Abstract
Inhibition of the metabolic regulator AMP-activated protein kinase (AMPK) is increasingly being investigated for its therapeutic potential in diseases where AMPK hyperactivity results in poor prognoses, as in established cancers and neurodegeneration. However, AMPK-inhibitory tool compounds are largely limited to compound C, which has a poor selectivity profile. Here we identify the pyrimidine derivative SBI-0206965 as a direct AMPK inhibitor. SBI-0206965 inhibits AMPK with 40-fold greater potency and markedly lower kinase promiscuity than compound C and inhibits cellular AMPK signaling. Biochemical characterization reveals that SBI-0206965 is a mixed-type inhibitor. A co-crystal structure of the AMPK kinase domain/SBI-0206965 complex shows that the drug occupies a pocket that partially overlaps the ATP active site in a type IIb inhibitor manner. SBI-0206965 has utility as a tool compound for investigating physiological roles for AMPK and provides fresh impetus to small-molecule AMPK inhibitor therapeutic development.
Keywords: AMP-activated kinase (AMPK), cell metabolism, drug development, inhibition mechanism, signaling, structure-function, appetite suppression, neuroprotection
Introduction
The ability to maintain energy homeostasis during acute or chronic periods of nutrient shortfall is an essential characteristic of all living organisms. A direct molecular link between nutrient supply and energy demand is provided by AMP-activated protein kinase (AMPK),5 a key regulator of cellular and whole-body metabolism (1, 2). AMPK is an evolutionarily conserved serine/threonine kinase that senses, and is activated by, low adenylate charge (elevated AMP/ATP and ADP/ATP ratios) resulting from energetically demanding processes such as muscle contraction or reduced energy supply caused by hypoxia or nutrient deprivation. AMPK redirects cellular metabolism by down-regulating numerous ATP-consuming anabolic processes (e.g. protein, cholesterol, and fatty acid synthesis) and up-regulating similarly diverse ATP-producing catabolic processes (e.g. fat oxidation, glycolysis, and autophagy) to restore energy balance. It does this by direct phosphorylation, either modulating the activities of rate-limiting enzymes in multiple metabolic processes or regulating transcriptional activities of the factors governing their expression. For example, AMPK phosphorylation of cytosolic acetyl-CoA carboxylase 1 (ACC1) and mitochondrial-associated ACC2, inhibits de novo lipogenesis and promotes fat oxidation, respectively. AMPK signaling has also been associated with a range of nonmetabolic regulatory roles (e.g. circadian rhythm, mitochondrial fission, and appetite control).
AMPK is an αβγ heterotrimeric complex, consisting of a catalytic α subunit (isoforms α1 and α2) and regulatory subunits β (isoforms β1 and β2) and γ (isoforms γ1, γ2, and γ3). The α subunit contains a canonical, bi-lobed kinase domain at the N terminus, followed by autoinhibitory (AID) and scaffolding domains, and a Ser/Thr-rich loop region (ST loop). The β subunit contains a mid-molecule carbohydrate-binding module (CBM) and C-terminal scaffolding domain, whereas the γ subunit consists of four cystathionine β-synthase domains and three adenine nucleotide binding sites (termed γ-sites 1, 3, and 4) that endow AMPK with its energy-sensing capabilities. AMPK signaling is dynamically and tightly controlled by numerous activating/inhibiting and localization mechanisms. Spatially, AMPK is targeted to various subcellular organelles and compartments in response to leptin and circadian rhythm (nucleus) (3, 4), glucose starvation (lysosomes, plasma membranes) (5, 6), mitochondrial damage (mitochondria), and increased autophagic flux (autophagosomes) (7, 8). Activation is triggered primarily by binding of AMP or ADP to exchangeable γ-sites 1 and 3, stimulating phosphorylation of the kinase activation loop residue Thr-172 (pThr-172) by upstream kinases LKB1 and Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) and co-localizing AMPK with substrates on intracellular membranes (6, 9). pThr-172–independent activity has been demonstrated, although the contribution this makes to overall AMPK metabolic control is presently unclear (8, 10). AMPK signaling is negatively regulated by multiple mechanisms, including exchange of AMP/ADP for ATP, AID-mediated autoinhibition, pThr-172–dephosphorylating actions of phosphatases, phosphorylation of suppressive regulatory sites in the α-ST loop, and a variety of nutrients (glucose, amino acids, and lipids), hormones (insulin, leptin, resistin), and cytokines (tumor necrosis factor α, ciliary neurotrophic factor, and interleukin-6) (11).
The metabolic dimensions associated with major human diseases, such as type 2 diabetes, cancer, and inflammatory disorders, have encouraged efforts to develop small-molecule AMPK activators. Patented examples now number in the hundreds. One of the first pharmacological AMPK activators discovered was 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) (12). AICAR is taken up by cells via the adenosine transport system (13) and converted to the monophosphorylated derivative ZMP, which functions as an AMP-mimetic. Other AMPK agonists can be broadly classified as direct activators (those that bind to drug sites located either between the kinase domain and β-CBM (e.g. A-769662, salicylate, 991, and PF-937 (14–17)) or within the γ subunit (e.g. C2 (18, 19)) or as indirect activators (those that commonly induce energy imbalance through mitochondrial toxicity, including metformin, xenobiotics, and other natural products).
Pharmacological AMPK inhibition provides potential strategies to treat obesity (appetite suppression), type 2 diabetes (enhanced insulin secretion), and stroke (neuroprotection) (11), and AMPK hyperactivity has also been linked to pathogenesis of neurodegeneration (20). The role of AMPK in cancer is complex; whereas initial studies demonstrated a tumor-suppressive role, AMPK signaling also contributes to the metabolic adaptations associated with tumor growth (e.g. increased glycolytic flux (the Warburg effect) and maintenance of ATP and NADPH) and promotes anchorage-independent proliferation (21–23). AMPK also promotes autophagic processes, via phosphorylation of ULK1, to maintain homeostasis in the neoplastic cell (24). Thus, AMPK is considered pro-tumorigenic under certain circumstances, underpinning the attraction of AMPK inhibition as a strategy for cancer treatments.
Current availability of small-molecule AMPK inhibitors, either for clinical application or as research tools to delineate AMPK's physiological roles, is extremely limited. By far the most widely applied AMPK inhibitor, the pyrazolopyrimidine derivative compound C (dorsomorphin), was originally selected from a high-throughput screen and used to confirm AMPK-dependent effects of AICAR and metformin in cultured hepatocytes (25). Compound C is an ATP-competitive inhibitor and binds to the highly conserved active site of AMPK (26). However, in vitro screening has shown that compound C is promiscuous, inhibiting multiple kinases with similar or greater potency than AMPK (27). Numerous studies have since described off-target or AMPK-independent cellular effects, including inhibition of bone morphogenetic protein type I receptors ALK2, ALK3, and ALK6 (28), hypoxia-induced HIF-1 activation (29), preadipocyte proliferation (30), and macrophage chemotaxis (31). Compound C also blocks AICAR cellular uptake through competition for adenosine transporter binding sites, which largely accounts for its suppressive effects on AICAR-mediated AMPK activation (13). In light of these undesirable properties, conclusions drawn from using compound C as an AMPK inhibitor are now viewed with extreme caution, and recommendations are to avoid its application altogether (27). Other characterized direct antagonists include MT47-100, a low-potency, β2-AMPK allosteric inhibitor that intriguingly also activates β1-complexes (32), and SU6656, that paradoxically stimulates net cellular AMPK signaling by promoting phosphorylation of Thr-172 by LKB1 (33).
Here, we report the discovery of the small-molecule SBI-0206965 as a direct, type IIb AMPK inhibitor that demonstrated improved potency and kinase selectivity relative to compound C. A crystal structure of the AMPK α2 kinase domain/SBI-0206965 complex revealed a binding pocket that partially overlapped the ATP site; however, SBI-0206965 displayed mixed-competitive kinetics. SBI-0206965 inhibited AMPK signaling in a variety of cell types. Our study also highlights limitations pertaining to SBI-0206965 when used in conjunction with AICAR.
