Summary
Nup133 belongs to the Y-complex, a key component of the nuclear pore complex (NPC) scaffold. Studies on a null mutation in mice previously revealed that Nup133 is essential for embryonic development but not for mouse embryonic stem cell (mESC) proliferation. Using single pore detection and average NE-fluorescence intensity, we find that Nup133 is dispensable for interphase and postmitotic NPC scaffold assembly in pluripotent mESCs. However, loss of Nup133 specifically perturbs the formation of the nuclear basket as manifested by the absence of Tpr in about half of the NPCs combined with altered dynamics of Nup153. We further demonstrate that its central domain mediates Nup133's role in assembling Tpr and Nup153 into a properly configured nuclear basket. Our findings thus revisit the role of the Y-complex in pore biogenesis and provide insights into the interplay between NPC scaffold architecture, nuclear basket assembly and the generation of heterogeneity among NPCs.
Keywords: Nuclear pore, nucleoporin, NPC scaffold, nuclear basket, biogenesis, dynamics, mouse embryonic stem cells
Introduction
Nuclear pore complexes (NPCs), massive protein assemblies spanning the double membrane of the nuclear envelope (NE), serve as the primary conduit for nucleocytoplasmic transport. NPCs and their constituent nucleoporins (Nups) also contribute to multiple transport-independent functions, notably gene regulation, chromatin organization, genome integrity, and cell cycle control (reviewed in Bukata et al., 2013; Raices and D'Angelo, 2017; Hezwani and Fahrenkrog, 2017). The NPC, an eightfold rotationally symmetric structure, consists of ~ 30 distinct Nups each present in a multiple of eight copies (reviewed in Beck and Hurt, 2017; Hoelz et al., 2016). Over the past decade, the application of cryo-electron tomography, biochemistry and X-ray crystallography has produced highly refined 3D-maps of the NPC scaffold. It consists of an inner ring that sits embedded in the nuclear pore membrane between the symmetrical cytoplasmic and nuclear rings. In vertebrates, the latter rings are each composed of 16 copies of the evolutionarily conserved Y-complex (also known as "Nup107-160 complex" or "coat nucleoporin complex") that contains 9 distinct Nups (Beck and Hurt, 2017; Hoelz et al., 2016). These three rings anchor additional, non-scaffolding Nups that comprise the central channel, cytoplasmic filaments and nuclear basket. Key among the non-scaffolding Nups is a subset of proteins featuring unfolded phenylalanine–glycine (FG)-repeat domains, which are the primary determinants of the permeability and selectivity properties of the NPC (Schmidt and Gorlich, 2016). These same Nups also facilitate NPC biogenesis and stabilize pore structure (Onischenko et al., 2017). Different peripheral structures emanating from the cytoplasmic and nuclear rings confer asymmetry to the NPC. Pertinent to our findings, the nuclear basket-containing Tpr (a long coiled-coil 267 kDa protein) and Nup153 and Nup50 (two highly mobile FG-Nups)-play a pivotal role in transport-dependent and -independent functions of the NPC (Cordes et al., 1997;Rabut et al., 2004;Griffis et al., 2004; Snow and Paschal, 2014). Despite established interactions between Nup153 and the basket Nups Tpr and Nup50, as well as with the Y-complex (Hase and Cordes, 2003; Makise et al., 2012; Vasu et al., 2001), little is known about how the nuclear basket assembles and integrates with the NPC scaffold.
Assembly of new pores is a complex process involving the organization of ~1,000 proteins into a single NPC. In metazoan cells, NPC assembly occurs at two distinct phases of the cell cycle: at the end of mitosis, concomitant with NE reformation, and during interphase when the NE is intact. Previous studies, using Xenopus extracts or RNAi depletion in human cancer cell lines, revealed that the Y-complex plays a key role in both post-mitotic and interphase NPC assembly (Walther et al., 2003; Harel et al., 2003; D'Angelo et al., 2006). However, these studies did not evaluate the function of individual Y-complex subunits in NPC formation. For example, RNAi-mediated depletion of the Nup133 subunit of the Y-complex induced destabilization of multiple Y-complex Nups and led to major NPC assembly defects (Walther et al., 2003). In contrast, our previous work on a null allele of mouse Nup133 (merm) revealed that Nup133, although required for embryonic development beyond gastrulation stages, is dispensable for mouse embryonic stem cell (mESC) proliferation (Lupu et al., 2008). Notably, analyses performed on Nup133merm/merm mESCs and tissues detected no major alteration in the levels or subcellular location of Nups that would be suggestive of impaired NPC assembly (Lupu et al., 2008). However, the assays used in that study lacked sufficient sensitivity and resolution to assess whether the absence of Nup133 compromises the efficiency and/or accuracy of distinct steps during NPC assembly. Additionally, Doucet et al. (2010) demonstrated a critical function of an amphipathic helix located within the Nup133 N-terminal domain for interphase NPC assembly in human U2OS cells (Doucet et al., 2010). Concluding that the role played by Nup133 during NPC assembly merited re-evaluation in mESCs, we undertook quantitative approaches to assess NPC number and composition in wild-type (WT) and Nup133-deficient mESCs.
Results
Nup133 is not required for interphase NPC assembly in mESCs
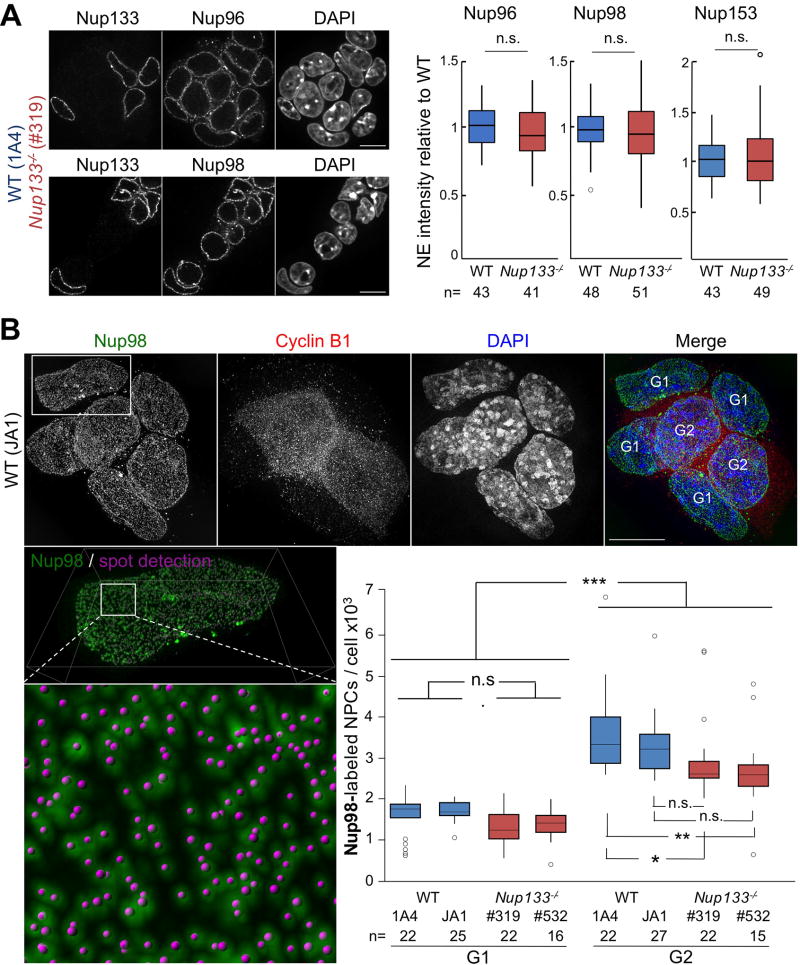
As a first approach to assess NPC composition and density in WT and Nup133-deficient mESCs, we quantified the average fluorescence intensity at the NE for three different Nups: Nup96, a component of the Y-complex; Nup98, a symmetric FG-Nup, and Nup153, a FG-Nup of the nuclear pore basket (Beck and Hurt, 2017). As depicted in Fig 1A, we observed no significant differences between WT and Nup133merm/merm (hereafter named Nup133−/−) mESCs in the level of fluorescence intensity measured for Nup96, Nup98 and Nup153, thereby suggesting that NPC density remains largely unaltered in the absence of Nup133 in mESCs.
