ABSTRACT
The DNA damage response ddrI gene encodes a transcription regulator belonging to the cAMP receptor protein (CRP) family. Cells devoid of the DdrI protein exhibit a pleiotropic phenotype, including growth defects and sensitivity to DNA-damaging agents and to oxidative stress. Here, we show that the absence of the DdrI protein also confers sensitivity to heat shock treatment, and several genes involved in heat shock response were shown to be upregulated in a DdrI-dependent manner. Interestingly, expression of the Escherichia coli CRP partially compensates for the absence of the DdrI protein. Microscopic observations of ΔddrI mutant cells revealed an increased proportion of two-tetrad and anucleated cells in the population compared to the wild-type strain, indicating that DdrI is crucial for the completion of cell division and/or chromosome segregation. We show that DdrI is also involved in the megaplasmid MP1 stability and in efficient plasmid transformation by facilitating the maintenance of the incoming plasmid in the cell. The in silico prediction of putative DdrI binding sites in the D. radiodurans genome suggests that hundreds of genes, belonging to several functional groups, may be regulated by DdrI. In addition, the DdrI protein absolutely requires cAMP for in vitro binding to specific target sequences, and it acts as a dimer. All these data underline the major role of DdrI in D. radiodurans physiology under normal and stress conditions by regulating, both directly and indirectly, a cohort of genes involved in various cellular processes, including central metabolism and specific responses to diverse harmful environments.
IMPORTANCE Deinococcus radiodurans has been extensively studied to elucidate the molecular mechanisms responsible for its exceptional ability to withstand lethal effects of various DNA-damaging agents. A complex network, including efficient DNA repair, protein protection against oxidation, and diverse metabolic pathways, plays a crucial role for its radioresistance. The regulatory networks orchestrating these various pathways are still missing. Our data provide new insights into the crucial contribution of the transcription factor DdrI for the D. radiodurans ability to withstand harmful conditions, including UV radiation, mitomycin C treatment, heat shock, and oxidative stress. Finally, we highlight that DdrI is also required for accurate cell division, for maintenance of plasmid replicons, and for central metabolism processes responsible for the overall cell physiology.
KEYWORDS: Deinococcus radiodurans, CRP family, cAMP signaling, transcription regulator, heat shock response, plasmid maintenance
INTRODUCTION
The molecular mechanisms contributing to the impressive capacity of Deinococcus radiodurans to withstand very harmful genotoxic stresses, including ionizing and UV radiation, desiccation, and reactive oxygen species (ROS), are now well documented (for recent reviews, see references 1–4). These include very efficient DNA repair mechanisms encoded by D. radiodurans to reassemble an intact genome from hundreds of DNA fragments (5–7) and a compact structure of the nucleoid maintained after irradiation which may favor DNA double-strand break (DSB) repair by avoiding dispersion of DNA fragments (8, 9). D. radiodurans also possesses very efficient ROS detoxification pathways, including both enzymatic and nonenzymatic systems (for reviews, see references 2 and 10) and protects its genome by limiting iron import and concomitant Fenton reactions (11). In addition, the radiation-resistant Deinococcus species share an upregulation of specific DNA damage response (ddr) genes following irradiation or desiccation (12). Recently, a screening of a transposon insertion mutant library exposed to different genotoxic stresses has identified a combination of various metabolic and structural functions involved in the extreme radioresistance of D. radiodurans (13). Nevertheless, the regulatory networks orchestrating these various pathways remain to be elucidated.
