Abstract
This article contains the peptide data obtained while performing disulfide bond mapping of the recombinant Plasmodium falciparum protein, Pfs25, produced from the baculovirus expression system. Pfs25 is a malaria transmission-blocking vaccine candidate, with a compact and complex structure including 22 cysteines. This supplementary data is related to the research “Disulfide bond mapping of Pfs25, a recombinant malaria transmission blocking vaccine candidate” (Lee et al., 2018) [1]. In brief, Pfs25 was digested with trypsin/Lys-C and derived peptides separated by High Performance Liquid Chromatography (HPLC) and analyzed by mass spectrometry (MS) by MSE fragmentation. The theoretical peptides and their respective masses along with disulfide bond locations with linked peptides are presented here alongside the mass spectrometry analysis. The raw mass spectrometry data is made available through the Mass Spectrometry Interactive Virtual Environment (MassIVE) with identifier: MSV000081982.
Keywords: Pfs25, Disulfide, Mass spectrometry, Malaria, LC-MSMS
Specifications Table
| Subject area | Chemistry, Biology, |
| More specific subject area | Disulfide bond analysis by liquid chromatography and mass spectrometric analysis |
| Type of data | Tables, figures |
| How data was acquired | Data was generated using liquid chromatography (Waters 2695 Separations Module and Waters 2489 UV/Vis Detector) and mass spectroscopy (Waters QTOF Premier mass spectrometer) |
| Data format | Collated data from analysis with Waters BiopharmaLynx 1.3 and MassLynx |
| Experimental factors | Recombinant Pfs25 digested with 20 µg of trypsin/Lys-C at 37 °C overnight and subsequent further digestion by additional 20 µg of trypsin/Lys-C for 3–4 hours at 37 °C |
| Experimental features | Identification of the proper pairing of 11 disulfide bonds in Pfs25 through digestion of peptides and LC-MS/MS |
| Data source location | Mass spectrometry data acquired in Middleton, WI, USA |
| Data accessibility | Data is provided within this article and RAW MS files have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) with identifier: MSV000081982 (ftp://massive.ucsd.edu/MSV000081982). MassIVE is a member of the ProteomeXchange Consortium |
Value of the data
-
•
The derived peptides and mass spectrometry data is provided here for further details from the disulfide bond analysis of Pfs25.
-
•
The disulfide bond locations and linked peptides are discussed alongside the mass spectrometry analysis here and data made accessible to the scientific community.
-
•
Pfs25 is a compact and complex 17.9 kDa protein with 22 cysteines (11 disulfide bonds) that has presented difficulty in prior disulfide bond analysis
-
•
A method was developed to map the disulfide bonds of a complex and compact protein, which may be applicable to other proteins, an important step in recombinant protein development for vaccines.
1. Data
The Pfs25 disulfide bond mapping peptides are discussed in further detail in this manuscript to further support the elucidation of disulfide bonds of Pfs25 as discussed in [1]. Further, the mass spectrometry RAW files have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE). Theoretical peptides, produced from Trypsin/Lys-C digestion of Pfs25, are presented in Table 1.
Table 1.
Theoretical Fragments for Trypsin/Lys-C Digestion of Pfs25.
| Peptides | Position | Peptide Label | Theoretical Mass (Da) |
|---|---|---|---|
| DAK | 1–3 | T1 | 332.17 |
| VTVDTVCK | 4–11 | T2 | 863.44 |
| R | 12-12 | T3 | 174.11 |
| GFLIQMSGHLECK | 13–25 | T4 | 1461.71 |
| CENDLVLVNEETCEEK | 26–41 | T5 | 1865.80 |
| VLK | 42–44 | T6 | 358.26 |
| CDEK | 45–48 | T7 | 493.18 |
| TVNKPCGDFSK | 49–59 | T8 | 1194.57 |
| CIK | 60–62 | T9 | 362.20 |
| IDGNPVSYACK | 63–73 | T10 | 1165.54 |
| CNLGYDMVNNVCIPNECK | 74–91 | T11 | 2027.86 |
| QVTCGNGK | 92–99 | T12 | 805.38 |
| CILDTSNPVK | 100–109 | T13 | 1088.55 |
| TGVCSCNIGK | 110–119 | T14 | 980.44 |
| VPNVQDQNK | 120–128 | T15 | 1040.53 |
| CSK | 129–131 | T16 | 336.15 |
| DGETK | 132–136 | T17 | 548.24 |
| CSLK | 137–140 | T18 | 449.23 |
| CLK | 141–143 | T19 | 362.20 |
| ENETCK | 144–149 | T20 | 722.29 |
| AVDGIYK | 150–156 | T21 | 764.41 |
| CDCK | 157–160 | T22 | 467.15 |
| DGFIIDQESSICTHHHHHH | 161–179 | T23 | 2248.98 |
Utilizing Biopharmalynx 1.3 the mass spectral data was analyzed and compared to the theoretical peptides to obtain the localization of the 11 disulfide bonds present in the recombinant Pfs25. The disulfide bond locations and linked peptides (including theoretical and observed) masses are presented in Table 2. Each disulfide bond (referenced by nomenclature SS#) is further presented with the peptide information and mass spectrometry (MS) and MSMS data obtained during the analysis in the subsequent figures and tables presented in this manuscript.
Table 2.
Disulfide bond locations for Pfs25 including theoretical and observed masses of fragments.
| Disulfide Bond | Linked Cysteines | Tryptic Peptide Label | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (ppm) |
|---|---|---|---|---|---|
| SS1 | Cys10-Cys24 | T2+T4 | 2323.1375 | 2323.1152 | 9.6 |
| SS2 | Cys26-Cys38 | T5 | 1863.7866 | 1863.7752 | 6.1 |
| SS3 | Cys45-Cys60 | T7+T9 | 853.3674 | 853.3656 | 2.1 |
| SS4 | Cys54-Cys72 | T8+T10 | 2358.0670 | 2358.0799 | 5.5 |
| SS5 | Cys74-Cys85 | T11+T13 | 3112.3796 | 3112.3534 | 8.4 |
| SS6 | Cys90-Cys100 | ||||
| SS7 | Cys95-Cys113 | T12+T14+T16 | 2117.9326 | 2117.9182 | 6.8 |
| SS8 | Cys115-Cys129 | ||||
| SS9 | Cys137-Cys148 | T18+T20 | 1169.5056 | 1169.4970 | 7.3 |
| SS10 | Cys141-Cys157 | T19+T22+T23 | 3074.3003 | 3074.2795 | 6.8 |
| SS11 | Cys159-Cys172 |
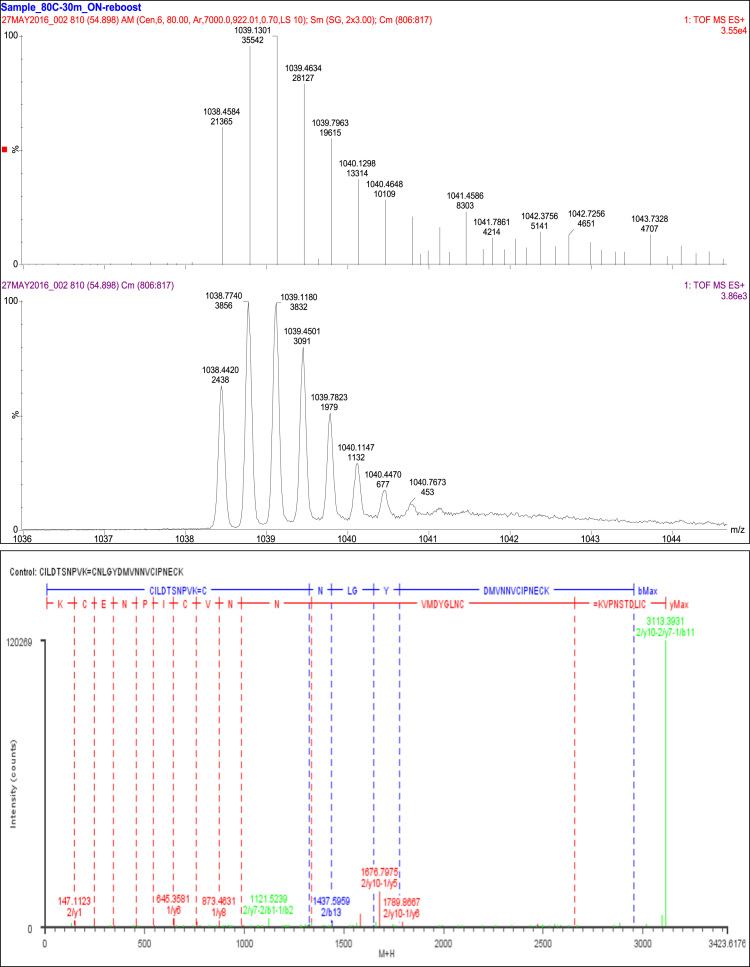
2. Disulfide bond SS1
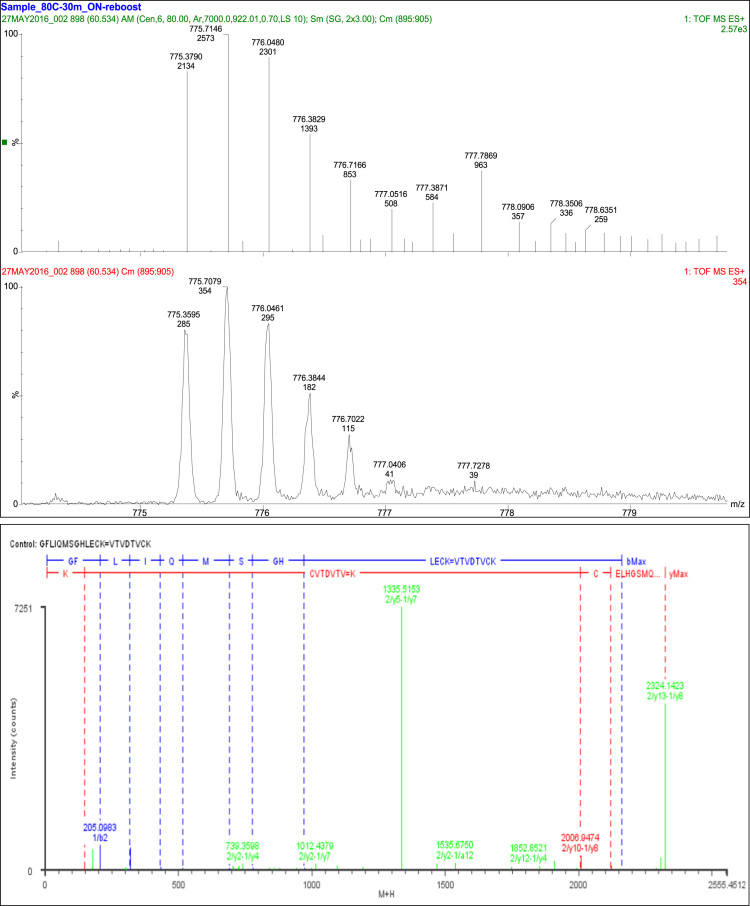
A total of 30 fragments were observed, with 20 fragment ions of this peptide consistent with the linkage of Cys10 and Cys24. The remaining ten fragment ions were consistent with constituent peptides (Table 3, Fig. 1).