Figure 1. NPC assembly appears unaltered in Nup133-deficient mESCs.
A. Left; representative spinning disk images (one z-section) of co-cultured WT (1A4) and Nup133−/− (#319) mESCs stained for DAPI, Nup133 and Nup96 (top) or Nup98 (bottom); bar 10µm. Right; Signal intensities for Nup96, Nup98 and Nup153 at the NE in WT (1A4) and Nup133−/− (#319) mESCs were quantified for n cells from ⪰ two independent experiments and normalized to the average for WT nuclei in the same field. B. Top; representative 3D-SIM images of WT mESCs stained for Nup98, Cyclin B1 and DAPI, bar 10µm. Bottom left; enlarged views of the NE surface of a G1 cell show Nup98-labeled NPCs and the corresponding spots defined as individual NPCs by Imaris. Bottom right; Imaris-based quantification of the total number of Nup98-labelled NPCs in WT (1A4, JA1) and Nup133−/− (#319, #532) mESCs in G1 and G2 (see Methods).
To refine this analysis and identify potential cell cycle–specific defects in NPC assembly, we applied structured illumination microscopy (SIM) to quantitate the number of individual nuclear pores during G1 and G2 in independently derived WT (1A4 and JA1) and Nup133−/− (#319 and #532) mESC lines. As detailed in "Experimental procedures" section, we labeled NPCs using a Nup98 antibody and classified cells as G1 or G2 based on the absence or presence, respectively, of strong cyclin B1 fluorescence (Fig 1B). We then used Imaris software to identify and count NPCs on SIM-acquired images of G1 and G2 cells. In WT mESCs this analysis measured 1.7 (± 0.4) 103 and 3.8 (± 0.9) 103 NPCs in G1 and G2, respectively (Fig 1B). These values fall within the range reported in previous studies (Dultz and Ellenberg, 2010; Maeshima et al., 2010; Maul et al., 1972) and indicate an approximate doubling of pore number during interphase in mESCS. Quantitation of single NPCs in Nup133−/− mESCs detected 1.3 (± 0.4) 103 and 2.8 (± 0.9) 103 NPCs per nucleus during G1 and G2, respectively. Compared to pore numbers measured in the WT counterparts, this represents a small decrease, with statistical significance only in G2. More importantly, this analysis documented a ~two-fold increase in NPC number between the G1 and the G2 phases of the cell cycle in Nup133−/− mESCs (Fig 1B). Our data thus indicate that Nup133 is largely dispensable for the assembly of new NPCs during interphase in pluripotent mESCs.
Assembly of Tpr within the basket of all NPCs requires Nup133
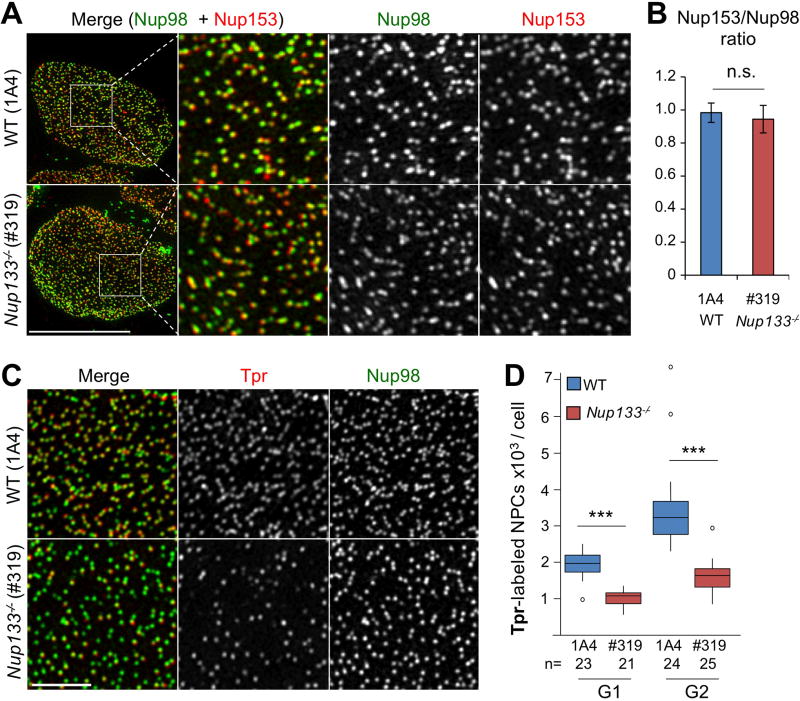
Concomitant with our studies quantitating Nup98-labeled pores, we performed dual color high resolution imaging on WT and Nup133−/− mESCs using, in addition to Nup98, antibodies specific for Nup153 or Tpr, two components of the NPC basket. SIM-acquired images revealed a widespread overlap in the patterns of fluorescence for Nup98 and Nup153 in both Nup133−/− and WT mESCs, validating the identification of individual NPCs (Fig 2A). Applying Imaris to SIM-acquired images of co-stained mESCs, we found a comparable Nup153/Nup98 ratio of pore numbers in WT and Nup133−/− mESCs (Fig 2B). Likewise, WT mESCs displayed extensive overlap of Nup98 and Tpr staining at the NE. In contrast, Nup133−/− mESCs contained multiple NPCs stained by Nup98 but not by Tpr (Fig 2C).
Figure 2. 3D SIM images reveal a widespread overlap in the patterns of fluorescence for Nup98 and Nup153 in WT and Nup133−/− mESCs but lack of Tpr in about half of the NPCs in Nup133-deficient mESCs.
A. Representative SIM Images of WT (1A4) and Nup133−/− (#319) mESCs double labeled with anti-Nup98 (green) and anti-Nup153 (red) antibodies. The square insets depict the region shown at higher magnification in the three panels to the right; the individual and merged images for Nup98 and Nup153. Bar 10 µm. B. Relative numbers of Nup98 and Nup153-labelled NPCs in WT (1A4) and Nup133−/− (#319) mESCs. Numbers of Nup98- and Nup153-labelled NPCs were independently quantitated for 11 WT (1A4) and 12 Nup133−/− (#319) mESCs and the ratio of Nup153 to Nup98-containing NPCs determined for each cell. The average ± SDs are presented. C. Representative SIM microscopy images of the NE surface of WT (1A4) and Nup133−/− (#319) mESCs stained for Nup98 and Tpr. Bar 2µm. D. Quantification of the total number of Tpr-labeled NPCs in WT and Nup133−/− mESCs during G1 and G2. See also Fig S1A.
To quantitate the reduction in numbers of Tpr-containing NPCs in Nup133−/− versus WT mESCs, we counted single pores labeled by Tpr antibodies in G1 and G2 cells. This analysis revealed an approximate two-fold reduction in the number of Tpr-labeled NPCs in Nup133−/− compared to WT mESCs at both G1 and G2 (Figs 2D and S1A). These results indicate that NPCs formed in the complete absence of Nup133 vary in their subunit composition: approximately one-half fail to assemble and/or maintain a Tpr-containing nuclear basket. Of note, the number of Tpr-stained NPCs doubles between G1 and G2 in both WT and Nup133−/− mESCs. Thus the observed heterogeneity in the nuclear basket of Nup133-deficient pores is unlikely to solely reflect a defect specific to interphase NPC assembly.
