Abstract
Sepsis and metabolic syndrome (MetS) are both inflammation-related entities with high impact for human health and the consequences of concussions. Both represent imbalanced parasympathetic/cholinergic response to insulting triggers and variably uncontrolled inflammation that indicates shared upstream regulators, including short microRNAs (miRs) and long non-coding RNAs (lncRNAs). These may cross talk across multiple systems, leading to complex molecular and clinical outcomes. Notably, biomedical and RNA-sequencing based analyses both highlight new links between the acquired and inherited pathogenic, cardiac and inflammatory traits of sepsis/MetS. Those include the HOTAIR and MIAT lncRNAs and their targets, such as miR-122, −150, −155, −182, −197, −375, −608 and HLA-DRA. Implicating non-coding RNA regulators in sepsis and MetS may delineate novel high-value biomarkers and targets for intervention.
Keywords: metabolic syndrome, miRs, ncRNAs, sepsis, SNPs
Significance Statement
Sepsis and metabolic syndrome (MetS) both carry a high toll on human health and modern healthcare. Sepsis is the reaction to overwhelming infection in the organism, usually addressed with systemic antimicrobial therapy and systemic supporting measures, whereas MetS is an amalgam of interlinked metabolic disturbances such as obesity, diabetes, hypertension and associated cardiac comorbidities. These seemingly unrelated entities may be regulated by nervous system-mediated interlinked molecular pathways, which can explain the accompanying mal-reaction to pathogens and metabolic challenges. Novel molecular/genomic assessment tools can offer potential new intervention points to consider in future approaches, foreseeing new management modalities.
Sepsis and MetS Present Escalating Inter-Related Challenges
Sepsis and MetS represent an inter-related escalating disease burden for modern healthcare systems. In spite of increasingly structured approaches, sepsis remains a leading cause of death in hospitals; and MetS is evolving to become a global epidemic, characterized as a combination of obesity, dyslipidemia, hypertension, insulin resistance, and increased thrombosis (Grundy et al., 2004; de Simone et al., 2007). MetS increases the risk for adverse cardiac outcomes such as coronary artery disease, heart failure, cardiac fibrosis and intracardiac malconduction leading to dysfunction and arrhythmias (affected by sympathetic imbalance, changed tissue impedance and changed cardiac electrophysiologic properties; Wilson et al., 2005; Ingelsson et al., 2006; Grassi et al., 2007; de Simone et al., 2007; Suzuki et al., 2008; Wang et al., 2010; Sardu et al., 2014a, 2017). Additionally, MetS elevates the risk for complications of sepsis, possibly relating in part to these maladaptive cardiometabolic alterations (Angus and van der Poll, 2013; Abbasi, 2017). One such dreaded complication of sepsis is intracardiac malconduction and altered electrical properties, exposing patients to life-threatening arrhythmias (Shahreyar et al., 2018). These complications are increasingly being recognized as part of MetS, and may lead to specific device therapies in the future, with discrete molecular pathology-and-response profiles (Marfella et al., 2013; Sardu et al., 2017). Inflammation is key to both sepsis and MetS, with established descriptions of inflammation in the former, and more recent integration into contemporary clinical definitions of the latter (Grundy et al., 2004). Therefore, novel scientific tools are being sought for evolving insights into the underlying biological mechanisms, with impressive attempts at new therapies.
Both sepsis and MetS show increasing interactions with the complex networks of long non-coding RNAs (lncRNAs), microRNAs (miRs) and their targets, compatible with the escalating importance of non-coding RNAs in various pathologies in the neurodegenerative and cardiometabolic arenas (Marfella et al., 2013; Meydan et al., 2016; Salta et al., 2016). Also, the apparent roles for lncRNAs and miRs in immune regulation (Morchikh et al., 2017) highlight their inflammatory association and may lead to the development of novel molecular tools co-regulating these discrete medical entities. Identification of signaling pathways involving environmental and nutritional influences may also pave the way for new interventions as well as shed light on existing therapeutic strategies (Wellen and Thompson, 2012). Sepsis involves an overwhelming systemic inflammatory reaction to infection and represents a complex joint action of pro-inflammatory and anti-inflammatory agents, driven by combined pathogen-specific and host-specific features and the interactions between them (Rittirsch et al., 2008; van der Poll and Opal, 2008). It is characterized by central nervous system (CNS), hemodynamic, cardiac and renal malfunction; adrenergic, glycemic, and coagulation deregulation; and cholinergic/inflammatory imbalance (Angus and van der Poll, 2013), which are largely amplified with underlying comorbidities, and impose high mortality even in modern healthcare settings. A recent call-to-arms by the Global Sepsis Alliance demonstrates this ongoing challenge (Reinhart et al., 2017), and the underlying mechanisms of sepsis have increasingly drawn attention over the last few decades.
Viewing sepsis and MetS as a reflection of acutely imbalanced neuroimmune reactions has spurred an investigation into the specific mechanisms acting jointly with immune machinery and their application in other contexts, such as brain function, which may decline during acute sepsis or recovery (Hanisch et al., 2008; Jackson et al., 2014). This decline involves cortisol decreases in the circulation, due to inhibition of the cholinergic hypothalamic-pituitary-adrenal axis by anxiety and/or changes to associated molecular machinery (Lebow et al., 2012; Galatzer-Levy et al., 2017). Dysfunction of this axis is a key characteristic of both sepsis and MetS (see Supplementary Material). Genomic polymorphisms in cholinergic elements predispose to other mental pathologies, such as drug addictions; therefore, brain malfunction during sepsis is likely to reflect this genomic pathway as well. That cholinergic imbalance is paramount to inflammatory reactions strengthens this assumption (Li and Burmeister, 2009), while raising the issue of personalized cholinergic reinforcement for intervention (Maskos et al., 2005).
In comparison to sepsis, MetS shows a sharply rising prevalence, with a steadily widening presence in many countries, and a risk for coronary and cerebrovascular diseases (Mottillo et al., 2010; O’Neill and O’Driscoll, 2015). Longstanding evidence defines MetS as an inflammatory entity which combines obesity, dyslipidemia, insulin resistance, and hypertension (Ridker et al., 2005), albeit with incompletely understood mechanisms. MetS further complicates the treatment of sepsis, including obesity-related restrictive lung disease and supply-related myocardial ischemia (Type II myocardial infarction), with putative inter-related mechanisms. In this aspect, MetS may present a differently-paced decline of immune-related functions, which parallel those that fail in septic events. This calls for exploring inter-related regulatory pathways controlling these two entities, including non-coding RNA controllers of inflammation that operate well under healthy conditions but may fail in sepsis and MetS. Combining biomedical and RNA-sequencing based evidence indicates that sepsis and MetS (although differing in their clinical nature) share malfunctioning biological domains. This implicates shared miRs and lncRNAs as key cellular regulators of immune-related and other conditions, and as plausible effectors of some of the common clinical and molecular features of sepsis and MetS.
Several miRs are evident in the pathologies of both MetS and sepsis; MiR-122 regulates lipid synthesis and oxidation, and also serves as a biomarker in hepatic ischemia, viral hepatitis and sepsis; MiR-150 is downregulated in sepsis and correlates with its severity as assessed by clinical scoring, but is upregulated in dysglycemia and diabetes mellitus; MiR-182 regulates glucose levels and its metabolism, and is also upregulated in sepsis; and miR-197, −375, −155 and −132 show dual roles in both infection control and MetS (Supplementary Material). An important advantage of miRs is their small size and “druggable” nature, i.e., antisense-oriented oligonucleotides for functional suppression, and vector-based approaches generating “sponge molecules” which act as decoys or miR-mimics for potentiating their activities (Li and Rana, 2014). These strategies may serve as impetus for further elucidation of the roles and possible applications of miRs as direct targets for therapeutics or for exerting regulatory effects on other targets. Several miR-based therapeutics are already in development in the cardio-metabolic context, including an antisense inhibitor of miR-208a, which offers improved recovery from heart failure in animal models (Miragen Therapeutics Inc., Boulder, CO, USA; Montgomery et al., 2011); and preclinical trials for a miR-195 inhibitor, also for heart failure (Servier, miRagen; Li and Rana, 2014).
Recent reports demonstrate a tilted axis of inflammatory mediators and cholinergic machinery which accompany the systemic derangements both in sepsis and MetS. Here, we explore the context of possibly shared elements driving these seemingly distinct clinical spheres. Such discussion becomes increasingly relevant as new molecular and intracellular mechanisms are rapidly being discovered, characterized and harnessed for clinical applications. In turn, new understanding of lncRNAs and miRs, delineation of their interactions, and uncovering their functions in various clinical scenarios can contribute to both the basic and translational research levels of these two conditions. We hope that the current study would spur new initiatives in these contexts, which continue to frustrate modern healthcare systems.
The lncRNA-miR Link with Inflammation in Sepsis and MetS
MiRs are non-coding regulatory RNA molecules, 100-fold smaller than coding RNAs. Several thousand miRs were identified, many of which are primate-specific (Barbash et al., 2014). LncRNAs as well have been subjected to active evolutionary pressure, adapting them to their diverse functional capacity towards multiple molecular targets and multi-leveled systemic mechanisms (Berezikov, 2011; Barbash et al., 2014). MiRs primarily attach to complementary motifs in target transcripts, thus halting translation of coding RNAs sharing such motifs and exacerbating their degradation. Specific miRs may control shared pathological processes; for example, 11 different miRs were reported to associate with both anxiety-spectrum clinical entities and with MetS, with transcripts of the cholinergic signaling pathway predictably targeted by all of those via the inflammatory triggers shared by these syndromes (Meydan et al., 2016).
