Abstract
Fragile X syndrome (FXS) is an X-chromosome linked intellectual disability and the most common known inherited single gene cause of autism spectrum disorder (ASD). Building upon demonstrated deficits in neuronal plasticity and spatial memory in FXS, we investigated how spatial information processing is affected in vivo in an FXS mouse model (Fmr1-KO). Healthy hippocampal neurons (so-called place cells) exhibit place-related activity during spatial exploration, and their firing fields tend to remain stable over time. In contrast, we find impaired stability and reduced specificity of Fmr1-KO spatial representations. This is a potential biomarker for the cognitive dysfunction observed in FXS, informative on the ability to integrate sensory information into an abstract representation and successfully retain this conceptual memory. Our results provide key insight into the biological mechanisms underlying cognitive disabilities in FXS and ASD, paving the way for a targeted approach to remedy these.
Introduction
The most common known inherited single gene cause of autism spectrum disorder (ASD) is fragile X syndrome (FXS)1,2: an intellectual disability in which expression of the fragile X mental retardation protein (FMRP) is silenced or affected by loss of function mutations, resulting in disturbed neuronal communication3,4. Due to its simple genetic etiology, FXS shows promise for understanding neuropsychiatric disease from genes, to circuits, to cognitive impairment5. Particularly affected in FXS human patients and animal models is the hippocampus6,7, a brain structure essential for consolidating experiences into conceptual and spatial memory8–11. When healthy humans and animals explore a space, hippocampal neurons (so-called place cells) exhibit place-related activity12,13. These cells tend to stably maintain their firing fields over time, forming a spatial representation of the environment14,15. It follows that anomalous place cell activity in disease models may be characteristic of cognitive impairment in neurological disorders. We used a spatial exploration paradigm to investigate in an FXS mouse model (Fmr1-KO)16,17 how spatial information processing is affected in vivo by recording hippocampal place cell activity.
Results and Discussion
Unaffected exploratory behavior in Fmr1-KO mice
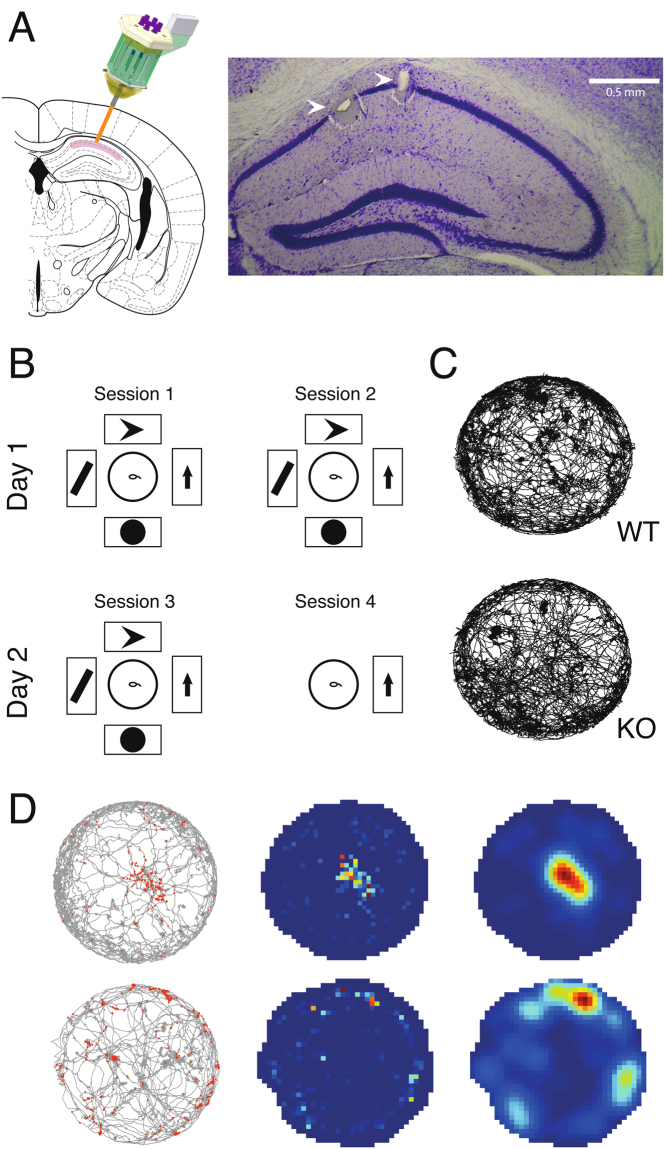
We recorded neuronal activity in hippocampal CA1 (Fig. 1A) during four subsequent spatial exploration sessions (across two days) in five Fmr1-KO mice and five WT control mice (Fig. 1B). The first two recording sessions (on the morning and afternoon of the first day) and the third recording sessions (on the morning of the second day) were done with a complete set of four visual cues marking the environment (“Full cue” sessions). For the fourth session (on the afternoon of the second day), three of these visual cues were removed from the room, leaving an incomplete set of cues by which the animal could localize itself (“Probe” session). Our recordings yielded 124 WT and 141 Fmr1-KO putative pyramidal cells that exhibited spatially modulated activity (place cells). The numbers of place cells recorded per animal and their contribution to the total number of cells recorded per genotype are provided in Table S1.
