ABSTRACT
The rRNA genes of Borrelia (Borreliella) burgdorferi are unusually organized; the spirochete has a single 16S rRNA gene that is more than 3 kb from a tandem pair of 23S-5S rRNA operons. We generated an rnc null mutant in B. burgdorferi that exhibits a pleiotropic phenotype, including decreased growth rate and increased cell length. Here, we demonstrate that endoribonuclease III (RNase III) is, as expected, involved in processing the 23S rRNA in B. burgdorferi. The 5′ and 3′ ends of the three rRNAs were determined in the wild type and rncBb mutants; the results suggest that RNase III in B. burgdorferi is required for the full maturation of the 23S rRNA but not for the 5S rRNA nor, curiously, for the 16S rRNA.
IMPORTANCE Lyme disease, the most common tick-borne zoonosis in the Northern Hemisphere, is caused by the bacterium Borrelia (Borreliella) burgdorferi, a member of the deeply branching spirochete phylum. B. burgdorferi carries a limited suite of ribonucleases, enzymes that cleave RNA during processing and degradation. Several ribonucleases, including RNase III, are involved in the production of ribosomes, which catalyze translation and are a major target of antibiotics. This is the first study to dissect the role of an RNase in any spirochete. We demonstrate that an RNase III mutant is viable but has altered processing of rRNA.
KEYWORDS: Borrelia burgdorferi, RNA processing, endoribonuclease III, ribosomal RNA, spirochetes
INTRODUCTION
Borrelia (Borreliella) burgdorferi is a spirochete that causes Lyme disease (1–3). The bacterium possesses a unique, segmented genome composed of a 910-kb linear chromosome with hairpin ends, as well as a couple dozen or so linear and circular plasmids (4–6). The chromosome harbors most of the housekeeping genes, while the plasmids, which have more genetic variation, predominantly carry genes encoding factors that mediate interactions with the vertebrate host and tick vector (5–11). B. burgdorferi is maintained in nature in an enzootic cycle, where it passes between a tick and a vertebrate (8, 10, 12). The B. burgdorferi genome has lost genes over its evolutionary history as the spirochete has become restricted to an obligate parasitic lifestyle (7, 9, 13–16). Likely as a consequence of this genome reduction, the rRNA genes have been repositioned into an idiosyncratic arrangement (5, 17–22).
In most bacteria, the three rRNAs (16S, 23S, and 5S) are produced as a single polycistronic transcript. Many microorganisms carry multiple copies of the rRNA operon (seven in Escherichia coli and 10 in Bacillus subtilis). Each operon almost always contains the three rRNA genes in the canonical 16S-23S-5S order, with minimal intergenic spacing. Various tRNA genes may be inserted between the rRNA genes. The polycistronic transcript is cotranscriptionally processed by endoribonucleases to separate each individual rRNA (23–27). Bacterial species that are host or niche restricted, with a reduced genome size and low GC content, generally possess fewer rRNA copies. Presumably, some operons are lost through recombination events during genome reduction; this can result in unusual rRNA gene organization, regulation, and chromosomal distribution (13, 28). B. burgdorferi and closely related Lyme disease species possess only a single copy of the 16S rRNA gene, and it is separated by more than 3 kb from the tandemly duplicated 23S-5S rRNA operons (5, 17–22, 29). The relapsing fever Borrelia species also have a split ribosomal operon, with additional genes inserted, but have only one copy of each rRNA gene (30). Although split ribosomal operons and uneven numbers of rRNA genes have been described (13, 28, 31–33), to our knowledge, the specific rRNA gene organization of B. burgdorferi has not been observed in any other genome.
Bacteria typically encode a suite of ribonucleases (RNases) that perform a variety of functions, including ribosome processing. RNase III, an endoribonuclease encoded by the rnc gene, acts as a homodimer that specifically recognizes and cleaves double-stranded RNA (34–36). The enzyme catalyzes the initial processing step of the 16S and 23S rRNAs, separating them from the polycistronic primary transcript prior to ribosome assembly (37–39). During transcription, stems form between the complementary 5′ and 3′ flanking regions of each rRNA and serve as double-stranded substrates for RNase III (40). RNase III cleaves both strands simultaneously, releasing the individual rRNAs. Exonucleases, and a few endonucleases, further process the remaining stem to generate the mature rRNAs (27, 35, 36). RNase III also processes mRNA to regulate transcript levels (35, 41–44). B. burgdorferi encodes an RNase III homolog with 44% identity to the B. subtilis enzyme and 35% identity to the E. coli enzyme. The 5S rRNA is processed by either RNase E in E. coli or RNase M5 in B. subtilis, following its liberation from the 30S transcript by RNase III cleavage (45, 46); B. burgdorferi possesses a homolog of RNase M5 (BB0626).
RNase III is not essential in many bacteria, including E. coli, because alternative RNA processing pathways can be utilized, although these rnc mutants have a growth phenotype (25, 47–50). The phenotype is likely due to aberrant ribosomes; the precursor 30S and pre-23S rRNA species accumulate in rnc null mutants (37, 39, 47, 49–52). mRNA and noncoding RNA (ncRNA) processing defects might also contribute to the phenotype, due to the role of RNase III in regulating levels of these RNA species (35, 41, 42, 44). However, RNase III is essential in B. subtilis (39) because it cleaves several prophage-encoded double-stranded toxin-antitoxin duplexes, preventing these mRNAs from being translated (53).
RNase III has not been previously investigated in spirochetes. We now demonstrate that RNase III is not essential in B. burgdorferi, but an rncBb mutant has a growth and morphological phenotype. The 23S rRNA is processed, as expected, by RNase III. However, RNase III is not required to generate wild-type 16S rRNA ends, which may be related to the distinct rRNA arrangement in Lyme disease Borrelia species.
RESULTS
Generation of an RNase III (rncBb) null mutant.