Table 3.
Disulfide bond SS1 (Cys10-Cys24) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 30 Fragments | Constituent Peptides 10 Fragments | 1/a2 | 173.129 | 173.0568 | 0.0722 | 38 | VT |
| 1/b5 | 516.267 | 516.2606 | 0.0063 | 69 | VTVDT | ||
| 1/y1 | 147.1133 | 147.0415 | 0.0718 | 19 | K | ||
| 2/a2 | 177.1028 | 177.1026 | 0.0002 | 567 | GF | ||
| 2/b2 | 205.0977 | 205.0983 | −0.0006 | 691 | GF | ||
| 2/b3 | 318.1818 | 318.1831 | −0.0013 | 634 | GFL | ||
| 2/b4 | 431.2658 | 431.1857 | 0.0801 | 82 | GFLI | ||
| 2/b6 | 690.3649 | 690.3612 | 0.0037 | 40 | GFLIQM | ||
| 2/b7 | 777.3969 | 777.4837 | −0.0868 | 34 | GFLIQMS | ||
| 2/b9 | 971.4773 | 971.4425 | 0.0348 | 52 | GFLIQMSGH | ||
| Cys10 and Cys24 20 Fragments | 1/b7-2/y3 | 1094.486 | 1094.477 | 0.0095 | 103 | VTVDTVC=ECK | |
| 1/b7-2/y9 | 1747.782 | 1747.827 | −0.0454 | 86 | VTVDTVC=QMSGHLECK | ||
| 1/y2-2/a12 | 1535.717 | 1535.675 | 0.0422 | 196 | CK=GFLIQMSGHLEC | ||
| 1/y2-2/y4 | 739.3483 | 739.3598 | −0.0115 | 163 | CK=LECK | ||
| 1/y2-2/y5 | 876.4072 | 876.3971 | 0.0101 | 68 | CK=HLECK | ||
| 1/y2-2/z4 | 722.3217 | 722.2222 | 0.0995 | 26 | CK=LECK | ||
| 1/y3-2/y3 | 725.3326 | 725.3351 | −0.0025 | 101 | VCK=ECK | ||
| 1/y3-2/y4 | 838.4167 | 838.3221 | 0.0946 | 56 | VCK=LECK | ||
| 1/y4-2/y12 | 1852.912 | 1852.852 | 0.0603 | 119 | TVCK=FLIQMSGHLECK | ||
| 1/y5-2/y3 | 941.4072 | 941.3948 | 0.0125 | 50 | DTVCK=ECK | ||
| 1/y5-2/y5 | 1191.55 | 1191.505 | 0.0454 | 74 | DTVCK=HLECK | ||
| 1/y5-2/y7 | 1335.604 | 1335.515 | 0.0884 | 7251 | DTVCK=SGHLECK | ||
| 1/y5-2/y8 | 1466.644 | 1466.662 | −0.0175 | 177 | DTVCK=MSGHLECK | ||
| 1/y7-2/y10 | 1907.903 | 1907.823 | 0.0797 | 268 | TVDTVCK=IQMSGHLECK | ||
| 1/y7-2/y2 | 1012.481 | 1012.438 | 0.0428 | 181 | TVDTVCK=CK | ||
| 1/y8-2/y10 | 2006.971 | 2006.947 | 0.0239 | 390 | VTVDTVCK=IQMSGHLECK | ||
| 1/y8-2/y11 | 2120.055 | 2120.046 | 0.0098 | 112 | VTVDTVCK=LIQMSGHLECK | ||
| 1/y8-2/y13 | 2324.145 | 2324.142 | 0.0029 | 4580 | VTVDTVCK=GFLIQMSGHLECK | ||
| 1/y8-2/z10 | 1989.945 | 1989.963 | −0.0178 | 63 | VTVDTVCK=IQMSGHLECK | ||
| 1/z7-2/y2 | 995.4542 | 995.5009 | −0.0468 | 47 | TVDTVCK=CK | ||
Fig. 1.
Disulfide bond SS1.
3. Disulfide bond SS2
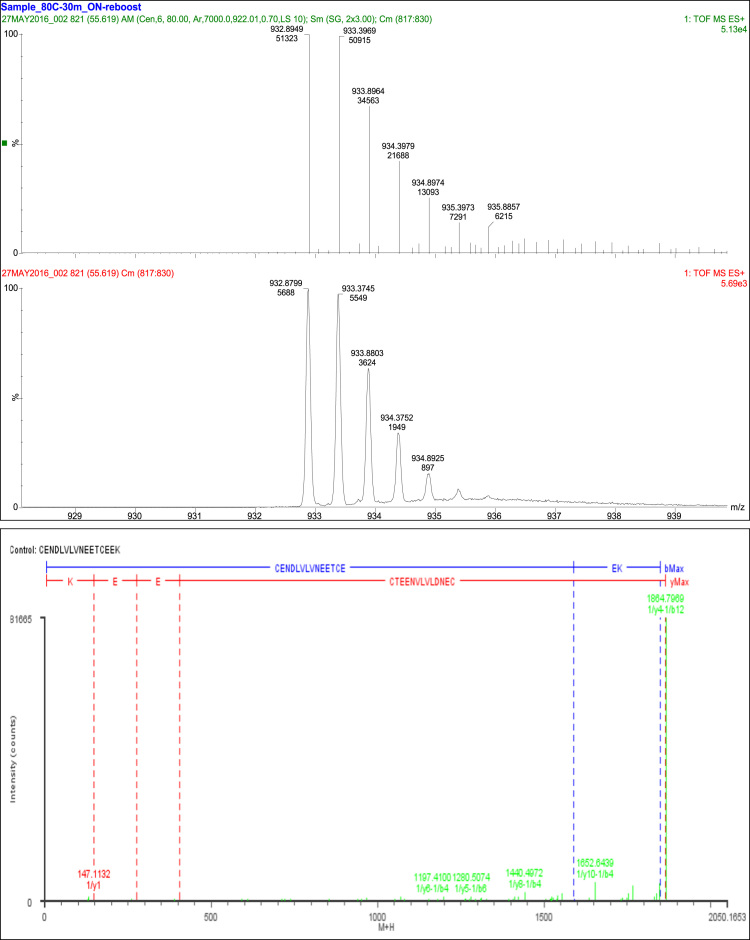
A total of 39 fragment ions of this peptide were observed with 36 fragment ions consistent with the linkage of Cys26 and Cys38. Three additional fragments were consistent with constituent peptides (Table 4, Fig. 2).
Table 4.
Disulfide bond SS2 (Cys26-Cys38) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 39 Fragments | Const. Pep.3 Fragments | 1/y1 | 147.1133 | 147.1132 | 0.0001 | 2129 | K |
| 1/y2 | 276.1559 | 276.159 | −0.0031 | 1082 | EK | ||
| 1/y3 | 405.1985 | 405.1972 | 0.0013 | 825 | EEK | ||
| Cys26 and Cys38 36 Fragments | 1/b14 | 1589.646 | 1589.614 | 0.0321 | 335 | CENDLVLVNEETCE (Internal) | |
| 1/y10-1/a3 | 1509.62 | 1509.584 | 0.0366 | 87 | LVNEETCEEK=CEN | ||
| 1/y10-1/b3 | 1537.615 | 1537.644 | −0.0291 | 246 | LVNEETCEEK=CEN | ||
| 1/y10-1/b4 | 1652.642 | 1652.644 | −0.002 | 5544 | LVNEETCEEK=CEND | ||
| 1/y10-1/b5 | 1765.726 | 1765.722 | 0.0043 | 4497 | LVNEETCEEK=CENDL | ||
| 1/y11-1/b4 | 1751.71 | 1751.709 | 0.0017 | 2109 | VLVNEETCEEK=CEND | ||
| 1/y12-1/b1 | 1506.682 | 1506.587 | 0.0948 | 66 | LVLVNEETCEEK=C | ||
| 1/y12-1/b2 | 1635.725 | 1635.631 | 0.0941 | 586 | LVLVNEETCEEK=CE | ||
| 1/y4-1/a12 | 1836.8 | 1836.787 | 0.0127 | 2176 | CEEK=CENDLVLVNEET | ||
| 1/y4-1/a2 | 710.249 | 710.2508 | −0.0018 | 210 | CEEK=CE | ||
| 1/y4-1/a6 | 1151.471 | 1151.5 | −0.0284 | 109 | CEEK=CENDLV | ||
| 1/y4-1/a7 | 1264.555 | 1264.47 | 0.0858 | 499 | CEEK=CENDLVL | ||
| 1/y4-1/b1 | 609.2012 | 609.2489 | −0.0477 | 393 | CEEK=C | ||
| 1/y4-1/b10 | 1634.704 | 1634.629 | 0.0752 | 999 | CEEK=CENDLVLVNE | ||
| 1/y4-1/b12 | 1864.794 | 1864.797 | −0.0024 | 81665 | CEEK=CENDLVLVNEET | ||
| 1/y4-1/b2 | 738.2438 | 738.2711 | −0.0273 | 341 | CEEK=CE | ||
| 1/y4-1/b3 | 852.2868 | 852.3066 | −0.0198 | 172 | CEEK=CEN | ||
| 1/y4-1/b4 | 967.3137 | 967.3153 | −0.0016 | 992 | CEEK=CEND | ||
| 1/y4-1/b5 | 1080.398 | 1080.422 | −0.024 | 317 | CEEK=CENDL | ||
| 1/y4-1/b6 | 1179.466 | 1179.442 | 0.024 | 380 | CEEK=CENDLV | ||
| 1/y4-1/b7 | 1292.55 | 1292.497 | 0.053 | 258 | CEEK=CENDLVL | ||
| 1/y4-1/b8 | 1391.619 | 1391.581 | 0.0372 | 224 | CEEK=CENDLVLV | ||
| 1/y5-1/a5 | 1153.451 | 1153.433 | 0.0179 | 274 | TCEEK=CENDL | ||
| 1/y5-1/b4 | 1068.362 | 1068.366 | −0.004 | 1172 | TCEEK=CEND | ||
| 1/y5-1/b6 | 1280.514 | 1280.507 | 0.0065 | 1279 | TCEEK=CENDLV | ||
| 1/y5-1/b8 | 1492.666 | 1492.607 | 0.059 | 72 | TCEEK=CENDLVLV | ||
| 1/y6-1/a6 | 1381.562 | 1381.568 | −0.0067 | 109 | ETCEEK=CENDLV | ||
| 1/y6-1/b4 | 1197.404 | 1197.41 | −0.0061 | 1266 | ETCEEK=CEND | ||
| 1/y6-1/b5 | 1310.488 | 1310.488 | 0.0001 | 885 | ETCEEK=CENDL | ||
| 1/y6-1/b6 | 1409.557 | 1409.556 | 0.0009 | 1232 | ETCEEK=CENDLV | ||
| 1/y7-1/a1 | 940.3392 | 940.3529 | −0.0137 | 271 | EETCEEK=C | ||
| 1/y7-1/b4 | 1326.447 | 1326.445 | 0.0021 | 764 | EETCEEK=CEND | ||
| 1/y7-1/b6 | 1538.599 | 1538.576 | 0.0226 | 1457 | EETCEEK=CENDLV | ||
| 1/y8-1/a5 | 1525.579 | 1525.593 | −0.014 | 1104 | NEETCEEK=CENDL | ||
| 1/y8-1/b4 | 1440.49 | 1440.497 | −0.0077 | 2676 | NEETCEEK=CEND | ||
| 1/y8-1/b5 | 1553.574 | 1553.575 | −0.0016 | 2348 | NEETCEEK=CENDL | ||
Fig. 2.
