Abstract
The bacterial type VI secretion system (T6SS) delivers effectors into eukaryotic host cells or toxins into bacterial competitor for survival and fitness. The T6SS is positively regulated by the threonine phosphorylation pathway (TPP) and negatively by the T6SS-accessory protein TagF. Here, we studied the mechanisms underlying TagF-mediated T6SS repression in two distinct bacterial pathogens, Agrobacterium tumefaciens and Pseudomonas aeruginosa. We found that in A. tumefaciens, T6SS toxin secretion and T6SS-dependent antibacterial activity are suppressed by a two-domain chimeric protein consisting of TagF and PppA, a putative phosphatase. Remarkably, this TagF domain is sufficient to post-translationally repress the T6SS, and this inhibition is independent of TPP. This repression requires interaction with a cytoplasmic protein, Fha, critical for activating T6SS assembly. In P. aeruginosa, PppA and TagF are two distinct proteins that repress T6SS in TPP-dependent and -independent pathways, respectively. P. aeruginosa TagF interacts with Fha1, suggesting that formation of this complex represents a conserved TagF-mediated regulatory mechanism. Using TagF variants with substitutions of conserved amino acid residues at predicted protein–protein interaction interfaces, we uncovered evidence that the TagF–Fha interaction is critical for TagF-mediated T6SS repression in both bacteria. TagF inhibits T6SS without affecting T6SS protein abundance in A. tumefaciens, but TagF overexpression reduces the protein levels of all analyzed T6SS components in P. aeruginosa. Our results indicate that TagF interacts with Fha, which in turn could impact different stages of T6SS assembly in different bacteria, possibly reflecting an evolutionary divergence in T6SS control.
Keywords: bacterial genetics, gene regulation, protein secretion, protein phosphorylation, protein-protein interaction, Agrobacterium tumefaciens, antibacterial activity, post-translational regulation, Pseudomonas aeruginosa, type VI secretion system, TagF, PppA, Fha
Introduction
The type VI secretion system (T6SS)4 is a versatile weapon deployed by many bacterial species to deliver diverse effector proteins into eukaryotic host cells or bacterial competitors. The major targets of T6SS antibacterial effectors include the membrane, cell wall, or nucleic acid; some of these are shared by eukaryote-targeting effectors (e.g. membranes), whereas the latter may have additional targets (e.g. actin cytoskeleton). The delivery and activity of these T6SS toxins and effectors have a clear impact in interbacterial competition and/or pathogenesis during eukaryotic host infection (1, 2). The T6SS apparatus relies on ∼13–14 conserved core components to build a contractile phage tail-like structure anchored to the bacterial cell envelope. To initiate the T6SS assembly, a TssJLM (or TssLM) transmembrane complex (3–6) serves as a docking site for the TssAEFGK baseplate complex (7, 8), with TssK bridging the baseplate and the membrane complex. On the baseplate, Hcp is polymerized in a tail tube-like structure and wrapped around by a TssB-TssC outer sheath. In some studies, it was proposed that TssA is responsible for initiating sheath polymerization (9). Upon contraction of the sheath, the Hcp tube, tipped by the VgrG-PAAR puncturing device, and the T6SS effectors associated with it (10, 11) are propelled across the cell envelope.
T6SS is regulated at multiple levels (12–15). A subset of T6SS gene clusters encode orthologs of serine/threonine kinase PpkA, the cognate phosphatase PppA, and the forkhead-associated (FHA) domain–containing proteins (16), which suggests the involvement of a threonine phosphorylation (TPP) regulatory pathway in these bacteria (17–20). Pseudomonas aeruginosa H1-T6SS is post-translationally regulated, positively by PpkA and negatively by the cognate phosphatase PppA. Such control occurs via threonine phosphorylation at the Thr-362 residue on an FHA domain–containing protein, Fha1, in P. aeruginosa (21) and Serratia marcescens (20). Remarkably, the T6SS inner-membrane protein TssL and not Fha was identified as the substrate of PpkA first in Agrobacterium tumefaciens (22) and recently in Vibiro alginolyticus (23). In A. tumefaciens, TssL forms a stable complex with TssM (4, 6, 24), which exhibits ATPase activity (5). Phosphorylated TssL recruits Fha to the TssM-TssL complex for T6SS activation (22). In P. aeruginosa, four type VI secretion–associated genes, namely tagQRST, participate in post-translational regulation and act upstream of PpkA to promote kinase activity and subsequent T6SS-dependent secretion (25, 26). Remarkably, the P. aeruginosa H1-T6SS can also be activated independently of Fha1 phosphorylation and TPP activity by inactivating a negative regulator, TagF (19), yet the molecular basis underlying the regulatory mechanisms of TagF-mediated T6SS repression has not been addressed.
A. tumefaciens harbors one T6SS that is activated at both transcriptional and post-translational levels when sensing acidity (22, 27, 28). Three T6SS effectors, including one peptidoglycan amidase (Tae) and two DNases (Tde1 and Tde2), conferring antibacterial activity, have been identified in this bacterium (29). Autointoxication is prevented through the production of cognate immunity proteins. In A. tumefaciens, TagF and PppA are encoded as a fusion protein named TagF-PppA, the overexpression of which abolishes Hcp secretion (22). In this study, we investigated the TagF-PppA suppression mechanism and how this affects T6SS-dependent secretion and antibacterial activity in A. tumefaciens. Our data indicate that the TagF domain alone is sufficient to post-translationally repress the T6SS independently of TPP. By performing protein–protein interaction studies, we identified that the cytoplasmic T6SS core component Fha is the binding target for TagF. Using structural modeling, we identified conserved TagF amino acid residues and demonstrated their importance in Fha interaction and TagF-mediated repression of T6SS activity. Remarkably, these conserved residues are also required for P. aeruginosa TagF in repressing the H1-T6SS activity and interaction with Fha1. TagF may have evolved while keeping the Fha protein as a target for controlling and preventing T6SS assembly.
Results
Both TagF and PppA domains can repress type VI secretion and antibacterial activity at post-translational levels in A. tumefaciens
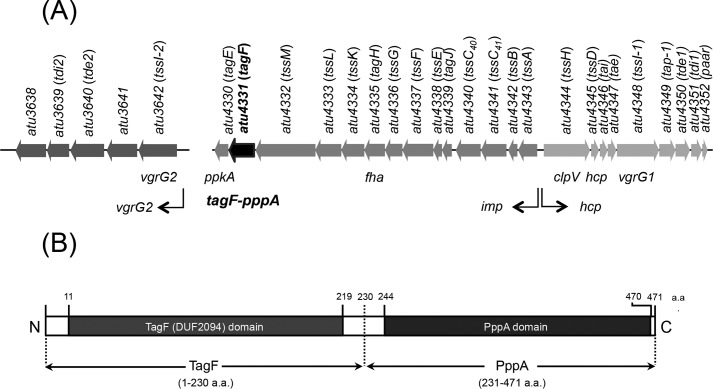
TPP regulation of T6SS has been demonstrated in only a few bacteria, including A. tumefaciens strain C58 (22). PpkA (Atu4330), TagF-PppA (Atu4331), and Fha (Atu4335), the three major components of the TPP pathway, are all encoded within the imp operon (Fig. 1A) (22, 27, 30). Atu4331 is a fusion protein, which contains N- and C-terminal domains homologous to TagF (DUF2094) (aa 11–219) and PppA (aa 244–470), respectively (thus named TagF-PppA) (Fig. 1B) (22). We previously reported that TagF-PppA plays a negative role in regulating Hcp secretion when TagF-PppA is overexpressed in A. tumefaciens (22). However, the molecular mechanism underlying this regulation is unknown. Thus, we first investigated the impact and respective role of individual domain from the TagF-PppA chimera on the TagF-PppA–mediated repression activity on T6SS activity. In A. tumefaciens strain C58, we separately produced the TagF domain located at the N terminus (aa 1–230) and the C-terminal region (aa 231–471) encompassing the PppA domain (Fig. 1B). Overexpression of TagF-PppA, TagF, and tagged TagF-Strep in C58 completely abolished type VI secretion (Hcp, Tae, and Tde1) (Fig. 2A). The T6SS antibacterial activity is also shut down, as shown by counting surviving Escherichia coli target cells. The number of survivors is indeed similar to that when the attacker is an A. tumefaciens T6SS mutant, ΔtssL (Fig. 2B). We also performed an in planta interbacterial competition assay with an A. tumefaciens prey strain lacking the three T6SS toxins (Tae, Tde1, and Tde2) and cognate immunity proteins, Δ3Tis (29). Previous observation showed a reduced number of viable prey cells when co-infected with WT C58 (31). Similarly, here, the survival of Δ3TIs was lower after co-infection with WT C58 harboring the vector pTrc200 (V). This was not seen when co-infecting with a T6SS inactive mutant ΔtssL-carrying pTrc200 (V) or C58 overexpressing the T6SS repressor TagF (i.e. TagF-PppA, TagF, or TagF-Strep) (Fig. 2C). Interestingly, overexpression of PppA alone reduced but did not abolish Hcp secretion (Fig. 2A). The antibacterial activity of the PppA overexpression strain was also not completely abolished, whereas modest antibacterial activity was detected with our in planta interbacterial competition assay (Fig. 2C). In addition, the protein levels of all analyzed T6SS components encoded within the imp (including TssM, TssL, TssK, Fha, TssC41, TssB, and TssA) and hcp (including ClpV and VgrG1) operons remained the same in all backgrounds tested, including overexpression of TagF-PppA, TagF, TagF-Strep, or PppA (Fig. 2A). Taken together, these data show that in A. tumefaciens, both TagF and PppA domains contribute the repressor function of TagF-PppA on T6SS effector secretion and antibacterial activity via a post-translational regulatory control.
Figure 1.
A. tumefaciens C58 t6ss gene clusters and TagF-pppA domain organization. A, the imp operon (atu4343 to atu4330), hcp operon (atu4344 to atu4352), and vgrG2 in A. tumefaciens strain C58 were designated tss or tag based on nomenclature proposed by Shalom et al. (34) and specific names derived from Lin et al. (30) and Bondage et al. (31). B, TagF-PppA domain organization according to information from the NCBI conserved domain database. TagF-PppA is predicted as a cytoplasmic protein (aa 1–471) with an N-terminal conserved TagF (DUF2094) domain (aa 11–219) and a C-terminal PppA (PP2Cc) domain (aa 244–470).
