Abstract
Introduction
Cystic fibrosis (CF) is a multi-organ disorder characterized by chronic sino-pulmonary infections and inflammation. Many patients with CF suffer from repeated pulmonary exacerbations that are predictors of worsened long-term morbidity and mortality. There are no reliable markers that associate with the onset or progression of an exacerbation or pulmonary deterioration. Previously, we found that the Mirc1/Mir17–92a cluster which is comprised of 6 microRNAs (Mirs) is highly expressed in CF mice and negatively regulates autophagy which in turn improves CF transmembrane conductance regulator (CFTR) function. Therefore, here we sought to examine the expression of individual Mirs within the Mirc1/Mir17–92 cluster in human cells and biological fluids and determine their role as biomarkers of pulmonary exacerbations and response to treatment.
Methods
Mirc1/Mir17–92 cluster expression was measured in human CF and non-CF plasma, blood-derived neutrophils, and sputum samples. Values were correlated with pulmonary function, exacerbations and use of CFTR modulators.
Results
Mirc1/Mir17–92 cluster expression was not significantly elevated in CF neutrophils nor plasma when compared to the non-CF cohort. Cluster expression in CF sputum was significantly higher than its expression in plasma. Elevated CF sputum Mirc1/Mir17–92 cluster expression positively correlated with pulmonary exacerbations and negatively correlated with lung function. Patients with CF undergoing treatment with the CFTR modulator Ivacaftor/Lumacaftor did not demonstrate significant change in the expression Mirc1/Mir17–92 cluster after six months of treatment.
Conclusions
Mirc1/Mir17–92 cluster expression is a promising biomarker of respiratory status in patients with CF including pulmonary exacerbation. Published by Elsevier B.V. on behalf of European Cystic Fibrosis Society.
Keywords: Cystic fibrosis, MicroRNA, Mir17–92a, Biomarker, Correlation, Pulmonary exacerbation
1. Introduction
Cystic fibrosis (CF) is a multi-organ disease caused by mutations of the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel. Patients with CF characteristically suffer from repeated episodes of acute worsening in respiratory symptoms and decline in lung function termed a pulmonary exacerbation (PEx) [1]. Many definitions of PEx have been proposed [2], but regardless of the designation used, these clinical events are associated with progressive long-term deterioration of lung function and heightened morbidity and mortality [3,4]. While a number of biomarkers based on inflammatory markers in CF have been suggested, relevant biomarkers associated with exacerbation or decline in lung function have not been developed for clinical use. Current efforts are attempting to predict and identify PEx earlier to be able to more promptly provide treatment with antimicrobials and anti-inflammatory agents [5]. Yet, there are no reliable diagnostic or prognostic risk-related markers associated with PEx. Therefore, identifying biomarkers that can reflect the existence of an exacerbation at an early stage would provide an invaluable opportunity for early detect PEx, to avoid further physiologic and immunologic injury, and prevent the need for costly medical care, such as hospitalization.
Biomarkers are anatomical, physiological, biochemical or imaging features that can be used for diagnosis, monitoring disease progression, or response to treatment. An ideal biomarker should be stable and easy to measure, cost efficient and consistent across gender and ethnic groups. To date, none of the available biomarkers in CF satisfy all of these criteria. The major limitations of markers are low specificity, sensitivity, and false positive results.
MicroRNAs (Mirs) are endogenous, evolutionarily conserved small non-coding RNA that have been shown to be effective tools to study the biology of diseases and to have great potential as novel diagnostic and prognostic biomarkers with high specificity and sensitivity [6]. Circulating Mirs are short non-coding RNAs involved in biological and pathological processes of every cell type. Mirs have many necessary features of ideal biomarkers. Mirs are stable in various biological fluids, such as plasma, serum, saliva, milk, cerebrospinal fluid. Particularly, the expression of serum Mirs is firmly linked to various diseases. Mirs are considered potential biomarkers for several chronic disorders due to their stability in the circulation, and are both disease-and tissue-specific, which makes them attractive circulatory biomarkers [7,8]. In addition, the quantity of Mirs can be easily estimated by various methods, such as qRT-PCR, microarray, hybridization and deep-sequencing. qRT-PCR (quantitative real time polymerase chain reaction) is the most common, reliable and available, inexpensive method used for quantifying the small amount of miRNAs with the highest sensitivity and specificity [9,10].
Mirs act primarily through degradation of target mRNA with subsequent decrease or loss of expression of encoded proteins. The Mir17–92 family maps to human chromosome 13 and encodes for the Mirc1/Mir17–92 cluster (Mir17, Mir18a, Mir19a, Mir20a, Mir19b-1, Mir92a) and two paralogs (Mir106a, Mir106b) [11]. We have recently found that the expression of Mir17 and Mir20a within the Mirc1/Mir17–92 cluster is elevated in CF macrophages from F508del mice and humans. These two Mirs target several essential autophagy proteins such as Atg5, Atg12, Atg7 and Atg16 [12,13]. We have confirmed that the upregulation of Mir17 and Mir20a is associated with down-regulation of their predicted autophagy targets contributing to weak autophagy activity in murine macrophages. Restoring the level of Mir17 and Mir20a to normal levels in vivo by intra-tracheal administration of specific antagomirs to live CF mice improves autophagy, controls infection, and reduces pneumonia [13]. Notably, in vitro, Mir17 and Mir20a antagomirs ameliorate CFTR function via the activation of autophagy [13]. Our published data suggest that targeting Mir17 and Mir20a improves several clinical symptoms in patients with CF. However, it is not clear if these Mirs are elevated in human samples and if their level correlates with clinical symptoms and change in response to treatment. Hence, in the current study, we determined that the Mirc1/Mir17–92 cluster is expressed broadly in human cells including neutrophils and in biological fluids including plasma and sputum. In addition, the most striking finding is that Mir cluster expression in sputum positively correlated with clinical symptoms such as PEx.
