Summary
Telomerase is an RNA-protein complex (RNP) that extends telomeric DNA at the 3’-ends of chromosomes using its telomerase reverse transcriptase (TERT) and integral template-containing telomerase RNA (TER). Its activity is a critical determinant of human health, affecting aging, cancer, and stem cell renewal. Lack of atomic models of telomerase, particularly one with DNA bound, has limited our mechanistic understanding of telomeric DNA repeat synthesis. We report the 4.8Å resolution cryo-electron microscopy structure of active Tetrahymena telomerase bound to telomeric DNA. The catalytic core is an intricately interlocked structure of TERT and TER, including a previously structurally uncharacterized TERT domain that interacts with TEN domain to physically enclose TER and regulate activity. This complete structure of a telomerase catalytic core and its interactions with telomeric DNA from the template to telomere-interacting p50-TEB complex provides unanticipated insights into telomerase assembly and catalytic cycle and a new paradigm for a reverse transcriptase RNP.
Keywords: telomere, IFD, replication protein A, non-coding RNA, group II intron, Penelope-like element, repeat addition processivity, CST, retrotransposase
Graphical abstract
INTRODUCTION
Telomerase is an RNA-protein complex (RNP) that extends the 3’ ends of linear chromosomes by repetitively synthesizing the short telomere repeat sequence (TTGGGG in ciliates and TTAGGG in humans) using an RNA template that is part of its essential telomerase RNA (TER) component and its specialized telomerase reverse transcriptase (TERT) (Blackburn and Collins, 2011). Telomerase activity is tightly regulated during development and oncogenesis (Shay, 2016). Telomerase insufficiency in stem and germline cells due to mutations in telomerase components is manifested as diseases including dyskeratosis congenita, aplastic anemia, and pulmonary fibrosis (Armanios and Blackburn, 2012), while mutations in the TERT promoter that result in activation of the normally silent promoter in somatic cells are the most common noncoding mutations in cancer (Heidenreich and Kumar, 2017).
All telomerases contain a catalytic core of TERT and TER, which are sufficient to reconstitute activity in vitro. Physiological function requires species-specific biogenesis and assembly protein(s) that contact TER (core RNP) (Chan et al., 2017) and additional proteins involved in localization, G-strand handling, recruitment of proteins for C-strand synthesis, and termination that associate with the core RNP either transiently or constitutively (Chan et al., 2017; Schmidt and Cech, 2015). TERT contains, in addition to the palm and fingers (reverse transcriptase, RT) and thumb (C-terminal element, CTE) domains found in other reverse transcriptases, a telomerase essential N-terminal domain (TEN) and a telomerase RNA binding domain (RBD) (Podlevsky et al., 2008). TER, a rapidly evolving non-coding (nc)RNA that varies greatly in size and structure between organisms, contains two distinct and essential conserved motifs for interaction with TERT: the template/pseudoknot (t/PK) domain and a stem-terminus element (STE) (Nelson and Shippen, 2015; Podlevsky and Chen, 2016).
A 9Å resolution cryo-electron microscopy (cryoEM) structure of Tetrahymena telomerase revealed the locations, identities, and interactions of its 9 proteins and most of the TER, defining its general path on TERT (Jiang et al., 2015). In Tetrahymena, the TERT-TER catalytic core is assembled with p65, which is required for biogenesis and for placement of the TER STE (Jiang et al., 2013; Stone et al., 2007). The other 7 proteins, p50 and two heterotrimeric Replication Protein A (RPA) (Sugitani and Chazin, 2015) related complexes, Teb1-Teb2/Rpa2-Teb3/Rpa3 (TEB) (Jiang et al., 2015; Upton et al., 2017) and p75-p45-p19 (Tetrahymena CTC1-STN1-TEN1 (CST)) (Jiang et al., 2015; Wan et al., 2015) are paralogous to human proteins that only transiently associate with telomerase (Chan et al., 2017; Chen and Lingner, 2013; Schmidt and Cech, 2015). p50-TEB binds TERT and greatly enhances telomerase activity in apparent homology to the activation activity of human telomere-associated proteins TPP1-POT1, which otherwise contribute to telomere end protection (Wang et al., 2007). The single-stranded telomeric DNA (sstDNA) binds to Teb1ABC OB-folds (Zeng et al., 2011) on its path out of TERT during telomeric repeat synthesis, similar to POT1 (Lei et al., 2004).
The TER template is typically complementary to 1.5–1.8 telomere repeats, for sstDNA alignment and telomere repeat synthesis (Greider and Blackburn, 1987). Multiple steps of sstDNA template binding/realignment, nucleotide addition, strand separation, and template translocation are required for synthesis of a single telomere repeat and telomere repeat addition processivity (RAP) (Wu et al., 2017b). These activities require both TERT and TER elements in a manner that has largely eluded structural characterization. Several TERT and TER domain structures have been determined (reviewed in (Chan et al., 2017), but the only structure of a (putative) TERT is the unusual Tribolium castaneum (flour beetle) TERT (Gillis et al., 2008; Mitchell et al., 2010), which showed that the RBD-RT-CTE formed a ring structure that has guided many structure-function studies of telomerase (Wu et al., 2017a; Xie et al., 2010) as well as modeling of TERT into low resolution electron microscopy (EM) maps (Jiang et al., 2015; Jiang et al., 2013; Sauerwald et al., 2013). However, there are indications that this is not a true TERT, as no cognate TER for Tribolium TERT has been identified; it intrinsically lacks two regions unique to almost all other TERTs that are required for RAP, the TEN (Jacobs et al., 2006; O'Connor et al., 2005) and a large insertion in the RT (Lingner et al., 1997; Lue et al., 2003); and genome sequencing of Tribolium castaneum suggests that its telomeres are not maintained by telomerase (Tribolium Genome Sequencing et al., 2008). Here we present the cryoEM structure of Tetrahymena telomerase with bound sstDNA midway through a telomere repeat synthesis at 4.8Å resolution. The structure reveals how the sstDNA and TERT-specific domains, including a previously uncharacterized domain, interact with TER and each other to regulate the catalytic cycle and how telomere-interacting proteins bind to TERT, and it explains decades of biochemical and mutagenesis data on ciliate and human telomerase.
RESULTS AND DISCUSSION
CryoEM Reconstruction of Tetrahymena Telomerase with Bound DNA
Endogenously expressed telomerase was purified from Tetrahymena as described (Jiang et al., 2015) (see Methods). To obtain telomerase with bound telomeric DNA, we added a three telomeric repeat DNA [d(GTTGGG)2GTLTLGLGLG], where superscript L indicates locked-nucleic acid (LNA) nucleotides that stabilize the RNA-DNA duplex (Vester and Wengel, 2004), plus dGTP, to form a 6 bp template-DNA duplex plus 13 nts sstDNA at the 5’-end (Jiang et al., 2015). Direct telomerase activity assays with dGTP on the purified telomerase confirmed that the DNA binds and a single dG is added in the expected register (Figure S1). As observed for DNA-free telomerase, sstDNA bound telomerase is conformationally heterogeneous (Jiang et al., 2015; Jiang et al., 2013) (see Methods). We collected a dataset of over 3 million particles and developed a custom protocol for 2D and 3D classification (Figure S2). Focused 3D classification and refinement of the particles was carried out using a soft mask that excludes the dynamic CST complex (Jiang et al., 2015) and N-terminal domains of p65 (Figure S2). The remaining subunits TERT, TER, TEB, p50, and sstDNA (~260 kDa) within the mask still exhibited substantial dynamics, ultimately limiting our resolution to 4.8Å (Figure S2). For comparison, we obtained a structure of DNA-free telomerase from a somewhat smaller dataset to 6.4Å resolution (Figure S3). The cryoEM maps of the sstDNA bound and substrate free telomerase (Figure S3) are globally similar, indicating that no large-scale changes in the positions and fold of protein and TER domains are required to convert the substrate-free into this active enzyme. The better resolution of the DNA-bound telomerase may be partially attributed to a potentially more rigid core around the DNA-RNA duplex.
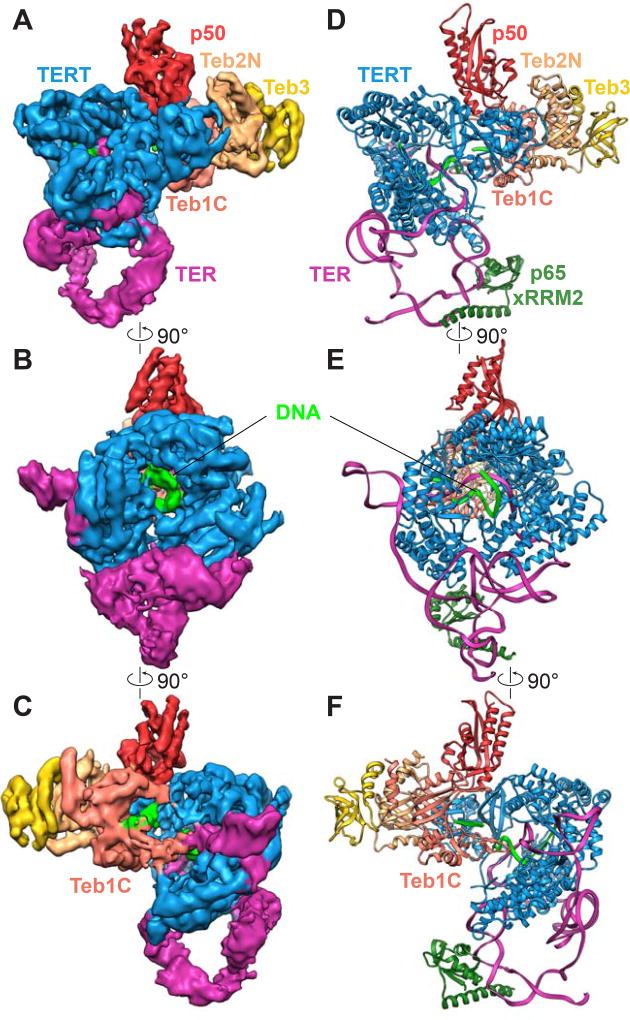
The cryoEM map of DNA-bound telomerase is shown in Figure 1A–1C. Protein and nucleic acid helices and most β-strands can be resolved throughout, the path of single-stranded RNA and most of the DNA clearly traced, and density from a few amino acid side chains and nucleic acid bases is visible (Figure S2 G–K). We obtained an almost complete atomic model of telomerase catalytic core with bound sstDNA, TEB, and p50 (Figures 1D–1F). The model was built by combining docking, density guided Rosetta modeling (Song et al., 2013), and manual rebuilding of crystal and NMR structures and homology models for TERT domains, TER domains, and TEB proteins (Jansson et al., 2015; Jiang et al., 2015) with the placement of the single-stranded regions of TER and secondary structure elements for the previously structurally uncharacterized p50 and a large TERT RT-specific insertion (see Methods) (Figures 1D–1F and S4A–S4K). The cryoEM structure shows intricate interactions between TERT and TER and between TERT, p50, and TEB. The DNA 3’ end of the template-DNA duplex is visible in the back view of the TERT ring formed by the RBD-RT-CTE domains (Figures 1B and 1E; see Figure 2A for domains), while the 5’ exiting DNA can be seen in the C-shaped cleft of Teb1C (Figure 1C and 1F).
Figure 1. CryoEM Reconstruction of Tetrahymena Telomerase with Telomeric DNA.
