Abstract
Immunomodulatory or immunosuppressive properties of bone marrow-derived mesenchymal stem cells (BM-MSCs) facilitate the treatment of acute respiratory distress syndrome and acute lung injury (ALI). Dysregulated miRNA (miRNA or miR) expression associated with the effects of BM-MSCs was assessed in a rat model of lipopolysaccharide (LPS)-induced ALI. The present study performed biochemical tests to assess five analytes, including alanine aminotransferase (ALT), aspartate aminotransferase (AST), lactate, blood urea nitrogen (BUN), and creatinine (CREA). Total cell count was assessed and the percentage of bronchoalveolar lavage neutrophil content was also examined. The results Histopathological examination of rat upper lobe lung tissue was then used to estimate lung injury score (LIS). The levels of AST, lactate, BUN and creatinine (excluding ALT), released into the circulation upon injury, were significantly lower in ALI rats treated with BM-MSCs than in ALI rats alone (P<0.05). BM-MSC rats exhibited a significantly decreased bronchoalveolar lavage neutrophil percentage and LIS compared with that of LPS treated rats alone (P<0.05). In addition, the miRNA expression profile was determined following treatment with BM-MSCs via microarray analysis. A total of 95/690 miRNAs were differentially expressed following the treatment of BM-MSCs in rats with ALI. Among the 95 miRNAs, 66 were upregulated and 29 were downregulated; 9 miRNAs were significantly upregulated (miR-1843-3p, miR-323-3p, miR-183-5p, miR-182 and miR-196b-3p) or downregulated (miR-547-3p, miR-301b-5p, miR-503-3p and miR-142-3p). A total of 3 miRNAs were inversely expressed in ALI treated with BM-MSCs compared with untreated ALI. Of these 3 miRNAs, the expression of miR-142-3p and miR-503-3p was upregulated in the LPS groups and downregulated in the BM-MSC groups. miR-196b-3p was downregulated in the LPS group and upregulated in the BM-MSC groups. miRNAs have a role in cell proliferation, immune response, inflammation and apoptosis, which may be associated with the therapeutic effects of BM-MSCs in ALI. In summary, BM-MSCs improved multi-organ damage and attenuated lung injury. Different miRNA profiles were expressed following BM-MSC treatment of ALI. These dysregulated miRNAs participated in BM-MSC-mediated immunomodulation of ALI.
Keywords: acute lung injury, expression profile, microRNA, mesenchymal stem cell
Introduction
Acute lung injury (ALI) and its more severe form, acute respiratory distress syndrome (ARDS), are generally diagnosed in critically sick patients characterized by widespread inflammation of the lung. The mortality rate for ARDS is as high as 36–44% (1). ALI and ARDS are caused by pneumonia, sepsis, severe trauma with shock, transfusion, drug toxicity, or aspiration of gastric contents. Lung inflammation, impaired gas exchange, destruction of the epithelium-capillary interface, and refractory hypoxemia are characteristic features of ARDS. There is currently no effective pharmacotherapy to improve survival of patients with ARDS (2,3).
Mesenchymal stem cells (MSCs) are multipotent progenitor cells that are isolated from various mesenchymal tissues, including umbilical cord, bone marrow (BM), placenta and adipose tissue (4). MSCs are attractive therapeutic candidates for the treatment of ARDS. The paracrine effects of MSCs modulate inflammation, endothelial injury, alveolar fluid clearance and apoptosis in ARDS. MSCs display anti-inflammatory, anti-apoptotic, neoangiogenic and immunomodulatory effects in various immune cells (5–9).
microRNAs (miRNAs or miRs) are short non-coding single-stranded RNA species approximately 19–25 nucleotides long. miRNAs modulate gene expression by translational inhibition, and are associated with diverse biological pathways, as diagnostic biomarkers and potential therapeutic targets (10–11). Altered miRNA expression levels have been associated with disease processes or therapeutic effects of different therapies (12). Previous studies have suggested that specific miRNAs are upregulated and others are downregulated in ALI and ARDS (13–16). Altered expression of miRNAs in the regulation of the inflammatory pathway and tissue repair in ALI and ARDS are correlated with inflammatory mediators and recruitment of B cells, T cells, and other immune cells in the lung (17,18).
In the present study, miRNA expression was profiled following treatment with BM-MSCs. Microarray analysis was used to investigate dysregulated miRNAs associated with the effects of BM-MSCs in a rat model of lipopolysaccharide (LPS)-induced ALI. To the best of our knowledge, the present study is the first attempt to estimate the miRNA expression profile in rat ALI following BM-MSC treatment.