2. Methods
2.1. Ethics statement
All human subjects were recruited as approved by the Institutional Review Board (IRB) of Nationwide Children’s Hospital (IRB15-00611 and IRB12-00405). All subjects underwent written consent for the procedures including all adult subjects provided informed consent, and a parent or guardian of any child participant provided informed consent on their behalf along with written assent from children.
2.2. Subjects
Male and female children older than 12 years and adult patients with CF were recruited from the CF clinic in either a state of baseline health or during the onset of a PEx. The diagnosis of CF was confirmed by the presence of two CF causing mutations on genetic blood testing and/or an elevated sweat chloride test. Data including patient demographics, medications, hospitalizations for PEx, and relevant clinical factors were collected in a database upon recruitment. Age and gender-matched healthy controls were recruited through Clinical Research Services. The demographics of subjects enrolled in this study are listed in Table 1. The control population was older and had more females, but subjects undergoing Ivacaftor/Lumacaftor treatment were not matched to healthy controls, and tended to be younger skewing the overall cohort. The CF subjects had moderate lung disease as shown by forced expiratory volume in 1 s (FEV1)% predicted based on American Thoracic Society criteria, [14] and the majority were pancreatic insufficient. The majority of the CF cohort had at least one copy of the F508del mutation.
Table 1.
Cohort demographics.
| CF | Controls | |
|---|---|---|
| n= | 76 | 49 |
| Females (%) | 40.9 | 66.7 |
| Age (years, st dev) | 26.6 ± 8.9 | 33.3 ± 10.3 |
| Caucasian (%) | 100 | 100 |
| Pancreatic insufficiency (%) | 93.0 | 0 |
| Genotype (%) | ||
| F508del homozygous | 80.9 | N/A |
| F508del heterozygous | 15.5 | N/A |
| FEV1 (% predicted, st dev) | 60.9 ± 22.2 | N/A |
| BMI (st dev) | 21.7 ± 3.7 | N/A |
2.3. Study parameters
Subject outcomes were stratified based upon Mirc1/Mir17–92 cluster expression. The occurrence of a PEx was the primary outcome measure which was defined by clinicians, and verified according to a previously published definition [15]. The outcome of lung function was measured FEV1 and forced vital capacity (FVC) on pulmonary function testing during routine clinical visits. Percent predicted measurements and z scores for FEV1 and FVC were derived from reference equations [16,17]. Nutritional status was determined by body mass index (BMI), which was calculated based on clinical measurements made by pulmonary dieticians during routine clinical visits.
2.4. Sample collection
Whole blood samples were obtained and plasma was isolated via centrifugation and frozen at −80 °C. Human neutrophils were then isolated and purified by negative selection. Briefly, blood was transferred to 50 ml conical tube and 50 μl of an antibody cocktail added per ml of blood (Stemcell Technologies, Vancouver, BC, Canada), plus 50 μl of magnetic beads per ml of blood. The sample was incubated 5 min at room temperature, then PBS-EDTA was added up to 50 ml, the tube was placed in the magnet for 10 min, the supernatant was transferred to a new tube and magnetic beads were added at the same amount of the previous step. The sample was placed 5 min in the magnet and the supernatant was collected. The cells were centrifuged at 600 ×g, and re-suspended in 1 ml of HBSS plus 1% of FBS before further experimentation. Sputum samples were obtained by spontaneous expectoration into sterile containers, aliquoted in Trizol, and frozen at −80 °C.
2.5. Quantitative real-time PCR (qRT-PCR) for expression of Mirs
Total RNA was isolated from neutrophils lysed in Trizol (Invitrogen Life Technologies, 15596-026) via chloroform (Fisher Scientific, 268320010), isopropanol (Fisher Scientific, BP2618-212), and glycogen (Fisher Scientific, 10814010). Total RNA from plasma and sputum samples was isolated using the miRNeasy Mini Kit (Qiagen, 217004). Expression of mature Mir17, Mir18a, Mir19a, Mir19b, Mir20a, Mir92a, Mir101, and SNORD48/RNU48 (as an endogenous control for human neutrophils) or cel-mir-39, cel-mir-54, and cel-mir238 (Thermo Fisher Scientific, 000200, 001361, and 000248, spike-in controls for plasma and sputum samples) were analyzed by first converting the RNA to cDNA using specific primers (Applied Biosystems, Assay ID 2308, 2422, 395, 396, 580, 431, 002253, 001232, respectively) with the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, 4366596). PCR was conducted according to the manufacturer’s guidelines. For qRT-PCR, cDNA was primed with specific TaqMan primers listed above and assayed using TaqMan Universal PCR MasterMix (Applied Biosystems, 4304437) and Applied Biosystems ABI 7900HT real-time PCR system. Expression was calculated as relative copy numbers. Ct values of each Mir were subtracted from the average Ct of the internal (SNORD48/RNU48) or spike-in (cel-mir-39, -54, and -238) control, and the resulting ΔCt was used in the equation: relative copy numbers = (2−ΔΔCt) [13,18].