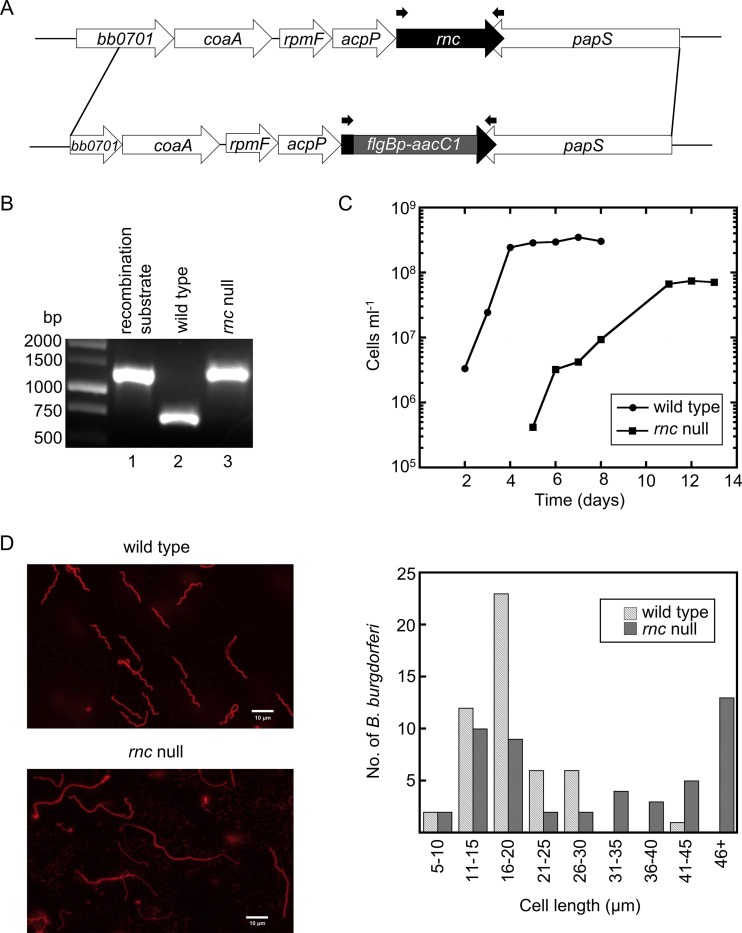
We hypothesized that the rRNA transcripts in B. burgdorferi, despite their unique organization, are processed as in other bacteria, including cleavage of the 16S and 23S rRNA by the endoribonuclease RNase III (37, 38, 39). The chromosomal rncBb gene was replaced through homologous recombination with the gentamicin resistance cassette flgBp-aacC1 (Fig. 1A). Three transformants were isolated in two B. burgdorferi strains (297 and B31-A3). The clones were examined for the presence of the flgBp-aacC1 insertion (Fig. 1B) by PCR, using primers specific for the flanking regions of the rncBb gene (Table 1; Fig. 1A). A second primer set specific for the flgBp-aacC1 cassette and flanking downstream rncBb region was also used to verify insertion of the resistance marker into the chromosome (data not shown). In addition, the absence of rncBb transcript in mutants was confirmed by reverse transcription-PCR (RT-PCR) (data not shown). Extensive attempts to complement the rncBb null mutants, either in trans with rncBb carried on a shuttle vector or in cis by genetic reconstitution, were unsuccessful; we speculate that our inability to transform the mutant was a consequence of an altered response to the competence preparation.
FIG 1.
A B. burgdorferi RNase III (rncBb) null mutant exhibits growth and morphological phenotypes. (A) Schematic of the genetic approach taken to construct the rncBb null mutant in B. burgdorferi. A gentamicin resistance cassette (flgBp-aacC1) replaced most of the chromosomal rncBb gene via allelic exchange by homologous recombination. Primers rnc 1F+NdeI and rnc 738R (Table 1), which were used to obtain PCR products shown in panel B, are indicated by the small black arrows above the rncBb gene and flgBp-aacC1 cassette. (B) Confirmation of an rncBb null mutant (in a B31 background). DNA from a B. burgdorferi transformant and controls were amplified by PCR, using primers flanking the insertion site, and the products were electrophoresed in a 1% agarose gel. Lane 1, cloned recombination substrate; lane 2, genomic DNA from wild-type B. burgdorferi; lane 3, genomic DNA from B. burgdorferi rnc null mutant. (C) B. burgdorferi wild type and the rncBb null mutant were inoculated in BSK II liquid medium at a cell density of 104 cell ml−1 and grown at 34°C until stationary phase. Cells were enumerated every 24 h, using a Petroff-Hausser counting chamber. (D) Microscopy images showing the cell morphology of the rncBb null mutant (lower panel) compared with that of wild-type B. burgdorferi (upper panel). Live B. burgdorferi cells were stained with a wheat germ agglutinin (WGA)-Alexa Fluor 594 conjugate and assayed by fluorescence microscopy. The length of 50 cells for each strain was measured as described by Lybecker et al. (54) and sorted into bins of 5-μm increments.
TABLE 1.
Oligonucleotides used in this study
| Purpose and name | Sequence (5′→3′) |
|---|---|
| rnc null mutant construction | |
| bb705 U1260F | TTTAAAGGTTGAAAATGAAG |
| bb705 92R+AatII+AgeI | ACCGGTCAAGACGTCTAAAGTCAATGCTCAAATT |
| bb705 617F+AatII | GACGTCTTTTTTGTGTGGAACTTTAT |
| bb705 D1945R+AgeI | ACCGGTATGAATCTAGGGAAAAACA |
| rnc null mutant screening | |
| rnc 1F+NdeI | CATATGAAAAAAAAATCTTCTGA |
| rnc 738R | TTAAAGGTTAATGTTTTCCA |
| pflgB5′+AgeI | ACCGGTACCCGAGCTTCAAGGAA |
| 5′ and 3′ RACE | |
| rrs 136R | CCCATCTCATAGGTAGATCATCCACGC |
| rrl 76R | GCTTTTCGCAGCTTACCACGACCTTC |
| rrl 198R | TTAGATGGTTCACTTCCCCTGGTATCGC |
| rrf 88R | CGAACTCGCAGTACCATCAGCGAATAAG |
| rrf 112R | CCCTGGCAATAACCTACTCTCCCGC |
| rrs 1365F | TGAATACGTTCTCGGGCCTTGTACACA |
| rrl 2770F | ACGTTCGGAAAGGATAACCGCTGAAAG |
| rrf 60F | CTTATTCGCTGATGGTACTGCGAGTTCG |
| rrf 49R | TGTGTTCGGAATGATAACAGGTGTTTCCTC |
| rrf 51R | TCTGTGTTCGGAATGATAACAGGTGTTTCC |
| rrf 8F | GGTTAAAGAAAAGAGGAAAC |
| rrl 76R | GCTTTTCGCAGCTTACCACGACCTTC |
| Northern blot probe: rnc 158R | CTAGATTTTTGATCCAACTCATTAGAATACGACGAATGACACAATG |
| Recombinant RNase III | |
| rnc 1F+NdeI | CATATGAAAAAAAAATCTTCTGA |
| rnc 735R+SapI | GCTCTTCCGCAAAGGTTAATGTTTTCCATAG |
| Artificial 23S rRNA substrate | |
| T7 prom | TAATACGACTCACTATAG |
| 23S stem+T7 | AGGAAGACAAAAATATGGCCAAAGTTGCCTTTGACCATATTTTTATCTTCCATCCTATAGTGAGTCGTATTA |
Characterization of the rncBb null mutant phenotype.
