Abstract
The spread of dengue (DENV) and Zika virus (ZIKV) is a major public health concern. The primary target of antibodies that neutralize DENV and ZIKV is the envelope (E) glycoprotein, and there is interest in using soluble recombinant E (sRecE) proteins as subunit vaccines. However, the most potent neutralizing antibodies against DENV and ZIKV recognize epitopes on the virion surface that span two or more E proteins. Therefore, to create effective DENV and ZIKV vaccines, presentation of these quaternary epitopes may be necessary. The sRecE proteins from DENV and ZIKV crystallize as native-like dimers, but studies in solution suggest that these dimers are marginally stable. To better understand the challenges associated with creating stable sRecE dimers, we characterized the thermostability of sRecE proteins from ZIKV and three DENV serotypes, DENV2–4. All four proteins irreversibly unfolded at moderate temperatures (46–53 °C). At 23 °C and low micromolar concentrations, DENV2 and ZIKV were primarily dimeric, and DENV3–4 were primarily monomeric, whereas at 37 °C, all four proteins were predominantly monomeric. We further show that the dissociation constant for DENV2 dimerization is very temperature-sensitive, ranging from <1 μm at 25 °C to 50 μm at 41 °C, due to a large exothermic enthalpy of binding of −79 kcal/mol. We also found that quaternary epitope antibody binding to DENV2–4 and ZIKV sRecE is reduced at 37 °C. Our observation of reduced sRecE dimerization at physiological temperature highlights the need for stabilizing the dimer as part of its development as a subunit vaccine.
Keywords: dengue virus (DENV), protein stability, virus assembly, thermodynamics, virus, dimerization, envelope protein, quaternary epitope antibody, temperature-dependent, Zika virus
Introduction
Dengue virus (DENV)3 infections have steadily been on the rise over the past 3 decades, with ∼390 million infections occurring each year (1, 2). A closely related flavivirus, Zika virus (ZIKV) has also become a major public health concern with the rapid spread of the virus throughout multiple countries and the discovery that diseases such as microcephaly in infants and Guillain-Barré syndrome in adults are associated with ZIKV infection (3). To date, there are no vaccines for ZIKV, with candidates still in clinical trials (4). The only licensed vaccine for DENV, Dengvaxia, is a tetravalent live-attenuated vaccine that has been shown to provide partial protection between DENV serotypes. Moreover, this vaccine is only recommended for use in people with partial immunity to DENVs because the vaccine appears to increase the risk of severe dengue disease when used in naive people (5–7). These facts highlight the need for alternative approaches for vaccine design and development against DENV and ZIKV (8).
DENV and ZIKV are closely related flaviviruses. Both viruses are structurally similar, displaying 180 copies of the envelope (E) glycoprotein as 90 anti-parallel homodimers organized in a herringbone pattern to cover the surface of the virion (9, 10). Truncation of the full-length E protein at the C terminus to remove the membrane-proximal and transmembrane domains results in a soluble ectodomain (aa 1–395) that is secreted from cells (11, 12). The structures of the soluble recombinant E proteins (sRecE) for DENV2–4 and ZIKV have been solved, revealing that sRecE adopts an intricate multidomain, primarily β-stranded structure consisting of three domains (DI, DII, and DIII) and crystallizes as a dimer, similar to the conformation of E dimers displayed on the virion surface (12–16).
DENV- and ZIKV-infected patients develop neutralizing and protective Abs that target epitopes on the E protein, which has led researchers to test whether sRecE or domains of the E protein can serve as effective vaccines for DENV (11, 17–19). Vaccine formulations containing E protein subunits have been shown to elicit neutralizing antibodies in humans and nonhuman primates, but unfortunately the titers were low and decreased quickly over time (11, 17, 18, 20). Recently, it has been discovered that many of the most potent neutralizing Abs isolated from human patients for DENV and ZIKV bind to quaternary epitopes that span across multiple E protein chains on the surface of the virion (21–24). Some DENV serotype–specific or ZIKV type–specific and strongly neutralizing Abs bind to quaternary epitopes that only require the formation of the homodimer (22, 25). A new class of human quaternary epitope Abs, named the E dimer epitope (EDE) Abs, bind to a conserved epitope across the E dimer and, strikingly, are broadly neutralizing against all four serotypes of DENV and ZIKV (16, 26). These results suggest that to create effective subunit vaccines, it will be important to engineer antigens that present these newly discovered quaternary epitopes. The dimer-specific epitopes are potentially the most straightforward to create with recombinant protein, and it has been proposed that sRecE dimers may function as effective vaccines (8, 27). For sRecE dimers to elicit EDE antibodies, it will be critical that the dimers be stable during vaccine formulation and long-term storage, as well as under physiological conditions during patient immunization (28).
Both DENV and ZIKV sRecE proteins crystallize as dimers, indicating that at high concentrations, the dimer is the dominant state. DENV2 and ZIKV sRecE monomers and dimers have been observed in solution, with the DENV2 sRecE being primarily monomeric at higher-salt conditions (16, 29–31). Also, EDE Abs do not bind to sRecE that is directly coated to hydrophobic or Ni-NTA ELISA plates but can bind to intact virion, further suggesting that the sRecE dimer epitopes are not maintained at low protein concentrations (21, 27).
Physiological temperature has been shown to induce “viral breathing”, attributed to increased E protein dynamics on the surface of flavivirus virions (32). Further investigation of DENV2 virions incubated at 37 °C revealed substantial structural E protein rearrangement and reduced E protein dimer contacts on the virion surface (33–36). These temperature effects have only recently been probed in the context of sRecE. Slon Campos et al. (37) reported improved DENV and ZIKV sRecE secretion in HEK293T cells by reducing the expression temperature from 37 to 28 °C and/or engineering a disulfide across the sRecE dimer interface, attributing secretion success to sRecE dimer formation. A recently published study from our laboratory (27) observed that the binding of DENV2 and ZIKV sRecE to quaternary Abs was temperature-dependent.
Here, we use a variety of biophysical methods and additional quaternary Ab-binding experiments to probe the thermostability of DENV2–4 and ZIKV sRecE monomers and dimers. We show that all of the sRecE proteins irreversibly unfold at moderate temperatures (46–53 °C) and that dimer formation is highly dependent on temperature. In-depth analyses of DENV2 sRecE reveals that the steep temperature dependence for dimerization is due to a large and exothermic enthalpy of binding (−79 kcal/mol).
Results
Expression and purification of DENV2–4 and ZIKV sRecE
DENV2–4 and ZIKV sRecE (aa 1–395) were constructed with prM for mammalian expression and secretion in EXPI293 (DENV2–3 and ZIKV) and DG44 CHO (DENV4) cells containing an N-terminal IL-2 secretion peptide and genetically encoded C-terminal His6 tag for purification (Fig. S1). As in other studies conducted with soluble recombinant E proteins, including antibody-binding studies, our construct does not include the C-terminal membrane-proximal and transmembrane domains (8). Relatively high expression yields were observed for DENV4 sRecE (11.5 mg/liter) and DENV2 sRecE (∼4 mg/liter), and lower yields were observed for DENV3 and ZIKV sRecE (∼1 mg/liter for each). High sRecE protein purity was achieved via nickel affinity chromatography followed by gel filtration (Fig. S2). CD spectral scans produced negative banding at 213–218 nm, indicating that the purified DENV2–4 and ZIKV sRecE were properly folded (Fig. S3).
