Abstract
RAS genes (HRAS, KRAS, and NRAS) comprise the most frequently mutated oncogene family in human cancer. With the highest RAS mutation frequencies seen with the top three causes of cancer deaths in the United States (lung, colorectal, and pancreatic cancer), the development of anti-RAS therapies is a major priority for cancer research. Despite more than three decades of intense effort, no effective RAS inhibitors have yet to reach the cancer patient. With bitter lessons learned from past failures and with new ideas and strategies, there is renewed hope that undruggable RAS may finally be conquered. With the KRAS isoform mutated in 84% of all RAS-mutant cancers, we focus on KRAS. With a near 100% KRAS mutation frequency, pancreatic ductal adenocarcinoma (PDAC) is considered the most RAS-addicted of all cancers. We review the role of KRAS as a driver and therapeutic target in PDAC.
Mutationally activated RAS genes (HRAS, KRAS, and NRAS) comprise the most frequently mutated gene family in cancer (27%; Catalogue of Somatic Mutations in Cancer [COSMIC] v80). KRAS is the predominant isoform mutated in cancer and the isoform mutated exclusively in pancreatic ductal adenocarcinoma (PDAC). With KRAS mutations found in nearly all PDAC, this cancer type is arguably the most RAS-addicted cancer. With substantial experimental evidence that mutant KRAS is essential for PDAC growth, the National Cancer Institute identified targeting KRAS as one of four major priorities for pancreatic cancer research. The current standards of care for PDAC consist of conventional cytotoxic drugs (Wolfgang et al. 2013). Although effective targeted therapies are now available for lung and colorectal cancer, no effective targeted therapies have been found for PDAC. With deaths caused by pancreatic cancer on the rise, the need for new therapies is now dire.
The KRAS small GTPase functions as a simple binary ON–OFF molecular switch, cycling between an active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate (GDP)-bound state (Vigil et al. 2010a). In normal quiescent cells, RAS is predominantly GDP-bound and inactive. Upon extracellular stimuli activation of receptor tyrosine kinases (RTKs) and other cell-surface receptors, there is rapid and transient formation of RAS-GTP, leading to engagement of effector proteins that then regulate a diversity of intracellular signaling networks (Cox and Der 2010) and thereby tightly control mitogenic processes. Cancer-associated RAS genes harbor missense mutations that encode single amino acid substitutions primarily (98%) at one of three mutational hot spots: glycine-12 (G12), glycine-12 (G13), or glutamine-61 (Q61). These mutations render RAS persistently GTP-bound and constitutively active independent of extracellular stimuli, resulting in overstimulation of effector signaling pathways to drive cancer growth. Thus, by analogy to the successful development of clinically effective adenosine triphosphate (ATP)-competitive inhibitors for protein kinases, small molecule GTP antagonists should provide a straightforward strategy to target mutant RAS. However, with picomolar affinity for GTP, and with millimolar GTP cellular concentrations, this approach has not been feasible. Furthermore, when the structure of RAS was determined, it did not reveal a surface topology amenable to the design of high-affinity small-molecule antagonists, deterring efforts to develop direct RAS inhibitors. Consequently, much of the past and current efforts have centered on indirect strategies. However, recent success in the identification of direct RAS-binding small molecules has fueled excitement that perhaps RAS is druggable after all. In this review, we first provide an overview of the role of KRAS in PDAC. We provide a snapshot of past and ongoing efforts and direct and indirect strategies to develop the long elusive anti-RAS drug for cancer treatment. We then focus on the development of inhibitors of KRAS effector signaling.
KRAS MUTATION AND PANCREATIC CANCER TUMORIGENESIS
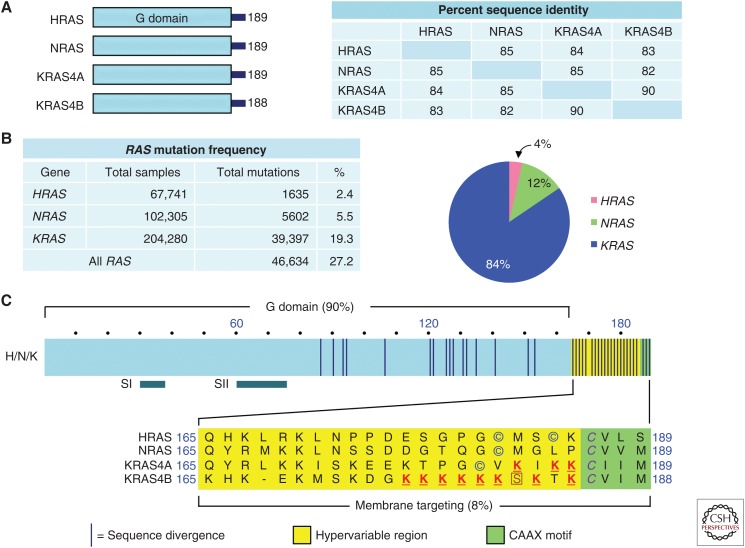
The three RAS genes encode four 188–189 amino acid proteins that share 82%–90% amino acid sequence identity and share near-identical structural and biochemical properties (Fig. 1A) However, they are differentially expressed and mutated with different frequencies in cancer (Prior et al. 2012; Cox et al. 2014). KRAS is the predominant mutated RAS gene in cancers (84% of all RAS missense mutations), followed by NRAS (12%), with HRAS rarely mutated (4%) (COSMIC v80) (Fig. 1B).
Figure 1.
RAS proteins. (A) Sequence identity of human RAS proteins. Amino acid sequence identity was determined by CLUSTALW multiple sequence alignment. (B) RAS mutation frequencies. Data were compiled from Catalogue of Somatic Mutations in Cancer (COSMIC) v80. (C) RAS domains. The amino-terminal amino acids (1–164) comprise the G domain involved in guanosine triphosphate (GTP) binding and hydrolysis and interaction with guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and effectors. RAS protein structure changes in the Switch I (SI; amino acids 30–38) and II (SII; amino acids 60–76) regions during guanosine diphosphate (GDP)–GTP cycling, with the GTP-bound form having higher affinity for effectors. Circled cysteine amino acids indicate sites of covalent modification by addition of a palmitate fatty acid. The boxed serine residue is phosphorylated by protein kinase C. The underlined lysine residues promote membrane targeting. The italicized cysteines in the CAAX motif are sites of covalent modification by addition of a farnesyl isoprenoid and carboxylmethylation.
