Abstract
Prions are protein-based infectious agents that autocatalytically convert the cellular prion protein PrPC to its pathological isoform PrPSc. Subsequent aggregation and accumulation of PrPSc in nervous tissues causes several invariably fatal neurodegenerative diseases in humans and animals. Prions can infect recipient cells when packaged into endosome-derived nanoparticles called exosomes, which are present in biological fluids such as blood, urine, and saliva. Autophagy is a basic cellular degradation and recycling machinery that also affects exosomal processing, but whether autophagy controls release of prions in exosomes is unclear. Our work investigated the effect of autophagy modulation on exosomal release of prions and how this interplay affects cellular prion infection. Exosomes isolated from cultured murine central neuronal cells (CAD5) and peripheral neuronal cells (N2a) contained prions as shown by immunoblotting for PrPSc, prion-conversion activity, and cell culture infection. We observed that autophagy stimulation with the mTOR inhibitor rapamycin strongly inhibited exosomal prion release. In contrast, inhibition of autophagy by wortmannin or CRISPR/Cas9-mediated knockout of the autophagy protein Atg5 (autophagy-related 5) greatly increased the release of exosomes and exosome-associated prions. We also show that a difference in exosomal prion release between CAD5 and N2a cells is related to differences at the level of basal autophagy. Taken together, our results indicate that autophagy modulation can control lateral transfer of prions by interfering with their exosomal release. We describe a novel role of autophagy in the prion life cycle, an understanding that may provide useful targets for containing prion diseases.
Keywords: prion, prion disease, exosome (vesicle), autophagy, CRISPR/Cas, Creutzfeldt–Jakob disease, extracellular vesicles, neurodegeneration, RT-QuIC, scrapie
Introduction
Prion diseases are fatal, infectious neurodegenerative disorders of man and animals that can be transmitted within and sometimes between species (1, 2). Examples are Creutzfeldt–Jakob disease, kuru and variant Creutzfeldt–Jakob disease in humans, scrapie in sheep and goat, bovine spongiform encephalopathy in cattle and other species, and chronic wasting disease in deer, elk, and moose (3–6). Loss of neurons is a key feature of prion diseases, accompanied by astrogliosis and mild microglia activation. This results in a progressive spongiform degeneration of the CNS,3 leading to ataxia, behavioral changes and, in humans, highly progressive loss of intellectual abilities (2, 7–9). The formation of the abnormally folded, protease-resistant isoform PrPSc of the host-encoded cellular prion protein (PrPC) is the underlying mechanism in prion disease pathogenesis (1, 2). PrPSc is derived post-translationally from PrPC by a profound conformational change (10, 11). The exact physiological function of PrPC is unclear (7), although roles in copper transport and neuroprotection are evident among the many potential roles attributed (12–14). PrPC was also described as receptor for oligomeric β-amyloid (15). Recently, a role of PrPC in regulating exosomal release was reported (16).
In prion-infected cultured neurons and in neurons of brain samples from prion disease affected humans, the appearance of multivesicular bodies (MVBs) and autophagic vacuoles has been reported (17–19). Interestingly, MVBs are known to give rise to exosomes. Exosomes are nanovesicles secreted by most cell types (20–23). They are formed intracellularly by inward budding of the limiting membrane of endocytic compartments. MVBs ultimately fuse with the plasma membrane, releasing their internal vesicles into the extracellular milieu (23). Exosomes are secreted into various body fluids such as plasma, urine, milk, amniotic fluid, and saliva, making them interesting candidates for novel biomarkers (24, 25). Exosomes are secreted by most cells in a constitutive manner and play a role in cell-to-cell communication (26–28).
It has been shown that exosomes can contain prions (29–31) and that they participate in delivering PrPSc to uninfected neighboring cells (32–34). We reported previously that cell-free cell culture media of prion-infected hypothalamic ScGT1 cells can induce PrPSc formation in recipient cells, indicating a cell-free transfer of prion infection (18). Exosomes also play a role in spreading the toxic forms of other aggregated proteins such as α-synuclein, β-amyloid, and misfolded SOD1 (35–38). A pivotal role of autophagy in the secretion of exosomes is emerging (39, 40). In macroautophagy (here referred to as autophagy), autophagosomes engulf cytosolic macromolecules and deliver them to lysosomes for degradation (41–43). The clearance of aggregation-prone proteins, such as mutant huntingtin fragments or mutant forms of α-synuclein causing Huntington's and Parkinson's disease, respectively, can be mediated by autophagy (44, 45). Animal models of Huntington's disease and of other proteinopathies revealed that treatment with rapamycin, a known inducer of autophagy, accelerates the clearance of toxic proteins (46–49). Of note, changes in autophagy level affect MVB formation and exosomal release. Upon stimulating autophagy, MVBs fuse more with autophagic vacuoles, resulting in inhibition of exosomal release (50, 51). The beneficial effect of up-regulated autophagy in prion disease models in vitro and in vivo has been described by us and others (52–58).
The impact of autophagy on exosomal release of prions is not known. In this work, we address the link between autophagy, exosomes and release of prion infectivity in two different cell culture models. Persistently prion-infected ScCAD5 cells are used as neuronal cells of CNS origin; ScN2a cells represent peripheral nervous system neuronal cells. Upon stimulating autophagy using rapamycin, we found a decrease in exosomal release and exosome-associated prions. On the other hand, disruption of the autophagic machinery by knocking out Atg5 using CRISPR/Cas9 technology resulted in increased release of exosomes and increase in exosome-associated PrPSc. Similarly, inhibition of autophagy using wortmannin (phosphatidylinositol 3-kinase inhibitor) enhanced the secretion of exosomes and of exosomal PrPSc. In summary, our work shows that autophagy modulation controls exosomal release and, consequently, secretion of prions in exosomes. These findings help to better understand how autophagy can control cellular prion infection, not only via regulation of autophagosomal-lysosomal prion clearance but also by controlling exosomal release of prions.
Results
Isolation and characterization of exosomes from prion infected ScCAD5 and ScN2a cells
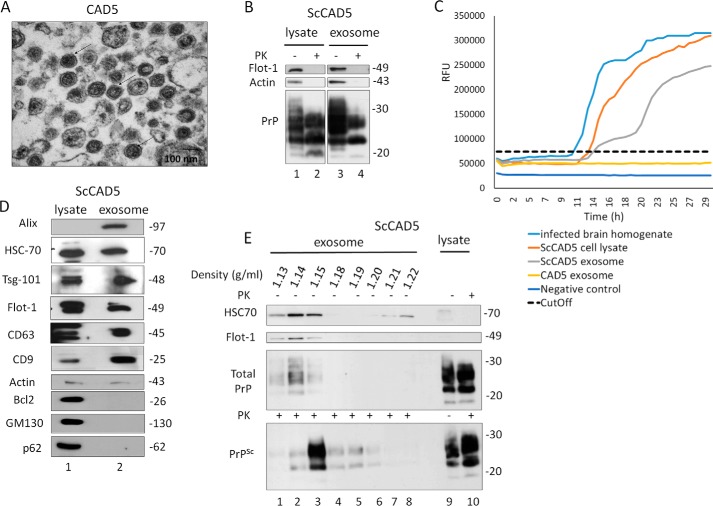
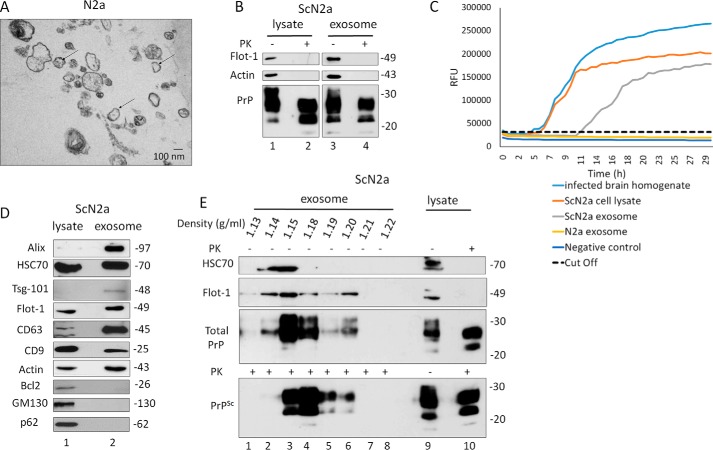
To study the effect of autophagy on exosomal release of prions, we used two prion-infected cell culture models. The murine cell lines ScCAD5 (CNS, catecholaminergic/neuronal) and ScN2a (peripheral nervous system, neuroblastoma/neuronal) are both persistently infected with the mouse-adapted scrapie prion strain 22L (59, 60). Exosomes isolated from cell culture media of ScCAD5 and ScN2a cells were characterized as shown in Figs. 1 and 2, respectively. The protein markers for exosomes flotillin-1, Alix, Tsg101, CD63, CD9, and HSC70 were detected in isolated exosomes from both cell lines using immunoblotting (Figs. 1D and 2D). The purity of isolated exosomes was confirmed by lack of Golgi (GM130), mitochondrial (Bcl2), and nuclear (nucleoporin p62) contaminants (Figs. 1D and 2D). Transmission EM (TEM) imaging of exosomal preparations showed vesicles with approximate diameters from 80 to 150 nm, and the morphology of vesicles was consistent with the previously reported cup shape of exosomes (Figs. 1A and 2A) (61, 62). To address in these cell models the role of exosomes in the release of prion proteins (both cellular and pathological isoforms) into the extracellular milieu, exosomes isolated from cell-free culture media of ScCAD5 and ScN2a cells were subjected to proteinase K digestion, and then both nontreated (−PK) and treated (+PK) fractions were analyzed in immunoblot for PrP using the monoclonal anti-PrP antibody 4H11. As expected, total PrP (−PK) and PrPSc (+PK) were detected in exosomes, which confirms the packaging of both PrP isoforms into exosomes in these cell lines (Figs. 1B and 2B). For further characterization of the exosomes, we analyzed the fractionation of 100,000 × g exosome isolate in a continuous sucrose gradient. As expected, the exosomes reside in fractions with density ranged from 1.13 to 1.18 g/ml (Figs. 1E and 2E). This fraction's density range has been previously reported to contain the exosomes, which indicates the high purity of our exosome isolates (23, 28). Interestingly, both total PrP and PrPSc were enriched in the same density gradient fractions as the exosomes, indicating that secreted PrPSc may be associated with the exosomes in both cell lines (Figs. 1E and 2E). Of note, some PrPSc was found in the collected pellets at all steps of the differential centrifugation (2,000 × g and 10,000 × g), indicating a considerable amount of PrPSc released into the culture media in both cell lines (Fig. S1, A and B).
