Abstract Abstract
The development of avian cytogenetics is significantly behind that of mammals. In fact, since the advent of cytogenetic techniques, fewer than 1500 karyotypes have been established. The Barbary partridge Alectoris barbara Bonnaterre, 1790 is a bird of economic interest but its genome has not been studied so far. This species is endemic to North Africa and globally declining. The Chukar partridge Alectoris chukar Gray, 1830 is an introduced species which shares the same habitat area as the Barbary partridge and so there could be introgressive hybridisation. A cytogenetic study has been initiated in order to contribute to the Barbary partridge and the Chukar partridge genome analyses. The GTG, RBG and RHG-banded karyotypes of these species have been described. Primary fibroblast cell lines obtained from embryos were harvested after simple and double thymidine synchronisation. The first eight autosomal pairs and Z sex chromosome have been described at high resolution and compared to those of the domestic fowl Gallus domesticus Linnaeus, 1758. The diploid number was established as 2n = 78 for both partridges, as well as for most species belonging to the Galliformes order, underlying the stability of chromosome number in avian karyotypes. Wide homologies were observed for macrochromosomes and gonosome except for chromosome 4, 7, 8 and Z which present differences in morphology and/or banding pattern. Neocentromere occurrence was suggested for both partridges chromosome 4 with an assumed paracentric inversion in the Chukar partridge chromosome 4. Terminal inversion in the long arm of the Barbary partridge chromosome Z was also found. These rearrangements confirm that the avian karyotypes structure is conserved interchromosomally, but not at the intrachromosomal scale.
Keywords: Barbary partridge Alectoris barbara, chukar partridge Alectoris chukar, endemic species, banding cytogenetics, high resolution chromosomes, homologies, intrachromosomal rearrangements
Introduction
The Barbary partridge Alectoris barbara Bonnaterre, 1790 (Phasianidae) is the only native partridge naturally present in Algeria. This North African endemic species is found not only from Morocco to Egypt, but also in Gibraltar, Sardinia and the Canary Islands (Cramp and Simmons 1980, Madge and McGowan 2002). The Barbary partridge is a nesting sedentary bird found in different ecosystems: Mediterranean (coastal dunes and Atlas Mountains), Steppic and Saharian. This common game bird is prized for its meat; hence its overhunting leads to declining population size in some areas. Although the Barbary partridge is listed as Least Concern on the IUCN Red List (2015) (International Union of Conservation of Nature), it is nevertheless protected by several conventions. Indeed, the Barbary partridge was placed on the regulated species list protected by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES): Bird instruction 79/409 (Annex I, II / 2, III / 1). This bird is also protected by the Convention on the Conservation of European Wildlife and Natural Habitats (Bern Convention). Furthermore, the Barbary partridge has also a national scope of protection in commercialisation of some bird species on the French territory. Decline of the native population is mainly due to predation, to poaching (despite the law prohibiting hunting since 1991) and habitat degradation due to mechanisation of farming and urban proliferation (Madge and McGowan 2002). In Morocco, observations have also shown a sharp decrease in Barbary partridge populations, which could become alarming in the long term (Maghnouj 1991). Other factors such as excessive use of pesticides, hikers and stray animals could also disrupt the smooth conduct of breeding. All these factors are also responsible for the decline of partridge populations in Europe (Tejedor et al. 2007, Randi 2008).
In addition, introduction of the exotic Chukar partridge Alectoris chukar Gray, 1830 could also lead to introgression in the wild genome of native partridge and could give rise to infertile descendants. In fact, hybridisation may occur when isolating mechanisms break down naturally or as a result of human activity as in the Alectoris partridges (Barbanera et al. 2011). Several studies have recorded cases of artificial genetic pollution of Alectoris rufa Linnaeus, 1758 and Alectoris graeca Meisner, 1804 by the Alectoris chukar genome (Randi et al. 2003, Barbanera et al. 2005, Barilani et al. 2007, Tejedor et al. 2007). The Barbary partridge is the most phylogenetically divergent taxon in the genus Alectoris, while Alectoris chukar is the most recent gamebird (Randi 1996, Randi and Lucchini 1998, Kimball et al. 1999). Alectoris barbara and chukar lineages split from an ancestral species about 6 million years ago, at the Miocene-Pliocene boundary (Voous 1976, Randi et al. 1992).
Preservation of this endemic species is a priority, which has led to a restocking programme with captive-reared Barbary partridge carried out by the Centre Cynégétique de Zéralda (36°42'06"N, 2°51'47"E). The goal of this project is to obtain strains able to reproduce in captivity, and formulate demographic monitoring after repopulation. Although the Barbary partridge is the main game-bird species in North Africa, scarce research has been reported and it concerns the reproduction and ecology of this species (Alaoui 1992, Akil and Boudedja 2001). However, recent genetic studies have established genetic tests aiming to identify hybrid individuals (Rodríguez-García and Galián 2014). Actually, the only classical cytogenetics data reported on Alectoris genus concern red-legged and Chukar partridges whose karyotypes have been described by use of conventional staining (Arruga et al. 1996, Babak et al. 2014, Ishishita et al. 2014).
The Barbary partridge Alectoris barbara like the domestic fowl Gallus domesticus Linnaeus, 1758 belongs to the ancestral order of Galliformes which includes the most avian species whose genomes have been analysed. In fact, the domestic fowl is the best described one because of its economic importance. It is considered as a reference in phylogenetics and comparative genomics and represents the only standardised bird karyotype (Ladjali-Mohammedi et al. 1999). As a typical avian genome, the karyotype of the domestic fowl has 39 pairs of chromosomes represented by 10 pairs of autosomal macrochromosomes (1–10 chromosomes), 28 pairs of microchromosomes (11–38) and one pair of sex chromosomes. The male is the homogametic sex ZZ (equivalent to human XX), whereas the female is the heterogametic sex ZW (equivalent to human XY) (Masabanda et al. 2004). Despite their small physical size, microchromosomes are characterised by high gene density, high GC content (McQueen et al. 1996) and an early replicating pattern compared to macrochromosomes (Schmid et al. 1989, Ponce de Leon et al. 1992, Burt 2002). Due to the presence of high number of near-undistinguishable microchromosomes, most bird karyotypes are partial and confined to a few macrochromosomes (Shibusawa et al. 2004). However, the use of chicken probes has allowed identification of several microchromosomes in some bird species (Fillon et al. 1998, Nie et al. 2015, Galkina et al. 2017, Kretschmer et al. 2018).
On the other hand, the chicken is the first avian genome to have been sequenced (Hillier et al. 2004), followed by the zebra finch Taeniopygia guttata (Warren et al. 2010) and Turkey Meleagris gallopavo (Dalloul et al. 2010). The chicken genome assembly Gallus_gallus-4.0 covered 1.03 Gb or 96% of the total genome size, including the sequence of the 10 macrochromosomes, 19 microchromosomes and sex chromosomes (Schmid et al. 2015). Recently, coverage was improved by a gain of 183 Mb and three microchromosomes (30, 31 and 33) in the Gallus_gallus-5.0 assembly. However, 138 Mb are not yet assigned to chromosomes (Warren et al. 2017). Rapid advances in genome assembly software and technologies as Next Generation Sequencing (NGS) allowed entire genome sequencing of more than 57 birds (Dalloul et al. 2010, Jarvis et al. 2014). Among these species, 42 were a part of the Genome 10K Project which aims to facilitate the sequencing and analysis of 10.000 vertebrate genomes (http://genome10k.soe.ucsc.edu) (Genome 10K Community of Scientists 2009). The Avian Phylogenomics Consortium announced in 2015 a great project called B10K (web.bioinfodata.org/B10K) to generate draft genome sequences for all the 10.476 avian species within the next five years (until 2020). All these sequencing data corroborate the exceptional stability of avian karyotypes (Shibusawa et al. 2002, Derjusheva et al. 2004, Shibusawa et al. 2004). Indeed, the occurrence of interchromosomal rearrangements in birds is a relatively rare event estimated to 1.25 per million years, which is considerably lower than in mammals (Zhao and Bourque 2009, Romanov et al. 2014, Zhang et al. 2014). It is assumed that interchromosomal reshuffling could be the result of an adaptive response and a cause or consequence of speciation (King 1995, Griffin et al. 2007, Romanov et al. 2014).
Although avian high resolution mapping is well advanced, reported cytogenetic studies are nevertheless partial and fewer than those of mammals despite great contribution of this discipline. In fact, classical and banding cytogenetics highlighted important features of avian karyotype as interchromosomal stability (Tegelstrôm and Ryttman 1981, Belterman and De Boer 1984, Christidis 1990, Shibusawa et al. 2004) and intrachromosomal reshuffling in some macrochromosomes (Stock and Bunch 1982, Griffin et al. 2007, Hooper and Price 2017). Banding cytogenetics has also elucidated the process of karyotypic evolution in some orders of bird (Dobigny et al. 2004, Shibusawa et al. 2004, Nishida et al. 2008) and even in mammals (Di-Nizo et al. 2017).
The aim of the present study is to describe the chromosomes of Barbary partridge Alectoris barbara and Chukar partridge Alectoris chukar at high resolution level with morphological and dynamic banding techniques. Comparison of partridges and chicken banding patterns has been conducted in order to estimate the degree of conservation and rearrangements of these species during speciation.
Material and methods
Biological material
Barbary and Chukar partridge embryos were obtained from the Centre Cynégétique de Zéralda during the laying period (March to June). Four Barbary partridge and four Chukar partridge embryos were sampled after 5–6 days incubation at 37 °C, and kept under the same temperature and hygrometry conditions in the Laboratoire de Génétique du Développement (Faculté des Sciences Biologiques, USTHB) until at least 12 days old.
