution 1:1,000, Life) for 50 min at 25 °C. The nuclei were stained with DAPI (Sigma), and images were visualized with a Zeiss LSM 510 Meta inverted confocal microscope.
PtdIns(3,4,5)P3 phosphatase assay
Cell lysates from the indicated MEFs were immunoprecipitated and subjected to native elution. Prechilled 0.1 glycine (pH 2.5) was used for 10 min at 4 °C to elute immunocomplexes and further neutralized with 1 m Tris-HCl (pH 8). For phosphatase assays, a solution with 25 mm Tris-HCl (pH 7.5), 140 mm NaCl, 1 mm DTT, and 50 mm diC8-PtdIns(3,4,5)P3 (Echelon) was prepared, and the assay ran at 37 °C for 15 min. Free phosphate release was measured with malachite green reagent (Cell Signaling Technology, 12776S) according to the manufacturer's instructions.
RT-PCR and quantitative PCR
Total cell RNA was prepared using TRIzol reagent (Invitrogen) following the manufacturer's instructions. Five micrograms of total RNA was subjected to reverse transcription to synthesize complementary DNA using the First Strand cDNA Synthesis Kit (TOYOBO), and quantitative PCR was performed with the IQ5 system (Bio-Rad). PCRs were performed in 20-μl reaction volumes with SYBR Green PCR Master Mix (TOYOBO) and 0.2 μm specific primers. The primer sequences used for all qPCRs were as follows: PTEN, 5′-ACAGGCTCCCAGACATGACA-3′ and 5′-ATGCTTTGAATCCAAAAACCTTACT-3′; actin, 5′-TCCTAGCACCATGAAGATCAAGATC-3′ and 5′-CTGCTTGCTGATCCACATCTG-3′.
Author contributions
H. L., C.-P. C., and L. Z. conceptualization; H. L. and L. Z. resources; H. L. and P. Z. data curation; H. L., P. Z., Q. Z., and C. L. software; H. L. and P. Z. formal analysis; H. L. and L. Z. supervision; H. L., P. Z., Q. Z., and C. L. validation; H. L. and P. Z. investigation; H. L., P. Z., Q. Z., C. L., W. Z., Z. C., and C.-P. C. methodology; H. L., P. Z., and Q. Z. writing-original draft; H. L., C.-P. C., and L. Z. project administration; P. Z., Q. Z., C. L., and W. Z. visualization; C.-P. C. and L. Z. writing-review and editing; L. Z. funding acquisition.
This work was supported by National Natural Science Foundation of China (Grant 31330021) and Beijing Natural Science Foundation Project (Z151100003915083). The authors declare that they have no conflicts of interest with the contents of this article.
- PTEN
- phosphatase and tensin homolog
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI3K
- phosphatidylinositol 3-kinase
- KO
- knockout
- XIAP
- X-linked inhibitor of apoptosis protein
- Tg
- transgenic
- MEF
- mouse embryonic fibroblast
- EGF
- epidermal growth factor
- DKO
- double knockout
- E
- embryonic day
- P
- postnatal day
- MEM
- minimum Eagle's medium
- shRNA
- short hairpin RNA
- BAC
- bacterial artificial chromosome
- TRITC
- tetramethylrhodamine isothiocyanate
- DAPI
- 4′,6-diamidino-2-phenylindole
- ANOVA
- analysis of variance
- Ub
- ubiquitin.
References
- 1. Maehama T., and Dixon J. E. (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- 2. Shen W. H., Balajee A. S., Wang J., Wu H., Eng C., Pandolfi P. P., and Yin Y. (2007) Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128, 157–170 10.1016/j.cell.2006.11.042 [DOI] [PubMed] [Google Scholar]
- 3. Song M. S., Carracedo A., Salmena L., Song S. J., Egia A., Malumbres M., and Pandolfi P. P. (2011) Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144, 187–199 10.1016/j.cell.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liaw D., Marsh D. J., Li J., Dahia P. L., Wang S. I., Zheng Z., Bose S., Call K. M., Tsou H. C., Peacocke M., Eng C., and Parsons R. (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16, 64–67 10.1038/ng0597-64 [DOI] [PubMed] [Google Scholar]
- 5. Nelen M. R., van Staveren W. C., Peeters E. A., Hassel M. B., Gorlin R. J., Hamm H., Lindboe C. F., Fryns J. P., Sijmons R. H., Woods D. G., Mariman E. C., Padberg G. W., and Kremer H. (1997) Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum. Mol. Genet. 6, 1383–1387 10.1093/hmg/6.8.1383 [DOI] [PubMed] [Google Scholar]
- 6. Marsh D. J., Dahia P. L., Zheng Z., Liaw D., Parsons R., Gorlin R. J., and Eng C. (1997) Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat. Genet. 16, 333–334 10.1038/ng0897-333 [DOI] [PubMed] [Google Scholar]
- 7. Sutphen R., Diamond T. M., Minton S. E., Peacocke M., Tsou H. C., and Root A. W. (1999) Severe Lhermitte-Duclos disease with unique germline mutation of PTEN. Am. J. Med. Genet. 82, 290–293 10.1002/(SICI)1096-8628(19990212)82:4%3C290::AID-AJMG3%3E3.0.CO%3B2-0 [DOI] [PubMed] [Google Scholar]
- 8. Steck P. A., Pershouse M. A., Jasser S. A., Yung W. K., Lin H., Ligon A. H., Langford L. A., Baumgard M. L., Hattier T., Davis T., Frye C., Hu R., Swedlund B., Teng D. H., and Tavtigian S. V. (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 10.1038/ng0497-356 [DOI] [PubMed] [Google Scholar]