Figure 1.
Characterization of exosomes isolated from CAD5/ScCAD5 neuronal cells. A, representative TEM of exosomes isolated from CAD5 culture medium reveals a homogenous population of vesicles of 100 nm in diameter characteristic for exosomes (some denoted by black arrows). Scale bar, 100 nm. B, immunoblot of ScCAD5 cell lysate and exosome preparations probed for total PrP (−PK) and PrPSc (+PK) (anti-PrP mAb 4H11). Actin was used as loading control. Flotillin-1 (Flot-1) was used as an exosome marker. C, RT-QuIC of CAD5 exosome, ScCAD5 exosome, ScCAD5 cell lysate, 10% brain homogenate from terminally prion-sick mice (22L) or left unseeded (negative control). The average increase of thioflavin-T fluorescence of replicate wells is plotted as a function of time. The y axis represents RFU, and the x axis represents time (h). D, immunoblot analysis of ScCAD5 cell lysate and exosomes isolated from ScCAD5 culture medium. Exosome preparation is positive for exosome markers Alix, HSC70, Tsg-101, flotillin-1, CD63, and CD9 and negative for mitochondrial marker Bcl2, Golgi marker GM130, and nuclear marker nucleoporin p62. Actin was used loading control. E, ScCAD5 exosome pellet loaded on the top of a continuous sucrose gradient and ultracentrifuged. Fractions were analyzed by Western blotting and probed for HSC70, flotillin-1, and mAb 4H11 to detect PrP/PrPSc. Lanes 9 and 10 represent cell lysate before and after PK digestion, respectively.
Figure 2.
Characterization of exosomes isolated from N2a/ScN2a cells. A, representative TEM of exosomes isolated from N2a culture medium shows a population of vesicles of 100 nm in diameter (some denoted by black arrows). Scale bar, 100 nm. B, immunoblot of ScN2a cell lysate and exosome preparations probed for total PrP (−PK) and PrPSc (+PK) (anti-PrP mAb 4H11). Flotillin-1 (Flot-1) was used as exosome marker. Actin was used as loading control. C, RT-QuIC of N2a exosome, ScN2a exosome, ScN2a cell lysate, and 10% brain homogenate from terminally prion-sick mice (22L) or left unseeded (negative control). The average increase of thioflavin-T fluorescence of replicate wells is plotted as a function of time. The y axis represents RFU, and the x axis represents time in hours. D, immunoblot analysis of ScN2a cell lysate and exosomes isolated from ScN2a cell culture medium. Exosome preparation is positive for exosome markers Alix, HSC70, Tsg-101, flotillin-1, CD63, and CD9 and negative for mitochondrial marker Bcl2, Golgi marker GM130, and nuclear marker nucleoporin p62. Actin was used as loading control. E, ScN2a exosome pallet loaded on the top of a continuous sucrose gradient and ultracentrifuged. The fractions were analyzed by Western blotting and probed for HSC70, flotillin-1, and mAb 4H11 to detect total PrP and PrPSc. Lanes 9 and 10 are cell lysate before and after PK digestion, respectively.
There is mounting evidence that exosomes containing PrPSc are able to disseminate prions from cell to cell (29, 30). Notably, the exosomes isolated from the ScCAD5 cells could successfully infect naïve CAD5 cells as shown by Fig. S3. Even though partial resistance of PrP to PK digestion is a main determinant for prion infectivity, several reports have demonstrated that prion infectivity is not always related to PK resistance. This led us to test the infectivity of PrP packaged into exosomes using real-time quaking-induced conversion (RT-QuIC) assay. RT-QuIC is a highly sensitive in vitro amplification technique used for detection of prions and diagnosing prion diseases in various tissues and body fluids (63, 64). Prion infectivity of exosomes was assessed previously using prion-infected cell assay (34, 61) and protein misfolding cyclic amplification; however, detection of prion infectivity in exosomes using RT-QuIC is lacking. Exosomes isolated from ScCAD5 and ScN2a cells revealed significant conversion of recombinant mouse PrP into ThT-binding aggregates in RT-QuIC compared with exosomes from noninfected cells, indicating the presence of appreciable amounts of prion-seeding activity (Figs. 1C and 2C). In ScCAD5 cells, conversion started after 12 h (lag phase) and reached maximum relative florescence units (RFU) comparable to the corresponding cell lysate and brain homogenate of terminally prion-sick mice (used as positive control) (Fig. 1C). Exosomes isolated from ScN2a cells showed comparable prion-conversion activity, starting after 11 h (lag time) with maximum florescence equivalent to conversion induced by the cell lysate (Fig. 2C). Taken together, exosomes isolated from both cell lines contained PrPSc and were positive for prion-conversion activity, indicating release of bona fide prion infectivity.
Induction of autophagy reduces exosomal release of prions in ScCAD5 and ScN2a cells
Having established that both of our cell models released prions in exosomes, we wanted to study how manipulation of autophagy affects exosomal release of prions. Pharmacological induction of autophagy was reported to have anti-prion effects in vitro and in vivo by promoting the intracellular degradation of PrPSc in lysosomes (53, 54). There is growing evidence that the autophagy machinery and exosomal release are interconnected (65, 66). However, the cross-talk between autophagy and exosomes in modulating the spread of prion infection between cells is still elusive.
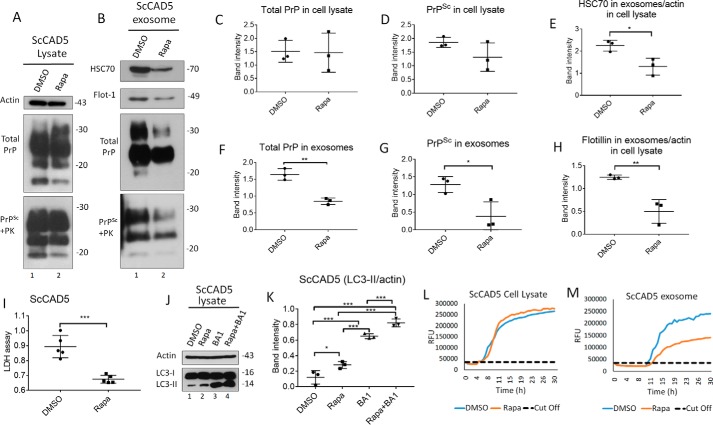
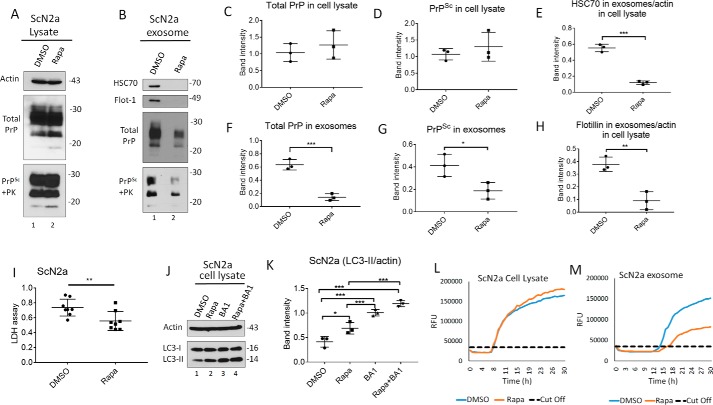
To study the role of autophagy induction in modulating secretion of exosomes and release of prions, we treated ScCAD5 and ScN2a cells with rapamycin, which is an mTOR-dependent autophagy stimulator. Rapamycin was first tested for potential toxicity in CAD5 and N2a cells. We found no significant toxicity for the concentrations used in this study when compared with vehicle-treated cells, yet the LDH release of rapamycin-treated cells was significantly lower than that of control cells, suggesting better survival with the rapamycin treatment (Figs. 3I and 4I). Additionally, rapamycin was confirmed to stimulate autophagy in both cell lines as shown by increase in LC3II signal (Figs. 3, J and K, and 4, J and K). In both cell lines, rapamycin treatment caused a significant decrease in exosome release (Figs. 3, B, E, and H, and 4, B, E, and H) and exosomal total PrP and PrPSc contents (Figs. 3, B, F, and G, and 4, B, F, and G). However, no change in total PrP and PrPSc amounts was found in the cell lysates (Figs. 3, A, C, and D, and 4, A, C, and D). Interestingly, the reduction in exosomal release caused by rapamycin treatment was as pronounced as that induced by GW 4869, which is widely used as an inhibitor for exosomal release (Fig. S1, C and D) (62). Next, we wanted to correlate observed effects on exosomal PrPSc to prion infectivity by testing prion-conversion activity of exosomes using RT-QuIC. Exosomes isolated from rapamycin-treated ScCAD5 and ScN2a cells showed less conversion activity compared with vehicle-treated controls for the same dilution of seed (Figs. 3, L and M, and 4, L and M). Taken together, our results demonstrate that induction of autophagy in prion-infected neuronal cells reduces the release of exosomes, paralleled by a marked reduction of prion infectivity in exosomes.