(A–C) 90° rotated views of the cryoEM map of Tetrahymena telomerase with bound DNA at 4.8Å resolution. The densities for the CST complex (p75-p45-p19) and p65 are invisible due to their dynamics and the soft mask that was used in the final 3D classification and refinement to exclude the CST complex and the majority of p65.(B) is back side of TERT ring.
(D–F) Views of the molecular model of TERT-TER-p65 RNP core, p50, and TEB, corresponding to A–C. The p65 xRRM2 is modeled in based on the crystal structure (PDB 4ERD) and the information from previous studies (Jiang et al., 2015; Jiang et al., 2013). Proteins and RNA are colored as follows: TERT (blue), TER (magenta), telomeric DNA (green), p65 xRRM (dark green), Teb1C (coral), Teb2N (tan), Teb3 (gold), p50 (red). See also Figures S1–S4 and Tables S1–S2.
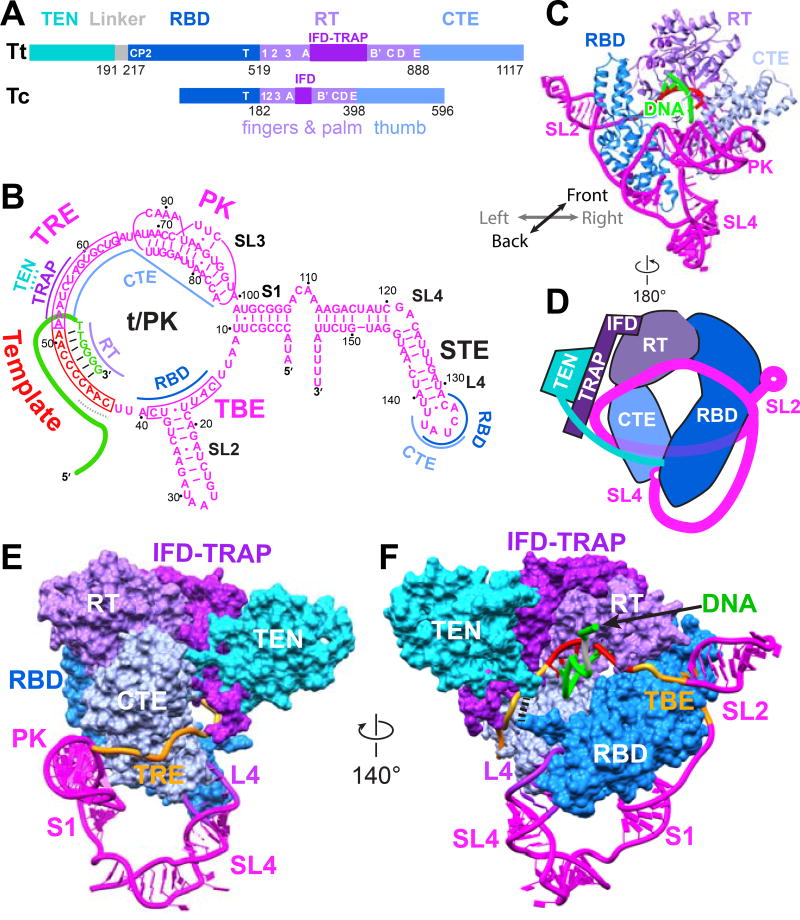
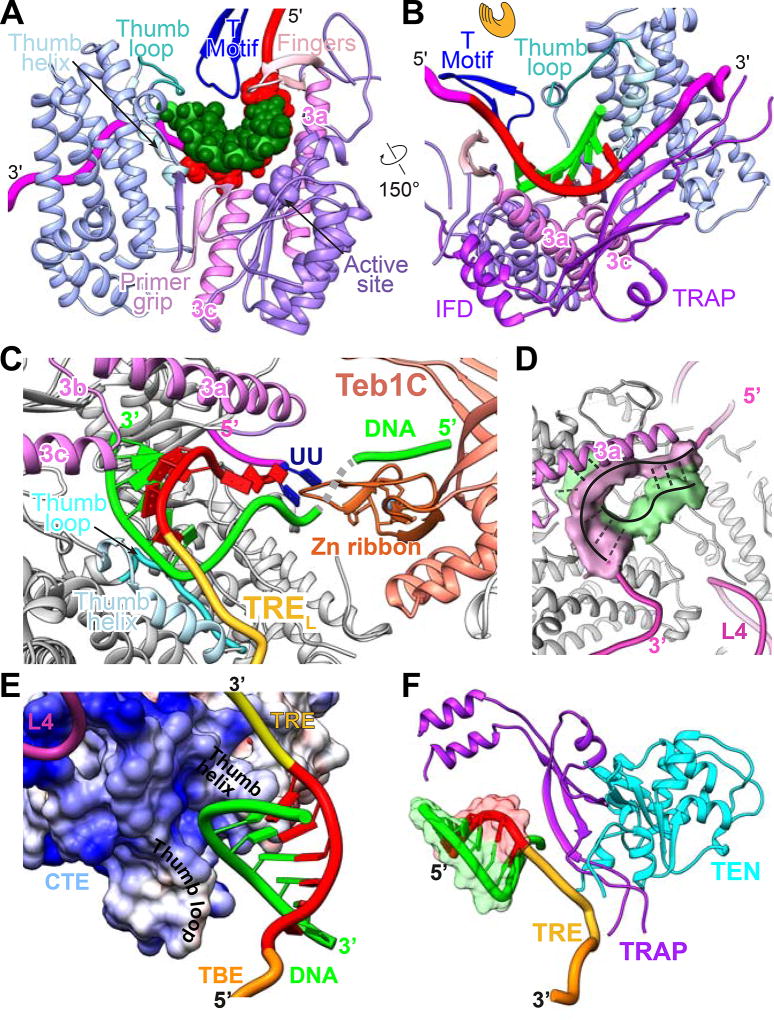
Figure 2. The TERT and TER Catalytic Core is an Interlocked Structure.
(A) Comparison of TERT domains of Tetrahymena (Tt) and Tribolium (Tc). For the RBD, the conserved CP2 (TFLY in vertebrates) motif that interacts with the TBE and T motif are annotated. In the RT domain, annotated motifs 1, 2, A, B’, C, D, E are common to reverse transcriptases; motif 3 and IFDa,c are common to TERT, Group II intron and PLE reverse transcriptases; and TRAP (between IFDa and IFDc) is unique to TERT.
(B) Schematic of TER secondary structure with bound DNA and sites of interaction with TERT domains.
(C) Structure of telomerase catalytic core, showing back view of TERT ring with pseudoknot, loop 4, and stem-loop 2. For clarity, only the DNA paired with template is shown.
(D) Schematic of TERT-TER structure illustrating the interlocking topology present in both the DNA bound and free telomerase.
(E–F) Structure of telomerase catalytic core with TERT as space-fill and TER as ribbon: (E) Side view showing TER between TRAP and CTE, (F) View corresponding to schematic in (D). Black dashes between TEN and RBD denote the linker between them. TERT domains are colored as follows: RT and IFD-TRAP (purple and violet), CTE (sky blue), RBD (blue), TEN (cyan); TER is magenta except template (red), and DNA is green. In E and F, TBE and TRE are orange.
See also Figure S4.
An Intricately Interlocked Catalytic Core
Our atomic model of telomerase with bound DNA reveals that the telomerase catalytic core has a unique architecture that interlocks TERT and TER (Figure 2). First, the TERT RBD-RT-CTE domains (Figure 2A) form a ring whose two ends (RBD and CTE) are anchored by TER loop 4 (L4), which acts as a molecular coupling (Figures 2C–2F). Second, the TER template/pseudoknot domain (t/PK) forms a circle, closed by stem 1 (Figure 2B), that wraps around the TERT ring, with the RNA contacting all three TERT ring domains (Figure 2C). The TER template boundary element (TBE), template, and template recognition element (TRE) are on the front side of the TERT ring (Figure 2F), while the pseudoknot is on the back over the CTE (Figures 2C and 2E). Third, within the RT domain there is an insertion in the fingers domain (IFD), containing a large previously structurally uncharacterized central region (IFDb; hereafter referred to as TRAP, see below) unique to telomerase, that extends above and over the t/PK circle (Figures 2D–2F). Last, the TERT TEN, which is connected to the RBD by a flexible linker which comes up from below the t/PK circle (Figures S4L–S4N), forms a structured interface on the outside of TRAP. The TEN-TRAP interaction thus forms a third, previously unknown, ring (TEN–RBD–RT-TRAP) that physically locks the t/PK circle onto the TERT ring, and sequesters the TRE between the CTE and TRAP (Figures 2D–2F). This intricately interlocked structure is important for function, as discussed below, and it explains why it is so difficult to assemble TERT and TER in vitro.
TRAP is a Newly Identified Structural Motif in TERT
The TERT RBD-RT-CTE ring is held together by L4, which forms a distinctive structure with stacked bases that has an extensive interface with the RBD on the minor groove side and the CTE on the major groove side (Figure 3A). The CTE (thumb) contains large insertions compared to Tribolium TERT (Gillis et al., 2008), which were identified in the modeling (Figure S4A). Most of the insertions in Tetrahymena CTE are helices on the outside of the TERT ring, while the interior is highly similar to crystal structures of Tribolium and human CTE (Hoffman et al., 2017) (Figure S5). The RBD adopts essentially the same conformation as in a Tetrahymena RBD–TBE crystal structure (Jansson et al., 2015) (Figure S5). The TERT RT domain is the most conserved subdomain among all species. It has two major insertions that have been identified as TERT specific domains, motif 3 (Xie et al., 2010) and the IFD (Lingner et al., 1997; Lue et al., 2003) (Figure 2A). In Tribolium TERT these motifs are parallel helices connected by short linkers (Gillis et al., 2008) (Figure 3). However, recent structures and sequence analysis have shown that these structural motifs are also found in other reverse transcriptases associated with group II introns (Qu et al., 2016; Stamos et al., 2017; Zhao and Pyle, 2016) and Penelope-Like Elements (PLE) (Gladyshev and Arkhipova, 2007) (Figure S6). In Tetrahymena, the linker (IFDb) (Lue et al., 2003; Podlevsky et al., 2008) between the two IFD helices (IFDa and IFDc) is 104 aa, 30% of the RT sequence (Figure 2A). Along with the TEN, a long IFD linker is unique to and found in almost all TERTs but not Tribolium, so to distinguish it from the other RT IFD linkers we name it TRAP, for its physical role in TRAPping TER (Figures 2D–2F and 3A–3B) as well as a functional role in regulating Telomerase RAP.
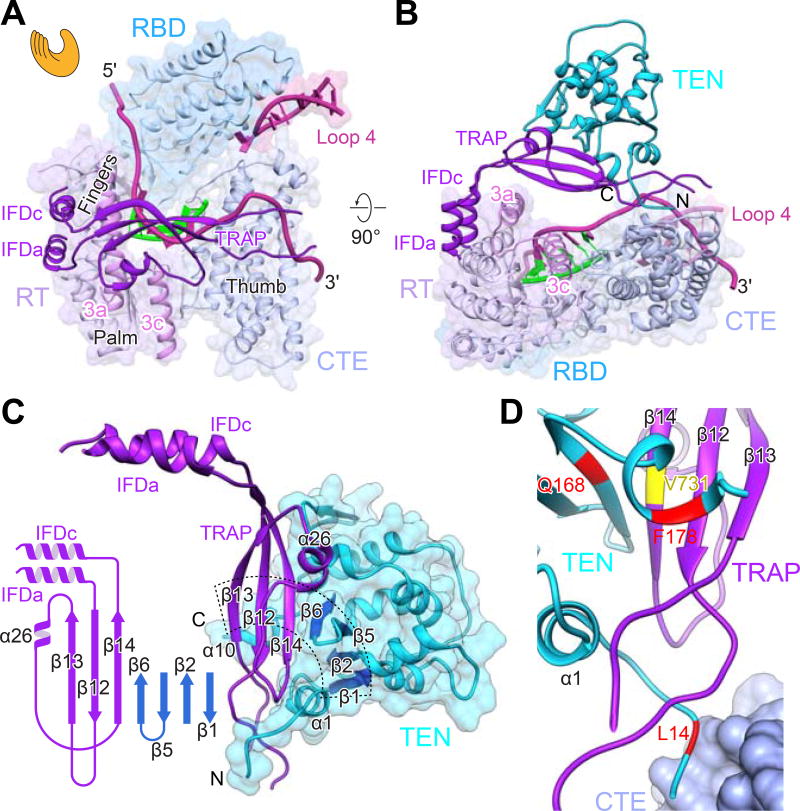
Figure 3. The TRAP-TEN Interaction is Unique to Telomerase.