Results
SBI-0206965: A potent, small-molecule AMPK inhibitor
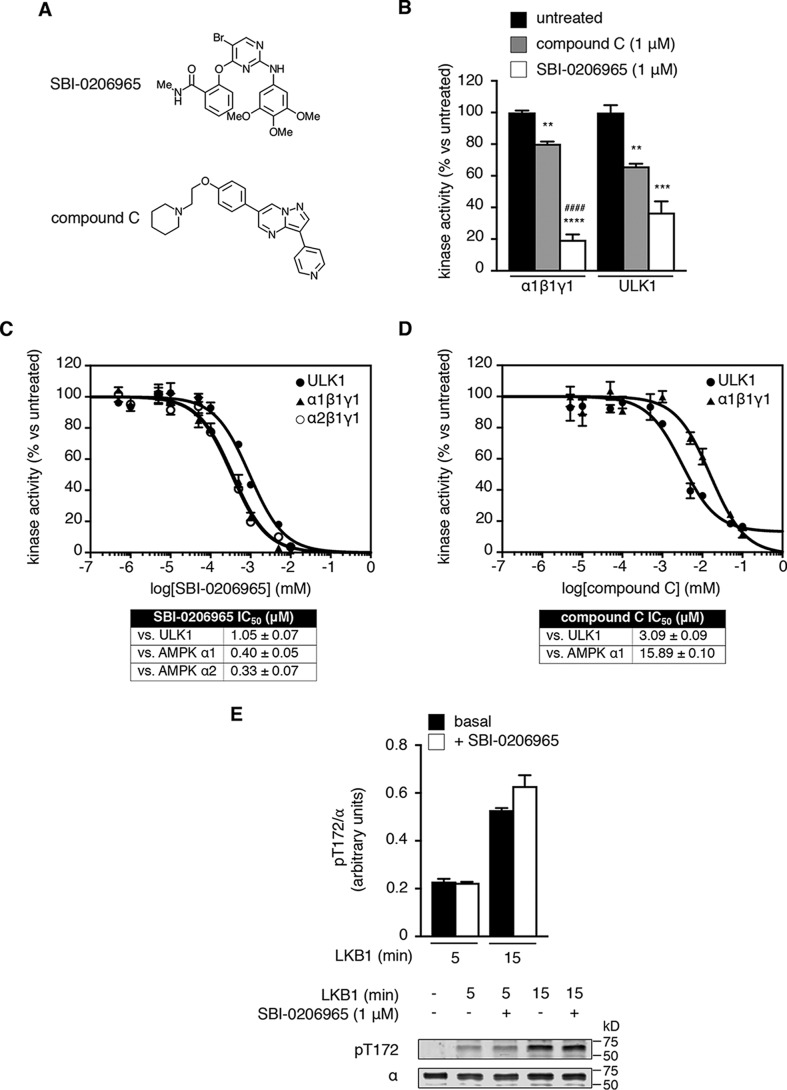
Using an active site competitive screen, Egan et al. (34) recently reported the 2-aminopyrimidine derivative SBI-0206965 (Fig. 1A) was a potent and highly selective inhibitor of the autophagy initiator ULK1. α1- and α2-AMPK complexes were also reported as hits in this screen. Subsequent studies in MEFs showed that SBI-0206965 (50 μm) produced a disconnect between AMPK-pThr-172 (elevated) and pACC (unchanged), indicative of direct inhibition of AMPK activity (34). Using cell-free phosphorylation of the AMPK and ULK1 synthetic peptide substrate S108tide as a measure of kinase activity (8), we found that SBI-0206965 (1 μm) was a more effective inhibitor of AMPK α1β1γ1 (80% inhibition) compared with ULK1 (63% inhibition) at 200 μm ATP (Fig. 1B). Conversely, compound C (Fig. 1A) was more effective as an inhibitor of ULK1 (36% inhibition) than AMPK (19% inhibition) at 200 μm ATP (Fig. 1B). Under our assay conditions, SBI-0206965 inhibited both α1β1γ1 and α2β1γ1 with nanomolar IC50 (Fig. 1C), whereas compound C inhibited both ULK1 and α1β1γ1 with micromolar IC50 (Fig. 1D). Using purified enzymes, we were unable to detect an effect of SBI-0206965 on the rate of LKB1-mediated phosphorylation of Thr-172 (Fig. 1E).
Figure 1.
Biochemical characterization of SBI-0206965. A, structures of SBI-0206965 and compound C. B, inhibition of AMPK α1β1γ1- and ULK1-phosphorylation of S108tide by SBI-0206965 and compound C. Error bars, mean percentage kinase activity versus untreated ± S.E. (error bars) (n = 3). **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 by one-way ANOVA with post hoc Dunnett's multiple-comparison test versus untreated kinase. ####, p < 0.0001 by one-way ANOVA with post hoc Dunnett's multiple-comparison test versus compound C–treated AMPK. Shown are dose-response curves for SBI-0206965 inhibition of ULK1, AMPK α1β1γ1, and AMPK α2β1γ1 (C) and compound C inhibition of ULK1 and AMPK α1β1γ1 (D). For C and D, assays were performed at fixed 200 μm ATP. IC50 values (μm) ± S.E. were calculated from triplicate experiments. E, Thr-172 phosphorylation assay. Bacterially expressed AMPK α2β1γ1 was incubated with LKB1 in the presence of SBI-0206965, and pThr-172 was measured by immunoblotting. Error bars, mean pThr-172/α (arbitrary units) ± S.E. (n = 3).
To assess kinase selectivity in terms of activity inhibition, we profiled SBI-0206965 (0.25 μm) against a diverse panel of 50 kinases (Fig. S1A). Compound C (2.5 μm) was screened in parallel for direct comparison (Fig. S1B). AMPK in this panel was assayed at 20 μm ATP, hence the need to profile inhibitors below the IC50 concentrations determined in our assays at 200 μm ATP (Fig. 1, C and D). Of the kinases in the panel, AMPK was inhibited to the greatest extent by both SBI-0206965 (22 ± 2% residual activity) and compound C (4 ± 1%); however, SBI-0206965 displayed a preferable selectivity profile, inhibiting activity of only five other kinases by >50% (compared with 23 kinases with compound C) and seven other kinases by >30% (compared with 31 kinases with compound C) (Fig. 2 and Fig. S1). SBI-0206965 inhibited LKB1 and CaMKK2 activities by 4 and 17%, respectively (Fig. 2).
Figure 2.

Kinase selectivity profile for SBI-0206965 (0.25 μm) and compound C (2.5 μm). Profiling (n = 2 per kinase, with or without inhibitor) was performed using the MRC-PPU Express Screen service. MKK1 (activated 180% by SBI-0206965) is excluded from the left-hand panel for clarity.
