Abstract
Neurodegenerative diseases share certain pathophysiological hallmarks that represent common targets for drug discovery. In particular, dysfunction of proteostasis and the resultant apoptotic death of neurons represent common pathways for pharmacological intervention. A library of aromatic carbamate derivatives based on the clinically available drug flupirtine was synthesized to determine a structure–activity relationship for neuroprotective activity. Several derivatives were identified that possess greater protective effect in human induced pluripotent stem cell-derived neurons, protecting up to 80% of neurons against etoposide-induced apoptosis at concentrations as low as 100 nM. The developed aromatic carbamates possess physicochemical properties desirable for CNS therapeutics. The primary known mechanisms of action of the parent scaffold are not responsible for the observed neuroprotective activity. Herein, we demonstrate that neuroprotective aromatic carbamates function to increase the Bcl-2/Bax ratio to an antiapoptotic state and activate autophagy through induction of beclin 1.
Graphical Abstract
INTRODUCTION
Neurodegenerative diseases are a group of disorders characterized by the loss of structure and/or function of neurons. At the molecular level, neurodegenerative diseases possess several similarities such as abnormal deposition of proteins (which in many disorders are misfolded),1 mitochondrial stress leading to the formation of reactive oxygen species (ROS),2 microglial activation and neuroinflammation,3 dysregulation of proteostasis involving the ubiquitin-proteasome pathway and autophagy-lysosome pathway,4 programmed cell death including apoptosis,5 and dysregulation of receptors involved in synaptic plasticity, memory, learning, and other functions.5,6 These features are evident in the most prevalent neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis (ALS), and Batten disease, the most common (yet rare) neurodegenerative disease of childhood. 1,5,7,8 Current treatment options for all of these disorders are symptomatic and do not slow or reverse disease progression. Pathophysiological similarities at the molecular level suggest that lessons learned from one neurodegenerative disease may be applicable to other diseases, leading to the design of pharmacological interventions with broad utility.9
Drug repurposing, employing a clinically approved agent for one indication in a different indication, has become a rich source of lead compounds.10 To this end, flupirtine (1, Figure 1), a nonopioid analgesic approved in the European Union for pain following surgery,11 has been shown to possess a range of additional pharmacological activities. The antiseizure activity of 1 has been ascribed to its indirect antagonism of the N-methyl-D-aspartate (NMDA) receptor.12 The compound is known to act as a potassium Kv7 channel (KCNQ) opener and reduces intracellular levels of calcium, which is related to its activity as an indirect NMDA receptor antagonist.13,14 Neuroprotective activity in a variety of neurodegenerative disease models has been reported for 1,14 with mechanisms of action including upregulation of the antiapoptotic protein B-cell lymphoma 2 (Bcl-2) and antioxidant activity via increased glutathione levels.11 Overall, in vitro, in vivo, and/or clinical trials have demonstrated protective properties in Alzheimer’s disease, Parkinson’s disease, epilepsy, Creutzfeldt–Jakob disease, glaucoma, age-related macular degeneration, ischemic stroke, prion disease, HIV related neuroinfection, multiple sclerosis, and Batten disease.11,13–15 However, chronic use of 1, as would be required to treat neurodegenerative diseases, may lead to mild hepatotoxicity that has led to restricted use in the European Union and precludes its use as a potential therapeutic for neurodegenerative disease.16
Figure 1.
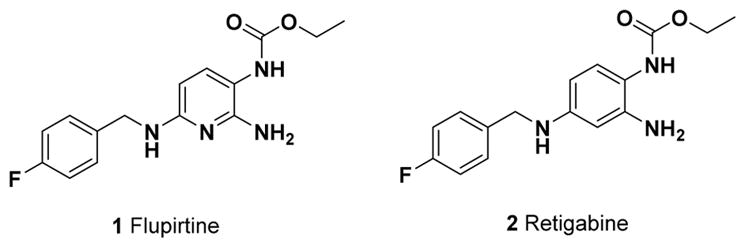
Structures of 1 and 2.
Retigabine (2, Figure 1), the phenyl bioisostere of 1, is approved by the Food and Drug Administration (FDA) for the treatment of seizures17 and is currently in phase II clinical trials for the treatment of ALS.18 In comparison with 1, 2 possesses greater potency as a NMDA receptor antagonist in neurons differentiated from ALS patient induced pluripotent stem cells (iPSCs).19 Like 1, 2 acts as a KCNQ opener and thus an indirect antagonist at the NMDA receptor20 and also possesses antioxidant properties.21
Given the noted neuroprotective effects of 1 across several neurodegenerative diseases, the recent progression of repurposing 2 to clinical trial for ALS and the currently ill-defined molecular targets of the two agents, we instigated a campaign to fully probe the unexplored structure–activity relationships (SARs) that define the neuroprotective properties of the aromatic carbamate scaffold. Even though libraries of both 1 and 2 derivatives have been reported in the literature, the focus was solely on NMDA antagonistic activity.12 Herein, we demonstrate the neuroprotective SAR of 1 violates its SAR for NMDA antagonism and directly show no correlation between neuroprotective activity and NMDA antagonism or antioxidant activity. We demonstrate the translational neuroprotective activity of designed aromatic carbamate derivatives in neurons differentiated from a human iPSC line, a model considered highly translatable and widely accepted for drug screening as well as repurposing molecules for neurodegenerative drug discovery.19,22 Further, we disclose the neuroprotective mechanism of action of structurally novel aromatic carbamates to be multimodal, acting to significantly upregulate the antiapoptotic protein Bcl-2 and induce autophagy by elevation of beclin 1.
CHEMISTRY
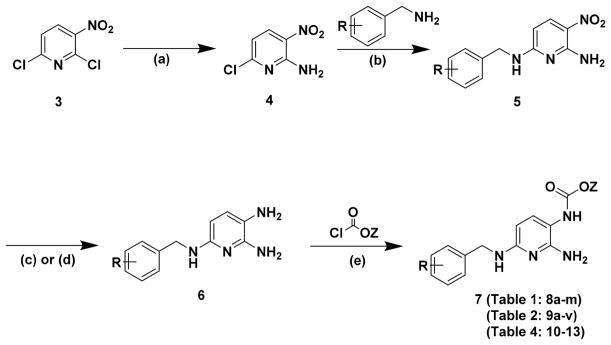
The synthetic route employed to access derivatives of 1 was adapted from Seydel et al.12 Commercially available 2,6-dichloro-3-nitropyridine (3) underwent selective amination at the 2-position to provide 6-chloro-3-nitropyridin-2-amine (4) in good yield (Scheme 1). Pyridine (4) underwent nucleophilic substitution with a suitably substituted benzylamine to afford the 6-amino-2-benzylamino-5-nitropyridine intermediate (5), which was subsequently reduced to provide triaminopyridine intermediate (6). Regioselective addition of a suitably functionalized chloroformate to the 3-amino position proceeded in good yield to afford the targeted aromatic carbamate of general structure (7).
Scheme 1a.
aReagents and conditions: (a) NH3, MeOH, BRSM-65%; (b) suitably funtionalized benzylamine, TEA, IPA; (c) SnCl2·2H2O, HCl; (d) Raney Ni, NH2NH2·2H2O, dioxane; (e) suitably functionalized chloroformate, dioxane.
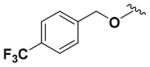
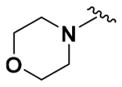
To access the corresponding ether bioisostere, commercially available 4-trifluoromethyl benzyl alcohol and pyridine intermediate (4) underwent a Williamson ether synthesis23 to afford the respective pyridine ether derivative (Supporting Information, Scheme S1). This intermediate was subjected to the same synthetic transformations as described in Scheme 1 to yield the 4-trifluorophenylmethyl ether analogue (14) (Table 2). The morpholino analogue (15) (Table 2) was synthesized based on the same synthetic route (Supporting Information, Scheme S2).
Table 2.
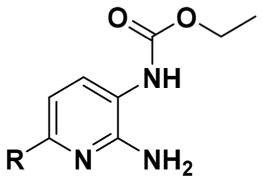
Protective Effect of Benzylamine Analogues of 1 to Ameliorate Etoposide-Induced Apoptosis in PC12 Cells (n = 3, Percentage Cell Viability Represented As Mean ± SEM)
| |||
|---|---|---|---|
| Compound | R | % Viability (3 μM) | MPO Score |
| Vehicle | 52.13 ± 1.13 | ||
| 1 |
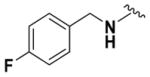
|
79.50 ± 3.22 | 4.50 |
| 9a |
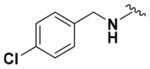
|
64.32 ± 2.47 | 4.44 |
| 9b |
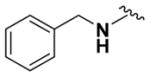
|
78.63 ± 6.27 | 4.50 |
| 9c |
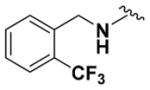
|
82.76 ± 2.15 | 4.30 |
| 9d |
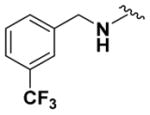
|
68.17 ± 3.05 | 4.30 |
| 9e |
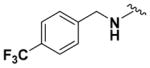
|
68.38 ± 3.67 | 4.30 |
| 9f |
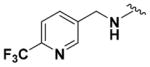
|
81.57 ± 6.54 | 4.10 |
| 9g |
|
80.59 ± 1.90 | 3.67 |
| 9h |
|
79.24 ± 13.63 | 3.56 |
| 9i |
|
135.53 ± 12.27 | 4.48 |
| 9j |
|
82.28 ± 12.82 | 4.48 |
| 9k |
|
79.68 ± 6.13 | 4.22 |
| 9l |
|
87.49 ± 6.13 | 4.22 |
| 9m |
|
72.94 ± 5.41 | 4.14 |
| 9n |
|
75.70 ± 3.69 | 3.41 |
| 9o |
|
78.04 ± 7.51 | 4.50 |
| 9p |
|
87.89 ± 4.66 | 4.23 |
| 9q |
|
48.88 ± 3.91 | 3.90 |
| 9r |
|
76.08 ± 7.37 | 3.96 |
| 9s |
|
57.59 ± 3.67 | 4.24 |
| 9t |
|
59.29 ± 5.17 | 4.50 |
| 9u |
|
49.96 ± 3.71 | 4.50 |
| 9v |
|
66.17 ± 2.76 | 4.50 |
| 14 |
|
69.94 ± 4.27 | 4.62 |
| 15 |
|
88.74 ± 5.53 | 5.00 |
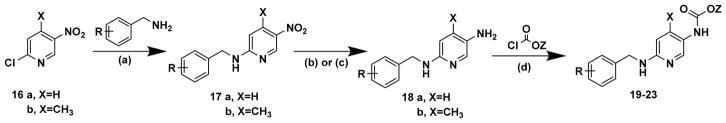
To achieve excision of the 2-amino moiety of 1, we employed either commercially available 2-chloro-5-nitropyridine (16a) or its 4-methyl counterpart (16b) as starting material and followed the same reaction sequence as delineated in Scheme 1 to obtain analogues (19, 21–23) and an analogue with a 4-methyl substitution on the pyridine ring (20) (Scheme 2).
Scheme 2a.
aReagents and conditions: (a) suitably funtionalized benzylamine, TEA, IPA; (b) SnCl2·2H2O, HCl; (c) Raney Ni, NH2NH2·2H2O, dioxane; (d) suitably functionalized chloroformate, dioxane.
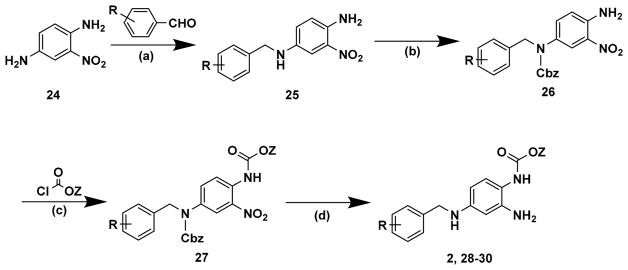
Synthetic access to structurally diverse derivatives of 2 was achieved by reductive amination of commercially available 2-nitro-1,4-phenyldiamine (24) with a suitably functionalized benzaldehyde (Scheme 3). The secondary amine was found to be more reactive compared to the primary amine and was thus protected as the benzylcarbamate (26). Addition of a suitably functionalized chloroformate afforded dicarbamate (27). Subsequent exposure of (27) to palladium on charcoal under a hydrogen atmosphere achieved concomitant benzylcarbamate deprotection and reduction of the 2-nitro moiety to the respective amine (2, 28–30) in one step.24
Scheme 3a.
aReagents and conditions: (a) (i) IPA, (ii) NaBH4; (b) benzyl chloroformate, DIPEA, IPA; (c) DIPEA, DMAP, dioxane; (d) H2, 10% Pd/C, MeOH.
RESULTS AND DISCUSSION
Screening
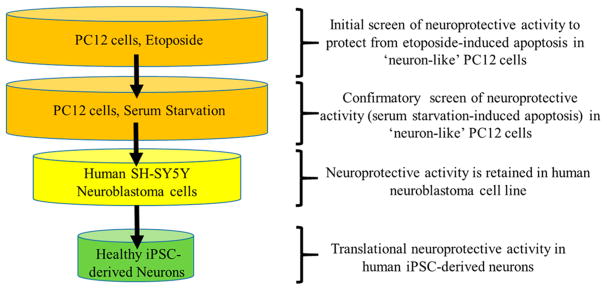
Our screening strategy (Figure 2) was designed to allow rapid initial evaluation of neuroprotective activity and identification and exclusion of toxic derivatives. To begin to understand the structural features of aromatic carbamates required for neuroprotective activity and to assign an initial SAR, we assayed their protective activity in “neuron-like” PC12 cells induced to apoptosis by exposure to etoposide.25 Etoposide is an anticancer agent that functions as a topoisomerase II inhibitor and is widely used to induce apoptosis in cells for screening of neuroprotective molecules.26 To ensure that screened compounds do not simply act to displace or block etoposide from its binding site, we conducted a secondary screen on selected derivatives to determine protective effect to ameliorate apoptotic cell death in PC12 cells exposed to serum starvation.25 Identical trends in activity were observed across both external insults; derivatives with greater protective effect than 1 to ameliorate etoposide-induced apoptosis also demonstrated greater protective effect to ameliorate serum starvation-induced apoptosis and vice versa (Supporting Information, Table S1). To rapidly assign a SAR, we compared protective activity at an aromatic carbamate concentration of 3 μM, with all compounds screened at 1, 3, 10, and 30 μM concentrations. The 3 μM concentration is the value at which hit compound 1 provided greatest protective activity in the primary etoposide-based assay and is an achievable value for translation to free brain concentration in vivo.27 Compounds which indicated greater protective activity than 1 were furthered screened at 0.1 and 0.3 μM to determine dose-dependent effects (Supporting Information, Tables S2–S5).
Figure 2.
Schematic representation of screening strategy to identify neuroprotective aromatic carbamates.
To determine protective effect in human cells, aromatic carbamates that showed greater activity than 1 in PC12 cells (and selected inactive compounds, compared to 1, to act as controls) were screened to determine their ability to ameliorate etoposide-induced apoptosis in human SH-SY5Y neuroblastoma cells. To determine the translational potential of aromatic carbamates that show protective effect in both PC12 and SH-SY5Y cells we screened the ability of these analogues (and selected inactive controls compared to 1) to ameliorate etoposide-induced apoptosis in human neurons differentiated from iPSCs. These iPSC-derived human neurons possess several advantages over immortalized neuronal cell lines and primary mouse neurons as they require less specialist isolation, can be produced in large quantities, and have no specialist licensing requirements.22
Structure–Activity Relationships
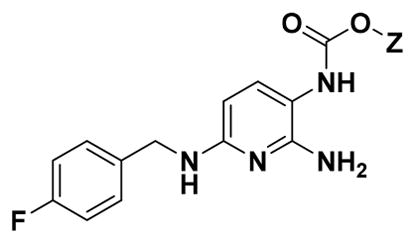
To provide a diverse library of aromatic carbamates, the structure of 1 was dissected in to three classes: analogues at the carbamate position (class I, Table 1), analogues of the benzylamine group (class II, Table 2), and analogues of the ring system (class III, Table 3). Further, hybrid analogues (Table 4) were also generated where modifications were made at more than one site. Multiparameter optimization (MPO) score was calculated for all synthesized analogues to allow further comparison of derivatives and identify predicted blood–brain barrier (BBB) penetration, an essential parameter for neuroprotective compounds.28 An MPO score ≥4 is predictive of brain penetration,29 therefore we designed our analogues such that the majority possess an MPO score ≥4 (Table 1–4, Supporting Information, Table S6).