Figure 2.
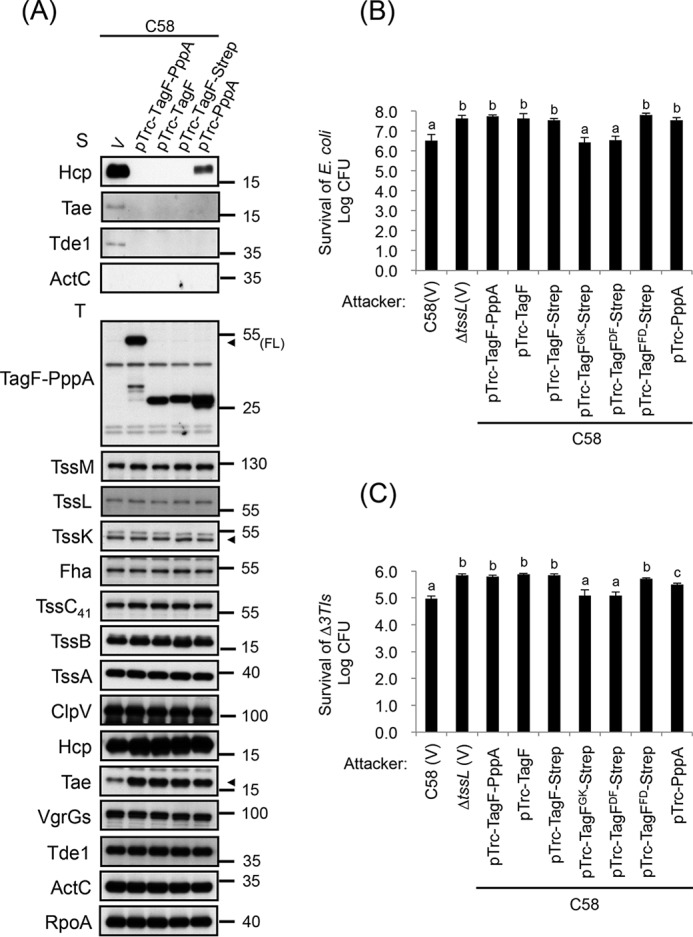
Both TagF and PppA domains can repress type VI secretion and antibacterial activity at post-translational levels in A. tumefaciens. A, type VI secretion analysis. Shown is Western blot analysis of total (T) and secreted (S) proteins isolated from WT C58 harboring the vector pTrc200 (V) or various overexpressing plasmids grown in AB-MES (pH 5.5) liquid culture with specific antibodies. The nonsecreted protein ActC and RNA polymerase α subunit RpoA were internal controls. The proteins analyzed and sizes of molecular weight standards are shown on the left and right, respectively, and indicated with arrowheads when necessary. FL, full-length TagF-PppA protein. B, A. tumefaciens antibacterial activity assay against E. coli. The A. tumefaciens WT C58 harboring the vector pTrc200 (V) or various overexpressed plasmids or ΔtssL mutant harboring the vector pTrc200 (V) was co-cultured on LB agar with E. coli strain DH10B cells harboring the plasmid pRL662. C, A. tumefaciens intraspecies competition in planta. The A. tumefaciens WT C58 harboring the vector pTrc200 (V) or various overexpressed plasmids or ΔtssL mutant harboring the vector pTrc200 (V) was used as attacker strain to mix with the target strain Δ3TIs harboring pRL662 and infiltrated into N. benthamiana leaves. B and C, data are mean ± S.D. of at least three biological replicates. Different letters above the bar indicate statistically significantly different groups of strains (p < 0.01 for B; p < 0.05 for C) based on cfu of the surviving target cells.
Both TagF and PppA domains repress T6SS activity independently of PpkA-mediated TssL phosphorylation
To explore the possible mechanisms of TagF-PppA-mediated T6SS repression at the post-translational level, we analyzed the impact on TssL phosphorylation. TagF-PppA, TagF, TagF-Strep, and PppA were overexpressed in the ΔtssL mutant also expressing His-tagged TssL (TssL-His). This TssL variant is functional and mediates Hcp, Tae, and Tde1 secretion (Fig. S1, A and B) (22). The TssL-His protein was purified by using Ni-NTA resins, and various A. tumefaciens strains were analyzed. The Phos-tag SDS-PAGE approach was used, which can detect two TssL-His protein bands, the lower band representing the unphosphorylated TssL-His and the upper band representing the phosphorylated TssL-His (p-TssL-His in Fig. 3A) (22). In all cases, the TssL-His protein displayed a similar phosphorylation pattern (Fig. 3A), which suggests that neither the full-length TagF-PppA nor any domain of TagF-PppA represses T6SS activity by controlling TssL phosphorylation.
Figure 3.
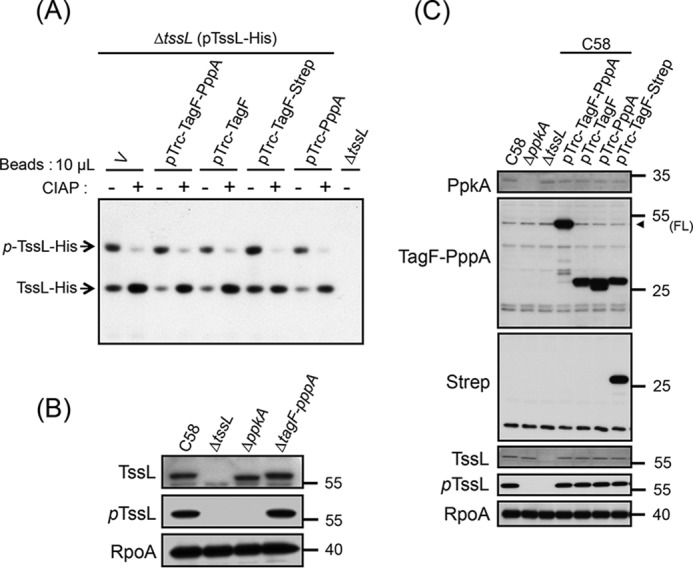
Both TagF and PppA domains repress T6SS activity independently of the PpkA-mediated TssL phosphorylation pathway in A. tumefaciens. A, Phos-tag SDS-PAGE analysis to detect the phosphorylation status of TssL-His. Shown is Western blot analysis of the same volumes of Ni-NTA resins (10 μl) associated with TssL-His from different strains treated with (+) or without (−) CIAP and examined by a specific antibody against His6. Total protein isolated from ΔtssL was a negative control. Phos-tag SDS-PAGE revealed the upper band indicating the phosphorylated TssL-His (p-TssL-His) and lower band indicating unphosphorylated TssL-His. B and C, Western blot analysis of the endogenous phosphorylation status of TssL (pTssL). Shown is Western blot analysis of total proteins isolated from WT C58, ΔppkA, ΔtssL, or C58 harboring the vector pTrc200 (V) or various overexpressing plasmids grown in AB-MES (pH 5.5) liquid culture with specific antibodies. The specific antibody for pTssL was generated against the 15-mer peptide (7SSWQDLPpTVVEITEE21), with phosphorylated Thr-14 of TssL underlined. RNA polymerase α subunit RpoA was an internal control. The proteins analyzed and molecular weight standards are on the left and right, respectively, and are indicated with an arrowhead when necessary. FL, full-length TagF-PppA proteins.
To determine whether the phosphorylation level detected from an overexpressed TssL-His is representative of an endogenous situation, we generated an antibody that specifically recognizes phosphorylated TssL (pTssL) (Fig. 3B). Phosphorylated TssL protein was detected as a single protein band and found with the same abundance when comparing WT C58 and strains overexpressing TagF-PppA, TagF, TagF-Strep, and PppA. However, the pTssL protein band was not detected in ΔtssL and ΔppkA mutant strains (Fig. 3C). We concluded that TagF-PppA represses T6SS activity independently of TssL phosphorylation in A. tumefaciens.
It is intriguing that overexpression of the PppA domain had no impact on TssL phosphorylation, which is mediated by PpkA, but still could repress T6SS activity. To confirm that the TPP-independent repression of both TagF and PppA domains is not caused by a secondary effect linked to overexpression, we also examined the T6SS secretion and antibacterial activity in a strain lacking the entire TPP (i.e. both ppkA and tagF-pppA). We generated a ΔppkAΔtagF-pppA strain and separately overexpressed the TagF or PppA domain in this mutant background. Interestingly, unlike ΔppkA, resulting in decreased type VI secretion (22), the ΔppkAΔtagF-pppA mutant retained type VI secretion activity comparable with that of the WT C58 (Fig. 4A). Overexpression of each of TagF or PppA domain in ΔppkAΔtagF-pppA abolished or greatly reduced the type VI secretion (Fig. 4B), suggesting that both TagF and PppA domains can inhibit type VI secretion in the absence of PpkA. As expected, the antibacterial activity was also abolished when the TagF or PppA domain was overexpressed in ΔppkAΔtagF-pppA, as shown by counting E. coli survivors and comparison with what is observed when the attacker is a T6SS mutant, ΔtssL (Fig. 4C). Interestingly, whereas the Hcp and effector (Tde1 and Tae) secretion levels in ΔppkAΔtagF-pppA were comparable with those of WT C58 (Fig. 4A), only partial antibacterial activity was observed (Fig. 4C). This phenotype is consistent with a previous observation in P. aeruginosa and S. marcescens, with ΔpppA showing reduced antibacterial activity despite elevated type VI secretion (20, 32, 33). These data indicate that both the TagF and PppA domains play a role in repressing the A. tumefaciens T6SS activity. This control is exerted at a post-translational level and is independent of PpkA and TssL phosphorylation.
Figure 4.