Assessing oligomeric state and homodimer affinity of sRecE proteins
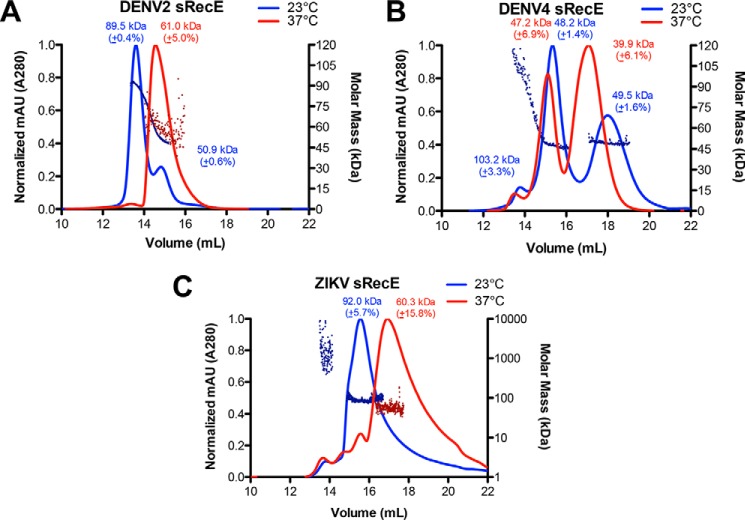
To assess the oligomeric state of sRecE proteins, size-exclusion chromatography with multiangle light scattering (SEC-MALS) experiments were performed at 23 °C with purified DENV2–4 and ZIKV sRecE. DENV2 sRecE eluted in two peaks, the major peak having a calculated molecular mass by MALS of 89.5 kDa and a minor peak mass of 50.9 kDa (Fig. 1A). The expected molecular mass for the monomer is 45.1 kDa; therefore, the data indicate that the protein is in equilibrium between a dimer and monomer with the dimer being more stable at these conditions. ZIKV sRecE eluted as two peaks, the major peak corresponding to a calculated molecular mass of 92.0 kDa, whereas aggregate was observed in a minor peak (Fig. 1C). The molecular mass of the monomer is 45.1 kDa, indicating that ZIKV sRecE is a dimer at these conditions, and unlike DENV2 sRecE, no monomer–dimer equilibrium was observed. Unexpectedly, the DENV4 sRecE elution profile contained three peaks, with a minor peak molecular mass calculated to be 103.3 kDa, followed by two consecutive major peaks with observed molecular masses at 48.2 and 49.5 kDa, which are near the expected mass for a monomer, 44.8 kDa (Fig. 1B). It is unclear why the DENV4 monomer elutes in two peaks from the gel filtration column. SEC analysis of deglycosylated DENV4 sRecE via peptide:N-glycosidase F treatment revealed similar traces observed in Fig. 1B, indicating that the presence of the two monomer peaks is not due to glycosylation (Fig. S4). One possibility is that the monomer is adopting two separate conformations with one of the conformations partially sticking to the column, which has been observed previously for other Class II fusion proteins (29, 38). When the data from the DENV4 SEC-MALS experiment are overlaid with the DENV2 SEC-MALS trace, it is apparent that the minor dimer peak from DENV4 coincides with the DENV2 dimer peak and that the first monomer peak from DENV4 coincides with the DENV2 monomer peak (Fig. S5B). Similar to DENV4, DENV3 sRecE also eluted with three peaks, with a minor peak molecular mass calculated as 83.7 kDa and the mass of the consecutive two major peaks as 49.3 and 52.7 kDa, respectively. Like DENV4 sRecE, DENV3 sRecE, with an expected molecular mass of 43.1 kDa, is mainly monomeric in solution and has a dimer–monomer equilibrium favoring the monomer (see Fig. S6A).
Figure 1.
Temperature dependence of sRecE oligomerization. A–C, SEC UV traces of DENV2 sRecE at 54 μm (A), DENV4 sRecE at 55 μm (B), and ZIKV sRecE at 21 μm (C) with MALS calculated molecular mass plotted on the right y axis. SEC-MALS analysis was performed at 23 °C (blue) and 37 °C (red). A Superdex 10/300 GL column was run using 1× PBS, pH 7.4 (see “Experimental procedures” for exact composition). For clarity, the MALS data are shown for DENV2 sRecE (A) at 23 °C and 37 °C, DENV4 sRecE (B) at 23 °C, and ZIKV sRecE (C) at both 23 and 37 °C. For DENV4 sRecE MALS data at 37 °C, see Fig. S5A.
We next probed how physiological temperature (37 °C) affects the equilibrium between DENV2 and ZIKV dimer and monomer. A SEC-MALS experiment with DENV2 and ZIKV sRecE was performed with the buffer, column, and all related equipment maintained at a temperature of 37 °C. DENV2 sRecE eluted predominantly as a single peak with a calculated molecular mass of 61.0 kDa and with the same retention time as the observed monomer in the 23 °C DENV2 sRecE experiment, indicating that at 37 °C, DENV2 sRecE is primarily monomeric (Fig. 1A). The observed molecular mass, 61.0 kDa, is higher than the mass of the monomer, 45.1 kDa, suggesting that at these conditions, there are still dimers in equilibrium with the monomer. Similar to DENV2, ZIKV sRecE eluted with a longer retention time at 37 °C with a calculated molecular mass of 60.3 kDa, indicating that ZIKV sRecE also was mostly monomer at 37 °C (Fig. 1C). SEC-MALS with DENV4 sRecE at 37 °C produced SEC traces similar to those observed at 23 °C. In contrast to the 23 °C experiment, at 37 °C, the final peak was the major eluting fraction instead of the monomer peak participating in the monomer–dimer exchange observed at 23 °C (Fig. 1B and Fig. S5A).
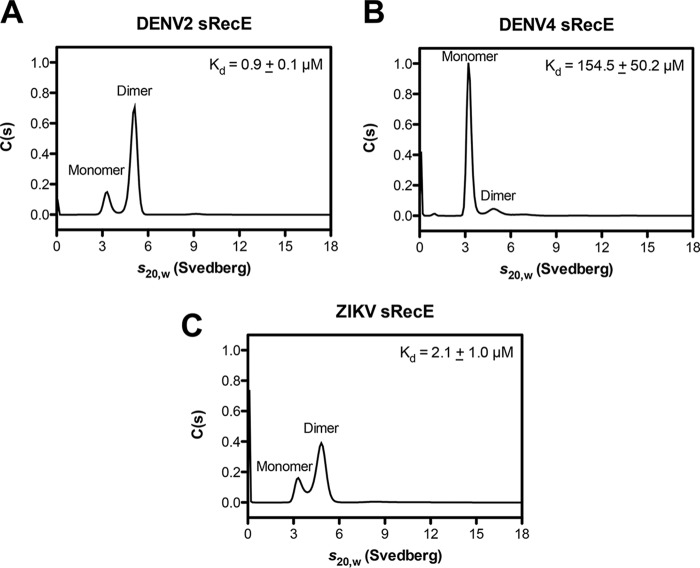
With the observed differences in DENV and ZIKV sRecE oligomeric state, we sought to measure the affinity of the sRecE homodimer and independently measure sRecE oligomeric state in solution by analytical ultracentrifugation (AUC) sedimentation velocity experiments. We performed each experiment at 21 °C at a concentration of 0.4 mg/ml (∼9 μm) for DENV2–4 and ZIKV sRecE. Both monomer and dimer conformations were observed for all sRecE tested (Fig. 2 (A–C) and Fig. S7). Consistent with our SEC-MALS data at 23 °C, DENV3 and DENV4 formed less stable dimers with Kd values for dimerization of 30.5 ± 10.6 and 154.5 ± 50.2 μm, respectively (Fig. 2B and Fig. S6C). Lower Kd values were observed for DENV2 sRecE (0.9 ± 0.1 μm) and ZIKV sRecE (2.1 ± 1.0 μm) (Fig. 2, A and C), supporting our observation of DENV2 and ZIKV sRecE dimer in the SEC-MALS experiments at 23 °C (Fig. 1, A and C). In summary, our SEC-MALS and AUC data indicate that whereas the DENV2 and ZIKV sRecE dimer is favored at 23 °C, at physiological temperature (37 °C), the equilibrium is shifted toward monomer, even at high concentrations. Both DENV3 and DENV4 sRecE are also in exchange between dimer and monomer; however, in contrast to DENV2 and ZIKV, at 23 °C, DENV3 and DENV4 sRecE are primarily monomers. All SEC-MALS experiments were performed with DENV protein concentrations of ∼2.5 mg/ml (∼55 μm) and ZIKV concentrations at ∼1 mg/ml (∼21 μm).
Figure 2.
sRecE oligomerization and homodimer affinity via AUC sedimentation velocity. Sedimentation velocity AUC experiments were performed with DENV2 (A), DENV4 (B), and ZIKV (C) sRecE at 0.4 mg/ml (∼9 μm) at 21 °C. Results representative of two independent experiments are plotted as the continuous sedimentation coefficient distribution (C(s)) versus the sedimentation coefficient (s20,w). Kd values of two independent experiments are reported as the mean ± S.D.