Table 1.
Protective Effect of Carbamate Analogues of 1 to Ameliorate Etoposide-Induced Apoptosis in PC12 Cells (n = 3, Percentage Cell Viability Represented As Mean ± SEM)
| |||
|---|---|---|---|
| Compound | Z | % Viability (3 μM) | MPO Score |
| Vehicle | 52.13 ± 1.13 | ||
| 1 |
|
79.50 ± 3.22 | 4.50 |
| 8a |
|
96.64 ± 5.97 | 4.50 |
| 8b |
|
46.66 ± 2.49 | 4.41 |
| 8c |
|
62.91 ± 1.72 | 4.22 |
| 8d |
|
42.17 ± 2.46 | 4.32 |
| 8e |
|
78.46 ± 6.52 | 3.73 |
| 8f |
|
84.87 ± 7.95 | 4.48 |
| 8g |
|
61.87 ± 4.01 | 4.50 |
| 8h |
|
88.94 ± 8.77 | 4.42 |
| 8i |
|
53.92 ± 3.68 | 4.50 |
| 8j |
|
77.25 ± 5.79 | 3.71 |
| 8k |
|
58.98 ± 3.97 | 4.02 |
| 8l |
|
81.21 ± 5.49 | 3.93 |
| 8m |
|
56.42 ± 5.19 | 2.82 |
Table 3.
Protective Effect of Ring Analogues of 1 to Ameliorate Etoposide-Induced Apoptosis in PC12 Cells (n = 3, Percentage Cell Viability Represented As Mean ± SEM
| Compound | Structure | % Viability (3 μM) | MPO Score |
|---|---|---|---|
| Vehicle | 52.13±1.13 | ||
| 1 |
|
79.50 ± 3.22 | 4.50 |
| 2 |
|
73.92 ± 8.03 | 4.50 |
| 19 |
|
109.51 ± 7.90 | 5.00 |
| 20 |
|
53.80 ± 1.14 | 4.79 |
Table 4.
Protective Effect of Hybrid Analogues of 1 to Ameliorate Etoposide-Induced Apoptosis in PC12 Cells (n = 3, Percentage Cell Viability Represented As Mean ± SEM)
| Compound | Structure | % Viability (3 μM) | MPO Score |
|---|---|---|---|
| Vehicle | 52.13 ± 1.13 | ||
| 1 |

|
79.50 ± 3.22 | 4.50 |
| 10 |
|
41.28 ± 5.84 | 4.48 |
| 11 |
|
107.25 ± 1.33 | 4.06 |
| 12 |
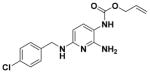
|
79.65 ± 3.06 | 4.25 |
| 13 |
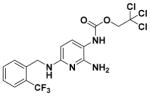
|
107.50 ± 4.23 | 2.99 |
| 21 |
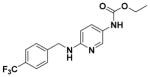
|
63.99 ± 3.21 | 4.68 |
| 22 |
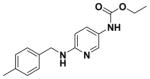
|
64.27 ± 7.19 | 4.87 |
| 23 |
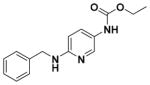
|
67.50 ± 7.60 | 5.00 |
| 28 |
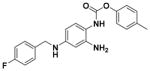
|
91.62 ± 6.35 | 3.71 |
| 29 |
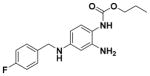
|
55.56 ± 4.28 | 4.39 |
| 30 |
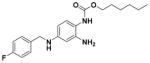
|
60.13 ± 6.59 | 3.73 |
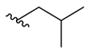
Within class I, a range of substituents could be tolerated at the carbamate moiety including electron-donating and electron-withdrawing groups (Table 1). To explore the effect of homologation within the carbamate moiety, analogues with 1, 3, 4, and 6 methylene groups, including linear and branched chains, were synthesized. The methyl (8a) and hexyl (8e) carbamate analogues demonstrated protective activity (cell viability of 97% and 78%, respectively). However, the hexyl derivative suffered a reduced MPO score <4. The isobutyl carbamate derivative (8c) showed a reduction in activity (63%), while the propyl (8b) and tert-butyl (8d) carbamates displayed no activity above vehicle control, demonstrating protection of cell viability at 47% and 42%, respectively, suggesting that branching alters binding to the target receptor and reduces activity, a commonly observed effect in medicinal chemistry, coupled with low levels of toxicity. The allyl carbamate (8f) was well tolerated (85%), while the alkyne carbamate (8g) displayed reduced activity (62%). Addition of electron-withdrawing groups influence protective effect based on their regiochemistry; 1-chloroethyl (8h) and 2,2,2-trichloroethyl (8j) analogues were well tolerated (89% and 77%, respectively) but with a reduced MPO score in the case of (8j), while the 2-chloroethyl analogue (8i) lost activity (54%), probably due to the accessible electrophilic site imparting toxicity. Phenyl (8k) and fluorenylmethyloxycarbonyl (Fmoc) (8m) derivatized carbamates also showed a reduction in activity (59% and 56%, respectively), further supporting that nonlinear steric bulk in this region of the scaffold engenders loss of activity. Interestingly, the benzyl analogue (8l) was equally tolerated when compared with allyl carbamate (8f) despite the clear steric differences between these two structures (81% and 85%, respectively).
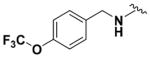
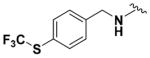
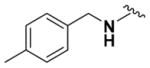
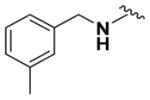
Within class II derivatives, the benzylamine moiety was found to be pharmacophoric with minor changes in substitution significantly affecting activity (Table 2). Substitution of chlorine at the para position (9a) reduced activity to 64% viable cells while hydrogen (9b) retains activity with 79% viable cells, suggesting this effect may be due to size rather than electronegativity. Fluorine and hydrogen have widely different electronegativities but comparable van der Waals radii. The 2-trifluoromethyl derivative (9c) retains activity, providing cell viability of 83%, while 3-trifluoromethyl (9d) and 4-trifluoromethyl (9e) both decrease activity to 68%. However, the 4-trifluoromethyl analogue (9e) at just 0.1 μM concentration increased cell viability to 84% (Figure 3), which was comparable to 1 at 3 μM concentration (80%). Bioisoteric replacement to incorporate a 3-(aminomethyl)-6-(trifluoromethyl)pyridine moiety (9f) showed similar cell viability to 1 at 3 μM concentration (82%). Similar to 4-trifluoromethyl (9e), 3-(aminomethyl)-6-(trifluoromethyl)pyridine (9f) also showed activity at concentrations as low as 0.1 μM with 74% viable cells. 4-Trifluoromethoxy benzylamine (9g) and its bioisostere 4-thio- (trifluoromethyl) benzylamine (9h) retain activity at 81% and 79%, respectively. However, their MPO scores are <4 at 3.67 and 3.56, respectively. Reduced concentration of 9g (0.1 μM) displays 77% cell viability, similar to 1 at 3 μM concentration. In comparison, 9h at 0.3 μM concentration increases cell viability to 93%. The 4-methyl analogue (9i) increases cell viability to 136%, suggesting an effect to enhance cell proliferation which would be advantageous in the treatment of neurodegenerative diseases. However, the derivative degrades in solvent upon storage for 4 days. The 3-methyl derivative (9j) retains activity comparable to 1 (82%) but is less active than its 4-substituted counterpart. The 4-methoxy derivative (9k) retains activity at 80%, while the 3-methoxy derivative (9l) increases activity to 87%. This suggests that both electron-withdrawing and electron-donating groups are well tolerated around the ring. Derivatives with electron-withdrawing 3,5-disubstitution (9m, 9n) enhance activity compared to their monosubstituted counterpart (9d). Derivatives with 2,4-disubstitution (9o, 9p) retain activity; this is particularly appreciable when electron-donating methyl groups are employed (88% cell viability). Adding steric bulk at the para-position in the form of a phenyl (9q) or thiophene (9r) reduces activity and induces toxicity at higher concentrations, along with reducing MPO score. Increasing steric bulk in the form of a naphthyl group (9s) reduces activity below that of 1. This effect is also seen when the carbon chain length is increased from methylene to ethylene (9t, 9u). These results combined may suggest that the benzylamine ring and its substitutions interact with a specific binding pocket of the protein target(s), and adding bulk or increasing linker length may disrupt this interaction. Adding an α-methyl group (9v) inactivates the molecule and imparts toxicity at higher concentrations. Bioisosteric replacement of the benzylamine with benzylether (14) results in reduced activity (70%) and, interestingly, the morpholino derivative (15) increases activity to 89%, which suggests that the amine may not be playing a role as a hydrogen bond donor.
Figure 3.
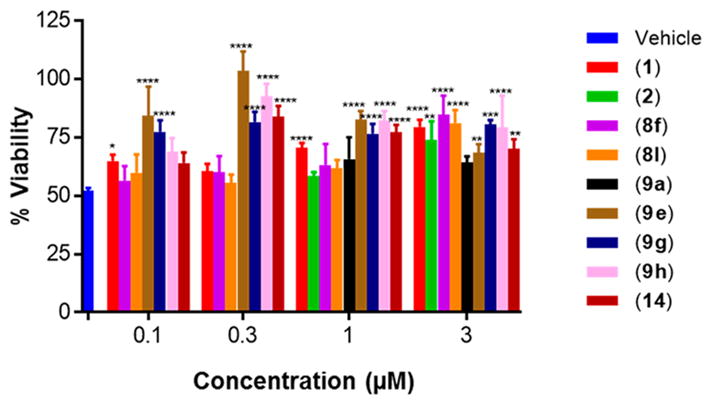
Summary of protective activity of selected aromatic carbamates to ameliorate etoposide-induced apoptosis in PC12 cells at low concentrations. Two-Way ANOVA, Tukey test, 95% confidence interval. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n = 3, % viability represented as mean ± SEM.
To understand the effect of substitutions on the pyridine ring (class III), ring analogues were synthesized (Table 3). The carbocyclic bioisostere of 1, 2 demonstrates similar protective activity (74% cell viability). Several analogues of 1 and 2 possessing the same substitution pattern were synthesized to further understand the role of the central ring in the activity of the molecule. Analogues of 2 were either equipotent with their 1 counterparts (1 vs 2, 8b vs 29) or less active (8e vs 30, which shows a decrease in cell viability protection by 20%). Excision of the 2-position amine (19) resulted in increased activity to 110% cell viability. Addition of a 4-methyl group to the central pyridine ring (20) resulted in no protective activity above vehicle control.
A number of hybrid analogues were synthesized to evaluate the cumulative effect on protective activity upon structural modification at more than one site (Table 4). The hybrid derivatives combining class I and class II analogues, demonstrated an additive effect on their activity (11 vs 9d and 8f; 12 vs 9a and 8f, and 13 vs 9c and 8j). As excision of the 2-position amine (19) resulted in enhanced cell proliferation, we sought to determine if further increases in cell viability can be achieved through combination with substituents that show high activity in other derivatives (21–23). However, all demonstrated reduced protective activity over their 2-amine counterpart.
A number of aromatic carbamates have been identified that possess significant protective activity to rescue PC12 cells from etoposide-induced apoptosis in our primary assay and demonstrate predicted (MPO score ≥4) BBB penetration. To determine the translational effect, a representative set of the most active compounds from all three classes, 1, 2, allyl carbamate (8f), benzyl carbamate (8l), 4-trifluoromethylbenzylamine (9e), 4-trifluoromethoxybenzylamine (9g), and 4-thio(trifluoromethyl) benzylamine (9h), and two less active control compounds, 4-chlorobenzylamine (9a) and benzylether (14), were screened in secondary assays. It should be noted that three compounds (8l, 9g, and 9h) possess MPO scores of 3.93, 3.67, and 3.56, respectively. However, these were included in this subset of compounds due to their much higher protective activity at concentrations as low as 0.1 μM (Figure 3). Further synthetic modifications to optimize BBB penetration are possible once lead compounds are identified.30
The ability of the representative set of compounds to ameliorate etoposide-induced apoptosis in human neuroblastoma SH-SY5Y cells was determined to evaluate if activity is retained upon transitioning from rat to human “neuron-like” cells. A similar trend in protective activity was observed in SH-SY5Y cells as in PC12 cells (Figure 4). The overall cell viability protection was lower in SH-SY5Y cells compared to PC12 cells (69% cell viability in SH-SY5Y for 1 at 3 μM vs 80% in PC12 cells), which may be attributed to different expression levels of the protein target(s) of our developed compounds between cell types.
Figure 4.
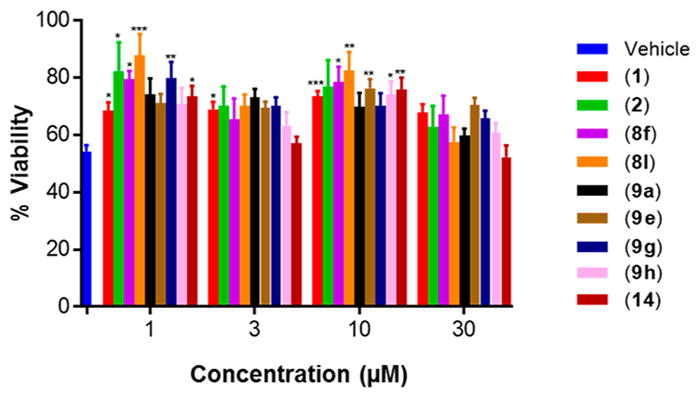
Selected aromatic carbamates are protective to ameliorate etoposide-induced apoptosis in human SH-SY5Y cells. Cell viability measured using PrestoBlue dye after treatment with 5 μM etoposide for 12 h. Two-Way ANOVA, Tukey test, 95% confidence interval. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n = 3, % viability represented as mean ± SEM.
Protective Effect in Human iPSC-Derived Neurons
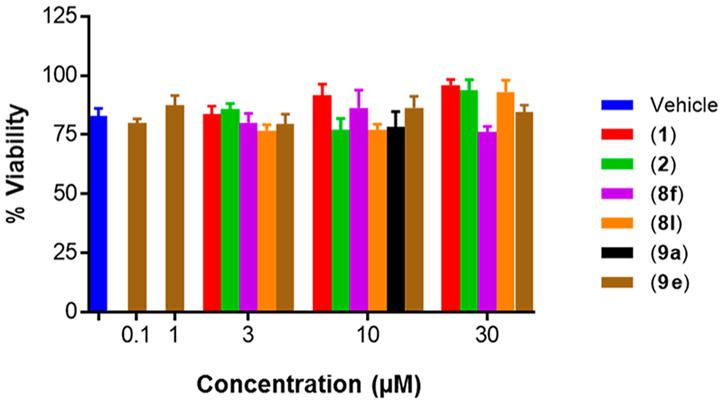
To determine the translational activity of selected analogues of 1 in the most representative model of terminally differentiated human neurons, aromatic carbamates were screened in neurons differentiated from IMR90-c4 iPSCs.22,31 These cells represent the best currently available in vitro model of human neurons for translational drug screening. The parent scaffold 1, benzyl carbamate analogue (8l), and 4-trifluoromethoxy analogue (9g) were active at 3, 10, and 30 μM concentrations, while 2 was active only at 10 and 30 μM concentrations (active defined as protective ability above that of vehicle control). Interestingly, aromatic carbamate 9a, inactive in the PC12 cell line, demonstrated low protective activity in the iPSC-derived neurons at 10 and 30 μM concentration but less so than 1 at these concentrations. Gratifyingly, the 4-trifluoromethyl benzylamine analogue (9e) retained significant protective activity from just 0.1 μM concentration through 3 μM (Figure 5). The electronegativity of the trifluoromethyl substituent may also have a role to deactivate the benzylamine, preventing formation of the quinone diimine reactive intermediate postulated to be responsible for the hepatotoxicity of 1.16 The ether bioisostere (14) provided 65% cell viability compared with 1 (70%) at 3 μM concentration. While this does represent a slight reduction in protective activity, the ether analogue is incapable of being metabolized to the quinone diimine reactive intermediate toxicophore, thus potentially offering an avenue to explore aromatic carbamates devoid of liver toxicity. This assay supports the translational potential of our preliminary screen as a similar trend in activity is observed in neurons differentiated from human iPSCs and rat “neuron-like” PC12 cells.
Figure 5.