Figure 3.
Autophagy stimulation decreases exosome secretion and consequently impacts exosomal PrPSc in ScCAD5 neuronal cells. A and B, Western blotting of cell lysate and exosomes from ScCAD5 cells treated with 500 nm of rapamycin (Rapa) or solvent only (DMSO). HSC70 and flotillin-1 (Flot-1) were used as exosome markers. PrP (−/+ PK) was probed with mAb 4H11. C and D, densitometric analysis for either total PrP or PrPSc, respectively, from ScCAD5 cell lysate normalized with actin (± S.D.) after treatment with 500 nm rapamycin or DMSO (n = 3 experiments). E and H, densitometric analysis for exosomal HSC70 and flotillin-1, respectively, normalized with actin in the corresponding cell lysate (± S.D.) after treatment with rapamycin or DMSO (n = 3 experiments). *, p < 0.05; **, p < 0.01. F and G, densitometric analysis for either total PrP or PrPSc, respectively, from ScCAD5 exosomes normalized with actin in the corresponding cell lysate (± S.D.) after treatment with rapamycin or DMSO (n = 3 experiments). **, p < 0.01. *, p < 0.05. I, lactate dehydrogenase (LDH) cytotoxicity assay (OD = 490 nm). Supernatant from ScCAD cells treated with rapamycin or DMSO for 6 days was tested to detect the level of LDH (± S.D.; n = 5 replicates). ***, p < 0.001. J, Western blotting of cell lysate of ScCAD5 cells treated with vehicle only (DMSO), 500 nm of rapamycin, 100 nm of bafilomycin A1 (BA1), or rapamycin + bafilomycin A1 for 4 h. LC3 was used to measure the autophagic flux, and actin was used as loading control. K, densitometric analysis for LC3-II protein levels normalized with actin (± S.D.; n = 3 replicates). *, p < 0.05; ***, p < 0.001. L and M, RT-QuIC for either ScCAD5 cell lysate or exosomes, respectively. The cells were treated with rapamycin or solvent only (DMSO).
Figure 4.
Autophagy stimulation mitigates exosome secretion and significantly decreases exosomal PrPSc in ScN2a cells. A and B, Western blotting of cell lysate and exosomes from ScN2a cells treated with 500 nm of rapamycin (Rapa) or solvent only (DMSO) for 6 days. HSC70 and flotillin-1 (Flot-1) were used as exosome markers. PrP (−/+ PK) was probed with mAb 4H11. Actin was used as a loading control for cell lysate. C and D, densitometric analysis for either total PrP or PrPSc, respectively, from ScN2a cell lysate normalized with actin (± S.D.) after treatment with rapamycin or DMSO (n = 3 experiments). E and H, densitometric analysis for exosomal HSC70 and flotillin-1, respectively, normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). ***, p < 0.001; **, p < 0.01. F and G, densitometric analysis for either total PrP or PrPSc, respectively, from ScN2a exosomes normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). ***, p < 0.001; *, p < 0.05. I, lactate dehydrogenase (LDH) cytotoxicity assay (OD = 490 nm). Supernatant from ScN2a cells treated with 500 nm rapamycin or DMSO for 6 days was tested to detect the level of LDH (± S.D.; n = 8 replicates). **, p < 0.01. J, Western blotting of cell lysate of ScN2a cells treated with vehicle only (DMSO), 500 nm of rapamycin, 100 nm of bafilomycin A1 (BA1), or rapamycin + bafilomycin A1 for 4 h. LC3 was used to measure the autophagic flux, and actin was used as loading control. K, densitometric analysis for LC3-II protein levels normalized with actin (± S.D.; n = 3 replicates). *, p < 0.05; ***, p < 0.001. L and M, RT-QuIC for either ScN2a cell lysate or exosomes; respectively; the cells were treated with rapamycin or solvent only (DMSO).
Inhibition of autophagy increases exosomal release of prions in ScCAD5 and ScN2a cells
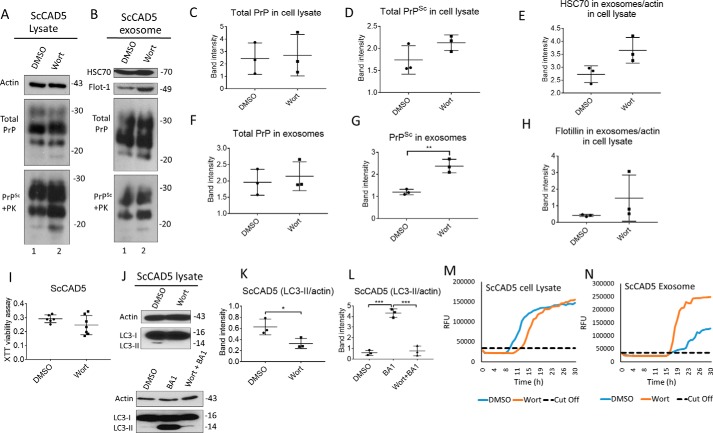
Given that increased autophagy reduces release of prions in exosomes, we next wanted to study whether a compromised autophagy would have opposite effects. To do so, autophagy was suppressed in ScCAD5 using wortmannin, a well known autophagy suppressor, which inhibits class III PI3K (67). The wortmannin concentration used in our study (4 nm) was nontoxic for CAD5 cells as shown by XTT assay (Fig. 5I). Although the basal autophagy in CAD5 cells is not very prominent, wortmannin was able to abolish the signal of LC3II in CAD5 cells indicating a successful inhibition of autophagy (Fig. 5, J–L). Interestingly, ScCAD5 cells with such compromised autophagy exhibited a slight increase in exosomes released (Fig. 5, B, E, and H), yet this increase in exosomal release was not statistically significant. Importantly, this was associated with significantly increased PrPSc amount in the exosomes (Fig. 5, B, F, and G). Additionally, wortmannin increased the infectivity of the ScCAD5 released exosomes as shown by RT-QuIC (Fig. 5, M and N). The effect of compromised autophagy on exosomal release was similar to the effects of treatment with monensin, a drug reported to stimulate the packaging of PrPSc into exosomes and the intercellular transfer of prion infectivity (34) (Fig. S1E).
Figure 5.
Inhibition of autophagy increases exosome release and exosomal PrPSc in ScCAD5 cells. A and B, Western blotting of cell lysate and exosomes from ScCAD5 cells, respectively, treated with 4 nm of wortmannin (Wort) for 48 h or solvent only treated (DMSO). HSC70 and flotillin-1 (Flot-1) were used as exosome markers. Actin was used as loading control for cell lysate. PrP (−/+ PK) was probed with mAb 4H11. C and D, densitometric analysis for either total PrP or PrPSc, respectively, from ScCAD5 cell lysate normalized with actin (± S.D.) after treatment with 4 nm of wortmannin or DMSO (n = 3 experiments). E and H, densitometric analysis for exosomal HSC70 and flotillin-1, respectively, normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). F and G, densitometric analysis for either total PrP or PrPSc, respectively, from ScCAD5 exosomes normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). **, p < 0.01. I, XTT viability assay. ScCAD5 cells were treated with 4 nm wortmannin or DMSO for 48 h; then cell viability was detected based on metabolic activity (± S.D.; n = 7 replicates). J, upper panel, Western blotting of cell lysate of ScCAD5 cells treated with vehicle only (DMSO) or 4 nm of wortmannin for 4 h. Lower panel, Western blotting of cell lysate of ScCAD5 cells treated with vehicle only (DMSO), 4 nm of wortmannin, or wortmannin with bafilomycin A1 (Wort + BA1) for 4 h. LC3 was used to measure the autophagic flux and actin was used as loading control. K and L, densitometric analysis for LC3-II protein levels normalized with actin (± S.D.; n = 3 replicates). *, p < 0.05; ***, p < 0.001. M and N, RT-QuIC for either ScCAD5 cell lysate or exosomes, respectively. The cells were treated with wortmannin or solvent only (DMSO).