Although D. radiodurans encodes two homologues of the Escherichia coli SOS response LexA repressor, LexA1 and LexA2, with self-protease activity stimulated by a mechanism requiring RecA, the physiological targets of these two potential repressors are still unidentified (14–16). Instead, another regulation pathway involving the IrrE metalloprotease and its cognate substrate, the DdrO repressor, has been shown to play a major regulatory role after exposure to ionizing radiation in both Deinococcus deserti (17) and D. radiodurans (18, 19). The IrrE metalloprotease from D. deserti cleaves the DdrO protein in response to gamma-ray exposure, allowing the induction of genes involved in DNA DSB repair (17). This pathway controls the radiation desiccation response (RDR) regulon, shared by several Deinococcus species (20, 21), and a regulatory RDR motif (RDRM) palindromic sequence was found upstream of the coding sequence of the RDR regulon members. In D. radiodurans, this regulon includes at least recA, gyrA, and Deinococcus-specific genes, like the ddrB, pprA, and ddrC genes. ddrB encodes an essential function for the single-strand annealing (SSA) repair mechanism (7, 22). All of these genes are repressed by DdrO in the absence of any genotoxic stress (18, 23, 24).
Another transcriptional regulator, DrRRA, crucial for the extreme resistance of D. radiodurans to gamma and UV irradiation, H2O2 exposure, and desiccation has been identified (25). It belongs to the response regulators of two-component systems, but its cognate histidine kinase is still unknown. Transcriptome analyses of the ΔdrRRA mutant revealed impaired induction of numerous genes after gamma-ray exposure, including recA, pprA, and uvr (ultraviolet radiation) DNA repair genes; katA and katE, encoding catalases; sodA and sodC, encoding superoxide dismutases; and genes encoding thioredoxin and ferritin/Dps-like proteins (25).
The ddrI gene (dr0997) was initially identified by transcriptome analyses of D. radiodurans as induced 4-fold following gamma-ray exposure (3 kilogray [kGy]) and 11-fold after desiccation (12). We have recently demonstrated that inactivation of ddrI by transposon insertion sensitizes D. radiodurans to gamma and UV irradiation, mitomycin C (MMC), and H2O2 (13). Yang et al. confirmed that a deletion of the ddrI gene sensitizes cells to these harmful conditions, suggesting an important role of DdrI in the cellular response of D. radiodurans to DNA damage and oxidative stress (26). Moreover, the absence of DdrI results in slow bacterial growth (26). The transcription of ddrI is induced by the response regulator DrRRA following gamma irradiation, and purified DrRRA binds to the ddrI promoter region (25). The DdrI protein exhibits typical features of the transcription factors belonging to the cAMP receptor protein (CRP) family, including an effector cyclic nucleotide monophosphate (cNMP)-binding domain at the N-terminal end, and a helix-turn-helix (HTH) DNA-binding domain at the C-terminal end (26). In E. coli, the expression of several dozen genes is regulated by CRP, including many genes involved in central carbon metabolism and transport of various metabolites (27). In D. radiodurans, it has been shown that the transcription of 18 genes encoding proteins mainly involved in metabolic pathways or DNA repair mechanisms is directly regulated by the DdrI protein (26).
In this study, we showed that the E. coli CRP can partially restore resistance to DNA-damaging agents and oxidative stress and complements the growth defect of D. radiodurans cells devoid of the DdrI protein. The in silico prediction of potential DdrI binding sites in the D. radiodurans genome suggests that hundreds of genes, belonging to several functional groups, may be regulated by DdrI. We showed that DdrI is involved in the heat shock response, regulation of cell division and/or chromosome segregation, stability of the megaplasmid MP1, and efficient plasmid transformation by facilitating the maintenance of the incoming plasmid in the cell. Moreover, we demonstrate that DdrI absolutely requires cAMP for in vitro binding to its specific DNA target and acts as a dimer. All these data underline the important regulatory role of DdrI in the adaptation of D. radiodurans to various stresses.
RESULTS
The ddrI locus is transcribed as a monocistronic unit.