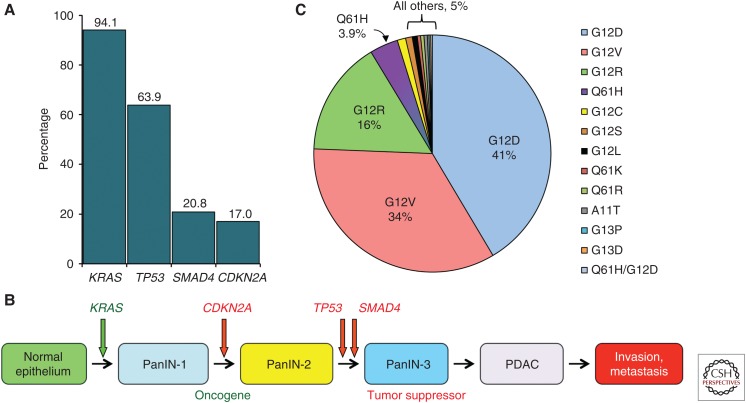
Significant cancer type preferences exist among the RAS genes (Cox et al. 2014). KRAS mutations predominate in lung, colorectal, and pancreatic cancer, whereas NRAS mutations predominate in cutaneous melanomas and acute myelogenous leukemia, and HRAS mutations are found in bladder and head and neck squamous cell carcinomas. Although already known in 1988 (Almoguera et al. 1988), subsequent comprehensive exome-wide deep sequencing verified that KRAS is mutationally activated in 94% of PDAC (Fig. 2A) (Jones et al. 2008; Biankin et al. 2012; Sausen et al. 2015; Waddell et al. 2015; Witkiewicz et al. 2015). These studies also verified an already well-established portrait of PDAC in which there are four major genetic alterations associated with the initiation and progression of PDAC, with the majority of gene alterations found in >10% of PDAC. In addition to KRAS, the TP53 (64%), SMAD4 (21%), and CDKN2A (17%) tumor suppressor genes are significantly mutated (Fig. 2A). Missense mutations and intragenic or homozygous deletion mutations and promoter methylation of CDKN2A (encoding p16INK4A and p19ARF), together with promoter silencing, result in a near universal loss of CDKN2A function in PDAC (Schutte et al. 1997).
Figure 2.
KRAS mutations in pancreatic cancer. (A) Frequency of mutations in the four major genes in pancreatic cancer. (B) Genetic progression of pancreatic cancer. (C) KRAS amino acid substitutions in pancreatic cancer. (Compiled from data in Jones et al. 2008, Biankin et al. 2012, Sausen et al. 2015, Waddell et al. 2015, and Witkiewicz et al. 2015.)
KRAS mutation is the initiating genetic event for PDAC. The progression of normal pancreatic tissue to PDAC involves a stepwise genetic transition projected to span 12 years (Iacobuzio-Donahue et al. 2012). Most commonly, pancreatic duct epithelium transitions to advancing stages of noninvasive microscopic ductal lesions, or pancreatic intraepithelial neoplasms (PanINs). In early-stage PanIN formation, flat pancreatic epithelial cells take on a cuboidal appearance, increase mucin production, and acquire atypical cytological and morphological features (Hruban et al. 2000, 2004; Distler et al. 2014). High-grade PanINs are usually characterized by a papillary morphology (Distler et al. 2014). Greater than 90% frequency of KRAS mutations are found in PanIN-1 lesions (Kanda et al. 2012).
Genetically engineered mouse models also support the initiating role of KRAS mutation in PDAC and additionally support the cooperating role of subsequent tumor suppressor loss in facilitating KRAS-driven progression to the malignant disease (Gopinathan et al. 2015; Lee et al. 2016). KrasG12D mutation alone resulted in PanIN formation and infrequent and protracted onset of PDAC (Aguirre et al. 2003; Hingorani et al. 2003). When coupled with mutational inactivation of Tp53 (Tp53R175H) (Hingorani et al. 2005), Cdkn2a (Aguirre et al. 2003; Bardeesy et al. 2006), or Smad4 (Kojima et al. 2007), PanIN formation was accelerated, progressing to rapid and high-frequency occurrence of metastatic PDAC.
It is currently not clear why PDAC is associated exclusively with KRAS mutations. One explanation for the isoform specificity of RAS-driven cancers could be that certain tissues are exposed to different carcinogens and environmental insults that lead to mutations in specific RAS genes. An alternative and intriguing possibility is that each isoform has specific functional properties that lead to these tissue preferences. One proposed basis for why KRAS is the preferred cancer isoform is protein expression levels. Although nearly identical on the amino acid level, KRAS, NRAS, and HRAS differ in the frequency of rare codon utilization, with the high frequency in KRAS contributing to a reduced rate of translation and lower RAS protein expression (Lampson et al. 2013). Elevated ectopic expression of mutant RAS in normal cells induces senescence rather than cell proliferation. Concurrent suppression of p53 function prevents senescence, allowing RAS-driven growth enhancement. With KRAS mutations, protein expression levels are hypothesized to be sufficiently high for tumor initiation but low enough to evade apoptosis and senescence. In contrast, mutant HRAS protein expression levels are high enough to elicit an apoptotic or senescent response. In support of this hypothesis, in vivo studies that show enriching the KRAS codons with more frequently translated HRAS codons, even though the amino acids are the same, resulted in reduced Kras-induced tumor formation in mice (Pershing et al. 2015).
Another unresolved issue is the role of the remaining KRAS wild-type (WT) allele in KRAS-mutant PDAC. There is evidence that suggests that the WT allele serves a paradoxical role as a tumor suppressor, with loss of the WT allele facilitating PDAC progression (Qiu et al. 2011). When evaluated in a KrasG12D-driven mouse model of PDAC, loss of the WT allele was associated with progression to metastatic disease. Loss of the KRAS WT allele is also seen in human PDAC. In contrast, a driver role for the remaining WT isoforms, HRAS and NRAS, has also been observed in KRAS-mutant pancreatic cell lines (Lim et al. 2008; Grabocka et al. 2014). However, whether their roles are simply quantitative or qualitatively distinct remains to be determined.
In addition to tissue-based isoform specificity, the frequency of different KRAS missense mutations can vary strikingly in different cancer types. For example, G12C mutations are rare in PDAC (1% of all KRAS mutations) but are the major KRAS mutation in lung adenocarcinoma (LAC) (43% of all KRAS mutations) (Fig. 2C) (Cox et al. 2014; Stephen et al. 2014; Hobbs et al. 2016). In contrast, G12R mutations are infrequently seen overall in cancer (<3%), yet comprise 16% of all KRAS mutations in PDAC. It is likely these different frequencies in KRAS mutations reflect, in part, distinct roles in cancer development and growth. Until recently, it was assumed that the different mutations cause essentially identical consequences on RAS function; clearly this is not the case, as it was recently shown that there are altered nucleotide exchange kinetics and effector binding abilities of different mutant KRAS proteins (Smith et al. 2013; Hunter et al. 2015). Thus, a shift away from the long-standing “all RAS mutations are created equal” paradigm has emerged, in which the study of specific RAS mutants is being pursued in search of mutation-selective functional vulnerabilities that can be exploited by different therapeutic strategies. The impact of this shift is evidenced by the mutation-specific inhibitors that have been developed to target G12C-mutant KRAS in LAC (Ostrem et al. 2013; Lim et al. 2014). There is limited evidence that mutant specificity also plays an important role in pancreatic cancer prognosis. One study suggests that KRAS mutations are associated with more aggressive cancers and a shorter survival time (Bournet et al. 2016). Another found that KRAS Q61 mutations were associated with improved survival compared with KRAS G12 mutant patients (Witkiewicz et al. 2015).
PANCREATIC DUCTAL ADENOCARCINOMA CHEMOTHERAPY
In 2016, pancreatic cancer surpassed breast cancer and became the third leading cause of cancer deaths in the United States (Siegel et al. 2016). With a continued increase in incidence, pancreatic cancer is projected to become the second leading cause of cancer death by around 2020 (Rahib et al. 2014). With no biomarkers for early detection, the deadly nature of PDAC is largely the result of late onset of symptoms when the cancer has already reached the metastatic state. The 5-year survival rate of pancreatic cancer is 8% (Siegel et al. 2016), giving this cancer the dubious distinction of being the deadliest cancer in the United States.