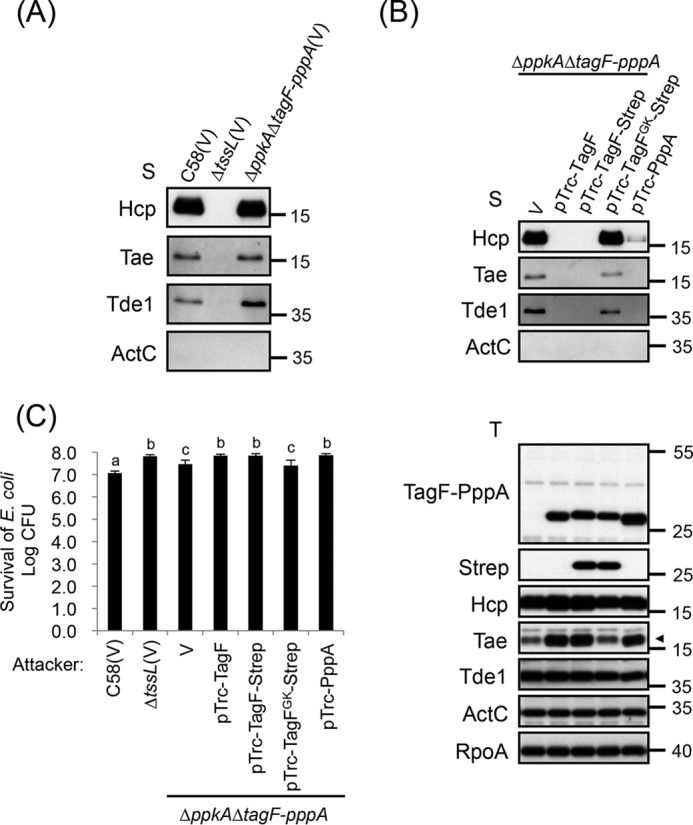
TagF represses T6SS activity independent of the TPP pathway in A. tumefaciens. A and B, type VI secretion analysis. Shown is Western blot analysis of total (T) and secreted (S) proteins isolated from WT C58 harboring the vector pTrc200 (V) or ΔtssL harboring the vector pTrc200 (V) or ΔppkAΔtagF-pppA harboring various overexpressing plasmids grown in AB-MES (pH 5.5) liquid culture with specific antibodies. The nonsecreted protein ActC and RNA polymerase α subunit RpoA were internal controls. The proteins analyzed and molecular weight standards are shown on the left and right, respectively, and indicated with an arrowhead when necessary. C, A. tumefaciens antibacterial activity assay against E. coli. The A. tumefaciens WT C58 harboring the vector pTrc200 (V), ΔtssL harboring the vector pTrc200 (V), or ΔppkAΔtagF-pppA harboring various overexpressing plasmids was co-cultured on LB agar with E. coli strain DH10B cells harboring pRL662. Data are mean ± S.D. (error bars) of at least three biological replicates. Different letters above the bar indicate statistically significantly different groups of strains (p < 0.01) based on cfu of the surviving target cells.
TagF interacts with the forkhead-associated protein Fha of A. tumefaciens and P. aeruginosa
Next, we investigated how TagF-PppA represses T6SS activity via a post-translational and TPP-independent pathway. We hypothesized that TagF-PppA may interact with the T6SS core component(s) to prevent T6SS activation. One plausible candidate is the forkhead-associated protein Fha because of its role in recruitment to a membrane-associated complex both independent of and dependent on TPP (19, 21, 22). Because TagF is the common repressor known to suppress T6SS independently of TPP in both A. tumefaciens and P. aeruginosa, we set up experiments to determine whether A. tumefaciens TagF-PppA and P. aeruginosa TagF can interact with their cognate Fha. Yeast two-hybrid (YTH) experiments revealed that both A. tumefaciens full-length TagF-PppA and the isolated TagF domain specifically interact with Fha (Fig. 5A). We also detected an interaction between the P. aeruginosa H1-T6SS–encoded TagF and Fha1 by using both YTH (Fig. 5B) and bacterial two-hybrid (BTH) assays (Fig. 5C). The data demonstrate that TagF directly interacts with Fha, probably interfering with its function and thereby preventing type VI secretion in both A. tumefaciens and P. aeruginosa.
Figure 5.
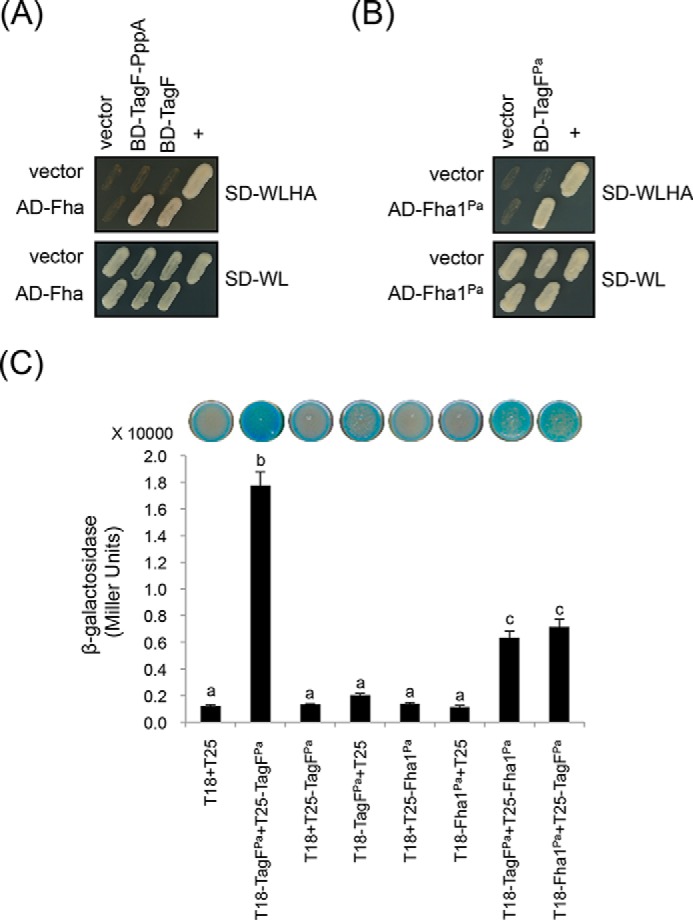
TagF directly interacts with Fha of A. tumefaciens and P. aeruginosa. A and B, yeast two-hybrid protein–protein interaction results. SD−WL medium (SD minimal medium lacking Trp and Leu) was used for the selection of plasmids. SD−WLHA medium (SD minimal medium lacking Trp, Leu, His, and Ade) was used for the auxotrophic selection of bait and prey protein interactions. The positive interaction was determined by growth on SD−WLHA medium at 30 °C for at least 2 days. The positive control (+) showing interactions of SV40 large T-antigen and murine p53 and negative control (vector) are indicated. C, bacterial two-hybrid analysis. Various combinations of recombinant pKT25 and pUT18C plasmids harboring P. aeruginosa TagFPa or Fha1Pa proteins were co-transformed into E. coli. A graphical representation of the β-gal activity from co-transformants is shown, the plasmid combinations are indicated below, and images of corresponding E. coli spots on LB agar plates containing X-gal are displayed at the top. The strength of the interaction was investigated by measuring the β-gal activity of cells. The average activity in Miller units is indicated. Experiments were carried out in duplicate, and data are mean ± S.D. (error bars) Different letters above the bar indicate statistically significantly different groups (p < 0.01). T18, empty vector pUT18C; T25, empty vector pKT25.
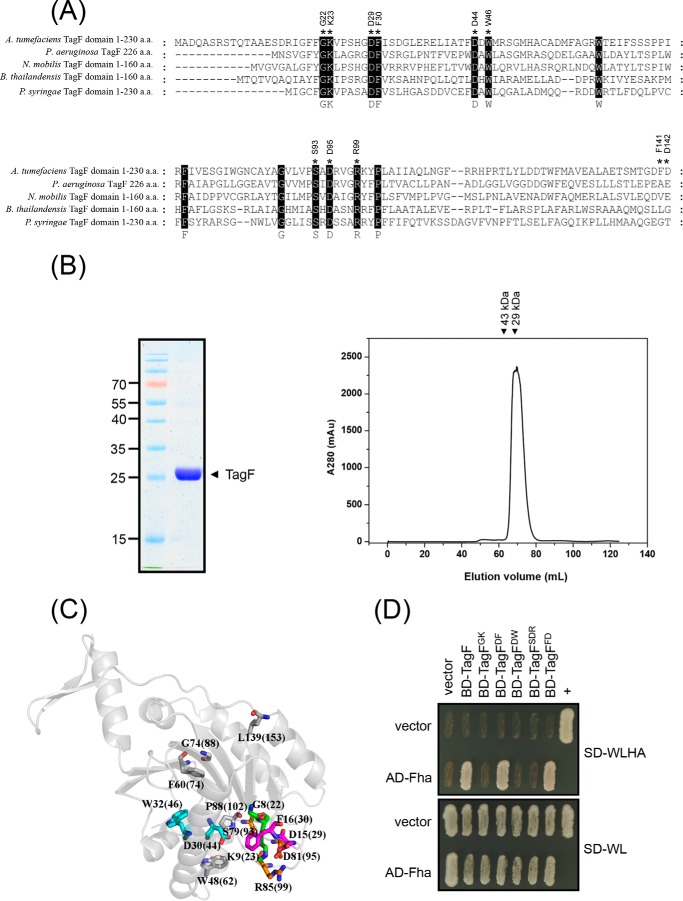
Conserved amino acid residues in TagF are critical for TagF–Fha interaction
Although Fha is the target for TagF in both A. tumefaciens and P. aeruginosa, the two TagF proteins share only limited amino acid similarity (Fig. 6A). Yet, 14 amino acid residues are highly conserved among various TagF orthologs (Fig. 6A and Fig. S3A). We hypothesized that these residues may play important roles in TagF function. The structure of P. aeruginosa TagF is presented as a homodimer (Protein Data Bank entry 2QNU). The self-interaction of P. aeruginosa TagF was also confirmed by BTH and YTH (Fig. 5C and Fig. S2A). In contrast, the A. tumefaciens TagF probably does function as a monomer, as supported by several lines of evidence. First, the TagF protein is mostly a monomer when analyzed by gel filtration in vitro (Fig. 6B). Second, A. tumefaciens TagF lacks the residues required for dimer formation in P. aeruginosa (Val-105 (V105), Leu-169 (L169), Leu-172 (L172), Ala-173 (A173), and Leu-195 (L195) in TagFPa) (Fig. S3A) (Protein Data Bank entry 2QNU). Finally, no self-interaction could be found for A. tumefaciens TagF using YTH (Fig. S2B). Other conserved residues are not involved in P. aeruginosa TagF homodimerization and were thus chosen for site-directed mutagenesis and characterization of the potential interface with other interacting proteins, namely Fha. A total of eight conserved residues (Gly-8 (G8), Lys-9 (K9), Asp-15 (D15), Phe-16 (F16), Ser-79 (S79), Asp-81 (D81), Arg-85 (R85), and Pro (P88) of P. aeruginosa TagF) form a specific loop, and the side chain of Asp-30 (D30) is outwardly exposed (Fig. 6C). Therefore, these are potential sites to interact with other proteins. In contrast, Phe-60 (F60), Gly-74 (G74), and Leu-139 (L139) are located inside the structure or the side chain wrapped in an internal structure to prevent interaction with other proteins. We generated four mutants with amino acid substitutions of alanine at GK (G8K9 in P. aeruginosa TagF; G22K23 in A. tumefaciens TagF), DF (D15F16 in P. aeruginosa TagF; D29F30 in A. tumefaciens TagF), DW (D30W32 in P. aeruginosa TagF; D44W46 in A. tumefaciens TagF), and SDR (S79D81R85 in P. aeruginosa TagF; S93D95R99 in A. tumefaciens TagF) (Fig. 6, A and C). We performed YTH analysis with the various A. tumefaciens TagF variants to determine the roles of the substituted residues in the TagF–Fha interaction. The interaction was completely lost in TagFGK, TagFDW, and TagFSDR mutants (Fig. 6D). In contrast, TagFDF and TagFFD with mutations in two randomly selected nonconserved residues (Phe-141 and Asp-142 (F141D142) in A. tumefaciens TagF) still retained full capacity to interact with Fha as compared with the WT TagF (Fig. 6D). Western blot analysis revealed that the protein abundance remained the same or was even slightly higher for all analyzed TagF variants as compared with the WT TagF (Fig. S3B), which indicates that conserved G22K23, D44W46, and S93D95R99 residues but not D29F30 of A. tumefaciens TagF are critical for the TagF–Fha interaction.