The ddrI locus dr0997 is located 178 bp upstream of a putative operon starting from dr0998 and ending at dr1000 (see Fig. S1 in the supplemental material), putatively encoding a member of the multiresistance and pH adaptation (MRP) family, a putative transcription regulator, and a 2′-5′ RNA ligase, respectively. Whereas a stem-loop structure forming a Rho-independent transcription terminator was found downstream of dr1000, there was no evidence for the presence of such a transcription terminator in the intergenic region between dr0997 and dr0998. To test if a ddrI deletion interferes with the expression of downstream genes, we assessed the presence of a putative operon from dr0997 to dr1000. The reverse transcription-PCR (RT-PCR) products for each coding sequence, as well as fragments overlapping dr0998 and dr0999 or dr0999 and dr1000, were amplified using cDNA synthesized from total D. radiodurans RNA as the template. No RT-PCR product was detected with one primer located in dr0997 and the other in dr0998, whereas an internal fragment of dr0997 was efficiently amplified (Fig. S1). Moreover, RT-PCR products beginning in dr0998 and ending in dr0999 or beginning in dr0999 and ending in dr1000 were amplified when dr0997 was deleted (Fig. S1). Therefore, dr0997 appeared to be transcribed as a monocistronic unit, upstream of the operon starting from dr0998 and ending at dr1000.
Expression of the E. coli CRP partially complemented growth defects and sensitivity to various stresses of the D. radiodurans ddrI deletion mutant.
As previously shown by Yang et al. (26), the ΔddrI null mutant exhibited a dramatic growth deficiency fully complemented by expression of the DdrI protein (Fig. 1). Interestingly, we show here that trans expression of the E. coli CRP in cells devoid of the DdrI protein resulted in a partial restoration of growth, with cells exhibiting a doubling time of 195 (±6) min compared to 304 (±10) min for the ΔddrI mutant strain and 120 (±9) min for the wild-type parental R1 strain when cultivated under standard growth conditions (Fig. 1). We also show that heterologous expression of the E. coli CRP in the ΔddrI mutant partially restored resistance to genotoxic agents, such as MMC and ultraviolet radiation (UVC) (Fig. 2A), and to oxidative stress, as shown by the diameters of the growth inhibition zones surrounding discs on which Paraquat (dimethyl-4,4′-bipyridinium dichloride) was spotted (Fig. 2B). These results strongly suggest that the E. coli CRP shared several common targets with the DdrI protein and regulated gene expression to restore efficient cell growth and resistance to harmful conditions.
FIG 1.

The ΔddrI mutant displays severe growth defects partially complemented by the expression of the E. coli CRP. GY9613 (wild type [WT]), GY13630 (ΔddrI mutant), GY13643 (ΔddrI/ddrI+ complemented strain), and GY13644 (ΔddrI/crp+ mutant) strains were grown at 30°C in TGY2X medium (supplemented with spectinomycin and 1 mM IPTG for the GY13644 strain). The A650 values of the cultures were measured and generation times calculated from 3 independent experiments.
FIG 2.
Expression of E. coli CRP in the ΔddrI mutant partially restores resistance to UVC, mitomycin C (MMC), and Paraquat. (A) Sensitivity assays of D. radiodurans to UVC and mitomycin C (MMC). GY9613 (wild type), GY13630 (ΔddrI mutant), GY13643 (ΔddrI/ddrI+ complemented strain), and GY13644 (ΔddrI/crp+ mutant) strains were grown to an A650 of 1, and appropriate dilutions (top) were spotted onto TGY agar plates exposed or not exposed to UVC rays (600 J/m2) or MMC (40 ng/ml). TGY agar plates were supplemented with spectinomycin and 1 mM IPTG for the GY13644 strain. (B) Sensitivity assays of D. radiodurans to oxidative stress generated by Paraquat. Exponential-phase cultures (A650, 0.5) of GY9613, GY13630, GY13643, and GY13644 were spread on TGY plates or TGY agar plates supplemented with spectinomycin and 1 mM IPTG for strain GY13644, and 10 μl of Paraquat (at the indicated concentrations, left) was spotted onto filter paper discs. After incubation at 30°C for 2 days, the diameters of the growth inhibition zones were measured.
In silico prediction of potential DdrI targets.
The complementation experiments described above suggest a high similarity between the binding sites of E. coli CRP and the D. radiodurans DdrI protein. Potential DdrI sites were therefore predicted using CRP sites determined experimentally. The E. coli CRP box is characterized by the 5′-TGTGA-N6-TCACA-3′ consensus sequence (the most conserved bases are underlined) (27). The DNA motifs recognized by CRP homologs are well conserved among a variety of bacterial species, including the GlxR factor of Corynebacterium glutamicum (28), the CRP of Mycobacterium tuberculosis (CRPMt) (29), and the CRP of Haemophilus influenzae (30) (Fig. S2). CRP and related regulators are able to recognize and bind to relatively divergent sites, reflected by the relatively low informational content of their consensus sequence.