Characterizing sRecE thermostability
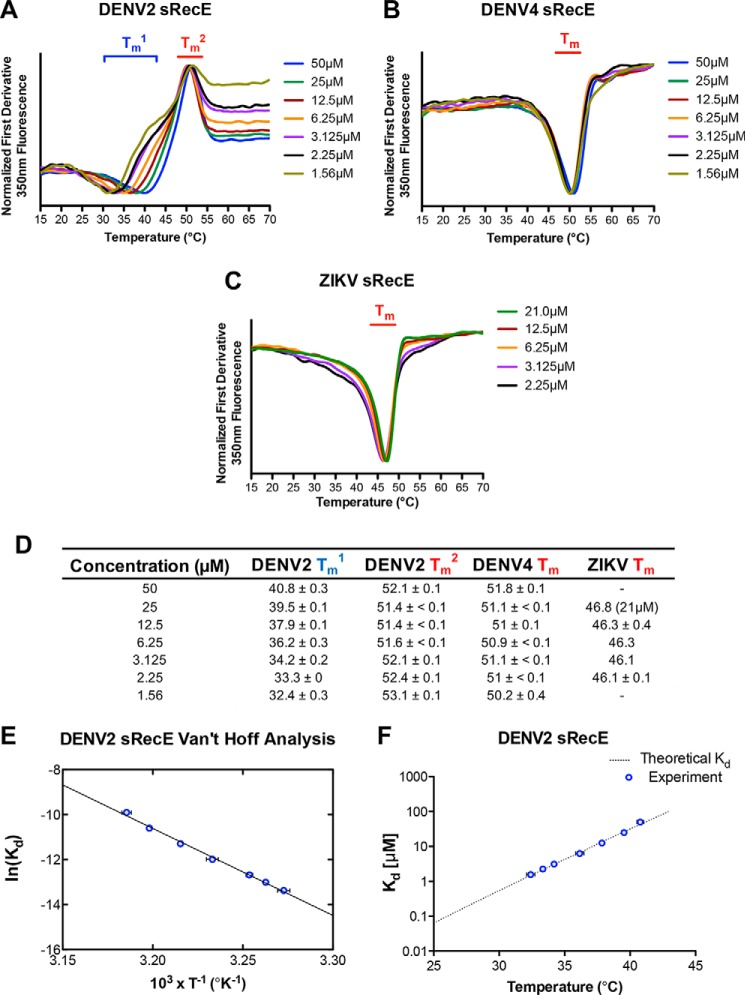
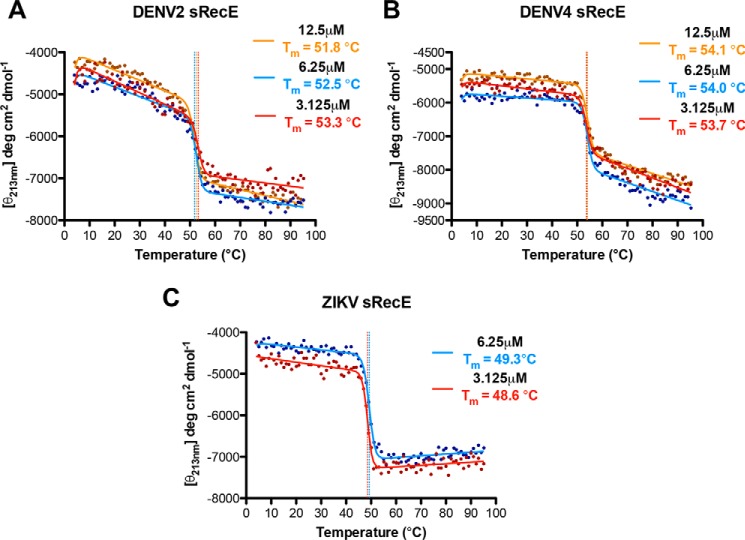
To more fully characterize changes in oligomeric state as a function of temperature and to probe the stability of sRecE monomers, we monitored temperature melts with nano-differential scanning fluorimetry (nanoDSF) and far-UV (213 nm) CD. During nanoDSF, the intrinsic fluorescence of aromatic groups in a protein is monitored as a function of temperature. Changes in fluorescence can be indicative of an unfolding event or changes in oligomerization state. We performed nanoDSF unfolding experiments with DENV2, DENV3, DENV4, and ZIKV sRecE at 2.25 μm from 15 to 95 °C. For DENV4 and DENV3 sRecE, single transitions were observed with midpoints (Tm) of 51.0 and 53.1 °C, respectively (Fig. 3B and Figs. S6B and S8C). We also observed a single transition for ZIKV sRecE, with a lower Tm, 46.0 °C, than DENV3 and DENV4 sRecE (Fig. S8D). Performing CD thermal melts with DENV3, DENV4, and ZIKV from 4 to 95 °C yielded Tm values similar to those observed with nanoDSF for DENV3, DENV4, and ZIKV (Fig. 4 (B and C), Fig. S6D, and Fig. 3D). Far-UV CD measures protein secondary structure, indicating that the Tm values observed for DENV3, DENV4, and ZIKV by nanoDSF and CD correspond with protein unfolding. NanoDSF was also used to probe the capacity of the sRecE proteins to refold. No refolding transitions were observed after cooling from 95 to 15 °C (Fig. S8, A–D).
Figure 3.
DENV2, DENV4, and ZIKV sRecE monomer and DENV2 sRecE dimer thermostability. Shown are NanoDSF fluorescence 350-nm first derivative plots of DENV2 sRecE (A), DENV4 sRecE (B), and ZIKV sRecE (C); thermal unfolding was monitored from 15 to 70 °C at concentrations ranging from 50 to 1.56 μm. D, calculated midpoint transitions (Tm) of DENV2, DENV4, and ZIKV sRecE from data represented in A–C, reported as mean ± S.D. of two independent experiments. E, Van't Hoff ln(Kd) versus T−1 plot of DENV2 sRecE Tm1, average of two independent experiments with error bars indicating S.D. Linear regression fit of the data goodness of fit (R2) was 0.9956. F, plot of DENV2 sRecE homodimer Kd values extrapolated from Van't Hoff analysis (E) at 25–43 °C.
Figure 4.
Thermostability of DENV2, DENV4, and ZIKV sRecE as measured with far-UV CD. Shown are temperature-induced changes in the molar ellipticity [θ] measured at 213 nm for DENV2 (A), DENV4 (B), and ZIKV sRecE (C) at 12.5 μm (orange), 6.25 μm (blue), and 3.125 μm (red). All sRecE samples were equilibrated on ice for ∼1 h before measurement.
In contrast, for DENV2 sRecE, we observed two transitions when monitoring unfolding with nanoDSF. With a protein concentration of 2.25 μm, the first transition had a midpoint at 33.1 °C (Tm1), and the second transition had a midpoint at 53.1 °C (Tm2) (Fig. S8A). Flavivirus E proteins contain a conserved tryptophan in the fusion loop that is buried at the E dimer interface between domain II and domain III (39, 40). Given the tryptophan at the dimer interface and the SEC-MALS data showing that DENV2 sRecE is predominantly a dimer at 23 °C and a monomer at 37 °C, we hypothesized that the Tm1 transition corresponds to DENV2 sRecE dimer dissociation. Consistent with this hypothesis, the Tm1 transition was reversible when heating DENV2 sRecE from 15 to 43 °C and cooling back down from 43 to 15 °C, but as expected, the protein irreversibly unfolded when heating it to 60 °C, just beyond Tm2 (Fig. S8, E and F). However, to rule out the possibility that the Tm1 transition is caused by a structural change in DENV2 sRecE, we performed a CD thermal melt with DENV2 sRecE from 4 to 95 °C, because CD monitors changes in protein secondary structure rather than the fluorescence of a limited number of side chains. As seen in Fig. 4A, only a single transition was observed with a Tm of 53.3 °C, consistent with Tm2 observed by nanoDSF (Fig. 3A and Fig. S8A).
