Abstract
Background
Poor prognosis of pancreatic cancer (PanCa) is associated with lack of an effective early diagnostic biomarker. This study elucidates significance of MUC13, as a diagnostic/prognostic marker of PanCa.
Methods
MUC13 was assessed in tissues using our in-house generated anti-MUC13 mouse monoclonal antibody and analyzed for clinical correlation by immunohistochemistry, immunoblotting, RT-PCR, computational and submicron scale mass-density fluctuation analyses, ROC and Kaplan Meir curve analyses.
Results
MUC13 expression was detected in 100% pancreatic intraepithelial neoplasia (PanIN) lesions (Mean composite score: MCS=5.8; AUC >0.8, P<0.0001), 94.6% of pancreatic ductal adenocarcinoma (PDAC) samples (MCS=9.7, P<0.0001) as compared to low expression in tumor adjacent tissues (MCS=4, P<0.001) along with faint or no expression in normal pancreatic tissues (MCS=0.8; AUC >0.8; P<0.0001). Nuclear MUC13 expression positively correlated with nodal metastasis (P<0.05), invasion of cancer to peripheral tissues (P<0.5) and poor patient survival (P<0.05; prognostic AUC=0.9). Submicron scale mass density and artificial intelligence based algorithm analyses also elucidated association of MUC13 with greater morphological disorder (P<0.001) and nuclear MUC13 as strong predictor for cancer aggressiveness and poor patient survival.
Conclusion
This study provides significant information regarding MUC13 expression/subcellular localization in PanCa samples and supporting the use anti-MUC13 MAb for the development of PanCa diagnostic/prognostic test.
Keywords: Pancreatic cancer, Mucin, MUC13, Diagnosis, Prognosis, Tumorigenesis
INTRODUCTION
By 2030, pancreatic cancer (PanCa) is projected to be the second leading cause of cancer related deaths 1. The early diagnosis of PanCa remains a clinical challenge, predominantly due the lack of effective diagnostic biomarkers. Therefore, the scientific community is making tremendous efforts toward identification of novel early diagnostic markers, as existing biomarkers, including CA19-9 have yielded only suboptimal efficacy in early stage detection 2.
Recent studies have suggested that high expression of MUC13, a newly identified epithelial cell surface mucin, in PanCa and the overexpression of MUC13 in PanCa cells leads to enhanced tumorigenic and metastatic phenotypes. These characteristics of PanCa cells are mediated by physical interactions between MUC13 and HER2/Neu 3–5. Additionally, MUC13 expression inversely correlates with the expression of the tumor suppressor microRNA-145 (miR-145) 5, while the restitution of miR-145 inhibits MUC13 expression and pancreatic tumor growth 6. In addition to our previous studies 3–5, the potential role of MUC13 in pancreatic pathophysiology has also been recognized by other institutions.
In this study, we have performed a comprehensive investigation of MUC13 expression using our newly generated anti-MUC13 monoclonal antibody (Mab) in a large cohort of human PanCa tissue samples. The expression pattern and subcellular localization of MUC13 was correlated with each patient’s clinical course to determine its diagnostic and/or prognostic significance. Based on our previous observations, we hypothesized that MUC13 expression could be detected in precursor lesions (i.e. PanIN).
MATERIALS AND METHODS
Human pancreatic cancer tissues
Tissue microarrays (TMAs) were obtained from multiple sources: TMA#Baptist-UTHSC (normal pancreatic tissue, tumor-adjacent normal pancreas, PanIN, pancreatic ductal adenocarcinoma [PDAC], other PanCa types) was obtained from Baptist Memorial Hospital (BMH), Memphis, TN; TMA#PA2012 (PDAC and other control organs) from the University of Nebraska Medical Center; TMA#642 (multiple types of invasive PanCa with normal pancreas) from Johns Hopkins; OD-CT-DgPan03-001 (PDAC), BIC14011 (PanIN, PanCa), PA241b (PDAC with matched normal adjacent tissue [NAT]), T141a (PanCa tissue), HPan-Ade150CS-01 (PDAC and matched NAT), PA961b (PDAC) and HPan-Ade180Sur-01 (PDAC with matched NAT) were obtained from US Biomax, Inc. (Derwood, MD, USA). Additionally, fresh pancreatic tissue samples were procured from BMH including PanCa with matched NAT. The institutional review boards at UTHSC and BMH approved this study.
PDAC cell lines and culture conditions
PDAC cells were obtained from ATCC (American Type Culture Collection). We generated MUC13 knock-down PDAC (sh-M13), MUC13 vector (sh-V) stable cell lines using HPAF-II cells; and ectopically expressing MUC13 (M13) and vector (V) stable cell lines using Panc-1 cells3. The cells were cultured in DMEM, RPMI or DME/F12 media supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere with 5% CO 2 in air at 37°C.
RT-PCR for MUC13 mRNA
RNA was extracted from fresh tissues using RNeasy Midi Kit (catalog number 75144, Qiagen). The relative expression levels of MUC13 mRNA were assessed by Semi - quantitative RT-PCR 8 using sequence-specific primers: MUC13 fwd: CCTTCGGTGTGATTATTATGGC, MUC13 rev: GCATCTGGCTGTCTCTGGAG.
Immunoblotting for MUC13 protein
The protein from tumor tissues was isolated as described before 8 and processed for immunoblotting as described previously 8, 9.
Immunohistochemistry for MUC13 expression
MUC13 expression was detected by immunohistochemical staining using a commercially available kit (Biocare Medical) as described before 5, 10, 11. Briefly, tissue sections were deparaffinized and rehydrated using different grades of alcohol followed by endogenous peroxidase quenching. Blocking and incubation with mouse monoclonal MUC13 antibody (produced in our lab) 3, 5, 12, 13 was then completed overnight. These antibodies exhibit high specificity and an intense reactivity with PDAC tissues in vitro and in vivo (xenograft mice-data not shown). The sections were then incubated with secondary MACH4 Universal HRP mouse polymer (#M4U534H) followed by staining with 3-diaminobenzidine (DAB) chromogen (#DB801) and counterstaining with hematoxylin.