The in silico prediction of potential DdrI targets in the D. radiodurans genome was performed using the Fast Investigation Tool for Bacterial and Archeal Regulons (FITBAR) Web tool (http://archaea.u-psud.fr/fitbar/) (31) dedicated to the identification of specific protein-DNA-binding sites on fully sequenced prokaryotic genomes. For this analysis, we used the “CRP-E. coli” matrix, which contains the set of 166 intergenic CRP target sites listed in RegulonDB (http://regulondb.ccg.unam.mx/), identified by genomic SELEX (27), and experimentally validated. The information carried by the compilation of these 166 CRP binding sites weighs only 7.29 bits, as calculated according to Schneider et al. (32).
A total of 176 sites located upstream of genes at distances <650 bp were found in intergenic regions (Table S1). These potential DdrI targets may regulate the expression of hundreds of genes, including monocistronic units or genes belonging to operons located downstream of these putative DdrI targets, and at least 68 divergent genes indicated in Table S1 (Hua and Hua [33]; GenBank accession numbers CP015081.1 to CP015081.4). These target genes are clustered into several functional categories, including DNA replication and repair, responses to oxidative stress and heat shock, signal transduction, carbohydrate metabolism, and transport, cell wall biogenesis, and lipid metabolism (Table S1). A matrix based on the set of these 176 potential DdrI sites was used in a second prediction to increase the informational content of the consensus sequence to 9.32 bits. With this additional information, 115 sites located upstream of genes at distances <650 bp were found in intergenic regions (Table S2). Among these 115 sites, 96 were among those present in Table S1, and 19 new sites were found when the 176 D. radiodurans binding sites were used to build the matrix.
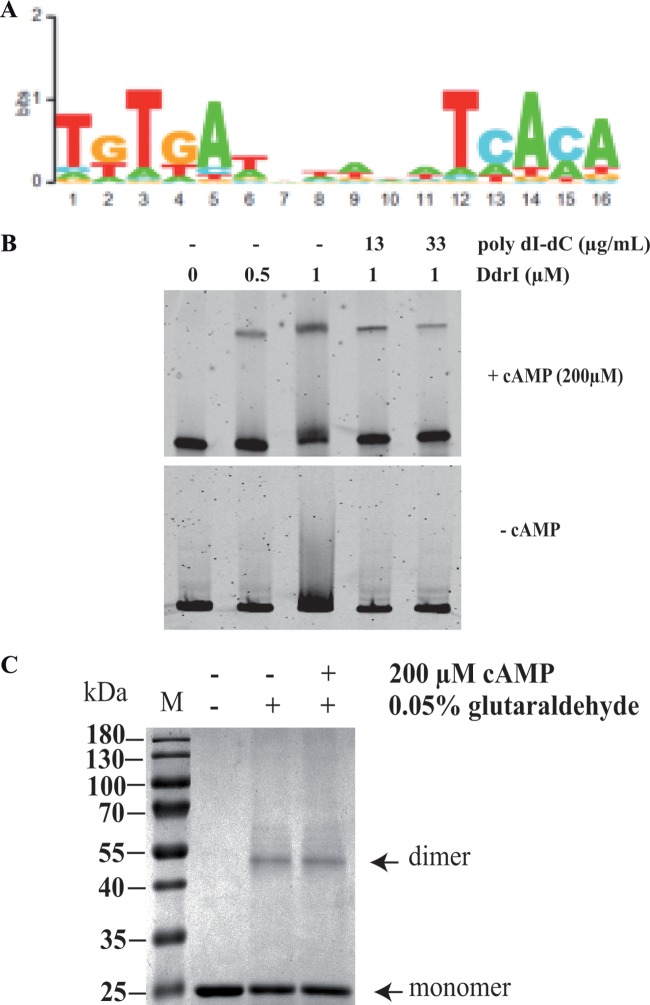
We then defined a D. radiodurans consensus sequence [5′-TGTGA(N6)TCACA-3′] (the palindromic sequence found in the consensus sequence is underlined) extrapolated by FITBAR from the 115 putative DdrI binding sites described in Table S2 (Fig. 3A). To assess the biological significance of this consensus sequence, we tested the ability of DdrI protein to bind to an 80-bp fragment containing, at position 33, the 5′-TGTGAAAAAACTCACA-3′ sequence related to the DdrI putative recognition sequence. For this purpose, we performed electrophoretic mobility shift assay (EMSA) analysis using a purified DdrI protein containing an N-terminal His6 tag, shown to be able to complement in vivo ΔddrI mutant deficiencies (Fig. S3). We found that a band shift is observed using the His6::DdrI protein, even in the presence of a large amount of a poly(dI-dC) competitor, but only when cAMP was added, as commonly observed for many E. coli CRP homologs (Fig. 3B). We also tested the binding of the His6::DdrI protein to a variant of this consensus sequence (5′-TGTTAAAAAACTTACA-3′) (the nucleotides different from those found in the consensus sequence are indicated in bold), which is present in the promoter region of the sodC (dr1546) gene, listed in Tables S1 and S2, and showed that the His6::DdrI protein also bound to this variant (Fig. S4).