Accelerated accumulation of publicly available RNA-sequencing datasets enables searching for inflammation-related miRs and lncRNAs and the changes occurring in them for sepsis and MetS separately as well as for their shared features (e.g., coagulation, adrenergic/cholinergic imbalance and diabetes, Supplementary Figures S1A,B, Supplementary Table S2). For example, the primate-specific miR-608 controls the MetS-related acetylcholinesterase (AChE; Hanin et al., 2014). A single nucleotide polymorphism (SNP) in the miR-608 sequence limits the risk of post-trauma sepsis (Zhang et al., 2015), indicating that this miR may be related to the aftermath of concussion (Bailes et al., 2013; Zaghloul et al., 2017) and possibly reflecting an apparent link between potential ncRNA controllers of sepsis and MetS at large. Since cholinergic signaling modulates inflammation (Tracey, 2010), ncRNA controllers of parasympathetic functioning are likely candidates for linking between these distinct inflammation-related pathologies and for playing a primary role in the clinical outcome of both sepsis and MetS. Figure 1 schematically presents these predicted inter-related ties between sepsis and MetS, their potential association with lncRNAs and miRs and their predicted mechanisms of action, which are far more diverse for lncRNAs as compared to miRs.
Figure 1.
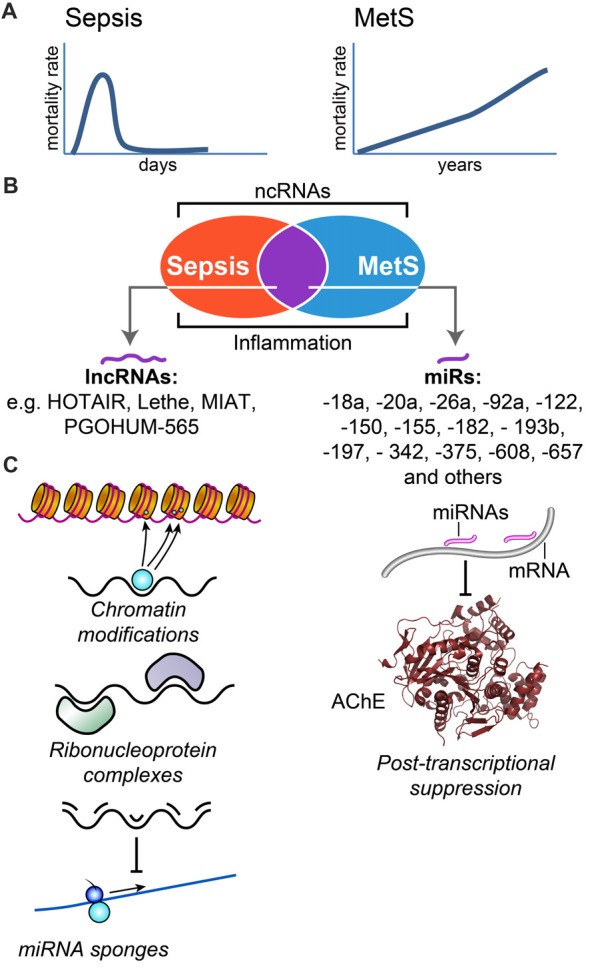
Sepsis and metabolic syndrome (MetS) share ncRNA controllers and inflammation characteristics. (A) Scheme of the impact over mortality rate for both sepsis and MetS. Sepsis involves rapid dynamics with mortality rate peaking in the scale of days, whereas entailed mortality from MetS related complications develops over many years. (B) Sepsis and MetS are both subject to ncRNA regulation, and share common microRNA (miR) and ncRNA regulators. (C) Long non-coding RNAs (LncRNAs; left pane, denoted as black line) exert diverse functions including chromatin modification, ribonucleoprotein complex formation or “sponge” activities, whereas miRs (right hand side, pink lines) primarily suppress their targets via post-transcriptional interaction with short sequence motifs on their target mRNAs, and lead to physiological changes (e.g., cholinergic changes, as a result of acetylcholinesterase (AChE) reduction).
In addition to their individual functions, lncRNAs cooperate with other ncRNAs, such as miRs, both mechanistically (as lncRNA “sponges” suppressing the reacting miRs) and as shared effectors, such as in vascular dysfunction (Yan et al., 2015). So far, 80 miRs were implicated in metabolic disorders (Zampetaki and Mayr, 2012; Arner and Kulyté, 2015). Those are involved in various adipose, lipid, glycemic and vascular impairments, including cardiometabolic clinical ramifications such as heart failure and atrial fibrillation (Marfella et al., 2013; Santulli et al., 2014; Sardu et al., 2015; Xu et al., 2016). In these contexts, miR regulators of organic function may respond to mechanical, electrical and biochemical stimuli, created by changing systemic environments. A notable example of these processes lies in cardiac remodeling, which has classically been viewed as a result of biophysical factors and insults; accumulating evidence implicates miRs as mediators and molecular effectors of this pathophysiology (Divakaran and Mann, 2008; Greco et al., 2012; Barwari et al., 2016). Atherosclerosis, cardiac regeneration, cardiac angiogenesis, cardiac electroconductance and lipid metabolism are additional MetS-related areas with miR involvement (Sardu et al., 2014b; Barwari et al., 2016), including their interaction with established mediators such as angiotensin-II (Pan et al., 2013; Sardu et al., 2016b), and their reaction to specific interventions. In this context, the effect of miRs on adaptive remodeling of cardiovascular organs has been studied, i.e., the association of different miRs’ levels with cardiomyocyte function, features and response to heart failure therapy (Sardu et al., 2014b, 2016a,b; Melman et al., 2015). Some of the metabolically-associated ncRNAs have an apparent role in sepsis as well (Table 1). Briefly, we hypothesized that lncRNA-miR interactions may reflect some of the shared symptoms observed clinically and the inflammatory processes driven by these shared mediators (Supplementary Material). Examples include miR-132, with established roles both in MetS and in the cellular response to viral infections (Lagos et al., 2010; Meydan et al., 2016), and miR-197, dysregulated in adipose tissue dysfunction, and in pulmonary and enteroviral infections (Tang et al., 2015). Such miR responses may both contribute to the host reaction towards infections while influencing seemingly distant clinical phenomena (such as MetS). Scientific tools such as genome wide association scale (GWAS) and RNA-sequencing analyses offer a glimpse towards the evolution of miRs and lncRNAs across different species (Ulitsky, 2016) and can identify associations encountered in current clinical practice, such as sepsis and MetS. Thus, both miRs and short-lived lncRNAs may play key roles in the dynamics and etiology of these conditions (Tani et al., 2012), affect the consequences of concussions and function cooperatively to exacerbate them or protect the body from the corresponding symptoms.
Table 1.
Non-coding RNA molecules associated with both sepsis and metabolic syndrome (MetS).