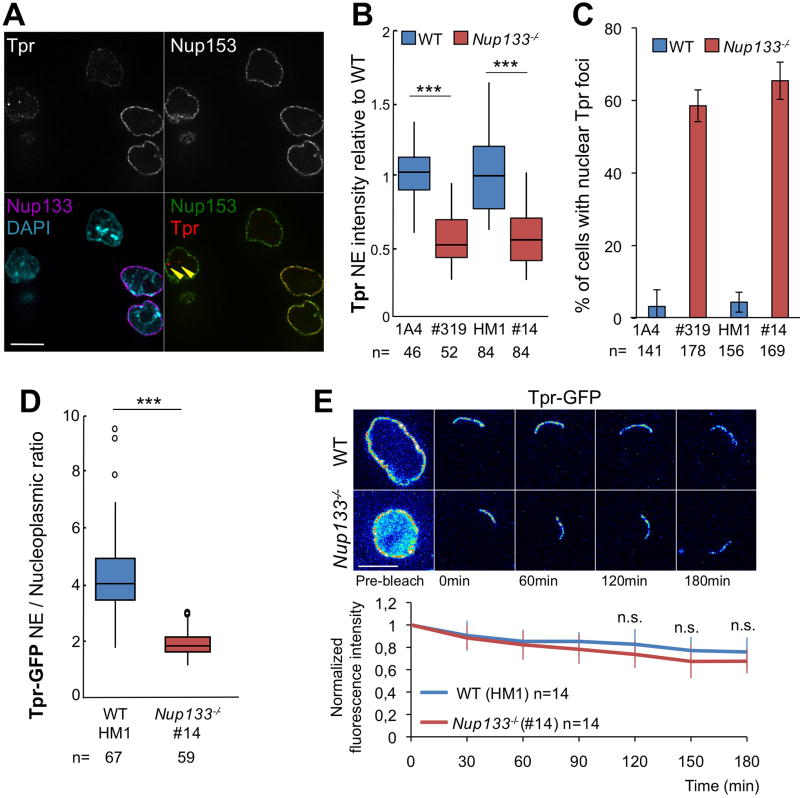
To independently confirm this observation, we quantified the levels of Tpr fluorescence at the NE. This analysis detected an approximate two-fold decrease in intensity of Tpr fluorescence in Nup133−/− (#319) compared to WT (1A4) mESCs (Fig 3A,B). We obtained an equivalent result in an independent Nup133−/− mESC line (#14), generated by CRISPR/Cas9 editing of WT-HM1 mESCs (Fig 3B; see Experimental procedures and Fig S1B,D). In contrast, mESCs co-stained for Nup153 and Tpr, showed similar levels of Nup153 fluorescence at the NE in the two distinct WT and Nup133−/− mESC lines (Fig S1C). These findings indicate that the Tpr-deficient NPC phenotype results from the loss of Nup133 rather than the strain background or other property specific to mESCs carrying the merm allele of Nup133.
Figure 3. The two-fold decrease in Tpr fluorescence intensity at the NE in Nup133-deficient compared to WT mESCs is not caused by its altered dynamics at NPCs.
A. Representative spinning-disk images (one z section) of WT (1A4) and Nup133−/− (#319) mESCs grown together and stained for Nup133, Nup153, Tpr and DAPI. Bar 10µm. Arrowheads point to intranuclear foci labeled by Tpr but not Nup153. B. Normalized Tpr signal intensity at the NE in WT (1A4 or HM1) and Nup133−/− (#319 or #14) mESCs. n: number of cells quantified from ⪰ two independent experiments; C. Quantification of the fraction of cells containing Tpr-positive and Nup153-negative intranuclear foci in WT and Nup133−/− mESCs. Average ± SD based on analysis of n cells from ⪰ two independent experiments. D. Tpr-GFP mislocalization in Nup133−/− mESCs was quantified based on the NE/Nucleoplasm intensity ratios acquired on individual live cells (n=) expressing Tpr-GFP. E. Top; Pseudocolored images of selected time frames (average projection of 3 z-sections used for quantitation) from representative iFRAP experiments performed on WT (HM1) or Nup133−/− (#14) mESCs expressing Tpr-GFP and treated with 50µM of cycloheximide. Time 0 was defined as the first time point after bleach. Scale bar, 10µm. Bottom; Plots of fluorescence decay kinetics from the unbleached region in WT (blue curves) and Nup133−/− (red curves) mESCs. Average normalized fluorescence signals ± SDs were quantified at each acquisition time point as detailed in Experimental procedures. See also Fig S1
In Nup133−/− mESCs we frequently detected a few bright Tpr-labeled nucleoplasmic foci that did not co-stain for Nup153 (Fig 3A). While rarely found in WT mESCs, such Tpr-positive/Nup153-negative foci were detected in a large fraction of Nup133−/− mESCs (Fig 3C). The absence of Nup153 distinguishes these foci from NPCs present on long NE invaginations, a common feature of nuclei in undifferentiated mESCs (Luo et al., 2009), and from intranuclear Nup clusters recently described in G0/G1 arrested cells (Perez-Garrastachu et al., 2017). However, this observation is reminiscent of the relocation of Tpr into intranuclear aggregates upon depletion of Nup153 in HeLa cells (Hase and Cordes, 2003) and suggests that a fraction of the Tpr protein not assembled into NPCs localizes to the nucleoplasm of Nup133−/− mESCs.
To validate these observations, we analyzed the subcellular location of human Tpr-GFP transiently transfected into WT and Nup133−/− mESCs under conditions that did not measurably alter total levels of Tpr at the NE (Fig S1E). Tpr-GFP localized at the NE in both WT and Nup133−/− mESCs, but in the latter, it was also detected within a few Nup153 negative intranuclear foci (Fig S1E). Moreover, in live transfected cells we found a significant decrease in the NE/nucleoplasmic ratio of Tpr-GFP in Nup133−/− compared to WT mESCs (Fig 3D), consistent with the reduction in NE-fluorescence observed in anti-Tpr stained Nup133−/− versus WT mESCs.
To determine if the observed mislocalization of Tpr reflects a change in its dynamics at NPCs, we applied inverse Fluorescence Recovery After Photobleaching (iFRAP) to mESCs transiently expressing Tpr-GFP (Rabut et al., 2004). Except for a small region of the NE, we photobleached the entire cell and measured the fluorescence decay of Tpr-GFP within the non-bleached region of the NE over 12 min (Fig S1F) or 3 hours (Fig 3E). The length of the non-bleached region of the NE of WT mESCs stayed fairly constant, indicating that NPCs did not diffuse within the plane of the NE over the time course of the assay. In addition, when normalized to non-bleached cells, the levels of Tpr-GFP fluorescence remained largely unchanged (Figs 3E and S1F), indicating that once assembled into NPCs, Tpr is quite stable in mESCs, a result in agreement with unpublished data reported in human cancer cell lines (Griffis et al., 2004). Most importantly, the iFRAP analysis found no significant difference in Tpr-GFP-dynamics between Nup133−/− and WT mESCs (Figs 3E and S1F), indicating that Tpr mislocalization in Nup133−/− mESCs is not the mere consequence of its increased dynamics at NPCs.