We further postulated that if Tm1 is due to DENV2 sRecE dimer dissociation, Tm1 should be concentration-dependent. NanoDSF experiments were conducted with DENV2 sRecE, DENV4 sRecE, and ZIKV sRecE with varying protein concentrations (1.56–50 μm). For DENV2 sRecE, we observed that increasing DENV2 sRecE concentration increased Tm1, whereas Tm2 remained unchanged (Fig. 3A). In contrast, increasing DENV4 sRecE concentration had no effect on the observed Tm (Fig. 3B), which is expected, as SEC-MALS and AUC identified DENV4 sRecE as mostly monomeric (Figs. 1B and 2B). Similar to DENV2 sRecE, ZIKV sRecE was observed as predominantly dimer at 23 °C, but unlike DENV2 sRecE, we only observed a single transition via nanoDSF for ZIKV sRecE at all concentrations tested (Fig. 3C). However, we do observe fluorescence peak broadening with ZIKV sRecE as concentration is reduced, suggesting that a Tm1 similar to DENV2 might be present but masked by protein unfolding due to the monomer's lower thermostability (46 °C for ZIKV compared with 52 °C for DENV2). Given these data, we concluded that the Tm1 transition is a measurement of the stability of the DENV2 sRecE dimer and that at Tm1, half of the protein is in the monomeric state and half in the dimeric state. This means that the protein concentration at which Tm1 is observed is the dissociation constant (Kd) at Tm1 for the dimer/monomer transition. Next, we used a Van't Hoff analysis to quantify the variance of the DENV2 sRecE homodimer Kd as a function of temperature (Fig. 3E) and calculate the change in enthalpy (ΔH0) for dimer formation. As expected for a two-state transition, the natural log of Kd varied linearly as a function of the reciprocal temperature, and binding was determined to be strongly exothermic with a ΔH0 of −77 kcal/mol (Figs. 3E and 5). From the analysis, we extrapolated the DENV2 sRecE homodimer Kd for temperatures ranging from 25 to 43 °C, revealing a Kd of 64.1 ± 2.3 nm at 25 °C increasing to 9.6 ± 0.4 μm at 37 °C, a ∼150-fold change, and to 102.7 ± 5.1 μm at 43 °C, a ∼1600-fold change (Fig. 3F). Note that the extrapolation used to estimate the Kd at 25 °C does not take into account that the ΔH0 of binding (ΔH°bind) will vary with temperature if there is a change in heat capacity upon binding. This is not expected to be a large effect over this temperature range, given the excellent fit of the data to the Van't Hoff equation, but may account for the difference between the dissociation constants measured at room temperature for DENV2 with AUC and the Van't Hoff analysis. Regardless, our data indicate that the stability of the DENV2 sRecE dimer is strongly dependent on temperature and is consistent with the reported reduction of DENV2 virion E dimer contacts at 37 °C (35).
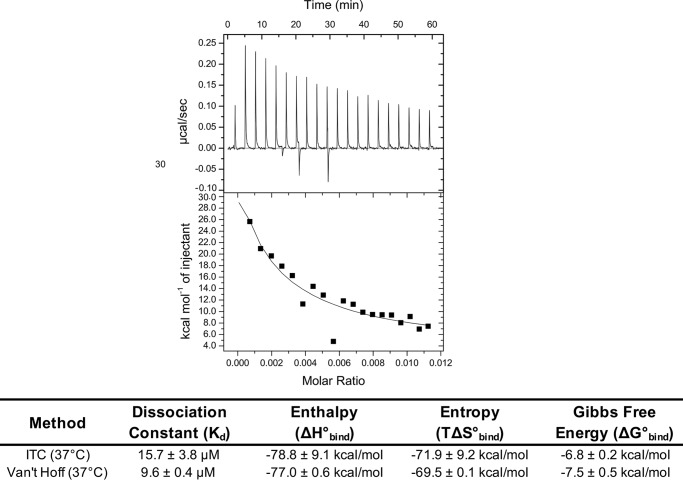
Figure 5.
Quantifying DENV2 sRecE dimer affinity using ITC. Upper, ITC isotherm of DENV2 sRecE dissociation at 37 °C at 67 μm (representative of two independent experiments). Lower, calculated DENV2 sRecE Kd and ΔH0 from homodimer dissociation model fit to integrated isotherm peaks and comparison with values obtained from Van't Hoff analysis of DENV2 sRecE (Fig. 3F). ITC ΔG0 and ΔS0 values were calculated using Equations 3 and 4 with the Kd and ΔH0 values obtained from homodimer dissociation model fit (see “Experimental procedures”). Values are reported as mean ± S.D.
Measuring DENV2 sRecE homodimer affinity at physiological conditions
To confirm the Van't Hoff analysis, we performed dissociation isothermal titration calorimetry (ITC) experiments by titrating concentrated DENV2 sRecE (54–67 μm) into buffer while maintaining the syringe and cell temperatures at 37 °C. From these experiments, the ΔH0bind and Kd of the sRecE dimer can be obtained from a dissociation model fit to the integrated isotherm (see “Experimental procedures”). At 37 °C, dissociation was observed for DENV2 sRecE in both replicates (Fig. 5 and Fig. S9) with a measured average Kd of 15.7 ± 3.8 μm, consistent with the predicted Kd of 9.6 ± 0.4 μm from the nanoDSF Van't Hoff analysis (Figs. 3F and 5). Our ITC experiment also detected a large ΔH°bind of −78.8 ± 9.1 kcal/mol, which was within 2 kcal/mol of ΔH0bind measured with the Van't Hoff analysis (Fig. 5). The large exothermic ΔH°bind probably reflects the large protein–protein interface between monomers in the DENV2 dimer (Fig. 6) (12, 13) and explains why dimer formation is so sensitive to temperature. Performing the same experiment at 67 μm with DENV2 sRecE at 23 °C did not yield a dissociation isotherm above background. This result is consistent with a Kd value at this temperature that is less than 1 μm, as the final diluted protein concentrations in the ITC cell were in the low micromolar range and therefore the protein was presumably still a dimer after dilution (Figs. 2A and 3F). Additionally, no dissociation isotherm was observed when testing DENV4 sRecE at 41.5 μm at 23 °C, consistent with the SEC-MALS and nanoDSF data indicating that DENV4 sRecE is monomeric at these conditions (Fig. S9B).
Figure 6.
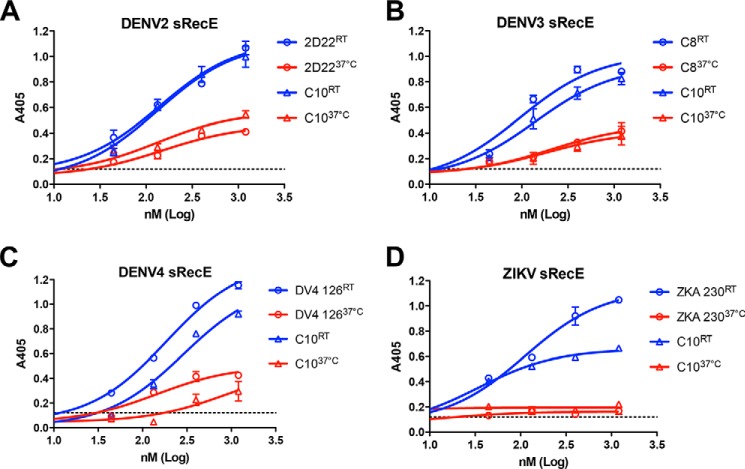
Temperature-dependent quaternary epitope Ab binding to sRecE. A–D, the effect of temperature on quaternary epitope Ab binding to DENV2 (A), DENV3 (B), DENV4 (C), and ZIKV (D) sRecE. sRecE at concentrations 0.04–1.2 μm was incubated with either type-specific (when available) or cross-reactive quaternary epitope Ab together at either room temperature (23 °C) or 37 °C for 1 h and loaded onto Ni2+-coated ELISA plates, and Ab-bound sRecE was detected. Values are reported as mean ± S.D. (error bars).