Scoring of MUC13 expression in stained pancreatic normal and cancerous tissues
After histochemical labeling, slides were digitally scanned and analyzed for MUC13 staining at the UTHSC Pathology Department. Blinded to the patient’s history, captured images were independently analyzed by three reviewers (N.Z, S.K, and S.C.C) and a consensus scoring was deduced as described earlier 14. The intensity of immunoreactivity of the MUC13 was graded on a 0 to 4 scale (0 for no staining, 1 for weak immunoreactivity; 2 for moderate immunoreactivity; 3 for strong immunoreactivity, and 4 for very strong immunoreactivity. The percentage of cells positive for MUC13 immunoreactivity within the tumor and adjacent normal tissue sections were scored as follows: 0–25% as 1, 26–50% as 2, 51–75% as 3 and 76–100% as 4. Acellular or stromal tissues were not scored, nor were neighboring acinar cells with negligible or no staining. Staining of uninvolved adjacent ducts was used to represent MUC13 expression in matched NAT for PDAC cases. The composite score (CS) values ranging from 0–16, were calculated by multiplying the value for staining intensity (0 – 4) and the percentage of immunoreactive cells (0 – 4) for each individual sample. Finally, the mean composite score (MCS) was determined by calculating an average of the composite scores of respective samples in each category.
Computational analysis for best feature selection
Computational analysis was utilized to better understand the role of MUC13 in predicting cancer aggressiveness and patient survival by defining strong determining features. The artificial intelligence techniques like ‘Best Fit’ filter method and the wrapper method of ‘Genetic Algorithm’ have been extensively described for best feature selection in the diagnosis of cancers15–17. Best Fit selects the first best attribute or feature set for further consideration whereas the Genetic Algorithm randomly searches every possible attribute or feature set with strong features. For evaluation of feature sets, we have additionally used a Neural Network classifier or a learner to identify the category or class to which an observation or instance belongs18, 19 depending upon the training validation and testing set provided to the classifier.
Quantification of spatial submicron scale mass density fluctuations associated with MUC13
We also studied the relationship between intracellular nanoscale morphological alterations reflected as submicron scale spatial mass-density fluctuation at different levels of MUC13 expression in PDAC by investigating the light localization property of cells 4, 20. Using confocal DAPI images of nuclei, we generated mass arrays and dielectric arrays in order to obtain normal modes of light waves to quantify the degree of the morphological distortion, Lmd with respect to the presence or absence of MUC13 staining in different areas within the samples or in contrast to the distinct samples as described before 20–22. Lmd stands for local mass distortion/variation m and lc for corresponding spatial correlation decay length23. For the statistical analysis, we took confocal micrographs of cells and tumor samples (n=20) from each type.
Statistical Analysis
The means of two groups were compared using a Students-t test, with P-values <0.05 considered statistically significant. Box plots were used to analyze data and generated using Sigma plot 12.0. For the computational analysis, the Mean Squared Error (MSE) was calculated as the average of the squares of the errors or deviations wherein the error is the difference between the expected and observed value. For immunostaining, MUC13 expression, represented by the MCS, was determined for the membrane, cytoplasm, nucleus, and overall (overall intensity x overall percentage staining). The MCS was compared among groups and the effects of MUC13 expression in conjunction with stage, grade, and long-term outcomes were studied. The ability of MUC13 to differentiate PDAC patients from the control groups (healthy control: HC and Chronic pancreatitis: CP patients for the initial study) was determined by analyzing the area under the curve (AUC; denotes the probability that the time-to-event is improved in one arm compared to the other) using ROC curve analysis. In multivariate analysis of these variables adjusted for overall (overall intensity x overall percentage staining) and MCS Nucleus, Cox Proportional Hazards model was used. Results were summarized with hazard ratios and confidence intervals. All analyses were done using SAS 9.4 (SAS institute Inc., Cary, NC) 24.
RESULTS
Progressive increase of MUC13 in PanIN lesions
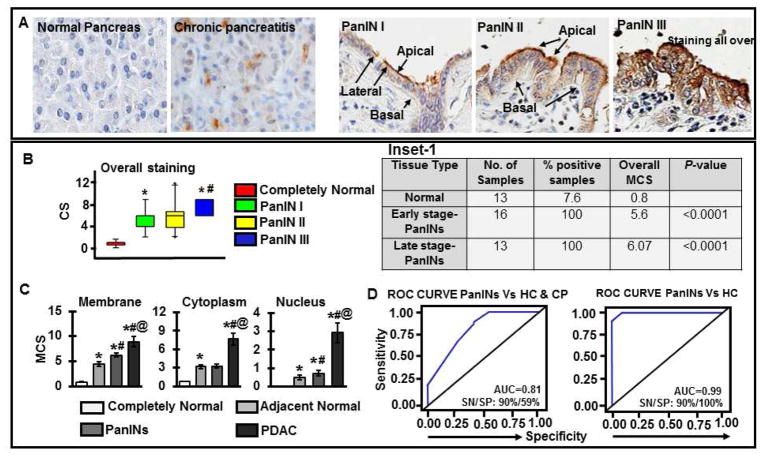
Tissues were immunostained for MUC13, including normal pancreas (n=13), chronic pancreatitis (n= 12), early-stage PanIN (n=16; PanIN-1A and PanIN-IB) and late-stage PanIN (n=13; PanIN-II and PanIN-III) (Supplementary Table 1). Normal pancreatic ducts showed no or very faint MUC13 expression, whereas, all of early (16 of 16) and late-stage PanINs (13 of 13) showed low to moderate MUC13 expression (Fig. 1A and Inset-1). MUC13 expression was predominantly at the apical side and less at the basal side of the epithelial cells in case of PanIN I/II, while, PanIN III samples showed staining all over the cell throughout lesions. Overall, MUC13 expression was significantly (p<0.0001) higher in PanINs compared to normal pancreatic ductal epithelium (Fig. 1B). Additionally, we analyzed changes in the subcellular localization of MUC13 in PanINs and observed a progressively increased MUC13 expression in the membrane, cytoplasm and nucleus, during the course of progression from PanIN-I to PanIN-III (Fig. 1C and Supplementary Figure 1A and B). We evaluated the lesion scores in two ways; Composite score (CS; taking in account of each case individually) and MCS (mean composite score) of each type of lesion. While assessing diagnostic significance of MUC13 using Receiver Operating Characteristic (ROC) analysis, MUC13 expression effectively differentiated PanINs (n=29) from healthy controls (HC; n=13) and chronic pancreatitis (CP; n=12) tissues (AUC: 0.99; and 0.81, respectively) (Fig 1D). The sensitivity (SN) and specificity (SP) differentiating PanINs from HC is SN: 90%, SP: 100% and that from HC and CP is SN: 90%, SP: 59%.