Nup133 indirectly contributes to limits on Nup153 dynamics at NPCs
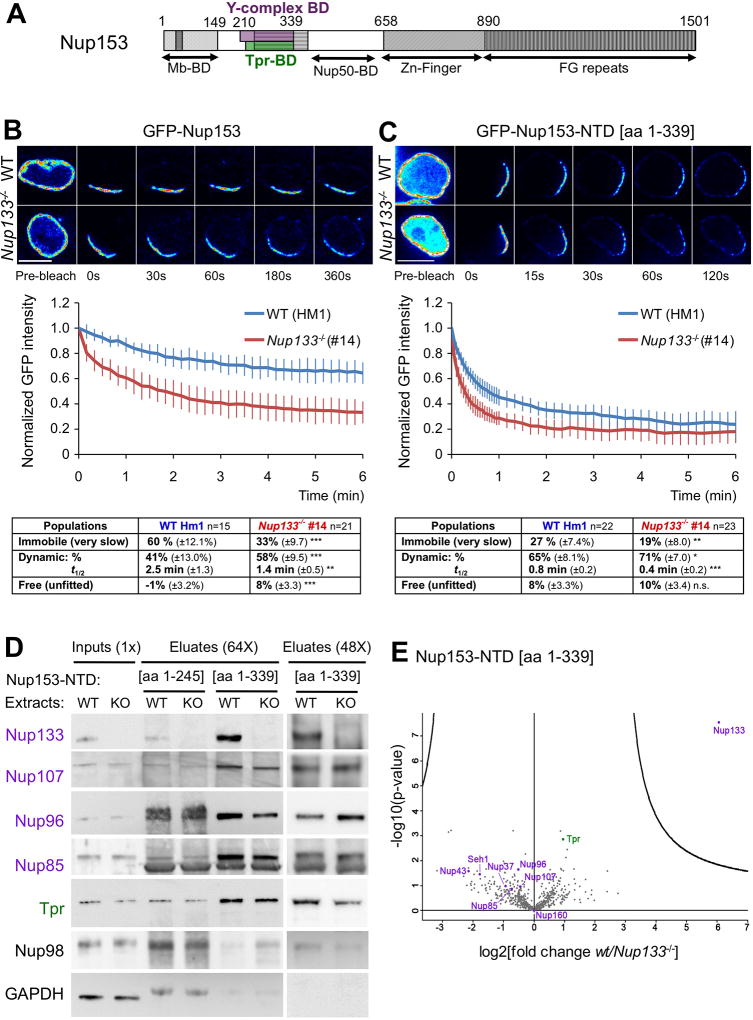
Nup153 is a large multifunctional protein that interacts with both the Y-complex and Tpr (Griffis et al., 2004; Hase and Cordes, 2003; Vasu et al., 2001; Vollmer et al., 2015) (Fig 4A). RNAi experiments previously demonstrated that long term depletion of Nup153 leads to the mislocalization of Tpr, implicating Nup153 in the assembly or maintenance of the nuclear basket (Hase and Cordes, 2003;Lussi et al., 2010; Mackay et al., 2010; Duheron et al., 2014). Our immunofluorescence data, showing largely unaltered levels of Nup153 at the NE, argue against reduced amounts of Nup153 as an explanation for Tpr-deficient pores in Nup133−/− mESCs (Figs 1A and S1C). However, these data do not take into account the dynamic properties of Nup153, notably the presence of at least two kinetically distinct populations of Nup153 at the NE (Griffis et al., 2004; Rabut et al., 2004). Therefore, we asked whether the absence of Nup133 perturbs the stability of Nup153 at NPCs by conducting iFRAP analyses on WT and Nup133−/− mESCs transiently transfected with GFP-Nup153. Consistent with previous studies, and in contrast to Tpr (Fig S1F), the iFRAP data depict Nup153 as quite dynamic in both WT and Nup133−/− mESCs lines (Fig 4B). However, the intensity of GFP-Nup153 fluorescence at the NE decreases at an appreciably faster rate in Nup133−/− mESCs as compared to their WT counterparts (Fig 4B). To rule out extraneous factors, we performed several control experiments. First, a comparison of the raw and normalized decay profiles, obtained from cells that express varying initial levels of GFP at the NE, showed that the amount of transfected Nup153 had no effect on its kinetics of dissociation (Fig S2A–B). Excluding strain and cell-line specific features, iFRAP experiments detected a similar increase in Nup153 dynamics at the NE when performed on an independently established WT-Nup133−/− mESC pair (Fig S2C). Finally use of a C-terminal rather than an N-terminal GFP-tagged Nup153 did not affect the iFRAP results (Fig S2D).
Figure 4. Nup133 deficiency increases Nup153 dynamics in vivo in mESCs but does not alter interactions between Nup153-NTD and the Y-complex or Tpr in vitro.
A. Schematic of Nup153 functional domains (BD, binding domain, Mb, membrane). Amino-acid numbers refer to human Nup153. See text for details. B–C. Top; pseudocolored images of selected time frames from representative iFRAP experiments performed on WT (HM1) or Nup133−/− (#14) mESCs expressing GFP-Nup153 (B) or GFP-Nup153-NTD (C). Time 0 was defined as the first time point after bleach. Scale bar, 10µm. Middle; Plots of fluorescence decay kinetics from the unbleached region in WT (blue curves) and Nup133−/− (red curves) mESCs. Average normalized fluorescence signals ± SDs were quantified at each acquisition time point as detailed in Experimental procedures. Cells were quantified from 3 (B) or 2 (C) independent experiments. Bottom; the fractions of the various GFP-Nup153 or GFP-Nup153-NTD populations exhibiting distinct dynamics at the NE and the half time of residency for the dynamic population (t1/2) were determined based on the fit of the normalized fluorescence signals from the indicated number of WT or Nup133−/− mESCs (see Experimental procedures). D. Pull-down experiments using as bait Nup153-NTD[aa1–245] or [aa1–339] and whole cell extracts from WT (HM1) or Nup133−/− (KO, #14) mESCs. Inputs (1× equivalent) and eluates (64× equivalent or 48× equivalent arising from a distinct experiment) were analyzed by western blot using the indicated Y-complex antibodies and Tpr. Anti-Nup98 and GAPDH were used as negative controls (original data are available as Mendeley dataset under doi:10.17632/w2bwx2528j.1). E. Volcano plot of quantitative mass spectrometry analysis of pull-down experiments using Nup153-NTD[aa 1–339] as bait and whole cell extracts from WT compared to Nup133−/− mESCs. The curves correspond to the 5% threshold cut-off. Note that except Nup133, Y-complex Nups (highlighted in purple) and Tpr (highlighted in green) are not significantly different between WT and Nup133−/−-purified fractions. See also Fig S2.
The decay kinetics of GFP-Nup153 in Nup133−/− mESCs did not fit with a single exponential function, consistent with multiple modes of interaction between Nup153 and NPCs (Griffis et al., 2004; Rabut et al., 2004). However, the curves did fit well with predictions of the reaction-dominant model (Sprague et al., 2004), which posits three distinct kinetic populations: an immobile (or slowly diffusing) subset stably anchored at the NPCs; a dynamic fraction; and an unfitted, "freely" diffusing class (see Experimental procedures). Our data find altered properties of GFP-Nup153 among these three populations in Nup133−/− versus WT mESCs: a decreased fraction of GFP-Nup153 behaves as immobile, while a larger fraction acts as freely diffusing molecules; in addition, the dynamic population shows more rapid dissociation from the NE, indicating a decreased affinity for NPCs (see tables within Figs 4B, S2C–D). Together these data demonstrate that the lack of Nup133 in mESCs alters the anchoring of Nup153 at the NPC by increasing its overall dynamics.
Because Nup153 interacts with the Y-complex and Tpr through its N-terminal NPC-targeting domain (NTD, [aa 1–339]) (Hase and Cordes, 2003; Vasu et al., 2001) (Fig 4A), the altered Nup153 dynamics observed in Nup133−/− mESCs may reflect perturbed interactions between its NTD and the NPC. To explore this possibility, we conducted iFRAP on WT and Nup133−/− mESCs transiently transfected with GFP-Nup153-NTD [aa 1–339].
GFP-Nup153-NTD localized at the NE; yet compared to GFP-Nup153, a larger fraction accumulated inside the nucleus (Fig 4C). In addition, and consistent with a previous study (Griffis et al., 2004), analysis of the post-bleach curves in WT mESCs revealed increased dynamic behavior of GFP-Nup153–NTD compared to that of GFP-Nup153 (See Tables within Figs 4B–C). Most importantly, the loss of Nup133 further enhanced the dynamic properties of GFP-Nup153-NTD in mESCs (Fig. 4C).
To explore the possibility that the interaction between Nup153-NTD and the Y-complex or Tpr requires Nup133, we performed pull down assays on whole protein extracts from WT and Nup133−/− mESCs using recombinant Nup153-NTD [aa 1–339] as bait. Nup153 [aa 1–245], lacking the binding domains for the Y-complex and Tpr (Griffis et al., 2004; Hase and Cordes, 2003; Vasu et al., 2001), served as control. Western blot and quantitative mass spectrometry analyses detected Nup133 only in WT samples and documented its specific enrichment, along with other tested Y-Nups and Tpr, in the Nup153-NTD [aa 1–339] bound fraction (Figs 4D and S2E). These analyses also revealed a similar enrichment of Tpr and Y-complex proteins in Nup153-NTD [aa 1–339]-purified extracts from either WT or Nup133−/− mESCs (Fig 4D–E). Thus, Nup133 does not represent the major Nup153-binding site to the Y-complex. These data show that in the context of whole cell lysates containing NPCs as dissociated subcomplexes, the lack of Nup133 neither precludes nor appreciably affects the biochemical interaction between Nup153-NTD and the Y-complex or Tpr. This differs from the in vivo situation that revealed a measurable impact of Nup133 on Nup153 dynamics and Tpr localization in the context of assembled NPCs. This apparent discrepancy between our in vitro and in vivo data indicates that the structural integrity of the NPC is likely required to visualize the contribution of Nup133 to the binding of Nup153 to Tpr or to the Y-complex.