Figure 1.
Experimental Setup. (A) Histology. Left, schematic of microdrive implantation target, modified from50. Right, coronal section showing the recording locations of two tetrodes in dorsal hippocampus CA1 (arrowheads). (B) Schematic of the behavioral protocol. On two consecutive days, in two sessions per day, animals freely explored a circular open field arena (middle). The arena was surrounded by four posters of geometric figures in the first three sessions, and only one poster in the fourth session. (C) Behavior. Accumulated trajectories of a WT and a KO animal exploring the arena during an example session. (D) Illustration of the three steps of firing map construction, for an example WT place cell (top row) and an example KO place cell (bottom row). Left, accumulated trajectory of animal exploration during the session, with spikes recorded from a single pyramidal cell superimposed in red. Middle, heat map of these spikes created by binning and normalizing this data. Right, smoothed heat map of these binned and normalized spikes.
Hippocampal place fields are thought to result from a complex process of integration of different types of information during exploration of an environment. Instantaneous multisensory inputs from the environment are combined into a ‘scaffold’ of internally generated representations, which depend to a large extent on self-motion information18,19. Therefore, we first controlled for parameters of exploratory behavior, which may act as a confounding factor for neuronal processing of spatial information (Fig. 1C). We found no difference between WT and Fmr1-KO running speed (WT median = 5.9 cm/s n = 30, KO median = 5.7 cm/s n = 33, Mann-Whitney U = 422, P = 0.32 two-tailed) and thigmotaxis (average distance from the center of the arena, WT median = 25.62 cm n = 38, KO median = 25.18 cm n = 32, Mann-Whitney U = 494, P = 0.18 two-tailed) across sessions. Additionally, there was no difference in the maximum and mean pyramidal cell firing rate across genotypes (Table S2).
Reduced spatial specificity of place cells in Fmr1-KO mice
Pyramidal cells of both genotypes exhibited spatially selective activity: place fields (Fig. 1D). We found no difference between WT and Fmr1-KO mice in the number of place fields per cell, or the spatial information that each spike carried. However, Fmr1-KO place fields were significantly larger than those of wildtype animals, based both on counting active pixels in the normalized map (WT median = 9448 n = 123, KO median = 9603 n = 141, Mann-Whitney U = 5666, P < 0.0001 two-tailed) and by comparing the place fields in the smoothed maps (Table S2).
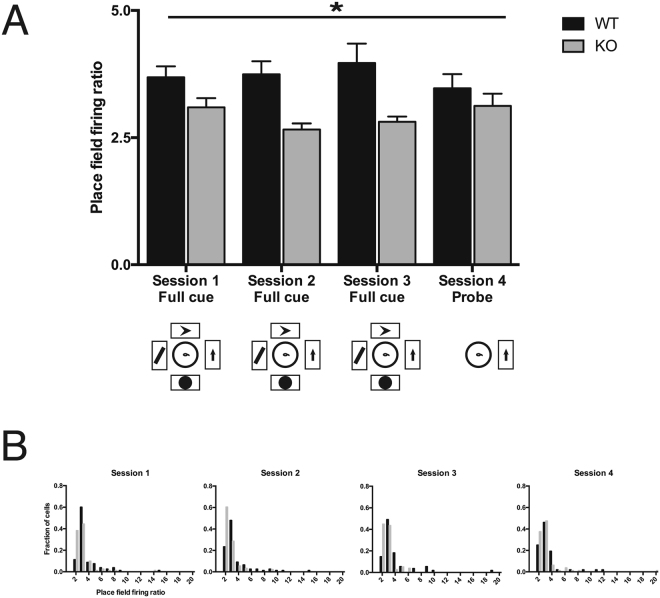
We determined spatial specificity of place cells as the firing rate increase of each cell within its field, relative to the firing rate of the same cell outside its field (Fig. 2). Spatial specificity of Fmr1-KO pyramidal cells was significantly reduced compared with WT (two-way genotype x session ANOVA, effect of genotype: F1,586 = 25.55; P < 0.0001). There was no effect of session and no interaction effect across cells.
Figure 2.
Spatial specificity of place cells per session. (A) Place field firing rate increase of WT (black) and Fmr1-KO (gray) pyramidal cells within their respective fields, relative to the firing rate of each cell outside its field (the place field firing ratio). Data are represented as mean ± SEM. Session 1: WT 80 cells, mean = 3.69, SEM = 0.22; KO 81 cells, mean = 3.09, SEM = 0.18. Session 2: WT 77 cells, mean = 3.75, SEM = 0.25; KO 91 cells, mean = 2.66, SEM = 0.12. Session 3: WT 55 cells, mean = 3.97, SEM = 0.38; KO 78 cells, mean = 2.81, SEM = 0.10. Session 4: WT 52 cells, mean = 3.47, SEM = 0.28; KO 80 cells, mean = 3.12, SEM = 0.24. (B) Distributions of place field firing rate increase of WT (black) and Fmr1-KO (gray) pyramidal cells within their respective fields, relative to the firing rate of each cell outside its field (the place field firing ratio). *P < 0.0001 main effect of genotype (two-way genotype x session ANOVA: F1,586 = 25.55). There was no effect of session and no interaction effect.