Figure 6.
Conserved amino acid residues of TagF are critical for TagF–Fha interaction in A. tumefaciens. A, amino acid sequence alignment of TagF or TagF domain orthologs from selected bacterial species. Conserved amino acid residues are highlighted in black, and those used for mutagenesis are indicated with an asterisk. Sequences were aligned and highlighted by use of ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) Part of the aligned result is shown here, and the fully aligned result and full information for bacterial strains and protein accession numbers are shown in Fig. S3A. B, Agrobacterium TagF protein is present as a monomer on gel filtration analysis in vitro. Purified His-tagged TagF domain (aa 1–214) was analyzed by SDS-PAGE. The proteins analyzed and molecular weight standards are shown on the right and left, respectively. His-tagged TagF proteins were further analyzed by use of a Superdex 75 16 × 60 column, and the elution profiles were recorded as absorbance at 280 nm showing that His-tagged TagF elutes as a single peak (∼26 kDa monomer). C, relative positions of the conserved amino acid residues in P. aeruginosa TagFPa protein revealed as a monomer with crystal structural information according to the X-ray crystal structure of P. aeruginosa TagF monomer (Protein Data Bank entry 2QNU). The corresponding conserved amino acid residues of A. tumefaciens TagF are indicated in parenthesis. D, yeast two-hybrid protein–protein interaction results with Fha and various TagF proteins. SD−WL medium (SD minimal medium lacking Trp and Leu) was used for selecting plasmids. SD−WLHA medium (SD minimal medium lacking Trp, Leu, His, and Ade) was used for the auxotrophic selection of bait and prey protein interactions. The positive interaction was determined by growth on SD−WLHA medium at 30 °C for at least 2 days. The positive control (+) showing interactions of SV40 large T-antigen and murine p53 and negative control (vector) are indicated.
Loss of TagF–Fha interaction upon site-directed mutagenesis abolishes the repression of T6SS activity in A. tumefaciens
To determine whether the TagF–Fha interaction is required for suppressing T6SS activity, we engineered the previously described mutations in the TagF-Strep variants and analyzed the impact of their overexpression on type VI secretion. As expected, overexpression of the WT controls, TagF-Strep or TagFFD-Strep, abolished Hcp, Tae, and Tde1 secretion. Remarkably, the secretion capacity remained high with overexpression of TagFGK-Strep, TagFDF-Strep, TagFDW-Strep, and TagFSDR-Strep in C58 (Fig. 7A). Upon Western blot analysis, the protein levels of all TagF variants were comparable with that of WT TagF-Strep, and overexpression did not affect the protein abundance of other T6SS components (Fig. 7A). These data suggest that the conserved residues GK, DW, and SDR of TagF are critical for TagF-mediated repression of type VI secretion via the TagF–Fha interaction. Intriguingly, TagFDF retained full binding capacity with Fha but lost the ability to repress type VI secretion, which suggests that D29F30 is not involved in binding Fha but is required for repressing T6SS activity. As expected, the type VI secretion activity of these TagF overexpression variants was consistent with their antibacterial activity (Fig. 2, B and C). The survival of E. coli was reduced to a level similar to that with the strain harboring the empty vector control when TagFGK-Strep was overexpressed in ΔppkAΔtagF-pppA (Fig. 4, B and C), which confirms that TagFGK-Strep lost the ability to repress T6SS-dependent antibacterial activity via a PpkA-independent pathway. To ensure that the overexpression phenotypes of the mutant alleles were also reflected when expressed at endogenous levels, we further generated chromosomal tagF-pppA alleles encoding the TagF amino acid substitution variants and further determined their T6SS suppression activity. All TagF amino acid substitution variants expressed from chromosomal alleles exhibited enhanced antibacterial activity (Fig. 7B), which is consistent with the derepressing T6SS activity demonstrated upon overexpression of these TagF variants (Fig. 2, B and C). However, our secretion assay could not detect a significant difference in secretion levels of Hcp and two effectors (Tae and Tde1) of all analyzed tagF-pppA mutants as compared with WT C58 (Fig. S4). Taken together, our data suggest that TagF negatively regulates T6SS via direct interaction with Fha in A. tumefaciens.
Figure 7.
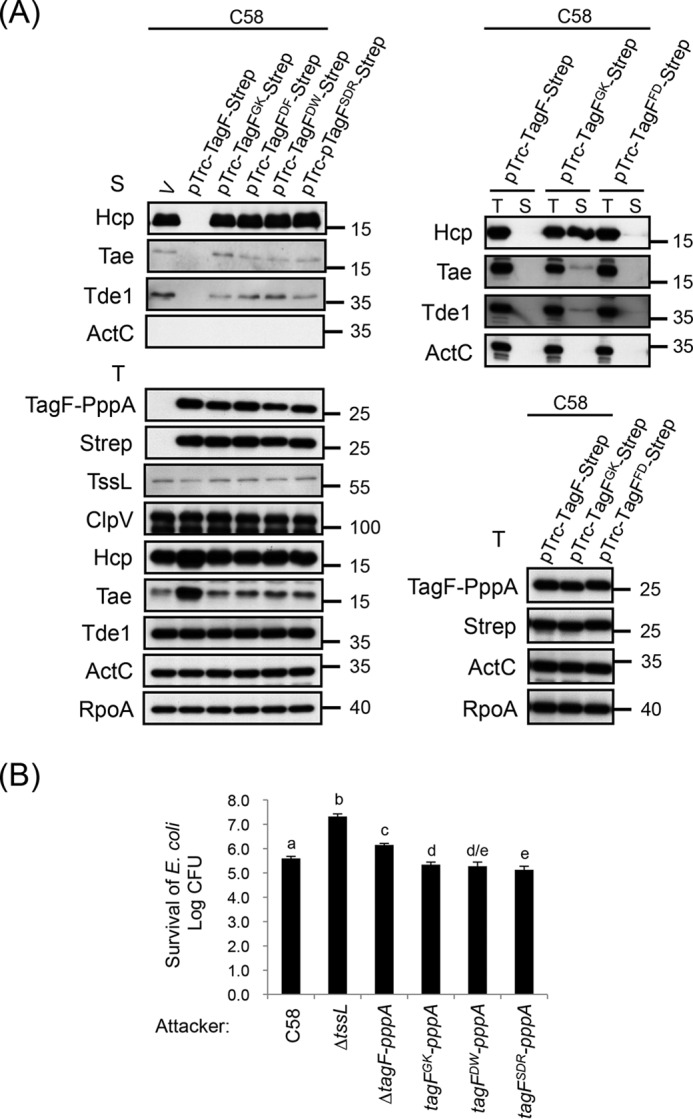
Conserved amino acid residues of TagF are required for repressing type VI activity in A. tumefaciens. A, Western blot analysis of total (T) and secreted (S) proteins isolated from WT C58 harboring the vector pTrc200 (V) or various TagF-Strep–overexpressing plasmids grown in AB-MES (pH 5.5) liquid culture with specific antibodies. The nonsecreted protein ActC and RNA polymerase α subunit RpoA were internal controls. The proteins analyzed and molecular weight standards are shown on the left and right, respectively. B, A. tumefaciens antibacterial activity assay against E. coli. The A. tumefaciens WT C58 or ΔtssL or chromosomally encoded tagF-pppA variants, including tagF-pppA with substitutions in the tagF domain (tagFGK-pppA, tagFDW-pppA, and tagFSDR-pppA), were co-cultured on LB agar with E. coli strain DH10B cells harboring the plasmid pRL662. Data are mean ± S.D. (error bars) of at least three biological replicates. Different letters above the bar indicate statistically significantly different groups of strains (p < 0.05) based on cfu of the surviving target cells.