(A) “Hand view” of TERT with template-DNA duplex and adjacent TRE and TBE shown. The RT (palm and fingers) and CTE (thumb) form the hand, with the RBD between fingers and thumb. L4 between RBD and CTE closes the TERT ring. The TRAP runs across the “hand” above the TER-DNA. TEN, above TRAP, is omitted in this view for clarity.
(B) 90° rotation of (A) showing back of the “hand view” of TERT to illustrate location of IFDTRAP and TEN relative to TERT ring. TER is in magenta and DNA is in green.
(C) IFD-TRAP interaction with TEN, showing complementary surface and an extended β-sheet formed between IFD-TRAP and TEN. Inset shows the secondary structure of IFD-TRAP and extended β-sheet surface that forms part of the TRAP-TEN interface. The part of the extended β-sheet in TEN is colored blue.
(D) Close-up view of TEN-TRAP interaction surface with some TEN residues (Q168, F178, L14) whose identities have been shown to be important for activity and RAP highlighted (Akiyama et al., 2015; Eckert and Collins, 2012; Jacobs et al., 2006; Zaug et al., 2008). L14 is also near the CTE. TRAP V731 (equivalent to human V791) is also at the TRAP-TEN interface (Chu et al., 2016a).
See also Figures S5 and S6.
The IFD helices are positioned similarly to those in group II intron and PLE reverse transcriptases on the periphery of the protein (Figure 3B). These helices form the short arm of an L-shaped structure where the TRAP is the long arm (Figure 3C). The corner of the L is buttressed by motif 3a helix, which inserts perpendicular to the IFDa,c helices (Figure 3B). The structure of TRAP is comprised primarily of β-strands and loops, and the backbone could be traced and tentative residue assignments made for all but a 23-residue highly acidic and apparently unstructured loop at the end of the L (Figures 3C–3D). The TEN (visible residues 11–181) straddles the TRAP with its N- and C-termini on either side, toward the CTE and RBD, respectively (Figures 3B–3C). Together, motif 3, IFD-TRAP, and TEN form a lid on the TERT ring that helps sequester and stabilize TER and the DNA (Figure 3B).
TRAP and TEN interface to form an extended β-sheet across the two domains (Figure 3C). Interaction with TRAP also orders or remodels structural elements of TEN at the interface compared to the TEN crystal structure (Jacobs et al., 2006) (Figures S4 and S5). TEN is required for RAP and has been extensively studied in both human and Tetrahymena telomerase. Mutagenesis and structural studies have suggested roles as a DNA anchor site, in TER binding, and in stabilization of short DNA-template duplexes (Akiyama et al., 2015; Eckert and Collins, 2012; Jacobs et al., 2006; Jurczyluk et al., 2011; Wyatt et al., 2009; Zaug et al., 2008). However, our structure reveals that TEN has no direct interactions with the DNA-template duplex or the exiting DNA, and only its C-terminus is close enough to contact TER. Several well studied TEN mutations (e.g. Q168A, F178A, L14A) whose identities affect activity and RAP (Akiyama et al., 2015; Eckert and Collins, 2012; Jacobs et al., 2006; Zaug et al., 2008) map to the TRAP-TEN (or TRAP-TEN-CTE for L14A) interface (Figure 3D). A human TRAP mutation, V791A (V731 in Tetrahymena) that affects activity, telomere length maintenance, and cell immortalization is also located at the TEN-TRAP interface (Chu et al., 2016a). The structure explains why TEN can be added in trans to the TERT ring and TER to reconstitute activity in vitro (Eckert and Collins, 2012; Robart and Collins, 2011). We conclude that TEN contributes indirectly to telomerase activity by forming a complex with TRAP that stabilizes its active fold, and that TRAP rather than TEN plays a direct role in RAP.
TER-TERT Interactions Regulate Function
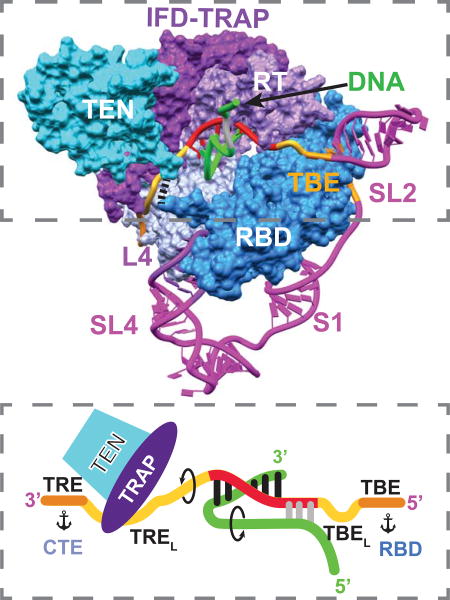
The cryoEM map provides a detailed view of TER interactions with TERT in a substrate bound complex (Figures 2E–2F and 4). In the t/PK circle, most of the TBE, template, and TRE (nts 15–64, except SL2) interact closely with TERT (Figures 4A–4F). In contrast, there are surprisingly few interactions on the back side of the TERT ring (Figures 1D and 1F). The pseudoknot has apparently only a few contacts to the highly basic surface of the CTE (Figure 4G), and there are no TERT contacts to S1 or the adjacent residues next to the TBE. In SL4, only the STE (L4) contacts TERT (Figure 4H). Comparison of the TER path in the DNA-free and DNA-bound telomerase shows that significant differences are confined to the template and 3’ and 5’ template adjacent residues in the TRE and next to the TBE, respectively (Figures 4B and S3).
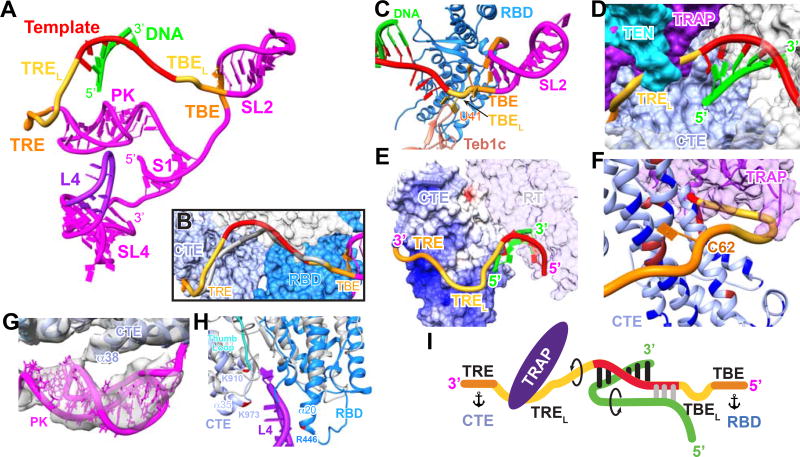
Figure 4. Role of TER in Telomerase Activity, Structure, and Assembly.
(A) Overall structure of TER, with regions of close interaction with TERT highlighted by colors.
(B) Comparison of TER location in DNA free and bound telomerase. Significant differences are seen only for the template and adjacent nucleotides. CTE-RBD surface shown is from the DNA-bound map.
(C–F) Close up views of the TBE–template–TRE traversing TERT: (C), Zoomed in view of TBE–RBD interface. TBEL U41U42 flip out and interact with the Zn ribbon of Teb1C. (D), TREL nucleotides in the channel formed by TRAP and CTE; (E) the TRE runs across the positively charged surface (shown as GRASP surface) of the CTE. TRAP and TEN are removed for clarity;
(F) TREL residues loop out to interact with the TRAP but not the CTE. TRE residues interact closely with a highly charged and aromatic rich surface of the CTE.
(G) Close-up view of the pseudoknot interaction with the back of the TERT ring.
(H) Zoomed in view of the RBD-L4-CTE interaction and comparison to Tribolium RBD-CTE (gray) interface which lacks RNA. Residues whose side chains appear to contact L4 are highlighted. RBD N-terminal residues 214–218 and the first two turns of helix α20 interact on the minor groove side and residues at the N-terminus of CTE helix α35 and connecting loop as well as the end of the thumb loop interact on the major groove side of L4.
(I) Cartoon of TRE-template-TBE interactions with TERT and sstDNA. Arrows indicate directions of rotation of TER and DNA during repeat synthesis, and anchors indicate TER anchor sites.
The TRE and TBE are essential for defining the 3’ and 5’ template boundaries, respectively (Lai et al., 2002; Miller and Collins, 2002). Discontinuities in these regions generally allow one round of telomere repeat synthesis but severely reduce RAP (Berman et al., 2011; Miller and Collins, 2002). In the cryoEM structure, the TBE is anchored around the outer edge of the RBD by specific interactions identical to those seen in the RBD-TBE crystal structure (Jansson et al., 2015) (Fig 4C). Notably, of the 3 residues (AUU) that link the template and TBE (here named TBEL for Linker between TBE and 5’ template) (Figure 4I) , two Us (41–42), conserved in ciliates and most vertebrates (Podlevsky et al., 2008), apparently flip out to interact with the Zn ribbon of Teb1C (Figure 4C); these residues were not observed in the crystal structure (Jansson et al., 2015). This unexpected interaction may explain why the Zn ribbon motif is required for high-RAP activity while it is not essential for telomeric DNA binding (Zeng et al., 2011). The 5’ end of the template extends over the RBD, with its bases facing outward (Figure 4C).
On the other side of the template, the TRE passes through a channel created by the CTE and TRAP–TEN (Figure 4D) and then traverses a basic surface on the CTE (Figure 4E). The 5’ end of the TRE (nts 52–59) (here named TREL, for 3’ template adjacent and residues that Loop out to bind TRAP) makes limited contacts with the CTE, but instead loops out to interact with the TRAP, consistent with a role for TRAP in regulating RAP (Figures 4F and 4I). On TEN, only the C-terminal residues around 180–181 are close enough to contact the TREL (Figure 4D). Residues 182–213 between TEN and RBD are missing in the density (Figure S4L–S4N). The 3’ end of the TRE (nts 60–68) closely interacts with the CTE, where there are numerous basic and aromatic groups, suggestive of a 3’ anchor point (Figure 4F).