Impaired short-term stability of spatial representation in Fmr1-KO mice
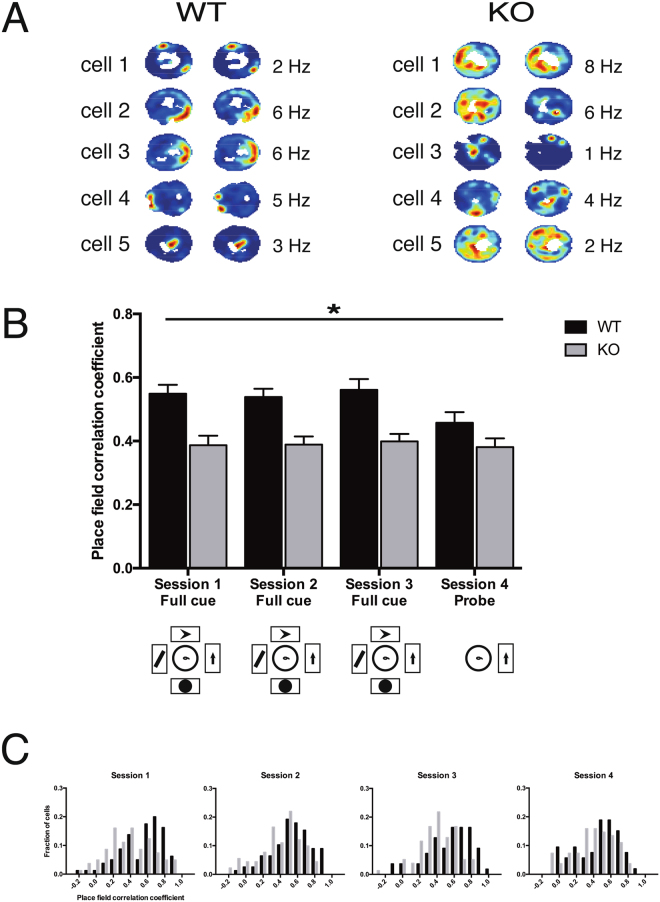
To assess the stability of place cell activity, we first evaluated the similarity of the firing rate maps between the halves of each session (Fig. 3). Pixel-based Pearson correlations between rate maps were significantly reduced in Fmr1-KO mice compared with WT (two-way genotype x session ANOVA, effect of genotype: F1,590 = 46.34; P < 0.0001). We found no effect of session and no interaction effect. Comparison between the quarters of each session yielded the same conclusion: correlations between rate maps were significantly reduced in Fmr1-KO mice compared with WT (two-way genotype x session ANOVA, effect of genotype: F1,1457 = 92.58; P < 0.0001) with no effect of session and no interaction effect (Table S3).
Figure 3.
Stability of firing rate maps within sessions. (A) Example WT and KO firing rate maps (selected across all mice and sessions), split between the first (left panels) and second (right panels) halves of each recording session, to illustrate the stability of each map. Each heat map is scaled by the maximum firing rate (indicated in Hz) of the cell within that session. Areas of the arena that were not visited during the recording session are marked in white. (B) Correlation of WT (black) and Fmr1-KO (gray) firing rate maps. Data are represented as mean ± SEM. Session 1: WT 80 fields, mean = 0.55, SEM = 0.028; KO 81 fields, mean = 0.39, SEM = 0.03. Session 2: WT 78 fields, mean = 0.54, SEM = 0.03; KO 91 fields, mean = 0.39, SEM = 0.03. Session 3: WT 55 fields, mean = 0.56, SEM = 0.03; KO 78 fields, mean = 0.40, SEM = 0.02. Session 4: WT 53 fields, mean = 0.46, SEM = 0.03; KO 82 fields, mean = 0.38, SEM = 0.03. (C) Distributions of the stability of WT (black) and Fmr1-KO (gray) pyramidal cells. *P < 0.0001 main effect of genotype (two-way genotype x session ANOVA: F1,590 = 46.34). There was no effect of session and no interaction effect.
Since each animal was used in multiple consecutive experiments in which only the wall cues were changed and the arena stayed the same, we assessed whether the novelty of the environment was diminished between experiments. We compared place field stability on the first (novel) day of each consecutive experiment and found no increase, allowing us to rule out the possibility that the mice were using subtle intra-maze cues to skew the stability of spatial representation between experiments.
Thus, the effects of the FXS mutation on spatial representation in Fmr1-KO mice are not attributable to behavioral differences or basic physiological properties such as pyramidal cell firing rate. However, the relative strength of firing of WT place cells within fields is greater than in Fmr1-KO mice, the latter shows increased size of place fields, and the location of place responses is less stable in Fmr1-KO mice than in WT controls in short intervals within recording sessions in the same environment.