2.6. Statistical analysis
All statistical analyses were performed using GraphPad Prism software (version 7.0) and R 3.4.1. Clinical outcomes included hospitalization for PEx, number of antibiotic courses, BMI, FEV1% predicted, and FVC% predicted. Mir expression was correlated with each clinical outcome using Spearman’s rank correlation coefficient (rho). Mann-Whitney U tests were used for comparisons of independent samples. Wilcoxon signed-rank tests were used for within patient comparisons of the pre-and post-Ivacaftor/Lumacaftor group. Statistical significance was defined as an unadjusted p value <0.05. Age and gender matched healthy controls were used as able in the analysis. Analysis was performed on negative dCT measurements using the average of Mir238, Mir54, and Mir39 as control measurements.
3. Results
3.1. The expression of Mirc1/Mir17–92 cluster is not significantly elevated in neutrophils derived from CF patients
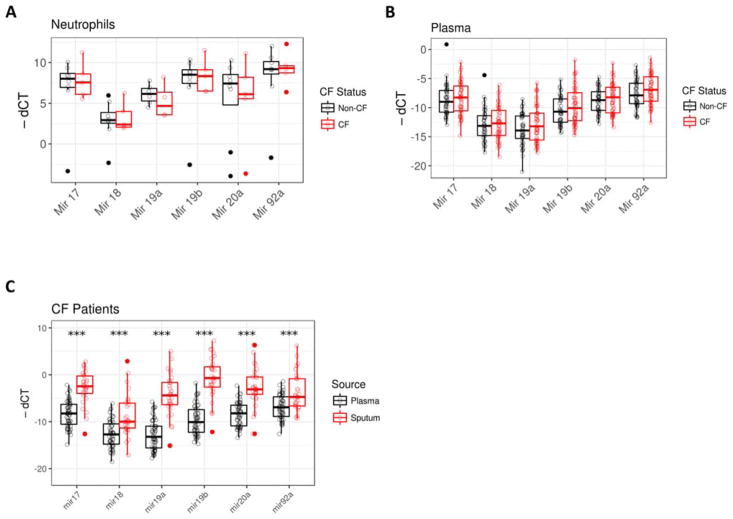
We have recently demonstrated that the Mirc1/Mir17–92 cluster is elevated in CF mouse model and in human CF macrophages [13]. To determine if this finding was specific to macrophages or more broadly expressed, we examined Mirc1/Mir17–92 cluster expression in neutrophils isolated from blood of 5 patients with CF and 8 age-matched, healthy controls. There was no significant difference in cluster expression between CF and non-CF neutrophils (Fig. 1A). Together, our published and recent findings herein demonstrate that the expression of specific Mirs within the Mirc1/Mir17–92 is significantly elevated in macrophages but not neutrophils from CF patients.
Fig. 1.
CF patients have elevated Mirc1/Mir17–92 cluster expression. A) Human neutrophils were isolated from peripheral blood samples from healthy controls and CF patients with at least one F508del mutation and a second class I or class II CFTR mutation. Mirc1/Mir17–92 cluster expression was determined by qRT-PCR, n = 8 for the control group and n = 5 for the CF group, significance determined with Mann-Whitney U tests. Mir17 p value 0.94, Mir18a p value 1, Mir 19a p value 0.47, Mir19b p value 1, Mir20a p value 0.83, and Mir92 p value 0.94. B) Human plasma was isolated from peripheral blood samples from CF patients and healthy controls. Mirc1/Mir17–92 cluster expression was determined by qRT-PCR, n = 55 for the CF group and n = 49 for the non-CF group. Mir17 p value 0.47, Mir18a p value 0.63, Mir 19a p value 0.22, Mir19b p value 0.22, Mir20a p value 0.43, and Mir92a p value 0.14. C) Sputum was obtained from 29 CF patients. Mirc1/Mir17–92 cluster expression was determined by qRT-PCR and expression levels compared to plasma levels from 1B. Mir17 p value < 0.0001, Mir18a p value < 0.0001, Mir 19a p value < 0.0001, Mir19b p value < 0.0001, Mir20a p value < 0.0001, and Mir92 p value < 0.0001. “*” = p value < 0.05, “**” = p value < 0.01, “***” = p value < 0.001.
3.2. CF patients have elevated Mirc1/Mir17–92 cluster expression in sputum and not plasma
Innate immune cells, such as macrophages from CF patients express high levels of Mirc1/Mir17–92 cluster and are associated with weak autophagy activity which is essential for CFTR regulation, offering a potential new biomarker for autophagy-related functions in immune cells. Although, the results with macrophages offer a reliable sample for testing the Mirc1/Mir17–92 cluster as a biomarker, the routine derivation of immune cells from blood samples can be laborious in a clinical setting. Therefore, we next determined the expression of the Mirc1/Mir17–92 cluster in readily available samples such as plasma and sputum. We measured the cluster expression in frozen plasma samples to determine if the Mirc1/Mir17–92 cluster could be detected in the absence of circulating phagocytes. Mirc1/Mir17–92 cluster expression was detected in low levels in both CF and non-CF plasma samples (Fig. 1B) with significant difference.
