Abstract
We provide a comprehensive diffusion MRI dataset acquired with a novel biomimetic phantom mimicking human white matter. The fiber substrates in the diffusion phantom were constructed from hollow textile axons (“taxons”) with an inner diameter of 11.8±1.2 µm and outer diameter of 33.5±2.3 µm. Data were acquired on the 3 T CONNECTOM MRI scanner with multiple diffusion times and multiple q-values per diffusion time, which is a dedicated acquisition for validation of microstructural imaging methods, such as compartment size and volume fraction mapping. Minimal preprocessing was performed to correct for susceptibility and eddy current distortions. Data were deposited in the XNAT Central database (project ID: dMRI_Phant_MGH).
Specifications Table
| Subject area | Neuroimaging |
|---|---|
| More specific subject area | Diffusion MRI |
| Type of data | MRI data |
| How data was acquired | 3 T MRI, Siemens CONNECTOM |
| Data format | NIfTI |
| Experimental factors | No special treatment was performed before the experiment. |
| Experimental features | Data were minimally preprocessed to correct for susceptibility and eddy current distortions. |
| Data source location | Boston, Massachusetts, USA |
| Data accessibility | Data is deposited in the XNAT Central database (https://central.xnat.org), Project ID: dMRI_Phant_MGH |
| Related research article | Fan, Q., Nummenmaa, A., Wichtmann, B., Witzel, T., Mekkaoui, C., Schneider, W., Wald, L.L., Huang, S.Y., Validation of diffusion MRI estimates of compartment size and volume fraction in a biomimetic brain phantom using a human MRI scanner with 300mT/m maximum gradient strength. Neuroimage. (2018). doi:10.1016/j.neuroimage.2018.01.004 |
Value of the data
-
•
The Connectome scanner at the MGH Martinos Imaging Center is equipped with up to 300 mT/m gradient strength for in vivo human imaging, which is the first of the three installed so far worldwide, and thus access to the system is extremely limited.
-
•
The Connectome system provides seven-fold higher gradient strength for diffusion encoding than state-of-art clinical scanners, which pushes upon the limits of diffusion resolution and provides unpreceded opportunity for microstructural imaging using diffusion MRI.
-
•
A biomimetic phantom mimicking human white matter was used to acquire the data, which to our best knowledge is the only existing prototype that is constructed with both intra- and extra-cellular compartments with known inner diameter, to provide a more realistic standard for diffusion MRI measurements of axon diameter and related microstructural metrics.
-
•
Our data was acquired with a comprehensive imaging protocol of about 40 hours of scanning time, which can be analyzed in multiple ways and allows great flexibility for other researchers to test their own algorithms.
-
•
Two datasets were provided, corresponding to Connectome and Prisma capabilities respectively, which can provide insight into the impact of gradient strength on the performance of diffusion MRI methods to be tested by other researchers.
1. Data
The motivation of this dataset was to validate estimates of compartment size and volume fraction using a biomimetic brain phantom constructed from hollow polypropylene yarns with known diameter. We acquired a comprehensive set of diffusion MRI measurements in the phantom using multiple gradient directions, diffusion times and gradient strengths on a human MRI scanner equipped with gradient strengths up to 300 mT/m.