Overexpression of TagF causes reduced T6SS protein accumulation and abolishes T6SS antibacterial activity in P. aeruginosa
We then assessed whether these specific conserved amino acid residues of TagF required for binding to Fha and T6SS repression in A. tumefaciens are also required in P. aeruginosa TagF for Fha1Pa interaction and T6SS activity. We generated two alanine substitution mutants in TagFPa, namely TagFPa-GK (Gly-8 and Lys-9) and TagFPa-SDR (Ser-79, Asp-81, and Arg-85). Consistent with the results obtained with A. tumefaciens, TagFPa-GK and TagFPa-SDR mutants lost the interaction with Fha1Pa, although their expression level was comparable with that of WT TagFPa in yeast (Fig. 8A and Fig. S3C). To determine whether the TagFPa–Fha1Pa interaction is critical for TagF-dependent H1-T6SS repression, WT TagFPa, Strep-tagged WT TagFPa, and the TagFPa-GK and TagFPa-SDR variants were expressed from pRL662 in the P. aeruginosa ΔretS mutant, a constitutively H1-T6SS active strain (35). As expected, Hcp1 and Tse3 were secreted into the culture medium of ΔretS harboring the vector pRL662 (V), but H1-T6SS secretion was greatly reduced when TagFPa or TagFPa-Strep was overexpressed (Fig. 8B). Furthermore, overexpression of TagFPa-GK-Strep or TagFPa-SDR-Strep did not repress Hcp1 and Tse3 secretion. Interestingly, in contrast to A. tumefaciens, where overexpressed TagF mutant variants are at a level comparable with the overexpressed WT form, in P. aeruginosa, we observed significantly higher levels of WT TagFPa-Strep as compared with TagFPa-GK-Strep or TagFPa-SDR-Strep (Fig. 8B). Furthermore, protein levels of all analyzed T6SS components (including secreted proteins Hcp1 and Tse3 and structural proteins TssB1 and TagJ1) were significantly reduced upon overexpression of WT TagF (TagFPa or TagFPa-Strep). Accordingly, protein levels of these T6SS components were restored to near WT levels in the presence of TagFPa-GK-Strep and TagFPa-SDR-Strep in P. aeruginosa (Fig. 8B).
Figure 8.
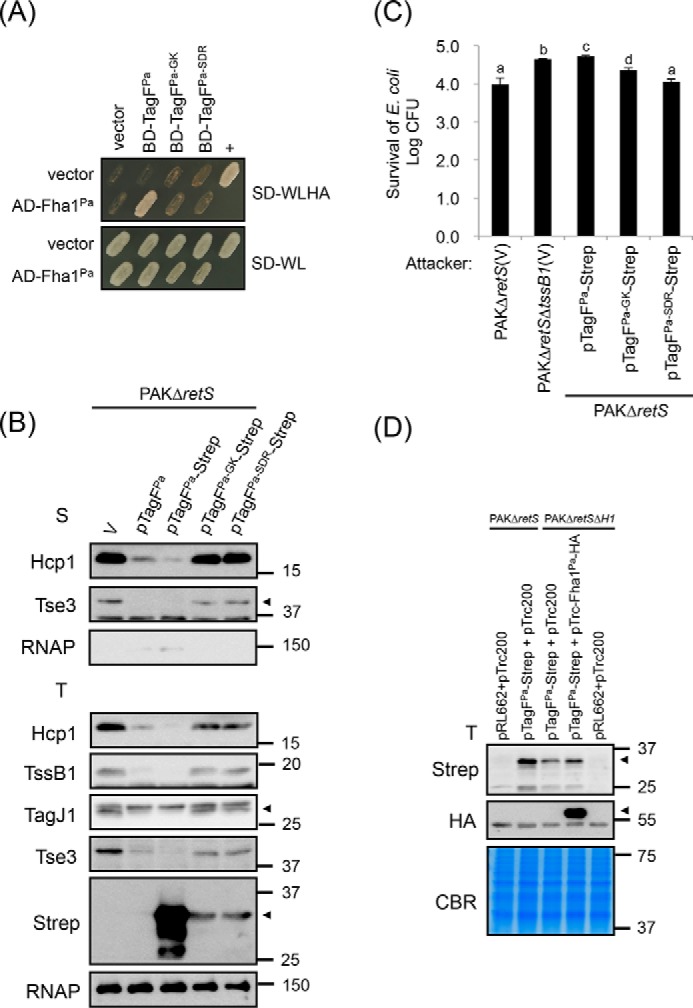
Conserved amino acid residues of TagFPa critical for TagFPa–Fha1Pa interaction are required for repressing H1-T6SS activity in P. aeruginosa. A, yeast two-hybrid protein–protein interaction results with P. aeruginosa Fha1 and various P. aeruginosa TagF proteins. SD−WL medium (SD minimal medium lacking Trp and Leu) was used for selecting plasmids. SD−WLHA medium (SD minimal medium lacking Trp, Leu, His, and Ade) was used for auxotrophic selection of bait and prey protein interactions. The positive interaction was determined by growth on SD−WLHA medium at 30 °C for at least 2 days. The positive control (+) showing interactions of SV40 large T-antigen and murine p53 and negative control (vector) are indicated. B, P. aeruginosa H1-T6SS secretion analysis. Shown is Western blot analysis of total (T) or secreted (S) proteins isolated from P. aeruginosa PAKΔretS (H1-T6SS–induced) harboring the vector pRL662 (V) or PAKΔretS harboring various overexpressed plasmids grown in TSB with specific antibodies. The nonsecreted RNA polymerase β subunit (RNAP) was an internal control. The proteins analyzed and molecular weight standards are shown on the left and right, respectively, and indicated with an arrowhead when necessary. C, P. aeruginosa H1-T6SS–mediated antibacterial assay against E. coli. Overnight cultures of P. aeruginosa PAKΔretS or PAKΔretSΔtssB1 (T6SS-defective strain) harboring the vector pRL662 (V) or various tagF-Strep–overexpressing plasmids were mixed with equivalent numbers of E. coli DH5α carrying a plasmid (pCR2.1) expressing β-gal. Data are mean ± S.D. of at least three biological replicates. Different letters above the bar indicate statistically significantly different groups of strains (p < 0.05) based on cfu of the surviving target cells. D, the presence of Fha1Pa increases the stability of TagFPa protein in P. aeruginosa. Shown is Western blot analysis of total (T) proteins isolated from P. aeruginosa PAKΔretS (H1-T6SS–induced) or PAKΔretSΔH1 (deletion of retS and H1-T6SS cluster) harboring various plasmid combinations grown in TSB with specific antibodies. All protein samples were analyzed by SDS-PAGE followed by Coomassie Blue staining (CBR) and served as an internal control. The proteins analyzed and molecular weight standards are shown on the left and right, respectively, and indicated with arrowheads when necessary.
Next, we performed an antibacterial activity assay and showed that when TagFPa-Strep was overexpressed in PAKΔretS, E. coli survival is similar to that for the T6SS-defective PAKΔretSΔtssB1 mutant. In contrast, the expression of TagFPa-GK-Strep and TagFPa-SDR-Strep conferred antibacterial activity similar to that of PAKΔretS (Fig. 8C). Because the reduced protein level/stability of both TagFPa-GK-Strep and TagFPa-SDR-Strep is associated with loss of ability to interact with Fha1Pa, we tested whether the presence or absence of Fha1Pa affected TagF abundance. Strikingly, TagFPa-Strep protein level was significantly reduced in PAKΔretSΔH1, which lacks endogenous Fha1Pa, as compared with PAKΔretS (Fig. 8D). TagFPa-Strep protein levels could be partially restored upon co-expression of TagFPa-Strep and Fha1Pa-HA in PAKΔretSΔH1 (Fig. 8D), which suggests that Fha1Pa may play a role in stabilizing TagFPa in P. aeruginosa. Taken together, our results indicate that the TagF domain represses T6SS activity via interaction with Fha but with distinct mechanisms when comparing A. tumefaciens and P. aeruginosa.
Discussion
In the present study, we have characterized the A. tumefaciens TagF-PppA and P. aeruginosa TagF and provide compelling evidence that TagF specifically interacts with Fha to repress type VI secretion and antibacterial activity independently of TPP. The loss of interaction with Fha is associated with loss of repression of T6SS activity in both cases, yet we observed differences in the nature and impact of TagF and its variants in repressing T6SS. This may have strategic consequences for how bacteria from various species communicate and respond to each other during T6SS-dependent bacterial warfare.
One remarkable difference is that TagF–Fha interaction has a different impact on T6SS protein abundance between A. tumefaciens and P. aeruginosa. A similar protein abundance of all TagFAt or TagFAt-Strep overexpressed (WT and all mutants) was observed in A. tumefaciens. In contrast, in P. aeruginosa, the protein level was significantly lower for TagFPa-GK-Strep or TagFPa-SDR-Strep than WT TagFPa-Strep (Figs. 2A, 7A, and 8B). Furthermore, all analyzed T6SS proteins accumulated to a similar level in the presence or absence of endogenous or overexpressed TagFAt in A. tumefaciens (Figs. 2A and 7A). However, the protein levels of P. aeruginosa H1-T6SS components (including secreted proteins Hcp1 and Tse3 and structural proteins TssB1 and TagJ1) were reduced with TagFPa or TagFPa-Strep overexpression but restored to nearly WT levels in the presence of mutant TagFPa-GK-Strep or TagFPa-SDR-Strep (Fig. 8B). Previous studies in P. aeruginosa suggested that TagFPa-mediated T6SS repression is mediated via a post-translational regulation mechanism (19). The evidence is based on the lack of influence on the expression of lacZ-translational fusion to fha1 or tssA1 and on the levels of two secreted proteins, Hcp and Tse1, in a ΔtagF mutant, which activates Hcp and Tse1 secretion. Instead of analyzing a tagF mutant here, we used strains overexpressing TagF, and our data suggest that the P. aeruginosa TagF-mediated post-translational repression occurs via influencing the protein stability of T6SS components when TagFPa is expressed in excess amounts. Taken together, Fha is a common target for TagF in repressing T6SS via post-translational regulation in both A. tumefaciens and P. aeruginosa, but each has a different strategy to exert this repression activity.
The role of Fha in activating T6SS in both TPP-dependent and -independent pathways may provide some clues to understand the mode of action by which TagF represses T6SS activity. Fha1 but not its phosphorylated form is required for ClpV1Pa foci formation and TagFPa-mediated derepression of type VI secretion in P. aeruginosa (19, 21). Also, Fha protein is a core component for T6SS in several bacteria lacking PpkA, PppA, and TagF (21, 22, 30, 36). Therefore, in addition to functioning as a scaffold protein specifically binding a phosphothreonine protein, Fha may also serve as a core T6SS component, probably via interaction with one or multiple T6SS components to activate T6SS assembly and secretion. TssM could be such a candidate because Fha1 foci formation is lost in a P. aeruginosa ΔicmF(tssM) mutant (37). In A. tumefaciens, Fha specifically interacts with TssL at phosphothreonine 14 to associate with the TssM-TssL inner membrane complex and recruit Hcp to interact with TssL for activating type VI secretion (4, 22). Together with our current knowledge of the T6SS assembly pathway (2, 10, 38), these studies indicate that Fha plays a key role in T6SS assembly at step(s) before recruitment of Hcp and TssB-TssC for tail polymerization. In A. tumefaciens, only WT TssL but not a TssL variant with the T14A amino acid substitution could interact with Fha, as assessed by a pulldown assay (22). Thus, Fha may interact with another core component(s) of the T6SS in addition to TssL or Fha itself, especially when PpkA is absent or not active. If so, TagF may compete with other T6SS core components for interacting with Fha and thereby prevent Fha from binding to a T6SS membrane–associated complex for stability (in P. aeruginosa) or activation of T6SS assembly (in A. tumefaciens).