SBI-0206965 is a mixed-type AMPK inhibitor
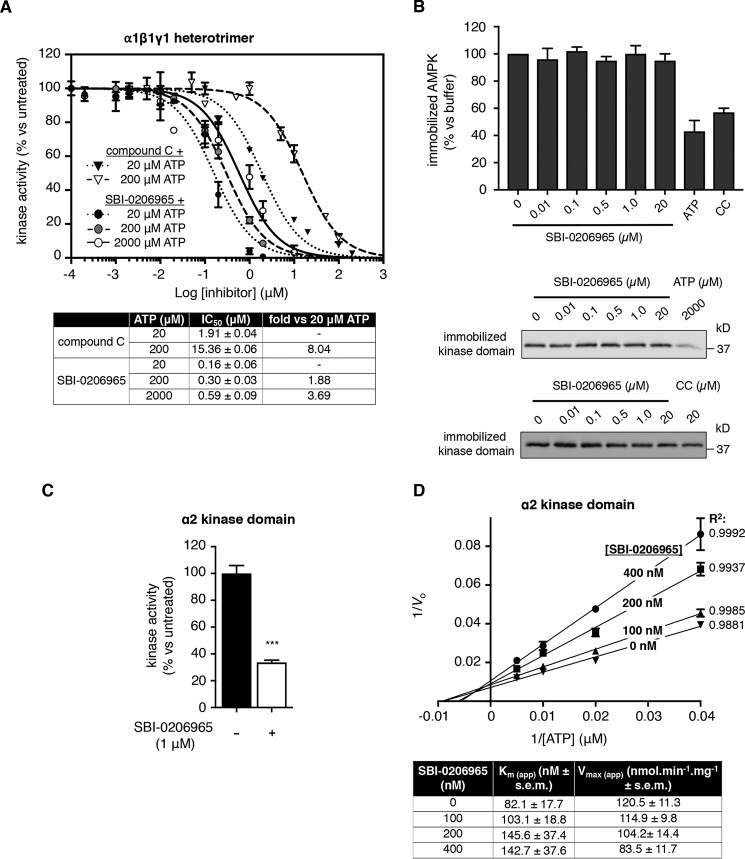
We investigated the inhibitory mechanism of SBI-0206965 by performing dose-response assays at different ATP concentrations using α1β1γ1 AMPK. SBI-0206965 IC50 increased <4-fold from 0.16 ± 0.06 to 0.59 ± 0.09 μm with 100-fold elevation in [ATP] (from 20 to 2000 μm), whereas compound C displayed conventional ATP-competitive inhibitor kinetics, with IC50 increasing 8-fold from 1.91 ± 0.04 to 15.36 ± 0.06 μm across a 10-fold elevation in [ATP] (from 20 to 200 μm) (Fig. 3A). Whereas ATP (2000 μm) and compound C (20 μm) blocked binding of Thr-172–phosphorylated, AMPK α2 kinase domain to ATP-agarose, SBI-0206965 (≤20 μm) was ineffective (Fig. 3B). However, SBI-0206965 (1 μm) inhibited the activity of phosphorylated α2 kinase domain to a similar extent as the AMPK heterotrimer at 200 μm ATP (Fig. 3C), confirming that the kinase domain alone encapsulates the SBI-0206965 binding site. We next tested the effect of varying ATP concentration on the activity of phosphorylated α2 kinase domain across a range of [SBI-0206965]. SBI-0206965 displayed kinetic properties indicative of mixed-type inhibitor modality, as the Ks for ATP increased and Vmax decreased with increasing [SBI-0206965] (Fig. 3D and Fig. S2, A and B). A secondary plot displaying the relationship between Km/Vmax (slopes on the Lineweaver–Burk plot (Fig. 3D)) and [SBI-0206965] generated a Ki of 261.4 nm (Fig. S2C). This is in contrast to the apparent αKi (also commonly termed Ki′) of 892.3 nm (Fig. S2D), which indicates that SBI-0206965 has a lower affinity for AMPK when ATP is bound. Equally, ATP has a lower affinity for AMPK when SBI-0206965 is bound (Ks(ATP) = 82.1 μm (Fig. 3D) versus αKs(ATP) =280.5 μm (derived from the equation, αKi/Ki = αKs/Ks)). In the context of AMPK regulation, these kinetic properties of SBI-0206965 are consistent with mixed-type inhibition.
Figure 3.
SBI-0206965 is an AMPK type II inhibitor. A, inhibition of AMPK α1β1γ1 phosphorylation of S108tide by SBI-0206965 and compound C at varying ATP concentrations (20, 200, and 2000 μm for SBI-0206965; 20 and 200 μm for compound C). Error bars, mean percentage kinase activity versus untreated ± S.E. (error bars) (n = 3). The table shows calculated IC50 values for each combination of inhibitor/ATP concentration and -fold increase in IC50 with increasing ATP concentration. B, ATP-agarose competitive binding assay. Thr-172–phosphorylated AMPK α2 kinase domain was incubated with ATP-agarose in the presence of SBI-0206965, ATP, or compound C (CC), as indicated. Immobilized AMPK was measured by immunoblot for α-subunit. Representative immunoblots of two individual experiments are shown. C, inhibition of α2 kinase domain phosphorylation of S108tide by SBI-0206965 (1 μm). Error bars, mean percentage kinase activity versus untreated ± S.E. (n = 3). ***, p < 0.001 by unpaired two-tailed Student's t test versus untreated kinase domain. D, Lineweaver–Burk (double reciprocal) plot showing inhibition of α2 kinase domain at four fixed SBI-0206965 concentrations (0–400 nm as indicated). Assays were performed in the presence of varying ATP concentrations (25–200 μm). The table shows apparent Km and Vmax values across SBI-0206965 dose range, calculated from data presented in Fig. S2A.
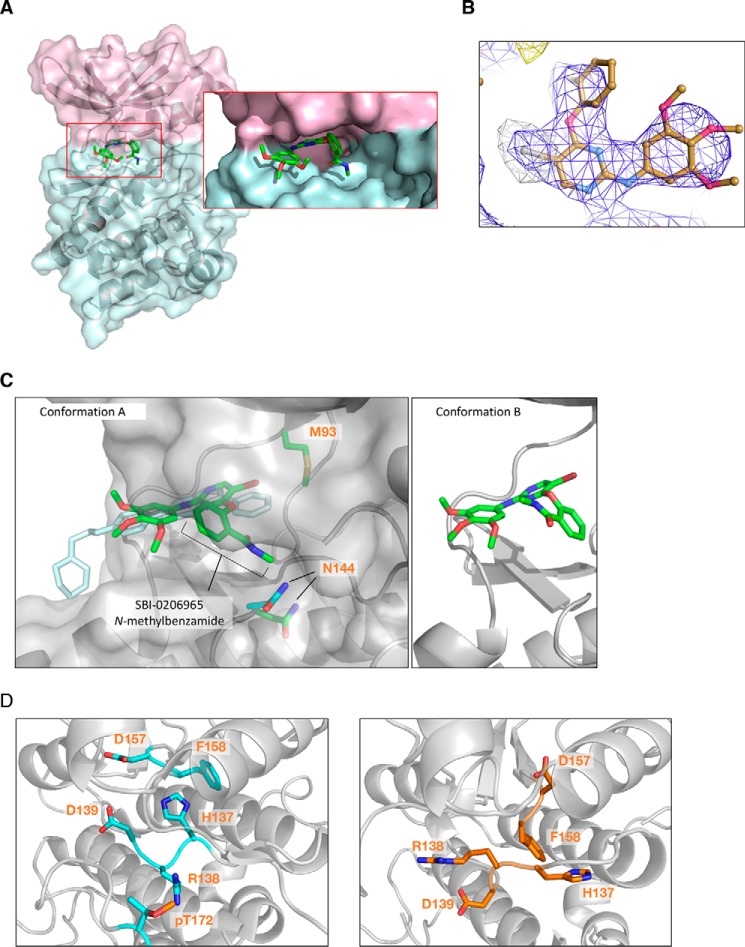
To confirm the binding mode of SBI-0206965, we solved a 2.9 Å resolution crystal structure of the inhibitor complexed to α2 kinase domain (residues 6–278), in which the activation loop Thr-172 was mutated to the phosphomimetic Asp (Table S1). A similar crystallization construct was used previously to visualize the compound C–binding pocket (26) (PDB entry 3AQV) and adopted a structure essentially identical to that of the Thr-172–phosphorylated WT kinase domain in the heterotrimeric complex (16, 19). SBI-0206965 was found to occupy a pocket located between the kinase N- and C-lobes and hinge region, which partially overlaps the compound C–binding site (Fig. 4A and Fig. S3A). Specifically, the trimethoxyphenyl and pyrimidinyl rings of SBI-0206965 lie approximately within the same plane as, and overlap substantially with, the phenyl and pyrazolo[1,5-a]pyrimidinyl rings, respectively, of the compound C core. The positioning of the electronegative bromine atom of SBI-0206965, probably contributing to increased potency relative to compound C, was confirmed by anomalous scattering at 13.6 keV (Fig. 4B) that revealed a large single peak for bromine (Table S1). The bromine moiety was found to occupy a large cavity bordered by residues Val-30, Lys-45, Ile-77, Met-93, Ala-156, and Asn-162.
Figure 4.
A, crystal structure of α2 kinase domain (residues 6–278) (T174D) complexed to SBI-0206965 in conformation A. Pink, N-lobe; blue, C-lobe. B, anomalous difference map at 3.0 Å of SBI-0206965 bromine contoured to 5.0 σ collected at the bromine edge (13.6 keV). The anomalous bromine peak (white mesh) strongly supports the placement of SBI-0206965 within the electron density (blue mesh). C, left, overlapping SBI-0206965 (green, conformation A) and compound C (cyan) binding sites. Surface represents SBI-0206965–bound α2. Gatekeeper residue α2-Met-93 and catalytic loop residue Asn-144 are shown (green, SBI-0206965 complex; cyan, compound C complex). Right, SBI-0206965 in conformation B. D, comparison of HRD motifs in the active α2 kinase conformation (PDB 4ZHX) (left) and α2 kinase domain complexed to SBI-0206965 (right).