Cell cultures
Primary fibroblast cell cultures were harvested from 6 to 12 days old embryos. The embryos were cleared from their annexes and totally ground in a trypsine solution (0.05%, Sigma). Cell suspension were incubated at 41 °C with an estimate concentration of 3×106 cells/ml in RPMI 1640 culture medium (20 mM HEPES, GIBCO) supplemented with 10% foetal calf serum (FCS, GIBCO), 1% L-Glutamine 200 mM (Sigma), 1% Penicillin, Streptomycin and Fungizone (Sigma). Trypsinisation of cells was realised to enhance division ability (adapted from Ladjali 1994, Ladjali et al. 1995).
Synchronisation of cell cultures
In order to increase the yield of metaphases and prometaphases cells, cultures were synchronised with a simple and double thymidine block during the S phase (Dutrillaux and Couturier 1981, Hayes et al. 1993, Ladjali et al. 1995). Cells were blocked for the first time during 16–18h with thymidine (final concentration: 10mg/ml, Sigma), and rinsed 2×15 min with BSS+ (Hank’s Balanced Salt Solution containing 5.6% NaHCO3 and 2 mM CaCl2) at 41 °C. Cells were incubated again in culture medium RPMI, and the day after, the step above was repeated for a second time to produce a double thymidine block. On the third day, when cells restarted division in RPMI with 5% FCS, an analogue of thymidine 5-Bromo-2-deoxyUridine (final concentration: 10 μg/ml, BrdU, Sigma) was incorporated into cultures. An hour after, 5-Fluoro-2-uridine (final concentration: 0.5 μg/ml, FdU, Sigma) was added to enhance BrdU incorporation. These treatments are required to prepare chromosomes for dynamic R-banding staining (Dutrillaux and Couturier 1981, Schmid et al. 1989, Hayes et al. 1993, Ladjali et al. 1995).
Cell harvest
The incorporation of BrdU into the S phase lasted 6–7 hours. Meanwhile cells were continuously observed by reversed microscope until the number of mitotic round cells peaked. Cells were trypsinysed (trypsine 0.05% + 0.02% EDTA, GIBCO) and harvested in a 15 ml tube with colchicine (final concentration: 0.05 μg/ml, Sigma). After centrifugation, hypotonic treatment was undertaken during 13 min at 37 °C with diluted newborn calf serum (1:5). Intracytoplasmic structures were prefixed with 1 ml of methanol/acetic acid (3:1) at 37 °C. Fixation was finally realised at 4 °C and after centrifugation, 1 ml was let in tubes until spreading. Slides were washed, rubbed and placed in cold water. A few drops from the cell suspension were spread at 10 cm of cold slide and left to dry until staining procedures occurred (adapted from Ladjali et al. 1995).
Banding staining
GTG-banding (G-bands obtained with Trypsin and Giemsa) was realised following the Seabright modified method (1971). Approximately; 3 to 4 days after spreading, slides were incubated for 8–10 seconds in a trypsine solution (final concentration: 0.25%, Sigma) at room temperature. Slides were rinsed twice in PBS- (Phosphate Buffered Solution, pH=6.8) and stained in 6% Giemsa for 8–10 minutes.
RBG-FPG banding (R-bands followed by fluorochrome-photolysis) procedure was undertaken following Ladjali et al. (1995). Slides were incubated in Hoechst 33258 solution (1 mg/ml) for 20 min. Slides were then rinsed and placed for 90 min in 2 × SSC buffer (Saline Sodium Citrate) at a distance of 15 cm from UV dark light (Mazdafluor OE TFWN 20). Slides are rinsed again and placed in Earle’s buffer at 87 °C for 10 min. Slides were washed and incubated for 20 min in 6% Giemsa staining solution.
RHG-banding (R-bands obtained by Heat and Giemsa) was realised on A. chukar spreads. Slides were incubated in Earle’s buffer (ph=5,8) at 87 °C for 20 minutes, then rinsed and stained in 6% Giemsa solution (containing phosphate buffer) (Dutrillaux and Leujeune 1971, Comings 1978).
Chromosome Classification
Slides were first observed with an optical microscope at objective magnification 10× to estimate the mitotic index (AxioZeiss Scope A1). Slides, showing a higher mitotic index, were analysed and prometaphases and metaphases, showing decondensed and dispersed chromosomes, were photographed (CoolCube1 Metasystems). The first eight macrochromosomes and Z sex chromosomes from Barbary partridge Alectoris barbara and Chukar partridge Alectoris chukar were classified in G- and R- banding as described in International System of Standardised Avian Karyotypes ISSAK (Ladjali-Mohammedi et al. 1999). Macrochromosomes pairs were classified according to decreasing size and centromere position (Shoffner 1974), whereas microchromosomes were not presented because of their small physical size making very difficult any classification or description. In order to avoid any ambiguity, nomenclature adopted in this article followed the ISSAK (Ladjali-Mohammedi et al. 1999) adapted from ISCN (1978).
Chromosome measurement
Analyses measurements of fifteen first pairs of chromosomes were undertaken using KARYOTYPE 2.0 software (Altinordu et al. 2016). Measured parameters were: Long (q) and short (p) arms, total chromosome length (p+q) and arm ratio r: Long/short. In the Results section below, morphometry will be presented of the first eight chromosomes and the Z chromosome, which have been compared to the domestic fowl. Other microchromosomes were physically too small and did not give significant values. Partridge’s karyotypes have been established manually, considering that software used in the present work was not adapted to birds.
Results
Primary fibroblasts cell lines were obtained a few hours after incubation and constituted a good source for obtaining chromosome preparations. The younger the embryos, the more mitotic divisions were obtained. The strict follow up of cell divisions after inhibition removal enabled the estimation of half cycle time to 7–8 hours for Barbary partridge Alectoris barbara and 6–7 hours for Chukar partridge Alectoris chukar. Important mitotic indices with high resolution chromosomes were obtained with simple synchronisation for A. barbara and double synchronisation for A. chukar during 18h. Furthermore, observation of cell cultures of both species showed that A. barbara cells were much more sensitive than A. chukar to the different drugs added during incubation. Trypsinisation and synchronisation steps caused important Barbary partridge cell death compared to Chukar partridge. In fact, we have incubated an average of 3×106 cells/ml (Ladjali 1994). After a confluence, we estimate that cells have divided four times (12.106 cells/ml). Following the trypsinisation, cells divided twice (24×106 cells/ml). A continuous observation of cultures after in vitro treatments shows an average decrease of 30% of live cells of Barbary partridge, equivalent to 7.2×106 cells/ml for all four embryos. Whereas, no diminution of mitotic power was observed in Chukar partridge regardless of trypsination, addition of BSS+, BrdU/FdU or colchicine.
Diploid numbers of Barbary partridge Alectoris barbara and Chukar partridge Alectoris chukar were estimated as 2n=78 from most metaphase plates (Fig. 1). Like most of birds, A. barbara and A. chukar karyotypes are composed of a few pairs of macrochromosomes and several microchromosomes with small physical size, which are very difficult to distinguish.
Figure 1.
Estimation of diploid number of Barbary and Chukar partridges. Major metaphase plates (10) displayed diploid number 2n= 78 chromosomes.
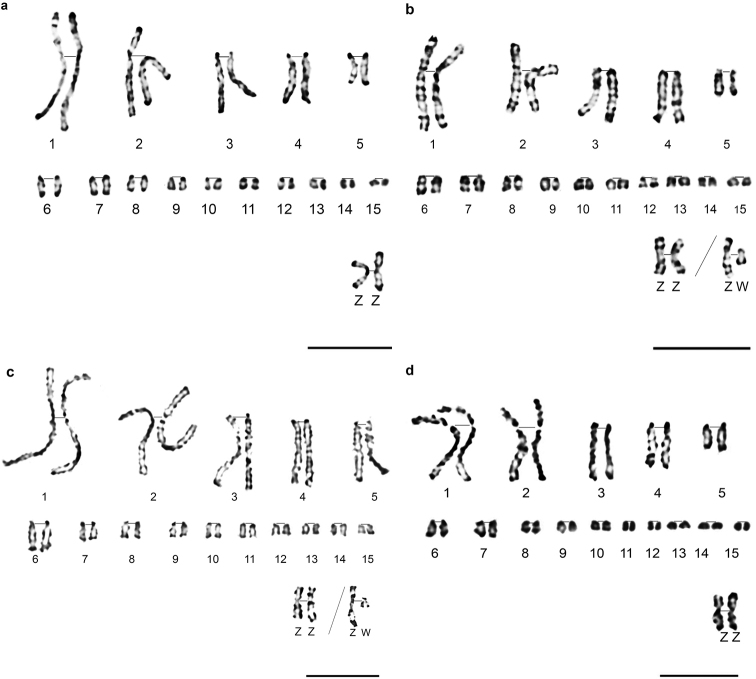
The authors proposed Alectoris barbara partial karyotype in GTG (Fig. 2a) and RBG banding (Fig. 2b), and Alectoris chukar partial karyotype in GTG (Fig. 2c) and RHG banding (Fig. 2d). Most metaphases show male genetic sex ZZ for both partridges, wherefore gonosome W was only described in RBG bands for A. barbara and GTG bands for A. chukar. The success of simple and double synchronisation resulted in high resolution chromosomes. Measurements show that chromosomes of A. chukar were more decondensed than those of A. barbara (Table 1). In fact, the size of the first eight macrochromosomes ranges from 14 µm to 3µm in A. chukar and from 9 µm to 2 µm for A. barbara. This is certainly due to the success of double synchronisation and extreme resistance of A. chukar cells to drugs added in vitro.
Figure 2.
Partial karyotypes of A. barbara in GTG bands (a), A. barbara in RBG bands (b), A. chukar in GTG bands (c), and A. chukar in RHG bands (d). Gonosomes Z W are classified apart. Scale bars: 5 µm.