Surgery, followed by adjuvant chemotherapy, is the only potentially curative treatment for PDAC, but only 15%–20% of patients are candidates for surgery. Chemotherapy for PDAC involves conventional cytotoxic drug combinations. The only targeted therapy approved for the treatment of PDAC, the epidermal growth factor receptor (EGFR) inhibitor erlotinib, when used in combination with gemcitabine, extended life by a statistically significant but clinically unimpressive 12 days more than gemcitabine alone (Moore et al. 2007). Since 1997, gemcitabine was the standard of care for PDAC. In 2011, the FOLFIRINOX (folinic acid, flurouracil, irinotecan, and oxaliplatin) chemotherapy regimen, an extremely toxic chemotherapy cocktail, was approved for PDAC. In 2013, the nab-paclitaxel (albumin-conjugated paclitaxel)-gemcitabine combination was found to increase life expectancy almost 2 months compared to gemcitabine and approved as a second standard of care for PDAC (Von Hoff et al. 2013). Therapeutic approaches as a whole have been largely unsuccessful in PDAC, with no treatment extending life longer than 1 year from diagnosis.
Although KRAS mutations are the initiating genetic step, continued mutant KRAS function is still required to maintain the growth of metastatic PDAC. RNA suppression studies in PDAC cell lines showed the strong requirement for tumorigenic growth (Brummelkamp et al. 2002; Lim and Counter 2005). Kras inactivation in KrasG12D-driven PDAC, in the continued presence of a Tp53 deficiency, showed rapid regression of primary (Collins et al. 2012a; Ying et al. 2012) and metastatic (Collins et al. 2012b) tumor growth. These studies support the significance of mutant KRAS as a therapeutic target in PDAC. Finally, although one study suggested that only a subset of KRAS-mutant PDAC cell lines were addicted to mutant KRAS (Singh et al. 2009), a subsequent study determined that all KRAS-mutant PDAC cell lines showed significant growth impairment on acute or sustained KRAS suppression (Hayes et al. 2016). The different conclusions from these two studies reflect the specific definition of “addiction” applied in each study. Thus, whereas there will likely exist intertumor variability in the specific mutant KRAS-driven cellular phenotypes, all KRAS-mutant PDAC will likely display KRAS addiction.
Although clearly a validated therapeutic target, KRAS-targeted therapies will also be subject to the limitations seen with targeted and conventional chemotherapy: the development of cancer-cell resistance. One mechanism by which PDAC loses its addiction to KRAS is through activation and expression of the Hippo pathway component YAP1 (Kapoor et al. 2014; Shao et al. 2014).
TARGETING KRAS: DO EFFECTOR INHIBITORS HOLD THE GREATEST PROMISE?
The main past and current strategies for developing therapeutics to block mutant KRAS function have focused on indirect approaches: to target proteins that support KRAS function and promote KRAS-driven cancer growth (Papke and Der 2017). These approaches have centered on targeting (1) proteins that promote KRAS association with the plasma membrane, (2) KRAS effector signaling, (3) components that support KRAS-dependent metabolic processes, and (4) synthetic lethal interactors critical for the growth of mutant but not WT KRAS cancer cells. More recently, RAS-binding small molecules that disrupt KRAS function have been identified, raising hope that RAS can be targeted directly. An ambitious goal of pancreatic cancer research is to double the rate of survival by the year 2020. With this urgency at hand, of these approaches, arguably inhibitors of KRAS effector signaling hold the greatest promise for the most immediate transition to the PDAC patient within the next several years. Therefore, for this review, we have focused on the development of inhibitors of KRAS effector signaling.
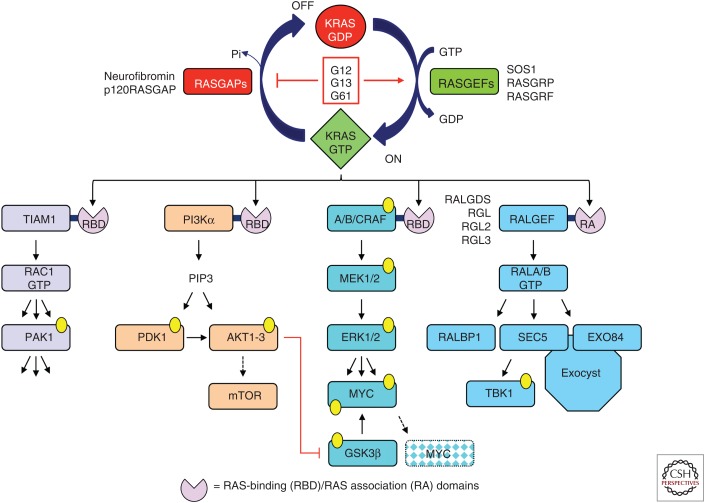
KRAS-GTP binds preferentially to downstream effectors that regulate a complex diversity of cytoplasmic signaling networks. The majority of effectors are characterized by the presence of a RAS-binding domain (RBD) or RAS association (RA) domain (Fig. 3). There are at least 11 different effector families with distinct catalytic functions. It is likely that KRAS drives tumorigenesis by the integrated result of multiple effector signaling pathways, with multiple KRAS effector pathways contributing to the tumor etiology. Thus, the concept of targeting KRAS effector signaling is not straightforward. Which effector pathway(s) are the best to target? Will concurrent inhibition of multiple effectors be required?
Figure 3.
KRAS guanosine diphosphate (GDP)–guanosine triphosphate (GTP) regulation and effector signaling. RAS-selective guanine nucleotide exchange factors (GEFs) and GTP-activating proteins (GAPs) regulate GDP–GTP cycling. Cancer associated mutations, found primarily (>99%) at residues G12, G13, or Q61 disrupt GDP–GTP cycling by impairing intrinsic and GAP-stimulated GTP hydrolysis and/or accelerating GDP–GTP exchange. KRAS effectors are characterized by a RAS-binding domain (RBD) or RAS association (RA) domains. AKT1 phosphorylates and inactivates GSK3β, preventing MYC phosphorylation at T58 and FBW7-mediated polyubiquitination and degradation.
There is substantial experimental evidence from cell culture and mouse model studies that validate driver roles for components of four KRAS effector signaling networks in cancer (Cox et al. 2014). However, some cautious interpretation is important when assessing the therapeutic value of effectors ablated by pharmacologic inhibitors. First, a majority of studies use genetic approaches to ablate effector expression and loss of protein expression, which may not accurately model target inhibition by pharmacologic approaches. Second, genetically engineered mouse model studies typically involve ablation of effector expression concurrently with induced expression of mutant RAS. Hence, they assess the role of effectors in tumor initiation and progression, and not maintenance of an already established metastatic PDAC. With these caveats in mind, we summarize the experimental evidence validating the therapeutic potential of targeting different effector signaling components, with a focus on the RAF effector signaling network.
The RACGEF-RAC1 Small GTPase Effector Signaling Network
The least validated and therapeutically tractable KRAS effector pathway implicated in oncogenesis is the RACGEF-RAC1 pathway (Fig. 3). Early studies in HRAS-transformed rodent fibroblasts provided the first indication of the role of RAC1 in RAS oncogenic functions (Bar-Sagi and Feramisco 1986; Khosravi-Far et al. 1995; Qiu et al. 1995). The subsequent identification of RAC1 missense mutations (P29S), primarily in cutaneous melanomas, provided strong evidence for a driver role for RAC1 activation in cancer (Hodis et al. 2012; Krauthammer et al. 2012). Further, Rac1-deficient mice exhibited reduced KrasG12D-driven pancreatic tumor formation and increased survival (Heid et al. 2011). However, constitutively active Rac1G12V alone was not sufficient to induce PanIN and PDAC formation (Eser et al. 2013).