The rncBb null mutants from both parental strains exhibited growth (Fig. 1C) and morphological (Fig. 1D) phenotypes. Wild-type B. burgdorferi cells reached mid-log-phase cell density within 1 day and stationary phase by 4 days. The rncBb mutant did not reach mid-log-phase cell density until about 5 days and the log phase was extended until about 11 days. Cell density reached by the rncBb null mutant at stationary phase was approximately a half log lower than that of the wild type. In addition to the growth phenotype, the rncBb null mutant cells are elongated compared to wild-type cells (Fig. 1D), which is a phenotype previously found in B. burgdorferi hfq mutants (54). A subpopulation of mutant cells has a wild-type length, resulting in a bimodal distribution (Fig. 1D).
Analyses of rRNA in the rncBb null mutant.
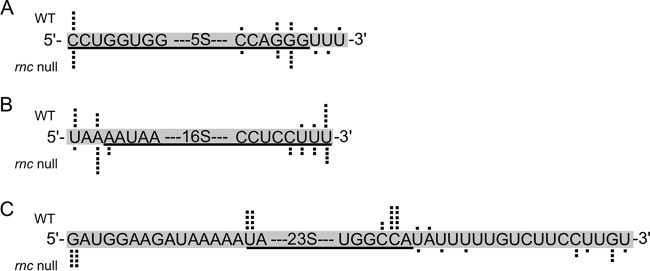
The ends of the three rRNAs in the rncBb mutant were determined by 5′ and 3′ rapid amplification of cDNA ends (RACE) to assay the function of RNase III in rRNA processing in B. burgdorferi. Total RNA was isolated from both B. burgdorferi rnc null mutants, as well as from the wild type. The 5′ and 3′ RACE PCR products for each rRNA were cloned and sequenced. The primary nucleotide sequence of the rRNA is shown with black dots above (wild-type sequences) and below (rncBb mutant sequences), indicating the experimentally determined location of the rRNA ends (Fig. 2). The ends of the 16S rRNA are the same in both the wild type and rncBb mutant, and map to the annotated ends, although the 3′ end is more variable (Fig. 2B). Therefore, RNase III is not essential for 16S rRNA processing in B. burgdorferi. However, the RACE data show that RNase III is required for full processing of the 23S rRNA (Fig. 2C). The mature 23S rRNA transcript is longer at both ends in an rncBb null mutant. The 5′ end of the 23S rRNA in the rncBb mutant is 34 nucleotides downstream from the predicted promoter. There is no difference in the 5S rRNA ends generated in the rncBb null mutant compared to those of the wild type, demonstrating that RNase III is not required for processing 5S rRNA (Fig. 2A), as expected (25–27, 45, 46).
FIG 2.
B. burgdorferi RNase III processes the 23S rRNA but not the 5S rRNA or the 16S rRNA. The 5′ and 3′ ends of the three rRNAs were examined by 5′ and 3′ RACE, respectively. Primary sequences for each rRNA 5′ end and 3′ end are shown, with the annotated rRNA sequence underlined. Individual sequencing events from multiple RACE PCR products for each rRNA are represented by black dots above the sequences for wild type (WT) and below the sequences for the rncBb mutant in a B31 background (rnc null). (A) 5S rRNA. (B) 16S rRNA. (C) 23S rRNA.
rncBb operon structure.
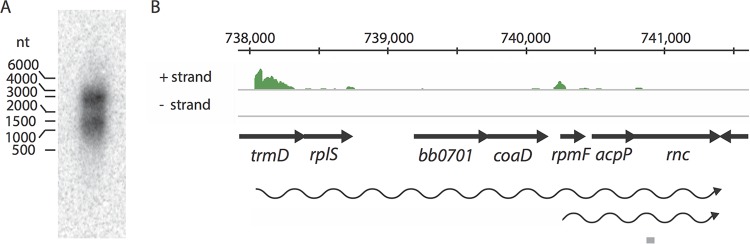
In B. burgdorferi, rnc is carried on the chromosome, in a region dense with genes related to translation. Northern blotting with an oligonucleotide probe to rncBb detected two polycistronic transcripts (Fig. 3A). The shorter mRNA, which may be either a primary or a processed transcript, begins with rpmF (encoding ribosomal protein L32), and the longer mRNA begins in trmD and includes rplS (encoding ribosomal protein L19). Examination of transcriptome data from wild-type B. burgdorferi (Fig. 3B) confirms the Northern blotting data. Junctional RT-PCR detected transcription throughout a larger 16-gene region (data not shown), beginning with bb0690 (bicA, dps, or napA), which may be due to either a larger polycistronic mRNA or pervasive transcription (55).
FIG 3.
rncBb is in an operon. (A) Northern blot analysis of 15 μg of total RNA fractionated on a formaldehyde-agarose gel, blotted to a nylon membrane, and hybridized with an oligonucleotide probe (gray bar in panel B). (B) Deep-sequencing results displayed as a coverage map. The chromosomal location and genomic context of the region around rncBb (bb0705) are depicted above and below the coverage map, respectively. The wavy arrows represent the predicted polycistronic mRNAs, and the gray bar illustrates the location of the probe used in the Northern blot.
Cleavage of an artificial 23S rRNA substrate.