| Non-coding RNA | Mets/Sepsis | Role | Reference | |
|---|---|---|---|---|
| microRNAs | miR-122 | MetS | Modulation of hepatocyte-associated proteins (ACC2, SCD1, ACLY, AMP-K) and lipid metabolism | Esau et al. (2006), Elmén et al. (2008) and Zampetaki and Mayr (2012) |
| Sepsis | Decreased in sepsis vs. healthy controls and non-septic SIRS, correlates with sepsis mortality | Caserta et al. (2016) | ||
| Increased in chronic liver infection with hepatitis C virus and involved in its pathogenesis | Janssen et al. (2013) | |||
| Marker for abnormal coagulation in sepsis | Janssen et al. (2013), Luna et al. (2015) and Correia et al. (2017) | |||
| miR-150 | MetS | Upregulated in adipose and hepatic tissues, and in insulin resistance | Karolina et al. (2012) | |
| Sepsis | Downregulated in sepsis vs. non-septic SIRS and healthy controls, correlates with SOFA score, predictive of mortality | Vasilescu et al. (2009) and Ma et al. (2017) | ||
| Correlates with proinflammatory cytokines (TNFα, IL-10, IL-18) | Vasilescu et al. (2009) | |||
| miR-182 | MetS | Implicated in insulin regulation and diabetes-associated muscle atrophy | Poy et al. (2004), Melkman-Zehavi et al. (2011) and Zhang et al. (2016) | |
| Sepsis | Upregulated in sepsis in GWAS | Vasilescu et al. (2009) | ||
| miR-197 | MetS | Upregulated in adipose tissue | Karolina et al. (2012) and Arner and Kulyté (2015) | |
| Sepsis | Upregulated in lung infections | Tang et al. (2015) | ||
| Decreased in chronic hepatitis B and enterovirus infections | Tang et al. (2015) | |||
| miR-375 | MetS | Suppresses insulin secretion | Zampetaki and Mayr (2012) | |
| Sepsis | Upregulated in hepatitis B virus infections | Li et al. (2010) | ||
| miR-155 | MetS | Involved in the pathogenesis of atherosclerosis and diabetes | Nazari-Jahantigh et al. (2012) and Lin et al. (2016) | |
| Sepsis | Involved in bacterial infections and hepatitis C-associated liver disease | Correia et al. (2017) | ||
| miR-608 | MetS | Involved in cholinergic signaling with implication for hypertension | Hanin et al. (2014) | |
| Sepsis | SNP is a prognostic marker for reduced risk of sepsis after major trauma | Zhang et al. (2015) | ||
| Interaction with IL-6 | Hanin et al. (2017) | |||
| Long non-coding RNAS (lncRNAs) | HOTAIR | MetS | Implicated in adipocyte differentiation | Wu et al. (2016) |
| Sepsis | Promotes TNFα production in cardiomyocites in sepsis, through NF-kB pathway | Wu et al. (2016) | ||
| Lethe | MetS | Leads to inflammatory effects in high-glucose environment | Zgheib et al. (2017) | |
| Sepsis | Regulates NF-kB and induced by IL-1β and TNFα | Rapicavoli et al. (2013) | ||
| NEAT1 | MetS | Regulates PPARg2 splicing during adipogenesis | Chen (2016) | |
| Mediates miR-140-induced adipogenesis | Gernapudi et al. (2016) | |||
| Sepsis | Induced by herpesvirus infections in a STAT3-dependant manner | Wang et al. (2017) | ||
| Upregulated in Hantavirus infections, downregulation in vitro causes impaired immune response, involved in innate immune response through RIG-I pathway | Ma et al. (2017) | |||
| DMRT2 | MetS | Suppressed in adipose tissue of obese humans (RNA sequencing) | Liu et al. (2014) | |
| Sepsis | Induced in vitro by LPS stimulation | Liu et al. (2014) | ||
| TP53I13 | MetS | Suppressed in adipose tissue of obese humans (RNA sequencing) | Liu et al. (2014) | |
| Sepsis | Induced in vitro by LPS stimulation | Liu et al. (2014) | ||
| Cox2 | Sepsis | Induced by Toll-like receptor activation | Carpenter et al. (2013) | |
| PACER | Sepsis | Involved in assembly of NF-kB | Krawczyk and Emerson (2014) | |
| Lnc-DC | Sepsis | Regulates dendritic cell differentiation | Wang et al. (2014) | |
| THRIL | Sepsis | Upregulates TNFα | Li et al. (2014) | |
| TNFAIP3 | Sepsis | Regulated by TNFα, coregulator of NF-kB | Vereecke et al. (2009) | |
Shown are lncRNAs and miRs reported to affect MetS and sepsis before and after concussions, including their predicted mechanism of action and associated molecular machinery (See Supplementary Material for additional citations).
Several lncRNAs play a role both in sepsis and in MetS. The Lethe lncRNA is involvedplay a role both in sepsis and in MetS in NF-kB regulation, and is co-induced with interleukin (IL)-1β and tumor necrosis factor (TNF)α, and downregulated by high-glucose environment, where it exacerbates the inflammatory cellular profile (Zgheib et al., 2017). This may affect the adverse outcomes of sepsis-induced hyperglycemia and the dysglycemia that accompanies the inflammatory driving force in MetS. In a mouse model of leptin deficiency and diet-induced insulin resistance, the E330013P06 lncRNA is both overexpressed in macrophages and correlated with inflammatory activity (Reddy et al., 2014). HOTAIR is another lncRNA which promotes the inflammatory response to pathogens through the NF-kB pathway (Wu et al., 2016), possibly by interacting with 23 different protein-coding transcripts correlated to adipocyte function. RNA-sequencing of 32 tissue types from 122 healthy human subjects revealed excessive levels of several highly expressed HOTAIR-associated transcripts (over 2-fold excess compared to tissue-wide average) in adipose tissue. These include the Kruppel-like factor 4 (KLF4), Notch homolog 3 (NOTCH3), Snail homolog 1 (SNAI1), Neurotrimin (NTM) and Vimentin (VIM). HOTAIR is also involved in multiple pathways, including the neuronal LSD1/CoREST/REST and PRC2 complexes, and regulates genes in the HOXD locus (Rinn et al., 2007; Tsai et al., 2010; Karpe and Pinnick, 2015). Correspondingly, HOTAIR plays an active role in adipocyte and adipose tissue development, with dispersion between central and peripheral tissues.
In search for putative functional relevance of HOTAIR, we investigated human peripheral RNA-sequencing data from the community acquired pneumonia and sepsis outcome diagnostic study, (CAPSOD, GSE accession number 63042), which enrolled 1152 subjects who presented to emergency care and were stratified into systemic inflammatory response syndrome (SIRS) and four septic groups with escalating severity: uncomplicated sepsis without organ dysfunction; severe sepsis with organ dysfunction; sepsis with hemodynamic collapse (shock); and those dying from sepsis within 28 days from admission. RNA sequencing identified 338 differentially-expressed liver genes between sepsis and non-sepsis SIRS, and 1238 differentially-expressed genes in the groups presenting increasing sepsis severity and outcome. We found several HOTAIR-associated genes to be upregulated in these septic patients, reaching 2–3 fold higher levels compared to non-infectious SIRS; stratifying according to sepsis severity highlighted angiopoietin-2, which is involved in atherosclerosis and in diabetic complications (Park et al., 2014), as increasing with the severity of sepsis, compatible with previous reports (Bopp et al., 2008; Aslan et al., 2017). This supports an inter-related impact for HOTAIR and other MetS-regulated genes, including cholinergic transcripts in the septic liver.
According to the Human Protein Atlas, the subset of MetS-modified genes is elevated 2.7-fold in adipose tissue compared to tissue average in healthy humans, and septic patients in the CAPSOD study show significant upregulation of such genes vs. non-infectious SIRS patients (Table 2). To expand the studied tissue scope, we searched for potential association between the levels of global MetS-modified blood cell coding and non-coding transcripts, and those of liver biopsied tissues from patients with increasing severity of sepsis symptoms. We predicted that transcripts differentially expressed in MetS blood cells would also be modified in the septic liver, and found that many of those transcripts which were changed in MetS blood cells were gradually decreased in septic liver samples with increasing symptoms severity (Supplementary Figure S2).
Table 2.
Molecular elements associated with the long non-coding RNA (lncRNA) HOTAIR in sepsis and MetS.
| Protein symbol | Human protein | Sepsis dataset of CAPSOD study, GSE63042 | |||||
|---|---|---|---|---|---|---|---|
| Protein name | Atlas, v16.1 | ||||||
| Tissue-wide average expression (TPM) | Adipose tissue expression (TPM) | Sepsis (mortality) vs. non-infectious SIRS, t-test | Septic shock vs. non-infectious SIRS, t-test | Severe sepsis vs. non-infectious SIRS, t-test | Uncomplicated sepsis vs. non-infectious SIRS, t-test | All sepsis vs. non-infectious SIRS, t-test | |
| SAV1 | 26.0 | 35.8 | 0.12 | 0.24 | 0.15 | 0.33 | 0.10 |
| Salvador homolog 1 | |||||||
| KLF4 | 44.2 | 101.0 | 0.09 | 0.61 | 0.51 | 0.15 | 0.67 |
| Kruppel-like factor 4 | |||||||
| EZH2 | 6.8 | 1.8 | 0.89 | 0.80 | 0.63 | 0.29 | 0.75 |
| Enhancer of zeste homolog 2 | |||||||
| SUZ12 | |||||||
| Suppressor of zeste 12 homolog | 19.4 | 13.9 | 0.40 | 0.89 | 0.94 | 0.74 | 0.78 |
| CHUK | |||||||
| Conserved helix-loop-helix ubiquitous kinase | 12.6 | 15.0 | 0.49 | 0.50 | 0.25 | 0.98 | 0.66 |
| IKBKB | |||||||
| Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | 8.9 | 7.4 | 0.56 | 0.22 | 0.09 | 0.04 | 0.17 |
| NOTCH3 | 20.6 | 55.8 | 0.15 | 0.15 | 0.03 | 0.21 | 0.16 |
| Notch homolog 3 | |||||||
| ELAVL1 | 36.4 | 33.2 | 0.91 | 0.21 | 0.23 | 0.13 | 0.21 |
| ELAV-like 1 (Hu antigen R) | |||||||
| KDM3A | 35.4 | 13.7 | 0.26 | 0.75 | 0.72 | 0.55 | 0.39 |
| Lysine Demethylase 3A | |||||||
| SNAI1 | 4.0 | 9.8 | * | * | * | * | * |
| Snail homolog 1 | |||||||
| TNF | 0.3 | 0.3 | 0.81 | 0.55 | 0.68 | 0.22 | 0.60 |
| Tumor necrosis factor | |||||||
| IGF2 | |||||||
| Insulin-like growth factor 2 (somatomedin A) | 147.6 | 102.0 | * | * | * | * | * |
| PCDH10 | 4.6 | 0.6 | * | * | * | * | * |
| Protocadherin 10 | |||||||
| FOXA1 | 9.2 | 0.3 | * | * | * | * | * |
| Forkhead box A1 | |||||||
| FOXM1 | 6.0 | 2.4 | 0.58 | 0.92 | 0.50 | 0.48 | 0.60 |
| Forkhead box M1 | |||||||
| CTNNB1 | |||||||
| Catenin (cadherin-associated protein), beta 1, 88 kDa | 154.3 | 156.3 | 0.44 | 0.07 | 0.02 | 0.14 | 0.047 |
| SETD2 | 12.2 | 10.0 | 0.44 | 0.07 | 0.02 | 0.14 | 0.047 |
| SET domain containing 2 | |||||||
| ASTN1 | 3.2 | 0.3 | * | * | * | * | * |
| Astrotactin 1 | |||||||
| PCDHA1 | 0.2 | 0.0 | 0.51 | 0.39 | 0.15 | 0.52 | 0.94 |
| Ptotocadherin alpha-1 | |||||||
| MUC5AC | |||||||
| Mucin 5AC, oligomeric mucus/gel-forming | 10.2 | 0.0 | * | * | * | * | * |
| NTM | 8.0 | 18.3 | * | * | * | * | * |
| Neurotrimin | |||||||
| PTK2B | 28.8 | 16.1 | 0.22 | 0.01 | 0.0004 | 0.01 | 0.0032 |
| Protein tyrosine kinase 2 beta | |||||||
| VIM | 955.0 | 2578.1 | 0.18 | 0.07 | 0.0005 | 0.01 | 0.01 |
| Vimentin | |||||||
*Unavailable data. Shown are the 23 known mediators associated with HOTAIR; their expression in adipose tissue relative to tissue-average, according to RNA sequencing of tissues from healthy human subjects (The Human Protein Atlas 16.1); and T-test values of their differential blood cells and liver expression levels in various sepsis conditions vs. non-septic SIRS (CAPSOD study, see text), in a descending order (mortality > shock > severe > uncomplicated). Significantly changed expression levels in the CAPSOD database are bolded (P value < 0.1).