FIG 3.
DdrI needs cAMP for binding a DNA fragment containing the predicted DdrI binding consensus. (A) The sequence logo of the putative DdrI binding site was deduced from the second round of DdrI target predictions by using a matrix based on the 115 potential DdrI binding sites found in the D. radiodurans genome (Table S2). (B) Electrophoretic mobility shift assays performed with an 80-bp DNA fragment containing the predicted DdrI binding site and the recombinant His6::DdrI protein (0.5 or 1 μM), in presence (+) or absence (−) of cAMP (200 μM). (C) DdrI protein (3 μg) was incubated with 0.05% glutaraldehyde in the presence or absence of cAMP at a final concentration of 200 μM. After migration, the gel was stained with Coomassie blue. M, molecular mass marker.
The putative DdrI recognition sequence being palindromic, we tested the ability of the DdrI protein to form dimers and any involvement of cAMP in the dimerization process. We observed that DdrI was able to form dimers without the requirement of cAMP (Fig. 3C).
The DdrI regulon includes genes involved in response to heat shock.
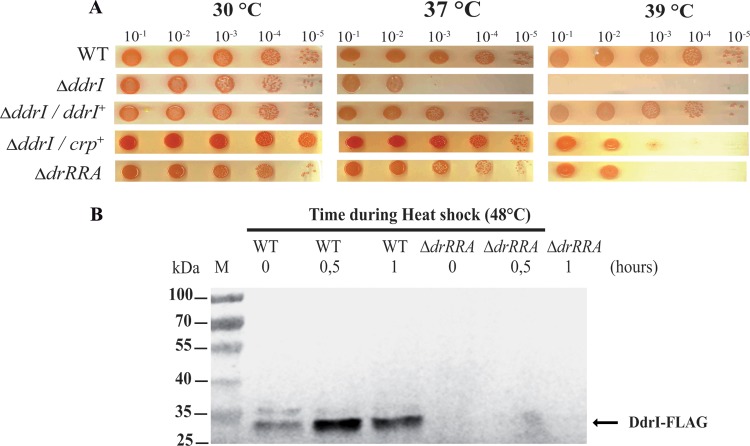
The ddrI gene was previously found among the 25 most highly upregulated genes upon heat shock treatment, as determined by microarray analysis (34). We therefore chose to investigate the effect of a ddrI deletion on the sensitivity of D. radiodurans to heat shock treatment. We found that cell viability decreased 1,000-fold in the absence of DdrI at 37°C, compared to wild-type cells grown under the same conditions, and cells devoid of DdrI were unable to grow at 39°C (Fig. 4A). The thermosensitive phenotype of the ΔddrI mutant was completely suppressed by the expression of a chromosomal ectopic ddrI gene and, to a lesser extent, by plasmid-based expression of the heterologous CRP from E. coli (Fig. 4A). Compared to the wild-type strain, the expression of the ddrI gene was shown to decrease in a drRRA mutant after DNA-damaging treatment (25). Therefore, the expression of DdrI was studied by Western immunoblot analyses of cell extracts from the R1 or ΔdrRRA mutant strain expressing the DdrI-FLAG-tagged protein when bacteria were exposed to heat shock. We also verified the functionality of the DdrI-FLAG protein to complement the ddrI deletion in vivo (Fig. S3). The expression of the DdrI-FLAG protein was induced in response to heat shock (0.5 or 1 h at 48°C) in the wild-type background, whereas in the ΔdrRRA mutant, the DdrI-FLAG protein was undetectable at 30°C, as well as after heat shock treatment, indicating a DrRRA-dependent expression of the DdrI protein (Fig. 4B). Thus, we tested the heat sensitivity of cells devoid of DrRRA and found that they also exhibited a thermosensitive phenotype at 39°C (Fig. 4A).