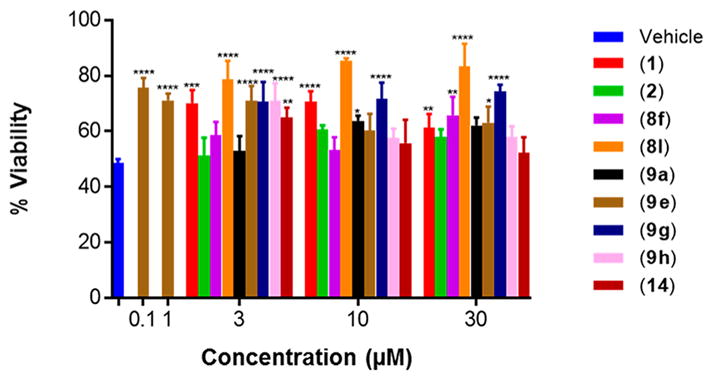
Selected aromatic carbamates ameliorate etoposide-induced apoptosis in human IMR90-c4 iPSC-derived neurons. Cell viability measured using PrestoBlue dye after treatment with 15 μg/mL etoposide for 36 h. Two-Way ANOVA, Tukey test, 95% confidence interval. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n = 3, % viability represented as mean ± SEM.
Mechanism of Action Determination
We sought to understand the mechanism by which the developed aromatic carbamates exert their neuroprotective activity to aid in future design of more active compounds. As antioxidant mechanisms of action are widely known to confer neuroprotective effect and both 1 and 2 are known to act to reduce reactive oxygen species (ROS) levels,32 we employed CellROX green dye which, by immunocytochemistry, correlates a proportional green intensity to the quantity of ROS present in a cellular system.
Exposure of PC12 cells to 10 μM of 1, 2, 8f, or 8l and staining with CellROX green dye indicated no amelioration of ROS upon treatment with the neuroprotective derivatives 8f or 8l, while 2 and 1 do ameliorate ROS induced by H2O2 as expected (Figure 6A). To further validate these results, PC12 cells were treated with aromatic carbamates in the concentration range of 1–30 μM. Introduction of ROS was generated using H2O2 and cell viability was measured after 4 h using PrestoBlue dye. Protection against cell death induced by H2O2 was observed with 1 and 2, while the more active analogues 8f, 8l, and 9e failed to show any activity above vehicle control (Figure 6B). Analogue 9a, which was less active compared to 1, was used as a negative control and showed no protective activity. Thus, synthesized aromatic carbamate derivatives with greater neuroprotective activity than 1 and 2 do not act as antioxidants.
Figure 6.
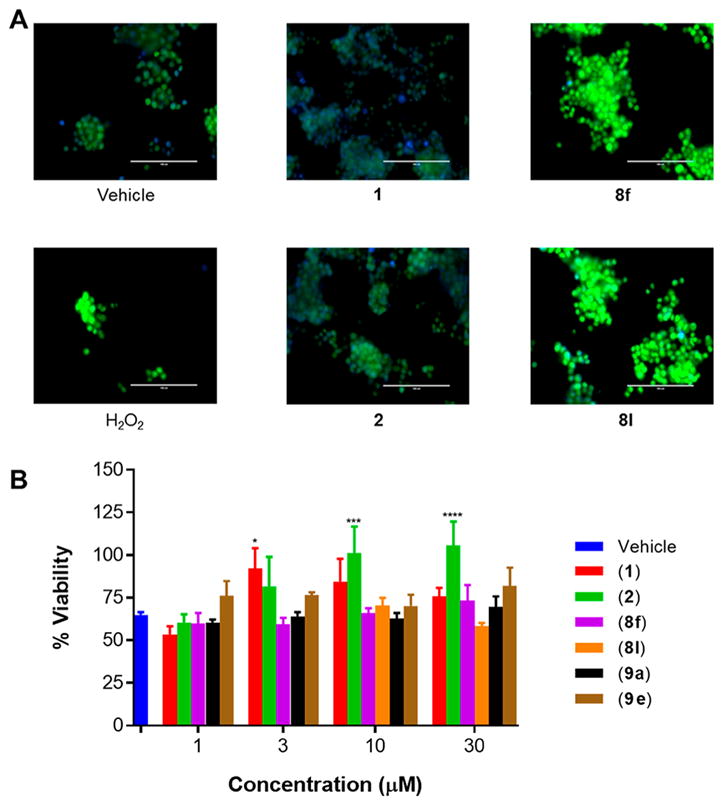
Antioxidant activity is demonstrated by 1 and 2. However, equipote0nt or greater potency protective derivatives 8f, 8l, and 9e do not act as antioxidants. (A) PC12 cells stained with CellROX and DAPI-blue nuclear stain. Greater intensity of green fluorescence indicates greater levels of ROS. Visualized through a fluorescence microscope (40× magnification, bar = 100 μm). (B) Measurement of cell viability using PrestoBlue dye after treating PC12 cells with 400 μM H2O2. Two-Way ANOVA, Tukey test, 95% confidence interval. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. n = 2, % viability represented as mean ± SEM.
Molecules acting as KCNQ openers, including 1 and 2, are known indirect antagonists of the NMDA receptor.33 We next sought to determine the activity of analogues of 1 to ameliorate NMDA induced excitotoxicity. NMDA receptors are expressed in PC12 cells, but the receptors are not functional.34 Thus, a mechanism of action involving NMDA receptor antagonism cannot be responsible for the observed protective activity. Human neurons differentiated from iPSCs, however, do express functional NMDA receptors,35 and we have shown translational protective effect of our aromatic carbamates against etoposide-induced toxicity (Figure 5). Thus, we employed neurons differentiated from human iPSCs (IMR90-c4) to evaluate our designed compounds in an NMDA challenge assay35 (Figure 7). No protective effect was observed with treatment of selected aromatic carbamates. However, a trend was observed where 1 and 2, at a higher concentration of 30 μM, protected against NMDA insult. The 4-trifluormethyl analogue (9e), active in the human iPSC-derived neuron assay at a low concentration of 0.1 μM, failed to show any NMDA antagonistic activity at all concentrations screened.
Figure 7.
Selected aromatic carbamates lack NMDA antagonistic activity when screened in human IMR90-c4 iPSC-derived neurons. Cell viability measured using PrestoBlue against 500 μM NMDA treatment. n = 1, 4 technical replicates. % viability represented as mean ± SEM.
Neuroprotective Derivatives Induce Bcl-2 and Increase Bcl-2/Bax Ratio
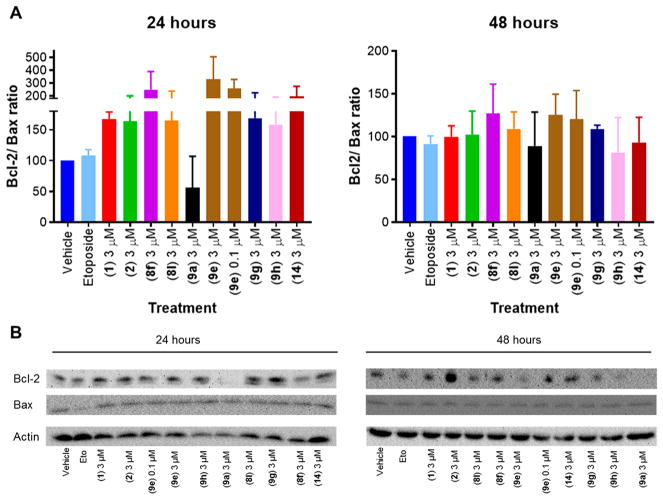
Upregulation of the antiapoptotic protein Bcl-2 is reported as a mechanism of action of 1, albeit at high concentration, 36 and thus we sought to understand the relationship of neuroprotective aromatic carbamates to the degree of Bcl-2 induction in PC12 cells. The ratio of Bcl-2 to Bcl-2-associated X protein (Bax), a pro-apoptotic protein, determines cell survival or cell death,37 and thus the ratio of Bcl-2 to Bax was determined. Cells were pretreated with the aromatic carbamates for 4 h before the addition of etoposide. After 24 h of treatment, 1 and 2 at 3 μM showed 68% and 64% increase in the Bcl-2/Bax ratio. Upon treatment with 8f at 3 μM, a 150% increase in the Bcl-2/Bax ratio was observed, while 9a, the reduced activity control, showed a 44% reduction. The 4-trifluoromethyl analogue (9e) at 3 and 0.1 μM showed 232% and 159% increase in the Bcl-2/Bax ratio, respectively. This was determined to be a transient effect as no change in the Bcl-2/Bax ratio was observed after 48 h of treatment (Figure 8A,B).
Figure 8.
Effect of selected aromatic carbamates to modulate the Bcl-2/Bax ratio in PC12 cells after 24 and 48 h of treatment. (A) Quantified results. n = 2, values represented as mean ± SEM. (B) Representative immunoblot for Bcl-2 and Bax.
Neuroprotective Derivatives Induce Autophagy
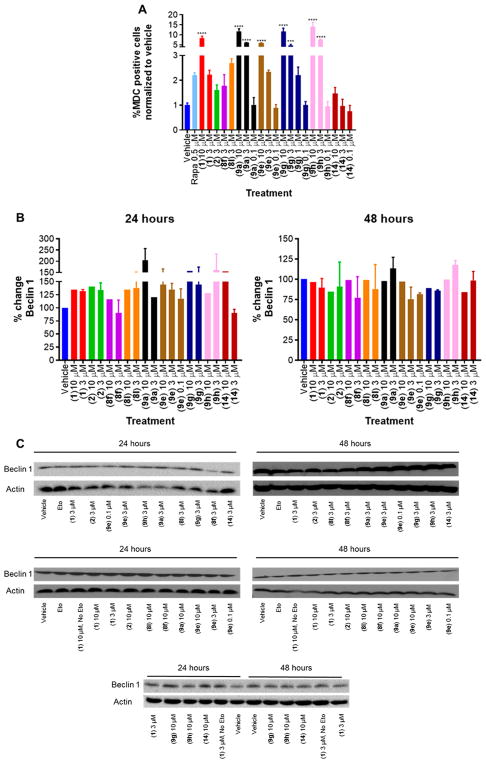
Mutations in genes regulating autophagy contribute to the pathogenesis of a wide range of neurodegenerative disorders. Activation of autophagy has been shown to be protective in several models of neurodegeneration.38,39 This effect results from enhanced clearance of abnormal or misfolded proteins from neurons.38,40 Thus, we evaluated the effect of analogues of 1 to induce autophagy. Induction of autophagy in PC12 cells was measured by monodansylcadaverine (MDC) staining using flow cytometry after 48 h of treatment.41 Rapamycin, which induces autophagy by mTOR inhibition,42 was employed as a positive control; at 500 nM concentration, a 2-fold increase in MDC staining was observed over vehicle. At 10 μM concentration, 1 showed an 8-fold increase in MDC staining, while 1, 2, and 8f at 3 μM show approximately 2-fold increase in MDC staining (Figure 9A). Derivative 8l at 3 μM resulted in approximately 3-fold increase in MDC staining. A dose-dependent induction in autophagy by MDC staining was observed with 9a, 9e, 9g, and 9h. Interestingly the inactive compound 9a, included as a negative control, demonstrated a significant effect to enhance autophagy, with a >5-fold increase at 3 μM concentration and >12-fold at 10 μM, suggesting that autophagy modulation alone is not sufficient for the aromatic carbamates to engender neuroprotective effect. Combination of Bcl-2 induction and autophagy activation is required (9a suppressed Bcl-2 expression (Figure 8A,B). Compound 9h (71% protection of cell viability of human iPSC-derived neurons from apoptosis) demonstrated 8-fold increase in autophagy at 3 μM, but comparison to the more protective derivative 8l (79% protection) at 3 μM reveals a 3-fold increase in autophagy. Perhaps suggesting a threshold value of autophagy modulation is required for protective activity that both compounds exceed to provide broadly similar activity, with the greater protective effect of 8l being attributed to slightly greater induction of Bcl-2 (Figure 8A).
Figure 9.
Effect of selected aromatic carbamates to modulate autophagy. (A) Effect of derivatives of 1 to induce autophagy as observed by MDC staining. One-Way ANOVA, Dunnett test, 95% confidence interval. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (B) Effect of aromatic carbamates on beclin 1 expression. Quantified results. n = 2, values represented as mean ± SEM. (C) Representative immunoblot for beclin 1.
A second experiment to determine autophagy modulation was conduted by measuring beclin 1 expression levels in PC12 cells by immunoblotting (Figure 9B,C). Activation of autophagy correlates with increased expression of beclin 1.43 Similar trends were observed to the MDC experiment. A 32% increase in beclin 1 expression was observed upon treatment with 1 at 3 μM. Interestingly, by comparison, aromatic carbamate 9a showed a reduced level of beclin 1 expression (20%) at 3 μM, although this did increase to 104% at 10 μM. A dose-dependent increase of beclin 1 expression was observed with aromatic carbamates 9e, 9g, and 14. The most protective derivative identified (9e), which confers 75% protection of human iPSC-derived neurons at 0.1 μM concentration, increases beclin 1 expression by 18% at this concentration (Figure 9B,C).
Upregulation of antiapoptotic Bcl-2 family proteins or down regulation of pro-apoptotic proteins of the Bcl-2 family has been widely shown in the literature as an effective strategy for neuroprotection. Small molecule inhibitors targeting Bax44 exert a neuroprotective effect.45 Likewise, modulation of autophagy by small molecules such as rapamycin results in a neuroprotective effect in several neurodegenerative diseases, including Parkinson’s disease, where rapamaycin enhances clearance of the pathological protein α-synuclein.46 Rapamycin functions to induce autophagy by binding FK506 binding protein 12 and then inhibiting phosphorylation of mTOR. In addition to inducing autophagy, rapamycin downregulates overall protein synthesis and cell proliferation due to its action on mTOR.47,48 Furthermore, rapamycin has poor BBB penetration, making it unsuitable as a therapeutic agent for neurodegenerative disease, making the identification of novel autophagy inducers an important pursuit.
CONCLUSIONS
The clinically available agent 1 has been known for years to possess moderate neuroprotective activity. However, the low propensity for neuroprotective effect even at high concentration, compounded by the known liver toxicity, precludes the therapeutic use of 1 for neurodegenerative diseases. Herein, we disclose the first detailed neuroprotective SAR of derivatives of 1, identifying a library of aromatic carbamates with enhanced neuroprotective effect. We disclose derivatives of 1 that engender protective activity to ameliorate etoposide- and serum starvation-induced apoptosis in “neuron-like” PC12 and SH-SY5Y cell lines. Further, we have identified a number of aromatic carbamates that demonstrate translational protective activity to ameliorate etoposide-induced apoptosis in human iPSC-derived neurons.
Mechanism of action determination shows that the antioxidant, NMDA antagonistic, and potassium channel opening activity of parent compounds 1 and 2 do not correlate to neuroprotective activity. We identify the mechanism of action of our developed aromatic carbamate derivatives of 1 to be multimodal: inducing Bcl-2, increasing the Bcl-2/Bax ratio to favor an antiapoptotic state, and increasing autophagy. Both of these actions are required to take place to confer neuroprotective effect with aromatic carbamate 9a increasing autophagy by 5-fold at 3 μM yet reducing Bcl-2/Bax ratio by 44%, which results in a compound with little neuroprotective effect in human iPSC-derived neurons above vehicle control. The most active derivative of 1, 4-trifluoromethane (9e), confers 76% cell viability in human iPSC-derived neurons at 0.1 μM concentration. This derivative engenders a 159% increase in the Bcl-2/Bax ratio and an approximate 18% increase in beclin 1 expression, indicating induction of autophagy at the same concentration, with both parameters increasing in intensity in a dose-dependent manner.
The correlation of neuroprotective activity to both induction of Bcl-2 and induction of beclin 1, used herein as a quantification of autophagy, may highlight another potential mechanism of action of the aromatic carbamates as disruptors of the Bcl-2:beclin 1 protein–protein interaction (PPI). The Bcl-2 protein is known to form a complex with beclin 1, where it functions to regulate autophagy by exerting an inhibitory effect.49 Thus, expression levels of Bcl-2 exert multiple influences on proteostasis and cell death in neurodegenerative diseases.40 Further studies to employ aromatic carbamates as chemical probes to investigate the target protein(s) are underway.
EXPERIMENTAL SECTION
General Synthetic Procedures
All reactions were carried out in oven- or flame-dried glassware under positive nitrogen pressure unless otherwise noted. Reaction progress was monitored by thin-layer chromatography (TLC) carried out on silica gel plates (2.5 cm × 7.5 cm, 200 μm thick, 60 F254) and visualized by using UV (254 nm) or by potassium permanganate and/or phosphomolybdic acid solution as indicator. Flash column chromatography was performed with silica gel (40–63 μm, 60 Å) using the mobile phase indicated or on a Teledyne Isco automated instrument (CombiFlash Rf 200 UV/vis). Commercial grade solvents and reagents were purchased from Fisher Scientific (Houston, TX) or Sigma-Aldrich (Milwaukee, WI) and were used without further purification except as indicated. Anhydrous solvents were purchased from Across Organics and stored under an atmosphere of dry nitrogen over molecular sieves.