The central domain of Nup133 is required for proper NPC basket assembly and dynamics
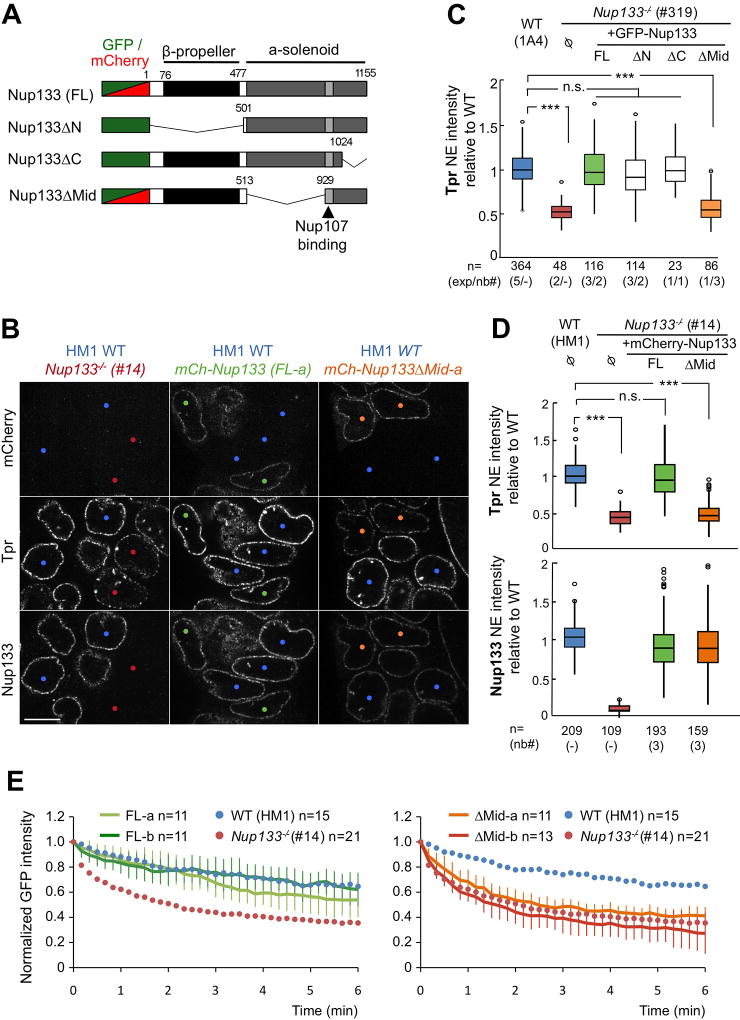
The data presented thus far on Nup133−/− mESCs indicate that Nup133 contributes to both Tpr recruitment and Nup153 dynamics at NPCs. To determine whether these two functions require the same domain of Nup133, we generated Nup133−/− mESC lines stably expressing GFP-or mCherry-tagged Nup133, either full-length (FL) or bearing one of three different deletions (Fig 5A): ΔN, a deletion of the N-terminal 500 aa that includes the first unfolded residues and β-propeller of Nup133; ΔMid, an α-solenoid domain deletion of aa 513–929 containing 17 α-helices located upstream of the Nup107 binding site; and ΔC, a second α-solenoid domain deletion of aa 1024–1155 comprising the last six C-terminal α-helices. All Nup133 fusion proteins were properly expressed and localized at the NE (Figs 5B and S3A–B), consistent with the presence of the Nup107-binding domain [aa 932–1024].
Figure 5. The Mid domain of Nup133 is required for NPC basket assembly.
A. Schematic representation of the GFP or mCherry-Nup133 constructs used in this study. B. Representative spinning-disk images of HM1 WT mESCs (blue dots) mixed and co-cultured with either Nup133−/− mESCs (#14, red dots, left panels), or with Nup133−/− mESCs stably expressing mCherry-Nup133 (mCh-Nup133 FL-a; green dots, middle panels) or mCherry-Nup133ΔMid (mCh-Nup133ΔMid-a; orange dots, right panels) and immunostained for Nup133 and Tpr. One z section of acquisition is shown. Bar, 10µm. C. Normalized Tpr signal intensity at the NE in WT (1A4) and Nup133−/− (#319) mESCs, either control (Ø) or stably expressing GFP-Nup133 (FL), GFP-Nup133ΔN or ΔMid or ΔC. The numbers of cells quantified (n), of independent experiments (exp) and when relevant, of independent clones analyzed (nb#) are indicated. D. Tpr and Nup133 normalized intensities at the NE in the indicated cell lines were quantified and plotted as in (C). E. Plots of GFP-Nup153 fluorescence decay kinetics from the unbleached region in Nup133−/− mESCs (#14) stably expressing either mCherry-Nup133 (FL-a and FL-b, left) or mCherry-Nup133ΔMid (ΔMid-a and ΔMid-b, right) at comparable level (see Fig. S3C). Average normalized fluorescence signals ± SDs are plotted at each time point and were quantified from the indicated number of cells (n=). Average values from iFRAP experiments on WT (HM1, blue dots) and Nup133−/− mESCs (#14, red dots) arising from Fig 4B are plotted on both graphs as references. See also Fig S3.
We then compared the levels of Tpr fluorescence detected at the NE of these various cell lines to those measured for WT and Nup133−/− mESCs (Fig 5C–D). Our findings show that expression of GFP- or mCherry-Nup133-FL efficiently complements the Tpr assembly defect at the NE of Nup133-deficient mESCs (Fig 5C–D). Expression of GFP-Nup133ΔN and GFP-Nup133ΔC also returned the intensity of Tpr fluorescence to WT levels at the NE, indicating that neither the N-terminal β-propeller of Nup133 nor its last C-terminal α-helices play critical roles in assembling Tpr into the NPC basket. In contrast, GFP- or mCherry-Nup133ΔMid expression failed to rescue Tpr localization to the NE (Fig 5B–D). Importantly, immunofluorescence and western blot analyses detected comparable levels of the Nup133-FL and -ΔMid fusions in transfected Nup133−/− mESC lines, eliminating potential impaired expression and localization of Nup133ΔMid as the reason for lack of complementation (Figs 5B, D and S3A, C, E–F). To identify the α-helices within the Nup133-Mid domain that mediate Tpr localization, we generated mCherry-Nup133ΔMid fusions for five overlapping deletions within this region (Fig S3D). None of these ΔMid deletions rescued Tpr localization, indicating that proper assembly of Tpr within the NPC involves the integrity of the entire Mid-domain of Nup133 (Fig S3F).
To determine if the lack of the Nup133-Mid domain also affects Nup153 dynamics, we performed iFRAP assays on Nup133−/− mESC clones stably expressing mCherry-Nup133 or mCherry-Nup133ΔMid and transiently transfected with GFP-Nup153 (Fig 5E). Nup133−/− mESCs expressing mCherry-Nup133-FL showed GFP-Nup153 dynamics at the NE very similar to those observed in WT mESCs, demonstrating rescue of the defect in Nup153 dynamics in Nup133−/− mESCs (Fig 5E, left). In contrast, Nup133−/− mESCs expressing mCherry-Nup133ΔMid, exhibited GFP-Nup153 dynamics resembling those found in Nup133−/− mESCs; thus proper dynamics of Nup153 at NPCs requires the Mid domain of Nup133 (Fig 5E, right).
Together, these data show that deletion of the Nup133-Mid domain recapitulates the two phenotypic defects in NPC basket assembly found in mESCs completely lacking Nup133; Tpr localization and Nup153 dynamics are thus closely correlated properties of NPCs.
Discussion
In this study, we have combined single NPC counting and measurements of fluorescence intensity at the NE to examine NPC assembly and dynamics in WT and Nup133-deficient mESCs. Consistent with our previous qualitative characterization of Nup133−/− (merm) embryos and mESCs (Lupu et al., 2008), these quantitative approaches reveal that Nup133 is dispensable for assembly of the NPC scaffold in pluripotent cells. This differs from previous Nup133 RNAi experiments performed in HeLa cells (Walther et al., 2003) likely because Y-complex Nups are not destabilized in Nup133−/− mESCs (Lupu et al., 2008 and Fig 4D).