As for any neural integration operation, self-localization is affected by the accumulation of errors, resulting in drift which increases with time20,21. Spatially informative sensory cues can realign the drifting map, therefore reducing error. In interpreting our current findings, one possibility is that the FXS mutation affects the sensory information-dependent updating of the self-motion based map, while leaving the map itself relatively spared.
Another possibility is that the Fmr1-KO mice are paying less attention to the cues, so that their information is not integrated properly into the spatial representation. As sensory cues may rapidly induce profound changes in the spatial map22, in an attention modulated way15, the increased instability we observe in Fmr1-KO mice may be due to a lower weight of sensory inputs in determining place cell firing. Indeed, FXS patients show defective attention and integration of new information23.
Fmr1-KO spatial representation does not reflect changes in environment
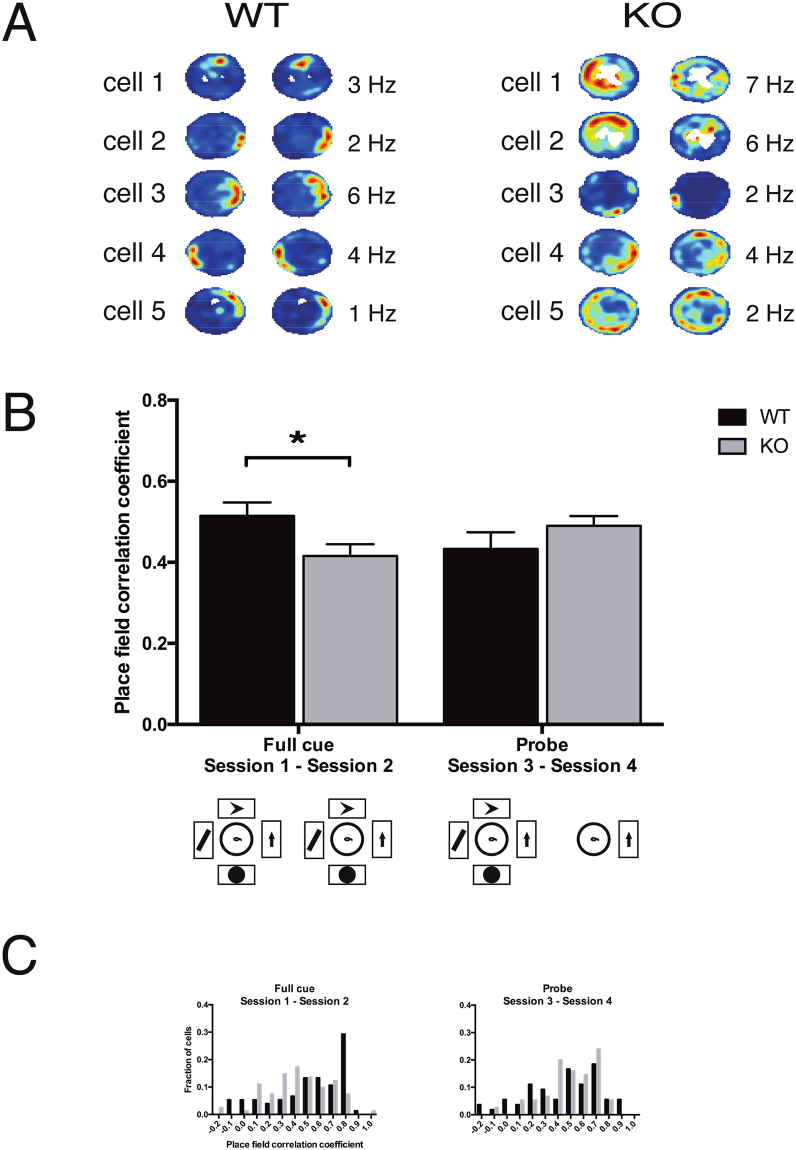
To determine whether Fmr1-KO mice are impaired in sensory information-dependent updating of their spatial representation, we assessed the stability of firing rate maps between “Full cue” and “Probe” sessions on the same day. Here, we found no direct effect of genotype or session, but we found a significant interaction effect (two-way genotype x session ANOVA followed by Bonferroni’s multiple comparisons test, interaction effect: F1,281 = 6.074; P = 0.0143). Specifically, Fmr1-KO pyramidal cells showed a significantly reduced spatial correlation of activity compared with WT between the “Full cue” sessions (i.e., sessions 1 and 2, in which the open field arena was surrounded by the same four visual cues). The firing rate map correlation between the “Full cue” (i.e., sessions 1 and 2) and the novel “Probe” (i.e., sessions 3 and 4, in between which three of these cues were removed) environment however, did not significantly differ between genotypes (Fig. 4), possibly signifying that constancy of external cues is not the main factor driving place cells in Fmr1-KO, which might rely on other (e.g. self-motion or internal24) sources of information.
Figure 4.