Given that sputum samples are non-invasively collected from CF patients on regular basis and yield considerable amounts of biological material, we determined if Mirc1/Mir17–92 cluster expression was detectable in 28 CF sputum samples. Control sputum from non-CF individuals was not available for comparison. All six cluster members were detectable in CF sputum (Fig. 1C). There was significantly higher expression of all cluster members in CF sputum compared to CF plasma (Fig. 1C). Therefore, although plasma samples do not demonstrate substantial differences between CF and non-CF individuals, sputum samples exhibit high levels of Mirc1/Mir17–92 cluster that may reflect disease prognosis.
3.3. Sputum Mirc1/Mir17–92 cluster expression correlates with lung function
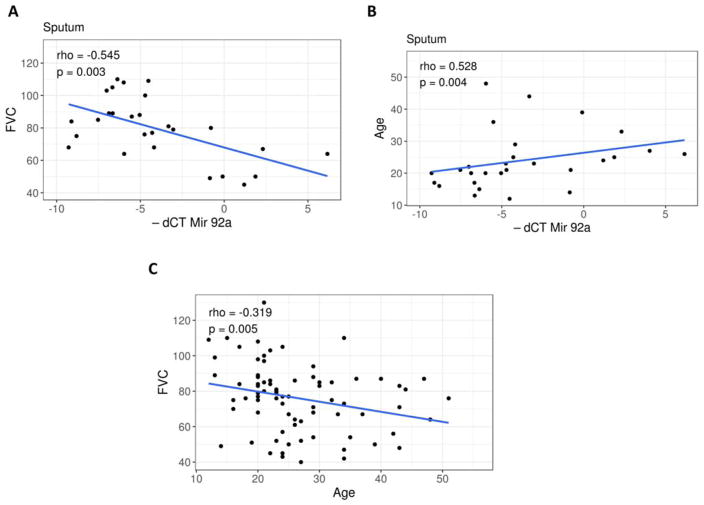
In order to determine if Mirc1/Mir17–92 cluster expression in sputum samples correlated with clinical outcomes, both plasma and sputum expression levels of Mirc1/Mir17–92 cluster were analyzed for their correlation with index lung function (FEV1, FVC), BMI, and age. Sputum concentrations for all six cluster members correlated with FVC (rho range −0.389–−0.545, Mir 92a) (Fig. 2A), FEV1 (rho range −0.392–−0.503), and age (rho range 0.44–0.528, Mir 92a) shown in Fig. 2B. Notably, there was a greater correlation of cluster expression with age, compared to FVC and age (Fig. 2C, rho = −0.319, p value 0.005).
Fig. 2.
Sputum Mirc1/Mir17–92 cluster expression correlates with lung function. A) Correlation plot for sputum Mir92a expression and forced vital capacity (FVC) from 27 patients with CF. Spearman’s rank correlation coefficient (rho) = −0.545, p value = 0.003. B) Correlation plot for sputum Mir92a expression and age from 28 patients with CF. Spearman’s rank correlation coefficient (rho) = 0.528, p value = 0.004. C) Correlation plot for FVC and age from 77 patients with CF. Pearson correlation coefficient (r) = −0.319, p value = 0.005.
3.4. Mirc1/Mir17–92 cluster expression correlates with the presence of pulmonary exacerbations
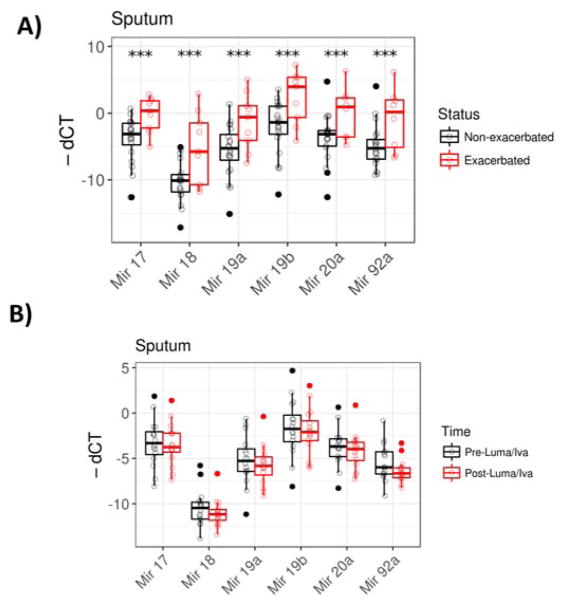
There are no reliable existing biomarkers in clinical use to detect early stages of PEx in patients with CF. PEx are associated with acute morbidity and rapid lung function decline, and hence, it is important to detect PEX at early stages to better prevent the deterioration of lung functions. Due to the wide spread of cluster expression in sputum observed (Fig. 1C), we determined if Mirc1/Mir17–92 cluster expression in sputum correlated with PEx status. Comparing exacerbated and non-exacerbated subjects, we found that all six members of the Mirc1/Mir17–92 cluster expression were significantly elevated in subjects at the start of a PEx compared to non-exacerbated subjects (Fig. 3A). The effect of exacerbation was still significant for all six members after controlling for sex and age using ANCOVA. These results indicate that the Mirc1/Mir17–92 cluster expression may be useful in a model for detecting PEx.