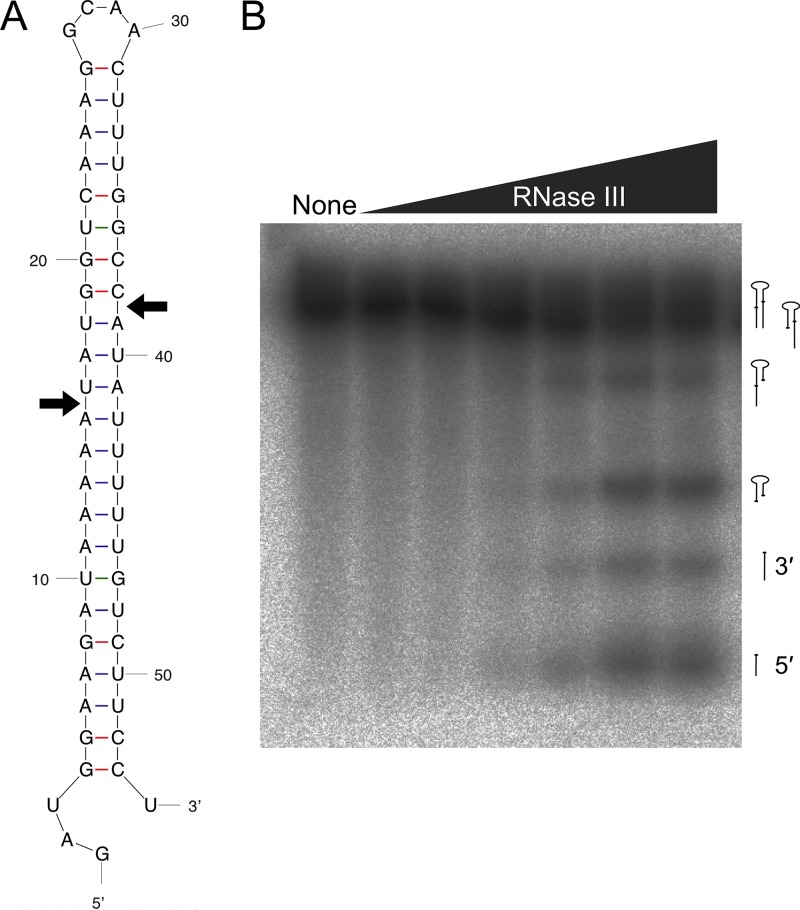
We biochemically assayed the activity of RNase III from B. burgdorferi using an in vitro cleavage reaction, with an artificial 23S rRNA stem-loop that serves as an RNase III substrate (56). The sequence is composed of the double-stranded stem portion of the B. burgdorferi 23S rRNA transcript with a loop of four unmatched nucleotides (Fig. 4A). RNase III from E. coli recognizes double-stranded stems and creates a staggered break with a 3′ two-nucleotide overhang (35, 36), although cleavage sites predicted from our RACE data (Fig. 2) result in a 5′ three-nucleotide overhang (Fig. 4A). The artificial 23S rRNA substrate was generated by in vitro transcription and radiolabeled with [32P]UTP. A molar excess of the substrate was incubated with recombinant B. burgdorferi RNase III protein as previously described (56, 57). Cleavage products were produced with increasing concentrations of RNase III, indicating that the stem-loop serves as a substrate (Fig. 4B).
FIG 4.
RNase III cleaves an artificial 23S rRNA substrate in vitro. (A) The double-stranded secondary structure of the artificial B. burgdorferi 23S rRNA substrate, predicted using Mfold (84). Substrate design was as described by Amarasinghe et al. (56). Arrows indicate cleavage sites predicted from RACE data (Fig. 2). (B) Phosphorimage showing the increase in [32P]UTP-labeled artificial 23S substrate cleavage products produced at 37°C after 5 min in the presence of increasing concentrations of recombinant RNase III (0 nM to 25 nM). The predicted species designations (right side) for each cleavage product are based on the work of Meng et al. (57) but have not been experimentally confirmed.
DISCUSSION
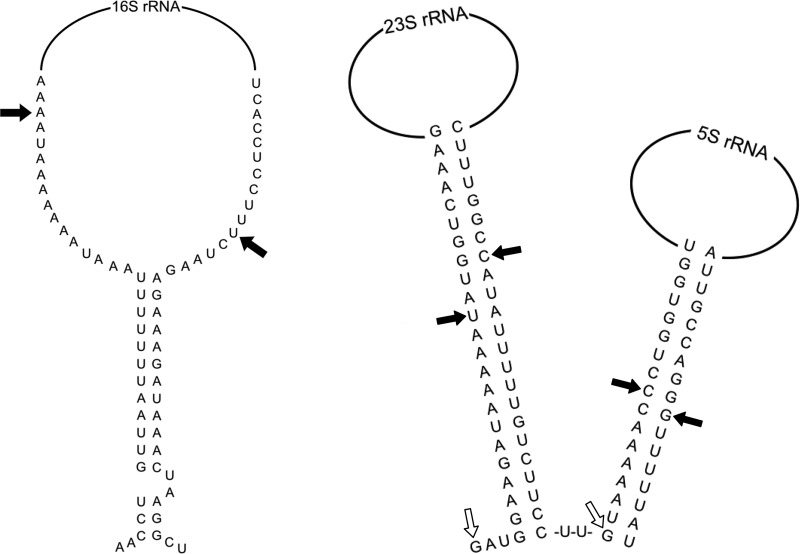
RNase III is a highly conserved endoribonuclease (35, 36, 42, 43, 58). This study is the first to characterize an RNase III homolog from a spirochete. Our results reveal that B. burgdorferi RNase III processes 23S rRNA, transcribed from the unusual tandem 23S-5S rRNA operons, as expected. However, the enzyme is not required to generate the mature 16S rRNA (Fig. 2), which is transcribed from a single gene separated from the 23S-5S rRNA operons. The experimentally determined 16S rRNA ends are not in the predicted double-stranded stem (Fig. 5), which may explain why these ends are not affected by the absence of RNase III.
FIG 5.
A model for rRNA processing in B. burgdorferi. Structures of the rRNA double-stranded stem regions are shown; the 16S rRNA structure (left side) was predicted using MacVector and Mfold (84), and the 23S and 5S rRNA structures (right side) are based on the work of Schwartz et al. (20). Experimentally determined 5′ and 3′ ends for the rRNAs from the RACE data (Fig. 2) are represented by black arrows (wild type) and open arrows (rncBb null mutant); 5′ and 3′ ends for 16S and 5S rRNAs are the same in the wild type and the rncBb mutant.
B. burgdorferi RNase III is more similar to that of the B. subtilis homolog than that of the E. coli homolog, and the palette of endoribonucleases in B. burgdorferi is more similar to the B. subtilis set. B. burgdorferi possesses a homolog of the 5S rRNA-processing enzyme RNase M5 from B. subtilis instead of RNase E from E. coli, in addition to an unannotated homolog of RNase Y (BB0504), which is also found in B. subtilis but not E. coli (M. L. Anacker, unpublished data) (59).
RNase III is essential in B. subtilis (39, 53), and rnc mutants of E. coli as well as those of S. aureus grow at a reduced rate, utilizing alternative RNA processing pathways (60, 61). The B. burgdorferi rnc mutant grows more slowly and reaches a lower cell density at stationary phase than the wild type (Fig. 1C). The growth phenotype may be due to defective ribosomes that are a consequence of incomplete 23S rRNA processing, or it may be due to defective processing of either a regulatory (44) or a housekeeping RNA. Abnormal 30S precursor ribosomal subunits containing partially processed 16S-23S-5S rRNAs are observed in rnc mutants of E. coli and B. subtilis (37, 39, 47, 50–52), and aberrantly long 23S rRNA can interfere with ribosome assembly (62). We were unable to complement the B. burgdorferi rnc mutants despite nearly exhaustive efforts, which may be another consequence of the growth defect; however, this is not uncommon, as complementation of mutants of other nucleic acid-interacting proteins has foundered in the spirochete (63–65).