Table 1.
A. barbara and A. chukar morphometry of the first eight macrochromosomes and gonosomes. Means are obtained at least from 10 prometaphases/metaphases (from 10 to 20). Chr: chromosome, q: long arm, p: short arm, t: total (p+q), r: ratio (q/p), lengths are given in micrometer (µm).
| A. barbara | A. chukar | |||||||
|---|---|---|---|---|---|---|---|---|
| Chr | p | q | t | r | p | q | t | r |
| 1 | 3.78 | 5.98 | 9.76 | 1.58 | 5.63 | 8.82 | 14.45 | 1.56 |
| 2 | 2.89 | 4.71 | 7.6 | 1.62 | 3.83 | 6.76 | 10.59 | 1.76 |
| 3 | 1.03 | 5.57 | 6.6 | 5.4 | 1.2 | 7.5 | 8.7 | 6.25 |
| 4 | 1.02 | 4.33 | 5.35 | 4.24 | 1.15 | 6.19 | 7.34 | 5.38 |
| 5 | 0.75 | 2.85 | 3.6 | 3.8 | 0.78 | 4.9 | 5.68 | 6.28 |
| 6 | 0.68 | 2.32 | 3 | 3.41 | 0.75 | 3.35 | 4.1 | 4.46 |
| 7 | 0.7 | 1.7 | 2.4 | 2.42 | 0.7 | 3 | 3.7 | 4.28 |
| 8 | 0.53 | 1.57 | 2.1 | 2.96 | 0.63 | 2.37 | 3 | 3.76 |
| Z | 2.5 | 3.1 | 5.6 | 1.24 | 3.1 | 3.5 | 6.6 | 1.12 |
| W | 0.75 | 1.03 | 1.78 | 1.37 | 0.93 | 1.37 | 2.3 | 1.47 |
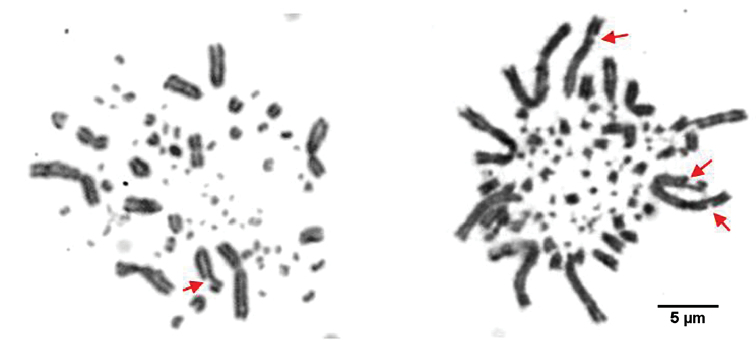
Observation of partridge’s spreads shows that in A. barbara an average of 45 metaphases /100 displayed break points. These breaks seem to appear in sub-terminal regions of macrochromosomes 1 and 3 (Fig. 3). None of A. chukar metaphases have shown this phenomenon. Furthermore, the same typical distribution of partridge’s chromosomes was observed. In fact, macrochromosomes are preferentially located towards the mitosis periphery, while microchromosomes are clustered within the mitosis interior (Fig. 3).
Figure 3.
Partridges’ metaphases showing spatial distribution of chromosomes (A. barbara on the left and A. chukar on the right). Macrochromosomes are located towards metaphases periphery, microchromosomes are confined to the central area. Arrows indicates break points in chromosomes. Bar = 5 µm.
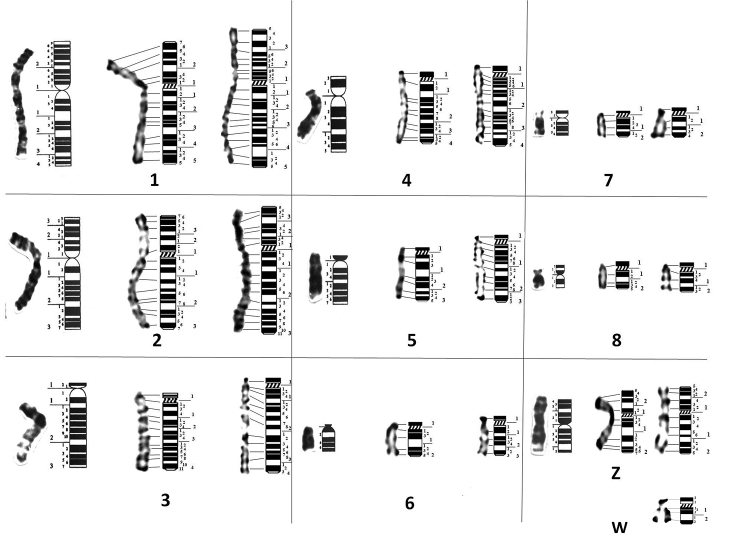
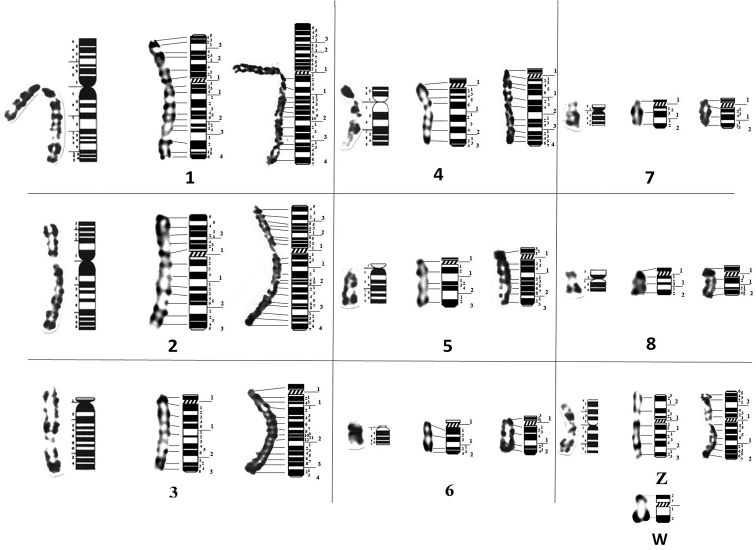
Partial ideograms of A. barbara and A. chukar were proposed on the basis of means of 20 metaphases plates following the International System of Standardised Avian Karyotypes (Ladjali-Mohammedi et al. 1999) (Fig. 4 and Fig. 5, Table 2 and 3).
Figure 4.
GTG partial ideograms of (from left to right) G. domesticus (Ladjali-Mohammedi et al. 1999), A. barbara and A. chukar. W chromosome is represented only in A. chukar. Horizontal traits indicate correspondence of positive bands between chromosomes and ideograms. Along ideograms: Large numbers indicate regions, smallest numbers indicate positive and negative bands.
Figure 5.
RBG and RHG partial ideograms of (from left to right) G. domesticus (Ladjali-Mohammedi et al. 1999), A. barbara and A. chukar. W chromosome is represented only in A. barbara. Horizontal traits indicate correspondence of positive bands between chromosomes and ideograms. Along ideograms: Large numbers indicate regions, smallest numbers indicate positive and negative bands.
Table 2.
Values summarized from partial ideograms of A. barbara and A. chukar described in GTG bands. Chr: chromosome, p: short arm, q: long arm, R: region, B: bands, LM: Landmark (all positions show negative landmarks except when (+) is added), empty boxes indicate that there is no particular landmark.
| Alectoris barbara | Alectoris chukar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | P arm | Q arm | P arm | Q arm | ||||||||
| R | B | LM | R | B | LM | R | B | LM | R | B | LM | |
| 1 | 2 | 11 | (21), (26) | 5 | 21 | (41) | 3 | 17 | (33) | 5 | 23 | (51) |
| 2 | 3 | 11 | (21) | 3 | 19 | (21), (31) | 3 | 13 | (31) | 3 | 21 | (31) |
| 3 | 1 | 3 | - | 4 | 23 | (13) (21) | 1 | 2 | - | 4 | 23 | (31), (41) |
| 4 | 1 | 1 | - | 4 | 19 | (21) | 1 | 2 | - | 4 | 25 | (26) + |
| 5 | 1 | 1 | - | 3 | 12 | (21) | 1 | 2 | - | 3 | 19 | (22) + |
| 6 | 1 | 1 | - | 2 | 8 | (21) | 1 | 2 | - | 3 | 9 | (22), (24) |
| 7 | 1 | 1 | - | 2 | 6 | - | 1 | 1 | - | 2 | 6 | (21) |
| 8 | 1 | 1 | - | 2 | 7 | - | 1 | 1 | - | 2 | 7 | - |
| Z | 2 | 7 | (21) | 2 | 11 | (21), (22)+ | 2 | 9 | - | 2 | 11 | (15), (21) |
| W | - | - | - | - | - | - | 1 | 2 | - | 2 | 5 | (11)+(22)+ |
Table 3.