Elevated temperatures reduce DENV and ZIKV quaternary epitope Ab binding to sRecE
Recent data suggest that the ability to induce quaternary epitope-binding Abs may be a promising vaccine strategy, as these Abs are potently neutralizing and define major antigenic sites targeted by the human Ab response (8, 27, 41, 42). sRecE has been identified as a potential vaccine antigen to elicit neutralizing Abs against DENV and ZIKV (26). In addition to their protective potential, previous reports have shown these Abs are capable of stabilizing sRecE dimers in solution (27, 29). We and others have previously shown that DENV2 and ZIKV sRecE binding to cross-reactive EDE1 C8 and C10 Ab and DENV2 type–specific 2D22 Ab in solution depends on temperature (27). To test whether this was also observed for DENV3 and DENV4, we incubated EDE1 C10 or serotype-specific quaternary Ab to DENV3 and DENV4 sRecE containing a C-terminal His6 tag in solution for 1 h at either 23 or 37 °C. Following incubation, the Ab-sRecE complex was immobilized to the Ni-NTA plate via the sRecE His tag, and Ab binding was measured using an anti-human IgG-AP–conjugated Ab. As observed in our previous studies, EDE1 C10 binding to DENV2 and ZIKV sRecE was observed at 23 °C (Fig. 6, A and D). Additionally, the recently discovered type-specific quaternary epitope Ab ZKA 230, specific to ZIKV sRecE (8), and 2D22, specific to DENV2 sRecE, also bound at the lower temperature (Fig. 6, A and D). However, when incubated at 37 °C, all Abs tested showed reduced binding to DENV2 sRecE and ZIKV sRecE (Fig. 6, A and D). For DENV3 and DENV4 sRecE, EDE1 C10 binding was also observed at 23 °C (Fig. 6, B and C). However, consistent with DENV2 and ZIKV sRecE, EDE1 C10 binding was reduced when incubated with DENV3 and DENV4 sRecE at 37 °C (Fig. 6, B and C). The observed Ab binding reduction at 37 °C to DENV4 sRecE was also observed with DENV4 126, a DENV4 type-specific Ab, and EDE1 C8 to DENV3 sRecE (Fig. 6, B and C). In contrast, Ab binding to either a conformational or linear fusion loop epitope present on sRecE monomers was temperature-independent, with observed binding to DENV2–4 and ZIKV sRecE at both 23 and 37 °C (Fig. S10). The observed reduction of these dimer conformation-specific Abs at 37 °C supports our biophysical data indicating that at physiological temperatures, the sRecE dimer is unstable, resulting in a shifted equilibrium toward the monomer.
Discussion
Our results indicate that temperature plays a significant role in DENV2–4 and ZIKV sRecE dimerization in solution. Consistent with our data, physiological temperatures have been shown to induce large rearrangements of the E protein and E protein dimer contacts on DENV2 virions (33–35), and physiological temperatures have been shown to increase E protein dynamics on the surface of flavivirus virions in West Nile Virus, all DENV serotypes, and ZIKV (23, 33, 43–47). These temperature-induced E protein dynamics, often referred to as “viral breathing,” have been identified through the discovery of neutralizing antibodies that bind to transiently exposed epitopes on the E protein at 37 °C, but not at lower temperatures, such as 4 °C (32). Further evidence has indicated that the E protein is the source of this viral breathing, as mutations within the E protein modulate this behavior (45). Our SEC-MALS and quaternary Ab-binding data provide evidence that physiological temperature induces sRecE protein dimer dissociation, which, we propose, is related to the E protein breathing observed on the virion; however, further experiments are needed to verify this claim, including consideration of the effects of the membrane-proximal and transmembrane domains on dimer stability, as these regions are not included in our constructs .
In agreement with our findings, previous studies have shown that the DENV2 and ZIKV sRecE monomer–dimer equilibrium is shifted toward dimer at room temperature (16, 30). In contrast, Rouvinski et al. (29) used SEC-MALS to characterize 1 mg/ml (22 μm) DENV2 sRecE made from S2 cells and found that it was monomeric at room temperature in 50 mm Tris buffer containing 500 mm NaCl at pH 8.0. A possible explanation for the inconsistency between this result and other studies, including our own, is that the higher salt concentration used in their experiment may reduce the ability of DENV2 sRecE to dimerize, as protein–protein interactions can be salt-dependent.
To gain insight into the strong temperature dependence we observed for DENV2 dimerization, we used both ITC and a Van't Hoff analysis to measure the enthalpy of dimerization. From the Gibbs–Helmhotz equation, we know that the natural logarithm of the dissociation constant (Kd) for a binding reaction is directly proportional to binding enthalpy and the inverse of temperature (see Equation 5). If binding is endothermic (ΔH0 > 0), then binding affinity increases as the temperature is raised. Endothermic binding events are typically driven by the hydrophobic effect, such as the binding between β-crystallins (48). If binding is exothermic (ΔH0 < 0), then binding affinity decreases as the temperature is raised. Exothermic binding events are driven by hydrogen bonding, salt bridges, and van der Waals interactions. The ΔH0 of dimerization for DENV2 is strongly exothermic, −78 kcal/mol. This result is consistent with the chemical composition of the dimer interface. We used the PISA server to identify interface residues from the DENV2 crystal structure (PDB code 1OAN), and of 49 nonglycine interface residues, 67% of the residues are polar or charged (49). This is a higher fraction of hydrophilic residues than is seen on average at protein–protein interfaces. In a large set of structures from the PDB, Yan et al. (50) observed that 51% of the nonglycine residues at protein–protein interfaces are polar or charged and that the percentage of polar and charged residues is even lower at homodimeric interfaces. One possibility for why the DENV2 interface is more polar than average is that some of the residues buried at the dimer interface become solvent-exposed when the protein switches to its trimeric form to promote viral fusion (51).
Our SEC-MALS experiments and AUC show that DENV3 and DENV4 sRecE are less stable as dimers than DENV2. In accordance with this finding, Luca et al. (30) used the crystal structures of these proteins to measure the change in accessible surface area (dSASA) upon binding and found that more contacts are made in the DENV2 dimer (dSASA = 1703–1929 Å2) than in the DENV3 dimer (dSASA = 1593 Å2). In contrast to our AUC data, which show monomer and dimer populations for DENV4 sRecE, another AUC study of DENV4 sRecE with sucrose gradients revealed that much larger DENV4 oligomers were present in solution (52). However, it is possible in the presence of sucrose that sRecE structural changes may occur. Previous studies suggest that glycosylation is a strong contributor to ZIKV sRecE dimerization, as Barba-Spaeth et al. (16) report that ZIKV sRecE produced in S2 cells forms a dimer at room temperature via SEC, and Dai et al. (31) report that bacterially expressed and refolded ZIKV sRecE is a monomer in solution.
The dynamic stability of E protein homodimers is probably related to the complex lifestyle of arboviruses. Highly stable dimers are unlikely to support the reorganization of E proteins from dimers to trimers required for viral fusion in endosomes to deliver the viral genome to the cytoplasm (51). These viruses have to infect a mosquito vector and primate host that maintain different body temperatures. The temperature-dependent differences in E protein homodimer stability are likely to be a result of viral adaptation to a dual-host lifestyle. In this study, we have analyzed a single representative E protein for DENV2–4 as well as ZIKV. As there is considerable E protein variation among DENVs within a serotype as well as some variation between different ZIKV strains, our results must be interpreted with caution. It is conceivable that large differences in dimer stability exist between viruses in the same serotype, and such differences may be linked to viral pathogenesis. Indeed, investigators have noted large differences in “viral breathing” between different DENV2 strains (53).
The effects of E protein dynamics, especially dimer stability, have been suggested to affect the type of Abs elicited during vaccination (8). Our data showing the reduced capability of DENV and ZIKV sRecE to dimerize at physiological temperatures are consistent with published data indicating that subunit vaccines based on the secreted E protein ectodomain mainly induce Abs to E monomer epitopes on EDIII or the undesired immunodominant conserved fusion loop E epitope (FLE) targeted by weakly neutralizing Abs (54). It has been proposed that effective DENV and ZIKV vaccines will require immunogens that present epitopes similar to those presented on the virion, in particular for the E protein, the quaternary and EDE epitopes, which have been shown to be potently neutralizing (21–23). Our data suggest that use of the sRecE as a vaccine immunogen requires methods that stabilize the sRecE protein as a dimer at physiological temperatures. The introduction of disulfides at the DENV2 sRecE E dimer DII/DIII (DII L107C and DIII A313C) (29) and DII/DII αB helix interface at position A259C (37) has been shown to stabilize sRecE dimers while retaining E dimer and quaternary epitope Ab binding. Additionally, the sRecE E dimer DII/DIII disulfide variant reduces the binding of FLE Abs, suggesting that using stabilized sRecE dimers in vaccinations may increase the probability of eliciting potently neutralizing quaternary epitope Ab rather than poorly neutralizing FLE Abs (8). By thorough analysis of the dimerization properties of E proteins from different flaviviruses, it should be possible to design stable and safer vaccine antigens that mimic the viral surface better than current subunit vaccines.