1H and 13C NMR spectra were recorded in the indicated solvent on a Bruker 400 MHz Avance III HD spectrometer at 400 and 100 MHz for 1Hand 13C, respectively, with TMS as an internal standard. Multiplicities are indicated by s (single), d (doublet), dd (doublet of doublets), t (triplet), q (quartet), m (multiplet), and br (broad). Chemical shifts (δ) are reported in parts per million (ppm) and coupling constants (J), in hertz. High-resolution mass spectroscopy was performed on a LC/MS IT-TOF (Shimadzu) using an ESI source conducted at the University of Texas at Arlington and Shimadzu Center for Advanced Analytical Chemistry. Low-resolution mass spectroscopy was performed on a LC/MS transmission quadrupole analyzer (Varian 1200) using an ESI source. High-pressure liquid chromatography was performed on a Gilson HPLC system with 321 pumps and 155 UV/vis detector using Trilution software v.2.1 with an ACE Equivalence 3 (C18, 3 mm, 4.6mm× 150 mm) column. All samples were determined to possess >95% purity, except where indicated otherwise.
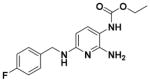
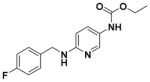
Ethyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)carbamate (1)
To a solution of N6-(4-fluorobenzyl)pyridine-2,3,6-triamine (265 mg, 1.14 mmol,) in 1,4-dioxane (10 mL) was added ethyl chloroformate (123 mg, 1.14 mmol). The reaction was stirred at room temperature overnight protected from light. The resultant precipitate was collected and washed with 1:1 dioxane:diethyl ether (40 mL) and diethyl ether (50 mL) and air-dried at room temperature to afford the title compound as a blue-green solid (82%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 4.8 Hz,CH2CH3), 4.06 (2H, q, J = 7.0 Hz, CH2CH3), 4.53 (2H, br s, CH2Ph), 5.92 (1H, d, J = 8.6 Hz, ArH), 7.20 (2H, t, J = 8.8 Hz, ArH,), 7.34 (1H, br s, NH), 7.43–7.50 (3H, m, ArH), 8.15 (1H, br s, NH), 8.52 (1H, br s, NH,), 13.06 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.93, 44.94, 60.97, 94.23, 107.16, 115.73 (d, J = 21.30 Hz), 130.04 (d, J = 8.21 Hz), 134.19 (d, J = 2.96 Hz), 143.26, 149.48, 155.50, 160.73, 163.14. HPLC: retention time, 5.94 min; purity, 99%. m/z (ESI) −305.1 (100%) [MH+]. HRMS (ESI): C15H17N4O2F requires 305.1408. Found: 305.1413.
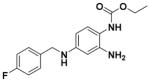
Ethyl (2-Amino-4-((4-fluorobenzyl)amino)phenyl)carbamate (2)
To a solution of benzyl (4-((ethoxycarbonyl)amino)-3-nitrophenyl) (4-fluorobenzyl)carbamate (180 mg, 0.39 mmol) in MeOH (5 mL) was added Pd/C (4 mg, 0.004 mmol) and stirred for 2 h at room temperature under an atmosphere of hydrogen. The reaction mixture was filtered through Celite washing with MeOH (50 mL) and evaporated in vacuo to provide the title compound as a purple solid (quant). 1H NMR (400 MHz, CDCl3) δH: 1.31 (3H, t, J = 7.24 Hz, CH2CH3), 3.61 (2H, br s, NH2), 4.21 (2H, q, J = 7.12 Hz, CH2CH3), 4.27 (2H, br s, CH2Ph), 6.02–6.09 (3H, m), 6.94 (1H, d, J = 8.40 Hz, ArH), 7.04 (2H, t, J = 8.68 Hz, ArH), 7.33 (2H, t, J = 8.61 Hz, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.54, 29.69, 61.58, 105.05, 107.94, 115.40 (d, J = 21.42 Hz), 117.74, 127.45, 129.87 (d, J = 7.50 Hz), 132.85, 139.37, 155.77, 160.99, 163.44. HPLC: retention time, 6.79 min; purity, 99%. m/z (ESI) −305.1 (100%) [MH+]. HRMS (ESI): C16H18N3O2F requires 304.1456. Found: 304.1454.
6-Chloro-3-nitropyridin-2-amine (4)
12To a solution of 2,6-dichloro-3-nitropyridine (3) (1 mol equiv) in MeOH (5 mL) was added a solution of 7N ammonia in MeOH (1 mol equiv), and the reaction was stirred overnight at room temperature. The reaction mixture was diluted with water (20 mL), and the insoluble portion was collected by filtration and crystallized from ethanol to obtain the title compound (based on recovery of starting material: 65%). 1HNMR(400 MHz, CDCl3) δ: 6.77 (1H, d, J = 8.57 Hz, ArH); 8.25 (2H, br s, NH2), 8.39 (1H, J = 8.56 Hz, ArH). 13C NMR (100 MHz, DMSO-d6) δ: 112.44, 126.57, 138.85, 153.92, 155.44. m/z (ESI) −174.1 (100%) [MH+]: 176.0 (40%).
General Synthetic Procedure for 6-Amino-2-benzylamino-5-nitropyridine Intermediates (5)
To a suspension of 6-chloro-3-nitropyridin-2-amine (4) (1.73 mmol) in IPA (5 mL) was added appropriately substituted benzylamine (2.07 mmol) and triethylamine (2.50 mmol) and the mixture refluxed for 3 h. The reaction mixture was allowed to cool to room temperature, and water (30 mL) was added slowly with stirring to provide a yellow solid. Extraction was performed using ethyl acetate (3 × 20 mL) and dried (Na2SO4), filtered, and evaporated in vacuo. Purification by column chromatography provided the title compound.
General Synthetic Procedure for Triaminopyridine Intermediate (6) Method A
SnCl2·2H2O (4.56 mmol) was dissolved in conc HCl (10 mL) and heated at 70 °C for 30 min until a colorless solution was formed. 6-Amino-2-benzylamino-5-nitropyridine intermediate (5) (1.14 mmol) was added portionwise and the solution heated at 70 °C for 3 h with a condenser equipped with subba seal and empty balloon. Water was added, and the orange solution was allowed to cool to room temperature. The solution was basified to pH 12 (2 M NaOH) and extracted with CH2Cl2 (3 × 20 mL), and the organic layer was washed with brine (50 mL), dried (Na2SO4), filtered, and evaporated in vacuo to provide the title compound. The title compound was taken forward without further purification.
General Synthetic Procedure for Triaminopyridine Intermediate (6) Method B
To a solution of 6-amino-2-benzylamino-5-nitropyridine intermediate (5) (1 mol equiv) in 1,4-dioxane (10 mL per mol equiv) was added Raney nickel (Al–Ni 50:50 wt %) (10× weight of 5) and hydrazine hydrate (10× volume of 5) and the suspension allowed to stir at room temperature overnight. The suspension was filtered through Celite washing with CH2Cl2 (100 mL) and evaporated in vacuo to provide the title compound. The title compound was taken forward without further purification.
General Synthetic Procedure for Pyridine Carbamate Derivatives (7)
To a solution of triaminopyridine (6) (1.14 mmol) in 1,4-dioxane (10 mL) was added the respective chloroformate (1.14 mmol). The reaction was stirred at room temperature overnight protected from light. The resultant precipitate was collected and washed with 1:1 dioxane:diethyl ether (40 mL) and diethyl ether (50 mL) and air-dried at room temperature to afford the title compound.
Methyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8a)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid. (85%). 1H NMR (400 MHz, DMSO-d6) δH: 3.61 (3H, s, CH3), 4.52 (2H, d, J = 5.16 Hz, CH2Ph), 5.91 (1H, d, J = 8.77 Hz, ArH), 7.21 (2H, t, J = 8.93 Hz, ArH), 7.34 (1H, br s), 7.37–7.47 (3H, m, ArH), 8.17 (1H, br s), 8.57 (1H, br s, NH), 13.03 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.95, 52.40, 94.10, 107.10, 115.77 (d, J = 21.32 Hz), 130.01 (d, J = 8.16 Hz), 134.19 (d, J = 2.80 Hz), 134.21, 143.42, 149.68, 155.96, 160.74, 163.15. 19F NMR (376 MHz, DMSO-d6) δF: −115.20. HPLC: retention time, 6.08 min; purity, 99%. m/z (ESI) −291.1 (100%) [MH+].
Propyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)-carbamate (8b)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (98%). 1H NMR (400 MHz, DMSO-d6) δH: 0.91 (3H, br s, CH2CH3), 1.59 (2H, br s, CH2), 4.00 (2H, t, J = 6.43 Hz, CH2Ph), 4.56 (2H, m, CH2), 5.97 (1H, t, J = 8.45 Hz, ArH), 7.19 (2H, t, J = 8.89 Hz, ArH), 7.42–7.52 (4H, m), 8.13 (1H, br s, NH), 8.44 (1H, br s, NH), 8.58 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 10.74, 22.31, 44.93, 66.52, 94.29, 95.49, 107.19, 115.73 (d, J = 21.32 Hz), 130.06 (d, J = 6.74 Hz), 134.22 (d, J = 2.99 Hz), 149.49, 160.73, 163.14. 19F NMR (376 MHz, DMSO-d6) δF: −115.23. HPLC: retention time, 5.18 min; purity, 99%. m/z (ESI) −319.6 (100%) [MH+].
Isobutyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8c)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (34%). 1H NMR (400 MHz, DMSO-d6) δH: 0.92 (6H, m,CH3), 1.88 (1H, br s, CH), 3.79 (2H, d, J = 6.35 Hz, CH2), 4.53 (2H, d, J= 5.11 Hz, CH2Ph), 5.92 (1H, d, J = 8.88 Hz, ArH), 7.20 (2H, t, J = 8.98 Hz, ArH), 7.36 (1H, br s, NH), 7.43–7.49 (3H, m), 8.19 (1H, br s, NH), 8.54 (1H, br s NH), 13.12 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 19.42, 27.98, 44.94, 66.81, 70.95, 94.09, 107.29, 115.77 (d, J = 21.30 Hz), 129.99 (d, J = 8.35 Hz), 134.20 (d, J = 2.53 Hz), 149.57, 160.73, 163.14. 19F NMR (376 MHz, DMSO-d6) δF: −115.23. HPLC: retention time, 5.42 min; purity, 99%. m/z (ESI) −333.1 (100%) [MNa+].
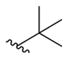
tert-Butyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8d)
According to the general procedure for pyridine carbamates, the title compound was obtained as a yellow-brown solid (25%). Rf = 0.24 (mobile phase 1:1 = hexane/EtOAc). 1H NMR (400 MHz, DMSO-d6) δH: 1.42 (9H, s, CH3), 4.35 (2H, d, J = 6.11 Hz, CH2Ph), 5.13 (2H, br s, NH), 5.68 (1H, d. J = 8.53 Hz, ArH), 6.50 (1H, t, J = 6.53 Hz, NH), 7.01 (1H, d, J = 7.35 Hz, ArH), 7.11 (2H, t, J = 8.97 Hz, ArH), 7.35 (2H, t, J = 6.67 Hz, ArH), 7.96 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 28.65, 44.10, 66.81, 78.71, 95.71, 107.48, 115.19 (d, J = 23.64 Hz), 129.49 (d, J = 8.02 Hz), 137.75 (d, J = 3.03 Hz), 153.25, 154.73, 155.82, 160.20, 162.60. HPLC: retention time, 5.82 min; purity, 99%. m/z (ESI) −333.1 (100%) [MH+].
Hexyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)-carbamate (8e)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (61%). 1HNMR(400 MHz, DMSO-d6) δH: 0.87 (3H, br s,CH3), 1.25–1.34 (6H, m, CH2), 1.58 (2H, br s, CH2), 4.00 (2H, q, J = 5.89 Hz, CH2CH3), 4.54 (2H, d, J = 4.72 Hz, CH2Ph), 5.92 (1H, d, J = 8.80 Hz, ArH), 7.20 (2H, t, J = 8.90 Hz, ArH), 7.38–7.47 (4H, m), 8.24 (1H, br s, NH), 8.53 (1H, br s, NH), 13.22 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.36, 22.49, 25.50, 28.90, 31.39, 44.93, 65.06, 94.11, 107.28, 115.77 (d, J = 21.32 Hz), 130.00 (d, J = 8.19 Hz), 134.16 (d, J = 3.18 Hz), 149.63, 155.59, 160.73, 163.15. 19F NMR (376 MHz, DMSO-d6) δF: −115.23. HPLC: retention time, 8.04 min; purity, 98%. m/z (ESI) −361.8 (100%) [MH+].
Allyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8f)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (78%). 1H NMR (400 MHz, DMSO-d6) δH: 4.53 (4H, m), 5.15–5.39 (2H, m), 5.91–5.98 (2H, m), 7.20 (2H, t, J = 8.31 Hz, ArH), 7.38–7.48 (4H, m), 8.21 (1H, br s, NH), 8.65 (1H, br s, NH), 13.15 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.94, 65.57, 91.40, 94.09, 107.08, 115.77 (d, J = 21.31 Hz), 117.97, 129.99 (d, J = 8.05 Hz), 133.71, 134.19 (d, J = 2.22 Hz), 155.25, 160.73, 163.15. 19F NMR (376 MHz, DMSO-d6) δF: −115.22. HPLC: retention time, 6.94 min; purity, 99%. m/z (ESI) −317.1 (100%) [MH+], 318.2 (20%). HRMS (ESI): C16H17N4O2F requires 317.1408. Found: 317.1411.
Prop-2-yn-1-yl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)- carbamate (8g)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (68%). 1H NMR (400 MHz, DMSO-d6) δH: 4.53 (2H, d, J = 4.00 Hz, CH2Ph), 4.69 (2H, s, CH2), 5.92 (1H, d, J = 8.85 Hz, ArH), 7.20 (2H, t, J = 8.90 Hz, ArH), 7.37–7.49 (4H, m), 8.21 (1H, br s, NH), 8.78 (1H, br s, NH), 13.13 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.94, 52.85, 78.13, 79.33, 94.12, 106.75, 115.78 (d, J = 21.48 Hz), 129.98 (d, J = 8.09 Hz), 134.18 (d, J = 3.46 Hz), 149.74, 154.70, 160.74, 163.16. 19F NMR (376 MHz, DMSO-d6) δF: −115.21. HPLC: retention time, 5.99 min; purity, 99%. m/z (ESI) −315.3 (100%) [MH+].
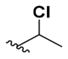
1-Chloroethyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8h)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (58%). 1H NMR (400 MHz, DMSO-d6) δH: 1.79 (3H, d, J = 5.71 Hz, CH3), 4.7 (2H, br s, CH2Ph), 5.96 (1H, d, J = 8.88 Hz, ArH), 6.57 (1H, q, J = 7.72 Hz, CH), 7.20 (2H, t, J = 8.89 Hz, ArH), 7.45–7.55 (3H, m), 8.43 (1H, br s, NH), 9.09 (1H, s, NH), 13.35 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 25.89, 44.96, 66.81, 84.37, 105.90, 115.77 (d, J = 21.46 Hz), 124.65, 130.12 (d, J = 8.06 Hz), 134.04 (d, J = 2.90 Hz), 143.44, 149.41, 160.76. m/z (ESI) −233.1 (100%) [fragmentation of carbamate group], 234.2 (15%), 339.1 (100%) [MH+], 341.0 (10%). HPLC: retention time, 6.12 min; purity, 99%.
2-Chloroethyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8i)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (3%). Rf = 0.72 (mobile phase 10:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, DMSO-d6) δH: 3.84 (2H, br s, CH2), 4.28 (2H, br s, CH2), 4.53 (2H, br s, CH2), 5.93 (1H, d, J = 8.72 Hz, ArH), 7.20 (2H, t, J = 8.32 Hz, ArH), 7.34 (1H, br s, NH), 7.43–7.49 (3H, m), 8.16 (1H, br s, NH), 8.74 (1H, s, NH), 13.05 (1H, br s, NH). 13C NMR (100 MHz, CDCl3) δC: 42.17, 45.71, 65.05, 96.16, 107.02, 115.39 (d, J = 21.31 Hz), 128.87 (d, J = 8.01 Hz), 134.87 (d, J = 3.02 Hz), 137.52, 153.46, 156.39, 160.78, 163.21. HPLC: retention time, 6.66 min; purity, 99%. m/z (ESI) −339.1 (100%) [MH+], 340.1 (25%), 341.1 (35%).