Fig. 1. MUC13 is progressively increased in PanINs.
(A) Images depicting the expression of MUC13 (brown) in completely normal pancreatic tissues, chronic pancreatitis and PanINs I–III. All images are captured at 400X. (B) Bars representing composite scores (CS) of overall MUC13 expression; *p-values<0.0001 compared to completely normal or #p-values<0.5 compared to panIN-I/II (Group means are indicated by solid lines); *p-values<0.5 compared to PanIN-I or #p-values<0.5 compared to panIN- II. Inset-1: Table showing MCS values for MUC13 expression in PanINs compared to normal pancreatic ductal epithelium (P-values <0.0001). (C) Comparative analysis of MUC13 expression and localization pattern in completely normal pancreatic tissues, adjacent normal, PanINs and PDAC. Mean composite score (MCS) representing subcellular localization of MUC13 in tissues; *p-values<0.0001 compared to completely normal, #p-values<0.5 compared to tumor adjacent normal and @ p-values<0.0001 compared to PanINs. Group means are indicated by solid lines. (D) For the diagnostic assessment of MUC13, ROC curves were generated examining the ability of MUC13 to differentiate between PanINs, healthy pancreas (HC) and chronic pancreatitis (CP).
Aberrant expression and subcellular localization of MUC13 in PDAC
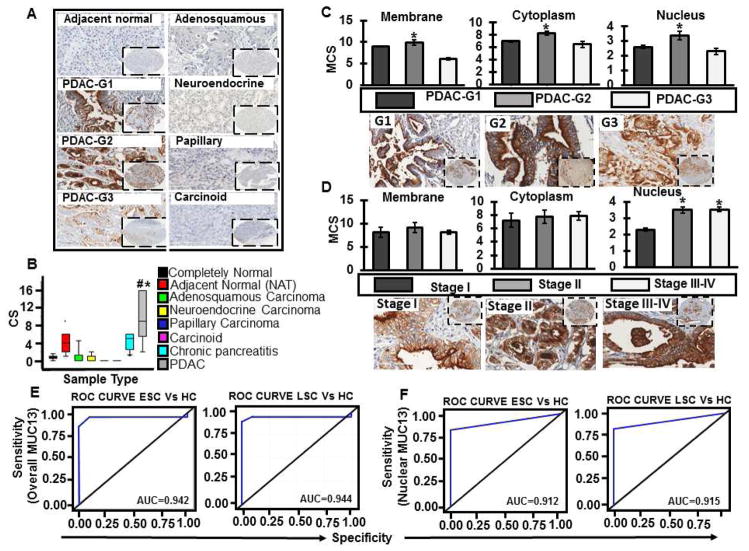
To determine involvement of MUC13 in PDAC, we performed MUC13 expression profile analysis in different histological PanCa types and normal pancreas. MUC13 expression was observed in 94.6% (p- value <0.0001) of PDAC samples (n=225, 95% positive, MCS=9.7; Fig. 2A and B) while, morphologically normal appearing tumor adjacent tissues (NAT) (n=100) showed faint to moderate staining in the ducts (n=100, 83% positive, MCS=4; Fig. 2A and B). A low to moderate MUC13 expression (MCS of 4) is observed in NAT, which is significantly (P-value<0.001) lower compared to PDAC (MCS of 9.7). Normal pancreatic ductal epithelia and acinar cells however, were largely negative for MUC13 expression (n=13, 7.6% positive, MCS=0.8; Fig. 2A and B). This data suggested some degree of molecular level alterations in NAT that are not morphologically apparent in these tissues. In contrast, faint or no staining was observed in adenosquamous carcinoma (n=18, 22% positive, MCS=1.2, P=0.001), neuroendocrine carcinoma (n=5, 20% positive, MCS=0.4, P=0.001), solid pseudo-papillary carcinoma and carcinoid tumors (n=6, 0% positive, MCS=0) (Fig. 2A and B). Completely normal ducts, acini or surrounding non-epithelial elements (lymphocytes or stroma) were typically not stained for MUC13 expression. Staining scores were recorded based on subcellular localization namely, membranous, cytoplasmic, nuclear and total staining and correlated with clinico-pathological parameters (Supplementary Table 2). Increased expression of MUC13 was observed only in PDAC samples sparing other PanCa types, suggesting its potential involvement in pancreatic ductal epithelial pathobiology.
Fig. 2. MUC13 expression profile analysis in normal pancreas and different pancreatic cancer types and its correlation with PDAC differentiation and stage.