Experimental procedures
Materials
DENV2–4 sRecE were cloned into the PαH mammalian expression vector constructed based on the WHO reference strains, DENV2 (aa 1–395 of S-16803), DENV3 (aa 1–395 of CH53489), DENV4 (aa 1–397 of TVP-376), and ZIKV (aa 1–404 of H/PF/2013). Expression and purification were performed according to our published protocol (27), using DG44 CHO cells for DENV4 sRecE and EXPI293 cells for DENV2–3 and ZIKV sRecE. All experiments were performed using 1× PBS at pH 7.4 (137 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4, 2 mm KH2PO4) buffer, following Thermo Scientific's recipe (catalog no. AM9625). Each sRecE stock, before each experiment, was thawed on ice and centrifuged at 10,000–13,000 rpm for 10–13 min at 4 °C, and supernatant was collected to prevent protein precipitates from interfering with experiments.
SEC-MALS
SEC-MALS experiments were performed using a GE Superdex 200 10/300 GL column–equipped Agilent FPLC system interfaced with a Wyatt DAWN HELEOS II light-scattering instrument, Wyatt T-rEX refractometer and a Wyatt dynamic light scattering module. Each experiment was conducted by loading 240–250 μg of ice-cold DENV2 sRecE (2.45 mg/ml), DENV3 sRecE (2.5 mg/ml), and DENV4 sRecE (2.47 mg/ml) and 100 μg of ZIKV sRecE (0.97 mg/ml). The entire experiment was performed at room temperature (∼23 °C) or 37 °C, at a flow rate of 0.5 ml min−1. For 37 °C experiments, all WYATT equipment was set to 37 °C, while maintaining buffer temperature using a temperature-controlled water bath and column temperature using THG thermal tape regulated by a digital thermostat. Before sample loading, the system was equilibrated with 1× PBS at pH 7.4 and 200 mg/liter sodium azide. MALS data were collected and analyzed using Wyatt ASTRA (version 6) software.
AUC sedimentation velocity
Sedimentation velocity experiments were performed in a ProteomeLab XL-I (Beckman Coulter) analytical ultracentrifuge. Protein samples at ∼0.4 mg/ml in 1× PBS were loaded in two-channel cells and spun in an An-50 Ti 8-place rotor at 30,000 rpm at 21 °C for 16 h. Absorbance at 280 nm was used for detection. Sedimentation velocity data were analyzed using Sedfit software (P. Schuck, NIBIB, National Institutes of Health).
CD
Measurements were conducted using a Jasco J-815 CD spectrometer. DENV2, DENV3, DENV4, and ZIKV sRecE proteins in 1× PBS, pH 7.4, with 10% glycerol, were diluted into 1× PBS at pH 7.4 to their designated concentrations and remained on ice before each experiment. Buffer controls with the appropriate glycerol dilutions were performed with each experiment to ensure that no significant signal was produced by the appropriately diluted buffer containing glycerol. CD spectral measurements from 250 to 200 nm were performed at 25 °C for DENV2, DENV4, and ZIKV sRecE in a 0.1-mm glass quartz cuvette at 25, 25, and 20 μm respective concentrations. DENV3 sRecE CD spectral measurements were performed in a 1-mm glass quartz cuvette at 3.125 μm concentration. All sRecE denaturation experiments were measured at 213 nm from 4 to 95 °C at a rate of 2 °C/min in a 1-mm glass quartz cuvette. The mean residue ellipticity was calculated using, using n values of 395, 393, 394, and 404 for DENV2, DENV3, DENV4, and ZIKV sRecE and fitted to Equation 1 of Ref. 55 to calculate the melting temperature (Tm) for each protein.
NanoDSF
nanoDSF experiments were performed using a Nanotemper Prometheus NT.48 system. DENV2, DENV3, DENV4, and ZIKV sRecE were diluted to the designated concentrations in 1× PBS at pH 7.4. Thermal denaturation unfolding and refolding experiments were performed for each sRecE at 2.25–12.5 μm from 15 to 95 °C (unfold) and from 95 to 15 °C (refold) at 1 °C/min. For concentration-dependent nanoDSF experiments with DENV2, DENV4, and ZIKV sRecE, proteins were diluted to 1.56–50 μm in 1× PBS, pH 7.4, and loaded into 10-μl capillary tubes. Thermal denaturation experiments were performed from 15 to 70 °C at 1 °C/min, measuring the intrinsic tryptophan fluorescence at 330 nm and its shift upon protein unfolding at 350 nm. The Tm for each experiment was calculated automatically by Nanotemper PR.Thermconstrol software. The fluorescence at 350-nm wavelength is reported for all sRecE nanoDSF experiments.
Van't Hoff analysis of DENV2 sRecE nanoDSF data
The sRecE protein homodimer interaction, assumed to be reversible, can be written as [M] + [M] ⇌ [D], where [M] and [D] are the concentrations of the monomer and dimer, respectively.
The equilibrium constant (Keq) of the sRecE homodimer can be defined as follows.
| (Eq. 1) |
This can also be written as the dissociation constant (Kd), which is more commonly used to describe protein–protein interactions.
| (Eq. 2) |
Assuming equilibrium conditions, the change in free energy is constant (ΔG = 0). Under these conditions and using the dissociation constant, the standard free energy of this reaction can be written as follows,
| (Eq. 3) |
where R is the ideal gas constant (1.987 cal·K−1·mol−1) and T represents temperature in K.
The standard free energy can also be described in terms of its enthalpy (ΔH0) and entropy (ΔS0) components.
| (Eq. 4) |
Combining Equations 3 and 4 yields the Van't Hoff equation, which describes how the dissociation constant varies with respect to temperature.
| (Eq. 5) |
An ideal Van't Hoff plot of ln(Kd) versus 1/T yields a straight line with a slope of −ΔH0/R and intercept of +ΔS0/R. Tm is defined as the point where the protein folded and unfolded fractions are equal (i.e. in equilibrium). Assuming that the Tm1 transition refers to the temperature when the DENV2 sRecE dimer dissociates, and because the Tm refers to equilibrium conditions, Tm1 can then represent when the amounts of dimer and monomer are equal to one another, instead of the fraction of folded protein. Therefore, the concentration at the observed Tm1 is equal to the dissociation constant of the sRecE homodimer at that given temperature equal to Tm1. From this, the values for Tm1 at the tested concentrations (1.56–50 μm) were used as the T values to generate the Van't Hoff plot in Fig. 3. The values from the fit were used to calculate the theoretical Kd values at different temperatures, and the slope Van't Hoff plot was used to calculate Hbind0.
ITC
DENV2 and DENV4 sRecE proteins were prepared for ITC by buffer exchange, with an Amicon 10,000 molecular weight cutoff concentrator or dialysis, 3000 molecular weight cutoff snakeskin tubing, into 1× PBS at pH 7.4, at 4 °C, to remove the glycerol concentration down to <10 nm (7.2 × 10−6% glycerol). DENV2 and DENV4 sRecE were concentrated to 54.1–67 μm and 41.5 μm, respectively, and incubated on ice for a minimum of 30 min before each dissociation experiment. The dissociation experiments were performed using a MicroCal Auto-ITC200 calorimeter at either 23 or 37 °C. For each experiment, a 2 μl × 19 injection protocol was used with an initial injection of 0.2 μl (this data point was discarded during analysis). The ITC cell (volume 200 μl) contained exchanged or dialyzed buffer. The stirring speed was kept at 750 rpm and the duration at 4 s. The data were fit to the dimer dissociation model using the Malvern/MicroCal ITC Origin version 7 analysis software to obtain enthalpy (ΔH0) and dissociation constant (Kd) values.