The locations of TBE–template–TRE provide a snapshot of how these elements are positioned during telomeric DNA synthesis midway through a telomere-repeat cycle. Anchoring of the TBE to the RBD has been proposed to define the 5’ template boundary by physically preventing nontemplate nucleotides from moving into the active site (Berman et al., 2011; Jansson et al., 2015; Jiang et al., 2015). The three TBEL residues could easily stretch (along with the flexible Zn motif, discussed below) to allow completion of a full repeat. An accordion model for telomere repeat synthesis has been proposed, with concerted expansion and compression of 5’ and 3’ template adjacent nucleotides (Berman et al., 2011). Here we find that TREL loops out, but rather than playing a passive role it is captured by TRAP (Figures 4F and 4I). In the DNA-free telomerase, the TBEL appears to be a little further out and the TREL is a little less looped out (Figure 4B). Differences observed in the DNA-free and DNA-bound telomerase density maps indicate that the TRAP-TREL interaction stabilizes and/or extends the TRAP-TEN interface (Figure S3E–S3F).
While the TRE–template–TBE play direct regulatory roles in sstDNA synthesis, other elements of TER in the catalytic core function in assembly, as structural scaffolds, or as linkers. The main interactions between the pseudoknot and the CTE are at A80U81 that form base triples (Cash and Feigon, 2017) and residues at the pseudoknot-TRE junction (Figure 4G). The pseudoknot has been proposed to be important for TER assembly with TERT (Jiang et al., 2015), forming only after the t/PK circle passes over the TERT ring (Cash and Feigon, 2017). An unfolded pseudoknot might be required for passage of the t/PK circle over the TRAP to bind the TERT ring.
L4 closes the TERT ring in both sstDNA-bound and -free telomerase, suggesting that it may have a structural role in addition to a role in assembly. The protein regions that interact with L4 are highly basic and mostly not present in Tribolium TERT (Figure 4H). Instead, Tribolium TERT has an extensive CTE–RBD interface that closes the TERT ring (Figure 4H) (Gillis et al., 2008; Mitchell et al., 2010) and would appear to preclude RNA binding. The TERT ring closure may be required in part to hold the RBD in place so that the TBE anchored to RBD does not move during telomere repeat synthesis.
DNA and TER Exit the Duplex in Opposite Directions
The 6 bp DNA-template duplex is cradled between the RT palm and the thumb (CTE) (Figure 5A), occupying a large cavity in the TERT ring (Figures 1B and 5A). On the back side of the TERT ring, the template-DNA duplex binding pocket contains the three catalytic aspartates, thumb loop, thumb helix, primer grip, and fingers found in other reverse transcriptases all of which interact with the DNA (Figures 5A–5B) (Ding et al., 1997; Mitchell et al., 2010; Stamos et al., 2017). The thumb helix lies along the minor groove and the thumb loop is near the DNA backbone of the first two base pairs of the template–DNA duplex. The telomerase RBD T-motif (Nakamura et al., 1997) β-hairpin loop (481–487) is sandwiched between the fingers domain and thumb loop on the major groove side of the template–DNA duplex, but does not appear to contact the DNA (Figure 5A). These 3 elements have been proposed to help retain the DNA strand during template translocation (Wu et al., 2017a). The last nucleotide in the template-DNA duplex is apparently in the active site, and the fingers’ β-hairpin stacks on the DNA 3’ end nucleotide (Figure 5A). Motif 3, on the front side of the TERT ring, has been proposed to facilitate template–DNA realignment (Xie et al., 2010) or influence the template conformation (Wu et al., 2017a) (Figures 5B–5D). The motif 3c helix interacts with the primer grip (Figure 5A), and the N-terminal end of motif 3c and linker 3b residues are in the minor groove of the template-DNA duplex (Figures 5A and 5C). In addition to buttressing the IFD-TRAP (Figure 3B), the N-terminal half of motif 3a helix cradles the 5’ end of the template (Figures 5B–5D). The TRAP, as discussed above, runs along and interacts with the TREL (Figure 5B).
Figure 5. The sstDNA and TER Exit the Template in Opposite Directions.
(A and B) Close-up views of interactions of TERT with the DNA-template from (A) back side highlighting the primer grip (orchid), thumb helix (light blue), thumb loop (light sea blue), active site aspartates (spheres) that interact with DNA, and RBD T-motif in major groove. Motif 3c is next to primer grip. (B) front side (“hand view”) highlighting interactions of motif 3 (3a, 3c helices) and TRAP with template and TRE.
(C) View of the template-DNA duplex in the active site and exiting TREL and sstDNA. No density is observed for the DNA after the region near the 3’ end of the template until the DNA binding cleft on Teb1C. The TBEL U41U42 contact Teb1C Zn ribbon.
(D) CryoEM map with regions corresponding to the template and DNA highlighted. There is continuous density assigned to sstDNA from the template-DNA duplex toward the 5’ end of the template where the densities of sstDNA and the last 3 nts of the template merge and appear to form a short helix. TERT is colored gray in C and D.
(E) Close up view of the sstDNA turn at the end of the template-DNA duplex, in the basic binding pocket between the thumb helix and thumb loop on the CTE.
(F) Side view of 6 bp template-DNA duplex exiting the TERT ring toward TRAP–TEN and TREL running along TRAP.
See also Figure S3.
The DNA-template duplex exits the TERT ring toward the TRAP, where the sstDNA and TREL continue in opposite directions (Figure 5C). After the last base pair, the sstDNA turns back toward the 5’ end of the template, while the TREL turns away from the duplex through the CTE-TRAP channel (Figures 5C and 4D). The sstDNA backbone turn is at a basic pocket between the thumb helix and the thumb loop (Figure 5E), and RBD F414 appears to stack on the first unpaired base (G13), which may facilitate the turn. The path of the sstDNA can then be traced to the 5’ template residues rC43A44A45 where the density for the two strands merges into what appears to be a 3 bp helix (Figures 5C–5D and 4I). Intriguingly, the location of the exiting DNA adjacent to the 5’ template nts suggests a possible role for the template in DNA handling. UV crosslinking studies have suggested that sstDNA can be near the TBE (Goldin et al., 2012). After the DNA nts near the template 5’-end there is a gap in the density for the sstDNA, corresponding to ~3 nt which are apparently flexible, until Teb1C (Figure 5C). The density assigned to the sstDNA on Teb1C is absent in the DNA-free telomerase (Figure S3G–S3H), confirming its location in the predicted Teb1C DNA-binding cleft based on mutagenesis (Zeng et al., 2011) and structural studies of related RPA-ssDNA complexes (Fan and Pavletich, 2012). A longer DNA would be expected to go from Teb1C to the Teb1AB domains (Figure 6), which are connected by flexible linkers and are invisible in the cryoEM maps (Jiang et al., 2015; Jiang et al., 2013).
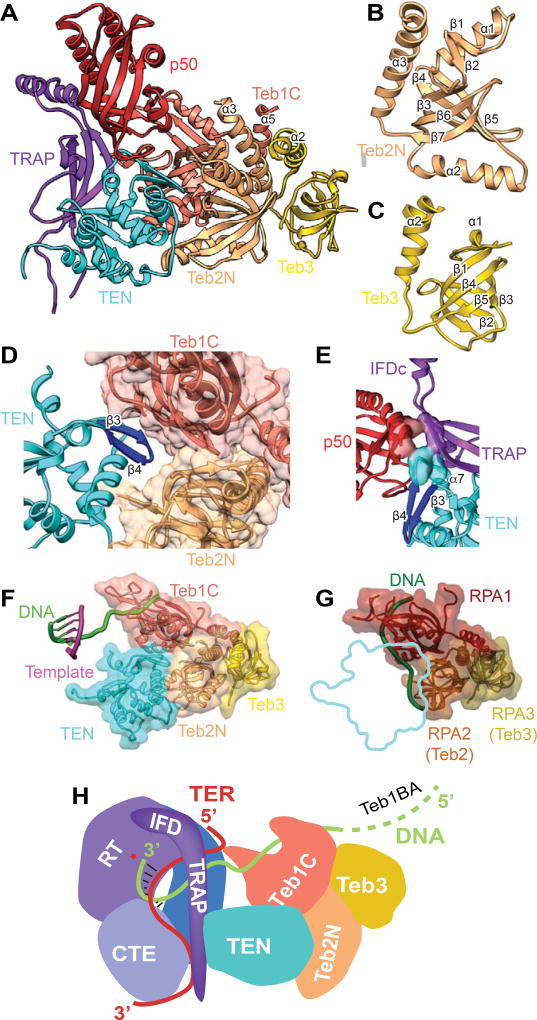
Figure 6. Structures and Interaction of p50 and TEB with TERT.
(A) Structure of IFD-TRAP-TEN-p50-TEB complex. TEN interacts with TRAP, p50, Teb1C, and Teb2N. TRAP interacts with p50 and TEN. Teb1C α5, Teb2N α3, and Teb3 α2 form a three-helix bundle. Teb1C structure is same as the crystal structure (PDB 3U50) except for the presence of α5 and the structure of the Zn ribbon motif (Figure 5C). The secondary structure of p50, modeled de novo, is a 6-stranded β-barrel with 4 α-helices.
(B) Structure of Teb2N/Rpa2N modeled from the cryoEM map. The structure is a 5-stranded β-barrel with 3 α-helices.
(C) Structure of Teb3/Rpa3 modeled from the cryoEM map. The structure is a 5-stranded β-barrel with 2 α-helices.
(D) Zoomed view of binding interface between Teb1C, Teb2N, and TEN. The TEN β3-β4 loop inserts between Teb1C and Teb2N.
(E) Close up view of the three-way interactions between p50, IFD-TRAP, and TEN, with interacting residues shown as surface. P50 and TRAP interact with TEN β4-α7 loop and α7, and p50 interacts with IFD-TRAP at the corner of the L linking them.
(F) TEN-TEB complex with sstDNA bound on Teb1C. The position of the sstDNA on Teb1C and the template-DNA duplex is shown relative to the TEN-TEB complex.
(G) RPA complex with DNA bound (PDB 4GNX) on RPA1 and RPA2 (equivalent to Teb2/Rpa2), with outline of TEN domain interaction in TEN-TEB complex to illustrate that TEN would occlude DNA binding surface on Teb2/Rpa2 in the Tetrahymena RPA complex.
(H) Cartoon illustrating the path of TRE-template-TBE on TERT and sstDNA from the active site of TERT to TEB. p50 is omitted for clarity.
See also Figures S4.
Biochemical studies of telomerase have provided evidence that a maximum length helix of 6–7 bp forms during telomere repeat synthesis (Forstemann and Lingner, 2005; Wang et al., 1998; Wu et al., 2017a), about the length of the duplex in our structure (Figure 5F). Here we see that the opposite paths of exiting sstDNA and TREL could limit the length of the helix to 6–7 bp by destabilizing the helix end. Furthermore, extension of the template-DNA duplex beyond ~7 bp with its adjoining sstDNA and TREL would also be sterically hindered by the TRAP (Figure 5F).
p50–TEB Structure and Recruitment
The telomerase structure reveals that IFD-TRAP–TEN also have roles in stabilizing the interaction of p50–TEB (Figure 6A). p50–Teb1 has been proposed to be structurally and functionally equivalent to the telomerase recruitment and activation activity of human TPP1–POT1(Jiang et al., 2015). In the p50–TEB complex (Figure 6A), only the structure of Teb1C was previously known (Zeng et al., 2011). We built an atomic model of the OB-fold proteins Teb2N and Teb3, and were able to trace the secondary structure elements of p50N, identifying an OB-fold comprising a six-stranded β-barrel with 3 distinct helices (Figures 6A–6C). p50 interacts with IFD-TRAP at the corner of the L, while TRAP and p50 interact with TEN at a loop and short helix (β4-α7) that are not structured in the free TEN (Figures 6A, 6E, and S5B). Human TPP1–POT1 transiently associates with telomerase TEN via a “TEL patch” on TPP1 to recruit it to telomeres. The orientation of the p50 OB-fold and interaction with TEN appear to mirror those in human TPP1–TEN (Nandakumar et al., 2012; Zhong et al., 2012), and these interactions are supported by mutational analysis in Tetrahymena (Jiang et al., 2015). The p50–IFD-TRAP interactions seen in Tetrahymena telomerase suggest that human TPP1 may interact with its IFD-TRAP in a comparable way. Indeed, mutations in human IFD-TRAP have recently been shown to affect telomerase recruitment to telomeres (Chu et al., 2016a; Chu et al., 2016b). The p50–IFD-TRAP–TEN interactions seen in Tetrahymena telomerase may also provide a rationale for why addition of p50 alone to the RNP core increases activity in vitro (Hong et al., 2013; Jiang et al., 2013), as it may stabilize the IFD-TRAP fold and/or TRAP–TEN interaction.