(A) Representative images of the tissues stained for MUC13 expression, depicted in color brown. (B) Bars representing composite scores of overall MUC13 expression in samples of normal pancreas and different histological pancreatic cancer types by IHC using MUC13 MAb. MUC13 expression was identified in 94.6% (p-value <0.0001) of pancreatic ductal adenocarcinomas; *#p-values<0.0001. Group means are indicated by solid lines. (C) Bars representing MCS of MUC13 with respect to subcellular localization in membrane, cytoplasm and nucleus in PDAC tissue samples; Grade 1 (G1) to Grade 3 (G3); and the representative images shown (D). Bars representing MCS of MUC13 with respect to subcellular localization in membrane, cytoplasm and nucleus in PDAC samples; stage I to stage IV; and the representative images shown. Group means are indicated by solid lines and *p-values<0.05. All images are captured at 400X and 40X. Diagnostic assessment of (E) overall MUC13 and (F) nuclear MUC13 using ROC curves in healthy controls, resectable ESC (Early-stage cancer) and unresectable LSC (Late-stage cancer).
Correlation of MUC13 expression with PDAC differentiation and stage
High MUC13 staining was observed in G2 (moderately differentiated) as compared to G1 (well differentiated) and G3 (poorly differentiated) (Fig. 2A and supplementary Figure 2A; p<0.05) samples. These results suggest progressive expression of MUC13 during PDAC from well to moderately differentiated tumors and the expression pattern changed with overall lower condensed MUC13 expression in the poorly differentiated tumors.
PDAC samples were further grouped into two categories based on the clinical stage of the tumor, early (stage I and II) and late (stage III and IV). The majority of samples in both the stages were positive for MUC13 staining with only 4.9 % and 5.3 % negative samples in early and late stage cancers, respectively. Although, the MUC13 subcellular localization was predominantly observed at M+C+N (membrane+cytoplasm+nucleus) in case of both early and late stage cancers, the percentage of cases positive for M+C+N varied and was higher (89.4%) in late stage cancers in comparison to early stage (81.6%). Overall, the analysis of subcellular localization patterns of MUC13 with respect to the tumor stages indicated increased nuclear expression of MUC13 in case of stage II–IV disease when compared to Stage I (Fig. 2D and Supplementary Figure 2B; p<0.05 and Supplementary Table 2). Supplementary Figure 2C is the representative image showing marked nuclear expression in stage IV. ROC curve analyses revealed that both overall (Fig. 2E) and nuclear (Fig. 2D) expression of MUC13 efficiently differentiated healthy controls (HC; n=13) from early stage (ESC; n= 182) and late stage cancers (LSC; n=19) (AUC>0.9).
Association of MUC13 expression with clinico-pathological parameters
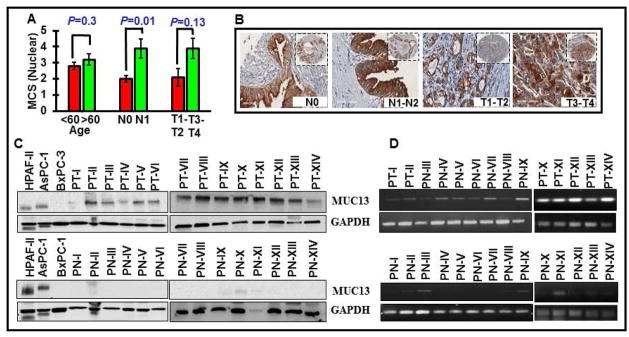
To establish association of MUC13 with clinico-pathological parameters, patient samples were grouped and analyzed for MUC13 positivity based on sex [130 males (96 % positive) and 74 females (100 % positive)], age [87 patients with age <60 (94.2 %) and 123 with age >60 (95.9 %)], and TNM staging [124 individuals with tumor located to pancreas (95.9 % positive) and 85 with extension beyond pancreas (94.1 % positive); 136 individuals (94.1 %) with no lymph node involvement (94.1 % positive) and 59 individuals with lymph node metastasis (96.6 % positive)]. The incidence of MUC13 positivity significantly correlated with advanced stage growth into adjacent organs or metastasis to lymph nodes (N1-N2); overall (MCS N0=8.9 Vs N1-N2=10.1; p=0.1), membrane (MCS N0=8.1 Vs N1-N2=9.1; p=0.5), Cytoplasm (MCS N0=6.9 Vs N1-N2=8.2; p=0.1) and nucleus (MCS N0=2.0 Vs N1-N2=3.9; p=0.01). Both the nuclear intensity and its percentage positivity in the tumor were higher in cases characterized to have increased nodal metastasis from 1– 4 lymph nodes (Fig. 3A, B, Supplementary Figure 4 and Table 3). MUC13 expression was lower in N0 disease compared with N1 while increased MUC13 staining was found in the nucleus with PDAC extending beyond pancreas (T3-T4) (MCS T1-T2=2.1 Vs T3-T4=3.9; p=0.13). These studies indicate noticeable differences in subcellular localization of MUC13 and association of its aberrant subcellular localization with the clinical disease parameters. Since, nuclear expression of MUC13 was considerably higher in T3/T4 or N1 disease (Fig. 3B), it suggests the clinical significance of nuclear MUC13 expression in pancreatic tumor invasion and metastasis.
Fig. 3. MUC13 expression and its association with clinico-pathological parameters.
(A) Analysis of nuclear MUC13 expression in PDAC tissues with respect to TNM staging (depth of invasion and lymph node metastasis). (B) Representative images are shown. (C and D) Immunoblotting and PCR analysis showing the expression of MUC13 in 14 fresh-frozen PDAC tissue (Upper panel; PT-I to PT-XIV) and 14 matched adjacent normal (NAT) (Lower panel; PN-I to PN-XIV).