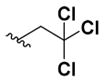
2,2,2-Trichloroethyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)carbamate (8j)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (63%). 1H NMR (400 MHz, DMSO-d6) δH: 4.57 (2H, d, J = 3.05 Hz, CH2Ph), 4.85 (2H, s, CH2), 5.95 (1H, d, J = 8.84 Hz, ArH), 7.20 (2H, t, J = 8.85 Hz, ArH), 7.42–7.50 (4H, m), 8.36 (1H, br s, NH), 9.12 (1H, br s, NH), 13.33 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.94, 66.81, 74.45, 94.36, 96.23, 106.24, 115.77 (d, J = 21.32 Hz), 129.99 (d, J = 8.19 Hz), 134.17 (d, J = 3.18 Hz), 149.60, 153.97, 160.74, 163.20. 19F NMR (376 MHz, DMSO-d6) δF: −115.19. HPLC: retention time, 6.43 min; purity, 99%. m/z (ESI) −408.9 (100%) [MH+], 409.8 (70%), 410.7 (25%).
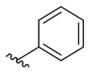
Phenyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)- carbamate (8k)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (37%). 1H NMR(400 MHz, DMSO-d6) δH: 4.38 (2H, d, J = 6.20 Hz, CH2Ph), 5.39 (2H, br s, NH2), 5.71 (1H, d, J = 8.31 Hz, ArH), 6.61 (1H, t, J = 6.30 Hz, NH), 7.08–7.24 (6H, m, ArH), 7.34–7.42 (4H, m, ArH), 8.78 (1H, br s, NH). 13C NMR (100 MHz, CDCl3) δC: 45.72, 96.39, 107.10, 115.41 (d, J = 21.34 Hz), 121.51, 125.63, 128.89 (d, J = 8.01 Hz), 129.37, 134.84 (d, J = 2.84 Hz), 137.30, 150.80, 153.14, 156.31, 160.80, 163.24. 19F NMR (376 MHz, DMSO-d6) δF: −115.62. HPLC: retention time, 6.63 min; purity, 99%. m/z (ESI) −353.4 (100%) [MH+].
Benzyl 6-(4-Fluorobenzylamino)-2-aminopyridin-3-ylcarbamate (8l)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (58%). 1H NMR (400 MHz, DMSO-d6) δH: 4.54 (2H, br s, CH2Ph), 5.09 (2H, br s, CH2Ph), 5.93 (1H, d. J = 8.74 Hz, ArH), 7.20(2H, t, J = 8.61 Hz, ArH), 7.34–7.51 (9H, m), 8.22 (1H, br s, NH), 8.70 (1H, br s, NH)), 13.18 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.96, 66.82, 94.20, 107.08, 115.76 (d, J = 21.32 Hz), 128.44, 128.84, 130.02 (d, J = 8.17 Hz), 134.19 (d, J = 2.50 Hz), 143.23, 149.63, 155.42, 160.74, 163.16. 19F NMR (376 MHz, DMSO-d6) δF: −115.22. HPLC: retention time, 6.10 min; purity, 99%. m/z (ESI) −367.1 (100%) [MH+], 368.2 (25%). HRMS (ESI): C20H19N4O2F requires 367.1565. Found: 367.1562.
(9H-Fluoren-9-yl)methyl (2-Amino-6-((4-fluorobenzyl)amino)- pyridin-3-yl)carbamate (8m)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (13%). Rf = 0.43 (mobile phase 20:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, DMSO-d6) δH: 4.22–4.38 (5H, m), 5.21 (2H, br s, NH2), 5.69 (1H, d, J = 8.21 Hz, ArH), 6.57 (1H, t, J = 6.20 Hz, NH), 7.00 (1H, d, J = 7.93 Hz, ArH), 7.11 (2H, t, J = 8.90 Hz, ArH), 7.34–7.45 (6H, m, ArH), 7.75–7.92 (4H, m, ArH), 8.41 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.10, 47.20, 66.09, 95.71, 115.23 (d, J = 21.23 Hz), 120.50, 120.59, 121.85, 125.77, 127.53, 127.75, 128.10, 129.50 (d, J = 8.02 Hz), 137.70, 137.73, 137.88, 139.87, 141.20, 144.32, 160.22, 162.62. HPLC: retention time, 7.25 min; purity, 80%. m/z (ESI) −455.3 (100%) [MH+].
Ethyl (2-Amino-6-((4-chlorobenzyl)amino)pyridin-3-yl)- carbamate (9a)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (65%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, br s, CH2CH3), 4.05 (2H, q, J = 6.50 Hz, CH2CH3), 4.55 (2H, br s, CH2Ph), 5.90 (1H, d, J = 8.73 Hz, ArH), 7.36–7.47 (6H, m), 8.22 (1H, br s, NH), 8.52 (1H, br s, NH), 13.25 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.93, 44.93, 60.97, 66.81, 94.04, 107.38, 128.94, 129.72, 132.39, 137.17, 149.65, 155.47. HPLC: retention time, 7.46 min; purity, 99%. m/z (ESI) −321.1 (100%) [MH+], 323.1 (30%).
Ethyl (2-Amino-6-(benzylamino)pyridin-3-yl)carbamate (9b)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (97%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21(3H, t, J = 6.68 Hz, CH2CH3), 4.05 (2H, q, J = 7.08 Hz, CH2CH3), 4.59 (2H, d, J = 3.04 Hz, CH2Ph), 5.94 (1H, d, J = 8.85 Hz, ArH), 7.29 (1H, m, ArH), 7.34–7.56 (6H, m), 8.40 (1H, br s, NH), 8.57 (1H, br s, NH), 13.45 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.94, 24.85, 45.72, 60.97, 94.21, 107.06, 127.94, 128.99, 137.94, 143.33, 149.59, 155.50. HPLC: retention time, 6.65 min; purity, 99%. m/z (ESI) −287.2 (100%) [MH+].
Ethyl (2-Amino-6-((2-(trifluoromethyl)benzyl)amino)pyridin-3-yl)- carbamate (9c)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (38%). 1H NMR (400 MHz, DMSO-d6) δH: 1.22 (3H, t, J = 7.94 Hz, CH2CH3), 4.06 (2H, q, J = 7.08 Hz, CH2CH3), 4.63 (2H, br s, CH2Ph), 5.79 (1H, d, J = 8.50 Hz, ArH), 7.44 (1H, br s, NH), 7.54 (1H, t, J = 7.78 Hz, ArH), 7.60 (1H, d, J = 7.83 Hz, ArH), 7.70 (1H, t, J = 7.72 Hz, ArH), 7.79 (1H, d, J = 7.98 Hz, ArH), 8.52 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.94, 43.10, 60.99, 93.39, 107.84, 124.81 (q, J = 274.19 Hz), 126.18 (q, J = 44.28 Hz), 127.11 (d, J = 30.09 Hz), 128.61, 129.60, 133.45, 136.02, 143.25, 149.73, 155.48. HPLC: retention time, 6.52 min; purity, 99%. HRMS (ESI): C16H17N4O2F3 requires 355.1376. Found: 355.1367.
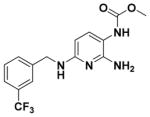
Ethyl 6-(3-(Trifluoromethyl)benzylamino)-2-aminopyridin-3-ylcarbamate (9d)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (32%). Rf = 0.56 (mobile phase 10:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, DMSO-d6) δH: 1.20 (3H, br s, CH2CH3), 4.03 (2H, q, J = 6.79 Hz, CH2CH3), 4.48 (2H, d, J = 5.45 Hz, CH2Ph), 5.36 (1H, br s, NH), 5.73 (1H, d, J = 8.34 Hz, ArH), 6.75 (1H, br s, NH), 7.06 (1H, br s, NH), 7.52–7.58 (2H, m, ArH), 7.63–7.68 (2H, m, ArH), 8.22 (1H, s, ArH). 13C NMR (100 MHz, DMSO-d6) δC: 15.04, 44.38, 60.44, 95.51, 107.31, 123.63 (q, J = 3.78 Hz), 124.02 (q, J = 3.76 Hz), 124.81 (q, J = 272.19 Hz), 129.36 (q, J = 31.46 Hz), 129.59, 131.81, 136.56, 143.08, 153.26, 155.56. HPLC: retention time, 5.98 min; purity, 99%. m/z (ESI) −355.1 (100%) [MH+].
Ethyl (2-Amino-6-((4-(trifluoromethyl)benzyl)amino)pyridin-3-yl)- carbamate (9e)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (53%). 1H NMR (400 MHz, DMSO-d6) δH: 1.22 (3H, t, J = 6.26 Hz, CH2CH3), 4.05 (2H, q, J = 6.98 Hz, CH2CH3), 4.67 (2H, d, J = 4.25 Hz, CH2Ph), 5.91 (1H, d, J = 8.80 Hz, ArH), 7.42–7.49 (2H, m), 7.61 (2H, t, J = 8.09 Hz, ArH), 7.74 (2H, t, J = 8.13 Hz, ArH), 8.37 (1H, br s, NH), 8.56 (1H, br s, NH), 13.43 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.93, 45.14, 60.99, 93.97, 107.54, 124.72 (q, J = 272.19 Hz), 125.84 (q, J = 3.86 Hz), 125.81, 128.40 (d, J = 31.55 Hz), 128.46, 143.13, 149.62, 155.49. 19F NMR (376 MHz, DMSO-d6) δF: −60.87. HPLC: retention time, 7.61 min; purity, 99%. HRMS (ESI): C16H17N4O2F3 requires 355.1376. Found: 355.1377.
Ethyl (2-Amino-6-(((6-(trifluoromethyl)pyridin-3-yl)methyl)- amino)pyridin-3-yl)carbamate (9f)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (84%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 6.43 Hz, CH2CH3), 4.06 (2H, q, J = 6.98 Hz, CH2CH3), 4.75 (2H, br s, CH2Ph), 5.97 (1H, d, J = 8.80 Hz, ArH), 7.42–7.56 (2H, m), 7.92 (1H, d, J = 8.07 Hz, ArH), 8.09 (1H, d, J = 7.11 Hz, ArH), 8.41 (1H, br s, NH), 8.58 (1H, br s, NH), 8.83 (1H, s, ArH), 13.44 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.92, 42.96, 61.01, 93.96, 107.87, 121.09 (q, J = 2.68 Hz), 122.19 (q, J = 273.69 Hz), 137.49, 138.02, 143.24, 145.88 (d, J = 33.71 Hz), 149.49 (d, J = 35.81 Hz), 149.92, 155.46. HPLC: retention time, 6.09 min; purity, 99%. m/z (ESI) −356.8 (100%) [MH+].
Ethyl (2-Amino-6-((4-(trifluoromethoxy)benzyl)amino)pyridin-3-yl)carbamate (9g)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (68%). 1H NMR (400 MHz, DMSO-d6) δH: 1.22 (3H, t, J = 7.94 Hz, CH2CH3), 4.06 (2H, q, J = 7.28 Hz, CH2CH3), 4.58 (2H, d, J = 4.00 Hz, CH2Ph), 5.91 (1H, d, J = 8.76 Hz, ArH), 7.30–7.53 (6H, m), 8.15 (1H, br s, NH), 8.53 (1H, br s, NH), 13.04 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.93, 44.91, 60.98, 94.01, 107.39, 120.53 (q, J = 256.88 Hz), 121.63, 129.77, 137.62, 143.26, 148.02, 149.63, 155.48. HPLC: retention time, 7.45 min; purity, 99%. HRMS (ESI): C16H17N4O3F3 requires 371.1326. Found: 371.1329.
Ethyl (2-Amino-6-((4-((trifluoromethyl)thio)benzyl)amino)- pyridin-3-yl)carbamate (9h)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (45%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 6.93 Hz, CH2CH3), 4.05 (2H, q, J = 7.11 Hz, CH2CH3), 4.66 (2H, d, J = 4.88 Hz, CH2Ph), 5.92 (1H, d, J = 8.81 Hz, ArH), 7.38–7.49 (2H, m, ArH, NH), 7.33 (2H, d, J = 8.23 Hz, ArH), 7.55 (2H, d, J = 8.13 Hz, ArH), 8.30 (1H, br s, NH), 8.55 (1H, br s, NH), 13.30 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.93, 45.10, 60.99, 93.92, 107.53, 122.19, 122.65, 129.26, 130.08 (q, J = 307.86 Hz), 136.91, 142.13, 149.64, 155.49. HPLC: retention time, 7.89 min; purity, 99%. HRMS (ESI): C16H17N4O2F3S requires 387.1097. Found: 387.1090.
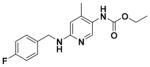
Ethyl (2-Amino-6-((4-methylbenzyl)amino)pyridin-3-yl)- carbamate (9i)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (8%). 1H NMR (400 MHz, MeOD-d4) δH: 1.29 (3H, br s, CH2CH3), 4.16 (2H, q, J = 7.13 Hz, CH2CH3), 7.10–7.16 (4H, m, ArH), 7.27 (1H, s, ArH). 13C NMR (100 MHz, DMSO-d6) δC: 14.90, 21.07, 33.16, 61.00, 66.80, 107.08, 107.19, 128.67, 129.49, 135.79, 136.31, 148.59. HPLC: retention time, 6.00 min; purity, 99%. HRMS (ESI): C16H20N4O2 requires 301.1659. Found: 301.1644.
Ethyl (2-Amino-6-((3-methylbenzyl)amino)pyridin-3-yl)- carbamate (9j)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (19%). Rf = 0.44 (mobile phase 10:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, CDCl3) δH: 1.26 (3H, br s,CH2CH3), 2.33 (3H, s,CH3), 4.15 (2H, q, J = 7.08 Hz, CH2CH3), 4.31 (2H, br s, CH2Ph), 4.92 (2H, br s, NH2), 5.38 (1H, br s, NH), 5.70 (1H, d, J = 8.45 Hz, ArH), 6.86 (1H, br s, NH), 7.06–7.23 (5H, m, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.57, 21.42, 46.42, 61.56, 95.19, 107.52, 124.27, 127.92, 128.00, 128.51, 138.25, 138.62, 152.48, 154.99, 155.86. HPLC: retention time, 7.09 min; purity, 99%. HRMS (ESI): C16H20N4O2 requires 301.1659. Found: 301.1652.
Ethyl (2-Amino-6-((4-methoxybenzyl)amino)pyridin-3-yl)- carbamate (9k)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (56%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 7.05 Hz, CH2CH3), 3.74 (3H, s, CH3), 4.05 (2H, q, J = 7.12 Hz, CH2CH3), 4.47 (2H, d, J = 4.91 Hz, CH2Ph), 5.92 (1H, d, J = 8.78 Hz, ArH), 6.93 (2H, d, J = 8.72 Hz, ArH), 7.32–7.47 (4H, m), 8.72 (1H, br s, NH), 8.53 (1H, br s, NH), 13.06 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.94, 45.21, 55.55, 60.98, 66.81, 94.18, 106.96, 114.40, 129.35, 129.69, 149.67, 155.54, 159.09. HPLC: retention time, 6.47 min; purity, 99%. m/z (ESI) −317.3 (100%) [MH+].
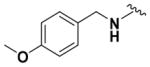
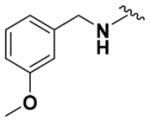
Ethyl (2-Amino-6-((3-methoxybenzyl)amino)pyridin-3-yl)- carbamate (9l)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (54%). 1H NMR (400 MHz, CDCl3) δH: 1.30 (3H, t, J = 6.64 Hz, CH2CH3), 1.77 (3H, s, CH3), 4.21 (2H, q, J = 7.10 Hz, CH2CH3), 4.44–4.48 (4H, m), 4.78 (1H, t, J = 6.21Hz,NH), 5.78 (1H, d, J =8.32Hz, ArH), 6.00 (1H, br s, NH), 7.11–7.17 (2H, m, ArH), 7.21 (1H, s, ArH), 7.28 (1H, ArH), 7.36 (1H, t, J = 7.83, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.92, 44.99, 60.99, 93.96, 107.53, 119.24, 120.29, 120.36, 121.79, 126.91, 130.98, 141.10, 148.97, 149.62, 155.48. HPLC: retention time, 6.23 min; purity, 99%. m/z (ESI) −317.3 (100%) [MH+].