Stability of firing rate maps between sessions. (A) Example WT and KO firing rate maps (selected across all mice and sessions), split between the first (left panels) and second (right panels) daily recording sessions, to illustrate the stability of each map. Each heat map is scaled by the maximum firing rate (indicated in Hz) of the cell within that session. Areas of the arena that were not visited during the recording session are marked in white. (B) Correlation of WT (black) and Fmr1-KO (gray) firing rate maps. Data are represented as mean ± SEM. Full cue session: WT 75 fields, mean = 0.51, SEM = 0.03; KO 81 fields, mean = 0.42, SEM = 0.03. Probe session: WT 54 fields, mean = 0.43, SEM = 0.04; KO 75 fields, mean = 0.49, SEM = 0.02. (C) Distributions of the stability of WT (black) and Fmr1-KO (gray) pyramidal cells. *P < 0.05 (Bonferroni’s multiple comparisons test).
Spatial representation impairments in Fmr1-KO provide biomarker for FXS deficits
Both animal8,9 and human10,11 studies link the hippocampus to spatial, contextual, and autobiographical memory. In the same way that place cells in animals exploring an environment can encode that space, the activity of hippocampal neurons in humans can encode abstract representations of multi-sensory perceptual information13,25. Hippocampal dysfunction is a critical component of intellectual pathologies such as FXS and ASD, in which impairments of conceptualization and memory are observed6,7,26. Here, the delicate system that allows the brain to carefully fine-tune which information it retains is disrupted, because of the devastating effect on activity-dependent synaptic plasticity that underlies learning and memory. This ultimately contributes to anomalous processing of social and environmental cues and associated deficits in memory and cognition27. Although they are equally affected neurologically, it has been difficult to assess these cognitive deficits in animal models with the same robustness as in human FXS patients28. Our findings take the middle ground by demonstrating on a cellular level in vivo that altered physiology in Fmr1-KO leads to impaired hippocampal information processing.
Indeed, one should note that place field disruption is not unequivocally correlated with a spatial and/or navigation memory deficit. In fact, it has been shown that even subtler distortions of hippocampal activity, for example ones affecting temporal coding, but leaving the place field map intact, may also correlate with learning and memory deficits29–31. On the other hand, reports from rat virtual reality experiments suggest that while place fields are disrupted, animals are still capable of learning spatial tasks in virtual space32. Similarly, mice with a deleted GluA1 subunit of the AMPA receptor have impaired place fields, but can solve a spatial memory task (while being impaired in a working memory task)33. Still, our finding of impaired stability of place fields in Fmr1-KO mice suggests deeply disrupted hippocampal function, which predicts cognitive deficits associated with FXS, especially as one moves to the more delicate, demanding cognitive abilities affected in human FXS patients.
There is a wide array of FXS physiological deficits which might underlie our results. Stability of spatial representation requires long-term potentiation (LTP) associated with NMDA receptor activity in hippocampal CA134–37. FMRP regulates subunit composition of hippocampal NMDA receptors38 and may therefore contribute to Fmr1-KO pathophysiology by affecting synaptic plasticity through altered subunit composition of NMDA receptors. Indeed, LTP deficits are observed in Fmr1-KO mice39–42. Additionally, Fmr1-KO mice show higher dendritic expression of the HCN1 gene in hippocampal CA143, which might limit certain aspects of spatial memory and plasticity in pyramidal neurons by affecting the ability of the entorhinal cortex to excite them44. The instability we find in Fmr1-KO firing rate maps may be interpreted within this context as an increase in HCN1-mediated control over CA1 pyramidal cell plasticity from entorhinal inputs through the perforant pathway, which affects the sensory information-dependent updating of the self-motion based map as described above. Finally, disrupted network mechanisms45 regulating the inflow of information between the hippocampus and entorhinal cortex46,47 may contribute to improper routing of sensory information to the hippocampus, or in the failure to elicit spike-timing dependent plasticity37,48,49. While the cognitive effects of these deficits have proven difficult to assess behaviorally in Fmr1-KO28, we find that they may contribute to disrupting neural mechanisms that establish associations between external cues and internally generated or self-motion dependent representations.
Hippocampal place cells are one of the best understood systems in the brain where we have reached an initial understanding of the relationship between neural dynamics, information encoding, and cognition. Here we have shown that they may provide a powerful tool in understanding intellectual disability and ASD in a mouse model of FXS, in which it has been surprisingly difficult to demonstrate consistent cognitive deficits despite its clear genetic etiology. We find impaired specificity and stability of CA1 place cell activity in Fmr1-KO mice, both within and across subsequent exploration sessions, while these mice show a relatively spared place field response and their behavior and firing-rate parameters do not significantly differ from WT mice. Our results link impaired physiology with cognition more deeply than possible with traditional behavioral of physiological assays, and offer a potential biomarker for testing of therapeutic strategies.
Methods
Subjects
We used five Fmr1-KO mice17 and five littermate wildtype (WT) control mice. All experiments were performed in accordance with Dutch National Animal Experiments regulations, were approved by the Universiteit van Amsterdam, and were carried out by certified personnel. Animals were received from the Erasmus Medisch Centrum Rotterdam breeding unit at an age of 8 weeks and group-housed until surgery. They were maintained on a regular 12-hour light-dark cycle (lights on: 8 am, lights off: 8 pm) and received standard food pellets and water ad libitum throughout the experiment. To minimize bias due to possible undetected changes in environmental conditions, Fmr1-KO and WT animals were always studied in pairs; both recordings were done on the same day and counterbalanced per genotype. Once habituated to the experimenter and handling, the mice underwent drive implantation surgery under buprenorphine-isoflurane anesthesia and were left to recover fully before the start of the experiment.