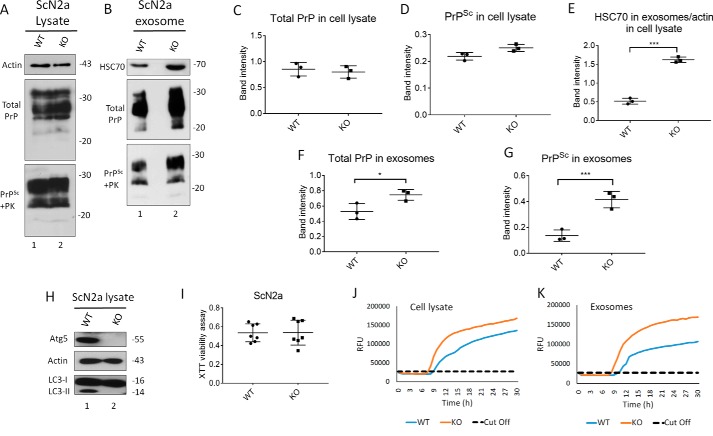
Next, we wanted to genetically compromise autophagy in ScCAD5 cells. The key autophagy protein Atg5 was knocked down using siRNA (siAtg5) (Fig. S2). We only achieved an incomplete transient knockdown of Atg5. Consequently, ScCAD5 cells with such compromised autophagy exhibited a minor increase in exosomes released (Fig. S2).
Using CRISPR/Cas9 technology, we generated Atg5-deficient ScN2a cells (KO), which are defective in autophagy (Fig. 6H). Notably, the Atg5-null ScN2a cells showed normal viability compared with their WT counterparts (Fig. 6I). Atg5-KO ScN2a cells revealed significantly higher exosome release compared with WT control cells, as shown by HSC70 signals in immunoblots (Fig. 6, B and E). Importantly, this was associated with strongly increased total PrP and PrPSc amounts in exosomes (Fig. 6, F and G, respectively). However, no change occurs in total PrP and PrPSc levels in the cell lysates (Fig. 6, A, C, and D). Testing prion-conversion activity by RT-QuIC demonstrated shorter lag time and higher maximum florescence (RFU) for exosomes from Atg5-KO cells, indicating higher prion-seeding activity compared with exosomes isolated from WT ScN2a cells (Fig. 6K). Notably, the effect of autophagy blockage on the induction of the exosome release in ScN2a-KO cells was comparable with the effect of treating ScN2a cells with monensin, which has been reported to increase exosomal release from neuronal cells (Fig. S1F). Taken together, blocking autophagy, either partially or completely, boosts release of prions in exosomes and consequently increases intercellular spread of prion infection.
Figure 6.
Knockout of Atg5 increases exosome release and exosomal PrPSc in ScN2a cells. A and B, Western blotting of cell lysate and exosomes from ScN2a cells respectively, either WT or KO for Atg5. HSC70 was used as exosomal markers. Total PrP and PrPSc were detected using mAb 4H11 antibody. C and D, densitometric analysis for either total PrP or PrPSc, respectively, from WT or KO ScN2a cell lysate normalized with actin (± S.D.; n = 3 experiments). E, densitometric analysis for exosomal HSC70 normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). ***, p < 0.001. F and G, densitometric analysis for either total PrP or PrPSc, respectively, from WT or KO ScN2a exosomes normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). ***, p < 0.001; *, p < 0.05. H, immunoblot showing complete knockout of Atg5 compared with WT ScN2a cells. Actin was used as loading control. Atg5 KO resulted in complete absence of LC3-II, confirming disruption of autophagy machinery. I, XTT viability assay. (OD = 490 nm). The cells were cultured for 48 h for XTT viability assay (n = 7 replicates). J and K, RT-QuIC for WT or KO ScN2a cells.
ScCAD5 cells release more prions in exosomes and have a lower level of autophagy than ScN2a cells
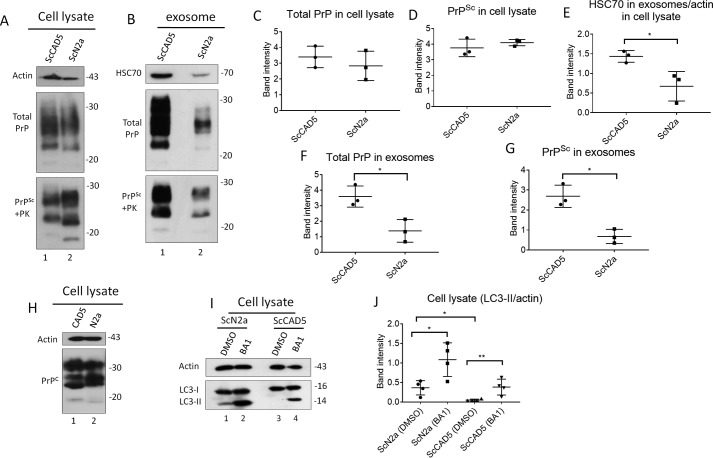
To explore the difference between CAD5 and N2a neuronal cells in terms of autophagy competence and exosomal release, we isolated exosomes from the same number of ScCAD5 and ScN2a cells. We found that the exosome yield obtained from ScCAD5 cells was markedly higher than that isolated from almost same number of ScN2a cells, as shown by HSC70 signal in immunoblot analysis (Fig. 7, B and E). Although ScCAD5 and ScN2a cells have comparable amounts of PrPtotal in the lysate, the PrP levels in ScCAD5 isolated exosomes are significantly higher than ScN2a exosomes (Fig. 7, A–D, F, and G). Then we wanted to correlate this to the level of basal autophagy in both cell types. Basal autophagic flux, as measured by LC3-II signals, was negligible in ScCAD5 cells compared with that in ScN2a cells (Fig. 7, I and J). A recent study reported that the level of PrPC plays a role in controlling exosomal release (16). This led us to test the difference in PrPC between uninfected CAD5 and N2a cells. We found that PrPC levels in CAD5 and N2a cells are not very different (Fig. 7H).
Figure 7.
ScCAD5 cells release more exosomes and PrPSc compared with ScN2a cells, which is inversely correlated to autophagy competence. Comparable numbers of ScN2a and ScCAD5 cells were used in this experiment. A and B, cell lysate and exosomes isolated from conditioned media from ScCAD5 and ScN2a cells were analyzed in immunoblot for HSC70 and PrP. C and D, densitometric analysis for either total PrP or PrPSc respectively from ScCAD5 or ScN2a cell lysate normalized with actin (± S.D.; n = 3 experiments). E, densitometric analysis for exosomal HSC70 normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). *, p < 0.05. F and G, densitometric analysis for either total PrP or PrPSc, respectively, from ScCAD5 or ScN2a exosomes normalized with actin in the corresponding cell lysate (± S.D.; n = 3 experiments). *, p < 0.05. H, immunoblot comparing PrPC levels between CAD5 and N2a cells using mAb 4H11. I, immunoblot comparing the level of LC3-I to LC3-II between ScN2a and ScCAD5 cells with and without treatment with bafilomycin A1 (BA1) for 4 h. Actin was used as a loading control. J, densitometric analysis for LC3-II protein levels normalized with actin (± S.D.; n = 4 replicates). *, p < 0.05, **, p < 0.01.
In summary, the lower level of basal autophagy could be an explanation for the higher exosomal release of prions from ScCAD5 cells. This may facilitate spreading of prions to neighboring cells and explain their high susceptibility to prion infection.
Discussion
Exosomes are nanovesicles secreted by almost all cell types into the extracellular space, which contains proteins, lipids, and nucleic acids that are derived from the parent cell. They play a crucial role in intercellular communication, both locally and systemically. This includes immune regulation, tissue repair, and neuronal and neuron–glia communication (39, 68–70). Exosomes originate from MVBs, which are late endosomes and hence a part of the endosomal system (23). Whereas exosomes are constitutively released from most cells under normal physiological conditions, their formation and release can be modulated by infection and cellular stress (71). The presence of exosomes with their cargo in body fluids makes them optimal platforms for intercellular transmission of diseases. This includes neurodegenerative disorders which involve prion and prion-like mechanisms (30, 72–75).
High prion-conversion activity in exosomes
Here, we use prion-infected cells to study the exosomal release of prions and the cellular programs involved. Notably, we detected significant prion-conversion activity in exosomes isolated from culture media of ScCAD5 and ScN2a cells using RT-QuIC, which is one of the most sensitive and specific techniques for detection of prion-seeding activity (63). Interestingly, infectivity in exosomes isolated from ScCAD5 cells was comparable with that of brain homogenates from mice with terminal prion disease and the cell lysates of ScCAD5 cells in terms of RT-QuIC curve kinetics. Considering that exosomes are highly diluted compared with prions as found in brain homogenate and cell lysate, the high prion-seeding activity of exosomes suggests that prion infectivity is very efficiently packaged into such extracellular vesicles. Interestingly, the exosomes isolated from the ScCAD5 cells efficiently infect naïve CAD5 cells (Fig. S3). This underscores the potential impact of exosomes in spreading prion infectivity to neighboring and more distant cells in the brain, in accordance with several previous reports (28, 30, 61).