Disulfide bond SS2.
4. Disulfide bond SS3
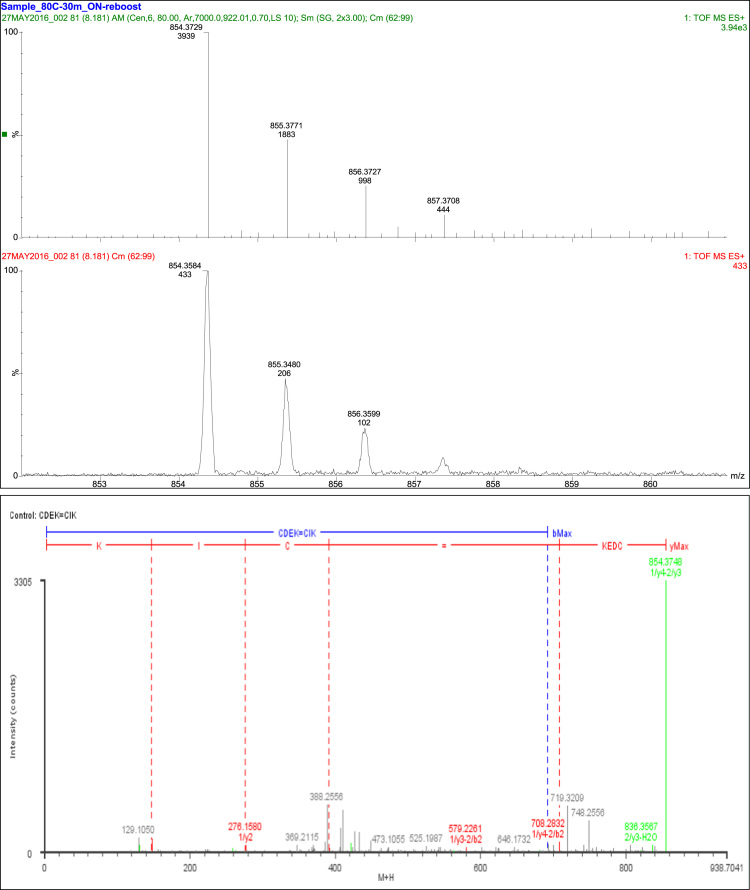
A total of nine fragments were observed, with six fragment ions of this peptide consistent with the linkage of Cys45 and Cys60. Three fragment ions were consistent with constituent peptides (Table 5, Fig. 3).
Table 5.
Disulfide bond SS3 (Cys45-Cys60) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 9 Fragments | Const. Pep.3 Fragments | 1/y1 | 147.1133 | 147.115 | −0.0017 | 163 | K |
| 1/y2 | 276.1559 | 276.158 | −0.002 | 84 | EK | ||
| 1/y3 | 391.1829 | 391.2075 | −0.0246 | 47 | DEK | ||
| Cys45 and Cys60 6 Fragments | 1/a1–2/a2 | 262.1048 | 262.0183 | 0.0865 | 22 | C=CI | |
| 1/b1–2/a3 | 421.0852 | 421.1291 | −0.0439 | 117 | C=CIK | ||
| 1/b2-2/y3 | 579.2271 | 579.2261 | 0.001 | 67 | CD=CIK | ||
| 1/y4-2/a2 | 680.2748 | 680.2768 | −0.002 | 23 | CDEK=CI | ||
| 1/y4-2/b2 | 708.2697 | 708.2832 | −0.0135 | 126 | CDEK=CI | ||
| 1/y4-2/y3 | 854.3752 | 854.3748 | 0.0004 | 3305 | CDEK=CIK | ||
Fig. 3.
Disulfide bond SS3.
5. Disulfide bond SS4
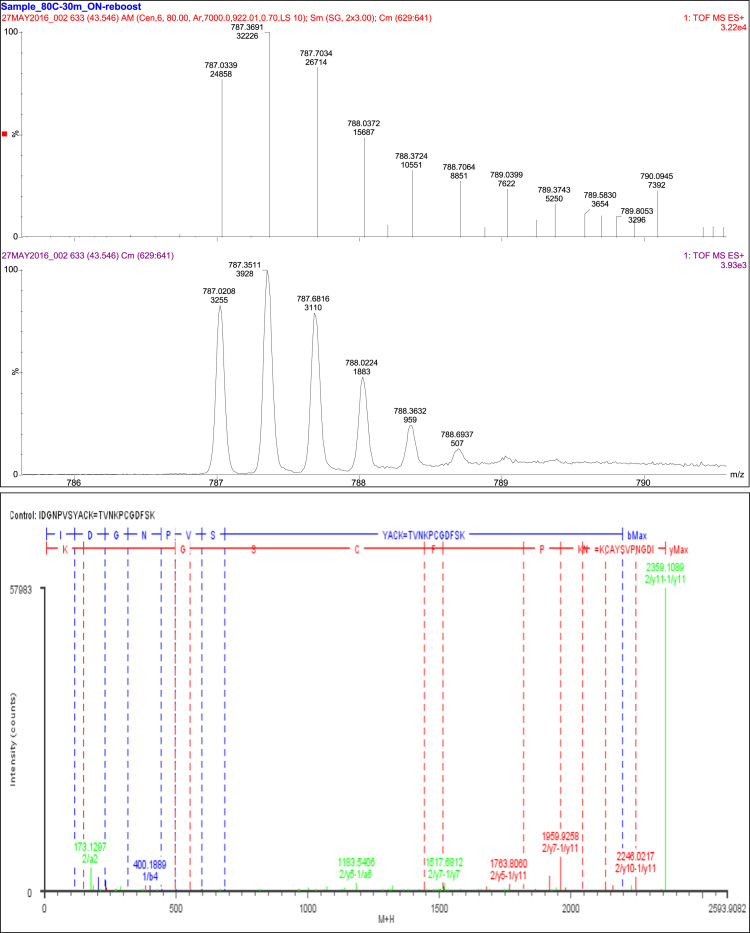
A total of 68 fragments were observed, with 45 fragment ions of this peptide consistent with the linkage of T8 to T10 through Cys54 and Cys72. The remaining 23 fragments were consistent with constituent peptides (Table 6, Fig. 4).
Table 6.