We could not unambiguously place the N-methylbenzamide group of SBI-0206965 within the electron density; therefore, we modeled two distinct conformations with equal occupancy (Fig. 4C). In one position (conformation A), the aromatic ring is rotated almost 80° with respect to the compound plane, and the N-methyl group extends into the space occupied by the catalytic loop, Mg2+-chelating residue Asn-144, and the ATP α-phosphate in structures of active kinases (35). In the alternate position (conformation B), the benzamide ring is rotated 180° relative to conformation A, and the N-methyl group is directed toward P-loop residues Leu-22 and Gly-23 (Fig. 4C). In both conformations, SBI-0206965 makes two electrostatic contacts with the main chain of Val-96 in the kinase hinge region; otherwise, drug binding is mainly stabilized by hydrophobic contacts with hinge (Tyr-95), N-lobe (Leu-22, Ala-43, and the gatekeeper residue Met-93), and C-lobe (Ile-77, Gly-99, Glu-100, Leu-146, and Ala-156) residues.
Comparisons with either apo- (PDB entry 2YZA), compound C-complexed (26), or activated (PDB entry 4ZHX) (19) kinase domain structures reveal the α2/SBI-0206965 complex contains many of the hallmarks of an unproductive kinase. In both compound C- and SBI-0206965-bound structures, the C-α-helix adopts a “swung-out” position, ensuring that the glutamate–lysine salt bridge, required for efficient phosphoryl transfer, is unformed (Fig. S3A). These residues (Lys-45 and Glu-64) are instead interdigitated by the activation loop residue Ser-161, hydrogen bonding with Glu-64. Glu-64 forms a further hydrogen bond with the backbone of Leu-160. Compared with the compound C-bound structure, the DFG motif (Asp157-Phe-Gly159) in the activation loop of our SBI-0206965-bound structure is displaced away from the ATP active site by a maximum of 2.9 Å (Phe-158 αC), presumably as a consequence of the nonplanar structure of SBI-0206965 relative to compound C. However, in common with the α2/compound C structure, this motif adopts a unique conformation that is neither “DFG-in,” in which Asp-157 is appropriately positioned to chelate Mg2+ ions required for coordination of ATP phosphate groups, nor classical “DFG-out,” in which the motif flips by 180° such that the Phe side chain now occludes the ATP pocket and Asp-157 is no longer able to coordinate Mg-ATP for catalysis (Fig. S3B). Instead, the inhibited α2 DFG motif adopts an intermediate conformation; Asp-157 is sterically hindered from adopting the “DFG-in” conformation by the compound N-methylbenzamide and is oriented toward the N-lobe (Fig. S3B), with Phe-158 vacating the regulatory R-spine to displace His-137 in the kinase HRD motif (Fig. 4D). A consequence of this displacement is rearrangement of the HRD backbone and positioning of the Arg-138 side chain to prevent coordination with the phosphate group of pThr-172.
SBI-0206965 inhibits cellular AMPK signaling
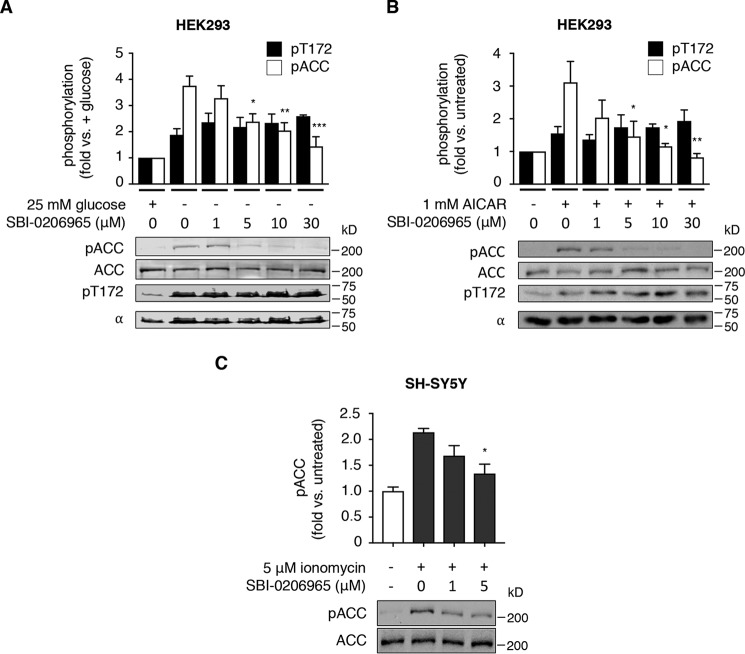
We explored the efficacy of SBI-0206965 to inhibit AMPK signaling in a range of cell lines. In HEK293 cells, glucose starvation triggered a 1.9-fold increase in pThr-172, indicative of energy stress, and this was accompanied by a 3.8-fold increase in pACC (Fig. 5A). The addition of SBI-0206965 to glucose-free medium significantly suppressed pACC at concentrations of ≥5 μm, probably due to direct AMPK inhibition because SBI-0206965 did not induce reductions in pThr-172, reflecting suppressed LKB1 activity (Fig. 5A). SBI-0206965 concentrations of ≥5 μm also blocked AMPK signaling in HEK293 cells treated with AICAR (Fig. 5B); however, this was probably due to inhibition of AICAR cellular uptake, reminiscent of compound C, because SBI-0206965 ≥ 5 μm also resulted in significantly reduced intracellular ZMP accumulation (Fig. S4A). SBI-0206965 ≤ 30 μm did not significantly affect ratios of adenine nucleotides in HEK293 cells, indicative of mitochondrial toxicity; however, we note a trend toward reduced adenylate energy charge at 30 μm (Fig. S4B). 5 μm SBI-0206965 also significantly suppressed pACC in the neuronal cell line SH-SY5Y, in which CaMKK2-mediated AMPK signaling was triggered by the addition of the calcium ionophore ionomycin (Fig. 5C).
Figure 5.
SBI-0206965 suppresses AMPK signaling in diverse cultured cell lines. Shown are representative immunoblots for pACC and pThr-172 from prepared lysates. HEK293 cells were incubated with glucose-free medium (A) and SBI-0206965 for 4 h or AICAR and SBI-0206965 for 1 h (B), as indicated. Error bars, mean -fold change in phosphorylation ± S.E. (error bars) (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001 by one-way ANOVA with post hoc Dunnett's multiple-comparison test versus glucose-starved (A) or AICAR-treated (B) cells. C, SH-SY5Y neuronal cells were co-incubated with CaMKK2 activator ionomycin and SBI-0206965 for 1 h, as indicated. Error bars, mean -fold change in phosphorylation ± S.E. (n = 3). *, p < 0.05 by one-way ANOVA with post hoc Dunnett's multiple-comparison test versus ionomycin-treated cells.
Discussion
SBI-0206965 was originally identified as an ATP-competitive inhibitor of the autophagy initiator kinase ULK1, with the ability to suppress cellular ULK signaling and ULK1-mediated survival of lung cancer and glioblastoma cells when coupled with nutrient stress (34). In this initial report, AMPK signaling in MEFs (evidenced as ACC phosphorylation) was unchanged following incubation with 50 μm SBI-0206965, despite increased Thr-172 phosphorylation, which may have arisen from fluctuations in adenine nucleotide ratios at high concentrations (Fig. S4B). SBI-0206965 has subsequently been shown to suppress non-small-cell lung cancer cell growth (36). We now show that SBI-0206965 can be repurposed as a potent and kinase-selective inhibitor of the metabolic coordinator AMPK, providing a useful alternative to the almost singularly used, yet promiscuous, compound C. Because AMPK is a major positive regulator of autophagy through direct phosphorylation of ULK1 (37) and other autophagy-associated proteins (e.g. VSP34 and Beclin-1), simultaneous inhibition of both AMPK and ULK1 signaling supports the application of SBI-0206965 as a highly effective suppressor of prosurvival autophagic responses in tumor cells. We previously used SBI-0206965 as an agent to confirm ULK1-mediated phosphorylation of the drug-sensitizing AMPK β1-subunit residue Ser-108, until recently regarded as an autophosphorylation site (8). Our demonstration now that SBI-0206965 also inhibits AMPK does not detract from our previous conclusions, because Ser-108 autophosphorylation is a cis event (10), and in our study, kinase-inactive AMPK enzyme was used as the cellular substrate.