RNase III also processes mRNA in order to globally regulate transcript levels (35, 41, 42, 66–68). An additional role for RNase III in S. aureus was found in tRNA and ncRNA, as well as mRNA regulation (68), and RNase III in Corynebacterium glutamicum regulates cell division via degradation of mraZ mRNA (44). Loss of mRNA and ncRNA regulation may also contribute to the growth and morphological phenotype of the B. burgdorferi rnc mutant. We hypothesize that the slow-growth and long-cell phenotype is due to delayed or improper cell division, caused by either inefficient translation or production of defective cell division machinery. Intriguingly, the rncBb mutant population also includes cells with a wild-type length, yielding a bimodal distribution (Fig. 1D).
RNase III has a fundamental role in processing of the 16S and 23S rRNAs during ribosome genesis, so we hypothesized that this would be conserved in B. burgdorferi. We assayed processing by determining the 5′ and 3′ ends of the mature rRNAs in the B. burgdorferi rnc null mutant. In other bacteria, endonucleolytic processing occurs cotranscriptionally, first releasing the 16S rRNA and then separating the 23S and 5S rRNAs from the polycistronic transcript (23).
Comparison of wild-type B. burgdorferi and the rncBb null mutant by 5′ and 3′ RACE revealed that the 23S and 5S rRNA transcripts exist in their canonical mature form, in spite of the tandem gene duplication (Fig. 2 and 5). The 23S rRNA is 15 nucleotides longer at the 5′ end and about 18 nucleotides longer at the 3′ end in the rncBb null mutant, which supports a role for RNase III in 23S rRNA processing. The 23S rRNA 5′ end in the rncBb null mutant maps to 34 nucleotides downstream of the predicted promoter, implying that this end is generated by minimal nucleolytic processing by an unknown RNase (Fig. 5). The 23S rRNA 3′ end in the rncBb null mutant maps to the 5′ end of the 5S rRNA unprocessed stem (Fig. 5), suggesting that an RNase, possibly RNase Y, is responsible for processing this end, following RNase M5 cleavage of the 5S rRNA. RT-PCR data showed an increase in the amount of RNA across the junction between the tandem 5S-23S rRNA operons in the rncBb null mutant (M. L. Anacker, A. F. Brinkworth, and D. S. Samuels, unpublished data), implying the presence of a large precursor transcript and a decrease in endonucleolytic processing, which is not without precedent, as a 30S unprocessed rRNA species is observed in both B. subtilis rncS and E. coli rnc null mutants (37, 39, 47, 50–52). On the other hand, there is no difference in the 5′ and 3′ ends of the 5S rRNA transcript between the wild type and rncBb null mutant, which is consistent with processing of 5S rRNA carried out by RNase E in E. coli and RNase M5 in B. subtilis (45, 46).
The 16S rRNA gene is spatially separated from the tandem 23S-5S rRNA operons on the chromosome in B. burgdorferi (5, 17–22). A truncated open reading frame (ORF) (bb0425) is present upstream, and a tRNAAla is downstream of the marooned 16S rRNA gene. RT-PCR data suggest that the 16S rRNA is cotranscribed with the two upstream genes (Anacker et al., unpublished), and Bugrysheva et al. (69) showed that the tRNAAla was cotranscribed with the 16S rRNA gene, yielding a large polycistronic transcript. Data from RACE analyses (Fig. 2) indicate that the 16S rRNA ends are the same in the rncBb null mutant as in the wild type, demonstrating that RNase III is not required for full processing of this rRNA. The experimentally determined ends of the 16S rRNA are not in the predicted double-stranded stem structure (Fig. 5) that potentially could be a cleavage site for RNase III. We propose the following model for generating the observed 16S rRNA 3′ end in the rncBb null mutant. RNase P processing of the tRNAAla 5′ end downstream of the 16S transcript would release a region of single-stranded RNA that undergoes subsequent processing by PNPase, a single-stranded exonuclease, and YbeY, a single-stranded endonuclease, as observed in E. coli (47, 70, 71). The mechanism of 16S rRNA 5′ end maturation is unknown in B. burgdorferi. Our data demonstrate that RNase III in B. burgdorferi processes the 23S rRNA, as expected; other endonucleases, possibly RNase Y and RNase M5, process the 5S rRNA; and a unique and currently undefined mechanism processes the 16S rRNA.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Low-passage-number B. burgdorferi strains B31-A3 (72) and 297 (BbAH130) (73) and the rncBb mutants, were cultivated at 34°C in Barbour-Stoenner-Kelly II (BSK II) liquid medium containing 6% rabbit serum (74) without gelatin (75). Cell density was assayed as previously described, by enumeration using a Petroff-Hausser counting chamber (76). E. coli Rosetta (DE3) (Novagen), used to overexpress recombinant B. burgdorferi RNase III, was grown at 37°C in LB medium containing 50 μg ml−1 ampicillin and 68 μg ml−1 chloramphenicol. E. coli TOP10F′ and DH5α cells, used for cloning, were grown at 37°C in LB medium containing 50 μg ml−1 kanamycin.
Generation of an rncBb null mutant in B. burgdorferi.
The rncBb gene (bb0705) was disrupted by replacement with flgBp-aacC1, which confers gentamicin resistance (72), as previously described (77, 78). Genomic regions encompassing 1.3 kb upstream and 1.3 kb downstream of rncBb were amplified by PCR using Taq polymerase (Sigma-Aldrich), cloned into pCR2.1-TOPO, confirmed by DNA sequencing, and ligated together. The gentamicin resistance cassette was ligated into a synthetic AatII site between the two flanking sequences. The plasmid was linearized with ScaI and electroporated into B. burgdorferi, as previously described (75, 79). Transformants were cloned in liquid BSK II medium in 96-well plates (80) containing 40 μg ml−1 gentamicin at 34°C in a 1.5% CO2 atmosphere and screened by PCR. At 34°C, mutant clones appeared in approximately 90 days, which is considerably longer than the 10 to 14 days typically required. Positive clones were confirmed by reverse transcription-PCR (RT-PCR). Total RNA was isolated using TRIzol (Ambion), and rncBb transcript was measured using a RETROscript kit (Ambion), as previously described (54, 77). The presence of endogenous plasmids was confirmed by PCR (81, 82). Two independent rncBb null mutants from B. burgdorferi strains B31-A3 and 297 were isolated.
Fluorescence microscopy.
Live B. burgdorferi cells were stained using a wheat germ agglutinin (WGA)-Alexa Fluor 594 conjugate (Invitrogen) (78). Briefly, a small volume of cells was washed once in prewarmed Dulbecco's phosphate-buffered saline (dPBS; 138 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, and 1.5 mM KH2PO4) plus 5 mM MgCl2 and resuspended in dPBS containing WGA-Alexa Fluor 594 at 1:200 dilution. The cells were incubated for 5 min at 37°C before pelleting by centrifugation. Cells were resuspended in a small volume of prewarmed dPBS and placed on precleaned slides with coverslips. Images were captured using an Olympus BX51 microscope through a 100× objective equipped with a DP72 digital camera, controlled by DP2-BSW software. Fluorescence images were processed and the length of 50 spirochetes for each strain was measured using ImageJ (National Institutes of Health; http://rsbweb.nih.gov/ij/) (54).