Materials and methods
Induction of ALI with LPS and administration of BM-MSCs
A total of 15 male Sprague-Dawley rats (age, 8–9 weeks; weight, 200–250 g) provided by Samtako Bio Korea (Osan, Korea) were used. All experimental procedures were approved by the Institutional Animal Care and Use Committee in Daejeon St. Mary's Hospital, Catholic University of Korea (Daejeon, Korea). All rats were housed under a 12 h light/dark cycle, a 50–60% humidity, and an ambient temperature of 22–24°C. In addition, rats received ad libitum access to food and water. All procedures were conducted by the same individual to minimize variation. In order to induce ALI, LPS extracted from Escherichia coli 055:B5 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) diluted in saline was used (20 mg/kg). ALI rats were injected intraperitoneally with LPS (5 mg/kg). In the control group, sham intervention was performed using the same amount of saline. Human MSCs were provided by The Catholic Institute of Cell Therapy (Seoul, Korea). The cells were preserved with Dulbecco's modified Eagle's medium containing 1,000 mg/l glucose, sodium bicarbonate, and pyridoxine (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere of 5% CO2 at 37°C. The cells at passages 3–4 were isolated for in vivo experiments. All 15 rats were assigned randomly to one out of the following three groups (n=5/group): Saline-treated controls, LPS-induced ALI with saline (ALI) and LPS-induced ALI with BM-MSC (LPS+BM-MSC). At 30 min following administration with LPS, BM-MSCs (2×106; 100 µl) or saline (100 µl) were injected slowly into the tail vein over 20 min.
Laboratory tests and histopathological examination
Rats were sacrificed at 6 h following administration of saline or BM-MSCs. Blood was harvested via cardiac puncture and plasma was centrifuged for 10 min at 3,000 × g at 37°C. Plasma samples were frozen at −70°C prior to analyze alanine aminotransferase (ALT), aspartate aminotransferase (AST), lactate, blood urea nitrogen (BUN), and creatinine (CREA) using an IDEXX VetTest® Chemistry Analyzer (IDEXX Laboratories, Inc., Westbrook, ME, USA).
The trachea was incised and bronchoalveolar lavage (BAL) fluid was obtained from the right lung. Total cells were counted using the LUNA automated cell counter (Logos Biosystems, Annandale, VA, USA) following the manufacturer's instructions. An aliquot of 200 µl of the diluted 500 µl pellet was cytospinned at a speed of 180 × g at 4°C, transferred to a slide, and stained with Wright-Giemsa stain at 24°C for 6 min. The 100-cell differential count was performed for estimating the percentage of neutrophils under a light microscope (magnification, ×400; Olympus Corporation, Tokyo, Japan) in 4 ideal slide zones. Rat left upper lobe lung tissue was fixed with 10% formalin overnight at 24°C, embedded in paraffin, and stained with hematoxylin and eosin at 24°C for 1 min. Each lung section was assessed independently by two clinical pathologists using microscopy (magnification, ×100) to evaluate the severity of lung injury. The lung injury score (LIS) comprises four components (hemorrhage, alveolar capillary congestion, inflammatory cells infiltrating the interstitium or airspace, and the alveolar wall thickness), with each component scored on a 5 point scale (0 = minimal damage, 1 = mild damage, 2 = moderate damage, 3 = severe damage, 4 = maximal damage). The LSI is the sum of all four component scores (9). The left lower lobe was frozen at −70°C prior to analysis of miRNA expression.
Microarray analysis and functional annotation
Total RNA was isolated from the rat lung tissue using TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH, USA) following the manufacturer's protocols. The total RNA pellet was dissolved in nuclease-free water and its quantity and yield was estimated using a 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). Rat miRNA expression profiling was performed using miRCURY LNA miRNA PCR Assays (Exiqon; Qiagen GmbH, Hilden, Germany). The seventh generation array included with this assay contains ~3,100 capture probes, covering all human, mouse and rat miRNAs annotated in miRBase (www.mirbase.org), as well as all viral miRNAs related to these species. Processed microarray slides were scanned using a G2565CA microarray scanner system (Agilent Technologies, Inc.) and imported using Feature Extraction software ver. 10.7.3.1 (Agilent Technologies, Inc.). The fluorescence intensities of each slide were quantified according to the Exiqon protocol. The results of miRNA expression were calculated as the mean ± standard error of the mean. Target prediction for functional estimation of the differentially expressed miRNAs was conducted using miRanda (34.236.212.39/microrna/home.do) and Targetscan ver. 7.0 (http://www.targetscan.org). The target lists of dysregulated miRNAs were submitted separately to the functional annotation tool provided by the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov), ver. 6.7. The predicted targets were annotated according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (19,20).
Visualization and analysis of dysregulated miRNAs
To visualize the predicted target genes associated with dysregulated miRNAs, the Network Analyzer plug-in (apps.cytoscape.org/apps/with_tag/networkanalysis) of Cytoscape 3.6 was used (21).
Statistical analysis
The Kruskal-Wallis test and a one-way analysis of variance for non-normally distributed data was used to test the median difference between each variable in the three groups. A post hoc Tamhane's T2 test was performed for pairwise comparison of subgroups. The box-and-whisker plot was used to present the data distribution in each figure. A line is drawn inside the box at the median and the box portion of the plot is defined by two lines at the 25th percentile and 75th percentile. The distance between the lower (25th percentile) and upper (75th percentile) lines of the box is defined as the inter-quartile range. The two whisker boundaries indicate the 10th (lower) and 90th (upper) percentiles. MedCalc Statistical Software Version 17.6 (MedCalc Software bvba, Ostend, Belgium) was used for statistical investigation. P<0.05 was considered to indicate a statistically significant difference.