Disulfide bond SS4 (Cys54-Cys72) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 68 Fragments | Constituent Peptides 23 Fragments | 1/a2 | 173.129 | 173.1297 | −0.0007 | 4507 | TV |
| 1/a3 | 287.1719 | 287.0969 | 0.075 | 897 | TVN | ||
| 1/a4 | 415.2669 | 415.2224 | 0.0445 | 28 | TVNK | ||
| 1/b2 | 201.1239 | 201.1235 | 0.0004 | 2754 | TV | ||
| 1/b3 | 315.1668 | 315.0896 | 0.0772 | 51 | TVN | ||
| 1/b4 | 443.2618 | 443.2626 | −0.0008 | 248 | TVNK | ||
| 1/y1 | 147.1133 | 147.0623 | 0.051 | 16 | K | ||
| 1/y2 | 234.1454 | 234.1455 | −0.0001 | 700 | SK | ||
| 1/y3 | 381.2138 | 381.2014 | 0.0124 | 1146 | FSK | ||
| 1/y4 | 496.2407 | 496.2404 | 0.0003 | 153 | DFSK | ||
| 1/y5 | 553.2622 | 553.2604 | 0.0018 | 564 | GDFSK | ||
| 2/a3 | 258.1454 | 258.1545 | −0.0092 | 78 | IDG | ||
| 2/a4 | 372.1883 | 372.1927 | −0.0044 | 300 | IDGN | ||
| 2/a5 | 469.2411 | 469.2278 | 0.0132 | 76 | IDGNP | ||
| 2/a6 | 568.3094 | 568.3065 | 0.0029 | 91 | IDGNPV | ||
| 2/a8 | 818.4048 | 818.3653 | 0.0396 | 129 | IDGNPVSY | ||
| 2/b1 | 114.0919 | 114.041 | 0.0509 | 18 | I | ||
| 2/b2 | 229.1188 | 229.119 | −0.0002 | 1962 | ID | ||
| 2/b3 | 286.1403 | 286.1415 | −0.0012 | 727 | IDG | ||
| 2/b4 | 400.1832 | 400.1889 | −0.0057 | 1203 | IDGN | ||
| 2/b5 | 497.236 | 497.2373 | −0.0013 | 252 | IDGNP | ||
| 2/b6 | 596.3044 | 596.3082 | −0.0038 | 205 | IDGNPV | ||
| 2/b7 | 683.3364 | 683.3206 | 0.0158 | 211 | IDGNPVS | ||
| Cys54 and Cys72 45 Fragments | 1/a6-2/y5 | 1183.5604 | 1183.541 | 0.0198 | 1608 | TVNKPC=SYACK | |
| 1/a6-2/y7 | 1379.6815 | 1379.604 | 0.0773 | 143 | TVNKPC=PVSYACK | ||
| 1/a8-2/y7 | 1551.73 | 1551.666 | 0.0636 | 108 | TVNKPCGD=PVSYACK | ||
| 1/a9-2/b10 | 1951.8682 | 1951.823 | 0.0454 | 93 | TVNKPCGDF=IDGNPVSYAC | ||
| 1/b10-2/a10 | 2038.9003 | 2038.99 | −0.09 | 43 | TVNKPCGDFS=IDGNPVSYAC | ||
| 1/b6-2/y3 | 961.4599 | 961.4065 | 0.0534 | 124 | TVNKPC=ACK | ||
| 1/b6-2/y8 | 1521.7194 | 1521.674 | 0.0449 | 161 | TVNKPC=NPVSYACK | ||
| 1/b7-2/y7 | 1464.6979 | 1464.675 | 0.0228 | 47 | TVNKPCG=PVSYACK | ||
| 1/b8-2/y10 | 1865.8162 | 1865.827 | −0.0106 | 68 | TVNKPCGD=DGNPVSYACK | ||
| 1/b8-2/y11 | 1978.9003 | 1978.891 | 0.0092 | 579 | TVNKPCGD= IDGNPVSYACK | ||
| 1/b8-2/y2 | 1062.4712 | 1062.441 | 0.0303 | 364 | TVNKPCGD=CK | ||
| 1/b8-2/y6 | 1482.6721 | 1482.651 | 0.021 | 137 | TVNKPCGD=VSYACK | ||
| 1/b8-2/y7 | 1579.7249 | 1579.69 | 0.0347 | 456 | TVNKPCGD=PVSYACK | ||
| 1/b9-2/y5 | 1530.6721 | 1530.667 | 0.0054 | 180 | TVNKPCGDF=SYACK | ||
| 1/b9-2/y7 | 1726.7932 | 1726.741 | 0.0527 | 66 | TVNKPCGDF=PVSYACK | ||
| 1/y11-2/y10 | 2246.0222 | 2246.022 | 0.0005 | 2740 | TVNKPCGDFSK=DGNPVSYACK | ||
| 1/y11-2/y11 | 2359.1062 | 2359.109 | −0.0027 | 57983 | TVNKPCGDFSK= IDGNPVSYACK | ||
| 1/y11-2/y2 | 1442.6771 | 1442.674 | 0.0034 | 677 | TVNKPCGDFSK=CK | ||
| 1/y11-2/y3 | 1513.7142 | 1513.711 | 0.0028 | 849 | TVNKPCGDFSK=ACK | ||
| 1/y11-2/y4 | 1676.7776 | 1676.777 | 0.0009 | 999 | TVNKPCGDFSK=YACK | ||
| 1/y11-2/y5 | 1763.8097 | 1763.806 | 0.0037 | 1320 | TVNKPCGDFSK=SYACK | ||
| 1/y11-2/y6 | 1862.8781 | 1862.893 | −0.0144 | 201 | TVNKPCGDFSK=VSYACK | ||
| 1/y11-2/y7 | 1959.9308 | 1959.926 | 0.005 | 6462 | TVNKPCGDFSK=PVSYACK | ||
| 1/y11-2/y9 | 2130.9951 | 2131.013 | −0.0173 | 418 | TVNKPCGDFSK=GNPVSYACK | ||
| 1/y11-2/z10 | 2228.9956 | 2228.999 | −0.0037 | 1038 | TVNKPCGDFSK=DGNPVSYACK | ||
| 1/y6-2/a10 | 1645.699 | 1645.775 | −0.0762 | 368 | CGDFSK=IDGNPVSYAC | ||
| 1/y6-2/y11 | 1819.7994 | 1819.795 | 0.0044 | 57 | CGDFSK= IDGNPVSYACK | ||
| 1/y6-2/y4 | 1137.4708 | 1137.472 | −0.0007 | 738 | CGDFSK=YACK | ||
| 1/y6-2/y7 | 1420.624 | 1420.613 | 0.0114 | 404 | CGDFSK=PVSYACK | ||
| 1/y7-2/b10 | 1770.7467 | 1770.722 | 0.0243 | 43 | PCGDFSK= IDGNPVSYAC | ||
| 1/y7-2/y11 | 1916.8523 | 1916.848 | 0.0039 | 3031 | PCGDFSK= IDGNPVSYACK | ||
| 1/y7-2/y2 | 1000.4232 | 1000.424 | −0.0004 | 627 | PCGDFSK=CK | ||
| 1/y7-2/y3 | 1071.4603 | 1071.461 | −0.0007 | 825 | PCGDFSK=ACK | ||
| 1/y7-2/y5 | 1321.5557 | 1321.558 | −0.0024 | 1193 | PCGDFSK=SYACK | ||
| 1/y7-2/y7 | 1517.6769 | 1517.681 | −0.0043 | 1667 | PCGDFSK=PVSYACK | ||
| 1/y7-2/y9 | 1688.7412 | 1688.747 | −0.0057 | 547 | PCGDFSK=GNPVSYACK | ||
| 1/y8-2/y11 | 2044.9471 | 2044.929 | 0.0182 | 64 | KPCGDFSK= IDGNPVSYACK | ||
| 1/y8-2/y2 | 1128.5182 | 1128.502 | 0.0162 | 491 | KPCGDFSK=CK | ||
| 1/y8-2/y4 | 1362.6187 | 1362.648 | −0.0288 | 120 | KPCGDFSK=YACK | ||
| 1/y8-2/y5 | 1449.6506 | 1449.647 | 0.0038 | 253 | KPCGDFSK=SYACK | ||
| 1/y8-2/y8 | 1759.8147 | 1759.8 | 0.0151 | 186 | KPCGDFSK=NPVSYACK | ||
| 1/y9-2/y11 | 2158.9902 | 2158.979 | 0.0112 | 1071 | NKPCGDFSK=IDGNPVSYACK | ||
| 1/y9-2/y3 | 1313.5981 | 1313.58 | 0.0184 | 279 | NKPCGDFSK=ACK | ||
| 1/y9-2/y4 | 1476.6615 | 1476.644 | 0.0173 | 144 | NKPCGDFSK=YACK | ||
| 1/z7-2/y2 | 983.3967 | 983.4453 | −0.0486 | 113 | PCGDFSK=CK | ||
Fig. 4.
Disulfide bond SS4.
6. Disulfide bonds SS5 and SS6
A total of 90 fragments were observed and four fragment ions (1/b12, 1/b13, 1/b15, and 1/b16) were consistent with an internal disulfide bond linkage between Cys74 and Cys85. Thirty-three fragment ions were consistent with the linkage of Cys90 and Cys100. An additional 44 fragments of this peptide were consistent with the combined linkages of Cys74 to Cys85 and Cys90 to Cys100 and nine fragments consistent with constituent peptides (Table 7, Fig. 5).
Table 7.