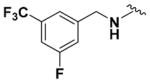
Ethyl (2-Amino-6-((3-fluoro-5-(trifluoromethyl)benzyl)amino)- pyridin-3-yl)carbamate (9m)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (65%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, br s, CH2CH3), 4.06 (2H, q, J = 6.80 Hz, CH2CH3), 4.68 (2H, br s, CH2Ph), 5.94 (1H, d, J = 8.76 Hz, ArH), 7.41–7.51 (2H, m), 7.60–7.68 (3H, m), 8.31 (1H, br s, NH), 8.56 (1H, br s, NH), 13.35 (1H, br s, NH). 13CNMR(100 MHz, DMSO-d6) δC: 14.92, 44.71, 60.99, 93.91, 107.85, 112.22 (d, J = 28.48 Hz), 119.08 (d, J = 23.84 Hz), 120.83 (q, J = 3.22 Hz), 121.05 (q, J = 274.68 Hz), 125.18, 131.29, 143.31, 149.72, 161.24, 163.69. 19F NMR (376 MHz, DMSO-d6) δF: −61.15, −110.51. HPLC: retention time, 6.28 min; purity, 99%. m/z (ESI) −373.2 (100%) [MH+].
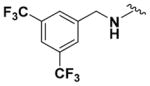
Ethyl (2-Amino-6-((3,5-bis(trifluoromethyl)benzyl)amino)pyridin-3-yl)carbamate (9n)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (22%). Rf = 0.30 (mobile phase 1:1 = hexane/EtOAc). 1H NMR (400 MHz, DMSO-d6) δH: 1.20 (3H, t, J = 6.72 Hz, CH2CH3), 4.03 (2H, q, J = 7.14 Hz, CH2CH3), 4.55 (2H, d, J = 6.30 Hz, CH2Ph), 5.25 (2H, br s, NH2), 5.73 (1H, d, J = 8.27 Hz, ArH), 6.78 (1H, t, J = 6.52 Hz, NH), 7.05 (1H, d, J = 7.97 Hz, ArH), 7.94 (1H, s, ArH), 8.02 (2H, s, ArH), 8.21 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 15.06, 44.03, 60.41, 95.65, 107.44, 120.68 (q, J = 3.97 Hz), 123.93 (q, J = 272.82 Hz), 128.47 (q, J = 3.21 Hz), 130.39 (d, J = 32.41 Hz), 136.18, 139.41. 145.66, 155.53. 19F NMR (376 MHz, DMSO-d6) δF: −61.23. HPLC: retention time, 5.19 min; purity, 99%. m/z (ESI) −423.1 (75%) [MH+], 445.1 (100%) [MNa+], 446.2 (20%).
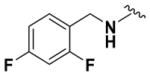
Ethyl (2-Amino-6-((2,4-difluorobenzyl)amino)pyridin-3-yl)- carbamate (9o)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (24%). Rf = 0.39 (mobile phase 1:1 = hexane/EtOAc). 1H NMR (400 MHz, CDCl3) δH: 1.26 (3H, br s, CH2CH3), 4.16 (2H, q, J = 7.09 Hz, CH2CH3), 4.38 (2H, br s, CH2Ph), 4.80 (2H, br s, NH2), 5.16 (1H, br s, NH), 5.72 (1H, d, J = 8.38 Hz, ArH), 6.58 (1H, br s, NH), 6.78 (2H, t, J = 8.43 Hz, ArH), 7.14 (1H, d, J = 7.16 Hz, ArH), 7.28 (1H, q, J = 8.10 Hz, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.54, 39.34, 61.54, 95.82, 103.66 (t, J = 25.24 Hz), 107.68, 111.28 (dd, J = 17.45, 3.66 Hz), 122.17 (dd, J = 11.09, 3.71 Hz), 130.16 (q, J = 3.36 Hz), 137.70, 153.34, 155.72, 160.13 (dd, J = 130.667, 11.84 Hz), 162.60 (dd, J = 129.95, 11.81 Hz). 19F NMR (376 MHz, DMSO-d6) δF: −111.89, −115.06. HPLC: retention time, 6.74 min; purity, 99%. m/z (ESI) −323.3 (100%) [MH+].
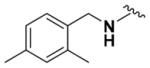
Ethyl (2-Amino-6-((2,4-dimethylbenzyl)amino)pyridin-3-yl)- carbamate (9p)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (33%). Rf = 0.36 (mobile phase 20:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, CDCl3) δH: 1.29 (3H, t, J = 6.29 Hz, CH2CH3), 2.32 (6H, s, CH3), 4.20 (2H, q, J = 7.11 Hz, CH2CH3), 4.33 (2H, d, J = 4.84 Hz, CH2Ph), 4.90 (2H, br s, NH2), 4.97 (1H, t, J = 5.15 Hz, NH), 5.77 (1H, d, J = 8.44 Hz, ArH), 6.25 (1H, s, ArH), 6.98–7.02 (2H, m, ArH), 7.18 (1H, d, J = 7.72 Hz, ArH), 7.22 (1H, br s, NH). 13C NMR (100 MHz, CDCl3) δC: 14.58, 18.84, 20.98, 44.46, 61.62, 95.66, 107.27, 126.73, 128.01, 131.28, 133.17, 136.02, 137.13, 138.31, 152.75, 155.44. HPLC: retention time, 6.11 min; purity, 99%. m/z (ESI) −315.3 (100%) [MH+].
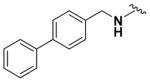
Ethyl (6-(([1,1′-Biphenyl]-4-ylmethyl)amino)-2-aminopyridin-3-yl)carbamate (9q)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (33%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 6.57 Hz, CH2CH3), 4.05 (2H, q, J = 6.71 Hz, CH2CH3), 4.61 (2H, d, J = 4.40 Hz, CH2Ph), 5.95 (1H, d, J = 8.83 Hz, ArH), 7.35–7.50 (7H, m, ArH, NH), 7.65–7.68 (4H, m, ArH), 8.29 (1H, br s, NH), 8.54 (1H, br s, NH), 13.27 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 19.94, 45.35, 60.97, 94.19, 107.19, 127.09, 127.31, 127.91, 128.48, 129.39, 137.30, 139.76, 140.25, 143.21, 149.85, 155.52. HPLC: retention time, 5.38 min; purity, 99%. m/z (ESI) −363.8 (100%) [MH+].
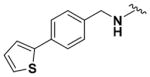
Ethyl (2-Amino-6-((4-(thiophen-2-yl)benzyl)amino)pyridin-3-yl)- carbamate (9r)
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (22%). 1H NMR (400 MHz, DMSO-d6) δH: 1.30 (3H, t, J = 7.34 Hz, CH2CH3), 4.15 (2H, q, J = 7.06 Hz, CH2CH3), 4.44 (2H, s, CH2Ph), 5.82 (1H, d, J = 8.35 Hz, ArH), 7.07–7.13 (2H, m, ArH), 7.34–7.44 (4H, m, ArH), 7.58 (2H, d, J = 8.30 Hz, ArH). 13C NMR (100 MHz, DMSO-d6) δC: 13.53, 44.95, 60.81, 95.82, 106.84, 122.55, 124.12, 125.33, 127.47, 127.64,132.97, 137.15,139.62, 143.97, 156.72, 156.86. HPLC: retention time, 6.41 min; purity, 99%. m/z (ESI) −369.6 (100%) [MH+].
Ethyl (2-Amino-6-((naphthalen-2-ylmethyl)amino)pyridin-3-yl)- carbamate (9s)
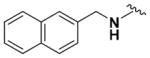
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (53%). Rf = 0.58 (mobile phase 1:1 = hexane/EtOAc). 1H NMR (400 MHz, MeOD-d4) δH: 1.28 (3H, t, J = 6.80 Hz, CH2CH3), 4.15 (2H, q, J = 7.07 Hz, CH2CH3), 4.84 (2H, s, CH2Ph), 5.82 (1H, d, J = 8.36 Hz, ArH), 7.12 (1H, d, J = 8.15 Hz, ArH), 7.38 (1H, t, J = 8.11 Hz, ArH), 7.45–7.51 (3H, m, ArH), 7.76 (1H, d, J = 8.19 Hz, ArH), 7.84–7.88 (1H, m, ArH), 8.05–8.09 (1H, m ArH). 13C NMR (100 MHz, CDCl3) δC: 14.62, 44.51, 61.50, 96.34, 107.38, 123.46, 125.50, 125.80, 126.31, 128.04, 128.74, 131.42, 133.80, 134.28, 137.34, 153.68, 155.58, 156.50. HPLC: retention time, 6.22 min; purity, 99%. m/z (ESI) −337.1 (100%) [MH+].
Ethyl (2-Amino-6-((4-fluorophenethyl)amino)pyridin-3-yl)- carbamate (9t)
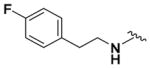
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (33%). 1H NMR (400 MHz, DMSO-d6) δH: 1.22 (3H, t, J = 6.78 Hz, CH2CH3), 2.86 (2H, t, J = 7.36 Hz, CH2), 3.56 (2H, q, J = 7.10 Hz, CH2), 4.06 (2H, q, J = 7.00 Hz, CH2CH3), 5.97 (1H, d, J = 8.79 Hz, ArH), 7.13 (2H, t, J = 8.93 Hz, ArH), 7.35–7.48 (4H, m, ArH, NH), 7.92 (1H, br s, NH), 8.53 (1H, br s, NH), 12.98 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.95, 34.00, 43.73, 60.96, 66.81, 94.27, 106.50, 115.47 (d, J = 20.77 Hz), 131.18 (d, J = 7.95 Hz), 135.19, 149.76 160.23, 162.63. HPLC: retention time, 5.62 min; purity, 99%. m/z (ESI) −319.6 (100%) [MH+].
Ethyl (2-Amino-6-(phenethylamino)pyridin-3-yl)carbamate (9u)
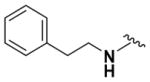
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue solid (15%). 1H NMR (400 MHz, CDCl3) δH: 1.28 (3H, t, J = 6.73 Hz, CH2CH3), 2.94 (2H, t, J = 7.32 Hz, CH2), 3.42 (2H, t, J = 7.33 Hz, CH2Ph), 4.19 (2H, q, J = 6.96 Hz, CH2CH3), 5.68 (1H, d, J = 8.90 Hz, ArH), 6.70 (2H, br s, NH2), 7.23–7.26 (3H, m, ArH), 7.31–7.44 (4H, m, ArH, NH), 13.76 (1H, br s, NH). 13C NMR (100 MHz, CDCl3) δC: 14.52, 35.00, 44.37, 62.09, 126.92, 128.77, 128.83, 137.71, 155.55. HPLC: retention time, 6.57 min; purity, 99%. m/z (ESI) −301.3 (100%) [MH+].
Ethyl (2-Amino-6-((1-phenylethyl)amino)pyridin-3-yl)carbamate (9v)
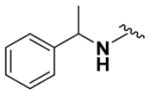
According to the general procedure for pyridine carbamates, the title compound was obtained as a blue-green solid (67%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 6.18 Hz,CH2CH3), 1.45 (3H, d, J = 7.06 Hz,CH3), 4.05 (2H, q, J = 7.31 Hz, CH2CH3), 4.17 (1H, q, J = 7.12 Hz, CHPh), 7.07–7.22 (4H, m), 7.26–7.32 (4H, m), 7.44 (1H, br s, NH), 8.55 (1H, br s, NH), 13.11 (1H, br s, NH). 13CNMR(100 MHz, DMSO-d6) δC: 14.91, 21.11, 35.77, 60.96, 107.56, 111.68, 126.80, 127.61, 128.85, 144.68, 147.98, 155.41. HPLC: retention time, 6.73 min; purity, 99%. m/z (ESI) −301.8 (100%) [MH+].
Methyl 6-(3-(Trifluoromethyl)benzylamino)-2-aminopyridin-3-ylcarbamate (10)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (30%). Rf = 0.53 (mobile phase 10:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, DMSO-d6) δH: 3.58 (3H, s, CH3), 4.48 (2H, d, J = 5.97 Hz, CH2Ph), 5.31 (1H, br s, NH), 5.71 (1H, d, J = 8.30 Hz, ArH), 6.73 (1H, br s, NH), 7.04 (1H, br s, NH), 7.52–7.67 (4H, m, ArH), 8.22 (1H, s, ArH). 13C NMR (100 MHz, DMSO-d6) δC: 44.35, 60.45, 95.46, 107.15, 118.77, 119.91,123.85 (q, J = 41.85 Hz), 124.12, 129.18, 129.61, 130.85 (q, J = 272.11 Hz), 131.82, 143.09, 156.03. 19F NMR (376 MHz, DMSO-d6) δF: −60.94. HPLC: retention time, 6.15 min; purity, 99%. m/z (ESI) −341.1 (100%) [MH+], 363.1 (30%) [MNa+].
Allyl 6-(3-(Trifluoromethyl)benzylamino)-2-aminopyridin-3-ylcarbamate (11)
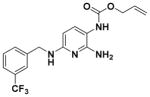
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (31%). Rf = 0.69 (mobile phase 10:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, CDCl3) δH: 4.49 (3H, m, CH2CHCH2), 4.64 (2H, d, J = 4.46 Hz, CH2Ph), 4.95 (1H, br s, NH), 5.30 (2H, m, CH2CHCH2), 5.76 (1H, d, J = 8.23 Hz, ArH), 5.97 (1H, br s, NH), 6.24 (1H, br s, NH), 7.16 (1H, d, J = 7.39 Hz, ArH), 7.44 (1H, m, ArH), 7.53 (2H, d, J = 7.54 Hz, ArH), 7.60 (1H, s, ArH). 13CNMR(100 MHz, CDCl3) δC: 45.91, 66.17, 96.28, 107.55, 118.26, 123.96, 124.12 (q, J = 272.20 Hz), 129.01, 130.56, 130.82 (d, J = 32.22 Hz), 132.43, 137.45, 140.55, 153.62, 155.20, 156.24. 19F NMR (376 MHz, DMSO-d6) δF: −60.95. HPLC: retention time, 6.60 min; purity, 99%. m/z (ESI) −367.1 (100%) [MH+], 368.2 (20%), 389.1 (50%) [MNa+]. HRMS (ESI): C17H17N4O2F3 requires 367.1376. Found: 367.1366.
Allyl (2-Amino-4-((4-chlorobenzyl)amino)phenyl)carbamate (12)
According to the general procedure for pyridine carbamates, the title compound was obtained as a green solid (61%). 1H NMR (400 MHz, DMSO-d6) δH: 4.47 (2H, d, J = 4.40 Hz, CH2), 4.54 (2H, d, J = 3.79 Hz, CH2), 5.22 (1H, d, J = 10.21 Hz, CH2), 5.35 (1H, d, J = 16.97 Hz, CH2), 5.80 (1H, d, J = 8.50 Hz, ArH), 5.95 (1H, m, CH), 6.31 (2H, br s, NH2), 7.24 (1H, d, J = 7.90 Hz, ArH), 7.38–7.42 (5H, m), 8.50 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 44.52, 65.27, 94.94, 107.09, 117.76, 128.69, 129.60, 131.81, 133.91, 139.04, 151.74, 153.09, 155.23. HPLC: retention time, 6.47 min; purity, 99%. m/z (ESI) −333.2 (100%) [MH+], 334.1 (30%).
2,2,2-Trichloroethyl (2-Amino-6-((2-(trifluoromethyl)benzyl)- amino)pyridin-3-yl)carbamate (13)
According to the general procedure for pyridine carbamates, the title compound was obtained as a gray solid (58%). 1H NMR (400 MHz, DMSO-d6) δH: 4.67 (2H, s, CH2), 4.86 (2H, br s, CH2Ph), 5.83 (1H, d, J = 8.75 Hz, ArH), 7.48–7.61 (3H, m, ArH), 7.70 (1H, t, J = 7.29 Hz, ArH), 7.79 (1H, d, J = 8.14 Hz, ArH), 8.07 (1H, br s, NH), 9.13 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 43.09, 74.47, 93.67, 96.24, 106.86, 126.68 (q, J = 3.11 Hz), 127.12 (d, J = 29.97 Hz), 127.53 (q, J = 273.54 Hz), 128.63, 129.63, 133.45, 136.01, 143.42, 149.81, 153.96. HPLC: retention time, 5.96 min; purity, 90%. HRMS (ESI): C16H14N4O2F3Cl3 requires 457.0207. Found: 457.0216.