The molecular and cellular mechanisms that influence the packaging and release of prions into exosomes are not fully understood. A recent study has demonstrated a role for the neutral sphingomyelinase pathway in packaging PrPSc into exosomes, albeit it did not exclude the involvement of other controlling pathways (62). In the current study, we demonstrate that autophagy, a basic cellular housekeeping and degradation machinery, controls the release of prions into exosomes.
Enhanced autophagy decreases exosomal release of prions
We show here that pharmacological induction of autophagy by rapamycin remarkably inhibits release of exosomes in ScCAD5 cells, a central nervous system cell line, and ScN2a cells, which are derived from the peripheral nervous system. This is paralleled by reduction of prion-conversion activity and PrPSc in such exosomes. Intriguingly, limiting the exosome release and hence the intercellular prion dissemination could be one of the ways used by autophagy to control the prion infection. At the cellular level, the effect of autophagy stimulation on exosome release can be explained by modulating the trafficking of MVBs. The latter fuse more with autophagosomes, the typical degradative and recycling organelles. This is in agreement with reports showing that induction of autophagy by starvation or LC3 overexpression enhances the fusion of MVBs with autophagic vacuoles (50, 51). There is a growing body of evidence showing that autophagy, MVBs, and exosome release are tightly interconnected (39). In addition to removal of harmful or unwanted material in exosomes in cellular stress situations, this mechanism has a general role in organizing unconventional secretion of proteins that are devoid of a signal peptide (40, 76–78). Despite efforts toward understanding the packaging of proteins and protein aggregates into exosomes, the factors that modulate whether fusion of MVBs with autophagic vacuoles results in release of exosomes or degradation in lysosomes are not fully elucidated.
Dysfunctional autophagy increases release of prions in exosomes
On the other side, inhibition of autophagy either by wortmannin in ScCAD5 cells or Atg5 knockout in ScN2a cells markedly promotes release of exosomes. Not unexpected, this was accompanied by release of prion infectivity in exosomes as revealed by our RT-QuIC results. This indicates that a dysfunctional autophagy may contribute to lateral spreading of prions. This finding is concordant with studies revealed that disrupted autophagy leads to exacerbated release of α-synuclein aggregates in exosomes that increased intercellular transmission of this detrimental protein to neighboring neurons (35, 79). Moreover, a dysfunctional autophagy is a characteristic of the elderly brain, associated with release of undigested or partially digested aggregates in exosomes (80). This could be a contributing factor for the high prevalence of neurodegenerative diseases in this age group.
CAD5 cells have less efficient autophagy and release more exosomes than N2a cells
Interestingly, our data show that PrPSc and prion-conversion activity released in exosomes isolated from ScN2a cells were miniscule compared with that released from ScCAD5 cells. One possible explanation is that the subcellular trafficking of PrPSc is different between ScN2a and CAD5 cells. It has been reported previously that PrPSc does not co-localize with flotillin in ScN2a cells, whereas CNS-derived ScGT1–7 cells show an abundance of flotillin-PrPSc–rich compartments (81).
On the other hand, this was not only true for PrPSc. It was also a general feature that CAD5 cells release higher amounts of exosomes than N2a cells do. The weak basal level of autophagy in CAD5 cells compared with N2a cells as shown by LC3-II signals could be one of the reasons making CAD5 cells better exosome releasers than N2a cells. However, the difference in exosome release in our two cell lines does not seem to be related to PrPC level. We propose that the difference in basal autophagy and consequently in exosomal spreading of prions between CAD5 and N2a cells could be a contributing factor for the difference in their response to prion infection (CAD5 is more susceptible to infection). However, further studies on different cell lines are required to test whether the weak basal autophagy is a general feature for the central neuronal cells, which show sever neuronal loss in response to prion infection
Taken together, there is an intricate cross-talk between autophagy and exosome release that profoundly influences lateral transmission of the prions. This interplay represents a novel target for developing therapeutic approaches to fight prion and prion-like diseases.
Experimental procedures
Reagents and antibodies
Unless otherwise indicated, all the reagents and chemical were obtained from Sigma–Aldrich. Proteinase K (PK; 03115879001) and Pefabloc inhibitor (11286700) were from Roche. Lipofectamine 2000 (Invitrogen, 1166807), Lipofectamine LTX transfection reagents (Invitrogen, 15338100), and On Target plus mouse Atg5 (11793) siRNA Smart Pool (GE Health Care, L-064838-00-0005) were acquired from the manufacturers indicated in parentheses.
Sources of the antibodies were as follows: anti-flotillin-1 (BD Transduction Laboratories, 610820), anti-Tsg101 (Santa Cruz Biotechnology, sc-7964), HSC70 (Thermo Scientific, PA5-27337), CD63 (Santa Cruz Biotechnology, SC-15363), Alix (Cell Signaling, 2171), anti-CD9 (Abcam, ab92726), Bcl2 (Cell Signaling, D17C4), Atg-5 (clone 7C6) (NanoTools, 0262-100), nucleoporin P62 (BD Transduction Laboratories, 610497), anti-GM130 (BD Transduction Laboratories, 610822), anti-β-actin mAb (Sigma–Aldrich, A5441), anti-LC3 (clone 2G6) (NanoTools, 0260-100), anti-PrP mAb 4H11 has been previously described (52). Peroxidase-conjugated Igs were from Jackson Immunoresearch Lab (goat anti-mouse HRP, 115-035-003; and goat anti-rabbit HRP, 111-035-45).
Maintenance of cell culture
The mouse neuroblastoma cell line N2a was obtained from ATCC (CCL-131), and they were cultured in Gibco Opti-MEM GlutaMAX medium (Gibco, 51985-34) containing 10% fetal bovine serum (Sigma, F1051), and penicillin/streptomycin in a 5% CO2 atmosphere. CAD5 cells are a central nervous system catecholaminergic cell line (82). The cells were subjected to single cell sub-cloning to be optimized for the prion infection (60). The cells were a generous gift from Dr. S. Mahal (Scripps Institute–Florida) and were cultured in Opti-MEM GlutaMAX medium containing 10% bovine growth serum (Hyclone, SH30541.03), and penicillin/streptomycin in a 5% CO2 atmosphere. The cells were infected with 10% brain homogenate of mouse-adapted scrapie strain 22L. For rapamycin treatment, the cells were treated with 500 nm of rapamycin or solvent only (DMSO) for 6 days. Firstly, rapamycin was added to complete media for 3 days; then complete medium was replaced with exosome-free medium, and rapamycin was added for another 3 days. For wortmannin treatment, the cells were cultured in complete medium for 48 h. Then complete medium was replaced with exosome-free medium, and the cells were treated with 4 nm of wortmannin for 48 h or solvent only–treated (DMSO). The cells were treated with 100 nm of bafilomycin A1 or vehicle only (DMSO) for 4 h.
Exosome isolation
Exosomes were isolated from five confluents (15-cm plates); each plate has ∼15 million cells. For the exosome collection, the culture medium was replaced by exosome-free medium 48 h after seeding the cells into 15-cm plates. The cells were left for 2 or 3 days as indicated, and the exosomes were isolated from culture medium by differential ultracentrifugation according to the Clayton group (23) using a Beckmann Coulter Optima XE-90 ultracentrifuge. Briefly, the culture supernatant was centrifuged at 2000 × g for 10 min; the pellet was discarded (dead cells); the supernatant was centrifuged again at 10,000 × g for 30 min; the pellet was discarded (cell debris); the supernatant was ultracentrifuged at 100,000 × g for 70 min; then the supernatant was discarded; and the pellet contains the exosomes. The pellet was washed one time with PBS and ultracentrifuged at 100,000 × g for 70 min. Exosome pellet was resuspended in 100 μl of PBS and then transferred to a continuous sucrose gradient, and 10 fractions were collected after ultracentrifugation at 285,000 × g for 50 min.
Knockout of Atg5 CRISPR/Cas9 in N2a cells
Expression vectors for guide RNA (sgRNA) were made targeting two exons of the Atg5 gene: exons 5 and 6. sgRNAs against exons 5 and 6 were purchased from Sigma. The following constructs were used: exon 5: target ID MM0000476061, vector U6gRNA-Cas9–2A-GFP, target sequence CCTCAACCGCATCCTTGGATGG; and exon 6, target ID MM0000476062, vector U6gRNA-Cas9–2A-RFP, target sequence GCCATCAACCGGAAACTCATGG. N2a-K21 cells were co-transfected with sgRNA and Cas9 expression vectors by Lipofectamine Plus LTX reagent according to the manufacturer's protocol. We targeted exons 5 and 6 together in double transfection. Transient GFP/RFP expression was used as transfection effectiveness control. Single cell clones were analyzed by immunoblotting for Atg5 and LC3-II and by DNA sequencing upon PCR amplification of genomic DNA.