The previously unnoticed association between MetS and sepsis transcripts is a striking and non-trivial one, both since we compared blood to liver transcripts and because SIRS may by itself lead to downregulation of some transcripts; indeed, certain MetS-modified transcripts (e.g., angiopoietin 2) showed elevated levels in SIRS compared to sepsis. Of note, testing for sepsis severity-related changes in genes that were reported to interact with HOTAIR showed no significant difference. However, concordant with the predicted cholinergic control of inflammation that may contribute to both conditions, we noted changes in several cholinergic genes. Specifically, the neuronal transcription regulator REST, the muscarinic receptor ChRM3, the nicotinic inflammation-blocking nAChR7 (chrna7) and the obesity-related nicotinic receptor nAChR3 receptor all showed a decline with sepsis severity. In contrast, both the epilepsy-related nicotinic receptor chrnb2 and the allosteric modulator of nicotinic reactions Lynx1 were increased with sepsis severity (Fedi et al., 2008). The apparent links between MetS and sepsis-related blood and liver transcripts provide further support for a functional relationship between the biomedical pathways affected in these two syndromes.
LncRNAs Involvement in Pancreatic Failure, Diabetes and Ischemic Myocardial Injury
In addition to identifying known disease-related genes, focused studies of specific tissue malfunctioning may detect sepsis and/or MetS-related changes in particular lncRNAs that were not yet defined as directly linked with these syndromes. In this context, pancreatic function is notoriously impaired in both sepsis and MetS. Transcript mapping of human pancreatic islets showed specific lncRNAs associated with beta-cell differentiation and maturation; some of those lncRNAs are glucose-regulated, and were therefore implicated in endocrine function, with two of those (KCNQ1OT1 and HI-LNC45) being dysregulated in type 2 diabetic humans compared to non-diabetics (Korostowski et al., 2012; Morán et al., 2012). Also, KCNQ1OT1 expression predicts heart failure following myocardial infarction, whereas the lncRNA SRA is involved in transactivation of the nuclear receptor PPARg, which contributes to adipocyte differentiation and function (Hubé et al., 2011). The CHAST lncRNA is upregulated in cardiomyocytes during hypertrophic heart disease (Viereck et al., 2016). In comparison, the circulating levels of LncRNA-p5549, -p21015 and -p19461 are downregulated in human obesity compared to non-obese individuals, showing negative correlation with clinical obesity parameters (BMI, waist circumference, waist-to-hip ratio); of these molecules, lncRNA-19461 also associates with glycemic laboratory markers, and is increased towards healthy-control values upon weight loss (Sun et al., 2016).
At the level of protein pathways, KEGG analysis revealed an association of lncRNA-p5549, -p21015 and -p19461 with 10 pathways, with the inflammatory pathway of the innate immune Toll-like receptor signaling being the most enriched (Sun et al., 2016). Also, lncRNA-BATE1 associated differentially in brown vs. white adipose tissue, by forming a functional ribonucleoprotein complex with hnRNP U (Alvarez-Dominguez et al., 2015), indicating functional relevance of RNA metabolism at large. Another brown fat-associated lncRNA molecule, Blnc1, is involved in adipogenesis and adipocyte differentiation and function, and interacts with hnRNP U and with the EBF2 transcription factor associated with adipocyte regulation (Zhao and Lin, 2015). Changes in each or all of these lncRNAs may hence associate with sepsis, MetS or both.
Ischemic myocardial injury is one of the most dreaded outcomes of MetS (Wilson et al., 2005). Elevated levels of the LIPCAR lncRNA associate with poor cardiac prognosis; and serum LIPCAR transcripts are altered in patients following myocardial infarction (Kumarswamy et al., 2014). Cardiac hypertrophy, a manifestation of advanced metabolic damage to the heart muscle also associates with elevated levels of the H19 lncRNA. The latter interacts with miR-675 and CaMKII-delta for its cardiac effects, and may form “sponges” for the let-7 miR (Kallen et al., 2013; Liu et al., 2016). Therefore, both pancreatic failure and ischemic cardiac injury may be susceptible for intricate patterns of ncRNA regulation in septic and MetS patients. Supplementary Table S1 lists further lncRNAs that were so far demonstrated to be involved with MetS, their possible mechanisms of action, and associated mediators.
Inflammatory Links with Sepsis and MetS-Mis-Regulated ncRNAs
Much of the reported associations between sepsis and MetS relate to their inflammatory links. In sepsis, major inflammatory reaction acutely recruits several systems, whereas in MetS, inflammation is chronic and subclinical, without the classic clinical inflammatory manifestation. Nevertheless, MetS has recently been earmarked as inflammatory in its nature, and shows inflammatory laboratory markers and associated activity of inflammatory mediators (such as CRP, TNFα, IFNγ, and various ILs). Apart from lncRNAs, the human genome includes 8000 lincRNAs (long intergenic non-coding RNAs) which may also contribute to immune system regulation (Carpenter et al., 2013; Rapicavoli et al., 2013). RNA-sequencing data shows that LPS modulates approximately 200 lincRNAs in blood and 60 in adipose tissue, and that those are enriched in NF-kB binding motifs as well as in TLR4 signaling targets, some of which (linc-DMRT2, linc-TP53I13) are expressed in adipose tissue. Also, TP53I13, which is suppressed in obese human subjects (Liu et al., 2014), is elevated in sepsis. In MetS, innate immune functions are co-modified with inflammatory elements: the MetS-upregulated lincRNA Cox2 modulates the expression of inflammatory-associated proteins, such as chemokines and interferon-stimulated genes (Carpenter et al., 2013), and the p50-associated lncRNA PACER leads the assembly of NF-kB transcription complexes, which are major promoters of inflammatory cellular processes (Krawczyk and Emerson, 2014).
Both lincRNAs and lncRNAs regulate inflammation on their own merit, and are themselves influenced by propagating inflammation via feedback mechanisms. The multi-directional nature of the lncRNA-inflammation axis is also evident in the interaction of NF-kB with Lethe, which has immunomodulatory roles over NFkB activities (Rapicavoli et al., 2013). Lnc-DC, another lncRNA governs the inflammatory activities of dendritic cells (antigen uptake, cytokine release) through the paramount STAT3 inflammatory activating pathway, whereas THRIL upregulates the inflammatory agent TNFα that by itself regulates TNFAIP3 (Vereecke et al., 2009; Li et al., 2014; Wang et al., 2014). Some lncRNAs participate in metabolic derangement by influencing other molecular elements involved in these pathologies (Table 2). The multi-leveled interactions of numerous lncRNAs, lincRNAs and miRs with sepsis and MetS highlight their importance for the inflammatory aspects of these syndromes.