Our biochemical characterization of SBI-0206965 as an AMPK antagonist highlights several limitations of use that demand consideration. Both Egan et al. (34) and our study here indicate a highly selective kinase profile for SBI-0206965; however, it must be noted that our combined, activity-based analyses cover only 57 kinases besides AMPK α1 and α2, representing just 11% of the human kinome. In light of our data implicating AMPK as being able to accommodate mixed-type inhibitors, we consider that activity, rather than ATP-competitive binding, is a more definitive metric of inhibitor selectivity. Besides ULK1 and -2, JAK3 and Src are flagged in both analyses as possible kinase targets for SBI-0206965, and we cannot exclude the possibility that others exist. We strongly advise against using SBI-0206965 in combination with AICAR when investigating cellular AMPK signaling. We also note that cellular incubations at high concentrations may induce Thr-172 phosphorylation through fluctuations in AMP/ATP and ADP/ATP ratios, rather than via stimulation of LKB1-mediated phosphorylation as with SU6656, which has the potential to confound interpretation of results.
Structural characteristics used to classify small-molecule kinase inhibitors label SBI-0206965 as a type IIb AMPK inhibitor (38, 39). Foremost, the SBI-0206965–binding pocket overlaps the ATP site in the α2 kinase domain, with the DFG motif positioned in a nonclassical “out” conformation. The Asp-157 side chain is not “in” (i.e. unable to coordinate phosphate-stabilizing Mg2+ ions), and Asn-144/Phe-158 (7.7 Å) and Glu-64/Phe-158 (10.1 Å) Cα atomic distances are consistent with a “DFG-out” classification (40). Additionally, the SBI-0206965 molecule does not extend into the back cleft of the α2 kinase domain, and the R-spine is distorted upon binding. Type IIb inhibitors bind to sites that incompletely overlap the ATP active site and are usually regarded as ATP-competitive. However, members of this inhibitor class have also been described as “ATP noncompetitive” (41, 42) and “indirectly ATP-competitive” (43), perhaps because comprehensive enzyme kinetic profiling is rarely reported alongside structural studies.
We have confirmed that SBI-0206965 partially shares its binding site on AMPK kinase domain with ATP but under our assay conditions is not ATP-competitive, instead displaying a mixed-type inhibition profile (Fig. 3D). Mixed-type inhibition, by definition, is a conceptual mixture of competitive (increased Km substrate) and uncompetitive inhibition (reduced Vmax), in which binding of the inhibitor affects substrate binding, and vice versa (44). From a structural perspective, mixed-type inhibition would appear incompatible with overlapping inhibitor/substrate binding sites. However, numerous studies have reported mixed-type inhibition by agents that have either been demonstrated to bind at the enzyme's active site or most likely do (i.e. transition state inhibitor analogues) (45–50). Specifically, a type II, active site binding inhibitor of insulin-like growth factor 1 receptor tyrosine kinase activity has been reported to display mixed-type kinetics with respect to ATP (45), whereas the tyrphostin inhibitor AG1296 displayed either competitive or mixed-type characteristics, depending on the activation state of its target kinase in platelet-derived growth factor receptor (46). Additionally, crystal structures of adenylate cyclase complexed to P-site inhibitors (adenine nucleosides/adenine nucleoside 3′-phosphates) show active site binding; these inhibitors display either noncompetitive (a special from of mixed inhibition) or uncompetitive kinetic properties (47). Active site binding by mixed inhibitors has also been inferred by direct methods (51, 52) or from close structural relationship to the substrate (53, 54). Type II kinase inhibitors such as SBI-0206965 probably stabilize the “DFG-out” conformation, an arrangement less favorable for ATP binding (55). Thus, the binding sites occupied by SBI-0206965 or ATP represent distinct conformational arrangements, each reducing the binding affinity of the competing ligand.
SBI-0206965 increased Km(ATP), indicating a preference for binding to the free enzyme rather than the enzyme-substrate complex. This is consistent with the location of the inhibitor-binding site resolved in our structure and our observation that SBI-0206965 cannot displace AMPK from ATP-agarose (Fig. 3C). This indicates that SBI-0206965 can only bind to an actively cycling AMPK, as the binding site must first become available following release of the ADP product from the active site. Furthermore, for the inhibitor-bound AMPK complexes, [ATP] in the activity assay may be below Ks ATP, precluding an increase of IC50 at the same rate as increase of [ATP], as dictated by the Cheng–Prusoff equation (56). This probably also explains why SBI-0206965 is very effective at inhibiting AMPK in cells; type II inhibitors typically display high cellular potency, whereas millimolar [ATP] often prevents type I inhibitors (e.g. compound C) from maintaining the potency observed in cell-free activity assays (43).
In summary, we show here that SBI-0206965 displays preferable characteristics, relative to compound C, as an AMPK inhibitor in vitro. Further studies are required to reveal its efficacy in vivo; however, biochemical and structural data provided in this study suggest that SBI-0206965 is a promising lead for the development of a new class of AMPK inhibitors with therapeutic potential.
Experimental procedures
Reagents
Antibodies for pan-AMPK α (catalog no. 2793, 1:1,000 dilution), and phosphospecific antibodies for α-pThr-172 (catalog no. 2535, 1:1,000 dilution) and pACC (catalog no. 3661, 1:1,000 dilution) were from Cell Signaling Technology. Streptavidin-IRDye 680RD (1:20,000 dilution) and anti-rabbit IgG IRDye 680 and anti-mouse IgG IRDye 800 fluorescent-labeled secondary antibodies (1:10,000 dilution) were from LI-COR Biosciences. GSH-Sepharose 4B (catalog no. 17075601) and streptavidin-Sepharose were from GE Life Sciences. Recombinant LKB1/MO25/STRAD was from Sigma (catalog no. SRP0246). SBI-0206965 was from ApexBio. ATP-agarose was from Innova Biosciences. cOmplete protease inhibitor mixture was from Roche Applied Science. DNA oligonucleotides and all other reagents were from Sigma.
Kinase inhibitor profiling
Inhibitor profiling (Express screen) was performed by the International Centre for Kinase Profiling, Medical Research Council Protein Phosphorylation and Ubiquitylation Unit (MRC-PPU), University of Dundee, UK.
Expression constructs
Assays
pET28b plasmid for expression of N-terminal His6 tag α2 kinase domain (residues 6–279) fusion protein was a generous gift from Dr. Dean Littler (Monash University). pET DUET constructs for Escherichia coli expression of AMPK heterotrimer α2β1γ1 have been described (10, 19).
Crystallization
pET21b plasmid for expression of α2 kinase domain (residues 6–278) incorporating T172D mutation, with a PreScission protease cleavable N-terminal His6 tag, was synthesized by General Biosystems. Constructs were sequence-verified, and expressed constructs were mass-verified by TOF MS.
Protein production
Heterotrimeric human AMPK GST-α1β1γ1 and GST-α2β1γ1, and human FLAG-ULK1 were produced in mammalian COS7 cells as described (8). AMPK and ULK1 were purified using GSH-Sepharose and anti-FLAG M2 affinity gel, respectively. FLAG-CaMKK2 was produced in Sf21 insect cells as described (6) and purified using anti-FLAG M2 affinity gel. Thr-172–phosphorylated AMPK His-α2(6–279) kinase domain was produced in bacteria as described (10). Briefly, the pET28b-α2(6–279) construct was transformed into E. coli strain Rosetta (DE3) and expressed by isopropyl 1-thio-β-d-galactopyranoside induction. Cells were ruptured using a precooled EmulsiFlex-C5 homogenizer (Avestin) and protein immobilized on nickel-Sepharose before phosphorylation with CaMKK2. Protein was eluted using 400 mm imidazole and buffer-exchanged with buffer A (50 mm Tris·HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 0.02% Tween 20) using a PD10 desalt column. A similar procedure was used to produce unphosphorylated AMPK α2β1γ1, except the CaMKK2 incubation step was omitted.