Of note, the conserved G22K23, D44W46, and S93D95R99 residues but not D29F30 of A. tumefaciens TagF are critical for the TagF–Fha interaction, but all are required for repressing T6SS activity. This result led us to propose that D29F30 may repress T6SS function independently of binding to Fha. TagF may target Fha in repressing T6SS activity in two steps. The first step is to bind Fha via an interface involving G22K23, D44W46, and S93D95R99. Upon binding, TagF interferes with Fha recruitment to the membrane-associated complex for T6SS activation via D29F30 residues. Alternatively, it is possible that TagF can also target other T6SS components in addition to Fha. Future work to elucidate the molecular details underlying how TagF–Fha interaction influences T6SS activity or identifying additional TagF-interacting partners may provide answers to distinguish between two possible mechanisms.
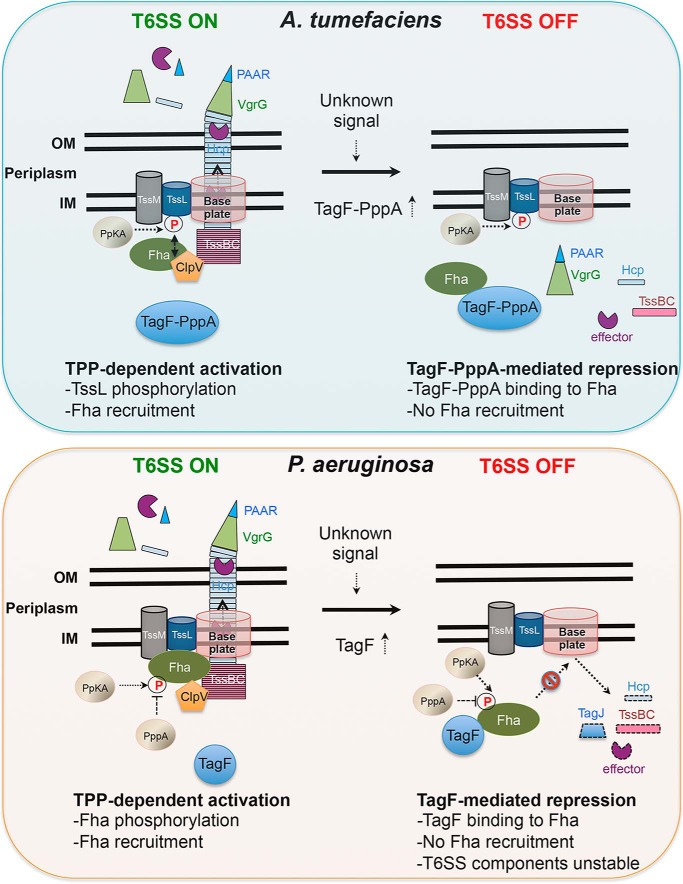
Combining previous (19, 21, 22) and current findings, we propose distinct models for TagF-mediated T6SS repression in A. tumefaciens and P. aeruginosa (Fig. 9). In WT A. tumefaciens when PpkA is active, the level of endogenous TagF-PppA is very low, and the protein does not bind to Fha, with no or little repression activity observed because Fha would then bind to pTssL for triggering T6SS assembly and secretion (T6SS ON shown in the top left panel of Fig. 9). On sensing an unknown signal, which may cause high accumulation of TagF-PppA or suppression of the TPP pathway, TagF-PppA can interact with Fha via its TagF domain to prevent it from binding to the membrane-associated complex and thus preventing T6SS activation (T6SS OFF shown in the top right panel of Fig. 9). Because TssM and TssL can form an inner-membrane complex in the absence of TssL phosphorylation (4, 22), the TssM-TssL complex and the associated baseplate complex probably remain properly assembled in the membrane when T6SS is suppressed by TagF-PppA. However, Hcp and TssBC may not be polymerized into the tail-like structure, and effector proteins are not loaded on the VgrG-PAAR spike for secretion. Because type VI secretion can be restored to the WT level in the ΔppkAΔtagF-pppA mutant (Fig. 4A), Fha probably also functions as a core T6SS component via interaction with one or multiple T6SS components to activate T6SS assembly and secretion in the absence of PpkA and TagF-PppA. This proposed mechanism also explains the previous observation that type VI secretion is highly attenuated but not completely abolished in the absence of PpkA (ΔppkA) (22, 30), because endogenous TagF-PppA, albeit at a low level, can bind Fha, and only a fraction of the Fha pool is available for recruitment to the T6SS core complex.
Figure 9.
Proposed models of TPP activation and TagF-mediated post-translational repression of type VI secretion in A. tumefaciens and P. aeruginosa. Proposed models of TPP activation (T6SS ON) and TagF-mediated repression (T6SS OFF) in A. tumefaciens (top) and P. aeruginosa (bottom) are illustrated. Key activation or repression events are summarized at the bottom of each model. Protein names are indicated in or near the designated molecules. IM, inner membrane; OM, outer membrane. For a detailed description of the proposed models, see “Discussion.”
In P. aeruginosa, type VI secretion is significantly enhanced in ΔpppA or ΔtagF as compared with the parental strain, and PppA phosphatase negatively regulates type VI secretion in a TPP-dependent manner, whereas TagF represses type VI secretion independently of TPP (19, 21). Thus, in the WT P. aeruginosa strain harboring both TPP components and TagF, type VI secretion remains at low levels, probably because of a series of phosphorylation and dephosphorylation events as well as TagF interaction with a fraction of the Fha1 pool (T6SS ON shown in the bottom left panel of Fig. 9). When TagF is expressed in excess amounts, TagF interacts with Fha to prevent it from binding to T6SS components and thus from activating T6SS assembly and subsequent secretion. Failure to recruit Fha1 to the membrane-associated T6SS complex may send out a signal to trigger degradation of cytoplasmic T6SS components and effectors (T6SS OFF shown in the bottom right panel of Fig. 9). Our current study may also provide an explanation for the enhanced T6SS secretion and antibacterial activity in a ΔtagF mutant in the presence or absence of PpkA (19, 21). Because Fha1 protein itself but not its phosphorylation is required for ClpV1Pa foci formation (19, 21), nonphosphorylated Fha1 may remain active in binding membrane-associated T6SS component(s), thus resulting in T6SS assembly and secretion in P. aeruginosa. With no TagF functionally available (i.e. in the absence of the protein or presence of a TagF mutant losing Fha1-binding activity), all Fha1 is available for activating T6SS assembly and secretion.
In conclusion, our proposed molecular model may provide answers to the longstanding question of how TagF mediates T6SS repression. We present compelling evidence suggesting that TagF specifically interacts with Fha and that such binding prevents Fha from being recruited to a T6SS membrane-associated complex. The TagF–Fha interaction has a different impact in different bacteria but ultimately prevents successful T6SS assembly. This study adds to our understanding of how bacteria deploy TPP-dependent activation and TPP-independent TagF-mediated repression mechanisms to control T6SS.
Experimental procedures
Bacterial strains, plasmids, and growth conditions
Strains, plasmids, and primer sequences used in this study are described in Tables S1 and S2. The growth conditions are described in detail in supporting information S1.
Plasmid construction and generation of in-frame deletion mutants
All in-frame deletion mutants were generated in A. tumefaciens C58 via double crossover using the suicide plasmid pJQ200KS (39) as described previously (4, 27). The detailed procedures for the construction of plasmids and mutant strains are described in supporting information S1.
Antibody production
The specific antibody for phosphorylated TssL (pTssL) was generated against the 15-mer peptide (7SSWQDLPpTVVEITEE21) containing the phosphorylated Thr-14 residue (22). The pTssL epitope located at the N-terminal region of TssL was used for polyclonal antibody production in rabbits.
A. tumefaciens type VI secretion and antibacterial competition assays
Type VI secretion assay was performed as described previously (4, 27, 29, 30). To study type VI secretion from A. tumefaciens grown in liquid medium, A. tumefaciens cells were grown in liquid 523 medium for 16 h at 25 °C. Cells were harvested, and A600 was adjusted to 0.1. A. tumefaciens cells continued to grow in liquid AB-MES medium (pH 5.5) (40) at 25 °C for 6 h. The cell suspension was centrifuged at 10,000 × g for 15 min at 4 °C; the resulting supernatant was concentrated by TCA precipitation, and Hcp, Tae, and Tde1 secretion was monitored as described previously (4, 27). An A. tumefaciens antibacterial activity assay using E. coli as target cells was performed as described previously (29). In brief, overnight grown A. tumefaciens and E. coli strains harboring appropriate plasmids were adjusted to A600 of 0.1 and incubated at 25 °C for 4–5 h before co-incubation. A. tumefaciens and E. coli cells were harvested, and A600 was adjusted, and then cells were mixed at a 1:30 ratio (A600 0.01:0.3) and spotted onto LB agar plates. Where applicable, the mixture was spotted onto an LB agar plate containing 0.5 mm isopropyl β-d-thiogalactopyranoside (IPTG) to induce expression from the pTrc200 plasmid. 16 h after incubation at 25 °C, the spots were harvested, serially diluted, and plated on LB agar plate containing appropriate antibiotic to quantify surviving E. coli cells by counting cfu. An in planta bacterial competition assay was performed as described previously (29). Briefly, A. tumefaciens strains were transformed with gentamycin, resistance conferred by the pRL662 plasmid, or spectinomycin, resistance conferred by the pTrc200 plasmid, for selecting surviving cells. The attacker (A600 of 5) and target (A600 of 0.5) strains were mixed in ½ Murashige and Skoog medium (pH 5.7) at a 10:1 ratio and infiltrated into leaves of 6–7-week-old Nicotiana benthamiana plants by use of a needleless syringe. After a 24-h incubation at room temperature, the infiltrated spot was punched out, ground in 0.9% NaCl, serially diluted, and plated in triplicates on LB agar containing appropriate antibiotic to select for the target cells. All assays were performed with at least two independent experiments, each with two biological replicates, or three independent experiments, each with one or two biological replicates.