Disulfide bond SS5 (Cys74-Cys85) and SS6 (Cys90-Cys100) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 90 Fragments | Cys74 and Cys85 4 Fragments | 1/b12 | 1324.513 | 1324.516 | −0.0031 | 1267 | CNLGYDMVNNVC (Internal) |
| 1/b13 | 1437.597 | 1437.596 | 0.0006 | 3004 | CNLGYDMVNNVCI (Internal) | ||
| 1/b15 | 1648.692 | 1648.71 | −0.0175 | 311 | CNLGYDMVNNVCIPN (Internal) | ||
| 1/b16 | 1777.735 | 1777.728 | 0.007 | 105 | CNLGYDMVNNVCIPNE (Internal) | ||
| Constituent Peptides 9 Fragments | 1/y1 | 147.1133 | 147.1123 | 0.001 | 2902 | K | |
| 2/y2 | 246.1818 | 246.1842 | −0.0024 | 789 | VK | ||
| 2/y3 | 343.2345 | 343.1509 | 0.0836 | 258 | PVK | ||
| 2/y4 | 457.2774 | 457.2771 | 0.0003 | 855 | NPVK | ||
| 2/y5 | 544.3095 | 544.3134 | −0.0039 | 711 | SNPVK | ||
| 2/y6 | 645.3572 | 645.3581 | −0.0009 | 3696 | TSNPVK | ||
| 2/y7 | 760.3841 | 760.3795 | 0.0046 | 2330 | DTSNPVK | ||
| 2/y8 | 873.4681 | 873.4631 | 0.0051 | 2651 | LDTSNPVK | ||
| 2/y9 | 986.5522 | 986.5494 | 0.0029 | 381 | ILDTSNPVK | ||
| Cys90-Cys100 33 fragments | 1/a17-2/a7 | 2569.065 | 2569.078 | −0.0127 | 433 | CNLGYDMVNNVCIPNEC=CILDTSN | |
| 1/y18-2/b2 | 2240.927 | 2240.983 | −0.0564 | 114 | CNLGYDMVNNVCIPNECK=CI | ||
| 1/y18-2/b4 | 2469.038 | 2469.068 | −0.03 | 1446 | CNLGYDMVNNVCIPNECK=CILD | ||
| 1/y18-2/b6 | 2657.118 | 2657.106 | 0.012 | 90 | CNLGYDMVNNVCIPNECK=CILDTS | ||
| 1/y2-2/a8 | 1063.492 | 1063.406 | 0.0859 | 167 | CK=CILDTSNP | ||
| 1/y2-2/b1 | 351.1161 | 351.204 | −0.0879 | 78 | CK=C | ||
| 1/y2-2/b2 | 464.2001 | 464.2095 | −0.0094 | 359 | CK=CI | ||
| 1/y2-2/b3 | 577.2842 | 577.284 | 0.0002 | 142 | CK=CIL | ||
| 1/y2-2/b4 | 692.3112 | 692.2997 | 0.0115 | 322 | CK=CILD | ||
| 1/y2-2/b7 | 994.4338 | 994.4278 | 0.006 | 837 | CK=CILDTSN | ||
| 1/y2-2/y10 | 1336.661 | 1336.634 | 0.027 | 664 | CK=CILDTSNPVK | ||
| 1/y3-2/b1 | 480.1587 | 480.1505 | 0.0081 | 107 | ECK=C | ||
| 1/y3-2/b4 | 821.3538 | 821.3419 | 0.0118 | 460 | ECK=CILD | ||
| 1/y3-2/b5 | 922.4014 | 922.3771 | 0.0244 | 283 | ECK=CILDT | ||
| 1/y3-2/b8 | 1220.529 | 1220.519 | 0.0098 | 340 | ECK=CILDTSNP | ||
| 1/y3-2/b9 | 1319.598 | 1319.584 | 0.0138 | 987 | ECK=CILDTSNPV | ||
| 1/y4-2/a1 | 566.2067 | 566.2282 | −0.0215 | 232 | NECK=C | ||
| 1/y4-2/a2 | 679.2908 | 679.3111 | −0.0203 | 202 | NECK=CI | ||
| 1/y4-2/a7 | 1209.524 | 1209.502 | 0.0221 | 927 | NECK=CILDTSN | ||
| 1/y4-2/b3 | 820.3697 | 820.3667 | 0.003 | 154 | NECK=CIL | ||
| 1/y4-2/b4 | 935.3967 | 935.3924 | 0.0043 | 481 | NECK=CILD | ||
| 1/y4-2/b5 | 1036.444 | 1036.448 | −0.0039 | 191 | NECK=CILDT | ||
| 1/y4-2/b8 | 1334.572 | 1334.572 | 0 | 1655 | NECK=CILDTSNP | ||
| 1/y4-2/y10 | 1579.746 | 1579.675 | 0.0706 | 5505 | NECK=CILDTSNPVK | ||
| 1/y5-2/b8 | 1431.625 | 1431.599 | 0.0256 | 644 | PNECK=CILDTSNP | ||
| 1/y5-2/y10 | 1676.799 | 1676.798 | 0.0012 | 15020 | PNECK=CILDTSNPVK | ||
| 1/y6-2/a1 | 776.3435 | 776.3327 | 0.0108 | 842 | IPNECK=C | ||
| 1/y6-2/a6 | 1305.618 | 1305.539 | 0.0798 | 217 | IPNECK=CILDTS | ||
| 1/y6-2/b2 | 917.4225 | 917.4044 | 0.0181 | 1033 | IPNECK=CI | ||
| 1/y6-2/b3 | 1030.507 | 1030.413 | 0.0941 | 390 | IPNECK=CIL | ||
| 1/y6-2/y10 | 1789.883 | 1789.867 | 0.0161 | 2137 | IPNECK=CILDTSNPVK | ||
| 1/z4-2/y10 | 1562.72 | 1562.661 | 0.0585 | 2069 | NECK=CILDTSNPVK | ||
| 1/z5-2/y10 | 1659.772 | 1659.854 | −0.0822 | 2401 | PNECK=CILDTSNPVK | ||
| Cys74 to Cys85 and Cys90 to Cys100 44 Fragments | 1/y10-1/a1–2/a2 | 1392.59 | 1392.579 | 0.0105 | 789 | NVCIPNECK=C=CI | |
| 1/y10-2/b9-1/a5 | 2594.151 | 2594.122 | 0.0295 | 85 | NVCIPNECK=CILDTSNPV=CNLGY | ||
| 1/y11-1/a4-2/a4 | 2003.918 | 2003.83 | 0.0873 | 148 | VNNVCIPNECK=CNLG=CILD | ||
| 1/y11-1/b4-2/b1 | 1718.712 | 1718.686 | 0.0265 | 115 | VNNVCIPNECK=CNLG=C | ||
| 1/y11-1/b6-2/b6 | 2526.077 | 2525.993 | 0.084 | 186 | VNNVCIPNECK=CNLGYD=CILDTS | ||
| 1/y12-2/a2-1/a5 | 2069.91 | 2069.88 | 0.0305 | 554 | MVNNVCIPNECK=CI=CNLGY | ||
| 1/y12-2/b5-1/a2 | 2093.895 | 2093.833 | 0.0625 | 298 | MVNNVCIPNECK=CILDT=CN | ||
| 1/y13-1/b1–2/a9 | 2492.075 | 2492.059 | 0.0166 | 490 | DMVNNVCIPNECK=C=CILDTSNPV | ||
| 1/y13-1/b2-2/a3 | 1992.847 | 1992.854 | −0.0063 | 826 | DMVNNVCIPNECK=CN=CIL | ||
| 1/y13-2/b3-1/b4 | 2190.948 | 2190.909 | 0.0388 | 313 | DMVNNVCIPNECK=CIL=CNLG | ||
| 1/y13-2/b5-1/a2 | 2208.922 | 2208.972 | −0.0498 | 102 | DMVNNVCIPNECK=CILDT=CN | ||
| 1/y14-1/a1–2/a8 | 2528.075 | 2527.986 | 0.0891 | 572 | YDMVNNVCIPNECK=C=CILDTSNP | ||
| 1/y14-2/b9-1/a3 | 2882.266 | 2882.24 | 0.0254 | 1679 | YDMVNNVCIPNECK=CILDTSNPV=CNL | ||
| 1/y15-1/a1–2/a9 | 2684.165 | 2684.074 | 0.0908 | 230 | GYDMVNNVCIPNECK=C=CILDTSNPV | ||
| 1/y15-1/b2-2/b9 | 2854.198 | 2854.166 | 0.0322 | 311 | GYDMVNNVCIPNECK=CN=CILDTSNPV | ||
| 1/y7-1/b1–2/b2 | 1121.425 | 1121.524 | −0.0988 | 3609 | CIPNECK=C=CI | ||
| 1/y7-2/a1-1/a2 | 1066.394 | 1066.391 | 0.0034 | 286 | CIPNECK=C=CN | ||
| 1/y7-2/a1-1/a8 | 1744.699 | 1744.779 | −0.0803 | 1284 | CIPNECK=C=CNLGYDMV | ||
| 1/y7-2/a2-1/a7 | 1758.715 | 1758.619 | 0.0957 | 156 | CIPNECK=CI=CNLGYDM | ||
| 1/y7-2/a3-1/a9 | 2084.91 | 2084.817 | 0.0925 | 164 | CIPNECK=CIL=CNLGYDMVN | ||
| 1/y7-2/a5-1/a3 | 1621.721 | 1621.642 | 0.0795 | 207 | CIPNECK=CILDT=CNL | ||
| 1/y7-2/a9-1/a11 | 2911.292 | 2911.285 | 0.0068 | 205 | CIPNECK=CILDTSNPV=CNLGYDMVNNV | ||
| 1/y7-2/a9-1/a8 | 2584.138 | 2584.076 | 0.062 | 990 | CIPNECK=CILDTSNPV=CNLGYDMV | ||
| 1/y7-2/b1-1/a3 | 1207.473 | 1207.5 | −0.027 | 508 | CIPNECK=C=CNL | ||
| 1/y7-2/b1-1/a6 | 1542.585 | 1542.666 | −0.0806 | 928 | CIPNECK=C=CNLGYD | ||
| 1/y7-2/b2-1/b4 | 1405.574 | 1405.526 | 0.0474 | 125 | CIPNECK=CI=CNLG | ||
| 1/y7-2/b3-1/b4 | 1518.658 | 1518.643 | 0.015 | 197 | CIPNECK=CIL=CNLG | ||
| 1/y7-2/b4-1/a10 | 2341.975 | 2341.988 | −0.0132 | 340 | CIPNECK=CILD=CNLGYDMVNN | ||
| 1/y7-2/b6-1/a10 | 2530.054 | 2529.985 | 0.0696 | 102 | CIPNECK=CILDTS=CNLGYDMVNN | ||
| 1/y7-2/b8-1/b3 | 1975.839 | 1975.88 | −0.0417 | 245 | CIPNECK=CILDTSNP=CNL | ||
| 1/y8-1/a1–2/a5 | 1493.663 | 1493.629 | 0.0336 | 182 | VCIPNECK=C=CILDT | ||
| 1/y8-2/b9-1/a3 | 2145.981 | 2145.931 | 0.0496 | 164 | VCIPNECK=CILDTSNPV=CNL | ||
| 1/y9-1/b1–2/b5 | 1663.695 | 1663.662 | 0.0337 | 104 | NVCIPNECK=C=CILDT | ||
| 1/y9-2/b1-1/a6 | 1755.696 | 1755.718 | −0.0212 | 658 | NVCIPNECK=C=CNLGYD | ||
| 1/y9-2/b1-1/b1 | 1221.453 | 1221.521 | −0.068 | 782 | NVCIPNECK=C=C | ||
| 1/y9-2/b8-1/a6 | 2496.067 | 2496.051 | 0.0159 | 634 | NVCIPNECK=CILDTSNP=CNLGYD | ||
| 1/y9-2/b9-1/a3 | 2260.023 | 2259.99 | 0.0334 | 668 | NVCIPNECK=CILDTSNPV=CNL | ||
| 2/y10-1/y12-1/b2 | 2665.192 | 2665.097 | 0.0942 | 258 | CILDTSNPVK=MVNNVCIPNECK=CN | ||
| 2/y10-1/y7-1/b10 | 3014.319 | 3014.31 | 0.0088 | 1407 | CILDTSNPVK=CIPNECK=CNLGYDMVNN | ||
| 2/y10-1/y7-1/b11 | 3113.388 | 3113.393 | −0.0056 | 120269 | CILDTSNPVK=CIPNECK=CNLGYDMVNNV | ||
| 2/y10-1/y7-1/b6 | 2556.124 | 2556.095 | 0.0291 | 1309 | CILDTSNPVK=CIPNECK=CNLGYD | ||
| 2/y10-1/y9-1/a4 | 2463.15 | 2463.093 | 0.0574 | 609 | CILDTSNPVK=NVCIPNECK=CNLG | ||
| 2/y10-1/y9-1/a5 | 2626.214 | 2626.114 | 0.0999 | 654 | CILDTSNPVK=NVCIPNECK=CNLGY | ||
| 2/y10-1/y9-1/b3 | 2434.124 | 2434.059 | 0.0645 | 571 | CILDTSNPVK=NVCIPNECK=CNL | ||
Fig. 5.