Values summarized from partial ideograms of A. barbara and A. chukar described in RBG/RHG bands. Chr: chromosome, p: short arm, q: long arm, R: region, B: bands, LM: Landmark (all positions show negative landmarks except when (+) is added), empty boxes indicate that there is no particular landmark.
| Alectoris barbara | Alectoris chukar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | P arm | Q arm | P arm | Q arm | ||||||||
| R | B | LM | R | B | LM | R | B | LM | R | B | LM | |
| 1 | 3 | 13 | (31) | 4 | 20 | (41) (42) | 3 | 18 | (21) (31) | 4 | 25 | (13), (31), (45) |
| 2 | 2 | 10 | (25) | 3 | 15 | (21) | 3 | 17 | (21) (31) | 4 | 25 | (31) |
| 3 | 1 | 2 | - | 3 | 16 | (22)+(31) | 1 | 2 | - | 4 | 27 | (31) |
| 4 | 1 | 2 | - | 3 | 14 | (21) (31) | 1 | 3 | - | 4 | 21 | (14)+, (16)+ (22)+ , (24)+ |
| 5 | 1 | 2 | - | 3 | 8 | (21) (31) | 1 | 3 | - | 3 | 15 | (12)+, (14)+ |
| 6 | 1 | 2 | - | 2 | 7 | (21) | 1 | 3 | - | 2 | 8 | (21) |
| 7 | 1 | 1 | - | 2 | 5 | (21) | 1 | 3 | - | 2 | 7 | (14)+ |
| 8 | 1 | 1 | - | 2 | 5 | (13)+ | 1 | 3 | - | 2 | 7 | (21) |
| Z | 2 | 7 | (21) | 3 | 9 | (24)+, (31) | 2 | 10 | (21) | 2 | 13 | (21) |
| W | 1 | 2 | (12)+ | 1 | 2 | (12)+ | - | - | - | - | - | - |
Partial ideograms of Alectoris barbara and Alectoris chukar described in GTG bands (Fig. 4, Table 2)
Chromosome 1
P arm
Barbary partridge: two regions. 11 G bands with a visible negative band (21) which divides the p arm into two regions. A large terminal positive band is also visible (26).
Chukar partridge: three regions. 17 G bands with a predominant terminal negative band (33).
Q arm
Barbary partridge: Five regions. 21 bands, four negative bands divide the q arm into four regions with one predominant negative band (41). The centromeric region is positively banded.
Chukar partridge: Five regions. 23 G bands, with a wide terminal negative band (51).
Chromosome 2
P arm
Barbary partridge: three regions. 11 G bands with a large negative proximal band (21).
Chukar partridge: three regions. 13 G bands with large negative terminal band (31).
Q arm
Barbary partridge: three regions. 19 G bands with two wide negative bands (21 and 31).
Chukar partridge: three regions. 21 G bands with a large negative subtelomeric band (31).
Chromosome 3
P arm
Barbary partridge: one region with 3 G bands.
Chukar partridge: one region with 2 G bands.
Q arm
Barbary partridge: four regions. 23 G bands with two wide proximal negative bands (13 and 21).
Chukar partridge: four regions. 23 G bands with two large negative bands (31 and 41).
Chromosome 4
P arm
Barbary partridge: one region.
Chukar partridge: one region with 2 G bands.
Q arm
Barbary partridge: four regions. 19 G bands with a wide proximal negative band (21).
Chukar partridge: four regions. 25 G bands with a visible central positive band (26).
Chromosome 5
Q arm
Barbary partridge: three regions. 12 G bands with a wide central negative band (21).
Chukar partridge: three regions. 19 G bands with a visible central positive band (22).
Chromosome 6
Q arm
Barbary partridge: two regions. 8 G bands with a wide central negative band (21).
Chukar partridge: two regions. 9 G bands with two central positive bands (22 and 24).
Chromosome 7
Q arm
Barbary partridge: two regions. 6 G bands.
Chukar partridge: two regions. 6 G bands with a visible central negative band (21).
Chromosome 8
Q arm
Barbary partridge: two regions. 7 G bands with a wide central negative band (21).
Chukar partridge: two regions. 7 G bands with a large terminal negative band (21).
Chromosome Z
P arm
Barbary partridge: two regions. 7 G bands showing a large negative band (21).
Chukar partridge: two regions. 9 G bands with a visible negative band (21).
Q arm
Barbary partridge: two regions. 11 G bands with a large negative band (21) and a positive land mark (22).
Chukar partridge: two regions. 11 G bands with two large negative bands (15 and 21).
Chromosome W
P arm
Chukar partridge: one region. 2 G bands with terminal positive band.
Q arm
Chukar partridge: two regions. 5 G bands with one positive subcentromeric band (11) and a telomeric positive band (22)
Partial ideograms of Alectoris barbara and Alectoris chukar described in RBG / RHG bands (Fig. 5, Table 3)
Chromosome 1
P arm
Barbary partridge: three regions. 13 RBG bands with a large terminal negative band (31).
Chukar partridge: Three regions. 18 RHG bands with two principal negative bands (21 and 31).
Q arm
Barbary partridge: Four regions. 20 bands with two wide terminal respectively negative and positive bands (41 and 42). The centromeric region is positively banded.
Chukar partridge: Four regions. 25 bands with three large negative bands which divided the q arm (13, 31 and 45).
Chromosome 2
P arm
Barbary partridge: two regions. 10 bands with a large negative telomeric band (25).
Chukar partridge: three regions. 17 bands with large negative proximal band (21).
Q arm
Barbary partridge: three regions. 15 bands with two wide negative bands (21 and 31).
Chukar partridge: four regions. 25 bands with a large negative telomeric band (31).
Chromosome 3
P arm
Barbary partridge: one region with 2 bands.
Chukar partridge: one region with 2 bands.
Q arm
Barbary partridge: three regions. 16 bands with a central positive band (22) and a telomeric negative band (31).
Chukar partridge: four regions. 27 bands with a large submedian negative band (31).
Chromosome 4
P arm
Barbary partridge: one region and 2 bands.
Chukar partridge: one region with 3 bands.
Q arm
Barbary partridge: three regions. 14 bands with two visible negative bands (21 and 31).
Chukar partridge: four regions. 21 bands with two proximal positive bands (14 and 16) and two central positive bands (22 and 24).
Chromosome 5
P arm
Barbary partridge: one region. 2 RBG bands.
Chukar partridge: one region. 3 RHG bands.
Q arm
Barbary partridge: three regions. 8 bands with two wide negative bands (21 and 31).
Chukar partridge: three regions. 15 bands with two large proximal positive bands (12 and 14).
Chromosome 6
P arm
Barbary partridge: one region showing 2 RBG bands.
Chukar partridge: one region presenting 3 RHG bands.
Q arm
Barbary partridge: two regions. 7 bands with a wide central negative band (21).
Chukar partridge: two regions. 8 bands and a large negative band (21).
Chromosome 7
P arm
Barbary partridge: one region.
Chukar partridge: one region with 3 RHG bands.
Q arm
Barbary partridge: two regions. 5 bands showing a large distal negative band (21).
Chukar partridge: two regions. 7 bands with a central positive band (14).
Chromosome 8
P arm
Barbary partridge: one region with one band.
Chukar partridge: one region with 3 RHG bands.
Q arm
Barbary partridge: two regions. 5 bands with a central positive band (13).
Chukar partridge: two regions. 7 bands and a central negative band (21).
Chromosome Z
P arm
Barbary partridge: two regions. 7 R bands and a wide terminal negative band (21).
Chukar partridge: two regions. 10 R bands with a large negative band (21).
Q arm
Barbary partridge: three regions. 9 R bands with a positive terminal land mark (24) and a large negative band (31).
Chukar partridge: two regions. 13 R bands with a visible terminal negative band (21).
W chromosome
P arm
Barbary partridge: one region. 2 RBG bands with terminal positive band.
Q arm
Barbary partridge: one region. 2 RBG bands with a large positive telomeric band. Centromeric region is negatively stained.
Alectoris barbara, Alectoris chukar and Gallus domesticus chromosome comparison
Comparison of morphological and dynamic G and R banding of A. barbara and A. chukar with domestic fowl (Ladjali-Mohammedi et al. 1999) shows a wide conservation of patterns in macrochromosomes. However, some rearrangements in partridges chromosomes 4 and Z were observed (Fig. 4 and Fig. 5). All centromeric regions of partridge chromosomes were positively stained in G and R banding. Chromosomes 1 and 2 are submetacentric in both Barbary and Chukar partridges, like in the domestic fowl. Despite the difference in chromosome 1 length, the arm ratio is quite similar (r = 1.58 / 1.56) (Table 1). On the other hand, the A. chukar long arm of chromosome 2 is longer than for A. barbara (r = 1.76 / 1.62). In both partridges, the centromere position is more submedian in chromosome 2 compared to chromosome 1. Chromosome 3 is acrocentric in partridges and domestic fowl. The banding pattern of the first three chromosomes is apparently widely conserved in all three species. Chromosome 4 is acrocentric in partridges and telocentric in chicken. The banding pattern is, however, conserved in A. barbara and G. domesticus, while in A. chukar, the subcentromeric region presents a different profile (Fig. 6). Chromosomes 5 and 6 are acrocentric in each species and present a similar pattern distribution, although, A. chukar presents higher number of bands due to decondensation. Chromosome 7 and 8 are acrocentric in both partridges and respectively, telocentric and submetacentric in the domestic fowl (Ladjali-Mohammedi et al. 1999). Surprisingly, the distribution of bands is conserved through these three Galliformes (comparison of chromosomes at the same decondensation stage). Sex chromosomes Z and W are submetacentric and morphologically conserved in all three species. However, the terminal region of the Z chromosome long arm presents a different pattern in A. barbara compared to that of A. chukar and the domestic fowl (Fig. 7). In the present work, we found in A. barbara a total of 145 G/123 R-bands and in A. chukar 173 G/187 R-bands only for the first eight chromosomes (Table 2 and 3).
Figure 6.
Schematic representation of a paracentric inversion in A. chukar (a), G. domesticus and A. barbara (b) chromosome 4. Corresponding bands are indicated by dashes. (ACH: A. chukar, GGA: G. domesticus and ABA: A. barbara).
Figure 7.