Our data also reveal that the complete absence of Nup133 does not prevent interphase NPC assembly in pluripotent mESCs. In contrast, a previous study conducted in U2OS cells concluded that interphase NPC assembly requires an amphipathic helix within the Nup133-β-propeller (ALPS motif) (Doucet et al., 2010). Notably, the Nup133 I254D ALPS mutant used in that study mislocalizes to the cytoplasm, whereas the deletion of the entire N-terminal domain does not affect Nup133 targeting to NPCs in either HeLa cells (Bolhy et al., 2011) or mESCs (this study, Fig S3B). This suggests that the negatively charged residue introduced into the Nup133 amphipathic helix by the I254D substitution may result in repulsive interactions with the NE. Thus Nup133 I254D, rather than causing loss-of-function, may generate an active inhibitor of NPC assembly, potentially explaining the apparent inconsistency between our findings and those of Doucet et al., 2010. Alternatively, a redundant pathway that bypasses the requirement for the Nup133 ALPS motif may be present in mESCs but absent in U2OS cells. Consistent with this possibility, Vollmer et al. identified an amphipathic helix in Nup153 that recruits the Y-complex to the inner side of the NE for interphase NPC assembly in Xenopus egg extracts (Vollmer et al., 2015).
While formation of the NPC scaffold proceeds largely unaltered in the absence of Nup133 or its central (Mid) domain, our results indicate that assembled NPCs vary in their subunit composition: approximately one-half lack a Tpr-containing nuclear basket. The iFRAP data showing a comparable off-rate for Tpr from NPCs in WT and Nup133−/− mESCs (Figs 3D and S1F) suggest that Tpr-deficient pores reflect a defect in assembly, rather than in stability, of the NPC basket. The key question then becomes why the absence of Nup133 specifically affects incorporation of Tpr into some NPC baskets. The implication of Tpr in the establishment of heterochromatin exclusion zones (Krull et al., 2010), as well as studies in budding yeast, suggest that alteration in chromatin structure might lead to assembly of Tpr-deficient pores. In WT yeast cells, the Mlp1/2 proteins (yeast Tpr orthologs) are excluded from NPCs located close to the nucleolus, possibly due to steric hindrance imposed by the nucleolus or by a peculiar chromatin conformation present beneath these NPCs (Galy et al., 2004). Considering the multiple links between nucleoporins and chromatin organization (Ptak and Wozniak, 2016), the absence of Nup133 may affect heterochromatin distribution, thereby locally preventing basket assembly by a similar hindrance-based mechanism. Nonetheless, this would have to occur at a local level, since no specific clustering of Tpr-less NPCs is apparent in the Nup133−/− mESCs (Fig 2C). However, we favour an alternative model in which the alteration imposed on the architecture of the NPC scaffold by the absence of Nup133, or its Mid domain, impairs or delays the basket assembly process. Indeed, a stereotypic organization of 16 Y-complexes defines the structures of the nuclear and cytoplasmic rings of the NPC scaffold; thus assembling a pore in the absence of one of the 9 Y-complex Nups will likely shift how the remaining 8 Y-subunits interact with each other, as well as with other NPC subcomplexes. In this model the co-existence of Tpr+ and Tpr− NPCs in Nup133−/− mESCs would reflect the existence of two types of NPC scaffolds, each conferring a distinct capability/fate on a NPC: unsuccessful or successful initiation and rapid completion of basket assembly.
We further demonstrate that the Mid domain, located upstream of the Nup107 binding site, mediates Nup133’s role in both Tpr localization to all NPCs and proper anchorage of Nup153 at NPCs (Figs 5 and S3). This indicates a mechanistic connection between these two properties of the nuclear basket. Because Nup153 directly interacts with Tpr (Hase and Cordes, 2003), Tpr-deficient NPCs might lack a subset of high affinity binding sites for Nup153, causing in turn, the overall increased mobility of Nup153 measured by iFRAP. Alternatively, the absence of Nup133 may primarily lead to the loss of high affinity binding sites for a specific kinetic subpopulation of Nup153 at NPCs, namely a stable pool required for the recruitment of Tpr. This hypothesis is consistent with the reported contribution of Nup153 to the assembly of Tpr within the nuclear basket (Hase and Cordes, 2003; Lussi et al., 2010; Mackay et al., 2010; Duheron et al., 2014).
In either case, our biochemical data (Figs 4D–E and S2E) indicate that the Mid domain of Nup133 does not represent the unique link between the Y-complex and either Tpr or Nup153. Consistently, functional links between the Tpr/Nup153 basket Nups and Y-complex Nups distinct from Nup133, specifically Nup107, are emerging. In vertebrates, a structural model of the NPC places Nup107 in close proximity to an electron optical density located on the nuclear side of the NPC scaffold; based on previous BioID experiments this density likely represents Tpr (von Appen et al., 2015; Beck and Hurt, 2017; Kim et al., 2014). In budding yeast, the Nup153 ortholog - yNup60- directly interacts in vitro with the heterodimer between Nup133 and the yeast Nup107 ortholog -yNup84. In vivo however, the loss of yNup84 -but not of yNup133-leads to altered localization of yNup60 (Nino et al., 2016). Evaluating our findings in the context of these data, we propose that the Mid domain of Nup133 contributes to proper NPC basket assembly by stabilizing a Nup107-dependent binding interface between the nuclear ring and the NPC basket. Noteworthy, mutations within NUP107 affecting Nup133 localization or protein levels were recently identified in patients with steroid resistant nephrotic syndromes (Miyake et al., 2015; Rosti et al., 2017). By providing insights into the interplay between NPC scaffold architecture and nuclear basket assembly, our study thus lays the foundation for future investigations into the role of scaffold Nup functions in normal and pathological conditions.
Experimental Procedures
Plasmids, cell lines, and oligonucleotides used in this study are provided in Supplemental Experimental Procedures as Tables S1 to S4, respectively. Classical procedures including details of conditions for cell growth and transfection, real-time quantitative PCR, and western blot analyses, are provided in Supplemental Experimental Procedures.
mESC lines
WT Nup133+/+ (1A4, JA1) and mutant Nup133merm/merm (#319, #532) mESC lines were generated from embryonic day 3.5 (E3.5) blastocysts recovered from 129.B6 Nup133merm/+ females mated to B6.129 Sox1gfp/+; Nup133merm/+ (JA1, 319, 532) or B6.129 Sox1gfp/+; Nup133+/+ (1A4) males following published procedures (Hogan et al., 1994; Nagy et al., 2003). Sox1gfp, a knockout-reporter allele, was previously crossed into the Nup133merm/+ strain to provide an early, specific marker of the neuroectoderm lineage (Aubert et al., 2003). mESCs homozygous for Nup133merm, an ENU-induced allele, express no detectable Nup133 protein (Lupu et al., 2008; Fig S1B,D) and are designated as Nup133−/− in this study. All mouse lines used for the generation of mESCs were housed and bred following procedures detailed in a protocol approved by the Memorial Sloan Kettering Institutional Animal Care and Use Committee.
Nup133−/− mESCs stably expressing GFP- or mCherry-tagged Nup133-FL, -ΔN, -ΔC or -ΔMid, were initially selected on 1µg/ml of puromicin (Invivogen) and subsequently grown in the presence of 0.5 µg/ml puromycin.
Nup133−/− (#14) mESCs were derived from HM1 cells by CRISPR/Cas9 editing and feature a large deletion of exons 2 through 6 on one allele, and a 35 bp deletion within exon 6 on the other allele; these lead to frameshifts and premature stop codons after aas 60 and 260, respectively. As a consequence of non-sense mediated mRNA decay, levels of the corresponding mRNAs are also strongly decreased (Fig. S1B).
Except for 3D-SIM imaging, mESCs were grown in serum/leukemia inhibitory factor (LIF)-containing stem cell medium on inactivated DR4-mouse embryonic fibroblasts (MEFs) (MEF-derived feeders).