Disulfide bonds SS5 and SS6.
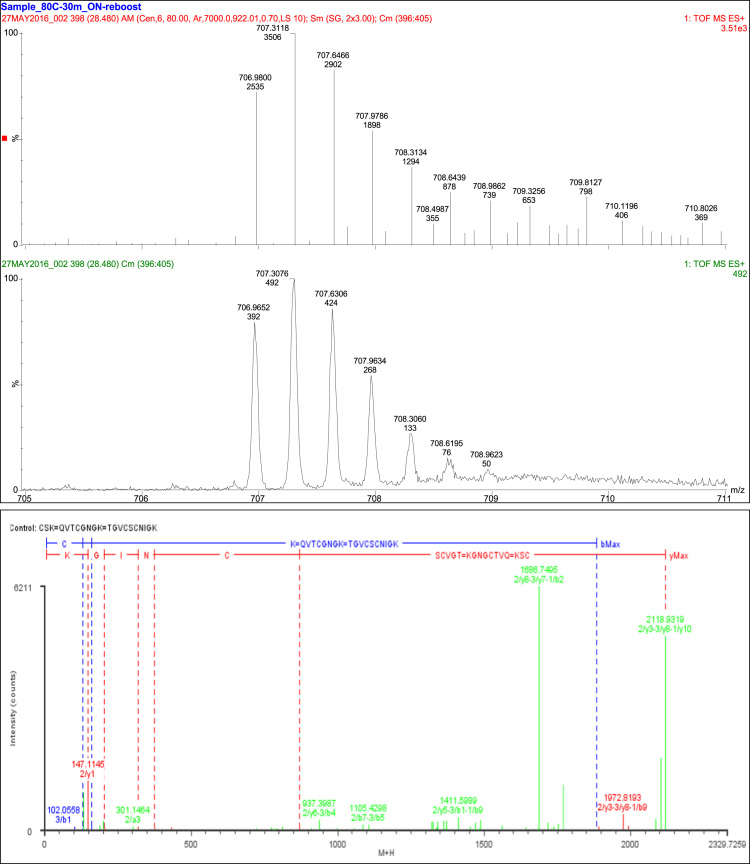
7. Disulfide bonds SS7 and SS8
A total of 39 fragments were observed. Three fragment ions (1/b7-2/b5, 1/y6-2/a4, and 1/y6-2/b4) were specific to the linkage between T12 and T14 and confirmed the Cys95 to Cys113 linkage. Four fragment ions (2/y5-3/b2, 2/y5-3/y3, 2/y6-3/a2, and 2/y6-3/b2) were specific to the linkage between T14 and T16 and confirmed the linkage of Cys115 to Cys129. A further 21 fragment ions were consistent with the linkage of T12, T14, and T16 and remaining 11 fragments consistent with constituent peptides (Table 8, Fig. 6).
Table 8.
Disulfide bond SS7 (Cys95-Cys113) and SS8 (Cys115-Cys129) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 39 Fragments | Constituent Peptides 11 Fragments | 1/a2 | 200.1399 | 200.141 | −0.0011 | 247 | QV |
| 1/a3 | 301.1876 | 301.1464 | 0.0412 | 100 | QVT | ||
| 1/b1 | 129.0664 | 129.0653 | 0.0011 | 33 | Q | ||
| 1/y1 | 147.1133 | 147.1145 | −0.0012 | 1258 | K | ||
| 1/y3 | 318.1777 | 318.1789 | −0.0012 | 47 | NGK | ||
| 1/y4 | 375.1992 | 375.2 | −0.0008 | 52 | GNGK | ||
| 2/a2 | 131.082 | 131.0274 | 0.0546 | 30 | T | ||
| 2/b1 | 102.0555 | 102.0558 | −0.0003 | 107 | T | ||
| 2/b2 | 159.077 | 159.0768 | 0.0002 | 140 | TG | ||
| 2/y2 | 204.1348 | 204.0705 | 0.0643 | 30 | GK | ||
| 2/y4 | 431.2618 | 431.2624 | −0.0006 | 72 | NIGK | ||
| Cys95 to Cys113 3 Fragments | 1/b7-2/b5 | 1105.441 | 1105.43 | 0.0109 | 157 | QVTCGNG=TGVCS | |
| 1/y6-2/a4 | 909.3923 | 909.4301 | −0.0378 | 58 | TCGNGK=TGVC | ||
| 1/y6-2/b4 | 937.3871 | 937.3987 | −0.0115 | 257 | TCGNGK=TGVC | ||
| Cys115 to Cys129 4 Fragments | 2/y5-3/b2 | 722.2966 | 722.3025 | −0.0059 | 45 | CNIGK=CS | |
| 2/y5-3/y3 | 868.4021 | 868.3544 | 0.0477 | 103 | CNIGK=CSK | ||
| 2/y6-3/a2 | 781.3337 | 781.4172 | −0.0835 | 21 | SCNIGK=CS | ||
| 2/y6-3/b2 | 809.3286 | 809.3259 | 0.0027 | 90 | SCNIGK=CS | ||
| T12, T14, and T16 linkage 21 Fragments | 1/a4-2/a6-3/a1 | 997.3728 | 997.3982 | −0.0254 | 39 | QVTC=TGVCSC=C | |
| 1/a4-2/a6-3/a2 | 1084.405 | 1084.434 | −0.0294 | 146 | QVTC=TGVCSC=CS | ||
| 1/a4-2/a8-3/b2 | 1339.527 | 1339.585 | −0.0582 | 210 | QVTC=TGVCSCNI=CS | ||
| 1/a4-2/y8-3/y3 | 1558.685 | 1558.64 | 0.0449 | 111 | QVTC=VCSCNIGK=CSK | ||
| 1/a5-2/a6-3/a1 | 1054.394 | 1054.432 | −0.0376 | 60 | QVTCG=TGVCSC=C | ||
| 1/a6-2/y8-3/y3 | 1729.749 | 1729.66 | 0.089 | 51 | QVTCGN=VCSCNIGK=CSK | ||
| 1/a7-2/a7-3/b1 | 1367.497 | 1367.472 | 0.0244 | 33 | QVTCGNG=TGVCSCN=C | ||
| 1/b5-2/a9-3/b2 | 1481.565 | 1481.493 | 0.0713 | 35 | QVTCG=TGVCSCNIG=CS | ||
| 1/b6-2/a7-3/b1 | 1338.47 | 1338.53 | −0.0596 | 72 | QVTCGN=TGVCSCN=C | ||
| 1/b6-2/b9-3/y3 | 1769.708 | 1769.651 | 0.0566 | 1155 | QVTCGN=TGVCSCNIG=CSK | ||
| 1/y5-2/b6-3/y3 | 1360.512 | 1360.561 | −0.0487 | 249 | CGNGK=TGVCSC=CSK | ||
| 1/y5-2/b9-3/b1 | 1411.523 | 1411.599 | −0.0762 | 348 | CGNGK=TGVCSCNIG=C | ||
| 1/y5-2/y8-3/a1 | 1371.564 | 1371.529 | 0.035 | 254 | CGNGK=VCSCNIGK=C | ||
| 1/y5-2/y8-3/b2 | 1486.591 | 1486.645 | −0.0536 | 270 | CGNGK=VCSCNIGK=CS | ||
| 1/y6-2/y10-3/y3 | 1891.814 | 1891.761 | 0.0521 | 105 | TCGNGK=TGVCSCNIGK=CSK | ||
| 1/y7-2/b8-3/b2 | 1641.649 | 1641.631 | 0.0188 | 78 | VTCGNGK=TGVCSCNI=CS | ||
| 1/y7-2/y10-3/y3 | 1990.882 | 1990.798 | 0.0837 | 124 | VTCGNGK=TGVCSCNIGK=CSK | ||
| 1/y7-2/y8-3/b2 | 1686.707 | 1686.75 | −0.0424 | 6211 | VTCGNGK=VCSCNIGK=CS | ||
| 1/y8-2/b8-3/b1 | 1682.676 | 1682.723 | −0.0466 | 32 | QVTCGNGK=TGVCSCNI=C | ||
| 1/y8-2/b9-3/y3 | 1972.835 | 1972.819 | 0.0156 | 407 | QVTCGNGK=TGVCSCNIG=CSK | ||
| 1/y8-2/y10-3/y3 | 2118.94 | 2118.932 | 0.0085 | 4955 | QVTCGNGK=TGVCSCNIGK=CSK | ||
Fig. 6.
Disulfide bonds SS7 and SS8.