TEB (Jiang et al., 2015) binds the exiting sstDNA on its Teb1 subunit (Figures 5C, 6F, and S3G–S3H). In Teb1C only the position of the Zn ribbon motif, where template adjacent U41U42 bind, differs substantially from the crystal structure (Zeng et al., 2011). The Zn ribbon motif has a lower resolution than the rest of Teb1C, consistent with dynamic positioning. TEB interacts with TEN primarily through the β3-β4 hairpin that acts as a lynchpin that anchors both Teb1C and Teb2N and is adjacent to the region that interacts with p50 (Figure 6D). The TEN interactions with Teb2 explain why Teb2-Teb3 are required in vitro with Teb1 for optimal activity and to recruit telomerase to telomeres (Jiang et al., 2015; Upton et al., 2017).
While Teb1 is unique to telomerase, Teb2 and Teb3 are shared subunits with Tetrahymena RPA (Rpa1-Teb2-Teb3) (Upton et al., 2017). The specific interaction of Teb2 with TEN is therefore somewhat surprising. In RPA complexes, single-stranded DNA binds to RPA2 along the equivalent surface of the β-barrel occluded by the TEN interaction with Teb2 (Fan and Pavletich, 2012) (Figure 6G). Thus, Teb2 apparently has a dual function in sequence non-specific singlestranded DNA binding in the RPA complex and specific interaction with TEN in telomerase. The sstDNA exits TERT to bind Teb1C and thence to Teb1B and A, in expected equivalency to the sstDNA binding to human POT1 (Figure 6H).
Implications for Mechanism and Human Telomerase
Structures of many RNPs, e.g. the ribosome and spliceosome, have shown that it is the RNA that does the catalysis, and the protein acts primarily as a scaffold; in contrast, many ncRNAs function as scaffolds for dynamic protein assemblies where the protein does the catalysis (Cech and Steitz, 2014). Our structure reveals that telomerase has uniquely evolved as a reverse transcriptase RNP where the protein catalyzes addition of individual nucleotides but the catalytic cycle of telomere repeat synthesis and processivity are dependent on TERT-unique domains and the ncRNA TER. All of the TERT domains have essential interactions with TER that together regulate template boundary, activity, RAP, translocation, and template-DNA helix length. The RBD interacts with TBE, the CTE binds the TRE and also interacts with the pseudoknot, the RT binds the template and also interacts with the TREL on the TRAP–TEN complex, and the RBD and CTE bind L4 to close the RBD-RT-CTE ring. Within the TERT ring, conserved RT and CTE motifs active site, fingers, primer grip, thumb loop, and thumb helix common to reverse transcriptases (Ding et al., 1997; Mitchell et al., 2010; Podlevsky and Chen, 2016; Stamos et al., 2017) interact with the DNA on the duplex, and the telomerase RBD T-motif is near the major groove (Figure 5). Motif 3, also found in group II intron and PLE reverse transcriptases (Qu et al., 2016; Xie et al., 2010; Zhao and Pyle, 2016) (Figure S6), interacts with the primer grip and minor groove of the duplex (motif 3c) and the 5’ end of the template and IFD-TRAP (motif 3a). Together, these TERT ring motifs function in template and sstDNA handling and single-telomere repeat synthesis.
The identification of the TRAP–TEN complex, above the TERT ring, and its interaction with TREL, together with the observed interactions of TBE and TRE with the TERT ring, and location of the exiting sstDNA provides a new window into mechanism (Figure 6H). We suggest the following as a working model of contributions of TBE-TBEL, TRE-TREL, and TRAP to telomere repeat synthesis. After sstDNA binds the 3’ end of the template (3 bp in Tetrahymena), DNA synthesis pulls the template DNA through the active site, until the maximal stretching of the TBEL residues adjacent to the TBE anchor halts the synthesis (Jansson et al., 2015; Jiang et al., 2015) and puts strain on the end of the duplex. As the growing RNA-DNA duplex begins to extrude through the large cavity in the TERT ring toward the TRAP, the TREL residues loop out to bind TRAP (Figures 4F and 4I). Duplex length is limited to 6–7 bp (Figure 5F) by destabilization of the helix end due to the TREL pulling away from the helix (Figure 5C) while the sstDNA turns toward the template at the CTE thumb helix-thumb loop binding pocket (Figure 5E), allowing the end bases to unpair. Additionally, longer duplexes would be sterically hindered by TRAP (Figure 5F). If the growing TREL loop is held tightly by the TRAP-TEN, then torsional stress would accumulate during synthesis of the telomere repeat (Figure 4I). The torsional stress and strain on the two ends of the duplex could be relaxed by release of the product DNA (strand separation), likely aided by DNA binding motifs on the TERT ring to help retain the DNA and possible rotation of the CTE (Wu et al., 2017a), allowing for subsequent template translocation (relaxation to its DNA-free state) (Figure 4I) and DNA product realignment. Whether an interaction between the exiting sstDNA and the template (Figures 4I and 5C) contributes to DNA handling, e.g. by restraining the sstDNA to exit to TEB only after each telomere repeat (Parks and Stone, 2014), remains to be determined.
The structure of telomerase bound to telomeric DNA reveals that TRAP and TEN are required together for processive sstDNA synthesis and also for recruitment of p50-TEB (TPP1-POT1 in human). The putative Tribolium TERT lacks these domains, and its smaller RBD lacks the CP2 (ciliates)/TFLY (vertebrates) motif that contributes to TBE binding (Akiyama et al., 2013; Harkisheimer et al., 2013; Jansson et al., 2015) as well as RBD and CTE elements that interact with L4, suggesting that it cannot function as a true TERT. Among other things, it could not processively synthesize multiple telomere repeats even if provided a template as there is no obvious mechanism for strand separation, template translocation, or duplex length limitation, and there is no TEN to bind telomere interacting proteins. Interestingly, the PLE telomere-associated retrotransposon reverse transcriptases (Gladyshev and Arkhipova, 2007) have a common domain organization with Tribolium TERT (Figure S6), suggesting they have a common function.
In contrast to Tribolium TERT, human TERT shares a common domain structure with Tetrahymena. Human TER diverges from ciliate TER in size and structure (Podlevsky and Chen, 2016) but its t/PK and STE element can be modeled to interact with TERT in a similar way (Wang et al., 2016). Indeed, the modeled TER in the catalytic core at 7.7Å resolution in a cryoEM model of human telomerase that was published while this paper was readied for production is consistent with these predictions (Nguyen, et al., 2018). While the human t/PK has a much larger pseudoknot with long double-stranded regions (Wang et al., 2016; Zhang et al., 2011), there are single-stranded and single-strand plus stem regions on either side of the template that we propose could function as TRE-TREL and TBE-TBEL, respectively, and an STE hairpin (P6.1 in the CR4/5 domain) that is predicted to act as the RBD–CTE coupler, inserting between the RBD and CTE in a manner similar to Tetrahymena L4 (Chan et al., 2017; Huang et al., 2014). Other regions of CR4/5 bind the RBD, likely stabilizing the interaction with TERT. Human telomerase has a longer TEN-RBD linker with low sequence complexity that has been implicated in the observed dimerization and multimerization of telomerase purified under certain conditions (Sauerwald et al., 2013; Wu et al., 2015). We suggest that dimers or multimers could result from non-physiological domain swapping of TEN in one particle to IFD-TRAP in another. Because the human TERT-TER catalytic core and IFD-TRAP–TEN–TPP1–POT1 interactions are expected to be similar to corresponding interactions in Tetrahymena (Chan et al., 2017; Jiang et al., 2015), this first complete model of a telomerase with bound DNA and sstDNA handling proteins also provides unprecedented insight into human telomerase mechanism, mutations that affect function, and recruitment to telomeres, and a framework for design of telomerase targeting therapeutics.
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed and will be fulfilled by the Lead Contact, Juli Feigon (feigon@mbi.ucla.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Tetrahymena thermophila CU522 TERT-FZZ strain with a replacement of the endogenous TERT gene with a C-terminally TAP-tagged (FLAG-TEV-ZZ) TERT (Min and Collins, 2009) (provided by Dr. Kathleen Collins) was used for telomerase purification. Details of the strain construction are given in (Min and Collins, 2009). TERT-FZZ cells were cultured in PPYS (1% proteose peptone, 0.15% yeast extract supplemented with 0.2% glucose and 30 µM FeCl3).
METHOD DETAILS
Telomerase sample preparation
Tetrahymena telomerase holoenzyme was purified as previously described (Jiang et al., 2013) with minor modifications. Fourteen liters of Tetrahymena TERT-FZZ cells were grown to ~400,000 cells/ml in PPYS. Cells were harvested by centrifugation, washed with 20 mM HEPES, pH 8.0, and lysed in detergent at 4 °C as described (Jiang et al., 2013). Lysate was clarified by ultracentrifugation and rabbit IgG-agarose resin (Sigma) was applied to supernatant. After binding, elution was performed by TEV protease. The TEV eluate was bound to anti-FLAG resin (Sigma) and eluted with 1.2 mL of 0.4 mg/ml FLAG peptide (Sigma) in buffer with 0.025% IGEPAL CA-630 (Sigma-Aldrich). To generate the DNA bound telomerase sample, the procedure described above was used with two modifications: 1µM of the DNA primer dGTTGGGGTTGGGGTLTLGLGLG (where superscript L is LNA) (Exiqon) and 10µM dGTP were added to the sample during the TEV elution step to saturate telomerase with DNA primer (Jiang et al., 2015). The final FLAG elution step was slightly modified for the cryoEM sample: a concentrated and detergent-free sample was generated by eluting from anti-FLAG resin using a small volume (25–50 µl) of 1mg/ml 3×FLAG peptide dissolved in elution buffer (20 mM HEPES•NaOH, pH 8.0, 50 mM NaCl, 1 mM MgCl2, 1 mM TCEP•HCl) supplemented with 50 µg/ml bacitracin.
CryoEM specimen preparation and data collection
For cryoEM, 2.5 µL of sample (~0.1–0.5 mg/ml estimated from silver stain gel) was applied to a glow-discharged Quantifoil R1.2/1.3 300-mesh holey carbon-coated grid. The grid was blotted with filter paper and flash-frozen in liquid ethane using an FEI Vitrobot Mark IV. Frozen-hydrated grids were loaded into an FEI Titan Krios electron microscope operated at 300 kV for automated image acquisition with Leginon (Suloway et al., 2005). Movies of dose-fractionated image frames were acquired with a Gatan K2 Summit direct electron detector operated in super-resolution mode at a calibrated magnification of 36,764× (pixel size of 0.68 Å on the sample level). A Gatan Imaging Filter (GIF) Quantum LS was installed between the electron microscope and the K2 camera and the slit width was set to 20 eV. The dose rate on the camera was set to ~2 e−/pixel/s and the total exposure time of each movie was 12 s fractionated into 48 frames of images with 0.25 s exposure time for each frame.