PK digestion and Western blotting
Immunoblot analysis was previously described (59). Briefly, confluent cells or exosome pellet was lysed in cold lysis buffer (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm EDTA, 0.5% Triton X-100, 0.5% sodium deoxycholate) for 10 min. Aliquots of lysates were incubated with PK for 30 min at 37 °C; PK was stopped by addition of proteinase inhibitors (0.5 mm Pefabloc) and directly precipitated with methanol. For samples without PK treatment proteinase inhibitors were added directly and precipitated with methanol. Precipitated proteins were resuspended in TNE buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA). For the gradient, the PK digestion was carried out after the fractionation. The samples were run on 12.5% SDS-PAGE, electroblotted on Amersham Biosciences Hybond P 0.45 PVDF (Amersham Biosciences, 10600023) and analyzed in immunoblot, using the Luminata Western Chemiluminescent HRP Substrates (Millipore, WBLUF0100).
Transmission electron microscopy
The exosome pellets were fixed with 2% paraformaldehyde (Electron Microscopy Sciences, 15700) and 2.5% glutaraldehyde (Electron Microscopy Sciences, 16220) in 0.1 m cacodylate buffer, pH 7.4, for 2 h. The fixation supernatants were discarded, and warm 1% agarose was added over the pellet. After the cooling of the agarose, the embedded pellet was cut into pieces and washed three times with same buffer. Then the pieces were postfixed in 1% osmium tetroxide (Electron Microscopy Sciences, 19100) in cacodylate buffer (Electron Microscopy Sciences, 11652) for 1 h, dehydrated through a graded series of ethanol, and embedded in Spurr resin (Electron Microscopy Sciences, 14300). Ultrathin sections were cut in a Leica EM UC7 ultramicrotome using a diamond knife and stained with 2% aqueous uranyl acetate and Reynolds's lead citrate. The sections were examined under a Hitachi H7650 transmission electron microscope at 80 kV. The images were acquired through an AMT 16000 CCD fixed on the microscope.
XTT cell viability assay
The cell viability was assessed by TACS® XTT cell proliferation Assay (Trevigen, 4891-025-K) according to the manufacturer's instructions. Absorbance was measured at 490 nm using BioTek Synergy HT. The data were expressed as percentages of viability compared with the corresponding control.
LDH cytotoxicity assay
The assay was conducted according to the manufacturer's protocol (Roche, 11644793001). Briefly, cell supernatant was centrifuged to remove cell debris. In a 96-well plate, 100 μl of cell supernatant and 100 μl of LDH reagent were mixed and incubated away from light for 10 min. The reaction was stopped using 50 μl of 1 n HCL. Measurement of the color intensity was done at 490 nm. At least six replicates were done for every condition.
Primary prion infection
Infection using cell homogenate
Either uninfected CAD5 or persistently infected ScCAD5 (22L strain) cells were harvested from one plate (10 cm). Culture medium was removed, followed by five washes with PBS, and then cells were mechanically detached by pipetting. The cell suspension was collected and centrifuged (200 × g, 5 min). Supernatant was discarded, and the cells were resuspended in PBS (180 μl). The cells were centrifuged again, the supernatant was discarded, and the pellet resuspended in PBS. The cells were transferred to cryogenic vials and stored at −20 °C for 2 h, thawed, and homogenized by passing through a 25-gauge needle 50 times.
Infection using brain homogenate
The mouse-adapted scrapie strain 22L was propagated in C57Bl/6 mice. The mice were euthanized when they were terminally sick. To prepare brain homogenates, brain was homogenized in PBS at a final concentration of 10% (w/v) using a gentle MACSTM dissociator at room temperature for 2 min, followed by centrifugation at 2,000 × g for 1 min, then aliquoted, and stored at −80 °C.
Infection using exosomes
Exosomes were collected from cell culture medium of 15 plates (10 cm) of either uninfected CAD5 or ScCAD5 (22L) cells. For primary infection, 1 × 105 cells were overlaid with 50 μl of cell homogenate, brain homogenate, or exosome preparation in 450 μl of serum-free culture medium. After 5 h of incubation, 500 μl of complete culture medium was added. Medium was removed 24 h later, and the cells were washed once with PBS before fresh culture medium was added to the cells. The cells were passaged several times and subjected to PK digestion for detection of PrPSc in immunoblot.
Real time quaking-induced conversion
Preparation of recombinant protein
The recombinant mouse PrP protein preparation was conducted according to Caughey group (63). Briefly, Escherichia coli has a pET41 vector with the mouse PrP sequence cultured in LB medium supplemented with kanamycin (0.05 mg/ml) and chloramphenicol (0.034 mg/ml), and the Overnight Express Autoinduction System 1 (Novagen, 71300) was used to induce the protein expression. The inclusion bodies were isolated from the pelleted bacteria using the Bug Buster Master Mix (Novagen, 71456) and stored at −20 °C. For purification of the recombinant PrP, the isolated inclusion bodies were dissolved into 8 m guanidine HCl in 100 mm sodium phosphate, pH 8.0, and then incubated on the rocker for 1 h at room temperature. Nickel–nitrilotriacetic acid Superflow resin beads (Qiagen, 1018401) were incubated in denaturing buffer (6 m guanidine HCl, 100 mm sodium phosphate, 10 mm Tris, pH 8.0) for 1 h at room temperature. Solubilized inclusion bodies were centrifuged at 7900 × g for 5 min, and the supernatant was added to the beads and incubated for 1 h with gentle rocking. The beads were then packed into a XK16 glass column (GE Healthcare Life Sciences; 28988937; length, 200 mm). Using an Amersham Biosciences AKTA Explorer FPLC running Unicorn software (version 5; GE Healthcare Life Sciences), the protein was refolded by a gradient from 100% denaturing buffer to 100% refolding buffer (100 mm sodium phosphate, 10 mm Tris, pH 8.0) over 4 h. Afterward, the column was washed for 30 min with refolding buffer. The protein was then eluted from the column using a linear gradient from 100% refolding buffer to 100% elution buffer (500 mm imidazole, 100 mm sodium phosphate, 10 mm Tris, pH 5.8). The central portions of the A280 UV peak were collected into dialysis buffer (10 mm sodium phosphate buffer, pH 5.8). The purified protein was then filtered with a 0.22-μm filter, transferred into Slide-A-Lyzer dialysis Cassette (molecular mass, 10 kDa; Thermo-Scientific, 87731), and placed in a 4-liter beaker of dialysis buffer overnight at 4 °C with continuous stirring. Following dialysis, the protein solution was filtered again with a prewashed 0.22-μm Argos syringe filter, measured using BCA protein assay (Thermo Scientific, 23227), aliquoted, and kept at −80 °C until use.
RT-QuIC assay
98 μl of the master mix containing a final concentration of 10 mm phosphate buffer (pH 7.4), 300 mm NaCl, 0.1 mg/ml rPrP, 10 μm ThT, and 1 mm of EDTA was loaded into each well of a black-walled 96-well plate with a clear bottom (Nunc, 165305), and reactions were seeded with 2 μl of the indicated test dilution for a final reaction volume of 100 μl. All reactions contained a final concentration of 0.002% SDS. The reactions were incubated at 42 °C and shaken every other minute at 700 rpm. Brain homogenate, cell, or exosomes lysate serial dilutions were prepared using 0.5-ml microtubes. The plates were sealed and incubated in FLUOstar Omega (BMG Labtech, Cary, NC) for 30 h with cycles of 60 s shaking and 60 s of rest throughout the incubation. ThT fluorescence measurements (450 nm excitation and 480 nm emission; bottom read) were taken every 15 min. To analyze the RT-QuIC, the data were averaged from four replicate wells and normalized to a percentage of the maximal fluorescence response of the instrument. The average values were plotted against the reaction times. The samples were scored positive if at least 50% of replicates reached a ThT fluorescence cut-off.
Statistical analysis
Statistical analysis was performed using GraphPad InStat (GraphPad Software Inc., version 3.05; Ralf Stahlman, Purdue University) using either the unpaired two-tailed t test for pair-wise comparisons or the one-way analysis of variance analysis with Tukey post test for multiple comparisons. Statistical significance was expressed as follows: ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001. The graphs were plotted using GraphPad Prism 7.
Author contributions
B. A. A., D. H. A., and H. M. S. conceptualization; B. A. A., D. H. A., and H. M. S. data curation; B. A. A., D. H. A., and H. M. S. formal analysis; B. A. A., D. H. A., and H. M. S. validation; B. A. A. and D. H. A. investigation; B. A. A., D. H. A., and H. M. S. methodology; B. A. A., D. H. A., and H. M. S. writing-original draft; D. H. A. and H. M. S. writing-review and editing; H. M. S. resources; H. M. S. supervision; H. M. S. funding acquisition; H. M. S. project administration.
Supplementary Material
Acknowledgments
We thank Wei-Xiang Dong (University of Calgary Microscopy and Imaging Facility) for help with TEM analysis. We also thank Dr. Sabine Gilch (University of Calgary) for help with RT-QuIC analysis.
This work was supported by NINDS, National Institutes of Health Grant R01 NS076853-01A1, Alberta Innovates–Health Solutions Grant 201200002, 201300020, and 201600010, and Canadian Natural Sciences and Engineering Research Council of Canada Grant RGPIN-2015-05130. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
- CNS
- central nervous system
- TEM
- transmission EM
- RT-QuIC
- real-time quaking-induced conversion
- MVB
- multivesicular body
- PK
- proteinase K
- RFU
- relative florescence unit(s)
- sgRNA
- guide RNA
- ThT
- thioflavin-T
- XTT
- 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide.