Electrophysiological techniques
Six independently moveable tetrodes were loaded into a custom-made microdrive37,50 and implanted over the dorsal hippocampus (AP: −2.0 mm, ML: −2 mm51; Fig. 1A). The tetrodes were lowered into the CA1 pyramidal cell layer guided by electrophysiological signals (sharp wave-ripple events) over the course of days following implantation surgery. Electrophysiological activity was recorded on a 27-channel analog Neuralynx data acquisition system at a 32 kHz sampling rate. Tetrode signals (bandpass filtered 0.6–6.0 kHz) were referred to a nearby tetrode which was targeted to a location devoid of single unit activity. Single-unit data were preprocessed with Klustakwik52 for automated spike clustering and the results were manually refined using Klusters53. The resulting spike trains were analyzed using custom-written MATLAB code. Animal tracking position was extracted from video footage by Ethovision XT software (Noldus, Wageningen, the Netherlands) which was synchronized with the electrophysiology data acquisition system. At the end of experiments, electrolytic lesions were made to verify tetrode placement. Brain tissue was fixed by transcardial perfusion and Nissl stained (Fig. 1A). Only animals with clear lesions in the CA1 pyramidal layer were included in the analysis.
Behavioral protocol
An experiment consisted of four sessions (two per day on two consecutive days) during which hippocampal neural ensemble activity was recorded as the mice freely explored (without foraging for food) a fully transparent, circular open field arena (diameter 64 cm) for 30 min. The arena was surrounded by black curtains and four large posters of geometric figures as visual cues (Fig. 1B). In the final (fourth) session, three of the visual cues were removed (“Probe” session); the same cues were removed for both genotypes. The two daily recording sessions were separated by a two-hour break, during which the animal rested in its home cage. Each animal was screened in its home cage in the experiment room for 30 min prior to each recording. Each animal was used for multiple (consecutive) experiments (on average 3 experiments per animal). A new set of visual wall cues was selected for each iteration: session 1 was always the first recording in the novel environment.
Neuronal analysis
Periods of inactivity (animal speed <3 cm/s) were excluded from analysis. Videotracking data were visually inspected, checked for accuracy, and corrected manually when necessary. Recording stability of individual clusters of spikes was examined; clusters whose first principal component drift exceeded more than three standard deviations across both sessions within a day were excluded from analysis. Classification of putative pyramidal cells was based on their firing rate and the mean of the autocorrelogram, as previously described by our lab37.
Place cell analysis
To create firing maps of individual neurons, spike data were (1) plotted on binned arena occupancy data (pixels: 2 × 2 cm), (2) normalized by the total time spent in each bin, and (3) smoothed (radius: 2). These three steps are illustrated for two example WT place cells recorded in two separate sessions in Fig. 1D. Bins that received insufficient sampling (<200 ms) were excluded from analysis. Only neurons that displayed place-related activity in at least one session were included in analysis. Place fields were defined as areas larger than 10 adjacent pixels where a pyramidal cell exhibited more than 30% of its maximum firing rate. Spatial information per spike was calculated as described in54. Spatial specificity (the place field firing ratio) was calculated as the firing rate increase of each cell within its field, relative to the firing rate of the same cell outside its field (in-field firing rate divided by out-field firing rate).
Data availability
The dataset generated and analyzed in the current study is available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank L. Noldus for the use of Ethovision XT software, K. Harris for the use of Klustakwik, and L. Hazan for the use of Klusters. Animals were kindly provided by Prof. Dr. R. Willemsen at the Department of Clinical Genetics, Erasmus MC in Rotterdam, The Netherlands. This work was supported by SenterNovem BSIK grant 03053, STW grant AET7613, and EU project 720270 – HBP SGA1 (Human Brain Project) to C.M.A.P.