P. aeruginosa type VI secretion and antibacterial competition assays
A P. aeruginosa type VI secretion assay was performed as described previously (41). In brief, P. aeruginosa strains harboring appropriate plasmids were grown in tryptone soy broth (TSB) overnight at 37 °C under agitation. Cells were harvested and subcultured to an A600 of 0.1, and then growth was continued in TSB to early stationary phase at 37 °C for 4–5 h to A600 of 5. Cells were separated from culture supernatants by centrifugation at 4000 × g at 4 °C. Cells were directly resuspended in 1 × SDS sample buffer. 10-Fold concentrated P. aeruginosa culture supernatant was prepared as follows. Proteins of the culture supernatant were precipitated using 6 m TCA (Sigma) at a final TCA concentration of 10%. Protein pellets were washed in 90% acetone, dried, and suspended in 1× SDS sample buffer; incubated at 96 °C for 20 min; and analyzed by SDS-PAGE. A P. aeruginosa antibacterial activity assay using E. coli as target cells was performed as described previously (42). In brief, overnight cultures of the indicated P. aeruginosa strains were incubated with overnight cultures of equivalent bacterial numbers of E. coli containing the plasmid pCR2.1 (carrying the lacZ gene) in a 1:1 ratio on LB agar for 5 h at 37 °C. In addition, P. aeruginosa and E. coli strains grown alone on LB agar for 5 h at 37 °C served as negative growth controls. Subsequently, patches of bacteria were collected and resuspended in LB broth, and dilution series ranging from 100 to 10−7 were plated in triplicate on LB supplemented with 100 mg/ml 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-gal; Invitrogen), allowing for colorimetric detection of lacZ-positive E. coli survivors. For quantitative analysis of the amount of E. coli survivors, the spots were harvested, serially diluted, and plated on LB agar plate containing X-gal and appropriate antibiotic to quantify surviving E. coli cells by counting. Data represent mean ± S.D. of all biological replicates.
Yeast two-hybrid assay
The Matchmaker yeast two-hybrid system was used as instructed (Clontech, Mountain View, CA) and as described previously (30). Each of the plasmid pairs was co-transformed into Saccharomyces cerevisiae strain AH109. The transformants were selected by their growth on synthetic dextrose (SD) minimal medium lacking tryptophan (Trp) and leucine (Leu) (SD−WL medium). The positive interaction of expressed fusion proteins was then determined by their growth on SD lacking Trp, Leu, adenine (Ade), and histidine (His) (SD−WLHA medium) at 30 °C for at least 2 days.
Total protein extraction from yeast
In brief, to prepare the total protein from yeast (43), overnight grown yeast strains harboring appropriate plasmids were subcultured at 28 °C in the same medium for further growth to A600 of 0.4–0.6. Yeast cells were harvested, 100 μl of protein extraction buffer containing 0.1% Nonidet P-40, 250 mm NaCl, 50 mm Tris-HCl (pH 7.5), 5 mm EDTA (pH 8.0), 1 mm DTT, 2× protease inhibitor mixtures (Roche Applied Science), 4 mm phenylmethylsulfonyl fluoride, and 50 μl of acid-washed glass beads (Sigma-Aldrich) were added. The cells were broken by vortex at the highest speed for 30 s, and then tubes were placed on ice for 30 s. The same procedure was repeated six times to make sure cells had been completely broken. The supernatant above the glass beads was collected (the first cell extract). Then 50 μl of protein extraction buffer was added to wash the tube containing broken cells and glass beads by vortexing at the highest speed for 30 s. The supernatant above the glass beads was collected again (the second cell extract). The two extracts were mixed together, and the final protein extract was centrifuged at 13,000 rpm for 5 min at 4 °C; the resulting supernatant was collected, and protein concentration was measured. An equal volume of 2× SDS loading buffer was added to the final protein sample, incubated at 96 °C for 20 min, and analyzed by SDS-PAGE.
Bacterial two-hybrid assay
The BTH assay was performed as described previously (44). In brief, DNA fragments encoding the protein of interest were amplified by PCR, adding appropriate restriction sites into the primers, using P. aeruginosa PAK genomic DNA. DNA fragments encoding the proteins or protein domains of interest were cloned into plasmids pKT25 and pUT18C, which each encode for complementary fragments of the adenylate cyclase enzyme, as described previously (44), resulting in constructs expressing an N-terminal fusion of the protein of interest with the T25 or T18 subunit of adenylate cyclase. Recombinant pKT25 and pUT18C plasmids were co-transformed into the E. coli DHM1 strain, which is devoid of adenylate cyclase, and transformants were spotted onto LB agar plates (Difco) supplemented with 40 mg/ml X-gal in the presence of 100 mg/ml ampicillin, 50 mg/ml kanamycin, and 1 mm IPTG. Positive interactions were identified as blue colonies on LB agar plates containing X-gal after a 48-h incubation at 30 °C. The experiments were performed at least in duplicate, and a representative result is shown.
β-gal assay
For quantitative analysis of BTH interactions, β-gal activity of co-transformants scraped from LB agar plates containing X-gal was measured as described previously, and activity was calculated in Miller units (45, 46).
Statistical analysis
Data represent mean ± S.E. of all biological replicates. Statistics were calculated by one-way analysis of variance and Tukey's honestly significance difference test (http://astatsa.com/OneWay_Anova_with_TukeyHSD/),5 and significant differences are indicated (p < 0.01 or p < 0.05).
Dephosphorylation and Phos-tag SDS-PAGE analyses
Dephosphorylation analysis by calf intestinal alkaline phosphatase (CIAP) was performed according to the user manual (New England Biolabs, Beverly, MA) with minor modifications as described previously (22). Equal amounts of Ni-NTA resins with purified TssL-His isolated from various A. tumefaciens strains were resuspended in 1× CIAP buffer containing 100 mm NaCl, 50 mm Tris-HCl (pH 7.9), 10 mm MgCl2, 1 mm DTT, and 1× protease inhibitor mixture (EDTA-free) with CIAP at 1 unit/μg of protein. The protein samples treated with or without CIAP were incubated at 37 °C for 90 min. An equal volume of 2× SDS loading buffer was added and incubated at 96 °C for 20 min and analyzed by Phos-tag SDS-PAGE. The Phos-tag SDS-PAGE analysis was performed according to the user manual for Phos-tag acrylamide AAL-107 (Wako Pure Chemical Industries, Osaka, Japan) with minor modifications as described previously (22). Protein samples were separated on 7% polyacrylamide gels containing 0.35 m BisTris-HCl (pH 6.8), 35 μm Phos-tag acrylamide AAL-107, and 100 μm ZnCl2, with electrophoresis conducted at 40 mA/gel under a maximum voltage of 90 V in a cold room. After electrophoresis, Phos-tag gels were washed with transfer buffer (25 mm Tris, 192 mm glycine, 20% methanol) containing 1 mm EDTA for 15 min with gentle shaking followed by a second wash in transfer buffer without EDTA for 15 min. The gels were washed with transfer buffer containing 1% SDS for 15 min before transfer to polyvinylidene difluoride membranes with a submarine blotting apparatus.
Protein purification and gel filtration analysis
N-terminal His-tagged TagF aa 1–214 proteins were expressed in E. coli BL21 (DE3) cells with the plasmid pET28a(+)-TagF 1–214. E. coli cells were grown in LB medium in the presence of kanamycin (20 μg/ml) at 37 °C until the cell density reached an A600 of 0.6–0.8. The cultures were induced with 1 mm IPTG for 16–20 h at 16 °C to induce production of His-tagged TagF aa 1–214 proteins. The cells were harvested by centrifugation followed by snap-freezing by liquid nitrogen and stored at −80 °C. Frozen bacterial pellets were resuspended with the lysis buffer (150 mm sodium chloride, 10 mm imidazole, and 50 mm Tris-HCl, pH 7.5), and then the cells were lysed by a microfluidizer. The cell lysate was clarified by centrifugation at 20,000 rpm for 40 min at 4 °C, and the supernatant was loaded onto an Ni-NTA column (GE Healthcare) pre-equilibrated with the lysis buffer. The column was washed with the washing buffer (150 mm sodium chloride, 80 mm imidazole, and 50 mm Tris-HCl, pH 7.5), and the bound protein was eluted by the elution buffer (150 mm sodium chloride, 300 mm imidazole, and 50 mm Tris-HCl, pH 7.5). The TagF aa 1–214 proteins were further analyzed by size-exclusion chromatography using a Superdex 75 16 × 60 column through a fast protein LC (FPLC) system (GE Healthcare). The column was equilibrated with 2 column volumes of buffer containing 50 mm Tris-HCl (pH 7.5) and 150 mm NaCl. To determine molecular weight, a parallel column was run with protein standards. The elution profiles were recorded as absorbance at 280 nm.
Western blot analysis
Western blot analysis was performed as described previously (40) with the primary polyclonal pTssL antibody produced in this study and those against proteins PpkA, TagF-PppA, TssK, Fha, TssC41, TssB, TssA, ClpV, Tae, VgrGs, and RpoA (30), TssL (4), TssM (4), Hcp (27), ActC (47), Tde1 (29), P. aeruginosa Hcp1, TssB1, TagJ1, and Tse3 (44), polyclonal antibodies against His (Sigma-Aldrich), monoclonal antibodies against HA (Sigma-Aldrich), the β subunit of RNA polymerase (Neoclone), c-Myc (Sigma-Aldrich), or Strep (IBA-Life Sciences, Goettingen, Germany), followed by incubation with a secondary antibody, horseradish peroxidase–conjugated goat anti-rabbit IgG (Chemichem) and detection by use of the Western Lightning System (PerkinElmer Life Sciences). Chemiluminescent bands were visualized on X-ray film (GE Healthcare).
Author contributions
J.-S. L. and E.-M. L. conceptualization; J.-S. L., P. P., and H.-H. W. data curation; J.-S. L., P. P., H.-H. W., M.-D. T., A. F., and E.-M. L. investigation; J.-S. L. and E.-M. L. writing-original draft; J.-S. L., P. P., H.-H. W., M.-D. T., A. F., and E.-M. L. writing-review and editing; M.-D. T., A. F., and E.-M. L. supervision; M.-D. T., A. F., and E.-M. L. funding acquisition.