As detailed below, we had to take a brute-force approach of obtaining a large dataset in order to improve the resolution. In total, 20,400 movies of the DNA-bound telomerase sample and 12,344 movies of the DNA-free telomerase sample were acquired for the structures presented in this paper. The dataset of 5,714 movies of the DNA-free telomerase sample that we collected previously (Jiang et al., 2015) were incorporated into the current datasets. Notably, as discussed below, most of the particles were considered as “bad” particles during 2D and 3D classifications and therefore discarded. For the DNA-bound telomerase, only 52,506 particles were selected from a total of 3.2 million for the final 3D refinement, which is only 1.64% (the actual percentage is slightly higher because some of the 3.2 million automatically boxed-out particles are not real particles, but rather ice contamination or empty background). Part of the problem might be due to the flexibility of the p75-p45-p19 (CST) complex, as demonstrated in our previous negative stain EM and cryoEM studies (Jiang et al., 2015; Jiang et al., 2013). In this study, focused 3D classification and refinement using a soft mask that excluded the dynamic CST complex indeed improved the resolution of the DNA-bound telomerase from 6.2 Å to 4.8 Å (Figure S2).
Comparison between negative stain EM images and cryoEM images of telomerase holoenzyme suggested a higher level of particle heterogeneity in cryoEM images and it was evident that some particles had fallen apart. This different level of particle heterogeneity between negative stain EM and cryoEM became more obvious when we used size exclusion chromatography to purify telomerase holoenzyme and remove IGEPAL CA-630 detergent and 3× FLAG peptide in the sample from affinity purification. This highly purified sample showed relatively good homogeneity of particles with a uniform shape in negative stain EM images, but very high heterogeneity in cryoEM images. It has been recognized in the cryoEM field that some complexes fall apart during preparation of cryoEM grids, likely due to exposure of the complex to the air/water interface. We tried to add detergent to the sample in order to reduce exposure of particles to hydrophobic environment; however, there were very few particles in holes of cryoEM grids in the presence of detergent (0.025% IGEPAL CA-630), suggesting that telomerase holoenzyme has affinity for the hydrophobic air surface. We also tried to concentrate the sample using Microcon centrifugal membrane filter in order to increase sample concentration for cryoEM with detergent, but it was not feasible due to insufficient sample quantity and because purified telomerase holoenzyme started to aggregate at higher concentrations. Finally, in the cryoEM samples used for data collection in this study, we used 50 µg/ml bacitracin to replace detergent in the last step of telomerase holoenzyme purification. Bacitracin can partially mimic detergent to protect exposure of telomerase particles to the air/water interface while keeping a sufficient number of particles in the holes of cryoEM grids for data collection. However, the problem of particle heterogeneity was only marginally improved by using bacitracin.
CryoEM image processing
All frames except for the first of each movie were 2x binned (final pixel size 1.36 Å) and aligned for correction of beam-induced drift using MotionCor2 (Zheng et al., 2017). Two average images were generated by MotionCor2 from each movie: one with dose weighting and the other one without. The average images without dose weighting were used for defocus determination and those with dose weighting were used for particle picking and further data processing, including image classification and 3D structure refinement. All average images after motion correction and their power spectra were visually inspected, and the images with poor quality were excluded. A total of 17,055 images of the DNA-bound telomerase sample and 10,708 images of the DNAfree telomerase sample were used for the following data processing.
The defocus values of the average images without dose weighting were determined by CTFFIND4 (Rohou and Grigorieff, 2015). Particles in the dose weighted average images were automatically picked with Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) using 17 projections from the previous cryoEM 3D reconstruction (Jiang et al., 2015). The dose weighted average images were corrected for contrast transfer function (CTF) by phase-flipping with the corresponding defocus and astigmatism values using Bsoft (Heymann and Belnap, 2007). Particles were extracted in 256×256 pixels from the phase-flipped images. The sum of the power spectrum of each particle was calculated. The particles with large or small sum of power spectrum mostly correspond to ice contamination or false particles, respectively, and were removed. A total of 3,200,000 DNA-bound telomerase particles and 2,300,000 DNA-free telomerase particles were selected for the following processing using RELION (Scheres, 2012). Because the particles preferred only a few orientations and were highly heterogeneous, particle selection using 2D and 3D classifications was carefully carried out in an iterative way (Figure S2), as detailed below. Notably, our procedure is different from the typical procedure recommended on the RELION website (https://www2.mrc-lmb.cam.ac.uk/relion/index.php/Recommended_procedures).
Our particle selection process for the DNA-bound telomerase is summarized in Figure S2, and a similar procedure was followed for the DNA-free telomerase. Briefly, particles were first classified into 6 subsets using 3D classification. No particles were discarded during this 3D classification step. The particles in each 3D class were then classified into 200 classes using 2D classification. The particles in each subset that are associated with the 2D class averages that showed uninterpretable or low-resolution features were removed. The reason for the above 3D and 2D classification steps is to avoid the loss of particles that are in the rare views. When we tried 2D classification with the whole data set, we found that particles in the rare views were often sorted into bad classes. The resulting “good” particles from all six subsets were combined for an additional 2D classification to further remove “bad” particles by selecting the 2D class averages with clear structural features. The selected particles were classified into only 1 class by 3D classification with a spherical mask to roughly align the particles (RELION option: -- healpix_order 3). The resulting particle orientation file was used as the input for another 3D classification of 6 classes with a small angular sampling (RELION options: --healpix_order 4 -- sigma_ang 2 --offset_range 5 --offset_step 1). The result shows 3D classes that have good density for p75-p45-p19 or lack this density. Comparison between these 3D classes suggests that the common parts (catalytic core, p50, and TEB) are similar between these 3D classes. We erased the poor density of the flexible N-terminus of p65 in the 3D map lacking the density of p75-p45-p19, and used this erased map to create a soft-edged mask that excludes the flexible parts. The particle orientation file from the above single-class 3D classification was used as the input for the subsequent 3D classification with this soft mask, and small angular sampling (RELION options: --healpix_order 4 --sigma_ang 2 --offset_range 5 --offset_step 1). The particles were classified into 6 classes and those in the best class were selected for the final 3D auto-refinement.
The 3D auto-refinement was performed using the same soft mask that was used in the last 3D classification. The resolution of the DNA-bound telomerase was estimated to be 4.8 Å by the relion_postprocess program using the ‘gold-standard’ Fourier Shell Correlation (FSC) at 0.143 criterion (Figure S3). The resolution of the DNA-free telomerase was estimated to be 6.4 Å using the same method. The cryoEM maps were sharpened with B-factor and low-pass filtered to the stated resolution using the relion_postprocess program. The local resolution was calculated by ResMap (Kucukelbir et al., 2014) using two cryoEM maps independently refined from halves of data. CryoEM data collection and processing statistics are given in Table S1.
Molecular modeling of TERT
Fitting of atomic coordinates into the cryoEM maps was performed using Coot (Emsley et al., 2010) and USCF Chimera (Pettersen et al., 2004). The TERT TEN, RBD, RT, and CTE domains were modeled as follows. For the TEN domain, modeling was initiated by docking of the crystal structure of Tetrahymena TEN (PDB 2B2A) (Jacobs et al., 2006) into the cryoEM map. We observed an overall good agreement of the template structure to the cryoEM density with differences confined to crystallographically unobserved loops, and to the N- and C-terminal portions of the protein, which participate in crystal contacts in the X-ray structure. Manual rebuilding in Coot resulted in a continuous protein model for residues 11–181, with no cryoEM density observed for the presumed flexible N-terminal tail (residues 1–10) and C-terminal linker (182–213).
The Tetrahymena RBD crystal structure in complex with SL2 (PDB 5C9H) (Jansson et al., 2015) showed excellent fit to the EM density, and was used without further modification except for the addition of four N-terminal amino acids (residues 214–217), which were not present in the crystallized construct. Altogether, RBD residues 214–249, and 293–509 are clearly observed in the EM density, and are followed by an apparently cryoEM-invisible flexible linker (residues 510–530).
Modeling of the RT domain was carried out using the structure of Tribolium TERT (PDB 3KYL) (Mitchell et al., 2010), which served as starting coordinates for rebuilding in RosettaCM (Song et al., 2013). 3000 models were calculated, and the top scoring hits were inspected in Coot, which revealed tight convergence for most of the RT sequence, including proper positioning of active-site residues (Asp618, Asp815, Asp816) for productive catalysis. However, for the TRAP (IFDb) (residues 638–742), which is not present in Tribolium TERT, no convincing models were obtained. A broad search for alternative TRAP template structures via HHpred (Zimmermann et al., 2017) resulted in no hits. Therefore, the EM map was hand-traced in Coot, revealing an internal structured domain of ~70 amino acids consistent with a twisted three-stranded antiparallel β-sheet that is N and C-terminally flanked by helical IFDa and IFDc regions, respectively. The IFDa and IFDc helices connected to either end of TRAP (IFDb) are well predicted, providing clear starting and ending points for the model, and the three β-strands and α-helix could be assigned with confidence. The only region of TRAP not visible in the cryoEM map is a region of low complexity at the periphery of the enzyme.
The CTE was modeled in RosettaCM using Tribolium (PDB 3KYL) and human CTE (PDB 5UGW) (Hoffman et al., 2017) template structures, yielding an atomic model that placed residues 891–1046. The presence of an additional ~50 residues of helical structure at the extreme C-terminus was clearly manifested in the cryoEM map, and subsequently manually built in Coot, providing a continuous CTE model out to amino acid position 1108, followed by a short, presumably unstructured, C-terminal tail (1109–1117).
Molecular modeling of TEB and p50
The heterotrimeric Tetrahymena TEB (Teb1C–Teb2N–Teb3) complex was modeled in RosettaCM using the template structures of Tetrahymena Teb1C crystal structure (PDB 3U50) (Zeng et al., 2011), with related human and fungal RPA complexes (PDB 1L1O, 4GNX) (Bochkareva et al., 2002; Fan and Pavletich, 2012) as additional templates and to provide relative orientations between protomers within the complex. The 25 highest scoring output structures (of 3000) were manually analyzed in Coot to select the best-fit model, covering Teb1C (residues 511–695), Teb2N (residues 29–180), and Teb3 (residues 1–121). Teb1NAB and Teb2C are connected by long linkers to Teb1C and Teb2N, respectively, and are not visible in the cryoEM maps due to positional flexibility (Jiang et al., 2015; Jiang et al., 2013). Modeling of p50 was carried out after the cryoEM densities for the other telomerase subunits had been accounted for. Idealized secondary structure elements (6 β-strands and 5 α-helices) were placed into the EM density using Coot and their lengths adjusted for optimal fit. Secondary structure elements were then connected in accordance with the expected topology of a six-stranded antiparallel β-barrel OB-fold. Hand tracing of the EM map was further supported by secondary structure predictions using Jpred (Drozdetskiy et al., 2015) and RaptorX (Wang et al., 2016).