One RACGEF, TIAM1, has been identified as a RAS effector (Lambert et al. 2002). Like the majority of KRAS effectors, TIAM1 contains an RBD that facilitates association with RAS-GTP. The importance of TIAM1 in RAS-driven tumorigenesis was shown by the impaired onset of Hras-driven skin carcinoma formation in Tiam1-deficient mice (Malliri et al. 2002). Phosphoinositide 3-kinase (PI3K) activation can also activate RACGEFs (e.g., PREX1/2). Although tool compound inhibitors of RACGEF activation of RAC1 have been described (e.g., NSC 23766), it has been difficult to advance clinically effective and selective inhibitors for cancer therapy.
Like KRAS, activated RAC1-GTP can interact with a spectrum of catalytically distinct effectors. Although the effectors critical to facilitate RAC1 as a cancer driver remain unclear, there is experimental support for the PAK1 serine/threonine kinase as an important effector in cancer (Baker et al. 2014). A Pak1 genetic deficiency or treatment with a pharmacologic inhibitor of Pak1 impaired KrasG12D-driven lung tumor formation (Chow et al. 2012). Another effector that will likely contribute to RAC1-dependent cancer growth is the p100β catalytic subunit of PI3K (Fritsch et al. 2013). Inhibitors of PAK1 and PI3Kβ are under preclinical and clinical development.
The RALGEF-RAL Small GTPase Effector Signaling Network
The next-best-validated RAS effector family is a family of four RA domain-containing RALGEFs (RALGDS, RGL, RGL2, and RGL3) (Fig. 3). RALGEFs act as guanine nucleotide exchange factors and activate RALA and RALB RAS-like small GTPases. Mice deficient in Ralgds showed impaired incidence of chemical carcinogen-induced Hras mutation and formation of squamous cell skin carcinomas (Gonzalez-Garcia et al. 2005). Rala/Ralb-deficient mice showed impaired KrasG12D-induced lung tumors (Peschard et al. 2012). Our studies found RGL2 overexpression in PDAC cell lines and patient tumors (Vigil et al. 2010b). Biochemical inhibition of RALGEF function as well as genetic suppression of RGL2 expression reduced PDAC cell line steady-state RAL activity, growth in soft agar, and Matrigel invasion. Interestingly, RGL2 exhibited both RAL-dependent and -independent roles in PDAC growth.
Like RAS, RAL proteins are activated by exchanging GDP in their active site for GTP. Although components of these effector pathways are not found mutated in cancer, sustained hyperactivation of this pathway and elevated levels of RAL-GTP were found more frequently in PDAC than the two more classical KRAS effector pathways, ERK mitogen-activated protein kinase (MAPK) and PI3K (Lim et al. 2006).
Interestingly, despite their strong sequence and biochemical identity, RALA and RALB have distinct roles in PDAC growth. RALA is necessary for PDAC anchorage-independent growth in vitro and tumorigenic growth (Lim et al. 2005). In contrast, RALB is dispensable during tumor initiation but important for PDAC invasion and metastases (Lim et al. 2006).
Active RAL-GTP can interact with a diversity of catalytically distinct effectors, several with demonstrated roles in supporting RAL-dependent cancer growth (Gentry et al. 2014). Of these, the best-characterized effectors are the EXO84 and SEC5 subunits of the octomeric exocyst complex that regulates vesicular trafficking. SEC5 can also act as a RALB effector independent of exocyst function, regulating the activity of the TBK1 serine/threonine kinase. Therefore, it was very intriguing when TBK1 was identified as a synthetic lethal interactor of mutant KRAS (Barbie et al. 2009). However, subsequent studies found that TBK1 function was not tightly linked with RAS-mutant cancer (Muvaffak et al. 2014). Another effector implicated in RALB-dependent PDAC invasion is RALBP1/RLIP76 (Neel et al. 2012), a multidomain protein that can act as a GTP-activating protein (GAP) for the Cdc42 and RAC1 RHO family small GTPases.
The PI3K-AKT-Mechanistic Target of Rapamycin (mTOR) Effector Signaling Network
There is substantial experimental evidence supporting the critical role of the catalytic subunits of class I PI3K (p110α/δ/γ) as critical effectors of mutant KRAS-driven oncogenesis (Fig. 3) (Castellano and Downward 2011). The p110 subunits contain amino-terminal RBDs and RAS-GTP binding promotes PI3K association with the cell membrane and additionally relieves autoinhibition. Mice harboring a Pik3ca locus that encoded a p110α variant with RBD mutations that impaired RAS-GTP binding showed strikingly impaired incidence of mutant KrasG12D-driven lung tumor or mutant Hras-induced skin tumor initiation and maintenance (Gupta et al. 2007; Castellano et al. 2013).
The important driver role for PI3K is supported by frequent activating PIK3CA mutations in cancer (10% all cancers; COSMIC v80). However, two studies reached different conclusions regarding the sufficiency of PI3K activation in driving PDAC development. One study found that the cancer-associated constitutively activated Pik3caH1047R mutant alone could not drive PanIN or PDAC development (Collisson et al. 2012). In contrast, a second study found that this mutant alone phenocopied KrasG12D and induced metastatic PDAC (Eser et al. 2013). A possible basis for these different findings may be the different pancreas cell types targeted.
PI3K phosphorylates phosphotidylinositol-4,5-bisphosphate (PIP2) and stimulates formation of plasma membrane–associated phosphotidylinositol-3,4,5-bisphosphate (PIP3) (Castellano and Downward 2011). PIP3 can then activate a multitude of proteins that include RACGEFs, PDK1, and the AKT1/2/3 serine–threonine kinases. Activated AKT can phosphorylate many other proteins that can then promote cell growth. The PTEN lipid phosphatase and tumor suppressor negatively regulates the pathway by dephosphorylating PIP3 to PIP2 and is an important tumor suppressor in PDAC (Ying et al. 2016) and PTEN loss is found commonly in cancer. However, PIK3CA and PTEN mutations are rare in PDAC. Further, whole-exome deep-sequencing data of pancreatic cancer patients shows that 93% of the rare PIK3CA mutations co-occur with a KRAS mutation, suggesting activated KRAS alone is not sufficient to effectively activate PI3K (Jones et al. 2008; Biankin et al. 2012; Sausen et al. 2015; Waddell et al. 2015; Witkiewicz et al. 2015). Supporting a limited linkage between KRAS and PI3K, KRAS suppression did not alter AKT activation levels in a majority of KRAS-mutant PDAC cell lines (Hayes et al. 2016).