Fig. 3.
Sputum Mirc1/Mir17–92 cluster expression correlates with pulmonary exacerbations. A) Sputum samples cluster expression levels were grouped according to the presence or absence of a pulmonary exacerbation in CF patients. Mir17 p value = 0.006, Mir18a p value = 0.034, Mir 19a p value = 0.023, Mir19b p value = 0.013, Mir20a p value = 0.043, and Mir92a p value = 0.049. B) Sputum from CF patients at baseline and 6 months-post-Lumacaftor/Ivacaftor initiation were obtained. Mirc1/Mir17–92 cluster expression was determined by qRT-PCR, n = 16 for each group. Expression levels were compared for each subject pre- and post-drug initiation. Mir17 p value = 0.59, Mir18a p value = 0.23, Mir 19a p value = 0.63, Mir19b p value = 0.63, Mir20a p value =0.37, and Mir92 p value = 0.05. *” = p value < 0.05, “**” = p value < 0.01, “***” = p value < 0.001.
3.5. Lumacaftor/Ivacaftor do not alter Mirc1/Mir17–92 cluster expression
Lumacaftor/Ivacaftor is a combination of CFTR modulators that was approved in July 2015 by the United States Food and Drug Administration for CF patients that are homozygous for the F508del mutation. Lumacaftor/Ivacaftor demonstrated modest increases in patient lung function and decreases in PEx in clinical trials [19]. Opinions regarding the clinical utility of this new drug combination remain guarded [20] and there is few existing data regarding its efficacy and immunologic effects in CF patients. We measured Mirc1/Mir17–92 cluster expression in sputum from 17 subjects before and 6 months after Lumacaftor/Ivacaftor initiation to determine if treatment initiation impacted cluster expression (Fig. 3B). There was an overall wide spread of data points for the other individual Mirs pre- and post-Lumacaftor/Ivacaftor initiation but there was no significant difference between pre-and post-treatment. Outliers were noted in subjects who were exacerbated at drug initiation, similar to sputum levels previously presented. Therefore, in congruence with modest clinical improvement observed in clinical trials, Lumacaftor/Ivacaftor treatment did not significantly affect the expression of the Mirc1/Mir17–92 cluster.
4. Discussion
There is a clear need for reliable, non-invasive and feasible methods to predict and detect PEx in CF patients beyond the limitations of the current practice of monitoring acute deterioration in clinical symptoms and pulmonary function, especially where timely intervention is critical for improved patient outcomes and to identify those at risk for accelerated deterioration in lung function. Finding biomarkers that are representative of overall prognosis and response to treatment will greatly improve the delivery of personalized care for patients with CF. Importantly; in this study we demonstrate that elevated expression of Mirc1/Mir17–92 cluster in sputum of CF patients is associated with PEx and increases with age.
Our understanding of Mirs and their role in different pathologies is increasing rapidly. During the past two decades, 2588 mature Mirs have been described in humans. The roles of Mirs have been described in several diseases, including cancers, [21–23] coronary diseases, [24,25] autoimmune diseases, [26–28] and viral infections [29,30]. Although several studies have emerged describing Mir dysregulation in CF (reviewed in [31]), Mir involvement in CF needs further investigation and elucidation. Several studies involving Mirs in CF have focused on their impact on CFTR expression. For instance, elevated Mir494 represses CFTR expression, [32,33] whereas that of Mir138 increases its level [34]. Other studies have assessed Mir-mediated regulation of inflammation in CF patients [35]. For instance Mir126 is highly expressed in the lung yet downregulated in CF bronchial epithelial cells which correlates with a significant upregulation of TOM1 mRNA [36] which modulates inflammatory response [37]. Mir17 overexpression in CF airway epithelial cells decreases interleukin-8 production [38]. Few studies reported the alteration of expression of Mirs affecting immune function and inflammation in CF [35,39].
Although Mirs predominantly reside intracellularly, they can stably exist in extracellular environments such as in serum, plasma, semen, cerebrospinal fluid and urine. This robust stability, together with demonstrated organ- and disease-specific expression, enables Mirs to act as potential non-invasive biomarkers for the detection and diagnosis of disease conditions. As revealed from the current study, the Mirc1/Mir17–92 cluster was expressed widely in both circulating CF cells and systemic and airway biologic samples, indicating its robustness as a marker of both systemic and local lung disease. As sputum may not be readily available in young or non-expectorating subjects, the optimal specimen for Mirc1/Mir17–92 cluster expression in CF patients where sputum is unable to be induced remains to be determined. Additionally, Mirc1/Mir17–92 cluster expression is minimally impacted by initiation of treatment with the CFTR modulator Ivacaftor/Lumacaftor. This finding corroborates with the lack of clinical improvement of this drug combination in several CF patients. Therefore, further studies are needed to determine if Mirc1/Mir17–92 cluster expression represents a prognostic marker of biologic response to current and newly developed CFTR modulators that offer significant improvement of clinical symptoms in the CF population. We believe this is highly likely as we have shown that therapeutic down-regulation of the Mirc1/Mir17–92 cluster improves CFTR function [13]. This effect is mediated by improvement of autophagy activity when the expression of the cluster is corrected [13]. Therefore, the level of expression of the Mirc1/Mir17–92 cluster may reciprocally correlate with the response to effective CFTR corrector therapies.