- 9. Li J., Yen C., Liaw D., Podsypanina K., Bose S., Wang S. I., Puc J., Miliaresis C., Rodgers L., McCombie R., Bigner S. H., Giovanella B. C., Ittmann M., Tycko B., Hibshoosh H., et al. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 10.1126/science.275.5308.1943 [DOI] [PubMed] [Google Scholar]
- 10. Di Cristofano A., Pesce B., Cordon-Cardo C., and Pandolfi P. P. (1998) Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355 10.1038/1235 [DOI] [PubMed] [Google Scholar]
- 11. Podsypanina K., Ellenson L. H., Nemes A., Gu J., Tamura M., Yamada K. M., Cordon-Cardo C., Catoretti G., Fisher P. E., and Parsons R. (1999) Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. U.S.A. 96, 1563–1568 10.1073/pnas.96.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stambolic V., Tsao M. S., Macpherson D., Suzuki A., Chapman W. B., and Mak T. W. (2000) High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/− mice. Cancer Res. 60, 3605–3611 [PubMed] [Google Scholar]
- 13. Knobbe C. B., Lapin V., Suzuki A., and Mak T. W. (2008) The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene 27, 5398–5415 10.1038/onc.2008.238 [DOI] [PubMed] [Google Scholar]
- 14. Garcia-Cao I., Song M. S., Hobbs R. M., Laurent G., Giorgi C., de Boer V. C., Anastasiou D., Ito K., Sasaki A. T., Rameh L., Carracedo A., Vander Heiden M. G., Cantley L. C., Pinton P., Haigis M. C., and Pandolfi P. P. (2012) Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 149, 49–62 10.1016/j.cell.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park K. K., Liu K., Hu Y., Smith P. D., Wang C., Cai B., Xu B., Connolly L., Kramvis I., Sahin M., and He Z. (2008) Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966 10.1126/science.1161566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun F., Park K. K., Belin S., Wang D., Lu T., Chen G., Zhang K., Yeung C., Feng G., Yankner B. A., and He Z. (2011) Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480, 372–375 10.1038/nature10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alimonti A., Carracedo A., Clohessy J. G., Trotman L. C., Nardella C., Egia A., Salmena L., Sampieri K., Haveman W. J., Brogi E., Richardson A. L., Zhang J., and Pandolfi P. P. (2010) Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 10.1038/ng.556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., and Jiang X. (2007) NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 10.1016/j.cell.2006.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trotman L. C., Wang X., Alimonti A., Chen Z., Teruya-Feldstein J., Yang H., Pavletich N. P., Carver B. S., Cordon-Cardo C., Erdjument-Bromage H., Tempst P., Chi S. G., Kim H. J., Misteli T., Jiang X., and Pandolfi P. P. (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156 10.1016/j.cell.2006.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fouladkou F., Landry T., Kawabe H., Neeb A., Lu C., Brose N., Stambolic V., and Rotin D. (2008) The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc. Natl. Acad. Sci. U.S.A. 105, 8585–8590 10.1073/pnas.0803233105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Themsche C., Leblanc V., Parent S., and Asselin E. (2009) X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 284, 20462–20466 10.1074/jbc.C109.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maddika S., Kavela S., Rani N., Palicharla V. R., Pokorny J. L., Sarkaria J. N., and Chen J. (2011) WWP2 is an E3 ubiquitin ligase for PTEN. Nature Cell Biol. 13, 728–733 10.1038/ncb2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahmed S. F., Deb S., Paul I., Chatterjee A., Mandal T., Chatterjee U., and Ghosh M. K. (2012) The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287, 15996–16006 10.1074/jbc.M111.321083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., and Mak T. W. (1998) Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39 10.1016/S0092-8674(00)81780-8 [DOI] [PubMed] [Google Scholar]
- 25. Zou W., Chen X., Shim J. H., Huang Z., Brady N., Hu D., Drapp R., Sigrist K., Glimcher L. H., and Jones D. (2011) The E3 ubiquitin ligase Wwp2 regulates craniofacial development through mono-ubiquitylation of Goosecoid. Nat. Cell Biol. 13, 59–65 10.1038/ncb2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fang L., Zhang L., Wei W., Jin X., Wang P., Tong Y., Li J., Du J. X., and Wong J. (2014) A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 10.1016/j.molcel.2014.06.018 [DOI] [PubMed] [Google Scholar]
- 27. Tawo R., Pokrzywa W., Kevei É., Akyuz M. E., Balaji V., Adrian S., Höhfeld J., and Hoppe T. (2017) The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell 169, 470–482.e13 10.1016/j.cell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]