Protein crystallization
His-α2(6–278) (T174D) was generated as described above for His-α2(6–279), except the CaMKK2 incubation step was omitted. His tag was removed with GST-tagged PreScission protease treatment (overnight, 4 °C) after the buffer exchange step. PreScission protease was removed using GSH-Sepharose and α2(6–279) (T174D) repurified by SEC. Concentrated α2(6–279) (T174D) (4 mg/ml) was incubated with 0.5 mm SBI-0206965 on ice for 30 min and centrifuged at 10,000 rpm for 3 min before setting crystallization experiments. Protein was mixed equally 1:1 at room temperature with a reservoir solution containing 9–14% ethanol, 5 mm MgCl2, 10 mm tris(2-carboxyethyl)phosphine, and 3–7 mm MnCl2. Diffraction quality crystals were obtained through streak seeding with a cat's whisker. Crystals appeared after 2–3 days and reached full size after 1–2 weeks. Crystals were then incubated in a cryoprotectant-containing reservoir solution with an addition of 25% ethylene glycol. Data were collected on both MX1 and MX2 beamlines at the Australian Synchrotron (Melbourne, Australia). Data were processed and integrated using XDS (57) and scaled using AIMLESS from the CCP4 suite (58). The structure was solved by molecular replacement using Phaser from the CCP4 suite (59) and 3AQV as the search model. Iterative rounds of model building and refinement were performed using Coot (60) and Buster (https://www.globalphasing.com/buster/)6 (61), respectively. Data for the bromine anomalous map was collected on the MX2 beamline at the Australian Synchrotron near the bromine edge at 13.6 KeV, calculated on the X-ray anomalous scattering website (http://skuld.bmsc.washington.edu/scatter/AS_form.html).6 An anomalous map was generated by Phenix.refine (62). SBI-0206965 molecular coordinates and restraints were generated using the PRODRG web server (63). Structural validation was performed using Molprobity (64). Omit maps were generated using Buster, and figures were created using PyMOL.
S108tide activity assay
AMPK and ULK1 activities were determined by phosphorylation of the AMPK β1 Ser-108 peptide (S108tide) using 100 μm S108tide, 5 mm MgCl2, and [γ-32P]ATP in a 25-μl reaction volume at 30 °C. Reactions were terminated after 10 min by spotting 15 μl onto P81 phosphocellulose paper (Whatman) and washing in 1% phosphoric acid. Radioactivity was quantified by scintillation counting.
LKB1 phosphorylation assays
200 ng of unphosphorylated His-α2β1γ1 was incubated with 20 ng of LKB1/STRADα/MO25 in the presence of buffer A supplemented with 2 mm MgCl2, 200 μm ATP, and compounds (final 1% DMSO) or 1% DMSO vehicle, as indicated, at 32 °C. Reactions were terminated by the addition of SDS sample buffer and immunoblotted for pThr-172.
Immunoblotting
Samples were separated by SDS-PAGE on a 12% gel (7% for ACC), previously enriched using streptavidin-Sepharose (10), and transferred to Immobilon-FL polyvinylidene difluoride membrane (EMD Millipore). Membrane was blocked in PBS + 0.1% Tween 20 (PBST) with 2% nonfat milk for 30 min at 22 °C and then incubated for either 1 h or overnight with primary antibodies (dilutions in PBST). After washes with PBST, membranes were incubated with anti-rabbit or anti-mouse IgG secondary antibodies, fluorescently labeled with IR680 or IR800 dye, for 1 h. Immunoreactive bands were visualized on an Odyssey® IR imaging system with densitometry analyses determined using ImageStudioLite software (LI-COR Biosciences).
ATP-agarose immobilization
20 μg of Thr-172–phosphorylated His-α2(6–279) kinase domain was incubated with ATP/compounds for 30 min at room temperature, before the addition of 10 μl of ATP-agarose suspended in 200 μl of buffer B (50 mm HEPES, pH 7.4, 100 mm NaCl, 10% glycerol, 5 mm MgCl2). Beads were incubated on a rotating wheel at 4 °C for 2 h, before washing three times with 500 μl of buffer A supplemented with 0.1% Tween 20. Beads were resuspended in 15 μl of SDS sample buffer, boiled, and immunoblotted for AMPK α.
Cell culture and incubations
HEK293 and SH-SY5Y mammalian cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and 1% penicillin/streptomycin antibiotics, at 37 °C with 5% CO2. Cells were incubated with fresh DMEM for 3 h, before simultaneous incubation with glucose-free DMEM and or/compounds as indicated. Glucose-starved cells were washed 4 h post-treatment with ice-cold PBS and harvested by rapid lysis using ice-cold lysis buffer (50 mm Tris·HCl, pH 7.4, 150 mm NaCl, 50 mm NaF, 1 mm sodium pyrophosphate, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1% Triton X-100, and cOmplete protease inhibitor mixture). AICAR/ionomycin-treated cells were harvested similarly, except this was performed 1 h post-treatment. Lysates were clarified by centrifugation (14,000 rpm; 3 min; 4 °C) and flash-frozen in liquid N2 until processed.
Nucleotide measurements
Adenine nucleotides and adenylate energy charge were measured by LC-MS from HEK293 perchlorate extracts, as described previously (32).
ZMP measurements
All ZMP measurements were acquired using LC-MS from perchlorate extracts, using a method similar to that described previously for nucleotides. Briefly, LC conditions were optimized for a 150-mm (length) and 0.5-mm (inner diameter) Hypercarb column (3 μm; Thermo Fisher Scientific Australia Pty. Ltd.). The LC solvent system was (A) 25 mm triethylammonium bicarbonate buffer at pH 7.8 and (B) acetonitrile with 0.1% TFA. ZMP was eluted at a flow rate of 500 μl/min in a gradient program consisting of 100% A (5 min), 0–25% B (10 min), 50–80% B (5 min), and 100% B (5 min). Data were analyzed with Multiquant 2.0.2, utilizing the area under the LC chromatogram for the corresponding ZMP peak. Calibration curves were obtained by linear regression of the peak area ratio of a ZMP standard. The MS conditions and MRM values for ZMP were optimized by separate infusion of 1 μg/ml solution in 25 mm triethylammonium bicarbonate buffer at a flow rate of 50 μl/min.
Statistical analysis
The data are presented as mean values ± S.E. of at least three independent experiments unless stated. Statistical analyses were performed using GraphPad Prism version 7.0c. Ordinary one-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test was used for all comparisons unless stated.
Author contributions
T. A. D., C. G. L., A. H., S. G., R. J. R., A. J. O., L. M. L., K. R. W. N., and N. X. Y. L. data curation; T. A. D., A. H., S. G., R. J. R., A. J. O., L. M. L., K. R. N., N. X. Y. L., B. E. K., J. W. S., and J. S. O. formal analysis; T. A. D., C. G. L., and A. H. methodology; T. A. D., C. G. L., J. W. S., and J. S. O. writing-original draft; L. F., J. W. S., and J. S. O. supervision; L. F., B. E. K., and J. S. O. funding acquisition; B. E. K., J. W. S., and J. S. O. conceptualization; J. S. O. project administration; J. S. O. writing-review and editing.
Supplementary Material
This work was supported by grants from the Australian Research Council (ARC), Cancer Australia, the National Health and Medical Research Council (NHMRC), and the Jack Brockhoff foundation (Grant JBF-4206, 2016). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
The atomic coordinates and structure factors (code 6BX6) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- AMPK
- AMP-activated protein kinase
- ACC
- acetyl-CoA carboxylase
- AID
- autoinhibitory
- CBM
- carbohydrate-binding module
- CaMKK2
- Ca2+/calmodulin-dependent protein kinase kinase 2
- AICAR
- 5-aminoimidazole-4-carboxamide ribonucleoside
- PDB
- Protein Data Bank
- DMEM
- Dulbecco's modified Eagle's medium
- ANOVA
- analysis of variance
- SBI-0206965
- 2-(5-bromo-2-(3,4,5-trimethoxyphenylamino)pyrimidin-4-yloxy)-N-methylbenzamide.