This is not the first report of differential expression of members of the Mirc1/Mir17–92 cluster. Another study demonstrated the diverse mature levels of individual Mirc1/Mir17–92 cluster members in different cell lines [40]. Together, our data and that of others suggest the existence of the molecular mechanisms responsible for differential maturation of individual Mir cluster members within one cluster [18,41]. In fact, the expression and function of Mirs are regulated at three levels: transcription, processing, and subcellular localization [42]. At the level of transcription, miRNA expression is controlled by many factors including chromatin modifications, DNA methylation, and activity of transcription factors [43]. Our data and that of others also suggest the presence of an alternative transcript to account for variable expression levels of individual miRNAs [44]. Additionally, Mir-related single nucleotide polymorphisms (SNPs) [45], including SNPs in Mir genes and target sites, function as regulatory SNPs to affect the phenotypes and disease susceptibility [46]. SNPs that affect miRNA binding and function are being increasingly reported [47].
Notably, several studies demonstrated that CF patients have an increased risk of cancer especially in the digestive tract [48,49]. Downregulation of CFTR mRNA gene expression was also included in a prognostic predictor gene set for poor prognosis in colorectal cancer [50]. Another study for early colon screening of adult CF patients revealed a high incidence of colon tumors, especially in males [51]. Mir17 is overexpressed in CF macrophages (this study) and epithelial cells [38], and is correlated with gastric cancer [52]. Interestingly, a recent meta-analysis study towards high-expression of Mirc1/Mir17–92 cluster has indicated poor prognosis of various cancers [53,54]. Therefore, our finding that Mirc1/Mir17–92 cluster is highly expressed in CF, may explain the increased incidence of adenomas and cancer in CF [55,56].
Our study shows that the expression of Mirs within the Mirc1/Mir17–92 cluster is increased with age. Given that high expression level of Mirc1/Mir17–92 cluster is a predictor of poor overall survival in patients with cancer [53,57], it is possible that the elevated level of Mirc1/Mir17–92 cluster correlates with poor prognosis of cancer in the CF population.
In summary, Mirc1/Mir17–92 cluster is highly expressed in CF cells and biologic fluids and correlates with respiratory status, age and PEx.
Acknowledgments
Studies in the Amer laboratory are supported by The Center for Clinical and Translational Science (CCTS), R01 HL094586 and R01 HL127651-01A1. BK is supported by 1K08AI108792-01A1, the Cystic Fibrosis Foundation (KOPP16I0), and UL1TR001070 from the National Center for Advancing Translational Sciences. KK is supported by Deutsche Forschungsgemeinschaft (DFG-German Research Foundation). The authors thank Melinda Smith and Mallory Rowell for assistance in sample collection. Human samples for this work were supplied by the C3F Live Cell Core at Nationwide Children’s Hospital, supported by a Research Development Program grant from the Cystic Fibrosis Foundation.
Footnotes
Conflict of interest
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. The authors declare that they have no conflict of interests. BK is on a CF advisory board for Vertex Pharmaceuticals.
References
- 1.de Boer K, Vandemheen KL, Tullis E, Doucette S, Fergusson D, Freitag A, et al. Exacerbation frequency and clinical outcomes in adult patients with cystic fibrosis. Thorax. 2011;66(8):680–5. doi: 10.1136/thx.2011.161117. [DOI] [PubMed] [Google Scholar]
- 2.Regelmann WE, Schechter MS, Wagener JS, Morgan WJ, Pasta DJ, Elkin EP, et al. Pulmonary exacerbations in cystic fibrosis: young children with characteristic signs and symptoms. Pediatr Pulmonol. 2013;48(7):649–57. doi: 10.1002/ppul.22658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.VanDevanter DR, Morris NJ, Konstan MW. IV-treated pulmonary exacerbations in the prior year: An important independent risk factor for future pulmonary exacerbation in cystic fibrosis. J Cyst Fibros. 2016;15(3):372–9. doi: 10.1016/j.jcf.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waters V, Ratjen F. Pulmonary Exacerbations in Children with Cystic Fibrosis. Ann Am Thorac Soc. 2015;12(Suppl 2):S200–6. doi: 10.1513/AnnalsATS.201502-098AW. [DOI] [PubMed] [Google Scholar]
- 5.Valletta EA, Rigo A, Bonazzi L, Zanolla L, Mastella G. Modification of some markers of inflammation during treatment for acute respiratory exacerbation in cystic fibrosis. Acta Paediatr. 1992;81(3):227–30. doi: 10.1111/j.1651-2227.1992.tb12209.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Yang PC. MicroRNA: a small molecule with a big biological impact. Microrna. 2012;1(1):1. doi: 10.2174/2211536611201010001. [DOI] [PubMed] [Google Scholar]