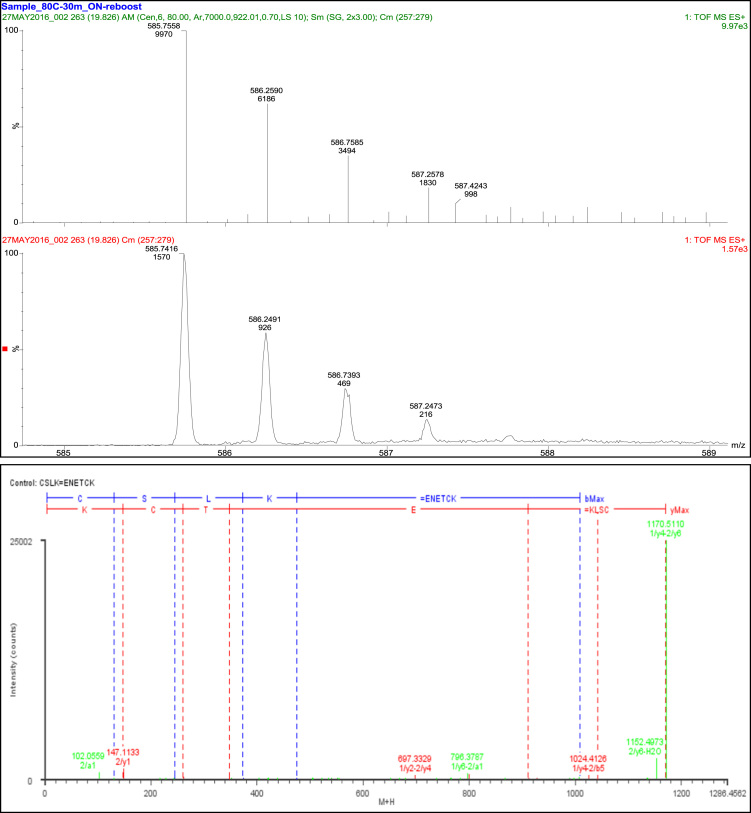
8. Disulfide bond SS9
A total of 35 fragment ions were observed with 25 fragment ions of this peptide consistent with the linkage of T18 to T20 through Cys137 and Cys148. Ten fragments were consistent with constituent peptides (Table 9, Fig. 7).
Table 9.
Disulfide bond SS9 (Cys137-Cys148) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 35 Fragments | constituent peptides 10 Fragments | 1/y2 | 260.1974 | 260.2041 | −0.0067 | 262 | LK |
| 1/y3 | 347.2294 | 347.2299 | −0.0005 | 129 | SLK | ||
| 2/a1 | 102.0555 | 102.0559 | −0.0004 | 754 | E | ||
| 2/a2 | 216.0984 | 216.1371 | −0.0387 | 65 | EN | ||
| 2/a4 | 446.1887 | 446.2249 | −0.0362 | 20 | ENET | ||
| 2/b1 | 130.0504 | 130.0469 | 0.0035 | 17 | E | ||
| 2/b2 | 244.0933 | 244.0929 | 0.0004 | 367 | EN | ||
| 2/b3 | 373.1359 | 373.1376 | −0.0017 | 171 | ENE | ||
| 2/b4 | 474.1836 | 474.1958 | −0.0122 | 88 | ENET | ||
| 2/y1 | 147.1133 | 147.1133 | 0 | 1131 | K | ||
| Cys137 and Cys148 25 Fragments | 1/a1–2/y2 | 323.1212 | 323.1008 | 0.0204 | 15 | C=CK | |
| 1/a1–2/y4 | 553.2114 | 553.2058 | 0.0056 | 53 | C=ETCK | ||
| 1/a1–2/y5 | 667.2544 | 667.2748 | −0.0204 | 59 | C=NETCK | ||
| 1/a2-2/y2 | 410.1532 | 410.1623 | −0.0091 | 36 | CS=CK | ||
| 1/a2-2/y4 | 640.2435 | 640.2652 | −0.0217 | 34 | CS=ETCK | ||
| 1/a3-2/y3 | 624.2849 | 624.2893 | −0.0044 | 27 | CSL=CKT | ||
| 1/a3-2/y5 | 867.3705 | 867.3508 | 0.0197 | 89 | CSL=NETCK | ||
| 1/b1–2/a5 | 650.1914 | 650.2377 | −0.0463 | 130 | C=ENETC | ||
| 1/b2-2/a5 | 737.2234 | 737.2545 | −0.0311 | 139 | CS=ENETC | ||
| 1/b1–2/y2 | 351.1161 | 351.1717 | −0.0556 | 207 | C=CK | ||
| 1/b1–2/y3 | 452.1638 | 452.1826 | −0.0188 | 23 | C=CKT | ||
| 1/b2-2/y2 | 438.1481 | 438.1512 | −0.0031 | 139 | CS=CK | ||
| 1/b2-2/y3 | 539.1958 | 539.2008 | −0.005 | 166 | CS=CKT | ||
| 1/b2-2/y5 | 782.2813 | 782.3434 | −0.0621 | 180 | CS=NETCK | ||
| 1/b3-2/y2 | 551.2322 | 551.2324 | −0.0002 | 219 | CSL=CK | ||
| 1/y4-2/y2 | 697.3377 | 697.3329 | 0.0048 | 465 | CSLK=CK | ||
| 1/y4-2/a5 | 996.413 | 996.4069 | 0.0061 | 130 | CSLK=ENETC | ||
| 1/y4-2/b5 | 1024.408 | 1024.4126 | −0.0046 | 502 | CSLK=ENETC | ||
| 1/y4-2/y3 | 798.3854 | 798.3802 | 0.0052 | 553 | CSLK=TCK | ||
| 1/y4-2/y4 | 927.428 | 927.4216 | 0.0064 | 223 | CSLK=ETCK | ||
| 1/y4-2/y5 | 1041.4709 | 1041.4377 | 0.0332 | 94 | CSLK=NETCK | ||
| 1/y4-2/y6 | 1170.5134 | 1170.511 | 0.0024 | 25002 | CSLK=ENETCK | ||
| 1/a1–2/y6 | 796.2969 | 796.3787 | −0.0818 | 646 | C=ENETCK | ||
| 1/b2-2/y6 | 911.3239 | 911.2963 | 0.0276 | 155 | CS=ENETCK | ||
| 1/y4-2/z3 | 781.3588 | 781.3229 | 0.0359 | 77 | CSLK=TCK | ||
Fig. 7.
Disulfide bond SS9.
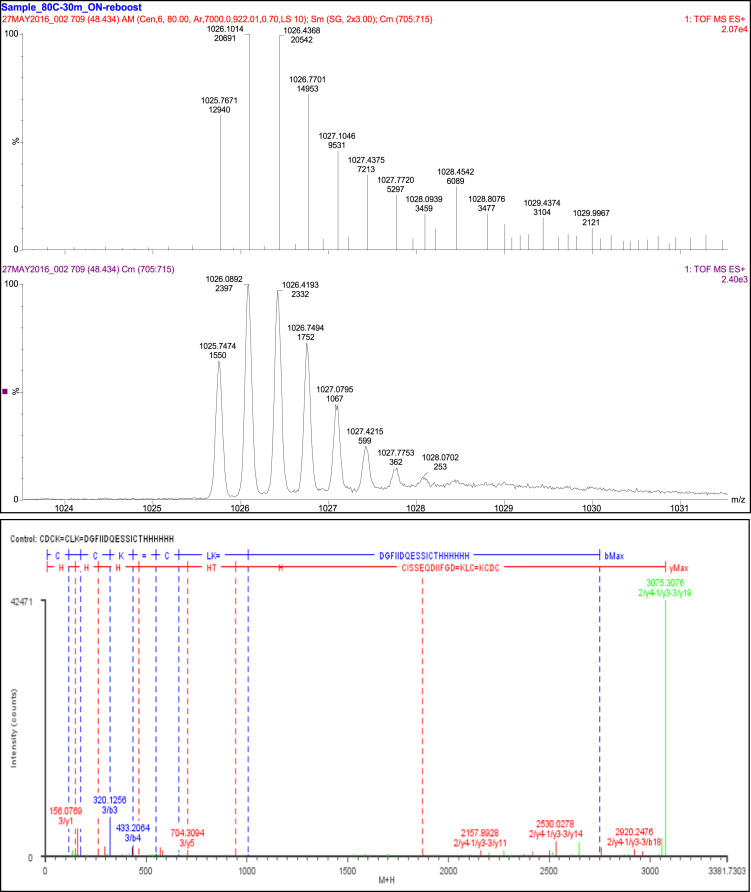
9. Disulfide bonds SS10 and SS11
A total of 65 fragments were observed. Eleven fragment ions were specific to the linkage of T19 and T22, and confirmed the linkage of Cys141 to Cys157. Three fragment ions were specific to the linkage between peptides T22 and T23 and confirmed the linkage of Cys159 to Cys172. Thirty-two fragment ions were consistent with the linkage of T12, T14, and T16 and an additional 19 fragments were consistent with constituent peptides (Table 10, Fig. 8).
Table 10.
Disulfide bond SS10 (Cys141-Cys157) and SS11 (Cys159-Cys172) peptides.