The RAF-MEK-ERK Signaling Network
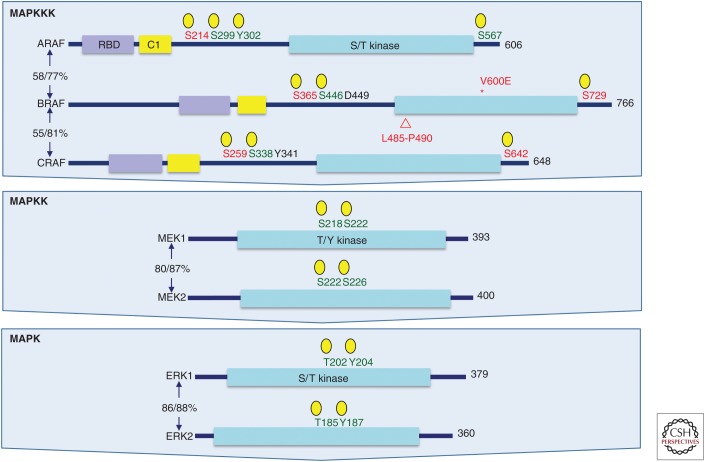
The three RAF serine-threonine kinases (ARAF, BRAF, and CRAF/RAF1) are the most significant effectors of KRAS-driven PDAC (Fig. 4). The importance of the extracellular regulated kinase (ERK) mitogen-activated protein kinase (MAPK) pathway in driving KRAS-dependent cancer growth is supported by the occurrence of BRAF mutations in cancer (18% all cancers; COSMIC v80). Although found infrequently in PDAC (<2%), BRAF-activating V600E mutations were mutually exclusive with KRAS mutations in PDAC (Witkiewicz et al. 2015). The widely studied BxPC-3 PDAC cell line, commonly used as a KRAS WT control in comparison with KRAS-mutant cell lines, harbors an in-frame genomic DNA deletion in BRAF that encodes a constitutively activated and transforming BRAF protein with a five-amino-acid deletion (Chen et al. 2016; Foster et al. 2016). Database analyses found that similar BRAF deletions were found in 4%–5% of KRAS WT PDAC and were mutually exclusive of BRAF missense mutations. Hence, BxPC-3 cells are not bona fide KRAS WT PDAC cells in that a major KRAS effector pathway is chronically activated in this cell line. Unlike BRAFV600E, BRAF deletion mutants were insensitive to the BRAF-selective inhibitor vemurafenib but sensitive to the pan-RAF inhibitor LY3009120.
Figure 4.
Components of the three-tiered extracellular regulated kinase (ERK) mitogen-activated protein kinase (MAPK) cascade. Each level of the ERK MAPK cascade is comprised of highly related isoforms. BRAF missense or deletion mutants (shown is the deletion in the BxPC-1 cell line) are found in pancreatic ductal adenocarcinoma (PDAC). RAF isoforms are regulated by positive (green) and negative (red) regulatory phosphorylation activities. Shown are the phosphorylation sites that activate MEK1/2 or ERK1/2. Domain topology was determined in simple modular architecture research tool (SMART) (smart.embl-heidelberg.de). Total protein and kinase domain sequence identities are indicated (%/%) as determined by CLUSTALW multiple sequence alignment. *, point mutation; ▵, deletion mutation.
Importantly, the predominant cancer-associated constitutive-activated BrafV600E mutant phenocopied KrasG12D and initiated PanIN formation in the pancreas of KRAS WT mice, and, when combined with a Tp53 mutation, resulted in lethal PDAC (Collisson et al. 2012). Moreover, a genetic deficiency in both Mek1 and Mek2 or both Erk1 and Erk2 blocked KrasG12D-driven mouse lung development (Blasco et al. 2011). Interestingly, genetic ablation of Raf1/Craf but not Braf also impaired KrasG12-driven mouse lung tumor development, suggesting that RAF1 is the critical RAF isoform involved in KrasG12D-induced lung tumor formation (Blasco et al. 2011; Karreth et al. 2011). However, surprisingly, a Raf1 deficiency did not impair KrasG12D-induced PDAC formation and progression and did not improve mouse survival, indicating tissue-specific utilization of effectors in KRAS-driven PDAC initiation (Eser et al. 2013). These observations argue that the RAF-MEK-ERK cascade is the key effector in supporting KRAS-dependent tumor initiation, progression, and maintenance.
GTP-bound RAS interacts with the amino-terminal RBD of RAF, increasing accumulation at the plasma membrane and leading to activating phosphorylation events, conformational changes and relief of autoinhibition, and RAF homo-/heterodimerization, promoting RAF kinase activation (Morrison 2012; Freeman et al. 2013). RAF subsequently phosphorylates and activates the MEK1 and MEK2 dual specificity kinases, which then phosphorylate and activate the ERK1 and ERK2 serine/threonine kinases. Activated ERK1/2 then phosphorylate >200 cytoplasmic and nuclear substrates, many of which are transcription factors that regulate the expression of genes involved in mitogenic signaling (McKay and Morrison 2007; Shaul and Seger 2007). Because the only well-established substrates of RAF are MEK1/2, and ERK1/2 are the only well-defined substrates of MEK1/2, the RAF-MEK-ERK protein kinase cascade is commonly depicted as a simple linear, unidirectional signaling pathway. However, with inputs and outputs at each level, with feedback regulatory mechanisms, and with a spectrum of scaffolds that regulate signaling input and output, this three-tier protein kinase cascade is the core of a complex signaling network.
With a diversity of substrates, the key substrates important for ERK-dependent cancer growth remain poorly defined. An important growth-promoting ERK substrate is the transcription factor MYC (Farrell and Sears 2014), an oncoprotein required for maintenance of RAS-driven tumors (Soucek et al. 2008). MYC protein levels are elevated in mouse and human PanIN and tumor tissue (Lin et al. 2013). Myc overexpression alone has been shown to drive PanIN and metastatic PDAC development. MYC protein is normally unstable with a short half-life (∼15 min). ERK stabilizes MYC by phosphorylation at residue S62, preventing FBXW7 E3 ligase-mediated protein degradation driven by GSK3β phosphorylation of MYC at T58 (Fig. 3) (Farrell and Sears 2014), and MYC protein half-life is prolonged in PDAC cell lines (Farrell et al. 2014; Hayes et al. 2016). Sensitivity to pharmacologic inhibitors of ERK was associated with MYC protein loss in growth-inhibited but not resistant PDAC cell lines (Hayes et al. 2016). ERK MAPK-directed regulation of MYC also drives the expression of genes that support the elevated glucose metabolism in Kras-mutant mouse PDAC cells in vivo (Ying et al. 2012).
Excessive ERK activation can drive senescence and growth cessation. To maintain a mitogenic level of ERK activity, the ERK MAPK cascade is also dynamically regulated by negative feedback signaling mechanisms that modulate the level of ERK activity (Ryan et al. 2015). Multiple ERK substrates can negatively regulate upstream signaling components that dampen the signaling strength of the cascade at different levels. For example, ERK phosphorylates CRAF at multiple negative regulatory sites that decrease RAS-driven RAF dimerization (McKay and Morrison 2007).
Substantial cross talk between KRAS effector pathways exists. This is particularly evident with the RAF and PI3K effector signaling networks. Inhibition of ERK signaling can cause compensatory activation of PI3K-AKT-mTORC1 signaling (Collisson et al. 2012). ERK inhibitor treatment stimulated increased AKT activation in sensitive but not resistant PDAC cell lines, and concurrent inhibition of PI3K synergistically enhanced ERK inhibitor growth suppression. Conversely, inhibition of PI3K signaling increased ERK signaling (Soares et al. 2015). The compensatory interrelationship between RAF and PI3K signaling contributes to the synergistic growth suppression seen in Kras-driven cancer models with concurrent pharmacologic inhibition of these two effector pathways. However, largely as a result of toxicity issues, such combinations have not shown the same promising antitumor activities in the clinical evaluation of KRAS-mutant cancers.