Molecular modeling of TER
The RNA was modeled starting with fitting of NMR and crystal structures of subdomains into the cryoEM maps using UCSF Chimera (Pettersen et al., 2004) and refining as follows. The solution NMR structures of SL2 (residues 19–37, PDB 2M22) (Richards et al., 2006) and the pseudoknot (residues 69–99, PDB 5KMZ) (Cash and Feigon, 2017) were recalculated by combining the cryoEM density map and NOE restraints using Xplor-NIH 2.45.6 (Bermejo et al., 2016). The bottom U-U base pair and single-stranded residues flanking SL2 (residues 14–18 and 38–39) were modeled from their positions in the crystal structure of Tetrahymena RBD-SL2 complex (PDB 5C9H) (Jansson et al., 2015). The structure of the central region of S4 (residues 119–125 and 144–148) was taken from the crystal structure p65 xRRM–S4 complex (PDB 4ERD) (Singh et al., 2012). The NMR structure of SL4 apical stem-loop (residues 125–144) (PDB 2FEY) (Chen et al., 2006) was manually adjusted to fit into the density using Coot (Emsley et al., 2010). Density corresponding to most of the loop residues was visible in the maps. An ideal 6 bp A-form helix for the DNA-RNA template duplex (RNA residues 46–51 and DNA residues 14–19) was fit into and refined to the density. Stem 1 (nts 4–10 and 101–107) was generated as an A-form helix by the online server RNAComposer (Popenda et al., 2012), and fit into cryoEM density that matches its shape, identifying two more base pairs than predicted. The remaining single-stranded regions of TER connecting the secondary structure elements discussed above were modeled into the cryoEM densities using Coot. Briefly, density corresponding to single-stranded RNA and DNA were identified, and fragments were created by Coot and placed into the density map. The bond angles of each nucleotide backbone were manually adjusted until the backbone fit into the density map, and then optimized by the “Regularize Zone” tool in Coot. Last, all the single-stranded regions and secondary structure elements were connected using Coot, and iteratively placed into the density map with the protein domains using UCSF Chimera.
Model Refinement
The combined molecular models for all nucleic acid (TER, telomeric DNA) and protein components (TERT, TEB, p50) were refined in real space using PHENIX (Adams et al., 2010). A three-stage refinement protocol (PHENIX options: rigid_body, global_minimization, adp) was carried out with secondary structure, Ramachandran, and rotamer restraints enforced throughout refinement. Validation statistics are given in Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analyses of electron microscopy data and the determination of structures by electron microscopy are integral parts of existing algorithms and software used which are described in Method Details.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The cryoEM density map has been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) under accession code EMDB: EMD-7820 (Tetrahymena telomerase without DNA), and EMD-7821 (Tetrahymena telomerase with DNA) and its associated model coordinates have been deposited in the Protein Data Bank (http://www.rcsb.org) under accession number PDB: 6D6V.
Supplementary Material
Reaction conditions are (1) d[GTTGGG]3 primer + dGTP; (2) d[GTTGGG]3 primer + dGTP & dTTP; (3) dGTTGGGGTTGGGGTLTLGLGLG primer [NL denotes LNA] + dGTP; (4) dGTTGGGGTTGGGGTLTLGLGLG + dGTP and dTTP. In (1), lack of dTTP in the reaction results in a product that fails to translocate properly but is still able to extend due to addition of dGTP and product slipping. In (2) a standard telomere addition is observed. (3) results in the addition of a single dG but product slippage is not observed due to LNA binding to template. (4) addition of GTTG occurs, but translocation is inhibited by LNA. Note that the LNA containing primer migrates slightly slower through the gel than non-modified DNA. Direct telomerase activity assays were performed as described (Jiang et al., 2015; Jiang et al., 2013) with 3 µM dGTP supplemented with dGTP-[α32P] for 30 min. RC is a recovery control.
(A) A cryoEM micrograph generated by averaging 48 motion-corrected frames in a movie recorded by K2 direct electron detector.
(B) The power spectrum of the micrograph in (A) showing the detectable data in the micrograph extending to spatial frequency of 1/3.8Å.
(C) CryoEM processing: Our custom cryoEM data processing method consists of multiple rounds of both 2D and 3D classification steps in order to sort out structurally homogeneous subsets from a huge dataset of over 3,000,0000 particles for improved resolution of the DNA-bound telomerase complexes. We first carried out Relion 3D classification directly on particle images to partition them into six classes. Several rounds of 2D classification selected 211,502 particles that were subjected to 3D classifications with small angular sampling using two different masking strategies, one with a spherical mask and the other with a soft mask, resulting in a 7.9 Å map with good CST density from 31,388 particles and a 4.8Å map without CST density from 52,506 particles, respectively. Please refer to the Methods for detailed description of the steps. The same workflow was used for the DNA free telomerase, starting from 2,300,000 particles.
(D) Plots of the gold-standard Fourier shell correlation coefficients as a function of spatial frequency for the DNA-bound and DNA-free telomerase complexes.
(E) Euler angle distributions of the particles in the final 3D auto-refinement.
(F) Local resolution evaluation of the cryoEM map of the DNA-bound telomerase generated by Resmap. The core region of the map has resolution near 4Å.
(G–K) Representative regions of the cryoEM density map (gray and mesh) superposed with select regions of our atomic models (color sticks and ribbons) of (G) RBD, (H) TEN, (I) RT, (J) IFD-TRAP, (K) the DNA-RNA duplex.
(A and B) Six rotated views of (A) DNA-bound telomerase and (B) DNA-free telomerase show that the two states are globally the same. For this comparison, both density maps are filtered to 6.4 Å resolution which is the lower resolution of them. Colors correspond to TERT (blue), TER (magenta), telomeric DNA (green), p65 xRRM (dark green), Teb1C (coral), Teb2N (tan), Teb3 (gold), p50 (red). Note that most of the density for p65 is invisible due to its flexibility.
(C and D) Region of the density map containing the TER template for telomerase with bound sstDNA (C) and for DNA-free telomerase (D). Magenta and green are cryoEM densities of RNA template and DNA, respectively. Grey is spherically masked cryoEM density of TERT. (E and F) Region of the density map containing at the TRAP–TEN–TRE interface in the DNA-bound telomerase (E) and in the DNA-free telomerase (F). The model of TRAP, TEN, and TRE in the telomerase–sstDNA complex is shown in E. In F the model of TRAP and TEN is from the telomerase–sstDNA complex to highlight the regions of TEN and TRAP that are no longer seen. The TRE (modeled into the DNA free map) also changes.
(G and H) Region of the density map containing Teb1C for telomerase with bound sstDNA (G) and for DNA-free telomerase (H). Note that both density maps are filtered to 6.4 Å resolution to match the resolution of the DNA-free telomerase.
(A) CryoEM map with atomic model of TERT ring, RBD-RT-CTE.
(B–D) TERT ring domain structures labeled with sequential assignment of helices and sheets: RBD (B), RT (C), CTE (D).
(E) CryoEM map with atomic model of TERT TEN–IFD-TRAP.
(F and G) TERT TEN and IFD-TRAP domain structures labeled with sequential assignment of helices and sheets: TEN (F), IFD-TRAP (G).
(H) CryoEM map with atomic model of TER and template bound DNA
(I) CryoEM map with p50 secondary structure elements. Amino acid sequence is not assigned.
(J) CryoEM map with atomic model of Teb1C and bound DNA
(K) CryoEM map with atomic model of Teb2N-Teb3.
(L) Cartoon representation of TERT-TER catalytic core showing that the TEN and RBD domains are connected by a flexible linker that crosses over the t/PK circle. Linker is represented by dotted line.
(M and N) Close-up views of the catalytic core highlighting the location of the (flexible, not visible) TEN-RBD linker (black dashed line) in the 4.8 Å cryoEM map. Linker length and start/end residue numbers are indicated.
(A) Tetrahymena TERT RBD-RT-CTE (blue, light purple, light blue) superimposed on Tribolium TERT (PDB 3KYL) (gray). For Tetrahymena TERT the IFD-TRAP (purple) is shown but the TEN is omitted for clarity.
(B) Tetrahymena TEN (cyan) superimposed on the crystal structure (PDB 2B2A) (gray). α7 and loop to β3 are not seen in the crystal structure.
(C) Tetrahymena Loop 4-RBD-TBE (blue and magenta) superimposed on RBD-TBE (gray) (PDB 5C9H).
(D) Tetrahymena CTE superimposed on human CTE (PDB 5UGW) (gray).
(A) Structures of HIV-1 RT (PDB 4PQU), G. stearothermophilus Group II Intron encoded RT (PDB 6AR1), Tribolium castaneum TERT (PDB 3KYL), and Tetrahymena thermophila TERT (this work), with template-primer duplexes.
(B) Comparison of conserved domains found in reverse transcriptases from HIV-1 and Group II introns, Penelope Like Element associated reverse transcriptase (PLE-RT) from A. vaga, Tribolium TERT, T. thermophila TERT, and human TERT. Note that the domain structure of Tribolium TERT appears more similar to PLE-RT than to Tetrahymena and human TERTs. Group II intron reverse transcriptase, PLE-RT, Tribolium TERT all have motif 3 and IFDac, and PLE-RT and Tribolium TERT have an RBD with a T-motif. The human and Tetrahymena TERTs (as well as TERTs from plants and yeasts) have the additional TRAP and TEN domains that are essential for RAP.
HIGHLIGHTS.
Structure of Tetrahymena telomerase with telomeric DNA provides mechanistic insights
Complete architecture of catalytic core reveals a new structural motif named TRAP
Detailed path of DNA from active site to telomere DNA-binding p50-TEB complex
Roles for telomerase RNA TRE-template-TBE in telomeric DNA synthesis
IN BRIEF.
The cryoEM structure of active Tetrahymena telomerase bound to telomeric DNA reveals unique insights into the catalytic core and DNA handling
Acknowledgments
This work was supported by NIH GM048123 and NSF MCB1517625 grants and award from National Center for Advancing Translational Science UCLA CTSI grant UL1TR001881 to J.F and NIH GM071940 to Z.H.Z. H.C. was supported in part by a Ruth L. Kirschstein National Research Service Award GM007185. We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and instrumentation grants NIH 1S10OD018111 and 1U24GM116792 and NSF DBI-1338135 and DMR-1548924.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes two tables and six figures can be found with this articleonline at *.