References
- 1. Steinberg G. R., and Kemp B. E. (2009) AMPK in health and disease. Physiol. Rev. 89, 1025–1078 10.1152/physrev.00011.2008 [DOI] [PubMed] [Google Scholar]
- 2. Hardie D. G., Schaffer B. E., and Brunet A. (2016) AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 26, 190–201 10.1016/j.tcb.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki A., Okamoto S., Lee S., Saito K., Shiuchi T., and Minokoshi Y. (2007) Leptin stimulates fatty acid oxidation and peroxisome proliferator-activated receptor α gene expression in mouse C2C12 myoblasts by changing the subcellular localization of the α2 form of AMP-activated protein kinase. Mol. Cell. Biol. 27, 4317–4327 10.1128/MCB.02222-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamia K. A., Sachdeva U. M., DiTacchio L., Williams E. C., Alvarez J. G., Egan D. F., Vasquez D. S., Juguilon H., Panda S., Shaw R. J., Thompson C. B., and Evans R. M. (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326, 437–440 10.1126/science.1172156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang C. S., Jiang B., Li M., Zhu M., Peng Y., Zhang Y. L., Wu Y. Q., Li T. Y., Liang Y., Lu Z., Lian G., Liu Q., Guo H., Yin Z., Ye Z., Han J., Wu J. W., Yin H., Lin S. Y., and Lin S. C. (2014) The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 20, 526–540 10.1016/j.cmet.2014.06.014 [DOI] [PubMed] [Google Scholar]
- 6. Oakhill J. S., Chen Z. P., Scott J. W., Steel R., Castelli L. A., Ling N., Macaulay S. L., and Kemp B. E. (2010) β-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK). Proc. Natl. Acad. Sci. U.S.A. 107, 19237–19241 10.1073/pnas.1009705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang J., Xu Z. X., Ding Z., Lu Y., Yu Q., Werle K. D., Zhou G., Park Y. Y., Peng G., Gambello M. J., and Mills G. B. (2015) Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance. Nat. Commun. 6, 7926 10.1038/ncomms8926 [DOI] [PubMed] [Google Scholar]
- 8. Dite T. A., Ling N. X. Y., Scott J. W., Hoque A., Galic S., Parker B. L., Ngoei K. R. W., Langendorf C. G., O'Brien M. T., Kundu M., Viollet B., Steinberg G. R., Sakamoto K., Kemp B. E., and Oakhill J. S. (2017) The autophagy initiator ULK1 sensitizes AMPK to allosteric drugs. Nat. Commun. 8, 571 10.1038/s41467-017-00628-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ross F. A., Jensen T. E., and Hardie D. G. (2016) Differential regulation by AMP and ADP of AMPK complexes containing different γ subunit isoforms. Biochem. J. 473, 189–199 10.1042/BJ20150910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott J. W., Ling N., Issa S. M., Dite T. A., O'Brien M. T., Chen Z. P., Galic S., Langendorf C. G., Steinberg G. R., Kemp B. E., and Oakhill J. S. (2014) Small molecule drug A-769662 and AMP synergistically activate naive AMPK independent of upstream kinase signaling. Chem. Biol. 21, 619–627 10.1016/j.chembiol.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 11. Viollet B., Horman S., Leclerc J., Lantier L., Foretz M., Billaud M., Giri S., and Andreelli F. (2010) AMPK inhibition in health and disease. Crit. Rev. Biochem. Mol. Biol. 45, 276–295 10.3109/10409238.2010.488215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corton J. M., Gillespie J. G., Hawley S. A., and Hardie D. G. (1995) 5-aminoimidazole-4-carboxamide ribonucleoside: a specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 229, 558–565 10.1111/j.1432-1033.1995.tb20498.x [DOI] [PubMed] [Google Scholar]
- 13. Fryer L. G., Parbu-Patel A., and Carling D. (2002) Protein kinase inhibitors block the stimulation of the AMP-activated protein kinase by 5-amino-4-imidazolecarboxamide riboside. FEBS Lett. 531, 189–192 10.1016/S0014-5793(02)03501-9 [DOI] [PubMed] [Google Scholar]
- 14. Cool B., Zinker B., Chiou W., Kifle L., Cao N., Perham M., Dickinson R., Adler A., Gagne G., Iyengar R., Zhao G., Marsh K., Kym P., Jung P., Camp H. S., and Frevert E. (2006) Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 3, 403–416 10.1016/j.cmet.2006.05.005 [DOI] [PubMed] [Google Scholar]
- 15. Hawley S. A., Fullerton M. D., Ross F. A., Schertzer J. D., Chevtzoff C., Walker K. J., Peggie M. W., Zibrova D., Green K. A., Mustard K. J., Kemp B. E., Sakamoto K., Steinberg G. R., and Hardie D. G. (2012) The ancient drug salicylate directly activates AMP-activated protein kinase. Science 336, 918–922 10.1126/science.1215327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao B., Sanders M. J., Carmena D., Bright N. J., Haire L. F., Underwood E., Patel B. R., Heath R. B., Walker P. A., Hallen S., Giordanetto F., Martin S. R., Carling D., and Gamblin S. J. (2013) Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4, 3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cokorinos E. C., Delmore J., Reyes A. R., Albuquerque B., Kjøbsted R., Jørgensen N. O., Tran J. L., Jatkar A., Cialdea K., Esquejo R. M., Meissen J., Calabrese M. F., Cordes J., Moccia R., Tess D., et al. (2017) Activation of skeletal muscle AMPK promotes glucose disposal and glucose lowering in non-human primates and mice. Cell Metab. 25, 1147–1159.e10 10.1016/j.cmet.2017.04.010 [DOI] [PubMed] [Google Scholar]
- 18. Hunter R. W., Foretz M., Bultot L., Fullerton M. D., Deak M., Ross F. A., Hawley S. A., Shpiro N., Viollet B., Barron D., Kemp B. E., Steinberg G. R., Hardie D. G., and Sakamoto K. (2014) Mechanism of action of compound-13: an alpha1-selective small molecule activator of AMPK. Chem. Biol. 21, 866–879 10.1016/j.chembiol.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langendorf C. G., Ngoei K. R., Scott J. W., Ling N. X., Issa S. M., Gorman M. A., Parker M. W., Sakamoto K., Oakhill J. S., and Kemp B. E. (2016) Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding. Nat. Commun. 7, 10912 10.1038/ncomms10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y. J., and Chern Y. (2015) AMPK-mediated regulation of neuronal metabolism and function in brain diseases. J. Neurogenet. 29, 50–58 10.3109/01677063.2015.1067203 [DOI] [PubMed] [Google Scholar]
- 21. Hardie D. G., and Alessi D. R. (2013) LKB1 and AMPK and the cancer-metabolism link: ten years after. BMC Biol. 11, 36 10.1186/1741-7007-11-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Popovics P., Frigo D. E., Schally A. V., and Rick F. G. (2015) Targeting the 5′-AMP-activated protein kinase and related metabolic pathways for the treatment of prostate cancer. Expert Opin. Ther. Targets 19, 617–632 10.1517/14728222.2015.1005603 [DOI] [PubMed] [Google Scholar]
- 23. Jeon S. M., and Hay N. (2015) The double-edged sword of AMPK signaling in cancer and its therapeutic implications. Arch. Pharm. Res. 38, 346–357 10.1007/s12272-015-0549-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galluzzi L., Pietrocola F., Bravo-San Pedro J. M., Amaravadi R. K., Baehrecke E. H., Cecconi F., Codogno P., Debnath J., Gewirtz D. A., Karantza V., Kimmelman A., Kumar S., Levine B., Maiuri M. C., Martin S. J., et al. (2015) Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880 10.15252/embj.201490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., and Moller D. E. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 10.1172/JCI13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Handa N., Takagi T., Saijo S., Kishishita S., Takaya D., Toyama M., Terada T., Shirouzu M., Suzuki A., Lee S., Yamauchi T., Okada-Iwabu M., Iwabu M., Kadowaki T., Minokoshi Y., and Yokoyama S. (2011) Structural basis for compound C inhibition of the human AMP-activated protein kinase α2 subunit kinase domain. Acta Crystallogr. D Biol. Crystallogr. 67, 480–487 10.1107/S0907444911010201 [DOI] [PubMed] [Google Scholar]