THERAPEUTICALLY TARGETING THE RAF EFFECTOR PATHWAYS
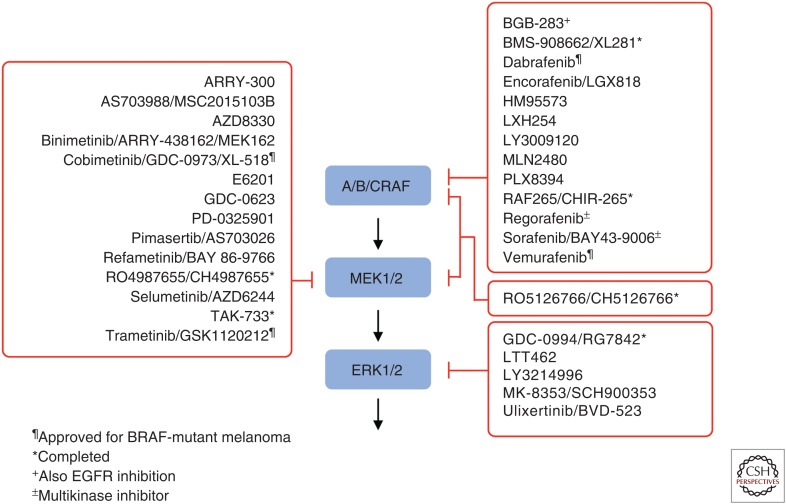
Because the ERK MAPK pathway is hyperactivated through mutations in RAS, BRAF, EGFR, NF1, and other critical cancer drivers, there has been intensive effort by the pharmaceutical industry to develop inhibitors of ERK MAPK signaling. Currently, there are more than 30 inhibitors under clinical evaluation, with two BRAF-selective (vemurafenib and debrafenib) and two MEK1/2-selective inhibitors (trametinib and cobimetinib) approved for use in BRAF-mutant melanoma (Fig. 5). In this section, we summarize the current status of inhibitors of each level of the cascade for the treatment of KRAS-mutant PDAC.
Figure 5.
Clinical candidate inhibitors of the RAF-MEK-extracellular regulated kinase (ERK) mitogen-activated protein kinase (MAPK) cascade. (Compiled from data in www.clinicaltrials.gov.)
RAF Inhibitors
The first-generation RAF inhibitors (vemurafenib and debrafenib) are BRAF-selective and have been effective in a subset of BRAF-mutant cancers, in particular, melanoma. They have not been effective with BRAF-mutant colorectal carcinoma due to up-regulation of EGFR signaling (Corcoran et al. 2012). Although there are cancer-type differences in the sensitivity to BRAF-selective inhibitors, the infrequent BRAFV600F mutant PDACs were responsive in vitro (Witkiewicz et al. 2015). However, PDAC with the atypical BRAF deletion mutants were surprisingly not responsive to vemurafenib (Peng et al. 2015).
In RAS-mutant cells, the BRAF-selective inhibitors cause paradoxical activation of ERK signaling (Hatzivassiliou et al. 2010; Heidorn et al. 2010; Poulikakos et al. 2010). This occurs through a mechanism involving RAS-GTP-mediated RAF dimerization, with the predominant dimer BRAF-CRAF heterodimers. BRAF but not CRAF is inhibited in the dimer, with the inactivated BRAF causing allosteric activation of the associated CRAF protein, leading to MEK-ERK activation. This reactivation of ERK MAPK signaling is associated with increased proliferation in cell culture and tumor formation in patients (Oberholzer et al. 2012; Su et al. 2012). Because KRAS mutation frequencies are found in PanIN-1 lesions (Witkiewicz et al. 2015), an obvious concern is that treatment with BRAF-selective inhibitors may accelerate PDAC development in BRAF-mutant cancer patients.
More recently developed second-generation RAF inhibitors are not limited by paradoxical activation. Pan-RAF inhibitors can overcome this paradoxical activation by blocking WT A-, B-, and CRAF in addition to BRAF V600E. Although dimer formation is induced with pan-RAF inhibitors, MEK activation is abrogated because each dimer molecule is inhibitor-bound and lacks kinase activity (Henry et al. 2015; Peng et al. 2015). The pan-RAF inhibitor LY3009120 was found to be active in RAS-mutant cancer cell lines, including KRAS-mutant PDAC. Several pan-RAF inhibitors are currently in phase I clinical trials. Another second-generation RAF inhibitor is the “paradox breaker” PLX8394 that also effectively inhibits CRAF and BRAF (Basile et al. 2014; Choi et al. 2014; Zhang et al. 2015). PLX8394 effectively blocked ERK activation and growth of RAS-mutant, vemurafenib-resistant melanoma cells.
MEK Inhibitors
MEK1/2-selective inhibitors have not shown potent activity in RAS-mutant cancers. This reduced potency is the result, in part, of vertical compensation mechanisms that lead to reactivation of ERK through up-regulation of upstream pathway components such as RTKs, BRAF, or KRAS (Little et al. 2011; Ryan et al. 2015). An additional basis of resistance involves MEK inhibitor treatment-induced activation of the PI3K-AKT-mTOR pathway. Further, reactivation of phospho-ERK on MEK inhibition with trametinib has been reported owing to compensatory mechanisms that involve kinome reprogramming of parallel pathways (Ryan et al. 2015), but these resistance mechanisms are still poorly understood (Samatar and Poulikakos 2014). Cell-culture analyses found that only a small subset of KRAS-mutant PDAC cell lines are sensitive to MEK inhibitor treatment (Witkiewicz et al. 2015; Hayes et al. 2016). Trametinib did not show efficacy in clinical evaluation with KRAS-mutant PDAC (Infante et al. 2014; Kasuga et al. 2015; Tolcher et al. 2015).
ERK Inhibitors
Because both RAF- and MEK-selective inhibitors are limited, in part, by mechanisms that cause reactivation of ERK downstream from the inhibitor block point, a logical approach to overcome this involve inhibitors that directly target ERK. Analyses with the ERK1/2-selective inhibitors SCH772984 or ulixertinib/BVD-523 found that ∼50% of PDAC cell lines were sensitive (Morris et al. 2013; Ryan et al. 2015; Hayes et al. 2016). ERK inhibitor treatment also blocked tumorigenic growth in PDAC cell lines and PDX tumor mouse models. BVD-523 (NCT02608 229) and LY3214996 (NCT0285 7270) are currently being evaluated in phase I clinical trials in PDAC in combination with nab-paclitaxel and gemcitabine.
Treatment-induced loss of MYC protein was seen in sensitive but not resistant cell lines, suggesting that ERK-independent mechanisms that prevent MYC degradation may be a mechanism of de novo resistance.
Concurrent Inhibition of the RAF and PI3K Effector Signaling Networks
Pharmacological inhibition of the PI3K pathway by using AKT inhibitors with single-agent strategies has produced negative results in vitro and in vivo, and activated PI3K cannot phenocopy mutant KRAS in PDAC mouse models. However, MEK inhibition leads to increased AKT signaling in vivo and combined inhibition of MEK and AKT resulted in robust synergistic inhibition of PDAC growth in vitro and in vivo (Collisson et al. 2012). Similarly, ERK inhibitor treatment also increased AKT activity in sensitive but not resistant PDAC cell lines, and cotreatment with a PI3K inhibitor synergistically enhanced ERK inhibitor-mediated growth suppression and caused apoptotic cell death (Hayes et al. 2016). Concurrent treatment with a PI3K inhibitor did not, however, overcome de novo resistance. Several completed or ongoing clinical trials have evaluated or are evaluating combinations of inhibitors of specific components of the RAF and PI3K effector pathways (www.clinicaltrials.gov).