Results
BM-MSCs reduce LPS-induced ALI
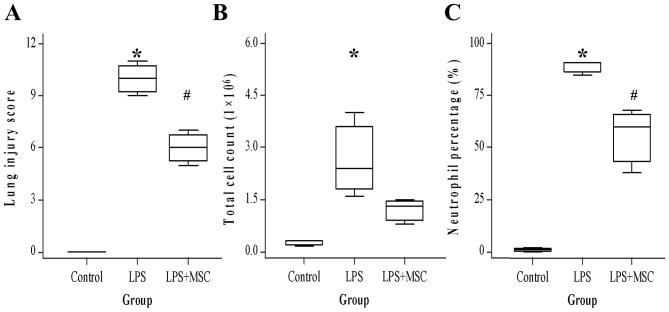
The presence of moderate pulmonary injuries including hemorrhage, congestive alveolar capillaries, inflammatory cell infiltration, and alveolar wall thickening in the LPS group were revealed via histopathological examination, compared with the LPS+BM-MSC group. The LIS was used to estimate the influence of BM-MSCs on lung injury. Similar to the histopathological examination, LIS (median, 10) in LPS rats was significantly higher than in controls (P<0.05). LIS (6) in BM-MSC rats was significantly lower than in LPS rats (P<0.05). The total cell count and neutrophil percentage in BAL fluid were counted to evaluate the protective role of BM-MSCs in LPS-induced ALI. As a result, BM-MSCs markedly reduced the number of total cell count (control, 0.3; LPS, 2.4; LPS+BM-MSC, 1.3), and significantly reduced the neutrophil percentage (Control, 1; LPS, 91; LPS+BM-MSC, 60; P<0.05) in the BAL fluid compared with the LPS group (Fig. 1).
Figure 1.
BM-MSCs reduce LPS-induced ALI. (A) LIS. Compared with controls, ALI rats exhibited significantly increased LIS. In contrast, BM-MSC rats exhibited decreased LIS compared with that of LPS. (B) The total cell count and (C) neutrophil percentage in the bronchoalveolar lavage fluid. Compared with the controls, ALI rats exhibited significantly increased total cell count (P<0.05) and neutrophil percentage (P<0.05). The BM-MSC group exhibited decreased neutrophil percentage compared with the LPS group (P<0.05). *P<0.05 vs. control; #P<0.05 vs. LPS. BM, bone marrow-derived; MSC, mesenchymal stem cell; LIS, lung injury score; ALI, acute lung injury; LPS, lipopolysaccharide.
BM-MSC treatment improves multi-organ damage induced by LPS
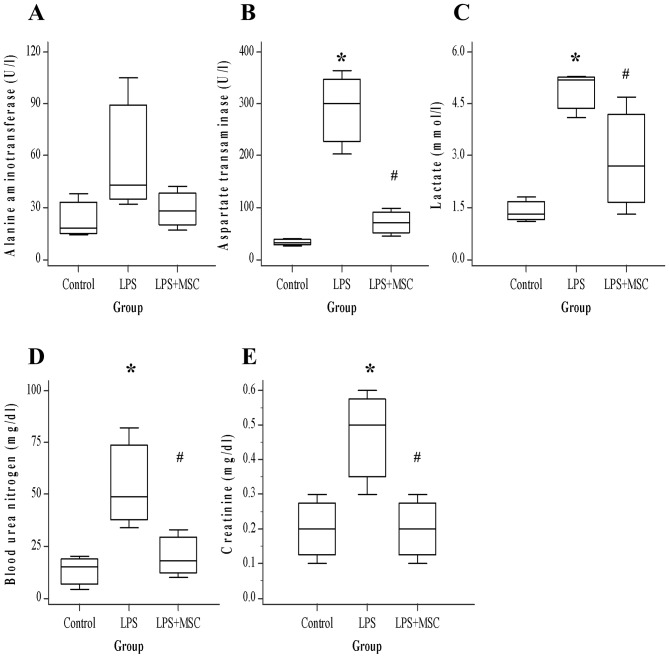
Organ damage was estimated by measuring serum biochemical indicators 6 h following administration with LPS. The levels of four analytes (excluding ALT) were significantly elevated by LPS (Fig. 2). The levels of liver enzymes ALT and AST released into circulation upon injury were lower in ALI treated with BM-MSCs than in ALI only (ALT, P=0.223; AST, P<0.05). In particular, AST was significantly decreased (control, 32; LPS, 300; LPS+BM-MSC, 70). The concentration of lactate, typically used as an indicator of tissue hypoperfusion, was lower (control, 1.3; LPS, 5.2; LPS+BM-MSC, 2.7) in ALI with BM-MSCs compared with ALI alone (P<0.05). In kidney injury, blood urea nitrogen (BUN; control, 15; LPS, 49; LPS+BM-MSC, 18) and creatinine (CREA; control, 0.2; LPS, 0.5; LPS+BM-MSC, 0.2) levels were also significantly lower in ALI with BM-MSCs, compared with ALI alone (BUN, P<0.05; CREA, P<0.05).
Figure 2.