- 7.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9(4):572–94. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 8.Faruq O, Vecchione A. microRNA: Diagnostic Perspective. Front Med (Lausanne) 2015;2(51) doi: 10.3389/fmed.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Planell-Saguer M, Rodicio MC. Detection methods for microRNAs in clinic practice. Clin Biochem. 2013;46(10–11):869–78. doi: 10.1016/j.clinbiochem.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Schmittgen TD, Lee EJ, Jiang J, Sarkar A, Yang L, Elton TS, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44(1):31–8. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy. 2011;7(11):1359–70. doi: 10.4161/auto.7.11.17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tazi MF, Dakhlallah DA, Caution K, Gerber MM, Chang SW, Khalil H, et al. Elevated Mirc1/Mir17-92 cluster expression negatively regulates autophagy and CFTR (cystic fibrosis transmembrane conductance regulator) function in CF macrophages. Autophagy. 2016;12(11):2026–37. doi: 10.1080/15548627.2016.1217370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beydon N, Davis SD, Lombardi E, Allen JL, Arets HG, Aurora P, et al. An official American Thoracic Society/European Respiratory Society statement: pulmonary function testing in preschool children. Am J Respir Crit Care Med. 2007;175(12):1304–45. doi: 10.1164/rccm.200605-642ST. [DOI] [PubMed] [Google Scholar]
- 15.Rosenfeld M, Emerson J, Williams-Warren J, Pepe M, Smith A, Montgomery AB, et al. Defining a pulmonary exacerbation in cystic fibrosis. J Pediatr. 2001;139(3):359–65. doi: 10.1067/mpd.2001.117288. [DOI] [PubMed] [Google Scholar]
- 16.Jones M, Castile R, Davis S, Kisling J, Filbrun D, Flucke R, et al. Forced expiratory flows and volumes in infants. Normative data and lung growth. Am J Respir Crit Care Med. 2000;161(2 Pt 1):353–9. doi: 10.1164/ajrccm.161.2.9903026. [DOI] [PubMed] [Google Scholar]
- 17.Castile R, Filbrun D, Flucke R, Franklin W, McCoy K. Adult-type pulmonary function tests in infants without respiratory disease. Pediatr Pulmonol. 2000;30(3):215–27. doi: 10.1002/1099-0496(200009)30:3<215::aid-ppul6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187(4):397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wainwright CE, Elborn JS, Ramsey BW. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med. 2015;373(18):1783–4. doi: 10.1056/NEJMc1510466. [DOI] [PubMed] [Google Scholar]
- 20.Jones AM, Barry PJ. Lumacaftor/ivacaftor for patients homozygous for Phe508del-CFTR: should we curb our enthusiasm? Thorax. 2015;70(7):615–6. doi: 10.1136/thoraxjnl-2015-207369. [DOI] [PubMed] [Google Scholar]
- 21.Aherne ST, Madden SF, Hughes DJ, Pardini B, Naccarati A, Levy M, et al. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer. 2015;15:329. doi: 10.1186/s12885-015-1327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, Wang ZX, Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther. 2010;10(12):1224–32. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Liu L, Xu Q, Wu P, Zuo X, Ji A. MicroRNA as a novel drug target for cancer therapy. Expert Opin Biol Ther. 2012;12(5):573–80. doi: 10.1517/14712598.2012.671293. [DOI] [PubMed] [Google Scholar]
- 24.Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394(3):792–7. doi: 10.1016/j.bbrc.2010.03.075. [DOI] [PubMed] [Google Scholar]
- 25.O’Sullivan JF, Neylon A, McGorrian C, Blake GJ. MicroRNA Expression in Coronary Artery Disease. Microrna. 2014;2(3):205–11. doi: 10.2174/22115366113026660018. [DOI] [PubMed] [Google Scholar]
- 26.Bernecker C, Halim F, Haase M, Willenberg HS, Ehlers M, Schott M. MicroRNA expressions in PMBCs, CD4+, and CD8+ T-cells from patients suffering from autoimmune Addison’s disease. Horm Metab Res. 2013;45(8):599–604. doi: 10.1055/s-0033-1341511. [DOI] [PubMed] [Google Scholar]
- 27.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32(3–4):189–94. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furer V, Greenberg JD, Attur M, Abramson SB, Pillinger MH. The role of microRNA in rheumatoid arthritis and other autoimmune diseases. Clin Immunol. 2010;136(1):1–15. doi: 10.1016/j.clim.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Roberts AP, Jopling CL. Targeting viral infection by microRNA inhibition. Genome Biol. 2010;11(1):201. doi: 10.1186/gb-2010-11-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7(8):e1002176. doi: 10.1371/journal.ppat.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKiernan PJ, Greene CM. MicroRNA Dysregulation in Cystic Fibrosis. Mediators Inflamm. 2015;2015:529642. doi: 10.1155/2015/529642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amato F, Seia M, Giordano S, Elce A, Zarrilli F, Castaldo G, et al. Gene mutation in microRNA target sites of CFTR gene: a novel pathogenetic mechanism in cystic fibrosis? PLoS One. 2013;8(3):e60448. doi: 10.1371/journal.pone.0060448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Megiorni F, Cialfi S, Dominici C, Quattrucci S, Pizzuti A. Synergistic post-transcriptional regulation of the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS One. 2011;6(10):e26601. doi: 10.1371/journal.pone.0026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran S, Karp PH, Jiang P, Ostedgaard LS, Walz AE, Fisher JT, et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A. 2012;109(33):13362–7. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya S, Balakathiresan NS, Dalgard C, Gutti U, Armistead D, Jozwik C, et al. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J Biol Chem. 2011;286(13):11604–15. doi: 10.1074/jbc.M110.198390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oglesby IK, Bray IM, Chotirmall SH, Stallings RL, O’Neill SJ, McElvaney NG, et al. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol. 2010;184(4):1702–9. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 37.Oglesby IK, Chotirmall SH, McElvaney NG, Greene CM. Regulation of cystic fibrosis transmembrane conductance regulator by microRNA-145, -223, and -494 is altered in DeltaF508 cystic fibrosis airway epithelium. J Immunol. 2013;190(7):3354–62. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- 38.Oglesby IK, Vencken SF, Agrawal R, Gaughan K, Molloy K, Higgins G, et al. miR-17 overexpression in cystic fibrosis airway epithelial cells decreases interleukin-8 production. Eur Respir J. 2015;46(5):1350–60. doi: 10.1183/09031936.00163414. [DOI] [PubMed] [Google Scholar]
- 39.Zhang PX, Cheng J, Zou S, D’Souza AD, Koff JL, Lu J, et al. Pharmacological modulation of the AKT/microRNA-199a-5p/CAV1 pathway ameliorates cystic fibrosis lung hyper-inflammation. Nat Commun. 2015;6:6221. doi: 10.1038/ncomms7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abasi M, Kohram F, Fallah P, Arashkia A, Soleimani M, Zarghami N, et al. Differential Maturation of miR-17 ~ 92 Cluster Members in Human Cancer Cell Lines. Appl Biochem Biotechnol. 2017;182(4):1540–7. doi: 10.1007/s12010-017-2416-5. [DOI] [PubMed] [Google Scholar]
- 41.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–14. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kishore A, Borucka J, Petrkova J, Petrek M. Novel insights into miRNA in lung and heart inflammatory diseases. Mediators Inflamm. 2014;2014:259131. doi: 10.1155/2014/259131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mullany LE, Wolff RK, Herrick JS, Buas MF, Slattery ML. SNP Regulation of microRNA Expression and Subsequent Colon Cancer Risk. PLoS One. 2015;10(12):e0143894. doi: 10.1371/journal.pone.0143894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vosa U, Esko T, Kasela S, Annilo T. Altered Gene Expression Associated with microRNA Binding Site Polymorphisms. PLoS One. 2015;10(10):e0141351. doi: 10.1371/journal.pone.0141351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullany LE, Herrick JS, Wolff RK, Slattery ML. Single nucleotide polymorphisms within MicroRNAs, MicroRNA targets, and MicroRNA biogenesis genes and their impact on colorectal cancer survival. Genes Chromosomes Cancer. 2017;56(4):285–95. doi: 10.1002/gcc.22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moszynska A, Gebert M, Collawn JF, Bartoszewski R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017;7(4) doi: 10.1098/rsob.170019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garg M, Ooi CY. The Enigmatic Gut in Cystic Fibrosis: Linking Inflammation, Dysbiosis, and the Increased Risk of Malignancy. Curr Gastroenterol Rep. 2017;19(2):6. doi: 10.1007/s11894-017-0546-0. [DOI] [PubMed] [Google Scholar]
- 49.Maisonneuve P, Marshall BC, Knapp EA, Lowenfels AB. Cancer risk in cystic fibrosis: a 20-year nationwide study from the United States. J Natl Cancer Inst. 2013;105(2):122–9. doi: 10.1093/jnci/djs481. [DOI] [PubMed] [Google Scholar]
- 50.Than BL, Linnekamp JF, Starr TK, Largaespada DA, Rod A, Zhang Y, et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene. 2016;35(32):4179–87. doi: 10.1038/onc.2015.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Billings JL, Dunitz JM, McAllister S, Herzog T, Bobr A, Khoruts A. Early colon screening of adult patients with cystic fibrosis reveals high incidence of adenomatous colon polyps. J Clin Gastroenterol. 2014;48(9):e85. doi: 10.1097/MCG.0000000000000034. [DOI] [PubMed] [Google Scholar]
- 52.Park D, Lee SC, Park JW, Cho SY, Kim HK. Overexpression of miR-17 in gastric cancer is correlated with proliferation-associated oncogene amplification. Pathol Int. 2014;64(7):309–14. doi: 10.1111/pin.12178. [DOI] [PubMed] [Google Scholar]
- 53.Li H, Wu Q, Li T, Liu C, Xue L, Ding J, et al. The miR-17-92 cluster as a potential biomarker for the early diagnosis of gastric cancer: evidence and literature review. Oncotarget. 2017;8(28):45060–71. doi: 10.18632/oncotarget.15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11(4):501–6. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez-Jimenez I, Fischman D, Cheriyath P. Colon cancer in cystic fibrosis patients: is this a growing problem? J Cyst Fibros. 2008;7(5):343–6. doi: 10.1016/j.jcf.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Audeh MW. Gastrointestinal cancer and the cystic fibrosis gene. N Engl J Med. 1995;333(2):129–30. doi: 10.1056/NEJM199507133330214. [DOI] [PubMed] [Google Scholar]
- 57.Xiao J, Yu W, Hu K, Li M, Chen J, Li Z. miR-92a promotes tumor growth of osteosarcoma by targeting PTEN/AKT signaling pathway. Oncol Rep. 2017;37(4):2513–21. doi: 10.3892/or.2017.5484. [DOI] [PubMed] [Google Scholar]