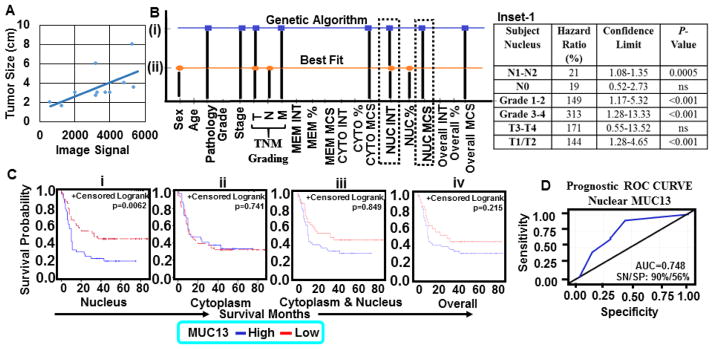
To validate MUC13 IHC results, we performed immunoblotting and RT-PCR experiments in a cohort of 14 matched fresh-frozen tumor (14 PDAC tumors; PT-I to PT-XIV) and adjacent tumor tissue samples (NAT; PN-I to PN-XIV). MUC13 expression was observed in all tumor samples (Fig. 3C and D, upper panels) whereas, faint (n=4) or no (n=10) expression was detected in NAT samples (Fig. 3C and D, lower panels). We also performed quantitative MUC13 expression analysis for its association with the tumor size (cm), which showed a positive correlation (R2=0.36) between increased MUC13 expression and the tumor size (Fig. 4A). Additionally, we performed genetic algorithm analysis using feature selector “best-fit attribute selector” and “classifier Neural Network” (Fig 4Bi and ii) softwares to determine association of different MUC13 expression patterns with clinical features. Analysis of this dataset was reduced to just 5 strong features namely sex, tumor invasion, lymph node involvement, nuclear intensity and nuclear percentage to predict clinical outcome/patient survival. On further applying genetic algorithm along with the same classifier Neural Network 25, the prediction of survival was more clear and accurate (Fig. 4Bi–ii and Supplementary Table 4). Our computational analysis suggests that the nuclear intensity of MUC13 (NUC INT) was the strongest attribute selected by “best-fit” and Genetic algorithm. However, percentage of area positive for MUC13 (NUC %) and its overall nuclear expression (NUC MCS) outperforms other attributes for patient survival. Our results show that Genetic algorithm along with Neural Network performed better compared to best fit with same classifier to predict patient’s prognosis (Supplementary Figure 5A–C). We have also shown the gradual reduction in error for 10 iterations, wherein Prediction Error Value is reduced after selecting strong features in each iteration using Genetic Algorithm and Neural Network. Further, we observed that overall nuclear expression (NUC MCS); attribute 17 with feature count value (FCV) as 10, is the strongest attribute in the feature set that can best predict the patient survival (Supplementary Figure 5B and C). Each time a feature is used in an iteration, its nuclear FCV increases. So the attribute with highest FCV is considered to be the strongest one, which means even less staining in nucleus, can predict patient’s poor prognosis. Nuclear MUC13 expression correlated with overall poor patient’s survival, nodal status (No-N1), depth of tumor invasion (T3-T4), tumor stage, and tumor grade compared with other subcellular localization patterns (Fig. 4C and Inset-1). A unit increase in nuclear MUC13 MCS resulted in a 21% (hazard ratio=1.20, 95% CI=1.08, 1.35, p=0.0005) increase in mortality. The tumors positive for high MUC13 expression or cytoplasm + nucleus also showed a trend toward shorter survival. These results suggest prognostic significance of nuclear expression of MUC13 in predicting the survival fate of PDAC patients in ROC curve (Fig. 4D; AUC>0.9).
Fig. 4. MUC13 association with tumor size and patient survival prediction.
(A) Stratified analysis of MUC13 expression and its association with tumor size in a pathologic specimen. Data was analyzed using Image Studio™ Lite and compared to total tumor size (cm) in the pathologic specimen. R2 = 0.36998. (B) Computational analysis for the selection of a strong attribute for prediction of survival in human pancreatic cancer patients. Graph shows the features selected by the algorithms, Genetic Algorithm (i), Best Fit (ii) along with the applied classifier Neural Network. (C) Kaplan-Meier survival curves compared using log-rank test were constructed to assess the role of subcellular distribution of MUC13 relative to patient survival: (i) nucleus, p = 0.006 (ii) cytoplasm, p = 0.74 (iii) cytoplasm and nucleus p = 0.84 and (iv) overall MUC13 expression p = 0.21. Inset-1: Proportional hazards regression was used for multivariable survival analyses. All P-values were computed two sided and defined as statistically significant when <0.05. (D) Prognostic ROC curve determining the potential of nuclear MUC13 expression to discriminate survived and diseased patients.
MUC13 expression leads to increased degree of disorder in PDAC
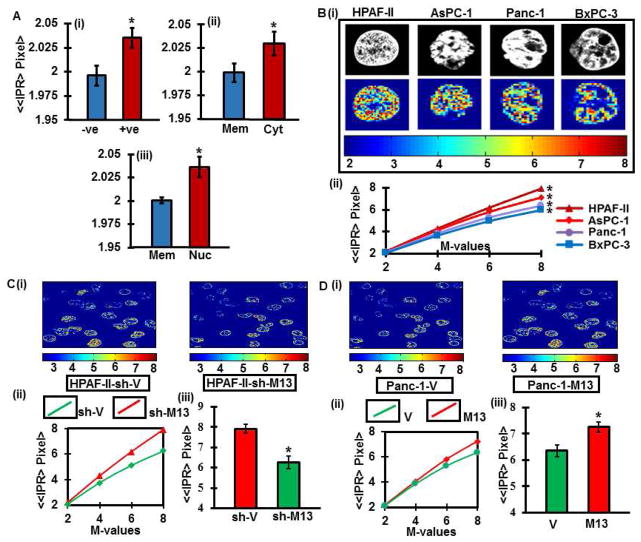
We sought to determine if MUC13 expression has an association with submicron scale morphological alterations occurring in PDAC tissues, as described earlier 4, 26. Enhanced morphological disorder (Lmd) in MUC13 positive tumors was observed compared to negative counterparts (p<0.001) (Fig. 5Ai). The order of morphological disorder was significantly higher (p<0.001) in nuclear positive areas, compared to being lowest in only membrane positive areas (Fig. 5Aii and iii).