Antibody–sRecE binding ELISA experiments
ELISAs were performed using our previously published protocol (27). For comparison of Ab binding to sRecE monomer and dimer epitopes, the following Abs were used: FLE Abs (4G2 and 1M7), quaternary epitope cross-reactive Abs (EDE1 C8 and C10), and quaternary epitope serotype-specific Abs (2D22, DENV4 126, and ZKA 230). sRecE was serially diluted 1:3 to 0–1.2 μm concentrations, to a total amount of 500 ng, and incubated at room temperature (23 °C) or 37 °C for 1 h. 500 ng of Ab at 10 ng/μl was added to each sRecE concentration and incubated on a rotator at room temperature or 37 °C for 1 h. Ni-NTA ELISA plates were then coated with the sRecE/Ab mixture for 2 h at room temperature or 37 °C. Wells were then blocked with 3% skim milk in TBS + 0.05% Tween 20 for 1 h at room temperature and washed with TBS + Tween 20. Wells containing 4G2 and 1M7 were incubated with a 1:1000 dilution of anti-mouse IgG conjugated with AP (Sigma) and with a 1:2500 dilution of anti-human IgG-AP (Sigma) for wells containing quaternary epitope Abs. After washing, wells were developed using AP-substrate (Sigma), and absorbance was measured at a wavelength of 405 nm.
Author contributions
S. T. K., L. P., S. W. M., A. M. d. S., and B. K. conceived and designed the experiments. S. T. K., L. P., S. W. M., A. M. P., A. A. B., and S. G. performed the experiments. S. T. K., L. P., S. W. M., A. M. d. S., A. T., and B. K. analyzed the data. A. T., M. J. M., S. G., and J. A. B. contributed reagents/materials/analysis tools.
Supplementary Material
Acknowledgments
We thank Frank Teets for help and ideas in designing the 37 °C SEC-MALS experiments. Production of DENV and ZIKV envelope proteins by the UNC Protein Expression and Purification core and use of the biophysical equipment in the UNC Macromolecular Interactions Facility were supported by NCI, National Institutes of Health, Grant P30CA016086.
This work was supported by NIGMS, National Institutes of Health (NIH), Grant R01GM073960; NIAID, NIH Grant U19 AI09784; and Centers for Disease Control and Prevention Grant 00HVCLJB-2017-04191. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S10.
- DENV
- dengue virus
- ZIKV
- Zika virus
- E
- envelope
- aa
- amino acid(s)
- sRecE
- soluble recombinant E
- Ab
- antibody
- EDE
- E dimer epitope
- Ni-NTA
- nickel-nitrilotriacetic acid
- SEC-MALS
- size-exclusion chromatography with multiangle light scattering
- AUC
- analytical ultracentrifugation
- nanoDSF
- nano-differential scanning fluorimetry
- dSASA
- change in accessible surface area
- FLE
- fusion loop epitope
- ITC
- isothermal titration calorimetry.
References
- 1. Bhatt S., Gething P. W., Brady O. J., Messina J. P., Farlow A. W., Moyes C. L., Drake J. M., Brownstein J. S., Hoen A. G., Sankoh O., Myers M. F., George D. B., Jaenisch T., and Wint G. R. W., Simmons C. P., et al. (2013) The global distribution and burden of dengue. Nature 496, 504–507 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization and the Special Programme for Research and Tropical Diseases (2009) Dengue guidelines for diagnosis, treatment, prevention and control. World Health Organization, Geneva: [PubMed] [Google Scholar]
- 3. Heinz F. X., and Stiasny K. (2017) The antigenic structure of Zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol. Mol. Biol. Rev. 81, e00055-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morabito K. M., and Graham B. S. (2017) Zika virus vaccine development. J. Infect. Dis. 216, S957–S963 10.1093/infdis/jix464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Y., Liu J., and Cheng G. (2016) Vaccines and immunization strategies for dengue prevention. Emerg. Microbes Infect. 5, e77 10.1038/emi.2016.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization (2016) Dengue vaccine: WHO position paper–July 2016. J. Wkly. Epidemiol. Rec. 91, 349–364 27476189 [Google Scholar]
- 7. Cohen J. (2017) If you haven't had dengue infection, don't use our vaccine, drug company warns. Science 80 10.1126/science.aar6371 [DOI] [Google Scholar]
- 8. Rey F. A., Stiasny K., Vaney M.-C., Dellarole M., and Heinz F. X. (2018) The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 19, 206–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X., Ge P., Yu X., Brannan J. M., Bi G., Zhang Q., Schein S., and Zhou Z. H. (2013) Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 10.1038/nsmb.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sirohi D., Chen Z., Sun L., Klose T., Pierson T. C., Rossmann M. G., and Kuhn R. J. (2016) The 3.8 Å resolution cryo-EM structure of Zika virus. Science 352, 467–470 10.1126/science.aaf5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coller B. A. G., Clements D. E., Bett A. J., Sagar S. L., and Ter Meulen J. H. (2011) The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine 29, 7267–7275 10.1016/j.vaccine.2011.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Modis Y., Ogata S., Clements D., and Harrison S. C. (2003) A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 100, 6986–6991 10.1073/pnas.0832193100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Zhang W., Ogata S., Clements D., Strauss J. H., Baker T. S., Kuhn R. J., and Rossmann M. G. (2004) Conformational changes of the flavivirus E glycoprotein. Structure 12, 1607–1618 10.1016/j.str.2004.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Modis Y., Ogata S., Clements D., and Harrison S. C. (2005) Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79, 1223–1231 10.1128/JVI.79.2.1223-1231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cockburn J. J. B., Navarro Sanchez M. E., Goncalvez A. P., Zaitseva E., Stura E. A., Kikuti C. M., Duquerroy S., Dussart P., Chernomordik L. V., Lai C. J., and Rey F. A. (2012) Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J. 31, 767–779 10.1038/emboj.2011.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barba-Spaeth G., Dejnirattisai W., Rouvinski A., Vaney M.-C., Medits I., Sharma A., Simon-Lorière E., Sakuntabhai A., Cao-Lormeau V.-M., Haouz A., England P., Stiasny K., Mongkolsapaya J., Heinz F. X., Screaton G. R., and Rey F. A. (2016) Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 536, 48–53 10.1038/nature18938 [DOI] [PubMed] [Google Scholar]
- 17. Govindarajan D., Meschino S., Guan L., Clements D. E., ter Meulen J. H., Casimiro D. R., Coller B. A. G., and Bett A. J. (2015) Preclinical development of a dengue tetravalent recombinant subunit vaccine: immunogenicity and protective efficacy in nonhuman primates. Vaccine 33, 4105–4116 10.1016/j.vaccine.2015.06.067 [DOI] [PubMed] [Google Scholar]
- 18. Manoff S. B., George S. L., Bett A. J., Yelmene M. L., Dhanasekaran G., Eggemeyer L., Sausser M. L., Dubey S. A., Casimiro D. R., Clements D. E., Martyak T., Pai V., Parks D. E., and Coller B. A. (2015) Preclinical and clinical development of a dengue recombinant subunit. Vaccine 33, 7126–7134 10.1016/j.vaccine.2015.09.101 [DOI] [PubMed] [Google Scholar]
- 19. Metz S. W., Tian S., Hoekstra G., Yi X., Stone M., Horvath K., Miley M. J., DeSimone J., Luft C. J., and de Silva A. M. (2016) Precisely molded nanoparticle displaying DENV-E proteins induces robust serotype-specific neutralizing antibody responses. PLoS Negl. Trop. Dis. 10, e0005071 10.1371/journal.pntd.0005071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delenda C., Frenkiel M. P., and Deubel V. (1994) Protective efficacy in mice of a secreted form of recombinant dengue-2 virus envelope protein produced in baculovirus infected insect cells. Arch. Virol. 139, 197–207 10.1007/BF01309465 [DOI] [PubMed] [Google Scholar]