AUTHOR CONTRIBUTIONS
J.J. performed cryoEM data collection and processing; Y.W., L.S., and J.J. performed model building, analyzed the data, and made figures; H.C. prepared telomerase samples; R.B. helped analyze TEB; Z.H.Z. supervised the cryoEM; J.F. supervised the project, analyzed the structure, and wrote the manuscript with Y.W., L.S., J.J., and Z.H.Z. All authors contributed to the final version.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama BM, Gomez A, Stone MD. A conserved motif in Tetrahymena thermophila telomerase reverse transcriptase is proximal to the RNA template and is essential for boundary definition. J Biol Chem. 2013;288:22141–22149. doi: 10.1074/jbc.M113.452425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama BM, Parks JW, Stone MD. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res. 2015;43:5537–5549. doi: 10.1093/nar/gkv406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AJ, Akiyama BM, Stone MD, Cech TR. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol. 2011;18:1371–1375. doi: 10.1038/nsmb.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo GA, Clore GM, Schwieters CD. Improving NMR Structures of RNA. Structure. 2016 doi: 10.1016/j.str.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harbor perspectives in biology. 2011;3:a003558. doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash DD, Feigon J. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res. 2017;45:482–495. doi: 10.1093/nar/gkw1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chan H, Wang Y, Feigon J. Progress in Human and Tetrahymena Telomerase Structure. Annu Rev Biophys. 2017;46:199–225. doi: 10.1146/annurev-biophys-062215-011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Lingner J. CST for the grand finale of telomere replication. Nucleus. 2013;4:277–282. doi: 10.4161/nucl.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TW, D'Souza Y, Autexier C. The Insertion in Fingers Domain in Human Telomerase Can Mediate Enzyme Processivity and Telomerase Recruitment to Telomeres in a TPP1-Dependent Manner. Mol Cell Biol. 2016a;36:210–222. doi: 10.1128/MCB.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TW, MacNeil DE, Autexier C. Multiple Mechanisms Contribute to the Cell Growth Defects Imparted by Human Telomerase Insertion in Fingers Domain Mutations Associated with Premature Aging Diseases. J Biol Chem. 2016b;291:8374–8386. doi: 10.1074/jbc.M116.714782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hughes SH, Arnold E. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers. 1997;44:125–138. doi: 10.1002/(SICI)1097-0282(1997)44:2<125::AID-BIP2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert B, Collins K. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem. 2012;287:12805–12814. doi: 10.1074/jbc.M112.339853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Lingner J. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep. 2005;6:361–366. doi: 10.1038/sj.embor.7400374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–9357. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin S, Kertesz Rosenfeld K, Manor H. Tracing the path of DNA substrates in active Tetrahymena telomerase holoenzyme complexes: mapping of DNA contact sites in the RNA subunit. Nucleic Acids Res. 2012;40:7430–7441. doi: 10.1093/nar/gks416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure. 2013;21:1870–1878. doi: 10.1016/j.str.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich B, Kumar R. TERT promoter mutations in telomere biology. Mutat Res. 2017;771:15–31. doi: 10.1016/j.mrrev.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hoffman H, Rice C, Skordalakes E. Structural Analysis Reveals the Deleterious Effects of Telomerase Mutations in Bone Marrow Failure Syndromes. J Biol Chem. 2017;292:4593–4601. doi: 10.1074/jbc.M116.771204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol. 2013;33:3962–3971. doi: 10.1128/MCB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol. 2014;21:507–512. doi: 10.1038/nsmb.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol. 2015;22:883–888. doi: 10.1038/nsmb.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O'Brien Johnson R, Collins K, Loo JA, et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science. 2015;350:aab4070. doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. The architecture of Tetrahymena telomerase holoenzyme. Nature. 2013;496:187–192. doi: 10.1038/nature12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes Dev. 2002;16:415–420. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci U S A. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell. 2009;36:609–619. doi: 10.1016/j.molcel.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol. 2010;17:513–518. doi: 10.1038/nsmb.1777. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AD, Shippen DE. Evolution of TERT-interacting lncRNAs: expanding the regulatory landscape of telomerase. Front Genet. 2015;6:277. doi: 10.3389/fgene.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature. 2018 doi: 10.1038/s41586-018-0062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks JW, Stone MD. Coordinated DNA dynamics during the human telomerase catalytic cycle. Nat Commun. 2014;5:4146. doi: 10.1038/ncomms5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Chen JJ. Evolutionary perspectives of telomerase RNA structure and function. RNA biology. 2016;13:720–732. doi: 10.1080/15476286.2016.1205768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang HW. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol. 2016;23:549–557. doi: 10.1038/nsmb.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Theimer CA, Finger LD, Feigon J. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res. 2006;34:816–825. doi: 10.1093/nar/gkj481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell. 2011;42:308–318. doi: 10.1016/j.molcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–460. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Cech TR. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6:584–593. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell. 2012;47:16–26. doi: 10.1016/j.molcel.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Lentzsch AM, Lambowitz AM. Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications. Mol Cell. 2017;68:926–939 e924. doi: 10.1016/j.molcel.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani N, Chazin WJ. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog Biophys Mol Biol. 2015;117:206–211. doi: 10.1016/j.pbiomolbio.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing C. Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Upton HE, Chan H, Feigon J, Collins K. Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes. J Biol Chem. 2017;292:217–228. doi: 10.1074/jbc.M116.763664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Wan B, Tang T, Upton H, Shuai J, Zhou Y, Li S, Chen J, Brunzelle JS, Zeng Z, Collins K, et al. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat Struct Mol Biol. 2015;22:1023–1026. doi: 10.1038/nsmb.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wang H, Gilley D, Blackburn EH. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc Natl Acad Sci U S A. 2016;113:E5125–5134. doi: 10.1073/pnas.1607411113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. Elife. 2015;4 doi: 10.7554/eLife.08363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Tam J, Collins K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017a doi: 10.15252/embj.201796887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Upton HE, Vogan JM, Collins K. Telomerase Mechanism of Telomere Synthesis. Annu Rev Biochem. 2017b;86:4.1–4.22. doi: 10.1146/annurev-biochem-061516-045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, Tsang AR, Lobb DA, Beattie TL. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One. 2009;4:e7176. doi: 10.1371/journal.pone.0007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res. 2010;38:1982–1996. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci U S A. 2011;108:20357–20361. doi: 10.1073/pnas.1113624108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kang M, Peterson RD, Feigon J. Comparison of solution and crystal structures of preQ1 riboswitch reveals calcium-induced changes in conformation and dynamics. J Am Chem Soc. 2011;133:5190–5193. doi: 10.1021/ja111769g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Pyle AM. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol. 2016;23:558–565. doi: 10.1038/nsmb.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reaction conditions are (1) d[GTTGGG]3 primer + dGTP; (2) d[GTTGGG]3 primer + dGTP & dTTP; (3) dGTTGGGGTTGGGGTLTLGLGLG primer [NL denotes LNA] + dGTP; (4) dGTTGGGGTTGGGGTLTLGLGLG + dGTP and dTTP. In (1), lack of dTTP in the reaction results in a product that fails to translocate properly but is still able to extend due to addition of dGTP and product slipping. In (2) a standard telomere addition is observed. (3) results in the addition of a single dG but product slippage is not observed due to LNA binding to template. (4) addition of GTTG occurs, but translocation is inhibited by LNA. Note that the LNA containing primer migrates slightly slower through the gel than non-modified DNA. Direct telomerase activity assays were performed as described (Jiang et al., 2015; Jiang et al., 2013) with 3 µM dGTP supplemented with dGTP-[α32P] for 30 min. RC is a recovery control.
(A) A cryoEM micrograph generated by averaging 48 motion-corrected frames in a movie recorded by K2 direct electron detector.
(B) The power spectrum of the micrograph in (A) showing the detectable data in the micrograph extending to spatial frequency of 1/3.8Å.
(C) CryoEM processing: Our custom cryoEM data processing method consists of multiple rounds of both 2D and 3D classification steps in order to sort out structurally homogeneous subsets from a huge dataset of over 3,000,0000 particles for improved resolution of the DNA-bound telomerase complexes. We first carried out Relion 3D classification directly on particle images to partition them into six classes. Several rounds of 2D classification selected 211,502 particles that were subjected to 3D classifications with small angular sampling using two different masking strategies, one with a spherical mask and the other with a soft mask, resulting in a 7.9 Å map with good CST density from 31,388 particles and a 4.8Å map without CST density from 52,506 particles, respectively. Please refer to the Methods for detailed description of the steps. The same workflow was used for the DNA free telomerase, starting from 2,300,000 particles.
(D) Plots of the gold-standard Fourier shell correlation coefficients as a function of spatial frequency for the DNA-bound and DNA-free telomerase complexes.
(E) Euler angle distributions of the particles in the final 3D auto-refinement.
(F) Local resolution evaluation of the cryoEM map of the DNA-bound telomerase generated by Resmap. The core region of the map has resolution near 4Å.
(G–K) Representative regions of the cryoEM density map (gray and mesh) superposed with select regions of our atomic models (color sticks and ribbons) of (G) RBD, (H) TEN, (I) RT, (J) IFD-TRAP, (K) the DNA-RNA duplex.
(A and B) Six rotated views of (A) DNA-bound telomerase and (B) DNA-free telomerase show that the two states are globally the same. For this comparison, both density maps are filtered to 6.4 Å resolution which is the lower resolution of them. Colors correspond to TERT (blue), TER (magenta), telomeric DNA (green), p65 xRRM (dark green), Teb1C (coral), Teb2N (tan), Teb3 (gold), p50 (red). Note that most of the density for p65 is invisible due to its flexibility.
(C and D) Region of the density map containing the TER template for telomerase with bound sstDNA (C) and for DNA-free telomerase (D). Magenta and green are cryoEM densities of RNA template and DNA, respectively. Grey is spherically masked cryoEM density of TERT. (E and F) Region of the density map containing at the TRAP–TEN–TRE interface in the DNA-bound telomerase (E) and in the DNA-free telomerase (F). The model of TRAP, TEN, and TRE in the telomerase–sstDNA complex is shown in E. In F the model of TRAP and TEN is from the telomerase–sstDNA complex to highlight the regions of TEN and TRAP that are no longer seen. The TRE (modeled into the DNA free map) also changes.
(G and H) Region of the density map containing Teb1C for telomerase with bound sstDNA (G) and for DNA-free telomerase (H). Note that both density maps are filtered to 6.4 Å resolution to match the resolution of the DNA-free telomerase.
(A) CryoEM map with atomic model of TERT ring, RBD-RT-CTE.
(B–D) TERT ring domain structures labeled with sequential assignment of helices and sheets: RBD (B), RT (C), CTE (D).
(E) CryoEM map with atomic model of TERT TEN–IFD-TRAP.
(F and G) TERT TEN and IFD-TRAP domain structures labeled with sequential assignment of helices and sheets: TEN (F), IFD-TRAP (G).
(H) CryoEM map with atomic model of TER and template bound DNA
(I) CryoEM map with p50 secondary structure elements. Amino acid sequence is not assigned.
(J) CryoEM map with atomic model of Teb1C and bound DNA
(K) CryoEM map with atomic model of Teb2N-Teb3.
(L) Cartoon representation of TERT-TER catalytic core showing that the TEN and RBD domains are connected by a flexible linker that crosses over the t/PK circle. Linker is represented by dotted line.
(M and N) Close-up views of the catalytic core highlighting the location of the (flexible, not visible) TEN-RBD linker (black dashed line) in the 4.8 Å cryoEM map. Linker length and start/end residue numbers are indicated.
(A) Tetrahymena TERT RBD-RT-CTE (blue, light purple, light blue) superimposed on Tribolium TERT (PDB 3KYL) (gray). For Tetrahymena TERT the IFD-TRAP (purple) is shown but the TEN is omitted for clarity.
(B) Tetrahymena TEN (cyan) superimposed on the crystal structure (PDB 2B2A) (gray). α7 and loop to β3 are not seen in the crystal structure.
(C) Tetrahymena Loop 4-RBD-TBE (blue and magenta) superimposed on RBD-TBE (gray) (PDB 5C9H).
(D) Tetrahymena CTE superimposed on human CTE (PDB 5UGW) (gray).
(A) Structures of HIV-1 RT (PDB 4PQU), G. stearothermophilus Group II Intron encoded RT (PDB 6AR1), Tribolium castaneum TERT (PDB 3KYL), and Tetrahymena thermophila TERT (this work), with template-primer duplexes.
(B) Comparison of conserved domains found in reverse transcriptases from HIV-1 and Group II introns, Penelope Like Element associated reverse transcriptase (PLE-RT) from A. vaga, Tribolium TERT, T. thermophila TERT, and human TERT. Note that the domain structure of Tribolium TERT appears more similar to PLE-RT than to Tetrahymena and human TERTs. Group II intron reverse transcriptase, PLE-RT, Tribolium TERT all have motif 3 and IFDac, and PLE-RT and Tribolium TERT have an RBD with a T-motif. The human and Tetrahymena TERTs (as well as TERTs from plants and yeasts) have the additional TRAP and TEN domains that are essential for RAP.