Fig. 5. Nanoscale morphological/architectural changes in human PDAC tissues.
(A) Histogram of ≪IPR>Pixel> for number of pixels in pancreatic tumor tissues that are MUC13 positive tumors compared to MUC13 negative counterparts (i), tissue areas positive for MUC13 membrane staining compared to the areas positive for MUC13 cytoplasmic staining (ii) and areas positive for nuclear staining (iii). Original magnification, 400X. *p<0.001. (B) Representatives of confocal grey scale images of MUC13 expressing (HPAF-II and AsPC-1) and MUC13 null (Panc-1 and BxPC-3) pancreatic cancer cell lines (Upper panel) and their corresponding IPR images (lower panel; IPR pixel values m=4) (i). Ensemble-averaged (n=20) values of ≪IPR(L)>Pixel> versus number of pixels for HPAF-II, AsPC-1, Panc-1 and BxPC-3 (*p<0.001) (ii). (C) Representative local Lmd distribution in two dimension images derived from the corresponding confocal image slices of nuclei for MUC13 expressing (sh-V) and MUC13 knockdown (sh-M13) HPAF-II cells (i). Ensemble-averaged (n=20) values of ≪IPR(L)>Pixel> versus number of pixels for sh-V and sh-M13 (*p<0.001) (ii). Histogram of ≪IPR>Pixel> for number of pixels m=4 (*p<0.001) (iii). (D) Representative local Lmd distribution in two dimension images derived from the corresponding confocal image slices of nuclei for MUC13 null (V) and ectopically expressing MUC13 (M13) Panc-1cells (i). Ensemble-averaged (n=20) values of ≪IPR(L)>Pixel> versus number of pixels for V and M13 (*p<0.001) (ii). Histogram of ≪IPR>Pixel> for number of pixels m=4 (*p<0.001) (iii).
Parallel results were corroborated in PDAC cell lines that endogenously express MUC13 (HPAF-II and AsPC-1) or are MUC13 null cells (Panc-1 and BxPC-3) (p<0.001) (Fig. 5B). Additionally, cells endogenously expressing MUC13 (HPAF-II-sh-V), show conspicuous morphological distortion (Lmd), while compared to MUC13 knockdown cells (HPAF-sh-M13) as depicted by the average IPR values (p<0.001) (Fig. 5C). However, ectopic expression of MUC13 in Panc-1 cells (MUC13 null) significantly (p<0.001) increased the degree of morphological distortion (Lmd) (Fig. 5D). This unique study suggest that MUC13 expression is associated with the enhanced mass-density variation, which further explains its role in tumor metastasis and aggressiveness 22, 23.
DISCUSSION
The high mortality observed for PanCa is mainly attributable to the often advanced stage of disease at diagnosis, thus the development of early diagnostic modalities for this disease should be a top priority and these modalities should be specific and sensitive enough to detect occult disease and precursor lesions. Our previous studies have suggested that MUC13, a recently identified membrane tethered glycoprotein, is highly overexpressed in PanCa while nearly undetectable in normal pancreatic tissue 3–5. However, due to the lack of availability of highly specific reagents related to MUC13, its utility as a PanCa biomarker has not been explored in a comprehensive manner. Considering this fact, our group has recently generated an array of anti-MUC13 MAbs that recognize MUC13 with high specificity. In the present study, using these novel reagents, we have established a MUC13 expression profile in different PanCa types. The results of this study demonstrate a potential role for MUC13 as a novel molecular biomarker for PanCa diagnosis/prognosis.
Our results demonstrate MUC13 positivity in PanIN lesions, with a progressive increase in expression from PanIN-I to PanIN-III suggesting its critical implications in PanCa progression. Undetectable MUC13 expression in normal pancreas; and significantly higher MUC13 expression in PDAC (as opposed to other PanCa subytypes), indicates that MUC13 expression is unique to PDAC and may be a useful marker for early PDAC. The diagnostic potential of MUC13 was further elaborated through a ROC analysis which discriminately differentiated early pancreatic lesions with controls (HC or BC or CP). We also found an increased MUC13 expression from well to moderately differentiated PDAC, although the expression declined in poorly differentiated PDAC, which might be the result of acquired epigenetic alterations in cancer cells. These epigenetic alterations following repression of MUC13 in poorly differentiated tumors are possibly required for successful seeding of tumor cells at sites of metastases. These underlying complex processes are a prerequisite to the dynamic regulation of the detachment and re-attachment of tumor cells at the metastatic site 8. We believe this phenomenon, involves shedding of the bulky proteins from the tumor cell surface to interact with and attach to metastatic proteins that aid tumor cells 27.
Staining of MUC13 in PDAC was observed in the cell membrane, cytoplasm as well as in the nucleus. Of note, high nuclear expression of MUC13 correlated with poor patient survival in PDAC, indicating its prognostic significance. Computational analysis of PDAC also confirmed nuclear expression as the strongest predictor of patient survival. Additionally, morphological distortion in pancreatic cancer cells and tissues suggested increased degree of disorder and aggressiveness with aberrant MUC13 expression (from membranous<cytoplasmic<nuclear). These results unequivocally indicate the importance of MUC13 expression and provide strong implications of nuclear MUC13 expression in PanCa progression. Additionally, cellular and molecular mechanisms that trigger aberrant MUC13 expression/localization is an interesting area for future research, as it can be helpful for developing new therapies to manage MUC13 driven PanCa progression and metastasis. Since MUC13 has a SEA domain (a cleavage site), therefore, it is also shed in the bloodstream and detected in sera of PanCa patients (Data not shown), efficiently discriminating PanCa patients with healthy individuals. Our hope is that this will lead to the development of a minimally invasive, cost-effective, and reproducible serum based detection system for PDAC. Moreover, our previous studies indicate a direct interaction of MUC13 with HER2 in PanCa cell lines and tissues 4, it would be exciting to investigate the association of MUC13 expression with clinical outcome in HER2+ PDAC.