2. Experimental design, materials, and methods
2.1. Phantom composition
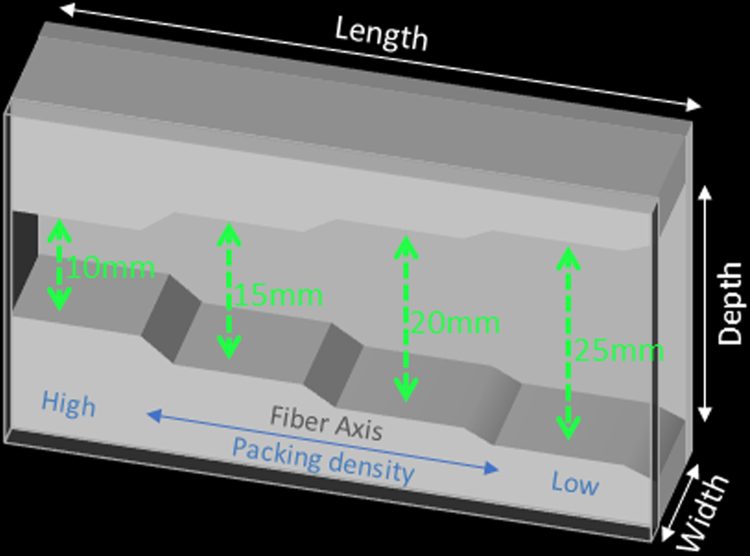
The fiber substrates in the diffusion phantom were constructed from hollow multifilament polypropylene yarns to generate textile axons (“taxons”) with an inner diameter of 11.8±1.2 µm and outer diameter of 33.5±2.3 µm [1] (Psychology Software Tools, Inc., Sharpsburg, Pennsylvania). The taxons were arranged in parallel within a 3D-printed taxon holder with rectangular chambers. Each of the chambers was built with fixed width and varying depths (i.e., 10 to 25 mm in Fig. 1) to achieve different fiber packing densities along the length of the fiber bundle. Fiber crossings of 90°, 45° and 30° were also created by interleaving the polypropylene filaments in separate 3D printed placeholders. See [2] for further information on phantom composition.
Fig. 1.
Illustration of the design of a representative taxon holder. The rectangular chamber has a fixed width but varying depth along the length of the chamber, such that the packing density decreases as the depth of the chamber increases.
2.2. MRI experiments
All scans were performed on the Siemens 3 T CONNECTOM MRI system with maximum gradient strength of 300 mT/m using a Siemens product 20-channel head-neck coil. A series of 2-mm isotropic resolution axial diffusion-weighted spin echo echo planar images (EPI) were acquired in the phantom using the following parameters: TR/TE = 4100/110 ms, gradient strength Gmax = 300 mT/m and 80 mT/m respectively for the Gmax=300 mT/m dataset and the Gmax=80 mT/m dataset, with a fixed diffusion gradient pulse duration δ = 8 ms. Diffusion times of Δ = 20, 30, 40, and 50 ms were sampled, which correspond to mean diffusion displacements of 9–15 μm, respectively, assuming an intrinsic diffusivity of 2.2×10-9 m2/s. Data were collected with 256 non-collinear diffusion-encoding gradient directions, which were evenly distributed on a sphere, with 20 interspersed b=0 images. For each diffusion time in the Gmax=300 mT/m dataset, the gradient strength linearly varied from 24 to 300 mT/m to produce b-values ranging between 50 to 18,250 s/mm2, and for the Gmax=80 mT/m dataset, the gradient strength ranges from 17 to 80 mT/m with a range of b-value of 50 to 1400 s/mm2 (Fig. 2, Table 1). Data were acquired in both anterior to posterior and posterior to anterior phase encoding directions to correct for image distortions due to susceptibility artifact. The total acquisition time was 38 h. Other imaging parameters included a field-of-view of 256×256 mm, partial Fourier of 6/8, and receive bandwidth of 1148 Hz/pixel. A regular non-accelerated EPI sequence was used to minimize parallel imaging artifact.
Fig. 2.
Exemplary diffusion weighted MR images. A representative axial slice through the center of the diffusion module of the phantom is shown. Diffusion-sensitizing gradients were applied perpendicular to the plane shown. The Gmax=300 mT/m dataset is shown in the upper panel and the Gmax=80 mT/m dataset in the lower panel.
Table 1.
Diffusion MR experiment parameters.