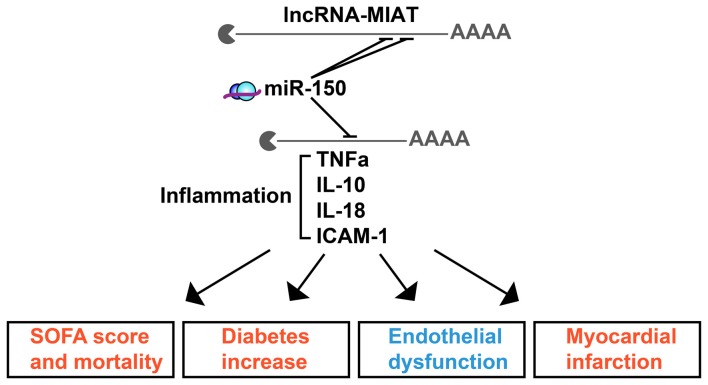
MIAT and miR-608 as Pan-Genomic Links of Sepsis with MetS and CNS Pathologies of Trauma and Mental Illness
The full scope of regulatory ncRNAs likely holds numerous yet-undiscovered connectors between diverse brain and body disease scenarios. One such example is the lncRNA MIAT, mis-regulation of which associates with endothelial dysfunction in diabetes. MIAT derives from chromosome 22 and carries recognition elements for 122 different miRs across numerous chromosomes. Of those, 41, 23 and 13 miRs associate with MetS, sepsis or both, respectively, creating lncRNA-miR-targets networks which may affect a number of related phenotypes (Figure 2). To assess the chromosomal origins and the relative impact of MIAT’s contributions to inflammation, we segregated the predicted MIAT miR targets according to their chromosomal origins and reported clinical associations (Figure 2), and identified a complex profile of MIAT’s contribution to MetS and sepsis. Segregated by their reported association with sepsis and/or MetS, the chromosomal loci of MIAT and its predicted miR targets spanned 11 different chromosomes. For example, in animal models, MIAT interacts with the chromosome 19-originated miR-150-5p; which is an established marker for sepsis staging and outcome in human patients, presumably via its regulation of several inflammation controlling cytokines including TNFα and ILs IL-10, IL-18 (Yan et al., 2015; Figure 2). MIAT’s interaction with miR-150-5p may explain its association with inflammation, severity score and mortality increase in diabetes as well as with the endothelial dysfunction and myocardial infarcts of septic patients, which likely extend to blood-brain barrier malfunctioning.
Figure 2.
The lncRNA MIAT targets multi-chromosome originated sepsis and MetS-related miRs. MIAT and miR-150 crosstalk exerts inflammatory impact in sepsis and MetS. Shown are selected inflammation-related targets of miR-150 which associate with the clinical characteristics of sepsis (red) or MetS (blue).
Molecular links in sepsis-MetS regulation and etiology predict that specific miRs may share targets (coding and non-coding) that are changed in both of these conditions. For example, a SNP in the miR-608-binding motif of the AChE gene associates with several MetS hallmarks (Hanin et al., 2014), whereas another SNP in the miR-608 gene itself limits the risk of sepsis in head injury patients (Zhang et al., 2015). The asymmetric impact of these two SNPs may hence indicate different consequences of AChE and miR-608 changes on MetS and sepsis. The mechanism of action explaining both associations may operate via cholinergic signaling, which can effectively block the inflammatory process via suppression by the homomeric alpha7 acetylcholine receptor of NF-kB-mediated production of pro-inflammatory cytokines (Tracey, 2002; Pavlov and Tracey, 2012). Several cholinergic-related miRs (“CholinomiRs”), including miR-132, miR-608 and miR-211, are functionally involved in inflammatory regulation (Meydan et al., 2016; Bekenstein et al., 2017). However, whether they impact sepsis, MetS or both has only scarcely been explored. Notably, miR-608 targets several inflammation-related transcripts as well as the histocompatibility antigen HLA-DRA (Barbash et al., 2017), which is a biomarker for immunosuppression in sepsis, predictive of survival, and a surrogate marker for GM-CSF experimental therapy of sepsis (Lekkou et al., 2004; Meisel et al., 2009; Cajander et al., 2013). Its association with miR-608 is hence of particular interest.
Previous reports revealed molecular cues that might explain the asymmetric impact of the SNP-mediated impairments of the AChE-miR-608 axis: the AChE SNP that interrupts the binding and functioning of miR-608 associates with elevated AChE, inflammation, trait anxiety and blood pressure while reducing cortisol levels in healthy volunteers (Hanin et al., 2014) and inducing hyper-alert reaction to stress in the pre-frontal cortex of minor allele carriers (Lin et al., 2016). The cholinergic imbalance caused by the AChE SNP is hence direct, but its impact is relatively limited. In comparison, the miR-608 SNP may modulate its interaction with both AChE and with numerous other inflammation-related transcripts. These include the amygdala-expressed inflammatory mediators IL-6, CD-44, CDC42 and TPP-1 (Hanin et al., 2014; Lin et al., 2016), which form a pathway resonant with the regulatory ncRNA-cholinergic-inflammatory network. Tilting this balance modulates cortisol levels, with an established role in both MetS and sepsis (Supplementary Material), compatible with targeting the histocompatibility antigen HLA-DRA gene, which is recognized as a hallmark of sepsis (Meisel et al., 2009; Barbash et al., 2017; Figure 3).
Figure 3.
Asymmetric impacts of single nucleotide polymorphisms (SNPs) interrupting miR-608/AChE interaction highlight the relevance for concussions. SNPs in the miR-608 and AChE genes may both interrupt the miR-608/AChE interaction, but the multi-target impact of miR-608 affects the risk of sepsis (red) whereas the AChE SNP relates to MetS (blue). Specifically, the miR-608 SNP (left) may modulate both HLA-DRA and inflammation by interrupting its cholinergic blockade. The AChE SNP (right) may likewise interfere with the cholinergic blockade of NFkB-mediated inflammation, while modulating anxiety, cortisol and blood pressure in MetS patients, which should be highly relevant for the consequences of concussions.
To find out the relevance of the imbalanced impact of ncRNAs to blood cells and/or liver transcripts, we sought for predicted targets of miR-608 that associate with MetS and/or sepsis reports, which highlighted its HLA-DRA target as uniquely related to both. Next, we re-searched the CAPSOD dataset for associations between individual transcript changes induced in miR-608 targets with MetS, sepsis or both. Strikingly, we identified parallel changes in the sepsis-related subset of those transcripts derived from patients’ liver tissues compared to healthy controls, and in MetS-derived RNA-sequencing profiles of patients’ nucleated blood cells with their appropriate controls. This is evident in the multi-chromosomal origins of miR-608’s predicted coding and noncoding RNA targets whose levels were modified in these two tissues, and demonstrated considerable similarities in the differential expression patterns of liver and blood cells between MetS and sepsis tissues (Figure 3). In both tissues, those chromosomal domains which showed pronounced response in the septic liver also showed prominent blood cell reactions to MetS, and vice versa, indicating that blood cells RNA-profiles may provide objective clinical assessment of symptoms severity.
Both MetS and sepsis also have mental implications (De Hert et al., 2011; Sonneville et al., 2013) which may relate to another class of lncRNAs, miR-reacting pseudogenes. These lncRNAs have been evolutionarily derived from coding genes, and carry miR recognition elements. SNPs in miR-responding pseudogenes associate with mental diseases such as schizophrenia, autism and bipolar disease, but their putative links to sepsis and MetS have not been explored to date. An example for the interaction between miR-608 and inflammation-regulating lncRNAs lies in the miRs-responding neuronal expressed pseudogene-PGOHUM00000243565 (PGOHUM-565; Barbash et al., 2017). Mechanistically, PGOHUM-565 shares eight different miR recognition elements with HLA-DRA and is predicted to bind the primate-specific miR-608 via its complementary “seed” motif. This may offset the action of miR-608 on numerous coding transcripts, including HLA-DRA and the cholinergic enzyme AChE. Imbalanced interactions of miR-608 with these two targets and others may both tilt the inflammatory response and be mechanistically related to the mental symptoms of sepsis (Cajander et al., 2013), once again highlighting the dissimilarity in the imbalanced ncRNA-target interactions that associate with sepsis and/or MetS.
Conclusions and Implications for Future Clinical Considerations
We identified genomic and clinical sepsis-related elements that associate with both sepsis and MetS, albeit at different progression rates and with different clinical ramifications. A deeper appreciation of the emerging ncRNAs underlying these shared elements found inflammation-associated molecular agents and cytokines that are affected in both sepsis and MetS. These add up on the established molecules such as Resistin, PAI-1, Adiponectin, Leptin and AGEs (Advanced Glycation End-products) that are prime examples underlying inflammation, putatively accounting for some of the apparent links between sepsis and MetS. The apparent interactions between MetS and the human response to pathogens are becoming evident across the spectrum of these processes. The gut microbiome, which functions mostly as a commensal pathogenic entity, impacts obesity and anti-obesity interventions (Liu et al., 2017). Consistent unfolding of the intricate links between MetS, inflammation and pathogens may further point at novel therapeutic avenues; one recent example is anti-IL-1β antibodies investigated for secondary prevention of myocardial infarction (Ridker et al., 2017). Highly relevant here is the aftermath of repeated concussions, for example in football players (Bailes et al., 2013; Zaghloul et al., 2017).
We suggest and provide evidence for miRs and lncRNAs as potent mediators of inflammatory regulation (including specific examples such as HOTAIR, Lethe, PGOHUM-565) in sepsis and MetS. Specifically, miR regulators of cellular mechanisms including miR-122, −150, −608 and −182 relate to both MetS and sepsis and affect anxiety as well (Supplementary Figures S1C). These lncRNA-miR interactions may form multi-directional regulatory networks, accounting for some of the shared clinical traits and molecular machinery involved. We conclude that altered RNA interactions may modify cellular and brain features in a parallel way to the impact of modified protein networks (Morais et al., 2014), adding a novel regulatory layer that needs to be clinically addressed.
Our current observations highlight upstream regulatory layers to MetS and sepsis progression and outcomes, and even more so, to the associations between them as evident in clinical traits. Based on our current insight, modulating the inflammatory pattern through relevant lncRNA/miR-based networks may be a prime target for sepsis, as an adjunct for antimicrobial and supportive care and for preventing complications after head injury. Hypothesis-driven exploration into the links between MetS and sepsis may hence promote the identification of biomarkers and novel therapies, which are urgently needed.