Blood chemistry levels (n=5 for each group). BM-MSC ameliorates LPS-induced aggravation in multi-organ damage. Evaluation of (A) ALT, (B) AST, (C) lactate, (D) BUN and (E) CREA. Compared with the control rats, the LPS group exhibited significantly increased AST, lactate, BUN and CREA. In contrast, the BM-MSC group exhibited significantly decreased AST, lactate, BUN and CREA (all P<0.05) compared with the LPS group. *P<0.05 vs. control; #P<0.05 vs. LPS. BM, bone marrow-derived; MSC, mesenchymal stem cell; LPS, lipopolysaccharide; ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CREA, creatinine.
miRNA expression profiles in ALI
miRNA expression profiling was performed to identify the alteration in miRNAs in the lungs of rats with LPS-induced ALI. A total of 128/690 rat miRNAs were expressed differently between the ALI and control groups. They included 68 upregulated and 60 downregulated miRNAs, respectively. Furthermore, 15 miRNAs were significantly upregulated or downregulated (fold-change ≥2) in the ALI group, compared with the control group (P<0.05). Five of these miRNAs (miR-760-3p, miR-223-3p, miR-449c-3p, miR-503-3p and miR-142-3p) were upregulated and 10 (miR-100-5p, miR-199a-5p, miR-99a-5p, miR-199a-3p, miR-181a-5p, miR-497-5p, miR-191a-5p, miR-28-5p, miR-3065-5p and miR-196b-3p) were downregulated following LPS treatment (Table I).
Table I.
miRNAs implicated in rats with ALI.
| miRNA name | Fold-Regulation | change direction | P-value |
|---|---|---|---|
| rno-miR-760-3p | 3.7 | Up | 0.030 |
| rno-miR-223-3p | 2.9 | Up | 0.048 |
| rno-miR-449c-3p | 2.1 | Up | 0.040 |
| rno-miR-503-3p | 2.1 | Up | 0.045 |
| rno-miR-142-3p | 2.0 | Up | 0.047 |
| rno-miR-100-5p | 2.4 | Down | 0.020 |
| rno-miR-199a-5p | 2.3 | Down | 0.030 |
| rno-miR-99a-5p | 2.3 | Down | 0.010 |
| rno-miR-199a-3p | 2.2 | Down | 0.030 |
| rno-miR-181a-5p | 2.1 | Down | 0.040 |
| rno-miR-497-5p | 2.1 | Down | 0.030 |
| rno-miR-191a-5p | 2.1 | Down | 0.010 |
| rno-miR-28-5p | 2.0 | Down | 0.030 |
| rno-miR-3065-5p | 2.0 | Down | 0.040 |
| rno-miR-196b-3p | 2.0 | Down | 0.046 |
miRNAs listed were significantly upregulated or downregulated in the lung tissue of rats with ALI compared with saline-treated controls, with P<0.05 and a fold-change ≥2. miRNA/miR, microRNA; ALI, acute lung injury; rno, Rattus norvegicus.
miRNA expression profiles in ALI following treatment with BM-MSCs
Among 690 rat miRNAs, 95 were differentially expressed between ALI in the BM-MSCs group and the control group. They included 66 upregulated and 29 downregulated miRNAs. Furthermore, nine miRNAs were significantly upregulated or downregulated in the ALI group, compared with the control group (fold-change ≥2; P<0.05). Among the nine miRNAs, five (miR-1843-3p, miR-323-3p, miR-183-5p, miR-182 and miR-196b-3p) were upregulated and four (miR-547-3p, miR-301b-5p, miR-503-3p and miR-142-3p) were downregulated following treatment with BM-MSCs (Table II). To investigate the effects of BM-MSCs in ALI, the inversely expressed miRNAs in ALI with BM-MSCs compared with ALI were selected. Three miRNAs were inversely expressed in the two groups. The expression of two of these miRNAs (miR-503-3p and miR-142-3p) was increased in the LPS group, and decreased in the BM-MSC group. The miR-196b-3p was downregulated in the LPS group, but upregulated in the BM-MSC group.
Table II.
Altered miRNAs in rats with ALI following BM-MSC treatment.
| miRNA name | Fold-Regulation | change direction | P-value |
|---|---|---|---|
| rno-miR-1843-3p | 4.5 | Up | 0.013 |
| rno-miR-323-3p | 3.8 | Up | 0.015 |
| rno-miR-183-5p | 3.7 | Up | 0.048 |
| rno-miR-182 | 2.9 | Up | 0.015 |
| rno-miR-196b-3p | 2.5 | Up | 0.043 |
| rno-miR-547-3p | 2.0 | Down | 0.010 |
| rno-miR-301b-5p | 2.1 | Down | 0.045 |
| rno-miR-503-3p | 2.0 | Down | 0.048 |
| rno-miR-142-3p | 2.0 | Down | 0.049 |
miRNAs listed were significantly upregulated or downregulated in the lung tissue of BM-MSC-treated rats with ALI compared with saline-treated controls, with P<0.05 and a fold-change ≥2. miRNA/miR, microRNA; ALI, acute lung injury; BM-MSC, bone marrow-derived mesenchymal stem cells; rno, Rattus norvegicus.
Pathway analysis of altered miRNAs in ALI following treatment with BM-MSCs
Gene ontology and KEGG pathway annotation analyses via DAVID ver. 6.7 revealed annotated KEGG pathways for the altered expressed miRNAs (Tables III and IV). The miRNA pathways were associated with inflammation, the immune response and cellular apoptosis.
Table III.