Immunostaining and quantification of fluorescence signals at the NE
mESCs grown on coverslips were washed with phosphate-buffered saline (PBS), then fixed using 3% paraformaldehyde (VWR) for 20 minutes, washed again with PBS, and permeabilized in PBS with 0.1% triton X-100 (Sigma), 0.02% Sodium Dodecyl Sulfate (SDS, Euromedex) and 10mg/ml bovine serum albumin (BSA, sigma). Antibody hybridizations and washes were also performed in this buffer. Primary and secondary antibodies were incubated for one hour at room temperature (antibodies used in this study are listed in Supplemental Experimental Procedures). Cells were then incubated 5 min with 1.10−4 mg/ml 4',6-diamidino-2-phenylindole (DAPI, Sigma) in PBS and mounted with Vectasheild (Vector). Images were acquired on either an inverted microscope (DMI8, Leica) with a CSU-W1 spinning disk head (Yokogawa, Japan) and a sCMOS Orca-Flash 4 V2+ camera (Hamamatsu) using 100x/1.4 oil objective or on an inverted microscope (Ti-Elipse, Nikon), with a CSUX1FW spinning disk head (Yokogawa, Japan) and EM-CCD camera (Hamamatsu) using 100x/1.4 oil objective.
To quantify fluorescent signals at the NE, the mESCs of interest were mixed with WT mESCs of the same genetic background (used as reference) and grown for 24h on coverslips prior to fixation and immunostaining. On each acquired image (≥ 3 independent images per condition), one z-section was selected; 8-pixels thick ROIs were drawn freehand on the NE of both WT and mutant mESCs. Following subtraction of background, the signal intensity at the NE for each cell was normalized to the average NE intensity measured for the WT mESCs in the same field.
The % of cells with Tpr-positive/Nup153-negative nuclear foci was determined in WT and Nup133−/− cells based on 3D-projections of 24 sections acquired with a z-step of 0.5µm and covering the entire depth of most nuclei.
High Resolution Imaging (3D SIM): immunostaining, image acquisition and processing (detailed procedures are provided in Supplemental Experimental Procedures)
For 3D SIM imaging, mESCs grown in 2i plus LIF medium were passaged onto fibronectin-coated cover slips (Zeiss) and grown for 2–3 days prior to staining. Cells were fixed in 4% paraformaldehyde for 15 minutes, washed in PBS, permeabilized with 0.2% Triton X-100 for 6 min, and saturated in IF Wash Buffer [PBS plus 3% IgG-free BSA and 0.1% Triton X-100] for 30 to 60 min. Antibody hybridizations and washes were also performed in this buffer. Following washes, cells were incubated with secondary antibodies, washed again and mounted with Prolong Gold (ThermoFisher).
For incubation with 2 rabbit antibodies (anti-Tpr, anti-Nup98; as in Fig 2C), initial antibody incubation and washes were with rabbit anti-Tpr, followed by Alexafluor 568-conjugated donkey anti-rabbit IgG plus DAPI. After incubation with IF wash buffer containing 3% normal rabbit serum, cells were incubated with rabbit anti-Nup98, preconjugated to AlexaFluor 488 (Cell Signaling Technology), followed by final washes and mounting.
3D-SIM super-resolution microscopy was performed using a DeltaVision OMX V4/Blaze 3D-SIM super-resolution microscope (GE Healthcare) housed in the Rockefeller University's Bio-Imaging Resource Center (BIRC). This OMX system was fitted with a 100x/1.40 UPLSAPO oil objective (Olympus); Evolve EMCCD cameras (Photometrics) were used in EM gain mode at a gain of 170, with 405nm, 488 nm and 568 nm laser lines for excitation, and 436/31, 528/48 and 609/37 nm emission filters, respectively. Optical sections were acquired at 125-nm intervals. Immersion oil refractive index (R.I.) was selected to optimize for the 568 nm channel and the ambient temperature. Structured illumination data sets were reconstructed using softWoRx software (GE Healthcare) as described (Demmerle et al., 2017). The effective pixel size of the reconstructed images is 40nm in xy.
3D-SIM images were processed using the Imaris software (Bitplane). Tpr, Nup98 and Nup153 antibodies selected for pore quantitation produced negligible intra and extra-nuclear staining, enabling visual definition of the nuclear boundary. To define fluorescence as a pore, the size of the “spots” function in Imaris was set at 80 nm and the intensity threshold at automatic. Under these conditions, the antibodies to Tpr, Nup98 and Nup153 generated a pixel intensity histogram with a prominent trough at the position where the automatic threshold was routinely placed. Such a curve of pixel intensities indicated that small changes in the threshold would not significantly impact the estimated total number of pores. Also, under these settings, the secondary antibody only controls produced few, if any, spots.
iFRAP experiments
For iFRAP experiments, mESCs were transiently transfected using plasmids expressing the indicated GFP-tagged protein. About 36h hours after transfection, mESCs were trypsinized and ~1.106 mESCs were seeded on microscopy-adapted 35mm dishes (µ-dish, 35 mm, high, Ibidi, Germany) coated with 0.1% gelatin and DR4-MEFs. Cells were used at least 4 hours after plating. Acquisitions were performed at 37°C and 5% of CO2 using an Axio Observer.Z1 inverted microscope (Zeiss, Germany) equipped with a 100x/1.4 oil objective, a CSU-X1 spinning disk head (Yokogawa, Japan) and a EMCCD QuantEM 512SC camera (Photometrics, AZ, USA) and a sCMOS PRIME 95B (Photometrics). The whole setup was driven with MetaMorph software (Molecular Devices, Sunnyvale, CA). Following pre-bleach acquisitions, the GFP signal within most of the cell excluding a small region of the NE was photobleached with a 473-nm line at 100% transmission (three iterations). 3z-sections with a step of 0.3µm were acquired at intervals adapted to the dynamics of the bleached protein, namely 10-s intervals for 6 min for GFP-Nup153 and Nup153-GFP; 3-s intervals for one min and 10-s intervals for 5 min for GFP-Nup153-NTD; 10-s intervals for 6 min and 30-s intervals for 6 min or 10-min intervals for 3 hours for Tpr-GFP. For the 3 hours-long experiments, 50µM of cycloheximide (Sigma) was added to prevent de novo protein synthesis. The raw data were processed using ImageJ software (National Institutes of Health, Bethesda, MD).
For quantification and representative illustrations, the average projection of the 3 z-sections was performed. For illustrations, pseudo colors “royal” were used to help the visualization of fluorescence intensities. To quantify the NE/Nucleoplasm intensity ratios of Tpr-GFP in live cells, ROIs were freehanded drawn on the NE and inside the nuclei (excluding the nucleoli and intranuclear foci) of individual z-sections before bleach, and areas outside of the cells were used for background subtraction. For iFRAP quantification, sequences were aligned using the StackReg registration plug-in on ImageJ. 5 pixels-thick freehand ROIs were freehanded drawn on the non-bleached part of the NE. After background subtraction, the fluorescence signals were subjected to double normalization using a manually defined region covering non-bleached cells to take into account the loss of total signal due to bleaching during post-bleach imaging. Raw data were then normalized to 1 at the first post-bleach point (time 0) to account for differences in fluorescence intensities in-between cells and experiments (Fig S2B). The resulting decay curves for each cell were fitted using the Matlab (The Math Works, Natick, MA) routine nlinfit, by the following model: Frap(t) = A1 + A2e−kt, where A1 and A2 are the fractions of non-diffusing (immobile) and reacting (dynamic) populations respectively, and k is the dissociation rate. In this model the loss of fluorescence is described by a non-diffusing and a reacting population. The assumption of this model is that the diffusion is very fast compared to binding and to the timescale of the FRAP measurement (Sprague et al., 2004). This “freely” diffusing population explains the difference between the theoretical curves resulting from the fit and the corresponding experimental data curves at t=0.
Graphs and Statistics
Box plots were generated using KaleidaGraph (Synergy Software): each box encloses 50% of the normalized values obtained, centered on the median value. The bars extending from the top and bottom of each box mark the minimum and maximum values within the dataset falling within an acceptable range. Values falling outside of this range are displayed as an individual point. Other graphs were generated using Excel. Statistical analyses were performed using two way ANOVA Tukey’s honest significant difference (HSD) test using the KaleidaGraph software except for Figs 2B, 3C and S1B where a t-test was performed using Excel. Standard conventions for symbols indicating statistical significance were used: ns, not significant: P> 0.05; *: P ≤ 0.05; ** :P ≤ 0.01, *** :P ≤ 0.001.