| Gmax = 300 mT/m |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffusion Time | Shell No. |
|||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||||||
| Δ = 20 ms | G = 25 mT/m | G = 62 mT/m | G = 101 mT/m | G = 140 mT/m | G = 176 mT/m | G = 215 mT/m | G = 253 mT/m | G = 291 mT/m | ||||||
| q = 0.009 μm-1 | q = 0.021 μm-1 | q = 0.034 μm-1 | q = 0.048 μm-1 | q = 0.060 μm-1 | q = 0.073 μm-1 | q = 0.086 μm-1 | q = 0.099 μm-1 | |||||||
| b = 50 s/mm2 | b = 300 s/mm2 | b = 800 s/mm2 | b = 1500 s/mm2 | b = 2450 s/mm2 | b = 3650 s/mm2 | b = 5050 s/mm2 | b = 6650 s/mm2 | |||||||
| Δ = 30 ms | G = 28 mT/m | G = 63 mT/m | G = 100 mT/m | G = 139 mT/m | G = 177 mT/m | G = 215 mT/m | G = 252 mT/m | G = 290 mT/m | ||||||
| q = 0.010 μm-1 | q = 0.022 μm-1 | q = 0.034 μm-1 | q = 0.047 μm-1 | q = 0.060 μm-1 | q = 0.073 μm-1 | q = 0.086 μm-1 | q = 0.099 μm-1 | |||||||
| b = 100 s/mm2 | b = 500 s/mm2 | b = 1250 s/mm2 | b = 2400 s/mm2 | b = 3900 s/mm2 | b = 5750 s/mm2 | b = 7950 s/mm2 | b = 10500 s/mm2 | |||||||
| Δ = 40 ms | G = 24 mT/m | G = 64 mT/m | G = 101 mT/m | G = 139 mT/m | G = 176 mT/m | G = 215 mT/m | G = 252 mT/m | G = 291 mT/m | ||||||
| q = 0.008 μm-1 | q = 0.022 μm-1 | q = 0.034 μm-1 | q = 0.047 μm-1 | q = 0.060 μm-1 | q = 0.073 μm-1 | q = 0.086 μm-1 | q = 0.099 μm-1 | |||||||
| b = 100 s/mm2 | b = 700 s/mm2 | b = 1750 s/mm2 | b = 3250 s/mm2 | b = 5300 s/mm2 | b = 7850 s/mm2 | b = 10850 s/mm2 | b = 14350 s/mm2 | |||||||
| Δ = 50 ms | G = 26 mT/m | G = 63 mT/m | G = 101 mT/m | G = 139 mT/m | G = 177 mT/m | G = 215 mT/m | G = 253 mT/m | G = 291 mT/m | ||||||
| q = 0.009 μm-1 | q = 0.021 μm-1 | q = 0.034 μm-1 | q = 0.047 μm-1 | q = 0.060 μm-1 | q = 0.073 μm-1 | q = 0.086 μm-1 | q = 0.099 μm-1 | |||||||
| b = 150 s/mm2 | b = 850 s/mm2 | b = 2200 s/mm2 | b = 4150 s/mm2 | b = 6750 s/mm2 | b = 9950 s/mm2 | b = 13800 s/mm2 | b = 18250 s/mm2 | |||||||
| Gmax = 80 mT/m | ||||||||||||||
| Diffusion Time | Shell No. |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||||||
| Δ = 20 ms | G = 25 mT/m | G = 36 mT/m | G = 50 mT/m | G = 62 mT/m | G = 71 mT/m | G = 80 mT/m | ||||||||
| q = 0.009 μm-1 | q = 0.012 μm-1 | q = 0.017 μm-1 | q = 0.021 μm-1 | q = 0.024 μm-1 | q = 0.027 μm-1 | |||||||||
| b = 50 s/mm2 | b = 100 s/mm2 | b = 200 s/mm2 | b = 300 s/mm2 | b = 400 s/mm2 | b = 500 s/mm2 | |||||||||
| Δ = 30 ms | G = 20 mT/m | G = 28 mT/m | G = 40 mT/m | G = 50 mT/m | G = 60 mT/m | G = 69 mT/m | G = 80 mT/m | |||||||
| q = 0.007 μm-1 | q = 0.010 μm-1 | q = 0.014 μm-1 | q = 0.017 μm-1 | q = 0.020 μm-1 | q = 0.024 μm-1 | q = 0.027 μm-1 | ||||||||
| b = 50 s/mm2 | b = 100 s/mm2 | b = 200 s/mm2 | b = 300 s/mm2 | b = 450 s/mm2 | b = 600 s/mm2 | b = 800 s/mm2 | ||||||||
| Δ = 40 ms | G = 17 mT/m | G = 30 mT/m | G = 38 mT/m | G = 51 mT/m | G = 59 mT/m | G = 70 mT/m | G = 80 mT/m | |||||||
| q = 0.006 μm-1 | q = 0.010 μm-1 | q = 0.013 μm-1 | q = 0.018 μm-1 | q = 0.020 μm-1 | q = 0.024 μm-1 | q = 0.027 μm-1 | ||||||||
| b = 50 s/mm2 | b = 150 s/mm2 | b = 250 s/mm2 | b = 450 s/mm2 | b = 600 s/mm2 | b = 850 s/mm2 | b = 1100 s/mm2 | ||||||||
| Δ = 50 ms | G = 22 mT/m | G = 30 mT/m | G = 40 mT/m | G = 50 mT/m | G = 60 mT/m | G = 70 mT/m | G = 81 mT/m | |||||||
| q = 0.007 μm-1 | q = 0.010 μm-1 | q = 0.014 μm-1 | q = 0.017 μm-1 | q = 0.020 μm-1 | q = 0.024 μm-1 | q = 0.027 μm-1 | ||||||||
| b = 100 s/mm2 | b = 200 s/mm2 | b = 350 s/mm2 | b = 550 s/mm2 | b = 800 s/mm2 | b = 1050 s/mm2 | b = 1400 s/mm2 | ||||||||