Key Concepts
Non-coding RNAs are active within genomic domains to alter protein expression and function, and across various clinical spectra, including MetS, sepsis, and head injury risks.
Cholinergic elements are neuronally-oriented molecular machinery transcripts with prominent roles in shaping inflammation as a controlled, proportional and adequate process.
The metabolic syndrome is an amalgam of chronic pathologic conditions which carry risks for cerebrovascular and other adverse sequelae, including coronary and mental disease.
Sepsis is a life-threatening pathological condition in which overwhelming inflammation is ensued in order to avert infections, with several major organic derangements occurring as a result.
Author Contributions
HS, CM and UB all participated in the information collection, data mining and manuscript design.
Conflict of Interest Statement
CM receives consultation fees from Raziel Therapeutics, Ltd. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- AChE
acetylcholinesterase
- AGEs
advanced glycation end-products
- CAPSOD
community acquired pneumonia and sepsis outcome diagnostic study
- CholinomiRs
cholinergic-related miRs
- CNS
central nervous system
- GWAS
genome wide association scale
- KLF4
Kruppel-like factor 4
- lncRNAs
long non-coding RNAs
- MetS
metabolic syndrome
- miRs
microRNAs
- NOTCH3
notch homolog 3
- NTM
neurotrimin
- SIRS
systemic inflammatory response syndrome
- SNAI1
snail homolog 1
- SNP
single nucleotide polymorphism
- VIM
vimentin.
Footnotes
Funding. This research was supported by the European Research Council (Advanced Award 321501), The Israeli Ministry of Science, Technology and Space, Grant No. 53140, The Israel I-Core Center of Excellence for Mass Trauma and The Legacy Heritage Science Initiative (LHSI) of The Israel Science Foundation Grant No. 817/13, The Austrian Research Promotion Agency (FFG Bridge1 project; Österreichische Forschungsförderungsgesellschaft, Grant number 853294) and the Edmond and Lily Safra Center for Brain Sciences (ELSC, to HS); UB was supported by the Howard and Diana Wendy pre-doctoral fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00189/full#supplementary-material
References
- Abbasi J. (2017). In treating sepsis, questions about timing and mandates. JAMA 318, 506–508. 10.1001/jama.2017.7997 [DOI] [PubMed] [Google Scholar]
- Alvarez-Dominguez J. R., Bai Z., Xu D., Yuan B., Lo K. A., Yoon M. J., et al. (2015). De novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. 10.1016/j.cmet.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus D. C., van der Poll T. (2013). Severe sepsis and septic shock. N. Engl. J. Med. 369, 840–851. 10.1056/NEJMra1208623 [DOI] [PubMed] [Google Scholar]
- Arner P., Kulyté A. (2015). MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 11, 276–288. 10.1038/nrendo.2015.25 [DOI] [PubMed] [Google Scholar]
- Aslan A., van Meurs M., Moser J., Popa E. R., Jongman R. M., Zwiers P. J., et al. (2017). Organ-specific differences in endothelial permeability-regulating molecular responses in mouse and human sepsis. Shock 48, 69–77. 10.1097/SHK.0000000000000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes J. E., Petraglia A. L., Omalu B. I., Nauman E., Talavage T. (2013). Role of subconcussion in repetitive mild traumatic brain injury. J. Neurosurg. 119, 1235–1245. 10.3171/2013.7.JNS121822 [DOI] [PubMed] [Google Scholar]
- Barbash S., Shifman S., Soreq H. (2014). Global coevolution of human microRNAs and their target genes. Mol. Biol. Evol. 31, 1237–1247. 10.1093/molbev/msu090 [DOI] [PubMed] [Google Scholar]
- Barbash S., Simchovitz A., Buchman A. S., Bennett D. A., Shifman S., Soreq H. (2017). Neuronal-expressed microRNA-targeted pseudogenes compete with coding genes in the human brain. Transl. Psychiatry 7:e1199. 10.1038/tp.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwari T., Joshi A., Mayr M. (2016). MicroRNAs in cardiovascular disease. J. Am. Coll. Cardiol. 68, 2577–2584. 10.1016/j.jacc.2016.09.945 [DOI] [PubMed] [Google Scholar]
- Bekenstein U., Mishra N., Milikovsky D. Z., Hanin G., Zelig D., Sheintuch L., et al. (2017). Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc. Natl. Acad. Sci. U S A 114, E4996–E5005. 10.1073/pnas.1701201114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E. (2011). Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 12, 846–860. 10.1038/nrg3079 [DOI] [PubMed] [Google Scholar]
- Bopp C., Bierhaus A., Hofer S., Bouchon A., Nawroth P. P., Martin E., et al. (2008). Bench-to-bedside review: the inflammation-perpetuating pattern-recognition receptor RAGE as a therapeutic target in sepsis. Crit. Care 12:201. 10.1186/cc6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajander S., Bäckman A., Tina E., Strålin K., Söderquist B., Källman J. (2013). Preliminary results in quantitation of HLA-DRA by real-time PCR: a promising approach to identify immunosuppression in sepsis. Crit. Care 17:R223. 10.1186/cc13046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Aiello D., Atianand M. K., Ricci E. P., Gandhi P., Hall L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792. 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta S., Kern F., Cohen J., Drage S., Newbury S. F., Llewelyn M. J. (2016). Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS). Sci. Rep. 6:28006. 10.1038/srep28006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. (2016). Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol. Metab. 5, 164–170. 10.1016/j.molmet.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C. N., Nalpas N. C., McLoughlin K. E., Browne J. A., Gordon S. V., MacHugh D. E., et al. (2017). Circulating microRNAs as potential biomarkers of infectious disease. Front. Immunol. 8:118. 10.3389/fimmu.2017.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hert M., Detraux J., van Winkel R., Yu W., Correll C. U. (2011). Metabolic and cardiovascular adverse effects associated with antipsychotic drugs. Nat. Rev. Endocrinol. 8, 114–126. 10.1038/nrendo.2011.156 [DOI] [PubMed] [Google Scholar]
- de Simone G., Devereux R. B., Chinali M., Best L. G., Lee E. T., Galloway J. M., et al. (2007). Prognostic impact of metabolic syndrome by different definitions in a population with high prevalence of obesity and diabetes: the strong heart study. Diabetes Care 30, 1851–1856. 10.2337/dc06-2152 [DOI] [PubMed] [Google Scholar]
- Divakaran V., Mann D. L. (2008). The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 103, 1072–1083. 10.1161/CIRCRESAHA.108.183087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Schütz S., Lawrence M., Petri A., Obad S., et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899. 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S. F., Yu X. X., Pandey S. K., Pear M., et al. (2006). miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98. 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Fedi M., Bach L. A., Berkovic S. F., Willoughby J. O., Scheffer I. E., Reutens D. C. (2008). Association of a nicotinic receptor mutation with reduced height and blunted physostigmine-stimulated growth hormone release. J. Clin. Endocrinol. Metab. 93, 634–637. 10.1210/jc.2007-1611 [DOI] [PubMed] [Google Scholar]
- Galatzer-Levy I. R., Ma S., Statnikov A., Yehuda R., Shalev A. Y. (2017). Utilization of machine learning for prediction of post-traumatic stress: a re-examination of cortisol in the prediction and pathways to non-remitting PTSD. Transl. Psychiatry 7:e1070. 10.1038/tp.2017.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernapudi R., Wolfson B., Zhang Y., Yao Y., Yang P., Asahara H., et al. (2016). MicroRNA 140 promotes expression of long noncoding RNA NEAT1 in adipogenesis. Mol. Cell. Biol. 36, 30–38. 10.1128/MCB.00702-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi G., Seravalle G., Quarti-Trevano F., Scopelliti F., Dell’Oro R., Bolla G., et al. (2007). Excessive sympathetic activation in heart failure with obesity and metabolic syndrome: characteristics and mechanisms. Hypertension 49, 535–541. 10.1161/01.hyp.0000255983.32896.b9 [DOI] [PubMed] [Google Scholar]
- Greco S., Fasanaro P., Castelvecchio S., D’Alessandra Y., Arcelli D., Di Donato M., et al. (2012). MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes 61, 1633–1641. 10.2337/db11-0952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy S. M., Brewer H. B., Jr., Cleeman J. I., Smith S. C., Jr., Lenfant C., American Heart Association et al. (2004). Definition of metabolic syndrome: report of the national heart, lung, and blood institute/american heart association conference on scientific issues related to definition. Circulation 109, 433–438. 10.1161/01.cir.0000111245.75752.c6 [DOI] [PubMed] [Google Scholar]