FIG 4.
The ΔddrI mutant exhibits a highly temperature-sensitive phenotype and induction of DdrI in response to heat shock that is dependent on the response regulator DrRRA. (A) Serial dilutions of GY9613 (wild type), GY13630 (ΔddrI mutant), GY13643 (ΔddrI/ddrI+ complemented strain), GY13644 (ΔddrI/crp+ mutant), and GY13681 (ΔdrRRA mutant) bacteria were spotted on TGY agar plates, and the plates were incubated at 30°C, 37°C, or 39°C for 3 to 5 days. (B) Exponential-phase cultures (A650, 0.45) of GY13663 (drRRA+ ddrI-FLAG mutant) and GY13679 (ΔdrRRA ddrI-FLAG mutant) bacteria grown at 30°C were transferred to 48°C for 0.5 or 1 h before Western blot analysis with anti-FLAG antibodies of their protein extracts (10 μg). Time zero corresponds to protein extracts prepared just before transfer to 48°C.
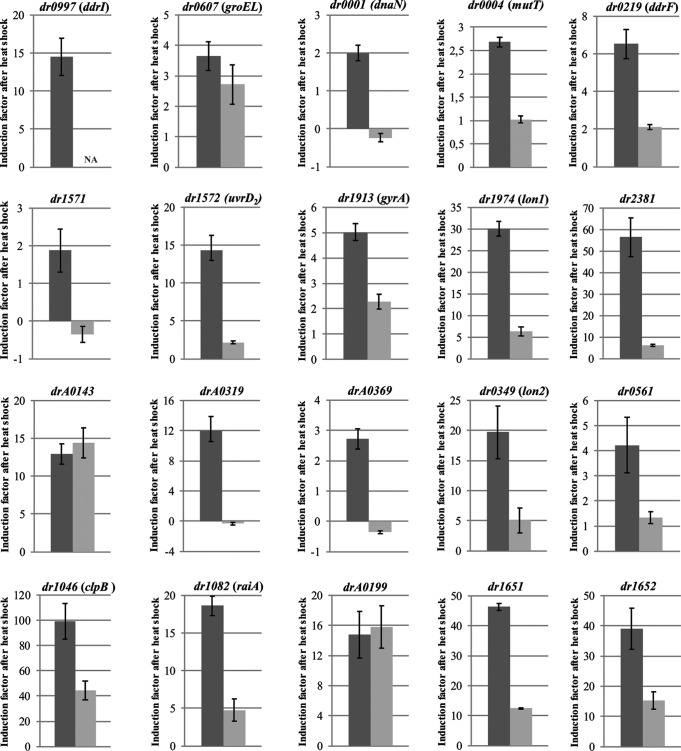
To test a possible DdrI-dependent expression of heat shock genes, we compared the target genes of DdrI predicted in silico (Tables S1 and S2) with the genes or proteins whose expression was found to be increased by heat shock (34, 35). We chose to study in wild-type and DdrI-deficient cells at 30°C and after 5 min of incubation of the cells at 48°C, by quantitative reverse transcription-PCR (RT-qPCR), the expression of genes that appeared from both approaches. We also included dr0004 in our study since many proteins of the Nudix family were found to be induced by heat shock treatment. We found that dr0001 (dnaN), dr0004 (mutT), dr0219 (ddrF), dr1571, dr1572 (uvrD2), dr1913 (gyrA), dr1974 (lon1), dr2381, drA0319, and drA0369 genes were induced by heat shock treatment in a DdrI-dependent manner (Fig. 5 and Table 1), suggesting that they may be direct targets of the DdrI regulatory protein (Tables S1 and S2). The dr0219 coding sequence, not annotated by Hua and Hua (33), was found 43 bp downstream of a putative DdrI binding site (5′-TGTTATGTTATATACG-3′) identified by FITBAR when the search was performed on the genome sequence published by White et al. (36). In contrast, the drA0143 gene predicted to be a DdrI target (Tables S1 and S2) was highly induced by heat shock treatment, but in a DdrI-independent manner (Fig. 5). However, in the ΔddrI mutant, the drA0143 gene was 2.5-fold less expressed than in the wild-type strain at 30°C and after heat shock. We did not observe any induction of dr0950 and dr0980 after heat shock treatment.
FIG 5.
DdrI is important for transcriptional heat shock induction of several genes. GY9613 (wild type, dark-gray bars) and GY13630 (ΔddrI mutant, light-gray bars) strains were grown to A650 of 0.5 at 30°C and transferred or not transferred to 48°C for 5 min before extraction of total RNA. Expression values at 48°C relative to 30°C were determined and were normalized to the expression values of reference genes tuf1 (dr0309) and recN (dr1477), as described in Materials and Methods. Error bars represent the standard deviations of the results from triplicate qRT-PCRs from three or four biological replicates.