- 21. Dejnirattisai W., Wongwiwat W., Supasa S., Zhang X., Dai X., Rouvinsky A., Jumnainsong A., Edwards C., Quyen N. T., Duangchinda T., Grimes J. M., Tsai W., Lai C., Wang W., Malasit P., et al. (2015) A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177 10.1038/ni.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fibriansah G., Ibarra K. D., Ng T.-S., Smith S. A., Tan J. L., Lim X.-N., Ooi J. S. G., Kostyuchenko V. A., Wang J., de Silva A. M., Harris E., Crowe J. E. Jr., and Lok S.-M. (2015) Dengue virus: cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91 10.1126/science.aaa8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fibriansah G., Tan J. L., Smith S. A., de Alwis R., Ng T. S., Kostyuchenko V. A., Jadi R. S., Kukkaro P., de Silva A. M., Crowe J. E., and Lok S. M. (2015) A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 6, 6341 10.1038/ncomms7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Alwis R., Smith S. A., Olivarez N. P., Messer W. B., Huynh J. P., Wahala W. M. P. B., White L. J., Diamond M. S., Baric R. S., Crowe J. E. Jr., and de Silva A. M. (2012) Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. U.S.A. 109, 7439–7444 10.1073/pnas.1200566109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hasan S. S., Miller A., Sapparapu G., Fernandez E., Klose T., Long F., Fokine A., Porta J. C., Jiang W., Diamond M. S., Crowe J. E. Jr., Kuhn R. J., and Rossmann M. G. (2017) A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun. 8, 14722 10.1038/ncomms14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M.-C., Kikuti C. M., Navarro Sanchez M. E., Dejnirattisai W., Wongwiwat W., Haouz A., Girard-Blanc C., Petres S., Shepard W. E., Desprès P., Arenzana-Seisdedos F., et al. (2015) Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520, 109–113 10.1038/nature14130 [DOI] [PubMed] [Google Scholar]
- 27. Metz S. W., Gallichotte E. N., Brackbill A., Premkumar L., Miley M. J., Baric R., and de Silva A. M. (2017) In vitro assembly and stabilization of dengue and Zika virus envelope protein homo-dimers. Sci. Rep. 7, 4524 10.1038/s41598-017-04767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scheiblhofer S., Laimer J., Machado Y., Weiss R., and Thalhamer J. (2017) Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev. Vaccines 16, 479–489 10.1080/14760584.2017.1306441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rouvinski A., Dejnirattisai W., Guardado-Calvo P., Vaney M. C., Sharma A., Duquerroy S., Supasa P., Wongwiwat W., Haouz A., Barba-Spaeth G., Mongkolsapaya J., Rey F. A., and Screaton G. R. (2017) Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nat. Commun. 8, 15411 10.1038/ncomms15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luca V. C., AbiMansour J., Nelson C. A., and Fremont D. H. (2012) Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 86, 2337–2346 10.1128/JVI.06072-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai L., Song J., Lu X., Deng Y. Q., Musyoki A. M., Cheng H., Zhang Y., Yuan Y., Song H., Haywood J., Xiao H., Yan J., Shi Y., Qin C. F., Qi J., and Gao G. F. (2016) Structures of the Zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704 10.1016/j.chom.2016.04.013 [DOI] [PubMed] [Google Scholar]
- 32. Kuhn R. J., Dowd K. A., Beth Post C., and Pierson T. C. (2015) Shake, rattle, and roll: impact of the dynamics of flavivirus particles on their interactions with the host. Virology 479, 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lim X. X., Chandramohan A., Lim X. Y. E., Bag N., Sharma K. K., Wirawan M., Wohland T., Lok S. M., and Anand G. S. (2017) Conformational changes in intact dengue virus reveal serotype-specific expansion. Nat. Commun. 8, 14339 10.1038/ncomms14339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Sheng J., Plevka P., Kuhn R. J., Diamond M. S., and Rossmann M. G. (2013) Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U.S.A. 110, 6795–6799 10.1073/pnas.1304300110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fibriansah G., Ng T.-S., Kostyuchenko V. A., Lee J., Lee S., Wang J., and Lok S.-M. (2013) Structural changes in dengue virus when exposed to a temperature of 37 °C. J. Virol. 87, 7585–7592 10.1128/JVI.00757-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang X., Sun L., and Rossmann M. G. (2015) Temperature dependent conformational change of dengue virus. Curr. Opin. Virol. 12, 109–112 10.1016/j.coviro.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Slon Campos J. L., Marchese S., Rana J., Mossenta M., Poggianella M., Bestagno M., and Burrone O. R. (2017) Temperature-dependent folding allows stable dimerization of secretory and virus-associated E proteins of dengue and Zika viruses in mammalian cells. Sci. Rep. 7, 966 10.1038/s41598-017-01097-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wengler G., Wengler G., and Rey F. A. (1999) The isolation of the ectodomain of the alphavirus E1 protein as a soluble hemagglutinin and its crystallization. Virology 257, 472–482 10.1006/viro.1999.9661 [DOI] [PubMed] [Google Scholar]
- 39. Wahala W. M. P. B., and Silva A. M. (2011) The human antibody response to dengue virus infection. Viruses 3, 2374–2395 10.3390/v3122374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai L., Wang Q., Qi J., Shi Y., Yan J., and Gao G. F. (2016) Molecular basis of antibody-mediated neutralization and protection against flavivirus. IUBMB Life 68, 783–791 10.1002/iub.1556 [DOI] [PubMed] [Google Scholar]
- 41. Gallichotte E. N., Widman D. G., Yount B. L., Wahala W. M., Durbin A., Whitehead S., Sariol C. A., Crowe J. E. Jr., de Silva A. M., and Baric R. S. (2015) A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. MBio 6, e01461-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nivarthi U. K., Kose N., Sapparapu G., Widman D., Gallichotte E., Pfaff J. M., Doranz B. J., Weiskopf D., Sette A., Durbin A. P., Whitehead S. S., Baric R., Crowe J. E. Jr., and de Silva A. M. (2017) Mapping the human memory B cell and serum neutralizing antibody responses to dengue virus serotype 4 infection and vaccination. J. Virol. 91, e02041-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kostyuchenko V. A., Chew P. L., Ng T.-S., and Lok S.-M. (2014) Near-atomic resolution cryo-electron microscopic structure of dengue serotype 4 virus. J. Virol. 88, 477–482 10.1128/JVI.02641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kostyuchenko V. A., Lim E. X. Y., Zhang S., Fibriansah G., Ng T. S., Ooi J. S. G., Shi J., and Lok S. M. (2016) Structure of the thermally stable Zika virus. Nature 533, 425–428 10.1038/nature17994 [DOI] [PubMed] [Google Scholar]
- 45. Goo L., VanBlargan L. A., Dowd K. A., Diamond M. S., and Pierson T. C. (2017) A single mutation in the envelope protein modulates flavivirus antigenicity, stability, and pathogenesis. PLoS Pathog. 13, e1006178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li J., Watterson D., Chang C. W., Che X. Y., Li X. Q., Ericsson D. J., Qiu L. W., Cai J. P., Chen J., Fry S. R., Cheung S. T. M., Cooper M. A., Young P. R., and Kobe B. (2018) Structural and functional characterization of a cross-reactive dengue virus neutralizing antibody that recognizes a cryptic epitope. Structure 26, 51–59.e4 10.1016/j.str.2017.11.017 [DOI] [PubMed] [Google Scholar]
- 47. Wu Y., Li S., Du L., Wang C., Zou P., Hong B., Yuan M., Ren X., Tai W., Kong Y., Zhou C., Lu L., Zhou X., Jiang S., and Ying T. (2017) Neutralization of Zika virus by germline-like human monoclonal antibodies targeting cryptic epitopes on envelope domain III. Emerg. Microbes Infect. 6, e89 10.1038/emi.2017.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sergeev Y. V., Hejtmancik J. F., and Wingfield P. T. (2004) Energetics of domain-domain interactions and entropy driven association of β-crystallins. Biochemistry 43, 415–424 10.1021/bi034617f [DOI] [PubMed] [Google Scholar]
- 49. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 50. Yan C., Wu F., Jernigan R. L., Dobbs D., and Honavar V. (2008) Characterization of protein-protein interfaces. Protein J. 27, 59–70 10.1007/s10930-007-9108-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Modis Y., Ogata S., Clements D., and Harrison S. C. (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 10.1038/nature02165 [DOI] [PubMed] [Google Scholar]
- 52. Hsieh S. C., Tsai W. Y., Nerurkar V. R., and Wang W. K. (2014) Characterization of the ectodomain of the envelope protein of dengue virus type 4: expression, membrane association, secretion and particle formation in the absence of precursor membrane protein. PLoS One 9, e100641 10.1371/journal.pone.0100641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lok S.-M. (2016) The interplay of dengue virus morphological diversity and human antibodies. Trends Microbiol. 24, 284–293 10.1016/j.tim.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 54. Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V. M., Malasit P., Rey F. A., Mongkolsapaya J., and Screaton G. R. (2016) Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat. Immunol. 17, 1102–1108 10.1038/ni.3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuhlman B., Boice J. A., Fairman R., and Raleigh D. P. (1998) Structure and stability of the N-terminal domain of the ribosomal protein L9: evidence for rapid two-state folding. Biochemistry 37, 1025–1032 10.1021/bi972352x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.