- Hanin G., Shenhar-Tsarfaty S., Yayon N., Yau Y. H., Bennett E. R., Sklan E. H., et al. (2014). Competing targets of microRNA-608 affect anxiety and hypertension. Hum. Mol. Genet. 23, 4569–4580. 10.1093/hmg/ddu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin G., Yayon N., Tzur Y., Haviv R., Bennett E. R., Udi S., et al. (2017). miRNA-132 induces hepatic steatosis and hyperlipidaemia by synergistic multitarget suppression. Gut 67, 1124–1134. 10.1136/gutjnl-2016-312869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch U. K., Johnson T. V., Kipnis J. (2008). Toll-like receptors: roles in neuroprotection? Trends Neurosci. 31, 176–182. 10.1016/j.tins.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Hubé F., Velasco G., Rollin J., Furling D., Francastel C. (2011). Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 39, 513–525. 10.1093/nar/gkq833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelsson E., Arnlöv J., Lind L., Sundström J. (2006). Metabolic syndrome and risk for heart failure in middle-aged men. Heart 92, 1409–1413. 10.1136/hrt.2006.089011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. C., Pandharipande P. P., Girard T. D., Brummel N. E., Thompson J. L., Hughes C. G., et al. (2014). Depression, post-traumatic stress disorder, and functional disability in survivors of critical illness in the BRAIN-ICU study: a longitudinal cohort study. Lancet Respir. Med. 2, 369–379. 10.1016/s2213-2600(14)70051-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H. L., Reesink H. W., Lawitz E. J., Zeuzem S., Rodriguez-Torres M., Patel K., et al. (2013). Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368, 1685–1694. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- Kallen A. N., Zhou X. B., Xu J., Qiao C., Ma J., Yan L., et al. (2013). The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52, 101–112. 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolina D. S., Tavintharan S., Armugam A., Sepramaniam S., Pek S. L., Wong M. T., et al. (2012). Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 97, E2271–E2276. 10.1210/jc.2012-1996 [DOI] [PubMed] [Google Scholar]
- Karpe F., Pinnick K. E. (2015). Biology of upper-body and lower-body adipose tissue—link to whole-body phenotypes. Nat. Rev. Endocrinol. 11, 90–100. 10.1038/nrendo.2014.185 [DOI] [PubMed] [Google Scholar]
- Korostowski L., Sedlak N., Engel N. (2012). The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 8:e1002956. 10.1371/journal.pgen.1002956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk M., Emerson B. M. (2014). p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife 3:e01776. 10.7554/eLife.01776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarswamy R., Bauters C., Volkmann I., Maury F., Fetisch J., Holzmann A., et al. (2014). Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575. 10.1161/CIRCRESAHA.114.303915 [DOI] [PubMed] [Google Scholar]
- Lagos D., Pollara G., Henderson S., Gratrix F., Fabani M., Milne R. S., et al. (2010). miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519. 10.1038/ncb2054 [DOI] [PubMed] [Google Scholar]
- Lebow M., Neufeld-Cohen A., Kuperman Y., Tsoory M., Gil S., Chen A. (2012). Susceptibility to PTSD-like behavior is mediated by corticotropin-releasing factor receptor type 2 levels in the bed nucleus of the stria terminalis. J. Neurosci. 32, 6906–6916. 10.1523/JNEUROSCI.4012-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkou A., Karakantza M., Mouzaki A., Kalfarentzos F., Gogos C. A. (2004). Cytokine production and monocyte HLA-DR expression as predictors of outcome for patients with community-acquired severe infections. Clin. Diagn. Lab. Immunol. 11, 161–167. 10.1128/cdli.11.1.161-167.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. D., Burmeister M. (2009). New insights into the genetics of addiction. Nat. Rev. Genet. 10, 225–231. 10.1038/nrg2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chao T. C., Chang K. Y., Lin N., Patil V. S., Shimizu C., et al. (2014). The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U S A 111, 1002–1007. 10.1073/pnas.1313768111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. M., Hu Z. B., Zhou Z. X., Chen X., Liu F. Y., Zhang J. F., et al. (2010). Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 70, 9798–9807. 10.1158/0008-5472.CAN-10-1001 [DOI] [PubMed] [Google Scholar]
- Li Z., Rana T. M. (2014). Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 13, 622–638. 10.1038/nrd4359 [DOI] [PubMed] [Google Scholar]
- Lin T., Simchovitz A., Shenhar-Tsarfaty S., Vaisvaser S., Admon R., Hanin G., et al. (2016). Intensified vmPFC surveillance over PTSS under perturbed microRNA-608/AChE interaction. Transl. Psychiatry 6:e801. 10.1038/tp.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Qin Y., Jia J., Lin T., Lin X., Chen L., et al. (2016). MiR-155 enhances insulin sensitivity by coordinated regulation of multiple genes in mice. PLoS Genet. 12:e1006308. 10.1371/journal.pgen.1006308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., An X., Li Z., Song Y., Li L., Zuo S., et al. (2016). The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 111, 56–65. 10.1093/cvr/cvw078 [DOI] [PubMed] [Google Scholar]
- Liu Y., Ferguson J. F., Xue C., Ballantyne R. L., Silverman I. M., Gosai S. J., et al. (2014). Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler. Thromb. Vasc. Biol. 34, 902–912. 10.1161/ATVBAHA.113.303123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Hong J., Xu X., Feng Q., Zhang D., Gu Y., et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868. 10.1038/nm.4358 [DOI] [PubMed] [Google Scholar]
- Luna J. M., Scheel T. K., Danino T., Shaw K. S., Mele A., Fak J. J., et al. (2015). Hepatitis C virus RNA functionally sequesters miR-122. Cell 160, 1099–1110. 10.1016/j.cell.2015.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Han P., Ye W., Chen H., Zheng X., Cheng L., et al. (2017). The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91:e02250-16. 10.1128/JVI.02250-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella R., Di Filippo C., Potenza N., Sardu C., Rizzo M. R., Siniscalchi M., et al. (2013). Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur. J. Heart Fail. 15, 1277–1288. 10.1093/eurjhf/hft088 [DOI] [PubMed] [Google Scholar]
- Maskos U., Molles B. E., Pons S., Besson M., Guiard B. P., Guilloux J. P., et al. (2005). Nicotine reinforcement and cognition restored by targeted expression of nicotinic receptors. Nature 436, 103–107. 10.1038/nature03694 [DOI] [PubMed] [Google Scholar]
- Meisel C., Schefold J. C., Pschowski R., Baumann T., Hetzger K., Gregor J., et al. (2009). Granulocyte-macrophage colony-stimulating factor to reverse sepsis-associated immunosuppression: a double-blind, randomized, placebo-controlled multicenter trial. Am. J. Respir. Crit. Care Med. 180, 640–648. 10.1164/rccm.200903-0363OC [DOI] [PubMed] [Google Scholar]
- Melkman-Zehavi T., Oren R., Kredo-Russo S., Shapira T., Mandelbaum A. D., Rivkin N., et al. (2011). miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 30, 835–845. 10.1038/emboj.2010.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman Y. F., Shah R., Danielson K., Xiao J., Simonson B., Barth A., et al. (2015). Circulating microRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study. Circulation 131, 2202–2216. 10.1161/CIRCULATIONAHA.114.013220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meydan C., Shenhar-Tsarfaty S., Soreq H. (2016). MicroRNA regulators of anxiety and metabolic disorders. Trends Mol. Med. 22, 798–812. 10.1016/j.molmed.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Montgomery R. L., Hullinger T. G., Semus H. M., Dickinson B. A., Seto A. G., Lynch J. M., et al. (2011). Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 124, 1537–1547. 10.1161/CIRCULATIONAHA.111.030932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán I., Akerman I., van de Bunt M., Xie R., Benazra M., Nammo T., et al. (2012). Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated and abnormally expressed in type 2 diabetes. Cell Metab. 16, 435–448. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais V. A., Haddad D., Craessaerts K., De Bock P. J., Swerts J., Vilain S., et al. (2014). PINK1 loss-of-function mutations affect mitochondrial complex I activity via NdufA10 ubiquinone uncoupling. Science 344, 203–207. 10.1126/science.1249161 [DOI] [PubMed] [Google Scholar]
- Morchikh M., Cribier A., Raffel R., Amraoui S., Cau J., Severac D., et al. (2017). HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387.e5–399.e5. 10.1016/j.molcel.2017.06.020 [DOI] [PubMed] [Google Scholar]
- Mottillo S., Filion K. B., Genest J., Joseph L., Pilote L., Poirier P., et al. (2010). The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J. Am. Coll. Cardiol. 56, 1113–1132. 10.1016/j.jacc.2010.05.034 [DOI] [PubMed] [Google Scholar]
- Nazari-Jahantigh M., Wei Y., Noels H., Akhtar S., Zhou Z., Koenen R. R., et al. (2012). MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Invest. 122, 4190–4202. 10.1172/JCI61716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S., O’Driscoll L. (2015). Metabolic syndrome: a closer look at the growing epidemic and its associated pathologies. Obes. Rev. 16, 1–12. 10.1111/obr.12229 [DOI] [PubMed] [Google Scholar]
- Pan W., Zhong Y., Cheng C., Liu B., Wang L., Li A., et al. (2013). MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS One 8:e53950. 10.1371/journal.pone.0053950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. W., Yun J. H., Kim J. H., Kim K. W., Cho C. H., Kim J. H. (2014). Angiopoietin 2 induces pericyte apoptosis via α3β1 integrin signaling in diabetic retinopathy. Diabetes 63, 3057–3068. 10.2337/db13-1942 [DOI] [PubMed] [Google Scholar]