Functional annotation of the altered microRNAs in rats with acute lung injury.
| Term | Count | P-value |
|---|---|---|
| hsa05200:Pathways in cancer | 35 | 0.004 |
| hsa04010:MAPK signaling pathway | 31 | 0.002 |
| hsa04810:Regulation of actin cytoskeleton | 24 | 0.013 |
| hsa04722:Neurotrophin signaling pathway | 22 | <0.001 |
| hsa04630:JAK-STAT signaling pathway | 21 | 0.002 |
| hsa04510:Focal adhesion | 21 | 0.040 |
| hsa04310:Wnt signaling pathway | 20 | 0.004 |
| hsa04360:Axon guidance | 19 | 0.001 |
| hsa04910:Insulin signaling pathway | 18 | 0.006 |
| hsa04530:Tight junction | 17 | 0.014 |
| hsa05211:Renal cell carcinoma | 14 | <0.001 |
| hsa05210:Colorectal cancer | 14 | 0.003 |
| hsa05322:Systemic lupus erythematosus | 14 | 0.012 |
| hsa04916:Melanogenesis | 14 | 0.012 |
| hsa04012:ErbB signaling pathway | 13 | 0.010 |
| hsa05215:Prostate cancer | 13 | 0.013 |
| hsa04520:Adherens junction | 12 | 0.011 |
| hsa05217:Basal cell carcinoma | 10 | 0.008 |
| hsa05212:Pancreatic cancer | 10 | 0.045 |
MAPK, mitogen-activated protein kinase; JAK-STAT, janus kinase-signal transducer and activator of transcription.
Table IV.
Functional annotation of the altered microRNAs in rats with acute lung injury following bone marrow-derived mesenchymal stem cell treatment.
| Term | Count | P-value |
|---|---|---|
| hsa04810:Regulation of actin cytoskeleton | 23 | <0.001 |
| hsa04010:MAPK signaling pathway | 22 | 0.005 |
| hsa04360:Axon guidance | 15 | 0.001 |
| hsa04310:Wnt signaling pathway | 14 | 0.013 |
| hsa04722:Neurotrophin signaling pathway | 12 | 0.018 |
| hsa04540:Gap junction | 10 | 0.014 |
| hsa04666:Fc gamma R-mediated phagocytosis | 10 | 0.021 |
| hsa04912:GnRH signaling pathway | 10 | 0.025 |
MAPK, mitogen-activated protein kinase; GnRH, gonadotropin-releasing hormone.
Pathway analysis of altered miRNAs in ALI following treatment with BM-MSCs
It was observed that miR-503-3p and miR-142-3p were associated with myeloid/lymphoid or mixed-lineage translocated to, 1; cyclin T2; and granzyme B gene is a serine protease with a notable role in the rapid induction of target cell apoptosis (22). It was also predicted that miR-503-3p and miR-196-3p were correlated with muscleblind-like protein 1 and DNA damage regulated autophagy modulator 1 (DRAM1) genes, whereas miR-142-3p and miR-196b-3p were associated with activator of heat shock protein ATPase 2, tyrosine-protein kinase ABL2 (ABL2), homeobox protein Nkx2-3 (Nkx2-3), Ras-responsive element-binding protein 1 (RREB1) and Musashi RNA binding protein 2.
Discussion
In the present study, the therapeutic effects of BM-MSCs were observed in an LPS-induced ALI rat model. The number of total inflammatory cells and neutrophil percentage in the BAL fluid were reduced in the ALI group treated with BM-MSCs, compared with the ALI group. AST/ALT (hepatic damage), BUN/CREA (renal damage) and lactate (tissue hypoperfusion) levels were measured to determine organ damage. BM-MSCs attenuated liver and kidney injury and improved tissue perfusion. Histological examination indicated that lung injury in the ALI group treated with BM-MSCs was less prominent than in the ALI group.
Immunomodulatory or immunosuppressive properties of BM-MSCs have been studied for many years (23–27). BM-MSCs have been considered as potential candidates for treatment of ALI and ARDS. Gupta et al (28) previously reported that intrabronchial infusion of BM-MSCs increased survival and reduced pulmonary edema. Improvement in lung histopathology was associated with decreased expression of pro-inflammatory cytokines including macrophage inflammatory protein-2, tumor necrosis factor-α (TNF-α) and elevation of anti-inflammatory cytokines, such as interleukin (IL)-1ra, IL-10, and IL-13 (28). Administration of BM-MSCs reduces not only systemic and pulmonary inflammation, but also organ damage, in mouse sepsis models (29). BM-MSCs mediate anti-inflammatory effects via concurrent downregulation of inflammation-related genes (IL-6 and IL-10) (29). Overall mortality in septic mice receiving MSCs was significantly decreased, likely due to decreased inflammation as evidenced by a reduction in protein and gene expression levels of pro-inflammatory cytokines, such as IL-6 (29).
miRNAs are non-coding small (~22 nucleotides) regulatory RNAs that affect the translation or stability of target mRNAs. The significance of miRNAs in various biological processes has been described previously (30). As miRNA regulation serves a crucial role in the immunomodulatory effects of MSCs, it may be associated with different miRNA expression patterns. MSCs suppress T cell proliferation via indoleamine 2,3-dioxygenase (IDO) (31) and prostaglandin E2, and act along with T-cells in inflammation (32). miR-181 is associated with T and B cell development (33), and enhances IL-6 and IDO expression when its expression is increased in MSCs (34). The expression of aberrant miRNAs associated with immune regulation was evaluated in ALI rats with or without BM-MSC treatment. It was demonstrated that 128 of the total of 690 miRNAs were expressed differently in ALI rats. This included 68 upregulated and 60 downregulated miRNAs. Significantly upregulated miRNAs included miR-760-3p, miR-223-3p, miR-449c-3p, miR-503-3p and miR-142-3p. Significantly downregulated miRNAs included miR-100-5p, miR-199a-5p, miR-99a-5p, miR-199a-3p, miR-181a-5p, miR-497-5p, miR-191a-5p, miR-28-5p, miR-3065-5p and miR-196b-3p.