Ethyl (2-Amino-6-((4-(trifluoromethyl)benzyl)oxy)pyridin-3-yl)- carbamate (14)
According to the general procedure for pyridine carbamates, the title compound was obtained as a white solid (44%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 6.68 Hz, CH2CH3), 4.08 (2H, q, J = 7.07 Hz, CH2CH3), 5.39 (2H, s, CH2), 5.88 (2H, br s, NH2), 6.19 (1H, d, J = 8.38 Hz, ArH), 7.58 (1H, d, J = 7.61 Hz, ArH), 7.67 (2H, d, J = 7.67 Hz, ArH), 7.76 (2H, d, J = 8.14 Hz, ArH), 8.67 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 14.97, 60.89, 67.66, 95.65, 113.07, 125.70, 125.78 (q, J = 3.72 Hz), 127.40 (q, J = 276.02 Hz), 128.72, 138.50, 142.03, 151.70, 155.15. HPLC: retention time, 6.57 min; purity, 97%. HRMS (ESI): C16H16N3O3F3 requires 356.1217. Found: 356.1224.
Ethyl (2-Amino-6-morpholinopyridin-3-yl)carbamate (15)
According to the general procedure for pyridine carbamates, the title compound was obtained as a brown solid (59%). 1H NMR (400 MHz, CDCl3) δH: 1.32 (3H, t, J = 7.00 Hz, CH2CH3), 3.64 (4H, t, J = 4.83 Hz, CH2), 3.87 (4H, t, J = 5.09 Hz, CH2), 4.23 (2H, q, J = 7.11 Hz, CH2CH3), 5.96 (1H, d, J = 8.74 Hz, ArH), 7.38 (2H, br s, NH2), 7.72 (1H, br s, ArH), 13.55 (1H, br s, NH). 13C NMR (100 MHz, CDCl3) δC: 14.54, 47.29, 62.10, 65.91, 67.09, 77.23, 94.86, 155.23. HPLC: retention time, 6.14 min; purity, 99%. m/z (ESI) −267.4 (100%) [MH+].
General Synthetic Procedure for 2-Benzylamino-5-nitropyridine Intermediates (17)
To a suspension of appropriately substituted 2-chloro-5-nitropyridine (16) (1.89 mmol) in IPA (5 mL) was added appropriately substituted benzylamine (2.27 mmol) and triethylamine (2.72 mmol) and the mixture refluxed for 3 h. The reaction mixture was allowed to cool to room temperature, and water (30 mL) was added slowly with stirring to provide a yellow solid. Extraction was performed using ethyl acetate (3 × 20mL), dried (Na2SO4), filtered, and evaporated in vacuo. Purification by column chromatography provided the title compound.
General Synthetic Procedure for Benzyl Diaminopyridine Intermediate (18) Method A
SnCl2·2H2O (4.87 mmol) was dissolved in conc HCl (10 mL) and heated at 70 °C for 30 min until a colorless solution was formed. 2-Benzylamino-5-nitropyridine intermediates (17) (1.21 mmol) was added portionwise and the solution heated at 70 °C for 3 h with a condenser equipped with subba seal and empty balloon. Water was added, and the orange solution was allowed to cool to room temperature. The solution was basified to pH 12 (2 M NaOH) and extracted with CH2Cl2 (3 × 20 mL), the organic layer was washed with brine (50 mL), dried (Na2SO4), filtered, and evaporated in vacuo to provide the title compound. The title compound was taken forward without further purification.
General Synthetic Procedure for Benzyl Diaminopyridine Intermediate (18) Method B
To a solution of 2-benzylamino-5-nitropyridine intermediates (17) (1 mol equiv) in 1,4-dioxane (10 mL per mol equiv) was added Raney nickel (Al–Ni 50:50 wt %) (10× weight of 17) and hydrazine hydrate (10× volume of 17) and the suspension allowed to stir at room temperature overnight. The suspension was filtered through Celite washing with CH2Cl2 (100 mL) and evaporated in vacuo to provide the title compound. The title compound was taken forward without further purification.
General Synthetic Procedure for Desamino Pyridine Carbamate Derivatives (19–23)
To a solution of benzyl diaminopyridine (18) (1.21 mmol) in 1,4-dioxane (10 mL) was added the respective chloroformate (1.21 mmol). The reaction was stirred at room temperature overnight protected from light. The resultant precipitate was collected and washed with 1:1 dioxane:diethyl ether (40 mL), diethyl ether (50 mL), and air-dried at roomtemperature to afford the title compound.
Ethyl (2-Amino-6-((4-fluorobenzyl)amino)pyridin-3-yl)carbamate (19)
According to the general procedure for desamino pyridine carbamates, the title compound was obtained as a brown solid (24%). 1H NMR (400 MHz, CDCl3) δH: 1.32 (3H, t, J = 6.79 Hz, CH2CH3), 4.23 (2H, q, J = 7.51 Hz, CH2CH3), 4.49 (2H, d, J = 4.31 Hz, CH2Ph), 5.53 (1H, br s, NH), 6.43–6.49 (2H, m), 7.04 (2H, t, J = 8.36 Hz, ArH), 7.34 (2H, t, J = 6.25 Hz, ArH), 7.71 (1H, br s, NH), 8.05 (1H, s, ArH). 13CNMR(100 MHz, DMSO-d6) δC: 15.00, 44.29, 60.61, 109.22, 115.26 (d, J = 21.05 Hz), 126.02, 129.67 (d, J = 8.05 Hz), 131.54, 136.75, 154.48, 160.35, 162.75. HPLC: retention time, 6.76 min; purity, 99%. HRMS (ESI): C15H16N3O2F requires 290.1299. Found: 290.1302.
Ethyl (6-((4-Fluorobenzyl)amino)-4-methylpyridin-3-yl)carbamate (20)
According to the general procedure for desamino pyridine carbamates, the title compound was obtained as a orange solid (46%). Rf = 0.37 (mobile phase 20:1 = CH2Cl2/MeOH). 1H NMR (400 MHz, CDCl3) δH: 1.26 (3H, br s,CH2CH3), 2.15 (3H, s,CH3), 4.17 (2H, q, J = 7.1 Hz, CH2CH3), 4.43 (2H, br s, CH2Ph), 5.62 (1H, br s, NH), 6.24 (1H, s, ArH), 6.56 (1H, br s, NH), 7.00 (2H, t, J = 8.62 Hz, ArH), 7.30 (2H, t, J = 8.20 Hz, ArH), 7.98 (1H, br s, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.56, 17.97, 45.66, 61.46, 107.73, 115.44 (d, J = 21.59 Hz), 123.68, 128.88 (d, J = 7.98 Hz), 134.50 (d, J = 3.00 Hz), 143.23, 156.24 160.81, 162.25. HPLC: retention time, 6.64 min; purity, 99%. m/z (ESI) −305.7 (100%) [MH+].
Ethyl (6-((4-(Trifluoromethyl)benzyl)amino)pyridin-3-yl)- carbamate (21)
According to the general procedure for desamino pyridine carbamates, the title compound was obtained as a green solid (61%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 7.09 Hz, CH2CH3), 4.07 (2H, q, J = 7.10 Hz, CH2CH3), 4.52 (2H, d, J = 6.09 Hz, CH2Ph), 6.50 (1H, d, J = 8.91 Hz, ArH), 7.02 (1H, t, J = 5.94 Hz, NH), 7.46–7.53 (3H, m), 7.66 (2H, d, J = 8.07 Hz, ArH), 7.96 (1H, s, ArH), 9.19 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 15.02, 44.44, 60.48, 108.28, 125.48 (q, J = 3.80 Hz), 126.24, 127.55 (d, J = 31.55 Hz), 127.60 (q, J = 271.79 Hz), 128.16, 139.33, 146.54, 154.54, 155.30. 19F NMR (376 MHz, DMSO-d6) δF: −60.69. HPLC: retention time, 5.54 min; purity, 99%. m/z (ESI) −340.1 (100%) [MH+].
Ethyl (6-((4-Methylbenzyl)amino)pyridin-3-yl)carbamate (22)
According to the general procedure for desamino pyridine carbamates, the title compound was obtained as a purple solid (38%). 1H NMR (400 MHz, DMSO-d6) δH: 1.21 (3H, t, J = 7.08 Hz,CH2CH3), 2.26 (3H, s, CH3), 4.07 (2H, q, J = 7.10 Hz, CH2CH3), 4.37 (2H, d, J = 6.07 Hz, CH2Ph), 6.45 (1H, d, J = 8.87 Hz, ArH), 6.79 (1H, t, J = 5.99 Hz, NH), 7.10 (2H, d, J = 7.86 Hz, ArH), 7.20 (2H, d, J = 7.97 Hz, ArH), 7.44 (1H, d, J = 7.55 Hz, ArH), 7.97 (1H, s, ArH), 9.16 (1H, br s, NH). 13C NMR(100MHz,DMSO-d6) δC: 15.03, 21.12, 44.68, 60.45, 108.04, 127.63, 129.15, 130.36, 135.84, 138.13, 139.48, 154.57, 155.67. HPLC: retention time, 6.16 min; purity, 99%. m/z (ESI) −306.7 (100%) [MK+].
Ethyl (6-(Benzylamino)pyridin-3-yl)carbamate (23)
According to the general procedure for desamino pyridine carbamates, the title compound was obtained as a purple solid (26%). 1H NMR (400 MHz, DMSO-d6) δH: 1.24 (3H, t, J = 7.09 Hz, CH2CH3), 4.14 (2H, q, J = 7.10 Hz, CH2CH3), 4.64 (2H, br s, CH2Ph), 7.14 (1H, d, J = 9.54 Hz, ArH), 7.30–7.34 (1H, m, ArH), 7.37–7.44 (4H, m, ArH), 7.88 (1H, dd, J = 7.45 Hz, 2.11 Hz, ArH), 8.11 (1H, s, ArH), 9.08 (1H, br s, NH), 9.92 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 19.02, 45.76, 56.47, 114.34, 126.81, 128.10, 128.21, 129.07, 136.82, 137.16, 149.89, 154.06. HPLC: retention time, 6.20 min; purity, 99%. m/z (ESI) −272.3 (100%) [MH+].
General Synthetic Procedure for 4-Amino-1-benzylamino-3-nitrobenzene Intermediate (25)
To a solution of 1,4-diamino-3-nitrobenzene (24) (3.27 mmol) in IPA (10 mL) was added the appropriately substituted benzaldehyde (3.43 mmol) and the solution stirred at 75 °C for 3 h. A solution of sodium borohydride (13.08 mmol) dissolved in 0.1% NaOH (1 mL) was added and the reaction mixture stirred for another 1 h. The mixture was allowed to cool to room temperature. The dark brown residue was filtered, washed with water, and air-dried at room temperature to obtain the title compound.
General Synthetic Procedure for Carboxybenzyl Protected 4-Amino-1-benzylamino-3-nitrobenzene Intermediate (26)
To a solution of 4-amino-1-benzylamino-3-nitrobenzene intermediate (25) (1.15 mmol) in IPA (10 mL) was added benzyl carbamate (1.38 mmol) and DIPEA (1.15 mmol) and the solution heated at 70 °C overnight. The orange solution was cooled to room temperature, extracted with CH2Cl2 (3 × 20 mL), and the organic layer was separated, washed with brine (50 mL), dried (Na2SO4), filtered, and evaporated in vacuo. Purification by column chromatography provided the title compound.
General Synthetic Procedure for Phenyl Carbamate Intermediate (27)
To a solution of carboxybenzyl protected 4-amino-1-benzylamino-3-nitrobenzene intermediate (26) (0.76 mmol) in 1,4-dioxane (5 mL) was added DMAP (0.38 mmol), DIPEA (3.8 mmol) and, dropwise, the appropriately substituted chloroformate (3.8 mmol), and the solution was heated at 90 °C overnight. The orange solution was cooled at room temperature, extracted with CH2Cl2 (3 × 20 mL), the organic layer separated, washed with brine (50 mL), dried (Na2SO4), filtered, and evaporated in vacuo. Purification by column chromatography provided the title compound.
General Synthetic Procedure for Benzene Carbamate Derivative (2, 28–30)
To a solution of intermediate (27) (0.43 mmol) in MeOH (5 mL) was added Pd/C (0.004 mmol) and stirred for 2 h at room temperature under an atmosphere of hydrogen. The reaction mixture was filtered through Celite by washing with MeOH (50 mL) and evaporated in vacuo to provide the title compound.
p-Tolyl (2-Amino-4-((4-fluorobenzyl)amino)phenyl)carbamate (28)
According to the general procedure for benzene carbamates, the title compound was obtained as a brown solid (18%). 1HNMR(400MHz, DMSO-d6) δH: 2.18 (3H, s,CH3), 4.19 (2H, d, J = 6.00Hz, CH2Ph), 5.88 (1H, t, J = 6.28 Hz, NH), 6.19–6.22 (2H, m, ArH), 6.61–6.66 (3H, m, ArH), 6.95 (2H, d, J = 8.06 Hz, ArH), 7.14 (2H, t, J = 8.91 Hz, ArH), 7.38 (2H, t, J = 8.62 Hz, ArH), 10.12 (2H, m). 13C NMR (100 MHz, DMSO-d6) δC: 20.56, 47.03, 94.31, 105.77, 109.30, 115.38 (d, J = 22.33 Hz), 121.21, 127.44, 129.37 (d, J = 8.02Hz), 130.13, 131.08, 137.18, 144.06, 155.74 (d, J = 32.16 Hz), 160.27, 162.67. HPLC: retention time, 6.55 min; purity, 99%. m/z (ESI) −386.2 (100%) [MK+].
Propyl (2-Amino-4-((4-fluorobenzyl)amino)phenyl)carbamate (29)
According to the general procedure for benzene carbamates, the title compound was obtained as a brown solid (36%). 1H NMR (400 MHz, DMSO-d6) δH: 0.90 (3H, br s, CH2CH3), 1.58 (2H, br s, CH2), 3.93 (2H, t, J = 5.69 Hz, CH2), 4.17 (2H, d, J = 4.68 Hz, CH2Ph), 4.53 (2H, br s, NH2), 5.81 (1H, d, J = 7.83 Hz, ArH), 5.92 (2H, m), 6.69 (1H, d, J = 7.30 Hz, ArH), 7.12 (2H, t, J = 8.53 Hz, ArH), 7.36 (2H, t, J = 6.61 Hz, ArH), 8.17 (1H, br s, NH). 13C NMR (100 MHz, DMSO-d6) δC: 10.75, 22.49, 40.40, 46.32, 65.83, 102.12, 113.47, 115.31 (d, J = 20.93 Hz), 129.29 (d, J = 8.11 Hz), 134.66, 137.25, 137.30, 147.79 160.24, 164.72. HPLC: retention time, 6.37 min; purity, 96%. m/z (ESI) −318.5 (100%) [MH+].
Hexyl (2-Amino-4-((4-fluorobenzyl)amino)phenyl)carbamate (30)
According to the general procedure for benzene carbamates, the title compound was obtained as a brown solid (quant). 1H NMR (400 MHz, CDCl3) δH: 0.90 (3H, t, J = 6.58 Hz, CH3), 1.33 (6H, m, CH2), 1.66 (2H, m, CH2), 3.72 (2H, br s, NH2), 4.13 (2H, t, J = 6.77 Hz, CH2), 4.24 (2H, br s, CH2Ph), 5.99 (1H, d, J = 2.26 Hz, ArH), 6.05 (1H, dd, J = 6.29 Hz, 2.15 Hz, ArH), 6.18 (1H, br s, NH), 6.91 (1H, d, J = 8.06 Hz, ArH), 7.03 (2H, t, J = 8.67 Hz, ArH), 7.31 (2H, t, J = 8.42 Hz, ArH). 13C NMR (100 MHz, CDCl3) δC: 14.04, 22.58, 25.53, 28.95, 31.48, 47.60, 65.61, 100.72, 104.32, 115.41 (d, J = 21.30 Hz), 128.96 (d, J = 7.84 Hz), 135.15 (d, J = 3.05 Hz), 142.99, 147.68, 155.71, 160.78, 163.21. HPLC: retention time, 6.75 min; purity, 74%. m/z (ESI) −360.3 (100%) [MH+].
Cell Culture
PC12 cells were obtained from ATCC (CRL-1721) and cultured in RPMI 1640 (Corning, 10-040-CV) supplemented with 10% heat inactivated horse serum (Corning, 35–030-CV), 5%fetal bovine serum (Corning, 35-010-CV), 1% penicillin–streptomycin (Corning, 30-002-CI), and maintained as monolayer cultures in a humidified atmosphere containing 5% CO2 at 37 °C. T-75 flasks and 96-well plates were coated with 100 μg/mL Rat Tail Collagen I (Corning, 354249) prepared in 0.02 N acetic acid solution.