References
- 1. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 2. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 10.1073/pnas.95.23.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilch S., Chitoor N., Taguchi Y., Stuart M., Jewell J. E., and Schätzl H. M. (2011) Chronic wasting disease. Top. Curr. Chem. 305, 51–77 10.1007/128_2011_159 [DOI] [PubMed] [Google Scholar]
- 4. Telling G. C. (2013) The importance of prions. PLoS Pathog. 9, e1003090 10.1371/journal.ppat.1003090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wadsworth J. D., Hill A. F., Beck J. A., and Collinge J. (2003) Molecular and clinical classification of human prion disease. Br. Med. Bull. 66, 241–254 10.1093/bmb/66.1.241 [DOI] [PubMed] [Google Scholar]
- 6. Watts J. C., Balachandran A., and Westaway D. (2006) The expanding universe of prion diseases. PLoS Pathog. 2, e26 10.1371/journal.ppat.0020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aguzzi A., and Polymenidou M. (2004) Mammalian prion biology: one century of evolving concepts. Cell 116, 313–327 10.1016/S0092-8674(03)01031-6 [DOI] [PubMed] [Google Scholar]
- 8. Collinge J. (2005) Molecular neurology of prion disease. J. Neurol. Neurosurg. Psychiatry 76, 906–919 10.1136/jnnp.2004.048660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weissmann C. (2004) The state of the prion. Nat. Rev. Microbiol. 2, 861–871 10.1038/nrmicro1025 [DOI] [PubMed] [Google Scholar]
- 10. Borchelt D. R., Scott M., Taraboulos A., Stahl N., and Prusiner S. B. (1990) Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 110, 743–752 10.1083/jcb.110.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caughey B., and Raymond G. J. (1991) The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 266, 18217–18223 [PubMed] [Google Scholar]
- 12. Brown D. R., Qin K., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., von Bohlen A., Schulz-Schaeffer W., Giese A., Westaway D., and Kretzschmar H. (1997) The cellular prion protein binds copper in vivo. Nature 390, 684–687 10.1038/37783 [DOI] [PubMed] [Google Scholar]
- 13. Roffé M., Beraldo F. H., Bester R., Nunziante M., Bach C., Mancini G., Gilch S., Vorberg I., Castilho B. A., Martins V. R., and Hajj G. N. (2010) Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc. Natl. Acad. Sci. 107, 13147–13152 10.1073/pnas.1000784107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zanata S. M., Lopes M. H., Mercadante A. F., Hajj G. N., Chiarini L. B., Nomizo R., Freitas A. R., Cabral A. L., Lee K. S., Juliano M. A., de Oliveira E., Jachieri S. G., Burlingame A., Huang L., Linden R., et al. (2002) Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J. 21, 3307–3316 10.1093/emboj/cdf325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., and Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 10.1038/nature07761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dias M. V., Teixeira B. L., Rodrigues B. R., Sinigaglia-Coimbra R., Porto-Carreiro I., Roffé M., Hajj G. N., and Martins V. R. (2016) PRNP/prion protein regulates the secretion of exosomes modulating CAV1/caveolin-1-suppressed autophagy. Autophagy 12, 2113–2128 10.1080/15548627.2016.1226735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liberski P. P., Sikorska B., Bratosiewicz-Wasik J., Gajdusek D. C., and Brown P. (2004) Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: from apoptosis to autophagy. Int. J. Biochem. Cell Biol. 36, 2473–2490 10.1016/j.biocel.2004.04.016 [DOI] [PubMed] [Google Scholar]
- 18. Schätzl H. M., Laszlo L., Holtzman D. M., Tatzelt J., DeArmond S. J., Weiner R. I., Mobley W. C., and Prusiner S. B. (1997) A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 71, 8821–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sikorska B., Liberski P. P., Giraud P., Kopp N., and Brown P. (2004) Autophagy is a part of ultrastructural synaptic pathology in Creutzfeldt–Jakob disease: a brain biopsy study. Int. J. Biochem. Cell Biol. 36, 2563–2573 10.1016/j.biocel.2004.04.014 [DOI] [PubMed] [Google Scholar]
- 20. Keller S., Sanderson M. P., Stoeck A., and Altevogt P. (2006) Exosomes: from biogenesis and secretion to biological function. Immunol. Lett. 107, 102–108 10.1016/j.imlet.2006.09.005 [DOI] [PubMed] [Google Scholar]
- 21. Simons M., and Raposo G. (2009) Exosomes: vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 22. Stoorvogel W., Kleijmeer M. J., Geuze H. J., and Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 10.1034/j.1600-0854.2002.30502.x [DOI] [PubMed] [Google Scholar]
- 23. Théry C., Amigorena S., Raposo G., and Clayton A. (2006) Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3:Unit 3.22 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 24. Properzi F., Logozzi M., and Fais S. (2013) Exosomes: the future of biomarkers in medicine. Biomark. Med. 7, 769–778 10.2217/bmm.13.63 [DOI] [PubMed] [Google Scholar]
- 25. Keller S., Ridinger J., Rupp A.-K., Janssen J. W., and Altevogt P. (2011) Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 9, 86 10.1186/1479-5876-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 27. Mittelbrunn M., and Sánchez-Madrid F. (2012) Intercellular communication: diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 13, 328–335 10.1038/nrm3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Février B., and Raposo G. (2004) Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16, 415–421 10.1016/j.ceb.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 29. Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., and Raposo G. (2004) Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U.S.A. 101, 9683–9688 10.1073/pnas.0308413101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vella L. J., Sharples R. A., Lawson V. A., Masters C. L., Cappai R., and Hill A. F. (2007) Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J. Pathol. 211, 582–590 10.1002/path.2145 [DOI] [PubMed] [Google Scholar]
- 31. Alais S., Simoes S., Baas D., Lehmann S., Raposo G., Darlix J. L., and Leblanc P. (2008) Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biol. Cell 100, 603–615 10.1042/BC20080025 [DOI] [PubMed] [Google Scholar]
- 32. Porto-Carreiro I., Février B., Paquet S., Vilette D., and Raposo G. (2005) Prions and exosomes: from PrPc trafficking to PrPsc propagation. Blood Cells Mol. Dis. 35, 143–148 10.1016/j.bcmd.2005.06.013 [DOI] [PubMed] [Google Scholar]
- 33. Bellingham S. A., Guo B. B., Coleman B. M., and Hill A. F. (2012) Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front. Physiol. 3, 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo B. B., Bellingham S. A., and Hill A. F. (2016) Stimulating the release of exosomes increases the intercellular transfer of prions. J. Biol. Chem. 291, 5128–5137 10.1074/jbc.M115.684258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Danzer K. M., Kranich L. R., Ruf W. P., Cagsal-Getkin O., Winslow A. R., Zhu L., Vanderburg C. R., and McLean P. J. (2012) Exosomal cell-to-cell transmission of α synuclein oligomers. Mol. Neurodegener. 7, 42 10.1186/1750-1326-7-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajendran L., Honsho M., Zahn T. R., Keller P., Geiger K. D., Verkade P., and Simons K. (2006) Alzheimer's disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. U.S.A. 103, 11172–11177 10.1073/pnas.0603838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sharples R. A., Vella L. J., Nisbet R. M., Naylor R., Perez K., Barnham K. J., Masters C. L., and Hill A. F. (2008) Inhibition of secretase causes increased secretion of amyloid precursor protein C-terminal fragments in association with exosomes. FASEB J. 22, 1469–1478 10.1096/fj.07-9357com [DOI] [PubMed] [Google Scholar]
- 38. Grad L. I., Yerbury J. J., Turner B. J., Guest W. C., Pokrishevsky E., O'Neill M. A., Yanai A., Silverman J. M., Zeineddine R., Corcoran L., Kumita J. R., Luheshi L. M., Yousefi M., Coleman B. M., Hill A. F., et al. (2014) Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 3620–3625 10.1073/pnas.1312245111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baixauli F., López-Otín C., and Mittelbrunn M. (2014) Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front. Immunol. 5, 403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang M., and Schekman R. (2013) Unconventional secretion, unconventional solutions. Science 340, 559–561 10.1126/science.1234740 [DOI] [PubMed] [Google Scholar]
- 41. Levine B., and Klionsky D. J. (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 42. Mizushima N., Levine B., Cuervo A. M., and Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mizushima N., and Klionsky D. J. (2007) Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 27, 19–40 10.1146/annurev.nutr.27.061406.093749 [DOI] [PubMed] [Google Scholar]
- 44. Ravikumar B., Duden R., and Rubinsztein D. C. (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 10.1093/hmg/11.9.1107 [DOI] [PubMed] [Google Scholar]
- 45. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., and Rubinsztein D. C. (2003) α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 10.1074/jbc.M300227200 [DOI] [PubMed] [Google Scholar]