- 27. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., and Peterson R. T. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 10.1038/nchembio.2007.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emerling B. M., Viollet B., Tormos K. V., and Chandel N. S. (2007) Compound C inhibits hypoxic activation of HIF-1 independent of AMPK. FEBS Lett. 581, 5727–5731 10.1016/j.febslet.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nam M., Lee W. H., Bae E. J., and Kim S. G. (2008) Compound C inhibits clonal expansion of preadipocytes by increasing p21 level irrespectively of AMPK inhibition. Arch. Biochem. Biophys. 479, 74–81 10.1016/j.abb.2008.07.029 [DOI] [PubMed] [Google Scholar]
- 31. Lee Y., Park B. H., and Bae E. J. (2016) Compound C inhibits macrophage chemotaxis through an AMPK-independent mechanism. Biochem. Biophys. Res. Commun. 469, 515–520 10.1016/j.bbrc.2015.12.015 [DOI] [PubMed] [Google Scholar]
- 32. Scott J. W., Galic S., Graham K. L., Foitzik R., Ling N. X., Dite T. A., Issa S. M., Langendorf C. G., Weng Q. P., Thomas H. E., Kay T. W., Birnberg N. C., Steinberg G. R., Kemp B. E., and Oakhill J. S. (2015) Inhibition of AMP-activated protein kinase at the allosteric drug-binding site promotes islet insulin release (2015) Chem. Biol. 22, 705–711 10.1016/j.chembiol.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 33. Ross F. A., Hawley S. A., Auciello F. R., Gowans G. J., Atrih A., Lamont D. J., and Hardie D. G. (2017) Mechanisms of paradoxical activation of AMPK by the kinase inhibitors SU6656 and Sorafenib. Cell Chem. Biol. 24, 813–824.e4 10.1016/j.chembiol.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Egan D. F., Chun M. G., Vamos M., Zou H., Rong J., Miller C. J., Lou H. J., Raveendra-Panickar D., Yang C. C., Sheffler D. J., Teriete P., Asara J. M., Turk B. E., Cosford N. D., and Shaw R. J. (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 59, 285–297 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng J., Trafny E. A., Knighton D. R., Xuong N. H., Taylor S. S., Ten Eyck L. F., and Sowadski J. M. (1993) A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 49, 362–365 10.1107/S0907444993000423 [DOI] [PubMed] [Google Scholar]
- 36. Tang F., Hu P., Yang Z., Xue C., Gong J., Sun S., Shi L., Zhang S., Li Z., Yang C., Zhang J., and Xie C. (2017) SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol. Rep. 37, 3449–3458 10.3892/or.2017.5635 [DOI] [PubMed] [Google Scholar]
- 37. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., Hansen M., and Shaw R. J. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dar A. C., and Shokat K. M. (2011) The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling, Annu. Rev. Biochem. 80, 769–795 10.1146/annurev-biochem-090308-173656 [DOI] [PubMed] [Google Scholar]
- 39. Roskoski R., Jr. (2016) Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 103, 26–48 10.1016/j.phrs.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 40. Vijayan R. S., He P., Modi V., Duong-Ly K. C., Ma H., Peterson J. R., Dunbrack R. L. Jr., and Levy R. M. (2015) Conformational analysis of the DFG-out kinase motif and biochemical profiling of structurally validated Type II inhibitors. J. Med. Chem. 58, 466–479 10.1021/jm501603h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blanc J., Geney R., and Menet C. (2013) Type II kinase inhibitors: an opportunity in cancer for rational design. Anticancer Agents Med. Chem. 13, 731–747 10.2174/1871520611313050008 [DOI] [PubMed] [Google Scholar]
- 42. Garuti L., Roberti M., and Bottegoni G. (2010) Non-ATP competitive protein kinase inhibitors. Curr. Med. Chem. 17, 2804–2821 10.2174/092986710791859333 [DOI] [PubMed] [Google Scholar]
- 43. Liu Y., and Gray N. S. (2006) Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2, 358–364 10.1038/nchembio799 [DOI] [PubMed] [Google Scholar]
- 44. Cleland W. W. (1963) The kinetics of enzyme-catalyzed reactions with two or more substrates or products. II. Inhibition: nomenclature and theory. Biochim. Biophys. Acta 67, 173–187 10.1016/0926-6569(63)90226-8 [DOI] [PubMed] [Google Scholar]
- 45. Lesuisse D., Mauger J., Nemecek C., Maignan S., Boiziau J., Harlow G., Hittinger A., Ruf S., Strobel H., Nair A., Ritter K., Malleron J. L., Dagallier A., El-Ahmad Y., Guilloteau J. P., et al. (2011) Discovery of the first non-ATP competitive IGF-1R kinase inhibitors: Advantages in comparison with competitive inhibitors. Bioorg. Med. Chem. Lett. 21, 2224–2228 10.1016/j.bmcl.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 46. Kovalenko M., Rönnstrand L., Heldin C. H., Loubtchenkov M., Gazit A., Levitzki A., and Böhmer F. D. (1997) Phosphorylation site-specific inhibition of platelet-derived growth factor β-receptor autophosphorylation by the receptor blocking tyrphostin AG1296. Biochemistry 36, 6260–6269 10.1021/bi962553l [DOI] [PubMed] [Google Scholar]
- 47. Dessauer C. W., Tesmer J. J., Sprang S. R., and Gilman A. G. (1999) The interactions of adenylate cyclases with P-site inhibitors. Trends Pharmacol. Sci. 20, 205–210 10.1016/S0165-6147(99)01310-3 [DOI] [PubMed] [Google Scholar]
- 48. Westmark P. R., Kelly J. P., and Smith B. D. (1993) Photoregulation of enzyme activity: photochromic, transition-state-analogue inhibitors of cysteine and serine proteases. J. Am. Chem. Soc. 115, 3416–3419 10.1021/ja00062a003 [DOI] [Google Scholar]
- 49. Marmor S., Petersen C. P., Reck F., Yang W., Gao N., and Fisher S. L. (2001) Biochemical characterization of a phosphinate inhibitor of Escherichia coli MurC. Biochemistry 40, 12207–12214 10.1021/bi015567m [DOI] [PubMed] [Google Scholar]
- 50. Thomson J. M., Prati F., Bethel C. R., and Bonomo R. A. (2007) Use of novel boronic acid transition state inhibitors to probe substrate affinity in SHV-type extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 51, 1577–1579 10.1128/AAC.01293-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davidson W., Frego L., Peet G. W., Kroe R. R., Labadia M. E., Lukas S. M., Snow R. J., Jakes S., Grygon C. A., Pargellis C., and Werneburg B. G. (2004) Discovery and characterization of a substrate selective p38R inhibitor. Biochemistry 43, 11658–11671 10.1021/bi0495073 [DOI] [PubMed] [Google Scholar]
- 52. Ghani U., and Ullah N. (2010) New potent inhibitors of tyrosinase: novel clues to binding of 1,3,4-thiadiazole-2(3H)-thiones, 1,3,4-oxadiazole-2(3H)-thiones, 4-amino-1,2,4-triazole-5(4H)-thiones, and substituted hydrazides to the dicopper active site. Bioorg. Med. Chem. 18, 4042–4048 10.1016/j.bmc.2010.04.021 [DOI] [PubMed] [Google Scholar]
- 53. Bdolah A., and Feingold D. S. (1968) Uridine diphosphate d-glucose dehydrogenase of Aerobacter aerogenes. J. Bacteriol. 96, 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Patel M. S. (1972) The effect of phenylpyruvate on pyruvate metabolism in rat brain. Biochem. J. 128, 677–684 10.1042/bj1280677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Knight Z. A., and Shokat K. M. (2005) Features of selective kinase inhibitors. Chem Biol. 12, 621–637 10.1016/j.chembiol.2005.04.011 [DOI] [PubMed] [Google Scholar]
- 56. Cheng Y., and Prusoff W. H. (1973) Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 10.1016/0006-2952(73)90196-2 [DOI] [PubMed] [Google Scholar]
- 57. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek P., Roversi P., Sharff A., Smart O., Vonrhein C., and Womack T. (2011) BUSTER version 2.10.0, Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 62. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schüttelkopf A. W., and van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- 64. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.