Author Contributions
T.A., C.M.A.P., and F.P.B. designed the experiments. T.A. performed the experiments. T.A. and F.P.B. analyzed the data. T.A., C.M.A.P., and F.P.B. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26853-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 2.Budimirovic DB, Kaufmann WE. What can we learn about autism from studying fragile X syndrome? Dev Neurosci. 2011;33:379–394. doi: 10.1159/000330213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pilpel Y, et al. Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of Fmr1 knockout mice. J Physiol. 2009;587:787–804. doi: 10.1113/jphysiol.2008.160929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: what a fly and mouse model can teach us. Learn Mem. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fung LK, Reiss AL. Moving Toward Integrative, Multidimensional Research in Modern Psychiatry: Lessons Learned From Fragile X Syndrome. Biol Psychiatry. 2016;80:100–111. doi: 10.1016/j.biopsych.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kates WR, Abrams MT, Kaufmann WE, Breiter SN, Reiss AL. Reliability and validity of MRI measurement of the amygdala and hippocampus in children with fragile X syndrome. Psychiatry Res. 1997;75:31–48. doi: 10.1016/S0925-4927(97)00019-X. [DOI] [PubMed] [Google Scholar]
- 7.Reiss AL, Lee J, Freund L. Neuroanatomy of fragile X syndrome: the temporal lobe. Neurology. 1994;44:1317–1324. doi: 10.1212/WNL.44.7.1317. [DOI] [PubMed] [Google Scholar]
- 8.Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science. 1992;256:675–677. doi: 10.1126/science.1585183. [DOI] [PubMed] [Google Scholar]
- 9.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 10.Manns JR, Hopkins RO, Squire LR. Semantic Memory and the Human Hippocampus. Neuron. 2003;38:127–133. doi: 10.1016/S0896-6273(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 11.Moscovitch M, Cabeza R, Winocur G, Nadel L. Episodic Memory and Beyond: The Hippocampus and Neocortex in Transformation. Annu Rev Psychol. 2016;67:105–134. doi: 10.1146/annurev-psych-113011-143733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Keefe J, Dostrovsky J. The hippocampus as a spatial map. Preliminary evidence from unit activity in the freely-moving rat. Brain Res. 1971;34:171–175. doi: 10.1016/0006-8993(71)90358-1. [DOI] [PubMed] [Google Scholar]
- 13.Ekstrom AD, et al. Cellular networks underlying human spatial navigation. Nature. 2003;425:184–188. doi: 10.1038/nature01964. [DOI] [PubMed] [Google Scholar]
- 14.Ziv Y, et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kentros CG, Agnihotri NT, Streater S, Hawkins RD, Kandel ER. Increased attention to spatial context increases both place field stability and spatial memory. Neuron. 2004;42:283–295. doi: 10.1016/S0896-6273(04)00192-8. [DOI] [PubMed] [Google Scholar]
- 16.Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell78, 23–33 (1994). [PubMed]
- 17.Mientjes EJ, et al. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol Dis. 2006;21:549–555. doi: 10.1016/j.nbd.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 18.McNaughton BL, Battaglia FP, Jensen O, Moser EI, Moser MB. Path integration and the neural basis of the ‘cognitive map’. Nat Rev Neurosci. 2006;7:663–678. doi: 10.1038/nrn1932. [DOI] [PubMed] [Google Scholar]
- 19.O’Keefe, J. & Nadel, L. The hippocampus as a cognitive map (Oxford University Press, Oxford, 1978).
- 20.Gothard KM, Skaggs WE, Moore KM, McNaughton BL. Binding of hippocampal CA1 neural activity to multiple reference frames in a landmark-based navigation task. J Neurosci. 1996;16:823–835. doi: 10.1523/JNEUROSCI.16-02-00823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabral HO, Fouquet C, Rondi-Reig L, Pennartz CM, Battaglia FP. Single-Trial Properties of Place Cells in Control and CA1 NMDA Receptor Subunit 1-KO Mice. J Neurosci. 2014;34:15861–15869. doi: 10.1523/JNEUROSCI.5320-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monaco, J. D., Rao, G., Roth, E. D. & Knierim, J. J. Attentive scanning behavior drives one-trial potentiation of hippocampal place fields. Nat Neurosci (2014). [DOI] [PMC free article] [PubMed]
- 23.Happé F, Frith U. The weak coherence account: detail-focused cognitive style in autism spectrum disorders. J Autism Dev Disord. 2006;36:5–25. doi: 10.1007/s10803-005-0039-0. [DOI] [PubMed] [Google Scholar]
- 24.Pastalkova E, Itskov V, Amarasingham A, Buzsáki G. Internally generated cell assembly sequences in the rat hippocampus. Science. 2008;321:1322–1327. doi: 10.1126/science.1159775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiroga RQ, Reddy L, Kreiman G, Koch C, Fried I. Invariant visual representation by single neurons in the human brain. Nature. 2005;435:1102–1107. doi: 10.1038/nature03687. [DOI] [PubMed] [Google Scholar]
- 26.Salmond CH, et al. The role of the medial temporal lobe in autistic spectrum disorders. Eur J Neurosci. 2005;22:764–772. doi: 10.1111/j.1460-9568.2005.04217.x. [DOI] [PubMed] [Google Scholar]
- 27.Happé F, Ronald A, Plomin R. Time to give up on a single explanation for autism. Nat Neurosci. 2006;9:1218–1220. doi: 10.1038/nn1770. [DOI] [PubMed] [Google Scholar]