TABLE 1.
Main putative DdrI targets, sorted by their function in DNA metabolism or stress responsesa
| Functional group and gene identified by Hua and Hua (33) | Gene identified by White et al. (36) | Function | FITBAR CRPb | FITBAR DdrIc | Heat shockd |
|---|---|---|---|---|---|
| Replication, recombination, and repair | |||||
| A2G07_00390 | DR0001 | DnaN, DNA polymerase III beta-subunit | X | X | X |
| A2G07_00380 | — | DdrC, DNA damage response protein | X | X | |
| A2G07_00375 | DR0004 | MutT, Nudix family protein | X | X | X |
| A2G07_00040 | DR0070 | DdrB, DNA damage response protein | X | X | |
| A2G07_13010 | DR0100 | SSB, single-stranded DNA-binding protein | X | X | |
| — | DR0219 | DdrF, DNA damage response protein | X | ||
| A2G07_11420 | DR0423 | DdrA, DNA damage response protein | X | X | |
| A2G07_05720 | DR1572 | UvrD2, DNA helicase | X | X | X |
| A2G07_04095 | DR1902 | RecD, exodeoxyribonuclease V subunit | X | ||
| A2G07_04045 | DR1913 | GyrA, DNA gyrase subunit A | X | X | X |
| A2G07_13525 | DRA0001 | ParA, chromosome-partitioning protein | X | X | |
| Stress response and chaperones | |||||
| A2G07_11785 | DR0349 | Lon2, ATP-dependent protease | X | ||
| A2G07_10695 | DR0561 | Periplasmic maltose-binding protein | X | ||
| A2G07_09290 | DR0844 | Thioredoxin domain-containing protein | X | ||
| A2G07_08295 | DR1046 | ClpB, ATP-dependent protease | X | ||
| A2G07_08110 | DR1082 | RaiA, ribosome-associated translation inhibitor | X | ||
| A2G07_06675 | DR1372 | Water stress, hypersensitive response domain-containing protein | X | X | |
| A2G07_05840 | DR1546 | SodC, superoxide dismutase | X | X | |
| A2G07_05725 | DR1571 | Peptide ABC transporter | X | X | X |
| A2G07_03765 | DR1974 | Lon1, ATP-dependent protease | X | X | |
| A2G07_01670 | DR2381 | Aldehyde dehydrogenase/enoyl-CoA hydratase | X | X | X |
| A2G07_14210 | DRA0146 | Catalase | X | X | |
| A2G07_14755 | DRA0259 | KatE2, catalase | X | ||
| A2G07_13265 | DRA0319 | Urease beta/gamma-subunit | X | X | X |
| A2G07_13520 | DRA0369 | Hypothetical protein | X | X | X |
| Regulation of transcription and signal transduction mechanisms | |||||
| A2G07_09790 | DR0743 | DNA-binding response regulator | X | X | |
| A2G07_09785 | DR0744 | Two-component sensor histidine kinase | X | ||
| A2G07_04605 | DR1797 | Transcription termination/antitermination protein NusA | X | X | |
| A2G07_04050 | DR1912 | Protein tyrosine phosphatase | X | X | |
| A2G07_00810 | DR2556 | Response regulator | X | ||
| A2G07_00435 | DR2629 | PadR family transcriptional regulator | X | X | |
| A2G07_13430 | DRA0350 | Histidine kinase | X | ||