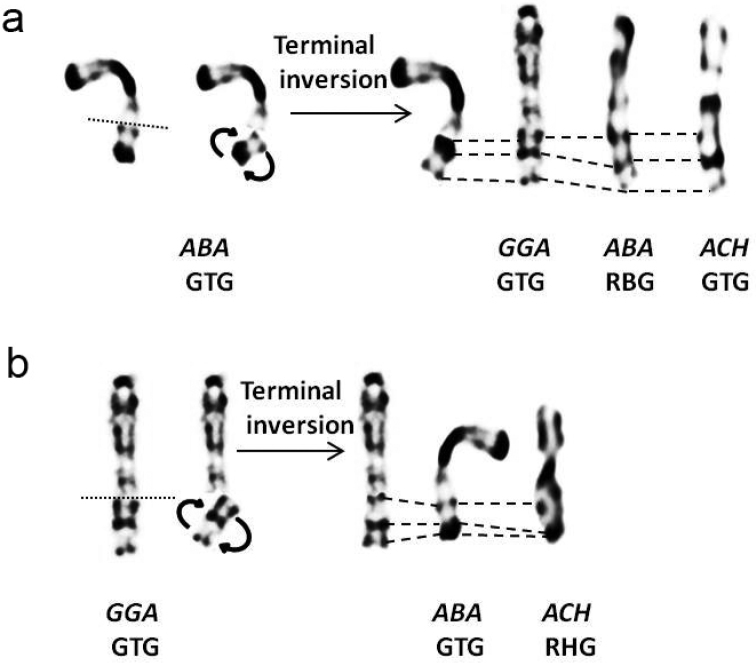
Schematic representation of a terminal paracentric inversion in chromosome Z of A. barbara in GTG (a) and chromosome Z of G. domesticus in GTG banding (b). Corresponding bands are indicated by dashes. (ACH: A. chukar, GGA: G. domesticus and ABA: A. barbara). Rearranged ABA Z in GTG corresponds to GGA Z and ACH Z in GTG. Rearranged GGA Z in GTG corresponds to ABA in GTG and ACH in RHG.
Discussion
Implementation of fibroblasts was observed in all cultures and confluence was quickly reached in all eight embryos, mainly in the youngest ones (6 days). This is due to the important mitotic power of cells at early embryonic stages (Ladjali et al. 1995). The high mortality in cell cultures of A. barbara (for the four embryos) is interestingly reflected in breeders’ observations regarding the Barbary partridge’s high vulnerability in breeding areas, unlike the Chukar partridge (personal communication of the Centre Cynégétique de Zéralda). Indeed, the Barbary partridge is a vulnerable endemic species, whereas Chukar partridge is usually introduced to reinforce the low local densities populations because of its easy practical prolificacy in captivity compared to other partridges (Rojas et al. 2011).
Distribution of partridges’ macrochromosomes and microchromosomes in metaphases is similar to that reported in several studies on chicken fibroblasts and neurons nucleis (Habermann et al. 2001, Federico et al. 2005) and mammalian fibroblasts nuclei (Cremer et al. 2000) (Fig. 3). In fact, it was reported that gene dense and early replicating chromatin, represented by microchromosomes (McQueen et al. 1996, Schmid et al. 1989, Ponce de Leon et al. 1992, Burt 2002, Skinner et al. 2009) were located in the nuclei central area, surrounded by gene-poor and later replicating chromatin (macrochromosomes) (Cremer et al. 2000). These results indicate that the radial position of chromosome territories is correlated with their size, their gene-density and replication timing (Habermann et al. 2001, Federico et al. 2005). Further, this specific distribution was assumed to be evolutionarily conserved in Galliformes (Maslova and Krasikova 2011) and also between mammals and birds despite their highly divergent karyotypes (Tanabe et al. 2002). The typical distribution of macro-and microchromosomes in metaphases could explain the particularly low rate of interchromosomal rearrangements in Galliformes (Shibusawa et al. 2002).
Fortuitously, 45% of A. barbara metaphase plates show breaks on some macrochromosomes which could be identified as fragile sites (Fig. 3). In birds, breakpoint regions of fragile sites are frequently associated with chromosomal rearrangements (Zlotina et al. 2010, Itoh et al. 2011, Skinner and Griffin 2012). Chromosomal fragile sites are loci prone to breakages within metaphase chromosomes (Fungtammasan et al. 2012). In mammals and birds, these breaks are assumed to occur in repetitive DNA clusters (Zlotina et al. 2010). Nevertheless, recent works in humans show that chromosomal rearrangements could appear in early replicating and actively transcribed gene clusters (Mortusewicz et al. 2013). It can be assumed that Barbary partridge chromosomes are particularly vulnerable to breakages, which could be favorable to intrachromosomal reshuffling. It would be very interesting to explore such genomic regions by molecular tools.
The diploid number of Alectoris barbara and Alectoris chukar was estimated as 2n = 78. This result is concordant with the exceptional stability of avian karyotype, i.e. about 65% of karyotyped birds displayed 76 to 82 chromosomes, including 7 to 8 pairs of macrochromosomes (Christidis 1990, Rodionov 1997). The diploid number of partridges emphasizes the conservation of karyotypes in the order of Galliformes (Stock and Bunch 1982, Shibusawa et al. 2002, Shibusawa et al. 2004). This is the case for the Chukar partridge described by Ishishita et al. (2014), as well as domestic fowl Gallus domesticus (Pollock and Fechheimer 1976, Ladjali-Mohammedi et al. 1999); Red-legged partridge Alectoris rufa (Arruga et al. 1996) and Japanese quail Coturnix japonica Temminck & Schlegel, 1849 (Stock and Bunch 1982). Interchromosomal conservation of partridges karyotype was also shown in previous studies. In fact, cross species painting using chicken macrochromosomes DNA probes (Zoo-FISH) has shown a perfect homology with, respectively, A. chukar and Alectoris rufa macrochromosomes (Kasai et al. 2003, Ishishita et al. 2014). Karyotypes of A. barbara and A. chukar show 8–10 pairs of macrochromosomes that have been measured and 30–28 pairs of microchromosomes whose morphology was difficult to determine despite obtaining high uncondensed chromosomes. Number of microchromosomes of partridges is quite similar to that of Galliformes (Stock and Bunch 1982, Shibusawa et al. 2002, Shibusawa et al. 2004). Microchromosomes were classified arbitrarily by decreasing size, their identification will be possible only by molecular cytogenetics (Zoo-FISH) using chicken microchromosomes specific markers (Fillon et al. 1998, Romanov et al. 2005).
Structural and dynamic R-bands obtained in the present work show similarities in pattern. However, dynamic RBG bands seem well delimited than morphological R-bands even if these latter present a higher number (Fig. 5). Pioneer studies have reported that RHG and RBG-bands are 75 to 85% congruent, and GTG and RHG-bands are 90% complementary, meaning that morphological G and R bands have a reverse pattern (Drouin et al. 1991). Dynamic and morphological R-bands are not totally stackable but correspond quite well and can be compared (Lemieux et al. 1990, Drouin et al. 1991).
Simple and double synchronisation of partridge cell cultures have offered the possibility to obtain important rate of prometaphasic chromosomes presenting high number of bands (Table 1 and 2). Comparatively, size of chicken macrochromosomes was ranged from 7 to 3 µm (Hammar 1966) and the first ten macrochromosomes of chicken haploid karyotype presented 209 G-bands and 182 R-bands (Ladjali et al. 1995). High resolution chromosomes allow detection of intrachromosomal changes that are not always visible at the metaphasic stage (Pollock and Fechheimer 1976, Ladjali et al. 1995, Ladjali-Mohammedi et al. 1999).
A. barbara and A. chukar chromosome 4 is acrocentric, while in G. domesticus it is telocentric. Furthermore, comparison of bands showed conservation of patterns in A. barbara and G. domesticus but not in A. chukar. This morphological difference could suggest repositioning of the centromere during the speciation event of partridges 6 million years ago (Randi et al. 1992). The difference in banding pattern in A. chukar could be explained by a paracentric inversion occurrence (4q11-4q31 in GTG) (Fig. 6). This result is supported by a previous study performed on red-legged partridge A. rufa chromosome 4, which is acrocentric (Arruga et al. 1996) and suggested that the morphological difference between A. rufa and G. domesticus was due to an inversion occurrence (Ramos et al. 1999). Later, Kasai et al. (2003) showed a perfect conservation of chicken BAC clones order on A. rufa chromosome 4 and introduced, for the first time in bird class, the term neocentromere (Kasai et al. 2003). Repositioning of the centromere or evolutionary new centromeres (ENC) is the movement of a centromere along the chromosome with the inactivation of the old one but without marker order alteration during evolution (Rocchi et al. 2012). Interestingly, this phenomenon is not so scarce and has been well described. In fact, several cases of de novo centromere formation have been reported in Japanese quail Coturnix japonica and Peking duck Cairina moschata Linnaeus, 1758 (Galkina et al. 2006, Skinner et al. 2009, Zlotina et al. 2012). Nevertheless, the hypothesis of double inversion occurrence should not be excluded as it was reported in the Japanese quail (Zlotina et al. 2012). High conservation of chromosome 4 in chicken and human over 300 million years has so far been reported (Chowdhary and Raudsepp 2000, Groenen et al. 2000). Conversely, the most common fusion reported in birds is between ancestral chromosome 4 and an ancestral microchromosome (Schmid et al. 2000, Shibusawa et al. 2002; 2004). In fact, in the chicken, whose karyotype is considered as the most similar to the ancestral bird karyotype, chromosome 4 is suggested to have arisen from a fusion of ancestral acrocentric chromosome 4 and ancestral microchromosome 10 (Belterman and De Boer 1984, Schmid et al. 2000, Griffin et al. 2007).
The morphological difference of chromosome 7 and 8 between partridges and the chicken, despite conservation of banding range, could be explained by repositioning of the centromere. However, double pericentric inversion cannot be excluded and only molecular investigations could elucidate such evolutionary events. Several studies show that chromosomes 7 and 8 are quite conserved in Galliformes and hybridize respectively to their homologous when using chicken chromosomal painting (Kasai et al. 2003). Exceptionally, in Guinea fowl Numida meleagris Linnaeus, 1758 belonging to Galliformes, Zoo-FISH with chicken DNA specific probes reveals a pericentric inversion in chromosome 7 which corresponds to chicken chromosome 8 (Shibusawa et al. 2002).