Identification of 5′ and 3′ ends of rRNAs.
The 5′ and 3′ ends of each of the rRNAs (16S, 23S, and 5S) were determined by 5′ and 3′ rapid amplification of cDNA ends (RACE). B. burgdorferi cells were grown to mid-log-phase cell density and lysed with TRIzol; RNA was extracted with chloroform, precipitated with isopropanol, and washed with 70% ethanol, before being resuspended in RNase-free water. Prior to 3′ RACE, total RNA was polyadenylated with a poly(A) tailing kit (Ambion). For the RACE protocol, nested primers and reverse transcriptase from the BD Smart RACE cDNA Amplification kit (Clontech) were utilized to generate cDNA from the total RNA. The cDNA was amplified by PCR, using a universal primer set and gene-specific primers for a sequence either downstream of the 5′ end or upstream of the 3′ end for each rRNA gene (Table 1). PCR products were analyzed on a 2% agarose gel, purified with a QIAquick gel extraction kit (Qiagen), and cloned into pCR2.1-TOPO. Clones were sequenced at the Murdock DNA Sequencing Facility (University of Montana) with an Applied Biosystems genetic analyzer (GeneScan), and sequence data were analyzed using MacVector.
RNA-seq and Northern blotting of rncBb operon.
Directional (strand-specific) RNA-seq cDNA libraries were constructed with a ligation-based protocol and analyzed as described previously (78). Operon predictions were made with DOOR2 (http://csbl.bmb.uga.edu/DOOR/) (83). For Northern blotting, total RNA was fractionated on a formaldehyde-agarose gel, blotted to a nylon membrane, and hybridized with oligonucleotide rnc 158R (Table 1), as previously described (54).
RNase III cleavage assay.
B. burgdorferi RNase III protein was purified using the IMPACT (intein-mediated purification with an affinity chitin-binding tag) kit (New England BioLabs). Briefly, cultures of E. coli carrying pTXB1 with the B. burgdorferi rnc gene were induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C and lysed by sonication. The lysate was centrifuged and loaded on chitin beads and washed with column buffer (20 mM Tris-HCl [pH 8.5], 500 mM NaCl, and 1 mM EDTA). Dithiothreitol (DTT) was added to the column to initiate self-cleavage of the intein tag. The unbound recombinant protein was then eluted from the column using column buffer.
An artificial 23S rRNA stem-loop that serves as an RNase III substrate was generated, as previously described (56); the sequence is composed of the 23S rRNA 5′ and 3′ complementary ends that form the stem portion of the rRNA transcript plus an intervening loop of four unmatched nucleotides. Oligonucleotides incorporating a T7 promoter (Table 1) were used to generate a template for transcribing a 32P-labeled RNA molecule. Oligonucleotides were annealed in water by heating for 5 min at 65°C and then quick cooling on ice. A MEGAscript (Ambion) in vitro transcription reaction with [α-32P]UTP (Perkin-Elmer) was incubated overnight at 37°C. Following subsequent DNase treatment (Turbo DNase), the substrate was resolved in a 15% Tris-borate-EDTA (TBE)–urea–polyacrylamide gel, extracted overnight using a gel elution buffer (0.5 M ammonium acetate, 1 mM EDTA, and 0.2% sodium dodecyl sulfate), and precipitated with ethanol. Detection of the substrate in the gel prior to extraction was performed using a phosphorimager (FLA-3000; Fujifilm). Following precipitation, RNA was resuspended in Tris-EDTA (TE) buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA).
The RNase III cleavage assay was performed as previously described (56, 57). The artificial 23S rRNA substrate was briefly heated at 100°C and rapidly cooled on ice to fold into a stem-loop structure. Substrate RNA (300 nM) and various levels of recombinant RNase III (0 to 25 nM) were combined in cleavage buffer (30 mM Tris-HCl [pH 8.0], 160 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT, and 0.1 mM tRNA) and incubated for 5 min at 37°C to bind enzyme and RNA. Addition of 10 mM MgCl2 initiated the cleavage reaction. Reactions were stopped after 5 min by adding gel loading buffer containing EDTA. Samples were resolved on a 15% (wt/vol) polyacrylamide gel containing 7 M urea and visualized on a phosphorimager.
ACKNOWLEDGMENTS
We thank David Bechhofer and Steve Lodmell for thoughtful and critical reading of the manuscript; David Bechhofer, Amanda Brinkworth, Mary Ellenbecker, Jean-Marc Lanchy, Steve Lodmell, Niko Popitsch, Philipp Rescheneder, Paula Schlax, and Kit Tilly for useful discussions; the Minnick lab for the use of their microscope; Patty McIntire for DNA sequencing; and Laura Hall for excellent technical assistance. We are profoundly grateful to the anonymous reviewers for constructive suggestions.
This work was supported by Public Health Service grants R01 AI051486 to D.S.S. and P20 GM103546 to Stephen R. Sprang (Center for Biomolecular Structure and Dynamics) from the National Institutes of Health. R.D.L. was supported by an Undergraduate Research Award from the Davidson Honors College, a scholarship through the Montana Space Grant Consortium under grant NNX10AJ83H from NASA, a Student Research Grant from the Montana Academy of Sciences, and an Interim Research Award through the Montana Integrative Learning Experience for Students (MILES) program under grant 52005905 from the Howard Hughes Medical Institute-Undergraduate Science Education Program.