In conclusion, our recently developed method for using our newly generated MAb may prove extremely useful for MUC13 detection in PanCa. While MUC13 expression could be used for early diagnosis of PanCa, its aberrant expression/subcellular localization (nuclear) is of high prognostic significance. Utilizing radionucleotide labeled MUC13 antibody, our aim is to develop a discriminatory, non-invasive imaging tools for identifying dysplasia in early PDAC and its precursor lesions. We will also employ a unique technique to probe MUC13 associated nanoscale morphological distortions appearing early at the initiation of PDAC. Additionally, we will formulate MUC13 targeted therapeutic strategies for PanCa patients. This will include designing of a novel nanoparticle technology for active tumor targeting via conjugation with MUC13 specific antibodies/aptamers for enhanced accrual of therapeutics at the tumor site. The largely, promising findings of this study are suggestive of future multi-center trials to assess the utility of MUC13 as a novel diagnostic/prognostic marker in pancreatic cancer.
Supplementary Material
NOVELTY AND IMPACT.
This study provides an evaluation of MUC13 expression and its subcellular localization in human PDAC tissues and its association with the clinical course of the disease progression. Our study represents the most extensive expression profiling of MUC13 for PDAC to date and supports its clinical development for diagnostic and prognostic purposes. Our newly generated anti-MUC13 MAb can be useful for the development of novel diagnostic/prognostic test(s).
Acknowledgments
The authors are thankful to Cathy Christopherson for editorial assistance.
Funding
This work was partially supported by grants from the Department of Defense (PC130870 to SCC and MJ), the National Institutes of Health (R01 CA206069, CA204552, CA210192, CA142736 to SCC, U01 CA162106A to SCC and MJ; R01 EB003682 to PP; K22CA174841 to MMY), the College of Pharmacy 2014 and 2015 Dean’s Seed/Instrument Grants of the University of Tennessee Health Science Center (to SCC, MJ and MMY) and Grants of the University of Memphis (to PP). Authors acknowledge the financial support of Herb Kosten Foundation.
Footnotes
Potential conflicts of interest
The authors declare that there are no financial and non-financial competing interests.
Contributors
- Study concept and design: Sheema Khan, Subhash C. Chauhan, Nadeem Zafar, Stephen W. Behrman.
- Acquisition of data: Sheema Khan, Nadeem Zafar, Stephen W. Behrman, Shabia S. Khan, Saini Setua, Zachary E Stiles, Hemendra Ghimire, Peeyush Sahay, Murali M. Yallapu, Tomoko Ise, Satoshi Nagata, Prabhakar Pradhan, Meena Jaggi, Subhash C. Chauhan.
- Analysis and interpretation of data: Sheema Khan, Nadeem Zafar, Stephen W. Behrman, Subhash C. Chauhan, Shabia S. Khan, Prabhakar Pradhan, Satoshi Nagata.
- Drafting of the manuscript: Sheema Khan and Subhash C. Chauhan.
- Statistical analysis: Lei Wang and Jim Y. Wan.
Competing interests
None declared.
Ethics approval
Institutional Review Board.
Data sharing statement
We are happy to share our data and resources upon request.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Can Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Kaur S, Baine MJ, Jain M, Sasson AR, Batra SK. Early diagnosis of pancreatic cancer: challenges and new developments. Biomark Med. 2012;6:597–612. doi: 10.2217/bmm.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chauhan SC, Ebeling MC, Maher DM, Koch MD, Watanabe A, Aburatani H, Lio Y, Jaggi M. MUC13 mucin augments pancreatic tumorigenesis. Mol Cancer Ther. 2012;11:24–33. doi: 10.1158/1535-7163.MCT-11-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan IA, Yoo BH, Masson O, Baron S, Corkery D, Dellaire G, Attardi LD, Rosen KV. ErbB2-dependent downregulation of a pro-apoptotic protein Perp is required for oncogenic transformation of breast epithelial cells. Oncogene. 2016;35:5759–5769. doi: 10.1038/onc.2016.109. [DOI] [PubMed] [Google Scholar]
- 5.Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, Thompson PA, Jaggi M, et al. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget. 2014;5:7599–609. doi: 10.18632/oncotarget.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setua S, Khan S, Yallapu MM, Behrman SW, Sikander M, Khan SS, Jaggi M, Chauhan SC. Restitution of Tumor Suppressor MicroRNA-145 Using Magnetic Nanoformulation for Pancreatic Cancer Therapy. J Gastrointest Surg. 2016;3:1–12. doi: 10.1007/s11605-016-3222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishii Y, Yamaguchi M, Kimura Y, Hasegawa T, Aburatani H, Uchida H, Hirata K, Sakuma Y. A newly developed anti-Mucin 13 monoclonal antibody targets pancreatic ductal adenocarcinoma cells. Int J Oncol. 2015;46:1781–7. doi: 10.3892/ijo.2015.2880. [DOI] [PubMed] [Google Scholar]