- Pavlov V. A., Tracey K. J. (2012). The vagus nerve and the inflammatory reflex—linking immunity and metabolism. Nat. Rev. Endocrinol. 8, 743–754. 10.1038/nrendo.2012.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P. E., et al. (2004). A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432, 226–230. 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- Rapicavoli N. A., Qu K., Zhang J., Mikhail M., Laberge R., Chang H. Y. (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2:e00762. 10.7554/eLife.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. A., Chen Z., Park J. T., Wang M., Lanting L., Zhang Q., et al. (2014). Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 63, 4249–4261. 10.2337/db14-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart K., Daniels R., Kissoon N., Machado F. R., Schachter R. D., Finfer S. (2017). Recognizing sepsis as a global health priority—a WHO resolution. N. Engl. J. Med. 377, 414–417. 10.1056/NEJMp1707170 [DOI] [PubMed] [Google Scholar]
- Ridker P. M., Cannon C. P., Morrow D., Rifai N., Rose L. M., McCabe C. H., et al. (2005). C-reactive protein levels and outcomes after statin therapy. N. Engl. J. Med. 352, 20–28. 10.1056/NEJMoa042378 [DOI] [PubMed] [Google Scholar]
- Ridker P. M., Everett B. M., Thuren T., MacFadyen J. G., Chang W. H., Ballantyne C., et al. (2017). Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131. 10.1056/NEJMoa1707914 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittirsch D., Flierl M. A., Ward P. A. (2008). Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8, 776–787. 10.1038/nri2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E., Sierksma A., Vanden Eynden E., De Strooper B. (2016). miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 8, 1005–1018. 10.15252/emmm.201606520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G., Iaccarino G., De Luca N., Trimarco B., Condorelli G. (2014). Atrial fibrillation and microRNAs. Front. Physiol. 5:15. 10.3389/fphys.2014.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C., Barbieri M., Rizzo M. R., Paolisso P., Paolisso G., Marfella R. (2016a). Cardiac resynchronization therapy outcomes in type 2 diabetic patients: role of microRNA changes. J. Diabetes Res. 2016:7292564. 10.1155/2016/7292564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C., Paolisso G., Marfella R. (2016b). Letter by sardu et al regarding article, “Circulating microRNA-30d is associated with response to cardiac resynchronization therapy in heart failure and regulates cardiomyocyte apoptosis: a translational pilot study”. Circulation 133:e388. 10.1161/CIRCULATIONAHA.115.018205 [DOI] [PubMed] [Google Scholar]
- Sardu C., Carreras G., Katsanos S., Kamperidis V., Pace M. C., Passavanti M. B., et al. (2014a). Metabolic syndrome is associated with a poor outcome in patients affected by outflow tract premature ventricular contractions treated by catheter ablation. BMC Cardiovasc. Disord. 14:176. 10.1186/1471-2261-14-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C., Marfella R., Santulli G., Paolisso G. (2014b). Functional role of miRNA in cardiac resynchronization therapy. Pharmacogenomics 15, 1159–1168. 10.2217/pgs.14.76 [DOI] [PubMed] [Google Scholar]
- Sardu C., Santamaria M., Funaro S., Sacra C., Barbieri M., Paolisso P., et al. (2017). Cardiac electrophysiological alterations and clinical response in cardiac resynchronization therapy with a defibrillator treated patients affected by metabolic syndrome. Medicine 96:e6558. 10.1097/MD.0000000000006558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardu C., Santamaria M., Paolisso G., Marfella R. (2015). microRNA expression changes after atrial fibrillation catheter ablation. Pharmacogenomics 16, 1863–1877. 10.2217/pgs.15.117 [DOI] [PubMed] [Google Scholar]
- Shahreyar M., Fahhoum R., Akinseye O., Bhandari S., Dang G., Khouzam R. N. (2018). Severe sepsis and cardiac arrhythmias. Ann. Transl. Med. 6:6. 10.21037/atm.2017.12.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Verdonk F., Rauturier C., Klein I. F., Wolff M., Annane D., et al. (2013). Understanding brain dysfunction in sepsis. Ann. Intensive Care 3:15. 10.1186/2110-5820-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ruan Y., Wang M., Chen R., Yu N., Sun L., et al. (2016). Differentially expressed circulating LncRNAs and mRNA identified by microarray analysis in obese patients. Sci. Rep. 6:35421. 10.1038/srep35421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Katz R., Jenny N. S., Zakai N. A., LeWinter M. M., Barzilay J. I., et al. (2008). Metabolic syndrome, inflammation, and incident heart failure in the elderly: the cardiovascular health study. Circ. Heart Fail. 1, 242–248. 10.1161/CIRCHEARTFAILURE.108.785485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. F., Huang R. T., Chien K. Y., Huang J. Y., Lau K. S., Jheng J. R., et al. (2015). Host microRNA miR-197 plays a negative regulatory role in the enterovirus 71 infectious cycle by targeting the RAN protein. J. Virol. 90, 1424–1438. 10.1128/JVI.02143-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Mizutani R., Salam K. A., Tano K., Ijiri K., Wakamatsu A., et al. (2012). Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 22, 947–956. 10.1101/gr.130559.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey K. J. (2002). The inflammatory reflex. Nature 420, 853–859. 10.1038/nature01321 [DOI] [PubMed] [Google Scholar]
- Tracey K. J. (2010). Understanding immunity requires more than immunology. Nat. Immunol. 11, 561–564. 10.1038/ni0710-561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. C., Manor O., Wan Y., Mosammaparast N., Wang J. K., Lan F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I. (2016). Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614. 10.1038/nrg.2016.85 [DOI] [PubMed] [Google Scholar]
- van der Poll T., Opal S. M. (2008). Host-pathogen interactions in sepsis. Lancet Infect. Dis. 8, 32–43. 10.1016/S1473-3099(07)70265-7 [DOI] [PubMed] [Google Scholar]
- Vasilescu C., Rossi S., Shimizu M., Tudor S., Veronese A., Ferracin M., et al. (2009). MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One 4:e7405. 10.1371/journal.pone.0007405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L., Beyaert R., van Loo G. (2009). The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 30, 383–391. 10.1016/j.it.2009.05.007 [DOI] [PubMed] [Google Scholar]
- Viereck J., Kumarswamy R., Foinquinos A., Xiao K., Avramopoulos P., Kunz M., et al. (2016). Long noncoding RNA chast promotes cardiac remodeling. Sci. Transl. Med. 8:326ra22. 10.1126/scitranslmed.aaf1475 [DOI] [PubMed] [Google Scholar]
- Wang Z., Fan P., Zhao Y., Zhang S., Lu J., Xie W., et al. (2017). NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell. Mol. Life Sci. 74, 1117–1131. 10.1007/s00018-016-2398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sarnola K., Ruotsalainen S., Moilanen L., Lepistö P., Laakso M., et al. (2010). The metabolic syndrome predicts incident congestive heart failure: a 20-year follow-up study of elderly Finns. Atherosclerosis 210, 237–242. 10.1016/j.atherosclerosis.2009.10.042 [DOI] [PubMed] [Google Scholar]
- Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., et al. (2014). The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313. 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]
- Wellen K. E., Thompson C. B. (2012). A two-way street: reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 13, 270–276. 10.1038/nrm3305 [DOI] [PubMed] [Google Scholar]
- Wilson P. W., D’Agostino R. B., Parise H., Sullivan L., Meigs J. B. (2005). Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation 112, 3066–3072. 10.1161/circulationaha.105.539528 [DOI] [PubMed] [Google Scholar]
- Wu H., Liu J., Li W., Liu G., Li Z. (2016). LncRNA-HOTAIR promotes TNF-α production in cardiomyocytes of LPS-induced sepsis mice by activating NF-κB pathway. Biochem. Biophys. Res. Commun. 471, 240–246. 10.1016/j.bbrc.2016.01.117 [DOI] [PubMed] [Google Scholar]
- Xu G., Cui Y., Jia Z., Yue Y., Yang S. (2016). The values of coronary circulating miRNAs in patients with atrial fibrillation. PLoS One 11:e0166235. 10.1371/journal.pone.0166235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Yao J., Liu J. Y., Li X. M., Wang X. Q., Li Y. J., et al. (2015). lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 116, 1143–1156. 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- Zaghloul N., Addorisio M. E., Silverman H. A., Patel H. L., Valdés-Ferrer S. I., Ayasolla K. R., et al. (2017). Forebrain cholinergic dysfunction and systemic and brain inflammation in murine sepsis survivors. Front. Immunol. 8:1673. 10.3389/fimmu.2017.01673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A., Mayr M. (2012). MicroRNAs in vascular and metabolic disease. Circ. Res. 110, 508–522. 10.1161/CIRCRESAHA.111.247445 [DOI] [PubMed] [Google Scholar]
- Zgheib C., Hodges M. M., Hu J., Liechty K. W., Xu J. (2017). Long non-coding RNA lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS One 12:e0177453. 10.1371/journal.pone.0177453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Q., Gu W., Zeng L., Zhang L. Y., Du D. Y., Zhang M., et al. (2015). Genetic variants of microRNA sequences and susceptibility to sepsis in patients with major blunt trauma. Ann. Surg. 261, 189–196. 10.1097/SLA.0000000000000687 [DOI] [PubMed] [Google Scholar]
- Zhang D., Li Y., Yao X., Wang H., Zhao L., Jiang H., et al. (2016). miR-182 regulates metabolic homeostasis by modulating glucose utilization in muscle. Cell Rep. 16, 757–768. 10.1016/j.celrep.2016.06.040 [DOI] [PubMed] [Google Scholar]
- Zhao X. Y., Lin J. D. (2015). Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem. Sci. 40, 586–596. 10.1016/j.tibs.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.