The Z chromosome in partridges shows a different terminal region. In fact, A. barbara Z gonosome presents an inversion of banding pattern in the terminus of long arm q compared to that of A. chukar and G. domesticus. Z gonosome of A. barbara in RBG corresponds to G. domesticus and A. chukar Z gonosome in GTG bands (Fig. 7). This result suggests occurrence of Z chromosome terminal inversion in the common ancestor of A. barbara, G. domesticus and A. chukar (Zq21 in GTG) (Fig. 7). The terminal region of Z chromosome in chicken is a characteristic heterochromatic band negatively stained in GTG (Ladjali-Mohammedi et al. 1999). Also, avian Z gonosome is particularly subjected to intrachromosomal rearrangements despite conservation of synteny in most species (Griffin et al. 2007, Nanda et al. 2008). In addition, total sequencing and assembly of chicken Z chromosome has confirmed low gene density (compared to autosomes) associated with high interspersed repeat content (Bellott et al. 2010), which is favorable to rearrangements (Völker et al. 2010).
In both partridges and chicken, the W chromosome is submetacentric and highly heterochromatic as reported in other studies on partridges (Ishishita et al. 2014, Arruga et al. 1996) (Fig. 2b, c). The W chromosome is ranked at the ninth position in A. barbara and A. chukar karyotypes. In different lineages of Neoaves, the W chromosome is supposed to have arisen by the accumulation of repetitive sequences and their conservation during evolution (Graves 2014, Schartl et al. 2016). A recent sequencing of chicken W chromosome has shown preservation of ancestral genes enriched for expressed dosage-sensitive regulators (Bellott et al. 2017). Therewith, it is well established that repetitive DNA polymorphism plays an important role in recombination, chromosomal instability and avian sex chromosome differentiation (Völker et al. 2010).
Conclusion
Banding cytogenetics performed on high resolution chromosomes allowed the precise description of Alectoris barbara Bonnaterre, 1790 and Alectoris chukar karyotypes. Comparative chromosomal mapping highlighted a large conservation with domestic fowl Gallus domesticus Linnaeus, 1758. However, rearrangements in acrocentric macrochromosomes 4, 7 and 8 were observed. Except for the Z chromosome, the partridge chromosomes share more similarities with the putative Galliform ancestral karyotype (Belterman and De Boer, 1984) than with chicken. Such cytogenetic studies could be of an important contribution to detect eventual chromosomal rearrangements in hybrids, given that A. barbara and A. chukar share an overlapping area. Obviously, more detailed molecular cytogenetic studies are necessary to refine the results of the present work. Indeed, we have selected clones from Wageningen chicken BAC (Bacterial Artificial Chromosomes) library (Zoorob et al. 1996, Fillon et al. 1998, Crooijmans et al. 2000) and hybridized them on Barbary partridge and Chukar partridge metaphases. The aim of this fluorescence In Situ Hybridization (FISH) is to confirm rearrangement events and individually identify each pair of microchromosomes (work in progress). This study shows that, despite the importance of molecular investigation, banding cytogenetics is still an important step that provides basic knowledge on evolution of avian karyotypes.
Funding
The present work has received financial support from the Ministère de l’Aménagement du Territoire et de l’Environnement MATE (project 223), Ministère de l’Enseignement Supérieur et de la Recherche Scientifique MESRS (project 209), Ministère de l’Intérieur, in the framework of Post Graduation Specialized: Empreintes génétiques en pratique judiciaire.
Acknowledgement
Special thanks to Dr Belhamra (University of Biskra, Algeria) for the interest and contribution to this work. We would like to thank collaborators from the Centre Cynégétique de Zéralda (Direction Générale des Forêts) for providing biological material, especially Miss Zemiti and Larinouna. We would like also to thank AJZ Engineering Algérie (Zeiss company), in particular Mister LAGUEL; for the precious help. Authors are deeply grateful to Dr Ruane John (Food and Agriculture Organization of the United Nations, FAO, Italy) for his linguistic revision.
Citation
Ouchia-Benissad S, Ladjali-Mohammedi K (2018) Banding cytogenetics of the Barbary partridge Alectoris barbara and the Chukar partridge Alectoris chukar (Phasianidae): a large conservation with Domestic fowl Gallus domesticus revealed by high resolution chromosomes. Comparative Cytogenetics 12(2): 171–199. https://doi.org/10.3897/CompCytogen.v12i2.23743
References
- Akil M, Boudedja S. (2001) Reproduction de la Perdrix gambra (Alectoris barbara) dans la région de Yakouren (Algérie). Game & Wildlife Science 18(3–4): 459–467. [Google Scholar]
- Alaoui MY. (1992) Ecologie de la ponte chez la perdrix gambra (Alectoris barbara) au Maroc. Gibier, Faune Sauvage 9: 405–415. [Google Scholar]
- Altınordu F, Peruzzi L Yu Y, He X. (2016) A tool for the analysis of chromosomes: KaryoType. Taxon 65(3): 586–592. https://doi.org/10.12705/653 [Google Scholar]
- Arruga MV, Tejedor MT, Villarroel MR, Heriz A, Ferreira E, Abenis FJ. (1996) Genetic studies of Alectoris rufa and A. graeca in Spain. Archivos de Zootecnia 45: 339–344. https://doi.org/10.1159/000075770 [Google Scholar]
- Babak A, Amirinia C, Gharahdaghi AA. (2014) Using cytogenetic analysis for detecting karyotype of Persian Chukar population. Indian Journal of Fundamental and Applied Life Sciences 4(3): 1407–1414. http://www.cibtech.org/sp.ed/jls/2014/03/JLS-186-S3-212-AFSHIN-USING-POPULATION.pdf [Google Scholar]
- Barbanera F, Negro JJ, Di Giuseppe G, Bertoncini F, Capppelli F, Dini F. (2005) Analysis of the genetic structure of red-legged partridge (Alectoris rufa, Galliformes) populations by means of mitochondrial DNA and RAPD markers: a study from central Italy. Biology Conservation 122: 275–287. https://doi.org/10.1016/j.biocon.2004.07.017 [Google Scholar]
- Barbanera F, Guerrini M, Bertoncini F, Cappelli F, Muzzeddu M, Dini F. (2011) Sequenced RAPD markers to detect hybridization in the Barbary partridge (Alectoris barbara, Phasianidae). Molecular Ecology Resources 11: 180–184. https://doi.org/10.1111/j.1755-0998.2010.02880.x [DOI] [PubMed] [Google Scholar]
- Barilani M, Bernard-Laurent A, Muccia N, Tabarronic C, Karkd S, Perez Garridoe JA, Randi E. (2007) Hybridisation with introduced chukars (Alectoris chukar) threatens the gene pool integrity of native rock (A. graeca) and red-legged (A. rufa) partridge populations. Biology Conservation 137: 57–69. https://doi.org/10.1016/j.biocon.2007.01.014 [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, Page DC. (2010) Convergent Evolution of Chicken Z and Human X Chromosomes by Expansion and Gene Acquisition. Nature 466(7306): 612–616. https://doi.org/10.1038/nature09172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Cho TJ, Brown L, Locke D, Chen N, Galkina S, Pyntikova T, Koutseva N, Graves T, Kremitzki C, Warren WC et al. (2017) Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nature Genetics 49(3): 387–394. https://doi.org/10.1038/ng.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belterman RHR, De Boer LEM. (1984) A karyological study of 55 species of birds, including karyotypes of 39 species new to cytology. Genetica 65(1): 39–82. https://doi.org/10.1007/BF00056765 [Google Scholar]
- Burt DW. (2002) Origin and evolution of avian microchromosomes. Cytogenetic and Genome Research 96: 97–112. https://doi.org/10.1159/000063018 [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T. (2000) HSA4 and G. domesticus 4: remarkable conservation despite 300-Myr divergence. Genomics 64: 102–105. https://doi.org/10.1006/geno.1999.6085 [DOI] [PubMed] [Google Scholar]
- Christidis L. (1990) Animal Cytogenetics 4: Chordata 3 B: Aves. Gebrüder Borntraeger, Berlin, 116 pp. [Google Scholar]
- Comings DE. (1978) Mechanisms of chromosome banding and implications for chromosome structure. Annual Review of Genetics 12: 25–46. https://doi.org/10.1146/annurev.ge.12.120178.000325 [DOI] [PubMed] [Google Scholar]
- Cramp S, Simmons KEL. (1980) Handbook of the birds of Europe, the middle East and North Africa. The birds of the Western Paleartic, vol II. Oxford University. Press, Oxford, 695 pp. [Google Scholar]
- Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. (2000) Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Critical Review in Eukaryotic Gene Expression 10(2): 179–212. https://doi.org/10.1615/CritRevEukarGeneExpr.v10.i2.60 [PubMed] [Google Scholar]
- Crooijmans RP, Vrebalov J, Dijkhof RJ, Van der Poel JJ, Groenen MAM. (2000) Two-dimensional screening of the Wageningen chicken BAC library. Mammalian Genome 11(5): 360–363. https://doi.org/10.1007/s003350010068 [DOI] [PubMed] [Google Scholar]
- Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K et al. (2010) Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biology 8(9): 1–21. https://doi.org/10.1371/journal.pbio.1000475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derjusheva S, Kurganova A, Habermann F, Gaginskaya ER. (2004) High chromosome conservation detected by comparative chromosome painting in chicken, pigeon and passerine birds. Chromosome Research 12(7): 715–723. https://doi.org/10.1023/B:CHRO.0000045779.50641.00 [DOI] [PubMed] [Google Scholar]
- Di-Nizo CB, Banci KRS, Sato-Kuwabara Y, Silva MJJ. (2017) Advances in cytogenetics of Brazilian rodents: cytotaxonomy, chromosome evolution and new karyotypic data. Comparative Cytogenetics 11(4): 833–892. https://doi.org/10.3897/CompCytogen.v11i4.19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobigny G, Ducroz JF, Robinson TJ, Volobouev V. (2004) Cytogenetics and cladistics. Systematic Biology 53(3): 470–84. https://doi.org/10.1080/10635150490445698 [DOI] [PubMed] [Google Scholar]
- Drouin R, Lemieux N, Richer CL. (1991) High-resolution R-banding at the 1250-band level. 111. Comparative analysis of morphologic and dynamic R-band patterns (RHG and RBG). Hereditas 114: 65–77. https://doi.org/10.1111/j.1601-5223.1991.tb00554.x [DOI] [PubMed] [Google Scholar]
- Dutrillaux B, Leujeune J. (1971) Sur une nouvelle technique d’analyse du caryotype humain. C.R. Académie des Sciences, Paris, 273: 2638–2640. [PubMed] [Google Scholar]
- Dutrillaux B, Couturier J. (1981) La pratique de l’analyse chromosomique. Paris, 87 pp.