| Assignment | Theoretical Mass (Da) | Observed Mass (Da) | Mass Error (Da) | Intensity (counts) | Identification | ||
|---|---|---|---|---|---|---|---|
| 65 Fragments | Constituent Peptides 19 fragments | 1/y1 | 147.1133 | 147.1126 | 0.0007 | 1153 | K |
| 1/y2 | 260.1974 | 260.2021 | −0.0047 | 167 | LK | ||
| 3/a2 | 145.0613 | 145.0646 | −0.0033 | 400 | DG | ||
| 3/a5 | 518.2979 | 518.296 | 0.0018 | 109 | DGFII | ||
| 3/a8 | 890.426 | 890.3521 | 0.0739 | 84 | DGFIIDQE | ||
| 3/a9 | 977.458 | 977.3666 | 0.0914 | 120 | DGFIIDQES | ||
| 3/b1 | 116.0348 | 116.0532 | −0.0184 | 28 | D | ||
| 3/b2 | 173.0562 | 173.0561 | 0.0001 | 645 | DG | ||
| 3/b3 | 320.1246 | 320.1256 | −0.001 | 6533 | DGF | ||
| 3/b4 | 433.2087 | 433.2064 | 0.0023 | 1906 | DGFI | ||
| 3/b5 | 546.2927 | 546.2884 | 0.0043 | 325 | DGFII | ||
| 3/b6 | 661.3197 | 661.3064 | 0.0133 | 295 | DGFIID | ||
| 3/b9 | 1005.453 | 1005.383 | 0.0699 | 237 | DGFIIDQES | ||
| 3/y1 | 156.0773 | 156.0769 | 0.0004 | 4578 | H | ||
| 3/y2 | 293.1362 | 293.1368 | −0.0006 | 1684 | HH | ||
| 3/y3 | 430.1951 | 430.1913 | 0.0038 | 1430 | HHH | ||
| 3/y4 | 567.254 | 567.2552 | −0.0012 | 1460 | HHHH | ||
| 3/y5 | 704.3129 | 704.3094 | 0.0035 | 1025 | HHHHH | ||
| 3/y7 | 942.4196 | 942.4053 | 0.0143 | 568 | THHHHHH | ||
| Cys141 to Cys157 11 Fragments | 1/a1–2/a1 | 149.0207 | 149.1165 | −0.0958 | 16 | C=C | |
| 1/a2-2/a1 | 262.1048 | 262.1324 | −0.0276 | 39 | CL=C | ||
| 1/a2-2/a2 | 377.1317 | 377.1469 | −0.0152 | 258 | CL=CD | ||
| 1/a2-2/b1 | 290.0997 | 290.1243 | −0.0246 | 45 | CL=C | ||
| 1/a2-2/b2 | 405.1266 | 405.1262 | 0.0005 | 230 | CL=CD | ||
| 1/y3-2/a2 | 551.2322 | 551.2063 | 0.0259 | 62 | CLK=CD | ||
| 1/b1–2/a2 | 292.0426 | 292.0312 | 0.0114 | 82 | C=CD | ||
| 1/b1–2/b1 | 205.0106 | 205.0966 | −0.086 | 154 | C=C | ||
| 1/b2-2/b1 | 318.0946 | 318.0872 | 0.0074 | 36 | CL=C | ||
| 1/y3-2/b1 | 464.2001 | 464.214 | −0.0139 | 41 | CLK=C | ||
| 1/y3-2/b2 | 579.2271 | 579.2289 | −0.0018 | 1027 | CLK=CD | ||
| Cys159 to Cys172 3 fragments | 2/y2–3/a12 | 1527.682 | 1527.653 | 0.0291 | 242 | CK=DGFIIDQESSIC | |
| 2/y2–3/y19 | 2497.089 | 2497.066 | 0.0227 | 994 | CK=DGFIIDQESSICTHHHHHH | ||
| 2/y3-3/a12 | 1642.709 | 1642.633 | 0.0763 | 69 | DCK=DGFIIDQESSIC | ||
| T12, T14, and T16 Linkage 32 Fragments | 1/a1–2/a3-3/a16 | 2156.8411 | 2156.815 | 0.0266 | 37 | C=CDC=DGFIIDQESSICTHHH | |
| 1/a1–2/a3-3/y10 | 1609.5782 | 1609.559 | 0.0193 | 132 | C=CDC=SICTHHHHHH | ||
| 1/a1–2/a3-3/y14 | 2068.7383 | 2068.799 | −0.0608 | 74 | C=CDC=DQESSICTHHHHHH | ||
| 1/a1–2/y4-3/y18 | 2673.0967 | 2673.159 | −0.0623 | 62 | C=CDCK=GFIIDQESSICTHHHHHH | ||
| 1/a1–2/y4-3/y8 | 1583.5625 | 1583.616 | −0.0533 | 363 | C=CDCK=CTHHHHHH | ||
| 1/a2-2/a3-3/a16 | 2269.925 | 2269.92 | 0.0051 | 921 | CL=CDC=DGFIIDQESSICTHHH | ||
| 1/a2-2/a3-3/a18 | 2544.043 | 2544.035 | 0.0076 | 58 | CL=CDC=DGFIIDQESSICTHHHHH | ||
| 1/a2-2/y4-3/a15 | 2306.9666 | 2306.904 | 0.0623 | 147 | CL=CDCK=DGFIIDQESSICTHH | ||
| 1/b1–2/b3-3/a12 | 1700.6064 | 1700.657 | −0.0505 | 100 | C=CDC=DGFIIDQESSIC | ||
| 1/b1–2/b3-3/y14 | 2124.7283 | 2124.776 | −0.0476 | 72 | C=CDC=DQESSICTHHHHHH | ||
| 1/b1–2/y4-3/a14 | 2084.8186 | 2084.862 | −0.0437 | 85 | C=CDCK=DGFIIDQESSICTH | ||
| 1/b1–2/y4-3/y17 | 2644.0703 | 2644.117 | −0.0471 | 2248 | C=CDCK=FIIDQESSICTHHHHHH | ||
| 1/b2-2/b3-3/a12 | 1813.6906 | 1813.709 | −0.0181 | 63 | CL=CDC=DGFIIDQESSIC | ||
| 1/b2-2/b3-3/b13 | 1942.7332 | 1942.833 | −0.0997 | 95 | CL=CDC=DGFIIDQESSICT | ||
| 1/b2-2/b3-3/y8 | 1578.536 | 1578.608 | −0.0721 | 228 | CL=CDC=CTHHHHHH | ||
| 1/b2-2/b3-3/y9 | 1691.62 | 1691.693 | −0.0729 | 105 | CL=CDC=ICTHHHHHH | ||
| 1/b2-2/y4-3/a14 | 2197.9026 | 2197.909 | −0.0066 | 671 | CL=CDCK=DGFIIDQESSICTH | ||
| 1/b2-2/y4-3/b17 | 2637.0742 | 2637.017 | 0.0576 | 233 | CL=CDCK=DGFIIDQESSICTHHHH | ||
| 1/b2-2/y4-3/y10 | 1924.7576 | 1924.81 | −0.0524 | 84 | CL=CDCK=SICTHHHHHH | ||
| 1/b2-2/y4-3/y16 | 2610.0859 | 2610.088 | −0.0017 | 407 | CL=CDCK=IIDQESSICTHHHHHH | ||
| 1/y3-2/y4-3/a18 | 2892.2439 | 2892.164 | 0.0803 | 403 | CLK=CDCK=DGFIIDQESSICTHHHHH | ||
| 1/y3-2/y4-3/b14 | 2372.0032 | 2372.038 | −0.0352 | 341 | CLK=CDCK=DGFIIDQESSICTH | ||
| 1/y3-2/y4-3/b18 | 2920.2388 | 2920.248 | −0.0088 | 1160 | CLK=CDCK=GFIIDQESSICTHHHHHH | ||
| 1/y3-2/y4-3/y11 | 2157.8953 | 2157.893 | 0.0024 | 922 | CLK=CDCK=SSICTHHHHHH | ||
| 1/y3-2/y4-3/y13 | 2414.9963 | 2415.007 | −0.011 | 897 | CLK=CDCK=QESSICTHHHHHH | ||
| 1/y3-2/y4-3/y14 | 2530.0232 | 2530.028 | −0.0046 | 2415 | CLK=CDCK=DQESSICTHHHHHH | ||
| 1/y3-2/y4-3/y16 | 2756.1914 | 2756.2 | −0.0088 | 1421 | CLK=CDCK=IIDQESSICTHHHHHH | ||
| 1/y3-2/y4-3/y18 | 2960.2812 | 2960.238 | 0.043 | 911 | CLK=CDCK=GFIIDQESSICTHHHHHH | ||
| 1/y3-2/y4-3/y19 | 3075.3081 | 3075.308 | 0.0005 | 42471 | CLK=CDCK=DGFIIDQESSICTHHHHHH | ||
| 1/y3-2/y4-3/y8 | 1870.7471 | 1870.768 | −0.0212 | 131 | CLK=CDCK=CTHHHHHH | ||
| 1/y3-2/y4-3/z14 | 2512.9968 | 2513.063 | −0.0657 | 102 | CLK=CDCK=DQESSICTHHHHHH | ||
| 1/y3-2/y4-3/z8 | 1853.7205 | 1853.793 | −0.0724 | 66 | CLK=CDCK=CTHHHHHH | ||
Fig. 8.
Disulfide bonds SS10 and SS11.
10. Experimental design, materials and methods
10.1. Sample preparation
Baculovirus Pfs25 [2] was denatured and digested as described in [1].
10.2. Chromatography
Digested peptides were separated with a 2695 Separations Module (Waters Corporation; Milford MA) and a 2489 UV/Vis Detector (Waters Corporation; Milford, MA) set at 214 nm. An XBridge (Waters Corporation; Milford, MA) BEH 300 C18 (2.1×250 mm, 5 µm) was used at a column temperature of 37 °C and gradient with 0.1% Triflouroacetic acid (TFA) in purified water (Mobile Phase A) and 0.1% TFA in acetonitrile (Mobile Phase B) as described in [1].
10.3. Mass spectrometry
MS analysis was done with a QTOF Premier mass spectrometer (Waters Corporation; Milford, MA) equipped with an electrospray source as described in [1]. MS data was acquired in MSE mode using MassLynx v4.1 (Waters Corporation; Milford, MA). RAW MS files have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) with identifier: MSV000081982.
10.4. Analysis of mass spectra
The mass spectral data was analyzed using BiopharmaLynx 1.3 (Waters Corporation; Milford, MA) as described in [1].
Acknowledgements
The authors thank Ashley Birkett and Merribeth Morin of PATH's Malaria Vaccine Initiative (MVI) for their support and assistance. The authors thank Steven Becht, Ying-Hua Chang and Jie Ding from PPD GMP Lab in Middleton, WI for work on the LC-MS/MS method for peptide mapping analysis. The authors thank the project team at Syngene International, a Biocon company, in Bangalore, India for cloning, production, and preliminary characterization of the baculovirus-expressed Pfs25. The work was supported by the Bill & Melinda Gates Foundation (OPP1108403). The views expressed herein are solely those of the authors and do not necessarily reflect the views of the Foundation.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2018.03.034.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Lee S.M., Plieskatt J.L., King C.R. Disulfide bond mapping of Pfs25, a recombinant malaria transmission blocking vaccine candidate. Anal. Biochem. 2018;542:20–23. doi: 10.1016/j.ab.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S.M., Wu C.K., Plieskatt J., McAdams D.H., Miura K., Ockenhouse C., King C.R. Assessment of Pfs25 expressed from multiple soluble expression platforms for use as transmission-blocking vaccine candidates. Malar. J. 2016;15:405. doi: 10.1186/s12936-016-1464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material