Supplementary Material
Acknowledgments
We thank the Lai and Filloux laboratory members for stimulating discussion. We also thank Sarah Wettstadt for technical assistance in P. aeruginosa experiments and the Genomic Technology Core Laboratory at the Institute of Plant and Microbial Biology, Academia Sinica, for DNA sequencing.
This work was supported by Ministry of Science and Technology of Taiwan (MOST) Grants 104-2311-B-001-025-MY3 (to E.-M. L.) and MOST Grants 105-0210-01-12-01 and 106-0210-01-15-04 and Taiwan Protein Project Grant 107-0210-01-19-02 (to M.-D. T.); Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/N002539/1 and Medical Research Council (MRC) Grant MR/N023250/1 (to A. F.); and MOST-BBSRC International Partnering Award (Grant 105-2911-I-001-503) (to E.-M. L.) and Grant BB/M02735X/1 (to A. F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2, Figs. S1–S4, and supporting information S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- T6SS
- type VI secretion system
- FHA
- forkhead-associated
- TPP
- threonine phosphorylation pathway
- aa
- amino acids
- Ni-NTA
- nickel-nitrilotriacetic acid
- pTssL
- phosphorylated TssL
- YTH
- yeast two-hybrid
- BTH
- bacterial two-hybrid
- LB
- lysogeny broth
- IPTG
- isopropyl β-d-thiogalactopyranoside
- cfu
- colony-forming units
- TSB
- tryptone soy broth
- X-gal
- 5-bromo-4-chloro-3-indolyl β-d-galactoside
- CIAP
- calf intestinal alkaline phosphatase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TagF
- type VI secretion–associated gene F.
References
- 1. Hachani A., Wood T. E., and Filloux A. (2016) Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93 10.1016/j.mib.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 2. Cianfanelli F. R., Monlezun L., and Coulthurst S. J. (2016) Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- 3. Aschtgen M. S., Bernard C. S., De Bentzmann S., Lloubès R., and Cascales E. (2008) SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531 10.1128/JB.00945-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma L. S., Lin J. S., and Lai E. M. (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 10.1128/JB.00029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma L. S., Narberhaus F., and Lai E. M. (2012) IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287, 15610–15621 10.1074/jbc.M111.301630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Felisberto-Rodrigues C., Durand E., Aschtgen M. S., Blangy S., Ortiz-Lombardia M., Douzi B., Cambillau C., and Cascales E. (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7, e1002386 10.1371/journal.ppat.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brunet Y. R., Zoued A., Boyer F., Douzi B., and Cascales E. (2015) The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11, e1005545 10.1371/journal.pgen.1005545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Planamente S., Salih O., Manoli E., Albesa-Jové D., Freemont P. S., and Filloux A. (2016) TssA forms a gp6-like ring attached to the type VI secretion sheath. EMBO J. 35, 1613–1627 10.15252/embj.201694024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zoued A., Durand E., Brunet Y. R., Spinelli S., Douzi B., Guzzo M., Flaugnatti N., Legrand P., Journet L., Fronzes R., Mignot T., Cambillau C., and Cascales E. (2016) Priming and polymerization of a bacterial contractile tail structure. Nature 531, 59–63 10.1038/nature17182 [DOI] [PubMed] [Google Scholar]
- 10. Basler M. (2015) Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150021 10.1098/rstb.2015.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brackmann M., Nazarov S., Wang J., and Basler M. (2017) Using force to punch holes: mechanics of contractile nanomachines. Trends Cell Biol. 27, 623–632 10.1016/j.tcb.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 12. Brunet Y. R., Bernard C. S., Gavioli M., Lloubès R., and Cascales E. (2011) An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7, e1002205 10.1371/journal.pgen.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernard C. S., Brunet Y. R., Gueguen E., and Cascales E. (2010) Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192, 3850–3860 10.1128/JB.00370-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leung K. Y., Siame B. A., Snowball H., and Mok Y. K. (2011) Type VI secretion regulation: crosstalk and intracellular communication. Curr. Opin. Microbiol. 14, 9–15 10.1016/j.mib.2010.09.017 [DOI] [PubMed] [Google Scholar]
- 15. Allsopp L. P., Wood T. E., Howard S. A., Maggiorelli F., Nolan L. M., Wettstadt S., and Filloux A. (2017) RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 114, 7707–7712 10.1073/pnas.1700286114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahajan A., Yuan C., Lee H., Chen E. S., Wu P. Y., and Tsai M. D. (2008) Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 1, re12 [DOI] [PubMed] [Google Scholar]
- 17. Cascales E. (2008) The type VI secretion toolkit. EMBO Rep. 9, 735–741 10.1038/embor.2008.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., and Attree I. (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10, 104 10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Silverman J. M., Austin L. S., Hsu F., Hicks K. G., Hood R. D., and Mougous J. D. (2011) Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol. Microbiol. 82, 1277–1290 10.1111/j.1365-2958.2011.07889.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fritsch M. J., Trunk K., Diniz J. A., Guo M., Trost M., and Coulthurst S. J. (2013) Proteomic identification of novel secreted anti-bacterial toxins of the Serratia marcescens Type VI secretion system. Mol. Cell. Proteomics 12, 2735–2749 10.1074/mcp.M113.030502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mougous J. D., Gifford C. A., Ramsdell T. L., and Mekalanos J. J. (2007) Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9, 797–803 10.1038/ncb1605 [DOI] [PubMed] [Google Scholar]
- 22. Lin J. S., Wu H. H., Hsu P. H., Ma L. S., Pang Y. Y., Tsai M. D., and Lai E. M. (2014) Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens. PLoS Pathog. 10, e1003991 10.1371/journal.ppat.1003991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Z., Zhou X., Ma Y., Zhou M., Waldor M. K., Zhang Y., and Wang Q. (2018) Serine/threonine kinase PpkA coordinates the interplay between T6SS2 activation and quorum sensing in the marine pathogen Vibrio alginolyticus. Environ. Microbiol. 20, 903–919 10.1111/1462-2920.14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng J., and Leung K. Y. (2007) Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206 10.1111/j.1365-2958.2007.05993.x [DOI] [PubMed] [Google Scholar]
- 25. Hsu F., Schwarz S., and Mougous J. D. (2009) TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol. Microbiol. 72, 1111–1125 10.1111/j.1365-2958.2009.06701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Casabona M. G., Silverman J. M., Sall K. M., Boyer F., Couté Y., Poirel J., Grunwald D., Mougous J. D., Elsen S., and Attree I. (2013) An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ. Microbiol. 15, 471–486 10.1111/j.1462-2920.2012.02816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu H. Y., Chung P. C., Shih H. W., Wen S. R., and Lai E. M. (2008) Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J. Bacteriol. 190, 2841–2850 10.1128/JB.01775-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu C. F., Lin J. S., Shaw G. C., and Lai E. M. (2012) Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS Pathog. 8, e1002938 10.1371/journal.ppat.1002938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma L. S., Hachani A., Lin J. S., Filloux A., and Lai E. M. (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104 10.1016/j.chom.2014.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin J. S., Ma L. S., and Lai E. M. (2013) Systematic dissection of the Agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS One 8, e67647 10.1371/journal.pone.0067647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bondage D. D., Lin J. S., Ma L. S., Kuo C. H., and Lai E. M. (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940 10.1073/pnas.1600428113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Basler M., Ho B. T., and Mekalanos J. J. (2013) Tit-for-Tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. LeRoux M., Kirkpatrick R. L., Montauti E. I., Tran B. Q., Peterson S. B., Harding B. N., Whitney J. C., Russell A. B., Traxler B., Goo Y. A., Goodlett D. R., Wiggins P. A., and Mougous J. D. (2015) Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. eLife 4, e05701 10.7554/eLife.05701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shalom G., Shaw J. G., and Thomas M. S. (2007) In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 10.1099/mic.0.2007/006585-0 [DOI] [PubMed] [Google Scholar]
- 35. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., and Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng J., Ho B., and Mekalanos J. J. (2011) Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876 10.1371/journal.pone.0023876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., and Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zoued A., Brunet Y. R., Durand E., Aschtgen M. S., Logger L., Douzi B., Journet L., Cambillau C., and Cascales E. (2014) Architecture and assembly of the Type VI secretion system. Biochim. Biophys. Acta 1843, 1664–1673 10.1016/j.bbamcr.2014.03.018 [DOI] [PubMed] [Google Scholar]
- 39. Quandt J., and Hynes M. F. (1993) Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
- 40. Lai E. M., and Kado C. I. (1998) Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180, 2711–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., and Filloux A. (2011) Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327 10.1074/jbc.M110.193045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hachani A., Lossi N. S., and Filloux A. (2013) A visual assay to monitor T6SS-mediated bacterial competition. J. Vis. Exp., e50103 10.3791/50103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin J. S., and Lai E. M. (2017) Protein-protein interactions: yeast two-hybrid system. Methods Mol. Biol. 1615, 177–187 10.1007/978-1-4939-7033-9_14 [DOI] [PubMed] [Google Scholar]
- 44. Lossi N. S., Manoli E., Simpson P., Jones C., Hui K., Dajani R., Coulthurst S. J., Freemont P., and Filloux A. (2012) The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel subcomplex in the bacterial type VI secretion system. Mol. Microbiol. 86, 437–456 10.1111/j.1365-2958.2012.08204.x [DOI] [PubMed] [Google Scholar]
- 45. Zhang X., and Bremer H. (1995) Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270, 11181–11189 10.1074/jbc.270.19.11181 [DOI] [PubMed] [Google Scholar]
- 46. Förster A., Planamente S., Manoli E., Lossi N. S., Freemont P. S., and Filloux A. (2014) Coevolution of the ATPase ClpV, the sheath proteins TssB and TssC, and the accessory protein TagJ/HsiE1 distinguishes type VI secretion classes. J. Biol. Chem. 289, 33032–33043 10.1074/jbc.M114.600510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu A. C., Shih H. W., Hsu T., and Lai E. M. (2008) A citrate-inducible gene, encoding a putative tricarboxylate transporter, is downregulated by the organic solvent DMSO in Agrobacterium tumefaciens. J. Appl. Microbiol. 105, 1372–1383 10.1111/j.1365-2672.2008.03874.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.