- 46. Qin Z.-H., Wang Y., Kegel K. B., Kazantsev A., Apostol B. L., Thompson L. M., Yoder J., Aronin N., and DiFiglia M. (2003) Autophagy regulates the processing of amino terminal huntingtin fragments. Hum. Mol. Genet. 12, 3231–3244 10.1093/hmg/ddg346 [DOI] [PubMed] [Google Scholar]
- 47. Berger Z., Ravikumar B., Menzies F. M., Oroz L. G., Underwood B. R., Pangalos M. N., Schmitt I., Wullner U., Evert B. O., O'Kane C. J., and Rubinsztein D. C. (2006) Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 15, 433–442 10.1093/hmg/ddi458 [DOI] [PubMed] [Google Scholar]
- 48. Shibata M., Lu T., Furuya T., Degterev A., Mizushima N., Yoshimori T., MacDonald M., Yankner B., and Yuan J. (2006) Regulation of intracellular accumulation of mutant huntingtin by beclin 1. J. Biol. Chem. 281, 14474–14485 10.1074/jbc.M600364200 [DOI] [PubMed] [Google Scholar]
- 49. Cortes C. J., and La Spada A. R. (2014) The many faces of autophagy dysfunction in Huntington's disease: from mechanism to therapy. Drug Discov. Today 19, 963–971 10.1016/j.drudis.2014.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fader C. M., and Colombo M. I. (2006) Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy 2, 122–125 10.4161/auto.2.2.2350 [DOI] [PubMed] [Google Scholar]
- 51. Fader C. M., Sánchez D., Furlán M., and Colombo M. I. (2008) Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250 [DOI] [PubMed] [Google Scholar]
- 52. Ertmer A., Gilch S., Yun S.-W., Flechsig E., Klebl B., Stein-Gerlach M., Klein M. A., and Schätzl H. M. (2004) The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J. Biol. Chem. 279, 41918–41927 10.1074/jbc.M405652200 [DOI] [PubMed] [Google Scholar]
- 53. Aguib Y., Heiseke A., Gilch S., Riemer C., Baier M., Schätzl H. M., and Ertmer A. (2009) Autophagy induction by trehalose counteracts cellular prion infection. Autophagy 5, 361–369 10.4161/auto.5.3.7662 [DOI] [PubMed] [Google Scholar]
- 54. Heiseke A., Aguib Y., Riemer C., Baier M., and Schätzl H. M. (2009) Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J. Neurochem. 109, 25–34 10.1111/j.1471-4159.2009.05906.x [DOI] [PubMed] [Google Scholar]
- 55. Cortes C. J., Qin K., Cook J., Solanki A., and Mastrianni J. A. (2012) Rapamycin delays disease onset and prevents PrP plaque deposition in a mouse model of Gerstmann—Straussler–Scheinker disease. J. Neurosci. 32, 12396–12405 10.1523/JNEUROSCI.6189-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yun S.-W., Ertmer A., Flechsig E., Gilch S., Riederer P., Gerlach M., Schätzl H. M., and Klein M. A. (2007) The tyrosine kinase inhibitor imatinib mesylate delays prion neuroinvasion by inhibiting prion propagation in the periphery. J. Neurovirol. 13, 328–337 10.1080/13550280701361516 [DOI] [PubMed] [Google Scholar]
- 57. Nakagaki T., Satoh K., Ishibashi D., Fuse T., Sano K., Kamatari Y. O., Kuwata K., Shigematsu K., Iwamaru Y., Takenouchi T., Kitani H., Nishida N., and Atarashi R. (2013) FK506 reduces abnormal prion protein through the activation of autolysosomal degradation and prolongs survival in prion-infected mice. Autophagy 9, 1386–1394 10.4161/auto.25381 [DOI] [PubMed] [Google Scholar]
- 58. Ishibashi D., Homma T., Nakagaki T., Fuse T., Sano K., Takatsuki H., Atarashi R., and Nishida N. (2015) Strain-dependent effect of macroautophagy on abnormally folded prion protein degradation in infected neuronal cells. PLoS One 10, e0137958 10.1371/journal.pone.0137958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gilch S., Winklhofer K. F., Groschup M. H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., and Schätzl H. M. (2001) Intracellular re-routing of prion protein prevents propagation of PrPSc and delays onset of prion disease. EMBO J. 20, 3957–3966 10.1093/emboj/20.15.3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahal S. P., Baker C. A., Demczyk C. A., Smith E. W., Julius C., and Weissmann C. (2007) Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. U.S.A. 104, 20908–20913 10.1073/pnas.0710054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coleman B. M., Hanssen E., Lawson V. A., and Hill A. F. (2012) Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 26, 4160–4173 10.1096/fj.11-202077 [DOI] [PubMed] [Google Scholar]
- 62. Guo B. B., Bellingham S. A., and Hill A. F. (2015) The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J. Biol. Chem. 290, 3455–3467 10.1074/jbc.M114.605253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Orrù C. D., Wilham J. M., Vascellari S., Hughson A. G., and Caughey B. (2012) New generation QuIC assays for prion seeding activity. Prion 6, 147–152 10.4161/pri.19430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zanusso G., Monaco S., Pocchiari M., and Caughey B. (2016) Advanced tests for early and accurate diagnosis of Creutzfeldt–Jakob disease. Nat. Rev. Neurol. 12, 427–427 10.1038/nrneurol.2016.85,10.1038/nrneurol.2016.92 [DOI] [PubMed] [Google Scholar]
- 65. Shrivastava S., Devhare P., Sujijantarat N., Steele R., Kwon Y.-C., Ray R., and Ray R. B. (2016) Knockdown of autophagy inhibits infectious hepatitis C virus release by the exosomal pathway. J. Virol. 90, 1387–1396 10.1128/JVI.02383-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tang Z., Ioja E., Bereczki E., Hultenby K., Li C., Guan Z., Winblad B., and Pei J.-J. (2015) mTor mediates Tau localization and secretion: implication for Alzheimer's disease. Biochim. Biophys. Acta 1853, 1646–1657 10.1016/j.bbamcr.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 67. Yang Y.-P., Hu L.-F., Zheng H.-F., Mao C.-J., Hu W.-D., Xiong K.-P., Wang F., and Liu C.-F (2013) Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol. Sin. 34, 625–635 10.1038/aps.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lai R. C., Arslan F., Lee M. M., Sze N. S., Choo A., Chen T. S., Salto-Tellez M., Timmers L., Lee C. N., El Oakley R. M., Pasterkamp G., de Kleijn D. P., and Lim S. K. (2010) Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 4, 214–222 10.1016/j.scr.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 69. Korkut C., Ataman B., Ramachandran P., Ashley J., Barria R., Gherbesi N., and Budnik V. (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139, 393–404 10.1016/j.cell.2009.07.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Robbins P. D., and Morelli A. E. (2014) Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Villarroya-Beltri C., Baixauli F., Gutiérrez-Vázquez C., Sánchez-Madrid F., and Mittelbrunn M. (2014) Sorting it out: regulation of exosome loading. Semin. Cancer Biol. 28, 3–13 10.1016/j.semcancer.2014.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Emmanouilidou E., Melachroinou K., Roumeliotis T., Garbis S. D., Ntzouni M., Margaritis L. H., Stefanis L., and Vekrellis K. (2010) Cell-produced α-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 30, 6838–6851 10.1523/JNEUROSCI.5699-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janas A. M., Sapoń K., Janas T., Stowell M. H., and Janas T. (2016) Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases. Biochim. Biophys. Acta 1858, 1139–1151 10.1016/j.bbamem.2016.02.011 [DOI] [PubMed] [Google Scholar]
- 74. Quek C., and Hill A. F. (2017) The role of extracellular vesicles in neurodegenerative diseases. Biochem. Biophys. Res. Commun. 483, 1178–1186 10.1016/j.bbrc.2016.09.090 [DOI] [PubMed] [Google Scholar]
- 75. Saman S., Kim W., Raya M., Visnick Y., Miro S., Saman S., Jackson B., McKee A. C., Alvarez V. E., Lee N. C., and Hall G. F. (2012) Exosome-associated Tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 287, 3842–3849 10.1074/jbc.M111.277061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Duran J. M., Anjard C., Stefan C., Loomis W. F., and Malhotra V. (2010) Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188, 527–536 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., and Deretic V. (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30, 4701–4711 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Qu Y., Franchi L., Nunez G., and Dubyak G. R. (2007) Nonclassical IL-1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 179, 1913–1925 10.4049/jimmunol.179.3.1913 [DOI] [PubMed] [Google Scholar]
- 79. Poehler A.-M., Xiang W., Spitzer P., May V. E., Meixner H., Rockenstein E., Chutna O., Outeiro T. F., Winkler J., Masliah E., and Klucken J. (2014) Autophagy modulates SNCA/α-synuclein release, thereby generating a hostile microenvironment. Autophagy 10, 2171–2192 10.4161/auto.36436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lipinski M. M., Zheng B., Lu T., Yan Z., Py B. F., Ng A., Xavier R. J., Li C., Yankner B. A., Scherzer C. R., and Yuan J. (2010) Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 107, 14164–14169 10.1073/pnas.1009485107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pimpinelli F., Lehmann S., and Maridonneau-Parini I. (2005) The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur. J. Neurosci. 21, 2063–2072 10.1111/j.1460-9568.2005.04049.x [DOI] [PubMed] [Google Scholar]
- 82. Qi Y., Wang J. K., McMillian M., and Chikaraishi D. M. (1997) Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17, 1217–1225 10.1523/JNEUROSCI.17-04-01217.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.