CONCLUSIONS AND FUTURE PERSPECTIVES
Despite the failures in developing anti-KRAS therapies, targeting KRAS remains one of the most promising directions in pancreatic cancer research. Although multiple directions continue to be pursued, with inhibitors already under clinical evaluation, targeting the key KRAS effector pathway—the RAF-MEK-ERK protein kinase cascade—arguably remains the most promising for short-term clinical success. However, the essential role of this pathway as a driver of KRAS-dependent PDAC growth has ensured that cancer cells have compensatory mechanisms that overcome the effectiveness of inhibitors when used as monotherapy. Instead, combination approaches will be needed to achieve a prolonged antitumor response that is less vulnerable to mechanisms of acquired resistance.
ACKNOWLEDGMENTS
We apologize to all colleagues whose work could not be cited because of space restrictions. Our research is funded by grants from the National Cancer Institute (CA042978, CA175747, CA199235, CA203657, and CA196510), the Department of Defense, the Pancreatic Cancer Action Network/American Association for Cancer Research, and the Lustgarten Foundation. The authors declare no financial interest.
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. 2003. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 17: 3112–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. 1988. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53: 549–554. [DOI] [PubMed] [Google Scholar]
- Baker NM, Yee Chow H, Chernoff J, Der CJ. 2014. Molecular pathways: Targeting RAC-p21-activated serine-threonine kinase signaling in RAS-driven cancers. Clin Cancer Res 20: 4740–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. 2006. Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci 103: 5947–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Sagi D, Feramisco JR. 1986. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 233: 1061–1068. [DOI] [PubMed] [Google Scholar]
- Basile KJ, Le K, Hartsough EJ, Aplin AE. 2014. Inhibition of mutant BRAF splice variant signaling by next-generation, selective RAF inhibitors. Pigment Cell Melanoma Res 27: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM Wu J, et al. 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, Baccarini M, Barbacid M. 2011. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell 19: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournet B, Muscari F, Buscail C, Assenat E, Barthet M, Hammel P, Selves J, Guimbaud R, Cordelier P, Buscail L. 2016. KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol 7: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. 2011. RAS interaction with PI3K: More than just another effector pathway. Genes Cancer 2: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, et al. 2013. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell 24: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Zhang Y, Van Horn RD, Yin T, Buchanan S, Yadav V, Mochalkin I, Wong SS, Yue YG, Huber L, et al. 2016. Oncogenic BRAF deletions that function as homodimers and are sensitive to inhibition by RAF dimer inhibitor LY3009120. Cancer Discov 6: 300–315. [DOI] [PubMed] [Google Scholar]
- Choi J, Landrette SF, Wang T, Evans P, Bacchiocchi A, Bjornson R, Cheng E, Stiegler AL, Gathiaka S, Acevedo O, et al. 2014. Identification of PLX4032-resistance mechanisms and implications for novel RAF inhibitors. Pigment Cell Melanoma Res 27: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow HY, Jubb AM, Koch JN, Jaffer ZM, Stepanova D, Campbell DA, Duron SG, O’Farrell M, Cai KQ, Klein-Szanto AJ, et al. 2012. p21-Activated kinase 1 is required for efficient tumor formation and progression in a Ras-mediated skin cancer model. Cancer Res 72: 5966–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. 2012a. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 122: 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MA, Brisset JC, Zhang Y, Bednar F, Pierre J, Heist KA, Galban CJ, Galban S, di Magliano MP. 2012b. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PloS ONE 7: e49707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP, Rabinovich BA, Hann B, Dankort D, et al. 2012. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov 2: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D, Hung KE, et al. 2012. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ. 2010. Ras history: The saga continues. Small GTPases 1: 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. 2014. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13: 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler M, Aust D, Weitz J, Pilarsky C, Grutzmann R. 2014. Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. Biomed Res Int 2014: 474905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, Hieber M, Arbeiter A, Klein S, Kong B, et al. 2013. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 23: 406–420. [DOI] [PubMed] [Google Scholar]
- Farrell AS, Sears RC. 2014. MYC degradation. Cold Spring Harb Perspect Med 4: a014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AS, Allen-Petersen B, Daniel CJ, Wang X, Wang Z, Rodriguez S, Impey S, Oddo J, Vitek MP, Lopez C, et al. 2014. Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res 12: 924–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Whalen DM, Ozen A, Wongchenko MJ, Yin J, Yen I, Schaefer G, Mayfield JD, Chmielecki J, Stephens PJ, et al. 2016. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29: 477–493. [DOI] [PubMed] [Google Scholar]
- Freeman AK, Ritt DA, Morrison DK. 2013. The importance of Raf dimerization in cell signaling. Small GTPases 4: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. 2013. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell 153: 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LR, Martin TD, Reiner DJ, Der CJ. 2014. Ral small GTPase signaling and oncogenesis: More than just 15 minutes of fame. Biochim Biophys Acta 1843: 2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. 2005. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 7: 219–226. [DOI] [PubMed] [Google Scholar]
- Gopinathan A, Morton JP, Jodrell DI, Sansom OJ. 2015. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis Model Mech 8: 1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabocka E, Pylayeva-Gupta Y, Jones MJ, Lubkov V, Yemanaberhan E, Taylor L, Jeng HH, Bar-Sagi D. 2014. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell 25: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. 2007. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell 129: 957–968. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al. 2010. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464: 431–435. [DOI] [PubMed] [Google Scholar]
- Hayes TK, Neel NF, Hu C, Gautam P, Chenard M, Long B, Aziz M, Kassner M, Bryant KL, Pierobon M, et al. 2016. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29: 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid I, Lubeseder-Martellato C, Sipos B, Mazur PK, Lesina M, Schmid RM, Siveke JT. 2011. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology 141: 719– 730, 730.e711–717. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. 2010. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 140: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JR, Kaufman MD, Peng SB, Ahn YM, Caldwell TM, Vogeti L, Telikepalli H, Lu WP, Hood MM, Rutkoski TJ, et al. 2015. Discovery of 1-(3,3-dimethylbutyl)-3-(2-fluoro-4-methyl-5-(7-methyl-2-(methylamino)pyrido[2,3- d]pyrimidin-6-yl)phenyl)urea (LY3009120) as a pan-RAF inhibitor with minimal paradoxical activation and activity against BRAF or RAS mutant tumor cells. J Med Chem 58: 4165–4179. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4: 437–450. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483. [DOI] [PubMed] [Google Scholar]
- Hobbs GA, Der CJ, Rossman KL. 2016. RAS isoforms and mutations in cancer at a glance. J Cell Sci 129: 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban RH, Goggins M, Parsons J, Kern SE. 2000. Progression model for pancreatic cancer. Clin Cancer Res 6: 2969–2972. [PubMed] [Google Scholar]
- Hruban RH, Takaori K, Klimstra DS, Adsay NV, Albores-Saavedra J, Biankin AV, Biankin SA, Compton C, Fukushima N, Furukawa T, et al. 2004. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am J Surg Pathol 28: 977–987. [DOI] [PubMed] [Google Scholar]
- Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. 2015. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res 13: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, Hruban RH. 2012. Genetic basis of pancreas cancer development and progression: Insights from whole-exome and whole-genome sequencing. Clin Cancer Res 18: 4257–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, Oh DY, Liu Y, Redhu S, Steplewski K, et al. 2014. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer 50: 2072–2081. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, et al. 2012. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142: 730–733.e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. 2014. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. 2011. C-Raf is required for the initiation of lung cancer by K-RasG12D. Cancer Discov 1: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga A, Nakagawa K, Nagashima F, Shimizu T, Naruge D, Nishina S, Kitamura H, Kurata T, Takasu A, Fujisaka Y, et al. 2015. A phase I/Ib study of trametinib (GSK1120212) alone and in combination with gemcitabine in Japanese patients with advanced solid tumors. Invest New Drugs 33: 1058–1067. [DOI] [PubMed] [Google Scholar]
- Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. 1995. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol 15: 6443–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Vickers SM, Adsay NV, Jhala NC, Kim HG, Schoeb TR, Grizzle WE, Klug CA. 2007. Inactivation of Smad4 accelerates KrasG12D-mediated pancreatic neoplasia. Cancer Res 67: 8121–8130. [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 44: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. 2002. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol 4: 621–625. [DOI] [PubMed] [Google Scholar]
- Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM. 2013. Rare codons regulate KRas oncogenesis. Curr Biol 23: 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Komar CA, Bengsch F, Graham K, Beatty GL. 2016. Genetically engineered mouse models of pancreatic cancer: The KPC model (LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre), its variants, and their application in immuno-oncology drug discovery. Curr Protoc Pharmacol 73: 14.39.1–14.39.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Counter CM. 2005. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell 8: 381–392. [DOI] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, Der CJ, Counter CM. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7: 533–545. [DOI] [PubMed] [Google Scholar]
- Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, Counter CM. 2006. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol 16: 2385–2394. [DOI] [PubMed] [Google Scholar]
- Lim KH, Ancrile BB, Kashatus DF, Counter CM. 2008. Tumour maintenance is mediated by eNOS. Nature 452: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SM, Westover KD, Ficarro SB, Harrison RA, Choi HG, Pacold ME, Carrasco M, Hunter J, Kim ND, Xie T, et al. 2014. Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl 53: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Rajbhandari N, Liu C, Sakamoto K, Zhang Q, Triplett AA, Batra SK, Opavsky R, Felsher DW, DiMaio DJ, et al. 2013. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res 73: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, Edwards PA, Smith PD, Cook SJ. 2011. A correction to the research article titled: “Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells” by Little A. S., Balmanno K., Sale M. J., Newman S., Dry J. R., Hampson M., Edwards P. A. W., Smith P. D., Cook S. J.. Sci Signal 4: er2. [DOI] [PubMed] [Google Scholar]
- Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. 2002. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature 417: 867–871. [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK. 2007. Integrating signals from RTKs to ERK/MAPK. Oncogene 26: 3113–3121. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, et al. 2007. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol 25: 1960–1966. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, et al. 2013. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 3: 742–750. [DOI] [PubMed] [Google Scholar]
- Morrison DK. 2012. MAP kinase pathways. Cold Spring Harb Perspect Biol 4: a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muvaffak A, Pan Q, Yan H, Fernandez R, Lim J, Dolinski B, Nguyen TT, Strack P, Wu S, Chung R, et al. 2014. Evaluating TBK1 as a therapeutic target in cancers with activated IRF3. Mol Cancer Res 12: 1055–1066. [DOI] [PubMed] [Google Scholar]
- Neel NF, Rossman KL, Martin TD, Hayes TK, Yeh JJ, Der CJ. 2012. The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol Cell Biol 32: 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, Roden C, Chalk CJ, Ardlie K, Palescandolo E, et al. 2012. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol 30: 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. 2013. K-RasG12C inhibitors allosterically control GTP affinity and effector interactions. Nature 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke B, Der CJ. 2017. Drugging RAS: Know the enemy. Science 355: 1158–1163. [DOI] [PubMed] [Google Scholar]
- Peng SB, Henry JR, Kaufman MD, Lu WP, Smith BD, Vogeti S, Rutkoski TJ, Wise S, Chun L, Zhang Y, et al. 2015. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 28: 384–398. [DOI] [PubMed] [Google Scholar]
- Pershing NL, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM. 2015. Rare codons capacitate Kras-driven de novo tumorigenesis. J Clin Invest 125: 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P, McCarthy A, Leblanc-Dominguez V, Yeo M, Guichard S, Stamp G, Marshall CJ. 2012. Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol 22: 2063–2068. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. 2010. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. 2012. A comprehensive survey of Ras mutations in cancer. Cancer Res 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu RG, Chen J, Kirn D, McCormick F, Symons M. 1995. An essential role for Rac in Ras transformation. Nature 374: 457–459. [DOI] [PubMed] [Google Scholar]
- Qiu W, Sahin F, Iacobuzio-Donahue CA, Garcia-Carracedo D, Wang WM, Kuo CY, Chen D, Arking DE, Lowy AM, Hruban RH, et al. 2011. Disruption of p16 and activation of Kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget 2: 862–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. 2014. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
- Ryan MB, Der CJ, Wang-Gillam A, Cox AD. 2015. Targeting RAS-mutant cancers: Is ERK the key? Trends Cancer 1: 183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatar AA, Poulikakos PI. 2014. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug Discov 13: 928–942. [DOI] [PubMed] [Google Scholar]
- Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay Li Q, et al. 2015. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 6: 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, et al. 1997. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res 57: 3126–3130. [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. 2014. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Seger R. 2007. The MEK/ERK cascade: From signaling specificity to diverse functions. Biochim Biophys Acta 1773: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2016. Cancer statistics, 2016. CA Cancer J Clin 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Neel BG, Ikura M. 2013. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci 110: 4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares HP, Ming M, Mellon M, Young SH, Han L, Sinnet-Smith J, Rozengurt E. 2015. Dual PI3K/mTOR inhibitors induce rapid overactivation of the MEK/ERK pathway in human pancreatic cancer cells through suppression of mTORC2. Mol Cancer Ther 14: 1014–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. 2008. Modelling Myc inhibition as a cancer therapy. Nature 455: 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. 2014. Dragging ras back in the ring. Cancer Cell 25: 272–281. [DOI] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. 2012. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 366: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolcher AW, Bendell JC, Papadopoulos KP, Burris HA III, Patnaik A, Jones SF, Rasco D, Cox DS, Durante M, Bellew KM, et al. 2015. A phase IB trial of the oral MEK inhibitor trametinib (GSK1120212) in combination with everolimus in patients with advanced solid tumors. Ann Oncol 26: 58–64. [DOI] [PubMed] [Google Scholar]
- Vigil D, Cherfils J, Rossman KL, Der CJ. 2010a. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer 10: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigil D, Martin TD, Williams F, Yeh JJ, Campbell SL, Der CJ. 2010b. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem 285: 34729–34740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, et al. 2013. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med 369: 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, et al. 2015. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 6: 6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. 2013. Recent progress in pancreatic cancer. CA Cancer J Clin 63: 318–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Dey P, Yao W, Kimmelman AC, Draetta GF, Maitra A, DePinho RA. 2016. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 30: 355–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Spevak W, Zhang Y, Burton EA, Ma Y, Habets G, Zhang J, Lin J, Ewing T, Matusow B, et al. 2015. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 526: 583–586. [DOI] [PubMed] [Google Scholar]