REFERENCES
- 1.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 2.Benach JL, Bosler EM, Hanrahan JP, Coleman JL, Bast TF, Habicht GS, Cameron DJ, Ziegler JL, Barbour AG, Burgdorfer W, Edelman R, Kaslow RA. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N Engl J Med 308:740–742. doi: 10.1056/NEJM198303313081302. [DOI] [PubMed] [Google Scholar]
- 3.Steere AC, Grodzicki RL, Kornblatt AN, Craft JE, Barbour AG, Burgdorfer W, Schmid GP, Johnson E, Malawista SE. 1983. The spirochetal etiology of Lyme disease. N Engl J Med 308:733–740. doi: 10.1056/NEJM198303313081301. [DOI] [PubMed] [Google Scholar]
- 4.Barbour AG, Garon CF. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 5.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J-F, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quakenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MK, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Casjens SR, Eggers CH, Schwartz I. 2010. Borrelia genomics: chromosome, plasmids, bacteriophages and genetic variation, p 27–53. In Samuels DS, Radolf JD (ed), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 7.Brisson D, Drecktrah D, Eggers CH, Samuels DS. 2012. Genetics of Borrelia burgdorferi. Annu Rev Genet 46:515–536. doi: 10.1146/annurev-genet-011112-112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker NS, Margos G, Blum H, Krebs S, Graf A, Lane RS, Castillo-Ramírez S, Sing A, Fingerle V. 2016. Recurrent evolution of host and vector association in bacteria of the Borrelia burgdorferi sensu lato species complex. BMC Genomics 17:734. doi: 10.1186/s12864-016-3016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caimano MJ, Drecktrah D, Kung F, Samuels DS. 2016. Interaction of the Lyme disease spirochete with its tick vector. Cell Microbiol 18:919–927. doi: 10.1111/cmi.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens SR, Gilcrease EB, Vujadinovic M, Mongodin EF, Luft BJ, Schutzer SE, Fraser CM, Qiu W-G. 2017. Plasmid diversity and phylogenetic consistency in the Lyme disease agent Borrelia burgdorferi. BMC Genomics 18:165. doi: 10.1186/s12864-017-3553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piesman J, Schwan TG. 2010. Ecology of borreliae and their arthropod vectors, p 251–278. In Samuels DS, Radolf JD (ed), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 13.Andersson SGE, Kurland CG. 1995. Genomic evolution drives the evolution of the translation system. Biochem Cell Biol 73:775–787. doi: 10.1139/o95-086. [DOI] [PubMed] [Google Scholar]
- 14.Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. doi: 10.1016/S0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 15.Batut B, Knibbe C, Marais G, Daubin V. 2014. Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol 12:841–850. doi: 10.1038/nrmicro3331. [DOI] [PubMed] [Google Scholar]
- 16.Martínez-Cano DJ, Reyes-Prieto M, Martínez-Romero E, Partida-Martínez LP, Latorre A, Moya A, Delaye L. 2015. Evolution of small prokaryotic genomes. Front Microbiol 5:742. doi: 10.3389/fmicb.2014.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson BE, MacDougall J, Saint Girons I. 1992. Physical map of the linear chromosome of the bacterium Borrelia burgdorferi 212, a causative agent of Lyme disease, and localization of rRNA genes. J Bacteriol 174:3766–3774. doi: 10.1128/jb.174.11.3766-3774.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukunaga M, Sohnaka M. 1992. Tandem repeat of the 23S and 5S ribosomal RNA genes in Borrelia burgdorferi, the etiological agent of Lyme disease. Biochem Biophys Res Commun 183:952–957. doi: 10.1016/S0006-291X(05)80282-7. [DOI] [PubMed] [Google Scholar]
- 19.Fukunaga M, Yanagihara Y, Sohnaka M. 1992. The 23S/5S ribosomal RNA genes (rrl/rrf) are separate from the 16S ribosomal RNA gene (rrs) in Borrelia burgdorferi, the aetiological agent of Lyme disease. J Gen Microbiol 138:871–877. doi: 10.1099/00221287-138-5-871. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz JJ, Gazumyan A, Schwartz I. 1992. rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. J Bacteriol 174:3757–3765. doi: 10.1128/jb.174.11.3757-3765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazumyan A, Schwartz JJ, Liveris D, Schwartz I. 1994. Sequence analysis of the ribosomal RNA operon of the Lyme disease spirochete, Borrelia burgdorferi. Gene 146:57–65. doi: 10.1016/0378-1119(94)90833-8. [DOI] [PubMed] [Google Scholar]
- 22.Marconi RT, Liveris D, Schwartz I. 1995. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol 33:2427–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gegenheimer P, Apirion D. 1975. Escherichia coli ribosomal ribonucleic acids are not cut from an intact precursor molecule. J Biol Chem 250:2407–2409. [PubMed] [Google Scholar]
- 24.Nomura M, Morgan EA, Jaskunas SR. 1977. Genetics of bacterial ribosomes. Annu Rev Genet 11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- 25.Apirion D, Gegenheimer P. 1981. Processing of bacterial RNA. FEBS Lett 125:1–9. doi: 10.1016/0014-5793(81)80984-2. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava AK, Schlessinger D. 1990. Mechanism and regulation of bacterial ribosomal RNA processing. Annu Rev Microbiol 44:105–129. doi: 10.1146/annurev.mi.44.100190.000541. [DOI] [PubMed] [Google Scholar]
- 27.Deutscher MP. 2009. Maturation and degradation of ribosomal RNA in bacteria. Prog Mol Biol. Transl Sci 85:369–391. doi: 10.1016/S0079-6603(08)00809-X. [DOI] [PubMed] [Google Scholar]
- 28.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct 4:13. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Criswell D, Tobiason VL, Lodmell JS, Samuels DS. 2006. Mutations conferring aminoglycoside and spectinomycin resistance in Borrelia burgdorferi. Antimicrob Agents Chemother 50:445–452. doi: 10.1128/AAC.50.2.445-452.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbour AG, Putteet-Driver AD, Bunikis J. 2005. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect Immun 73:6165–6168. doi: 10.1128/IAI.73.9.6165-6168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer SL, Flechtner VR, Johansen JR. 2001. Is the 16S-23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol 18:1057–1069. doi: 10.1093/oxfordjournals.molbev.a003877. [DOI] [PubMed] [Google Scholar]
- 32.Pei A, Li H, Oberdorf WE, Alekseyenko AV, Parsons T, Yang L, Gerz EA, Lee P, Xiang C, Nossa CW, Pei Z. 2012. Diversity of 5S rRNA genes within individual prokaryotic genomes. FEMS Microbiol Lett 335:11–18. doi: 10.1111/j.1574-6968.2012.02632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. 2015. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson HD, Webster RE, Zinder ND. 1968. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem 243:82–91. [PubMed] [Google Scholar]
- 35.Court DL, Gan J, Liang Y-H, Shaw GX, Tropea JE, Costantino N, Waugh DS, Ji X. 2013. RNase III: genetics and function; structure and mechanism. Annu Rev Genet 47:405–431. doi: 10.1146/annurev-genet-110711-155618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson AW. 2014. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip Rev RNA 5:31–48. doi: 10.1002/wrna.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikolaev N, Silengo L, Schlessinger D. 1973. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A 70:3361–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolaev N, Silengo L, Schlessinger D. 1973. A role for ribonuclease III in processing of ribosomal ribonucleic acid and messenger ribonucleic acid precursors in Escherichia coli. J Biol Chem 248:7967–7969. [PubMed] [Google Scholar]
- 39.Herskovitz MA, Bechhofer DH. 2000. Endoribonuclease RNase III is essential in Bacillus subtilis. Mol Microbiol 38:1027–1033. doi: 10.1046/j.1365-2958.2000.02185.x. [DOI] [PubMed] [Google Scholar]