Pull-down experiments and mass spectrometry analyses
His6-ProtA-Nup153-NTD-His6 recombinant proteins (aa 1–245 or 1–339) were produced from bacteria transformed with the pET28a-zz-hNup153 vectors (Vasu et al., 2001), purified on nickel beads, and incubated with mESC lysates as detailed in Supplemental Experimental Procedures. After incubation and extensively washes, the proteins were eluted either in 50µL of Laemmli for subsequent western blot analyses or washed with 20 mM (NH4)2CO3 digested on beads with trypsin and processed for mass-spectrometry analysis as detailed in Supplemental Experimental Procedures. Raw LC-MS/MS data were analyzed and quantified using MaxQuant (Cox et al., 2011) and MaxLFQ (Cox et al., 2014). Results are presented as Volcano-plots (Hubner and Mann, 2011) with a threshold set with an FDR < 0.05 and an S0 of 2.
Supplementary Material
Acknowledgments
We are grateful to I. Bouhlel for characterization of Nup133 antibody, to K. Taoud for help with characterizing Nup133ΔMid1-5 fusions, and to V. Heyer for generating the CRISPR/Cas9 vectors; we kindly thank B. Burke, V. Cordes, B. Fontoura and D. Forbes for sharing antibodies. We also acknowledge J. Collignon, C. Panayotou and V. Ribes for advices regarding mESCs culture and B. Palancade for critical reading of the manuscript. We thank A. North of the Rockefeller University Bio-Imaging Resource Center (BIRC) for expert guidance on acquisition of 3D-SIM imaging data; V. Boyko and S. Fujisawa of the Sloan Kettering Institute (SKI) Molecular Cytology Core for assistance on image analysis; and J. Liu and W. Mark of the SKI Mouse Genetics Core for assistance in ESC line derivation. We thank the proteomics core facility at the Institut Jacques-Monod, notably T. Léger and C. Garcia, for the LC-MS/MS experiments, Q. Defenouillere for MaxQuant analyses, and the Region Ile-de-France (SESAME), the Paris-Diderot University (ARS), and CNRS for funding part of the LC-MS/MS equipment. The BIRC OMX 3D-SIM system in the Rockefeller University Bio-Imaging Resource Center (BIRC) used in this study was supported by award S10RR031855 from the National Center for Research Resources. The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health. We also acknowledge the ImagoSeine core facility of the Institut Jacques Monod, member of IBISA and of the France-Bioimaging (ANR-10-INBS-04) infrastructures, notably G. Wentzinger and X. Baudin for help with cell sorting and spinning disk imaging, respectively. Work in the laboratory of VD is supported by the Centre national de la recherche scientifique (CNRS), the French National Research Agency under grant No. ANR-2012-BSV2-0008-01 to VD, the "Fondation pour la Recherche Médicale” (Foundation for Medical Research) under grant No DEQ20150734355, “Equipe FRM 2015" to VD and by the Labex Who Am I? (ANR-11-LABX-0071; Idex ANR-11-IDEX-0005-02). Support for work in the laboratory of EL came from NYSTEM C024333, Tri-Institutional Stem Cell Initiative UO1 Tri-I-Sci-EL, and the MSK CCSG P30 CA008748. BS was supported by "la Fondation ARC pour la recherche sur le Cancer", AB received PhD fellowships from the "Ministère de l’Enseignement Supérieur et de la Recherche" and the "Ligue Nationale contre le Cancer" and a “transition post-doc” grant from the Labex Who Am I?.
Footnotes
Accession Codes
The original 16-bit images and montages of the western blots used for Fig 4D are available as Mendeley dataset under doi:10.17632/w2bwx2528j.1
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2016) with the dataset identifier PXD000000.
Author Contributions
BS, EF, AB, BR, EL and VD conceived and designed the experiments; BS, EF, AB, and VA performed the experiments; BS, EF, AB, NA, EL and VD analyzed the data; BS, VD, and EL wrote the manuscript with contribution from all co-authors.
Declaration of interest
The authors declare that they have no conflict of interest.
References
- Aubert J, Stavridis MP, Tweedie S, O'Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, et al. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11836–11841. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol. 2017;18:73–89. doi: 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukata L, Parker SL, D'Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–386. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- D'Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- Demmerle J, Innocent C, North AJ, Ball G, Muller M, Miron E, Matsuda A, Dobbie IM, Markaki Y, Schermelleh L. Strategic and practical guidelines for successful structured illumination microscopy. Nat Protoc. 2017;12:988–1010. doi: 10.1038/nprot.2017.019. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duheron V, Chatel G, Sauder U, Oliveri V, Fahrenkrog B. Structural characterization of altered nucleoporin Nup153 expression in human cells by thin-section electron microscopy. Nucleus. 2014;5:601–612. doi: 10.4161/19491034.2014.990853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116:63–73. doi: 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezwani M, Fahrenkrog B. The functional versatility of the nuclear pore complex proteins. Semin Cell Dev Biol. 2017;68:2–9. doi: 10.1016/j.semcdb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Hoelz A, Glavy JS, Beck M. Toward the atomic structure of the nuclear pore complex: when top down meets bottom up. Nat Struct Mol Biol. 2016;23:624–630. doi: 10.1038/nsmb.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddinton R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hubner NC, Mann M. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC) Methods. 2011;53:453–459. doi: 10.1016/j.ymeth.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A. 2014;111:E2453–2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Gassman KL, Petell LM, Wilson CL, Bewersdorf J, Shopland LS. The nuclear periphery of embryonic stem cells is a transcriptionally permissive and repressive compartment. J Cell Sci. 2009;122:3729–3737. doi: 10.1242/jcs.052555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 2010;1:71–84. doi: 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J Cell Biol. 2010;191:923–931. doi: 10.1083/jcb.201007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima K, Iino H, Hihara S, Funakoshi T, Watanabe A, Nishimura M, Nakatomi R, Yahata K, Imamoto F, Hashikawa T, et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol. 2010;17:1065–1071. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem. 2012;287:38515–38522. doi: 10.1074/jbc.M112.378893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Maul HM, Scogna JE, Lieberman MW, Stein GS, Hsu BY, Borun TW. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Tsukaguchi H, Koshimizu E, Shono A, Matsunaga S, Shiina M, Mimura Y, Imamura S, Hirose T, Okudela K, et al. Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am J Hum Genet. 2015;97:555–566. doi: 10.1016/j.ajhg.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Nino CA, Guet D, Gay A, Brutus S, Jourquin F, Mendiratta S, Salamero J, Geli V, Dargemont C. Posttranslational marks control architectural and functional plasticity of the nuclear pore complex basket. J Cell Biol. 2016;212:167–180. doi: 10.1083/jcb.201506130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Tang JH, Andersen KR, Knockenhauer KE, Vallotton P, Derrer CP, Kralt A, Mugler CF, Chan LY, Schwartz TU, et al. Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell. 2017 doi: 10.1016/j.cell.2017.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garrastachu M, Arluzea J, Andrade R, Diez-Torre A, Urtizberea M, Silio M, Arechaga J. Nucleoporins redistribute inside the nucleus after cell cycle arrest induced by histone deacetylases inhibition. Nucleus. 2017:1–19. doi: 10.1080/19491034.2017.1320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak C, Wozniak RW. Nucleoporins and chromatin metabolism. Curr Opin Cell Biol. 2016;40:153–160. doi: 10.1016/j.ceb.2016.03.024. [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Raices M, D'Angelo MA. Nuclear pore complexes and regulation of gene expression. Curr Opin Cell Biol. 2017;46:26–32. doi: 10.1016/j.ceb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti RO, Sotak BN, Bielas SL, Bhat G, Silhavy JL, Aslanger AD, Altunoglu U, Bilge I, Tasdemir M, Yzaguirrem AD, et al. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J Med Genet. 2017;54:399–403. doi: 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Snow CJ, Paschal BM. Roles of the nucleoporin Tpr in cancer and aging. Adv Exp Med Biol. 2014;773:309–322. doi: 10.1007/978-1-4899-8032-8_14. [DOI] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B, Lorenz M, Moreno-Andres D, Bodenhofer M, De Magistris P, Astrinidis SA, Schooley A, Flotenmeyer M, Leptihn S, Antonin W. Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev Cell. 2015;33:717–728. doi: 10.1016/j.devcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.