The anti-inflammatory effects of miR-181a may be mediated via targeting of importin α3, and miR-181b may inhibit nuclear factor-κ-gene binding (NF-κB)-mediated inflammatory responses (35). miR-223 is hematopoietic-specific miRNA and is a key modulator of hematopoietic lineage differentiation. It is deregulated in various inflammatory disorders (36). miR-223 is also upregulated in autoimmune diseases such as inflammatory bowel disease and rheumatoid arthritis (37). Serum levels of miR-146a and miR-223 are significantly decreased in sepsis, compared with systemic inflammatory response syndrome (SIRS) and healthy populations. However, there were no significant differences in levels of miR-223 in SIRS, compared with the controls (38). Another study profiling serum miRNAs from 214 patients with sepsis (117 survivors and 97 non-survivors) reported that miR-223 levels were significantly decreased in patients with non-surviving sepsis compared with surviving sepsis (39). Various miRNAs modulated the expression of pro-inflammatory cytokines TNF-α and IL-6. The expression of miR-181 and miR-191 was associated with TNF-α, whereas miR-142, miR-223, miR-181 and miR-199 were associated with IL-6 (40).
In the present study, 95 out of 690 miRNAs were differentially expressed following the treatment of BM-MSCs in ALI rats. Of these 95 miRNAs, 66 were upregulated and 29 were downregulated. Among them, 9 miRNAs (upregulated 5 miRNAs: miR-1843-3p, miR-323-3p, miR-183-5p, miR-182 and miR-196b-3p; downregulated 4 miRNAs: miR-547-3p, miR-301b-5p, miR-503-3p and miR-142-3p) were significantly upregulated or downregulated.
Differently expressed miRNAs in ALI rats were associated with mitogen-activated protein kinase (MAPK), janus kinase-signal transducer and activator of transcription, Wnt and ErbB signaling pathways, which are controlled by altered levels of miRNAs in ALI rats. Altered miRNAs in these rats following treatment with BM-MSCs were likely associated with the MAPK and Wnt signaling pathways. miR323-3p has been implicated in the Wnt signaling pathway and the cadherin signaling pathway (41).
In particular, three miRNAs were significantly inversely expressed in ALI with BM-MSCs compared with ALI: The expression of two miRNAs (miR-503-3p and miR-142-3p) was increased in the LPS group and decreased in the BM-MSC group. miR-196b-3p was downregulated in the LPS group and upregulated in the BM-MSC group. miR-196b controls granulocytic colony numbers and suppresses granulocyte-colony stimulating factor-stimulated granulopoiesis (42). Therefore, miR-196b is a negative regulator of granulocytic differentiation (42). miR-196b significantly enhanced cell proliferation and partially inhibited the differentiation of mouse normal bone marrow precursors (43,44). miR-142 is expressed in hematopoietic or dendritic cells, and regulates immune response. It serves a critical role in LPS-induced endogeneous expression of IL-6, which is a significant component of LPS-induced endotoxemia (45). It was predicted that miR-142 mediated the regulation of apoptosis, which is a major metabolic process activated in the lungs of patients with ALI/ARDS. miR-503 inhibits cell proliferation, and induces cellular apoptosis and G0/G1 arrest by directly targeting E2F3, as an important transcription factor in proliferation and cell cycle distribution (46). These miRNAs are associated with cell proliferation, immune response, inflammation and apoptosis, and associated with the therapeutic effects of BM-MSCs in ALI.
Among the predicted target genes associated with dysregulated miRNAs, DRAM1 mediates autophagic defense against a broader range of intracellular pathogens, because the common bacterial endotoxin lipopolysaccharide induces DRAM1 expression (47). ABL2 suppresses fms-like tyrosine kinase 3 (FLT3)-internal tandem duplication-induced cell proliferation as negative regulator of signaling downstream of FLT3 by partially blocking FLT3-induced protein kinase B phosphorylation (48). Increased expression of Nkx2-3 at both RNA and protein level was demonstrated in intestinal specimens of Crohn's disease (49). RREB1 is activated by the MAPK pathway and negatively represses the miR-143/145 promoter through interaction with two Ras responsive elements and establishes complex network of regulation through which the miR-143/145 cluster is able to modulate KRAS signaling in colorectal cancer (50).
There are a few study limitations. First, the small sample size may render the result less powerful. Second, the temporal variation in the miRNA expression following LPS injection was not analyzed, which may be associated with discrepancies in miRNA expression levels in previous studies associated with ALI/ARDS. Third, dysregulated miRNAs following LPS injections or BM-MSC infusions were not quantified using reverse transcription-quantitative polymerase chain reaction because of small sample volumes.