- 40.Gegenheimer P, Apirion D. 1980. Precursors to 16S and 23S ribosomal RNA from a ribonuclease III− strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res 8:1873–1891. doi: 10.1093/nar/8.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 42.Kaberdin VR, Singh D, Lin-Chao S. 2011. Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci 18:23. doi: 10.1186/1423-0127-18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hui MP, Foley PL, Belasco JG. 2014. Messenger RNA degradation in bacterial cells. Annu Rev Genet 48:537–559. doi: 10.1146/annurev-genet-120213-092340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeda T, Tanaka Y, Takemoto N, Hamamoto N, Inui M. 2016. RNase III mediated cleavage of the coding region of mraZ mRNA is required for efficient cell division in Corynebacterium glutamicum. Mol Microbiol 99:1149–1166. doi: 10.1111/mmi.13295. [DOI] [PubMed] [Google Scholar]
- 45.Sogin ML, Pace B, Pace NR. 1977. Partial purification and properties of a ribosomal RNA maturation endonuclease from Bacillus subtilis. J Biol Chem 252:1350–1357. [PubMed] [Google Scholar]
- 46.Misra TK, Apirion D. 1979. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem 254:11154–11159. [PubMed] [Google Scholar]
- 47.Gegenheimer P, Watson N, Apirion D. 1977. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem 252:3064–3073. [PubMed] [Google Scholar]
- 48.King TC, Schlessinger D. 1983. S1 nuclease mapping analysis of ribosomal RNA processing in wild type and processing deficient Escherichia coli. J Biol Chem 258:12034–12042. [PubMed] [Google Scholar]
- 49.King TC, Sirdeshmukh R, Schlessinger D. 1984. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci U S A 81:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babitzke P, Granger L, Olszewski J, Kushner SR. 1993. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol 175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettijohn DE, Stonington OG, Kossman CR. 1970. Chain termination of ribosomal RNA synthesis in vitro. Nature 228:235–239. doi: 10.1038/228235a0. [DOI] [PubMed] [Google Scholar]
- 52.Dunn JJ, Studier FW. 1973. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease III. Proc Natl Acad Sci U S A 70:3296–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durand S, Gilet L, Condon C. 2012. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet 8:e1003181. doi: 10.1371/journal.pgen.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lybecker MC, Abel CA, Feig AL, Samuels DS. 2010. Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 78:622–635. doi: 10.1111/j.1365-2958.2010.07374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lybecker M., Bilusic I., Raghavan R. 2014. Pervasive transcription: detecting functional RNAs in bacteria. Transcription 5:e944039. doi: 10.4161/21541272.2014.944039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amarasinghe AK, Calin-Jageman I, Harmouch A, Sun W, Nicholson AW. 2001. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol 342:143–158. doi: 10.1016/S0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 57.Meng W, Nicholson RH, Nathania L, Pertzev AV, Nicholson AW. 2008. New approaches to understanding double-stranded RNA processing by ribonuclease III: purification and assays of homodimeric and heterodimeric forms of RNase III from bacterial extremophiles and mesophiles. Methods Enzymol 447:119–129. doi: 10.1016/S0076-6879(08)02207-6. [DOI] [PubMed] [Google Scholar]
- 58.Nicholson AW. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev 23:371–190. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 59.Archambault L, Borchert JS, Bergeron J, Snow S, Schlax PJ. 2013. Measurements of mRNA degradation in Borrelia burgdorferi. J Bacteriol 195:4879–4887. doi: 10.1128/JB.00659-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, Jacquier A, Vandenesch F, Romby P. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Resch A, Afonyushkin T, Lombo TB, McDowall KJ, Bläsi U, Kaberdin VR. 2008. Translational activation by the noncoding RNA DsrA involves alternative RNase III processing in the rpoS 5′-leader. RNA 14:454–459. doi: 10.1261/rna.603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gutgsell NS, Jain C. 2012. Gateway role for rRNA precursors in ribosome assembly. J Bacteriol 194:6875–6882. doi: 10.1128/JB.01467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dresser AR, Hardy PO, Chaconas G. 2009. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog 5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. 2009. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog 5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salman-Dilgimen A, Hardy PO, Dresser AR, Chaconas G. 2011. HrpA, a DEAH-box RNA helicase, is involved in global gene regulation in the Lyme disease spirochete. PLoS One 6:e22168. doi: 10.1371/journal.pone.0022168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gatewood ML, Jones GH. 2012. Expression of a polycistronic messenger RNA involved in antibiotic production in an rnc mutant of Streptomyces coelicolor. Arch Microbiol 194:147–155. doi: 10.1007/s00203-011-0740-7. [DOI] [PubMed] [Google Scholar]
- 67.Lim B, Sim S-H, Sim M, Kim K, Jeon CO, Lee Y, Ha N-C, Lee K. 2012. RNase III controls the degradation of corA mRNA in Escherichia coli. J Bacteriol 194:2214–2220. doi: 10.1128/JB.00099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lioliou E, Sharma CM, Caldelari I, Helfer A-C, Fechter P, Vandenesch F, Vogel J, Romby P. 2012. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8:e1002782. doi: 10.1371/journal.pgen.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bugrysheva JV, Godfrey HP, Schwartz I, Cabello FC. 2011. Patterns and regulation of ribosomal RNA transcription in Borrelia burgdorferi. BMC Microbiol 11:17. doi: 10.1186/1471-2180-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, RajBhandary UL, Walker GC. 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol 78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jacob AI, Köhrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98:12724–12729. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 75.Samuels DS, Drecktrah D, Hall LS. 2018. Genetic transformation and complementation, p 183–200. In Pal U, Buyuktanir O (ed), Borrelia burgdorferi: methods and protocols, vol 1690 Humana Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galbraith KM, Ng AC, Eggers BJ, Kuchel CR, Eggers CH, Samuels DS. 2005. parC mutations in fluoroquinolone-resistant Borrelia burgdorferi. Antimicrob Agents Chemother 49:4354–4357. doi: 10.1128/AAC.49.10.4354-4357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS, Drecktrah D. 2012. Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol Med Microbiol 66:157–165. doi: 10.1111/j.1574-695X.2012.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Drecktrah D, Lybecker M, Popitsch N, Rescheneder P, Hall LS, Samuels DS. 2015. The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. PLoS Pathog 11:e1005160. doi: 10.1371/journal.ppat.1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Drecktrah D, Hall LS, Hoon-Hanks LL, Samuels DS. 2013. An inverted repeat in the ospC operator is required for induction in Borrelia burgdorferi. PLoS One 8:e68799. doi: 10.1371/journal.pone.0068799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med 199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Labandeira-Rey M, Skare JT. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect Immun 69:446–455. doi: 10.1128/IAI.69.1.446-455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res 37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg120. [DOI] [PMC free article] [PubMed] [Google Scholar]