SH-SY5Y cells were obtained from ATCC (CRL-2266) and cultured in DMEM/F12 (Life Tech, 11320033) supplemented with 10% fetal bovine serum (Corning, 35-010-CV), 1% penicillin–streptomycin (Corning, 30-002-CI) and maintained as monolayer cultures in a humidified atmosphere containing 5% CO2 at 37 °C.50
IMR90-c4 iPSC line was derived from IMR90 fibroblasts31 obtained from WiCell (WiCell, USA). Undifferentiated iPSCs were maintained and passaged in E8Medium (Thermofisher, A1517001) and grown on growth-factor reduced Matrigel (Corning, 354277) for 4 days. iPSCs were differentiated into neurons using an adherent three-step differentiation method as previously described.51 Differentiation of these iPSCs into neural stem cells (NSCs) was induced using neural induction medium (Thermofisher, A1647801) for 9–11 days. After such induction period, NSCs were enzymatically dissociated by accutase (Corning, 25-058-CL) and seeded as single cells at a cell density of 100000 cells/cm2 in the presence of 10 μM Y-27632 (Tocris, 1254) on Matrigel-coated plates. Then, 24 h after seeding, NSCs were further differentiated into neural precursor cells (NPCs) by incubating them in the presence of neural differentiation medium [human pluripotent stem cell serum-free medium (Thermofisher, A1000701), supplemented with 2% bovine serum albumin, 1% Glutamax I (Thermofisher, 35050061), 10 ng/mL human recombinant brain-derived neurotrophic growth factor (Thermofisher, 10908010), and 10 ng/mL human recombinant glial-derived neurotrophic factor (Thermofisher, PHC7044) for 5 days. Differentiation of these NPCs into neurons was achieved by seeding 50000 cells/cm2 on tissue culture plastic surface-coated plates. Neurons were seeded on poly-D-lysine (2 μg/cm2; Sigma-Aldrich, P6407)/laminin (1 μg/cm2; Sigma-Aldrich, L2020)-coated plates and maintained in neuron maturation medium [NMM: Neurobasal-A medium, 2% B27 supplement (ThermoFisher, A1486701), and 1% CultureOne (ThermoFisher, A3320201)]. Cells were allowed to further differentiate for 21 days, with 50% medium replacement every 2 days.51
Cell Viability Assay
Briefly, PC12 cells were plated at a density of 50000 cells/well in 96-well plates coated with rat tail collagen and allowed to adapt overnight in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were pretreated (4 h) with the test compound and incubated with 15 μg/mL etoposide (Chem-Impex International, 28435). Cell viability was measured after 48 h using PrestoBlue at excitation 560 ± 9 nm and emission 590 ± 9 nm using a microplate reader (Biotek Synergy Mx, Reader model SMATBL) as previously reported.25
SH-SY5Y cells were plated at a density of 50000 cells/well in 96-well plates and allowed to adapt overnight in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were pretreated (4 h) with the test compound and incubated with 5 μg/mL etoposide. Cell viability was measured after 48 h using PrestoBlue at excitation 560 ± 9 nm and emission 590 ± 9 nm using a microplate reader (Biotek Synergy Mx, reader model SMATBL).
Neurons derived from IMR-90 iPSCs were plated at a density of 16000 cells/well in 96-well plates coated with poly D-lysine/laminin and allowed to mature in a humidified atmosphere containing 5% CO2 at 37 °C. The cells were pretreated (4 h) with the test compound and incubated with 15 μg/mL etoposide. Cell viability was measured after 36 h using PrestoBlue at excitation 560 ± 9 nm and emission 590 ±9 nm using a microplate reader (Biotek Synergy Mx, reader model SMATBL).
Immunoblotting
For Western blot experiments, cells were lysed at 4 °C using M-PER protein extraction reagent (Thermofisher, 78501) after 24 and 48 h of treatment with test compounds. Protein contents were estimated using BCA protein assay (ThermoFisher, 23225), and 50 μg of protein were electrophoresed on 10% SDS gel and then electrotransferred on nitrocellulose membranes. Blots were treated with 5% BSA solution and probed with Bcl-2 (CST, 34395; 1:1000 dilution), beclin 1 (CST, 37385; 1:2000 dilution), Bax (Santa Cruz, SC7480; 1:500 dilution), and actin (CST, 3700P; 1:5000 dilution) at 4 °C overnight. Blots were then washed six times with TBS-T (tris buffer saline with 0.1% Tween) for 1 h and incubated with horseradish peroxide conjugated antimouse or antirabbit IgG secondary antibody (1:5000) at room temperature for 1 h. After washing with TBS-T for 1 h, blots were exposed to chemiluminescence substrate to detect HRP activity by Versa Doc (BIO-RAD). The images were quantified using ImageJ software (National Institute of Health).
Flow Cytometry
Autophagy was analyzed by incubating PC12 cells with 0.5mMmonodansylcadaverine (Sigma-Aldrich, 30432) for 30 min at 37 °C. For flow cytometry analyses, cells were trypsinized and combined with floating cells from the medium. The cells were resuspended in PBS containing 1% FBS, and 10000 cells were analyzed using the FITC-A filter. Data was analyzed using FlowJo v10.2 (FlowJo, LLC) after removing cell debris.
Supplementary Material
Acknowledgments
SH-SY5Y cells were a kind gift from Dr. Thomas Abbruscato (Texas Tech University Health Sciences Center). Research reported in this publication was supported by Texas Tech University Health Sciences Center School of Pharmacy (P.C.T. and A.A.H.), the Rare Genomics Institute and Collaborative Drug Discovery under the BeHEARD challenge grant initiative (P.C.T.), and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award UL1TR001105 (P.C.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
ABBREVIATIONS USED
- ALS
amyotrophic lateral sclerosis
- Bax
Bcl-2-associated X protein
- BBB
blood–brain barrier
- Bcl-2
B-cell lymphoma 2
- CNS
central nervous system
- FDA
Food and Drug Administration
- Fmoc
fluorenylmethyloxycarbonyl
- iPSC
induced pluripotent stem cell
- KCNQ
potassium Kv7 channel
- MDC
monodansylcadaverine
- MPO
multiparameter optimization
- mTOR
mechanistic target of rapamycin
- NMDA
N-methyl-D-aspartate
- PPI
protein–protein interaction
- ROS
reactive oxygen species
- SAR
structure–activity relationship
Footnotes
Notes
The authors declare the following competing financial interest(s): A provisional patent application has been filed describing the compounds in this paper.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.7b01199.
Synthetic route for compounds 14 and 15, dose–response protection of selected compounds to ameliorate serum starvation-induced apoptosis in PC12 cells, dose–response of all compounds to ameliorate etoposide-induced apoptosis, calculations for MPO score, representative images for MDC staining, characterization data of all synthesized intermediates, 1H NMR and 13C NMR spectra of final products (PDF)
Molecular formula strings for all final products (CSV)
References
- 1.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 2.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria, oxidative stress and cell death. Apoptosis. 2007;12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 3.Xanthos DN, Sandkuhler J. Neurogenic neuroinflammation: Inflammatory CNS reactions in response to neuronal activity. Nat Rev Neurosci. 2014;15:43–53. doi: 10.1038/nrn3617. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 5.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: Mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am J Neurodegener Dis. 2013;2:145–175. [PMC free article] [PubMed] [Google Scholar]
- 6.Bredesen DE, Rao RV, Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovacs GG. Molecular pathological classification of neurodegenerative diseases: Turning towards precision medicine. Int J Mol Sci. 2016;17:189. doi: 10.3390/ijms17020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinarivala N, Trippier PC. Progress in the development of small molecule therapeutics for the treatment of neuronal ceroid lipofuscinoses (NCLs) J Med Chem. 2016;59:4415–4427. doi: 10.1021/acs.jmedchem.5b01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trippier PC, Jansen Labby K, Hawker DD, Mataka JJ, Silverman RB. Target- and mechanism-based therapeutics for neurodegenerative diseases: Strength in numbers. J Med Chem. 2013;56:3121–3147. doi: 10.1021/jm3015926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 11.Harish S, Bhuvana K, Bengalorkar GM, Kumar TN. Flupirtine: Clinical pharmacology. J Anaesthesiol Clin Pharmacol. 2012;28:172–177. doi: 10.4103/0970-9185.94833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seydel JK, Schaper KJ, Coats EA, Cordes HP, Emig P, Engel J, Kutscher B, Polymeropoulos EE. Synthesis and quantitative structure-activity relationships of anticonvulsant 2,3,6-triaminopyridines. J Med Chem. 1994;37:3016–3022. doi: 10.1021/jm00045a005. [DOI] [PubMed] [Google Scholar]
- 13.Schuster G, Schwarz M, Block F, Pergande G, Schmidt WJ. Flupirtine: A Review of its neuroprotective and behavioral properties. CNS Drug Rev. 1998;4:149–164. [Google Scholar]
- 14.Szelenyi I. Flupirtine, a re-discovered drug, revisited. Inflammation Res. 2013;62:251–258. doi: 10.1007/s00011-013-0592-5. [DOI] [PubMed] [Google Scholar]
- 15.Dhar S, Bitting RL, Rylova SN, Jansen PJ, Lockhart E, Koeberl DD, Amalfitano A, Boustany RM. Flupirtine blocks apoptosis in batten patient lymphoblasts and in human postmitotic CLN3- and CLN2-deficient neurons. Ann Neurol. 2002;51:448–466. doi: 10.1002/ana.10143. [DOI] [PubMed] [Google Scholar]
- 16.Siegmund W, Modess C, Scheuch E, Methling K, Keiser M, Nassif A, Rosskopf D, Bednarski PJ, Borlak J, Terhaag B. Metabolic activation and analgesic effect of flupirtine in healthy subjects, influence of the polymorphic NAT2, UGT1A1 and GSTP1. Br J Clin Pharmacol. 2015;79:501–513. doi: 10.1111/bcp.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stafstrom CE, Grippon S, Kirkpatrick P. Ezogabine (retigabine) Nat Rev Drug Discovery. 2011;10:729–730. doi: 10.1038/nrd3561. [DOI] [PubMed] [Google Scholar]
- 18.ClinicalTrialsgov. U.S. National Institutes of Health; Bethesda, MD: 2015. [accessed July 27, 2017]. Clinical trial of Ezogabine (retigabine) in ALS subjects. https://clinicaltrials.gov/ct2/show/NCT02450552. [Google Scholar]
- 19.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SSW, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH, Cudkowicz ME, Bean BP, Eggan K, Woolf CJ. Intrinsic membrane hyperexcitability of ALS patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn-Munro G, Dalby-Brown W, Mirza NR, Mikkelsen JD, Blackburn-Munro RE. Retigabine: Chemical synthesis to clinical application. CNS Drug Rev. 2005;11:1–20. doi: 10.1111/j.1527-3458.2005.tb00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boscia F, Annunziato L, Taglialatela M. Retigabine and flupirtine exert neuroprotective actions in organotypic hippocampal cultures. Neuropharmacology. 2006;51:283–294. doi: 10.1016/j.neuropharm.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- 23.Williamson A. Ueber die Theorie der Aetherbildung. Justus Liebigs Ann Chem. 1851;77:37–49. [Google Scholar]
- 24.Kinarivala N, Trippier PC. Exploration of relative chemo-selectivity in the hydrodechlorination of 2-chloropyridines. Tetrahedron Lett. 2014;55:5386–5389. [Google Scholar]
- 25.Kinarivala N, Shah K, Abbruscato TJ, Trippier PC. Passage variation of PC12 cells results in inconsistent susceptibility to externally induced apoptosis. ACS Chem Neurosci. 2017;8:82–88. doi: 10.1021/acschemneuro.6b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinarivala N, Suh JH, Botros M, Webb P, Trippier PC. Pharmacophore elucidation of phosphoiodyn A - Potent and selective peroxisome proliferator-activated receptor beta/delta agonists with neuroprotective activity. Bioorg Med Chem Lett. 2016;26:1889–1893. doi: 10.1016/j.bmcl.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 27.Bolo NR, Hode Y, Nedelec JF, Laine E, Wagner G, Macher JP. Brain pharmacokinetics and tissue distribution in vivo of fluvoxamine and fluoxetine by fluorine magnetic resonance spectroscopy. Neuropsychopharmacology. 2000;23:428–438. doi: 10.1016/S0893-133X(00)00116-0. [DOI] [PubMed] [Google Scholar]
- 28.Rankovic Z. CNS physicochemical property space shaped by a diverse set of molecules with experimentally determined exposure in the mouse brain. J Med Chem. 2017;60:5943–5954. doi: 10.1021/acs.jmedchem.6b01469. [DOI] [PubMed] [Google Scholar]
- 29.Wager TT, Hou X, Verhoest PR, Villalobos A. Moving beyond rules: The development of a central nervous system multiparameter optimization (CNS MPO) approach to enable alignment of druglike properties. ACS Chem Neurosci. 2010;1:435–449. doi: 10.1021/cn100008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trippier PC. Selecting good ‘drug-like’ properties to optimize small molecule blood-brain barrier penetration. Curr Med Chem. 2016;23:1392–1407. doi: 10.2174/0929867323666160405112353. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 32.Gassen M, Pergande G, Youdim MBH. Antioxidant properties of the triaminopyridine, flupirtine. Biochem Pharmacol. 1998;56:1323–1329. doi: 10.1016/s0006-2952(98)00126-9. [DOI] [PubMed] [Google Scholar]
- 33.Kornhuber J, Bleich S, Wiltfang J, Maler M, Parsons CG. Flupirtine shows functional NMDA receptor antagonism by enhancing Mg2+ block via activation of voltage independent potassium channels. Rapid communication. J Neural Transm (Vienna) 1999;106:857–867. doi: 10.1007/s007020050206. [DOI] [PubMed] [Google Scholar]
- 34.Edwards MA, Loxley RA, Williams AJ, Connor M, Phillips JK. Lack of functional expression of NMDA receptors in PC12 cells. NeuroToxicology. 2007;28:876–885. doi: 10.1016/j.neuro.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Zhang WB, Ross PJ, Tu Y, Wang Y, Beggs S, Sengar AS, Ellis J, Salter MW. Fyn Kinase regulates GluN2B subunit-dominant NMDA receptors in human induced pluripotent stem cell-derived neurons. Sci Rep. 2016;6:23837. doi: 10.1038/srep23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perovic S, Schroder HC, Pergande G, Ushijima H, Muller WE. Effect of flupirtine on Bcl-2 and glutathione level in neuronal cells treated in vitro with the prion protein fragment (PrP106–126) Exp Neurol. 1997;147:518–524. doi: 10.1006/exnr.1997.6559. [DOI] [PubMed] [Google Scholar]
- 37.Oltval ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 38.Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Füllgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC. Autophagy and neurodegeneration: Pathogenic mechanisms and therapeutic opportunities. Neuron. 2017;93:1015–1034. doi: 10.1016/j.neuron.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 39.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 40.Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M, Christoffersson J, Chaabane W, Moghadam AR, Kashani HH, Hashemi M, Owji AA, Los MJ. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Prog Neurobiol. 2014;112:24–49. doi: 10.1016/j.pneurobio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 42.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 43.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavathiotis E, Reyna DE, Bellairs JA, Leshchiner ES, Walensky LD. Direct and selective small-molecule activation of proapoptotic BAX. Nat Chem Biol. 2012;8:639–645. doi: 10.1038/nchembio.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niu X, Brahmbhatt H, Mergenthaler P, Zhang Z, Sang J, Daude M, Ehlert FGR, Diederich WE, Wong E, Zhu W, Pogmore J, Nandy JP, Satyanarayana M, Jimmidi RK, Arya P, Leber B, Lin J, Culmsee C, Yi J, Andrews DW. A small-molecule inhibitor of Bax and Bak oligomerization prevents genotoxic cell death and promotes neuroprotection. Cell Chem Biol. 2017;24:493.e5–506.e5. doi: 10.1016/j.chembiol.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 47.Wyttenbach A, Hands S, King MA, Lipkow K, Tolkovsky AM. Amelioration of protein misfolding disease by rapamycin: Translation or autophagy? Autophagy. 2008;4:542–545. doi: 10.4161/auto.6059. [DOI] [PubMed] [Google Scholar]
- 48.King MA, Hands S, Hafiz F, Mizushima N, Tolkovsky AM, Wyttenbach A. Rapamycin inhibits polyglutamine aggregation independently of autophagy by reducing protein synthesis. Mol Pharmacol. 2008;73:1052–1063. doi: 10.1124/mol.107.043398. [DOI] [PubMed] [Google Scholar]
- 49.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Kovalevich J, Langford D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol Biol. 2013;1078:9–21. doi: 10.1007/978-1-62703-640-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel R, Page S, Al-Ahmad AJ. Isogenic blood-brain barrier models based on patient-derived stem cells display inter-individual differences in cell maturation and functionality. J Neurochem. 2017;142:74–88. doi: 10.1111/jnc.14040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.