- 8.Sung HY, Park AK, Ju W, Ahn J-H. Overexpression of Mucin 13 due to Promoter Methylation Promotes Aggressive Behavior in Ovarian Cancer Cells. Yonsei Medical Journal. 2014;55:1206–13. doi: 10.3349/ymj.2014.55.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher DM, Khan S, Nordquist JL, Ebeling MC, Bauer NA, Kopel L, Singh MM, Halaweish F, Bell MC, Jaggi M, Chauhan SC. Ormeloxifene efficiently inhibits ovarian cancer growth. Cancer Lett. 356:606–12. doi: 10.1016/j.canlet.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S, Chauhan N, Yallapu MM, Ebeling MC, Balakrishna S, Ellis RT, Thompson PA, Balabathula P, Behrman SW, Zafar N, Singh MM, Halaweish FT, et al. Nanoparticle formulation of ormeloxifene for pancreatic cancer. Biomaterials. 2015;53:731–43. doi: 10.1016/j.biomaterials.2015.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S, Ebeling MC, Chauhan N, ATP, Gara Rk, Ganju A, Yallapu MM, Behrman SW, Zhao H, Zafar N, Singh MM, Jaggi M, et al. Ormeloxifene Suppresses Desmoplasia and Enhances Sensitivity of Gemcitabine in Pancreatic. Cancer Cancer Res. 2015;75:2292–04. doi: 10.1158/0008-5472.CAN-14-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, Bell MC, Koch MD, Aburatani H, Lio Y, Jaggi M. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009;69:765–74. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 13.Gupta BK, Maher DM, Ebeling MC, Stephenson PD, Puumala SE, Koch MR, Aburatani H, Jaggi M, Chauhan SC. Functions and regulation of MUC13 mucin in colon cancer cells. J Gastroenterol. 2014;49:1378–91. doi: 10.1007/s00535-013-0885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chauhan SC, Singh AP, Ruiz F, Johansson SL, Jain M, Smith LM, Moniaux N, Batra SK. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125) Mod Pathol. 2006;19:1386–94. doi: 10.1038/modpathol.3800646. [DOI] [PubMed] [Google Scholar]
- 15.Daelemans W, Hoste V, De Meulder F, Naudts B. Combined Optimization of Feature Selection and Algorithm Parameters in Machine Learning of Language. In: Lavrač N, Gamberger D, Blockeel H, Todorovski L, editors. Machine Learning: ECML 2003: 14th European Conference on Machine Learning; Cavtat-Dubrovnik, Croatia. September 22–26, 2003; Berlin, Heidelberg: Springer Berlin Heidelberg; 2003. pp. 84–95. Proceedings. [Google Scholar]
- 16.Kim K-j, Han I. Genetic algorithms approach to feature discretization in artificial neural networks for the prediction of stock price index. Expert Syst Appl. 2000;19:125–32. [Google Scholar]
- 17.Petricoin EF, III, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. The Lancet. 2002;359:572–7. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 18.Erguzel TT, Ozekes S, Tan O, Gultekin S. Feature Selection and Classification of Electroencephalographic Signals: An Artificial Neural Network and Genetic Algorithm Based Approach. Clin EEG Neurosci. 2015;46:321–6. doi: 10.1177/1550059414523764. [DOI] [PubMed] [Google Scholar]
- 19.Sudhakar M, Albert Mayan J, Srinivasan N. Intelligent Data Prediction System Using Data Mining and Neural Networks. In: Suresh PL, Panigrahi KB, editors. Proceedings of the International Conference on Soft Computing Systems: ICSCS 2015; New Delhi: Springer India; 2016. pp. 489–500. [Google Scholar]
- 20.Pradhan P, Damania D, Joshi HM, Turzhitsky V, Subramanian H, Roy HK, Taflove A, Dravid VP, Backman V. Quantification of nanoscale density fluctuations using electron microscopy: Light-localization properties of biological cells. Appl Phys Lett. 2010;97:243704. doi: 10.1063/1.3524523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradhan P, Damania D, Joshi HM, Turzhitsky V, Subramanian H, Roy HK, Taflove A, Dravid VP, Backman V. Quantification of nanoscale density fluctuations by electron microscopy: probing cellular alterations in early carcinogenesis. Phys Biol. 2011;8:026012. doi: 10.1088/1478-3975/8/2/026012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian H, Pradhan P, Liu Y, Capoglu IR, Li X, Rogers JD, Heifetz A, Kunte D, Roy HK, Taflove A, Backman V. Optical methodology for detecting histologically unapparent nanoscale consequences of genetic alterations in biological cells. PNAS. 2008;105:20118–23. doi: 10.1073/pnas.0804723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian H, Roy HK, Pradhan P, Goldberg MJ, Muldoon J, Brand RE, Sturgis C, Hensing T, Ray D, Bogojevic A, Mohammed J, Chang J-S, et al. Nanoscale Cellular Changes in Field Carcinogenesis Detected by Partial Wave Spectroscopy. Cancer Res. 2009;69:5357–63. doi: 10.1158/0008-5472.CAN-08-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DRC Regression Models and Life Tables. Series B Appl Stat. 1972;34:187–220. [Google Scholar]
- 25.Ahmad F, Mat Isa NA, Hussain Z, Osman MK, Sulaiman SN. A GA-based feature selection and parameter optimization of an ANN in diagnosing breast cancer. Pattern Anal Appl. 2015;18:861–70. [Google Scholar]
- 26.Damania D, Subramanian H, Tiwari AK, Stypula Y, Kunte D, Pradhan P, Roy HK, Backman V. Role of Cytoskeleton in Controlling the Disorder Strength of Cellular Nanoscale Architecture. Biophysical Journal. 2010;99:989–96. doi: 10.1016/j.bpj.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinn BV, von Minckwitz G, Denkert C, Eidtmann H, Darb-Esfahani S, Tesch H, Kronenwett R, Hoffmann G, Belau A, Thommsen C, Holzhausen HJ, Grasshoff ST, et al. Evaluation of Mucin-1 protein and mRNA expression as prognostic and predictive markers after neoadjuvant chemotherapy for breast cancer. Ann Oncol. 2013;24:2316–24. doi: 10.1093/annonc/mdt162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.