In spite of these limitations, the present study identified the miRNA expression profiles in ALI rats following BM-MSC treatment, and revealed that BM-MSCs improved multiorgan damage and attenuated lung injury. Furthermore, BM-MSC treatment of ALI rats dysregulated miRNA profiles. Dysregulated miRNAs mediated the immunomodulation of BM-MSCs in ALI. Further studies are required to elucidate the putative targets of dysregulated miRNAs.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Catholic Medical Center Research Foundation made in the program year of 2016 (grant no. 52015B000100173).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JP was involved in revising the manuscript and was responsible for the interpretation of general data and the statistical analysis. SJ contributed to the conception of the study. KP and KY made substantial contributions to the acquisition, analysis, and interpretation of the experimental data. SS collected the fund for this study, made substantial contributions to the conception and design of the study, revised it critically for important intellectual content, and gave final approval of the version to be published. All authors approved the final manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Daejeon St. Mary's Hospital, Catholic University of Korea (Seoul, Korea).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Phua J, Badia JR, Adhikari NK, Friedrich JO, Fowler RA, Singh JM, Scales DC, Stather DR, Li A, Jones A, et al. Has mortality from acute respiratory distress syndrome decreased over time?: A systematic review. Am J Respir Crit Care Med. 2009;179:220–227. doi: 10.1164/rccm.200805-722OC. [DOI] [PubMed] [Google Scholar]
- 2.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Zemans RL. The acute respiratory distress syndrome: Pathogenesis and treatment. Annu Rev Pathol. 2011;6:147–163. doi: 10.1146/annurev-pathol-011110-130158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlain G, Fox J, Ashton B, Middleton J. Concise review: Mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 5.Crisostomo PR, Markel TA, Wang Y, Meldrum DR. Surgically relevant aspects of stem cell paracrine effects. Surgery. 2008;143:577–581. doi: 10.1016/j.surg.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemeth K, Leelahavanichkul A, Yuen PS, Mayer B, Parmelee A, Doi K, Robey PG, Leelahavanichkul K, Koller BH, Brown JM, et al. Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat Med. 2009;15:42–49. doi: 10.1038/nm0409-462b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815–1822. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 8.Kinnaird T, Stabile E, Burnett MS, Lee CW, Barr S, Fuchs S, Epstein SE. Marrow-derived stromal cells express genes encoding a broad spectrum of arteriogenic cytokines and promote in vitro and in vivo arteriogenesis through paracrine mechanisms. Circ Res. 2004;94:678–685. doi: 10.1161/01.RES.0000118601.37875.AC. [DOI] [PubMed] [Google Scholar]
- 9.Mazhari R, Hare JM. Mechanisms of action of mesenchymal stem cells in cardiac repair: potential influences on the cardiac stem cell niche. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S21–S26. doi: 10.1038/ncpcardio0770. [DOI] [PubMed] [Google Scholar]
- 10.Staszel T, Zapala B, Polus A, Sadakierska-Chudy A, Kiec-Wilk B, Stepien E, Wybranska I, Chojnacka M, Dembinska-Kiec A. Role of microRNAs in endothelial cell pathophysiology. Pol Arch Med Wewn. 2011;121:361–366. [PubMed] [Google Scholar]
- 11.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 12.Luck ME, Muljo SA, Collins CB. Prospects for therapeutic targeting of micrornas in human immunological diseases. J Immunol. 2015;194:5047–5052. doi: 10.4049/jimmunol.1403146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie T, Liang J, Liu N, Wang Q, Li Y, Noble PW, Jiang D. Microrna-127 inhibits lung inflammation by targeting igg fcgamma receptor i. J Immunol. 2012;188:2437–2444. doi: 10.4049/jimmunol.1101070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai ZG, Zhang SM, Zhang Y, Zhou YY, Wu HB, Xu XP. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol. 2012;90:37–43. doi: 10.1139/y11-095. [DOI] [PubMed] [Google Scholar]
- 15.Vaporidi K, Vergadi E, Kaniaris E, Hatziapostolou M, Lagoudaki E, Georgopoulos D, Zapol WM, Bloch KD, Iliopoulos D. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;303:L199–L207. doi: 10.1152/ajplung.00370.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 17.Bhargava M, Wendt CH. Biomarkers in acute lung injury. Transl Res. 2012;159:205–217. doi: 10.1016/j.trsl.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y, Lyu YI, Tang J, Li Y. MicroRNAs: Novel regulatory molecules in acute lung injury/acute respiratory distress syndrome. Biomed Rep. 2016;4:523–527. doi: 10.3892/br.2016.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47 doi: 10.1002/0471250953.bi0813s47. 8.13.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman J. Granzyme A activates another way to die. Immunol Rev. 2010;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Nicola M, Carlo-Stella C, Magni M, Milanesi M, Longoni PD, Matteucci P, Grisanti S, Gianni AM. Human bone marrow stromal cells suppress T-lymphocyte proliferation induced by cellular or nonspecific mitogenic stimuli. Blood. 2002;99:3838–3843. doi: 10.1182/blood.V99.10.3838. [DOI] [PubMed] [Google Scholar]