- Federico C, Cantarella CD, Scavo C, Saccone S, Bed’Hom B, Bernardi G. (2005) Avian genomes: different karyotypes but a similar distribution of the GC-richest chromosome regions at interphase. Chromosome Research (2005) 13: 785–793. https://doi.org/10.1007/s10577-005-1012-7 [DOI] [PubMed]
- Fillon V, Morisson M, Zoorob R, Auffray C, Douaire M, Gellin J, Vignal A. (1998) Identification of 16 chicken microchromosomes by molecular markers using two-colour fluorescence in situ hybridization (FISH). Chromosome Research 6: 307–313. https://doi.org/10.1023/A:1009274925051 [DOI] [PubMed] [Google Scholar]
- Fungtammasan A, Walsh E, Chiaromonte F, Eckert KA, Makova KD. (2012) A genome-wide analysis of common fragile sites: What features determine chromosomal instability in the human genome? Genome Research 22: 993–1005. https://doi.org/10.1101/gr.134395.111 [DOI] [PMC free article] [PubMed]
- Galkina S, Deryusheva S, Fillon V, Vignal A, Crooijmans R, Groenen MAM, Rodionov A, Gaginskaya ER. (2006) FISH on avian lampbrush chromosomes produces higher resolution gene mapping. Genetica 128: 241–251. https://doi.org/10.1007/s10709-005-5776-7 [DOI] [PubMed] [Google Scholar]
- Galkina S, Fillon V, Saifitdinova A, Daks A, Kulak M, Dyomin A, Koshel E, Gaginskaya ER. (2017) Chicken Microchromosomes in the Lampbrush Phase: A Cytogenetic Description. Cytogenetic and Genome Research 152(1): 46–54. https://doi.org/10.1159/000475563 [DOI] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists. (2009) Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. Journal of Heredity 100: 659–674. https://doi.org/10.1093/jhered/esp086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. (2014) Avian sex, sex chromosomes, and dosage compensation in the age of genomics. Chromosome Research 22: 45–57. https://doi.org/10.1007/s10577-014-9409-9 [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LBW, Tempest HG, Skinner BM. (2007) The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenetic and Genome Research 117: 64–77. https://doi.org/10.1159/000103166 [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Cheng HH, Bumstead N, Benkel BF, Briles WE. (2000) A consensus linkage map of the chicken genome. Genome Research 10: 137–147. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC310508/pdf/x9.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I. (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Research 9(7): 569–584. https://doi.org/10.1023/A:1012447318535 [DOI] [PubMed] [Google Scholar]
- Hammar B. (1966) The Karyotypes of Nine Birds. Hereditas 55: 367–585. https://doi.org/10.1111/j.1601-5223.1966.tb02056.x [Google Scholar]
- Hayes H, Popescu P, Dutrillaux B. (1993) Comparative gene mapping of lactoperoxidase, retinoblastoma, and α-lactalbumin genes in cattle, sheep, and goats. Mammalian Genome 4(10): 593–597. https://doi.org/10.1007/BF00361391 [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME et al. (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. https://doi.org/10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Hooper DM, Price TD. (2017) Chromosomal inversion differences correlate with range overlap in passerine birds. Nature Ecology and Evolution 1: 1526–1534. https://doi.org/10.1038/s41559-017-0284-6 [DOI] [PubMed] [Google Scholar]
- ISCN (1978) An international system of human cytogenetic nomenclature: birth defects. Original Article Series, Vol. 14(8). The National Foundation, New York. Cytogenetics and Cell Genetics 21(6): 309–404. https://doi.org/10.1159/isbn.978-3-8055-8742-6747930 [Google Scholar]
- Ishishita S, Tsuruta Y, Uno Y, Nakamura A, Nishida C, Griffin DK, Tsudzuki M, Ono T, Matsuda Y. (2014) Chromosome size-correlated and chromosome size-uncorrelated homogenization of centromeric repetitive sequences in New World quails. Chromosome Research 22(1): 15–34. https://doi.org/10.1007/s10577-014-9402-3 [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kampf K, Arnold AP. (2011) Possible differences in the two Z chromosomes in male chickens and evolution of MHM sequences in Galliformes. Chromosoma 120: 587–598. https://doi.org/10.1007/s00412-011-0333-x [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P et al. (2014) Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346: 1320–1331. https://doi.org/10.1126/science.1253451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA. (2003) Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenetic and Genome Research 102(1–4): 326–30. https://doi.org/10.1159/000075770 [DOI] [PubMed] [Google Scholar]
- Kimball RT, Braun EL, Zwartjes PW, Crowe TM, Ligon JD. (1999) A molecular phylogeny of the pheasants and partridges suggests that these lineages are not monophyletic. Molecular Phylogenetics and Evolution 11: 38–54. https://doi.org/10.1006/mpev.1998.0562 [DOI] [PubMed] [Google Scholar]
- King M. (1995) Species Evolution: the Role of Chromosome Change. Cambridge University Press, Cambridge 578–580. https://doi.org/10.2307/2413666
- Kretschmer R, de Lima VLC, de Souza MS, Costa AL, O’Brien PCM, Ferguson-Smith MA, de Oliveira EHC, Gunski RJ, Garnero ADV. (2018) Multidirectional chromosome painting in Synallaxis frontalis (Passeriformes, Furnariidae) reveals high chromosomal reorganization, involving fissions and inversions. Comparative Cytogenetics 12(1): 97–110. https://doi.org/10.3897/CompCytogen.v12i1.22344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladjali K. (1994) Caryotype de la poule domestique « Gallus domesticus » et incidences des anomalies chromosomiques dans les troubles de la reproduction. Ph. D Dissertation, Institut National Agronomique Paris-Grignon, 179 pp. [Google Scholar]
- Ladjali K, Tixier-Boichard M, Cribiu P. (1995) High-Resolution Chromosome Preparation for G- and R- Banding in Gallus domesticus. Journal of Heredity 86: 136–139. https://doi.org/10.1093/oxfordjournals.jhered.a111543 [Google Scholar]
- Ladjali-Mohammedi K, Bitgood JJ, Tixier-Boichard M, Ponce De Leon FA. (1999) International System for Standardized Avian Karyotypes (ISSAK): Standardized banded karyotypes of the domestic fowl (Gallus domesticus). Cytogenetic and Cell Genetics 86: 271–276. https://doi.org/10.1159/000015318 [DOI] [PubMed] [Google Scholar]
- Lemieux N, Drouin R, Richer CL. (1990) High-resolution dynamic and morphologic G-bandings (GBG and GTG): a comparative study. Human Genetics 85: 261–266. https://doi.org/10.1007/BF00206742 [DOI] [PubMed] [Google Scholar]
- Madge S, McGowan P. (2002) Pheasants Partridges and Grouse A Guide to the Pheasants, Partridges, Quails, Grouse, Guineafowl, Buttonquails, and Sandgrouse of the World. A and C Black Ltd, London, 408 pp https://doi.org/10.1642/0004-8038(2003)120[0920:PPAGAG]2.0.CO;2 [Google Scholar]
- Maghnouj M. (1991) Perdreaux d’élevage: une reproduction naturelle satisfaisante de la perdrix gambra (Alectoris barbara). Annales de la Recherche Forestière au Maroc 25: 83–101. [Google Scholar]
- Masabanda JS, Burt DW, O’Brien PC, Vignal A, Fillon V et al. (2004) Molecular cytogenetic definition of the chicken genome: the first complete avian karyotype. Genetics 166: 1367–1373. https://doi.org/10.1534/genetics.166.3.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen HA, Fantes J, Cross SH, Clark VH, Archibald AL. (1996) CpG islands of chicken are concentrated on microchromosomes. Nature Genetics 12: 321–324. https://doi.org/10.1038/ng0396-321 [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Herr P, Helledaya T. (2013) Early replication fragile sites: where replication-transcription collisions cause genetic instability. Journal of the European Molecular Biology Organization 32(4): 493–495. https://doi.org/10.1038/emboj.2013.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. (2008) Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenetic and Genome Research 122: 150–156. https://doi.org/10.1159/000163092 [DOI] [PubMed] [Google Scholar]
- Nie W, O’Brien PCM, Fu B, Wang J, Su W, He K, Bed’Hom B, Volobouev V, Ferguson-Smith MA, Dobigny G, Yang F. (2015) Multidirectional chromosome painting substantiates the occurrence of extensive genomic reshuffling within Accipitriformes Bio Med Central Evolutionary Biology 15: 205. https://doi.org/10.1186/s12862-015-0484-0 [DOI] [PMC free article] [PubMed]
- Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, Griffin DK, Matsuda Y. (2008) Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Research 16(1): 171–181. https://doi.org/10.1007/s10577-007-1210-6 [DOI] [PubMed] [Google Scholar]
- Pollock DL, Fechheimer NS. (1976) The chromosome number of Gallus domesticus. British Poultry Science 17: 39–42. https://doi.org/10.1080/00071667608416247 [DOI] [PubMed] [Google Scholar]