- 28.Kazdoba TM, Leach PT, Silverman JL, Crawley JN. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizumori SJ, McNaughton BL, Barnes CA, Fox KB. Preserved spatial coding in hippocampal CA1 pyramidal cells during reversible suppression of CA3c output: evidence for pattern completion in hippocampus. J Neurosci. 1989;9:3915–3928. doi: 10.1523/JNEUROSCI.09-11-03915.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbe D, Buzsáki G. Alteration of theta timescale dynamics of hippocampal place cells by a cannabinoid is associated with memory impairment. J Neurosci. 2009;29:12597–605. doi: 10.1523/JNEUROSCI.2407-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middleton SJ, McHugh TJ. Silencing CA3 disrupts temporal coding in the CA1 ensemble. Nat Neurosci. 2016;19:945–51. doi: 10.1038/nn.4311. [DOI] [PubMed] [Google Scholar]
- 32.Cushman JD, et al. Multisensory control of multimodal behavior: do the legs know what the tongue is doing? PLoS One. 2013;8:e80465. doi: 10.1371/journal.pone.0080465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resnik E, McFarland JM, Sprengel R, Sakmann B, Mehta MR. The effects of GluA1 deletion on the hippocampal population code for position. J Neurosci. 2012;32:8952–68. doi: 10.1523/JNEUROSCI.6460-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHugh TJ, Blum KI, Tsien JZ, Tonegawa S, Wilson MA. Impaired hippocampal representation of space in CA1-specific NMDAR1 knockout mice [see comments] Cell. 1996;87:1339–1349. doi: 10.1016/S0092-8674(00)81828-0. [DOI] [PubMed] [Google Scholar]
- 35.Kentros C, et al. Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science. 1998;280:2121–2126. doi: 10.1126/science.280.5372.2121. [DOI] [PubMed] [Google Scholar]
- 36.Ekstrom AD, Meltzer J, McNaughton BL, Barnes CA. NMDA receptor antagonism blocks experience-dependent expansion of hippocampal “place fields”. Neuron. 2001;31:631–638. doi: 10.1016/S0896-6273(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 37.Cabral HO, et al. Oscillatory Dynamics and Place Field Maps Reflect Hippocampal Ensemble Processing of Sequence and Place Memory under NMDA Receptor Control. Neuron. 2014;81:402–415. doi: 10.1016/j.neuron.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godfraind JM, et al. Long-term potentiation in the hippocampus of fragile X knockout mice. American Journal of Medical Genetics. 1996;64:246–251. doi: 10.1002/(SICI)1096-8628(19960809)64:2<246::AID-AJMG2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Zhao MG, et al. Deficits in trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–7392. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang Y, et al. Fragile X mental retardation protein is required for chemically-induced long-term potentiation of the hippocampus in adult mice. J Neurochem. 2009;111:635–646. doi: 10.1111/j.1471-4159.2009.06314.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen T, et al. Pharmacological rescue of cortical synaptic and network potentiation in a mouse model for fragile X syndrome. Neuropsychopharmacology. 2014;39:1955–1967. doi: 10.1038/npp.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brager DH, Akhavan AR, Johnston D. Impaired dendritic expression and plasticity of h-channels in thefmr1(-/y) mouse model of fragile X syndrome. Cell Rep. 2012;1:225–233. doi: 10.1016/j.celrep.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nolan MF, et al. A behavioral role for dendritic integration: HCN1 channels constrain spatial memory and plasticity at inputs to distal dendrites of CA1 pyramidal neurons. Cell. 2004;119:719–732. doi: 10.1016/j.cell.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 45.Radwan B, Dvorak D, Fenton AA. Impaired cognitive discrimination and discoordination of coupled theta-gamma oscillations in Fmr1 knockout mice. Neurobiol Dis. 2016;88:125–138. doi: 10.1016/j.nbd.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colgin LL, et al. Frequency of gamma oscillations routes flow of information in the hippocampus. Nature. 2009;462:353–357. doi: 10.1038/nature08573. [DOI] [PubMed] [Google Scholar]
- 47.Lasztóczi B, Klausberger T. Layer-Specific GABAergic Control of Distinct Gamma Oscillations in the CA1 Hippocampus. Neuron. 2014;81:1126–1139. doi: 10.1016/j.neuron.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 48.Benchenane K, Tiesinga PH, Battaglia FP. Oscillations in the prefrontal cortex: a gateway to memory and attention. Curr Opin Neurobiol. 2011;21:475–485. doi: 10.1016/j.conb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Battaglia FP, Benchenane K, Sirota A, Pennartz CM, Wiener SI. The hippocampus: hub of brain network communication for memory. Trends Cogn Sci. 2011;15:310–318. doi: 10.1016/j.tics.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Battaglia FP, et al. The Lantern: an ultra-light micro-drive for multi-tetrode recordings in mice and other small animals. J Neurosci Methods. 2009;178:291–300. doi: 10.1016/j.jneumeth.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 51.Paxinos, G. & Franklin, K. B. J. The Mouse Brain in Stereotaxic Coordinates. 2nd ed (Academic Press, San Diego, 2001).
- 52.Harris KD, Henze DA, Csicsvari J, Hirase H, Buzsáki G. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J Neurophysiol. 2000;84:401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- 53.Hazan L, Zugaro M, Buzsáki G. Klusters, NeuroScope, NDManager: a free software suite for neurophysiological data processing and visualization. J Neurosci Methods. 2006;155:207–216. doi: 10.1016/j.jneumeth.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Skaggs, W. E., McNaughton, B. L., Gothard, K. G. & Markus, E. J. In Advances in Neural Information Processing Systems (eds Hanson, S. J., Cowan, J. D. & Giles, C. L.) 1030–1037 (Morgan Kaufmann Publishers, San Mateo, 1993).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated and analyzed in the current study is available from the corresponding author on reasonable request.