- 24.Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci USA. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002;30:215–222. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 26.Krampera M, Cosmi L, Angeli R, Pasini A, Liotta F, Andreini A, Santarlasci V, Mazzinghi B, Pizzolo G, Vinante F, et al. Role for interferon-gamma in the immunomodulatory activity of human bone marrow mesenchymal stem cells. Stem Cells. 2006;24:386–398. doi: 10.1634/stemcells.2005-0008. [DOI] [PubMed] [Google Scholar]
- 27.Le Blanc K. Immunomodulatory effects of fetal and adult mesenchymal stem cells. Cytotherapy. 2003;5:485–489. doi: 10.1080/14653240310003611. [DOI] [PubMed] [Google Scholar]
- 28.Gupta N, Su X, Popov B, Lee JW, Serikov V, Matthay MA. Intrapulmonary delivery of bone marrow-derived mesenchymal stem cells improves survival and attenuates endotoxin-induced acute lung injury in mice. J Immunol. 2007;179:1855–1863. doi: 10.4049/jimmunol.179.3.1855. [DOI] [PubMed] [Google Scholar]
- 29.Mei SH, Haitsma JJ, Dos Santos CC, Deng Y, Lai PF, Slutsky AS, Liles WC, Stewart DJ. Mesenchymal stem cells reduce inflammation while enhancing bacterial clearance and improving survival in sepsis. Am J Respir Crit Care Med. 2010;182:1047–1057. doi: 10.1164/rccm.201001-0010OC. [DOI] [PubMed] [Google Scholar]
- 30.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 31.Meisel R, Zibert A, Laryea M, Gobel U, Daubener W, Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood. 2004;103:4619–4621. doi: 10.1182/blood-2003-11-3909. [DOI] [PubMed] [Google Scholar]
- 32.Kroesen BJ, Teteloshvili N, Smigielska-Czepiel K, Brouwer E, Boots AM, van den Berg A, Kluiver J. Immuno-miRs: Critical regulators of T-cell development, function and ageing. Immunology. 2015;144:1–10. doi: 10.1111/imm.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat Immunol. 2009;10:1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Wang Y, Fan H, Zhao X, Liu D, Hu Y, Kidd AR, III, Bao J, Hou Y. MicroRNA-181a regulates local immune balance by inhibiting proliferation and immunosuppressive properties of mesenchymal stem cells. Stem Cells. 2012;30:1756–1770. doi: 10.1002/stem.1156. [DOI] [PubMed] [Google Scholar]
- 35.Sun X, He S, Wara AKM, Icli B, Shvartz E, Tesmenitsky Y, Belkin N, Li D, Blackwell TS, Sukhova GK, et al. Systemic delivery of microRNA-181b inhibits nuclear factor-kappaB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ Res. 2014;114:32–40. doi: 10.1161/CIRCRESAHA.113.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haneklaus M, Gerlic M, O'Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274:215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li YT, Chen SY, Wang CR, Liu MF, Lin CC, Jou IM, Shiau AL, Wu CL. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis Rheum. 2012;64:3240–3245. doi: 10.1002/art.34550. [DOI] [PubMed] [Google Scholar]
- 38.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients' mortality: A prospective observational study. PLoS One. 2012;7:e38885. doi: 10.1371/journal.pone.0038885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. Circulating micrornas as biomarkers for sepsis. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17010078. pii: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandis I, Ospelt C, Karagianni N, Denis MC, Reczko M, Camps C, Hatzigeorgiou AG, Ragoussis J, Gay S, Kollias G. Identification of microRNA-221/222 and microRNA-323-3p association with rheumatoid arthritis via predictions using the human tumour necrosis factor transgenic mouse model. Ann Rheum Dis. 2012;71:1716–1723. doi: 10.1136/annrheumdis-2011-200803. [DOI] [PubMed] [Google Scholar]
- 42.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 44.Georgantas RW, III, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, Malter C, Nieves E, Tawara I, Wang Y, et al. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood. 2011;117:6172–6183. doi: 10.1182/blood-2010-12-325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang SW, Yue J, Wang BC, Zhang XL. miR-503 inhibits cell proliferation and induces apoptosis in colorectal cancer cells by targeting E2F3. Int J Clin Exp Pathol. 2015;8:12853–12860. [PMC free article] [PubMed] [Google Scholar]
- 47.van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, Ottenhoff TH, Spaink HP, Meijer AH. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected] Cell Host Microbe. 2014;15:753–767. doi: 10.1016/j.chom.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Kazi JU, Rupar K, Marhall A, Moharram SA, Khanum F, Shah K, Gazi M, Nagaraj SR, Sun J, Chougule RA, Rönnstrand L. ABL2 suppresses FLT3-ITD-induced cell proliferation through negative regulation of AKT signaling. Oncotarget. 2017;8:12194–12202. doi: 10.18632/oncotarget.14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu W, Lin Z, Kelly AA, Hegarty JP, Poritz LS, Wang Y, Li T, Schreiber S, Koltun WA. Association of a Nkx2-3 polymorphism with Crohn's disease and expression of Nkx2-3 is up-regulated in B cell lines and intestinal tissues with Crohn's disease. J Crohns Colitis. 2009;3:189–195. doi: 10.1016/j.crohns.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.