- Ponce de Leon FA, Li Y, Weng Z. (1992) Early and late replicative chromosomal banding patterns of Gallus domesticus. Journal of Heredity 83: 36–42. https://doi.org/10.1093/oxfordjournals.jhered.a111154 [DOI] [PubMed] [Google Scholar]
- Randi E, Meriggi A, Lorenzini R, Fusco G, Alkon PU. (1992) Biochemical analysis of relationships of Mediterranean Alectoris partridges. The Auk 109(2): 358–367. https://doi.org/10.2307/4088204 [Google Scholar]
- Randi E. (1996) A mitochondrial cytochrome B phylogeny of the Alectoris partridges. Molecular Phylogenetics Evolution 6(2): 214–27. https://doi.org/10.1006/mpev.1996.0072 [DOI] [PubMed] [Google Scholar]
- Randi E, Lucchini V. (1998) Organization and evolution of the mitochondrial DNA control region in the avian genus Alectoris. Journal of Molecular Evolution 47(4): 449–462. https://doi.org/10.1007/PL00006402 [DOI] [PubMed] [Google Scholar]
- Randi E, Tabarroni C, Rimondi S, Lucchini V, Sfougaris A. (2003) Phylogeography of the rock partridge (Alectoris graeca). Molecular Ecology 12(8): 2201–2214. https://doi.org/10.1046/j.1365-294X.2003.01899.x [DOI] [PubMed] [Google Scholar]
- Randi E. (2008) Detecting hybridization between wild species and their domesticated relatives. Molecular Ecology 17(1): 285–93. https://doi.org/10.1111/j.1365-294X.2007.03417.x [DOI] [PubMed] [Google Scholar]
- Ramos PS, Dias D, Ponce de Leon FA. (1999) Molecular cytogenetic analysis of the chicken and red-legged partridge chromosome 4 repatterning. Animal Biotechnology 10: 123–126. https://doi.org/10.1080/10495399909525934 [DOI] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R. (2012) Centromere repositioning in mammals. Heredity 108: 59–67. https://doi.org/10.1038/hdy.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov AV. (1997) Evolution of avian chromosome and linkage groups. Russian Journal of Genetics 33: 605–617. [Google Scholar]
- Rodríguez-García MJ, Galián J. (2014) Rapid differentiation of Alectoris rufa L., 1758 and Alectoris chukar (Gray, 1830) (Galliformes: Phasianidae) by melting curve analysis of a parathyroid hormone gene SNP. Anales de Biología 36: 121–127. https://doi.org/10.6018/analesbio.36.19 [Google Scholar]
- Rojas M, González I, Pavón MÁ, Pegels N, Hernández PE, García T, Martín R. (2011) Mitochondrial and nuclear markers for the authentication of partridge meat and the specific identification of red-legged partridge meat products by polymerase chain reaction. Poultry Science 90: 211–222. https://doi.org/10.3382/ps.2010-00895 [DOI] [PubMed] [Google Scholar]
- Romanov MN, Daniels LM, Dodgson JB, Delany ME. (2005) Integration of the cytogenetic and physical maps of chicken chromosome 17. Chromosome Research 13: 215–222. https://doi.org/10.1007/s10577-005-1506-3 [DOI] [PubMed] [Google Scholar]
- Romanov MN, Farre M, Lithgow PE, Fowler KE, Skinner BM et al. (2014) Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BioMed Central Genomics 15: 1060. https://doi.org/10.1186/1471-2164-15-1060 [DOI] [PMC free article] [PubMed]
- Schartl M, Schmid M, Nanda I. (2016) Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma 125: 553–571. https://doi.org/10.1007/s00412-015-0569-y [DOI] [PubMed] [Google Scholar]
- Schmid M, Enderle E, Schindler D, Schempp W. (1989) Chromosome banding and DNA replication patterns in bird karyotypes. Cytogenetics and Cell Genetics 52: 139–146. https://doi.org/10.1159/000132864 [DOI] [PubMed] [Google Scholar]
- Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M et al. (2000) First Report on Chicken Genes and Chromosomes 2000. Cytogenetics and Cell Genetics 90: 169–218. https://doi.org/10.1159/000056772 [DOI] [PubMed] [Google Scholar]
- Schmid M, Smith J, Burt DW, Aken BL, Antin PB, Archibald AL, Ashwell C, Blackshear PJ, Boschiero C, Brown CT, Burgess SC et al. (2015) Third Report on Chicken Genes and Chromosomes. Cytogenetic and Genome Research 145: 78–179. https://doi.org/10.1159/000430927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright M. (1971) A rapid banding technique for human chromosomes. Lancet 2: 971–972. https://doi.org/10.1016/S0140-6736(71)90287-X [DOI] [PubMed] [Google Scholar]
- Shibusawa M, Nishida-Umehara C, Masabanda J, Griffin DK, Isobeb T, Matsuda Y. (2002) Chromosome rearrangements between chicken and guinea fowl defined by comparative chromosome painting and FISH mapping of DNA clones. Cytogenetic and Genome Research 98: 225–230. https://doi.org/10.1159/000069813 [DOI] [PubMed] [Google Scholar]
- Shibusawa M, Nishibori M, Nishida-Umehara C, Tsudzuki M, Masabanda J, Griffin DK, Matsuda Y. (2004) Karyotypic evolution in the Galliformes: an examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenetic and Genome Research 106: 111–119. https://doi.org/10.1159/000078570 [DOI] [PubMed] [Google Scholar]
- Shoffner RN. (1974) Chromosomes in birds. In: Busch H. (Ed.) The cell nucleus. Academic, San Diego, 223–261. https://doi.org/10.1016/B978-0-12-147602-1.50015-5
- Skinner BM, Robertson LB, Tempest HG, Langley EJ, Ioannou D, Fowler KE, Crooijmans RP, Hall AD, Griffin DK, Völker M. (2009) Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BioMed Central Genomics 10: 357. https://doi.org/10.1186/1471-2164-10-357 [DOI] [PMC free article] [PubMed]
- Skinner BM, Griffin DK. (2012) Intrachromosomal rearrangements in avian genome evolution: evidence for regions prone to breakpoints. Heredity 108: 37–41. https://doi.org/10.1038/hdy.2011.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AD, Bunch TD. (1982) The evolutionary implications of chromosome banding pattern homologies in the bird order Galliformes. Cytogenetics and Cell Genetics 34: 136–148. https://doi.org/10.1159/000131802 [DOI] [PubMed] [Google Scholar]
- Tanabe H, Habermann FA, Solovei I, Cremer M, Cremer T. (2002) Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutation Research 504(1–2): 37–45. https://doi.org/10.1016/S0027-5107(02)00077-5 [DOI] [PubMed] [Google Scholar]
- Tegelstrôm H, Ryttman H. (1981) Chromosomes in birds (Aves): evolutionary implications of macro- and microchromosome numbers and lengths. Hereditas 94: 225–233. https://doi.org/10.1111/j.1601-5223.1981.tb01757.x [Google Scholar]
- Tejedor MT, Monteagudo LV, Mautner S, Hadjisterkotis E, Arruaga MV. (2007) Introgression of Alectoris chukar Genes into a Spanish Wild Alectoris rufa Population. Journal of Heredity 98(2): 179–182. https://doi.org/10.1093/jhered/esm001 [DOI] [PubMed] [Google Scholar]
- Völker M, Backström N, Skinner BM, Langley EI, Bunzey SK, Ellegren H, Griffin DK. (2010) Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Research 20(4): 503–511. https://doi.org/10.1101/gr.103663.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voous KH. (1976) The birds of the tropical ‘Middle Seas,’ past and present. Proceedings of the XVIth International Ornithological Congress, Canberra, 12–17 August 1974. Canberra, 697–704.
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW et al. (2010) The genome of a songbird. Nature 464: 757–762. https://doi.org/10.1038/nature08819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W, Hillier LDW, Tomlinson C, Minx P, Kremitzki M et al. (2017) A New Chicken Genome Assembly Provides Insight into Avian Genome Structure. Genes, Genome, Genetics (Bethesda) 7(1): 109–117. https://doi.org/10.1534/g3.116.035923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM et al. (2014) Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. https://doi.org/10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Bourque G. (2009) Recovering genome rearrangements in the mammalian phylogeny. Genome Research 19: 934–942. https://doi.org/10.1101/gr.086009.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotina A, Galkina S, Krasikova A, Crooijmans RP, Groenen MAM, Gaginskaya ER, Deryusheva S. (2010) Precise centromere positioning on chicken chromosome 3. Cytogenetic and Genome Research 129: 310–313. https://doi.org/10.1159/000314923 [DOI] [PubMed] [Google Scholar]
- Zlotina A, Galkina S, Krasikova A, Crooijmans RP, Groenen MAM, Gaginskaya ER, Deryusheva S. (2012) Centromere positions in chicken and Japanese quail chromosomes: de novo centromere formation versus pericentric inversions. Chromosome Research 20(8): 1017–1032. https://doi.org/10.1007/s10577-012-9319-7 [DOI] [PubMed] [Google Scholar]
- Zoorob R, Billault A, Severac V, Fillon V, Vignal A, Auffray C. (1996) Two chicken genomic libraries in the PAC and BAC cloning systems: organization and characterization. Animal Genetics 27(Supplement 2): 69.