The shared data has been minimally preprocessed to correct for distortions due to gradient nonlinearity, susceptibility effects, B0 drift, and eddy currents [3], [4], [5]. Due to the length of the phantom scan (about a day), the static magnetic field (B0) drifts due to hardware heating during the scan resulted in small image displacements along the phase encoding direction, which mimicked head motion [6]. To address this issue, the first b=0 image of all individual shells (i.e., for each q-value) were concatenated and used for a joint field map estimate using TOPUP [7], [8], where image displacements due to B0 drift were modeled in the same way as head motion. Eddy current correction was performed using the EDDY [3] tool in FSL.
Acknowledgments
This work was funded by an NIH Blueprint for Neuroscience Research Grant: U01MH093765, as well as NIH funding from NCRR P41EB015896, NIBIB R01EB006847, NIBIB R00EB015445, NINDS K23NS096056, NHLBI R01HL131635, NHLBI R56HL125590, and Instrumentation Grants S10-RR023401, S10-RR023043, and S10-RR019307. Funding support was also received from Chronic Effects of Neurotrauma Consortium/Veterans Affairs Rehabilitation Research & Development project F1880, US Army 12342013 (W81XWH-12-2-0139), the American Heart Association Postdoctoral Fellowship Award (17POST33670452), a Radiological Sciences of North America Research Resident Grant number RR1427 and the MGH Executive Committee on Research Fund for Medical Discovery Fellowship Award.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2018.03.021.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Guise C., Fernandes M.M., Nóbrega J.M., Pathak S., Schneider W., Fangueiro R. Hollow polypropylene yarns as a biomimetic brain phantom for the validation of high-definition fiber tractography imaging. ACS Appl. Mater. Interfaces. 2016;8:29960–29967. doi: 10.1021/acsami.6b09809. [DOI] [PubMed] [Google Scholar]
- 2.Fan Q., Nummenmaa A., Wichtmann B., Witzel T., Mekkaoui C., Schneider W., Wald L.L., Huang S.Y. Validation of diffusion MRI estimates of compartment size and volume fraction in a biomimetic brain phantom using a human MRI scanner with 300 mT/m maximum gradient strength. Neuroimage. 2018 doi: 10.1016/j.neuroimage.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson J.L.R., Sotiropoulos S.N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkinson M., Beckmann C.F., Behrens T.E.J., Woolrich M.W., Smith S.M. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Fan Q., Witzel T., Nummenmaa A., Van Dijk K.R.A., Van Horn J.D., Drews M.K., Somerville L.H., Sheridan M.A., Santillana R.M., Snyder J., Hedden T., Shaw E.E., Hollinshead M.O., Renvall V., Zanzonico R., Keil B., Cauley S., Polimeni J.R., Tisdall D., Buckner R.L., Wedeen V.J., Wald L.L., Toga A.W., Rosen B.R. MGH–USC Human Connectome Project datasets with ultra-high b-value diffusion MRI. Neuroimage. 2016;124(Part B):1108–1114. doi: 10.1016/j.neuroimage.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haacke E.M., Thompson M.R., Venkatesan R., Brown R.W. J. Wiley & Sons; New York: 1999. Magnetic Resonance Imaging: Physical Principles and Sequence Design; p. 914. [Google Scholar]
- 7.Andersson J.L.R., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 8.Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E.J., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material