| A2G07_15105 | DRB0044 | Diguanylate cyclase | X | X |
—, not correctly annotated.
Genes proposed by FITBAR to be regulated by DdrI using as a matrix the 166 CRP binding sites identified in the E. coli genome (Table S1).
Genes proposed by FITBAR to be regulated by DdrI using as a matrix the 176 putative DdrI binding sites identified in the D. radiodurans genome (Table S2).
Genes induced by heat shock treatment in a DdrI-dependent manner (Fig. 5).
We also tested the expression of a panel of 6 genes (dr0997 [ddrI], dr0349 [lon2], dr0561, dr1046 [clpB], and dr1082 [lrpA, also called hpf or raiA], and drA0199) previously shown to be induced by heat shock a minimum of 5-fold in D. radiodurans (34), described as being not controlled by both Sig1 (an alternative sigma factor) and HspR (a negative regulator of the heat shock response) proteins (34, 35), and not found in Tables S1 and S2. As a control, we analyzed the expression of the groEL gene (dr0607), which is known to be induced by heat shock under the control of Sig1 and HspR (34, 35) and is not found in the list of putative genes controlled by DdrI (Tables S1 and S2). We found that groEL was induced by 3.5-fold after 5 min of incubation at 48°C, but in a DdrI-independent manner. We confirmed the upregulation of the ddrI gene after 5 min of incubation at 48°C (Fig. 5) (34), and we showed that drA0199 was induced independently of the DdrI protein, whereas dr0349, dr0561, dr1046, and dr1082 were induced by heat shock treatment in a DdrI-dependent manner (Fig. 5 and Table 1). The expression of these 4 genes may be indirectly regulated by the DdrI protein. The dr0178 gene, belonging to a transposable element of the IS200/IS605 family, was also reported to be upregulated 5.9-fold after heat shock treatment (34). As previously shown, the standard R1 ATCC 13939 strain used in our study only contains one active copy of an IS200/IS605 family member, called ISDra2F, while the second copy is inactive (37). This is in contrast with the first published complete genome sequence of D. radiodurans (36), in which 7 complete ISDra2 copies and one partial ISDra2 copy were reported (38). The gene corresponding to dr0178 in ISDra2F is dr1651 (tnpB) (39, 40). Thus, we tested, by RT-qPCR, the effect of DdrI deficiency on heat shock induction of both dr1651 (tnpB) and dr1652 (tnpA) genes belonging to the same operon and encoding a negative regulator of ISDra2 transposition and the ISDra2 transposase, respectively. We found that both ISDra2 genes were highly induced by heat shock treatment and that this induction required DdrI (Fig. 5).
Taken together, these results confirm the direct or indirect DdrI-dependent regulation of several key components of the heat shock regulon, in agreement with the thermosensitive phenotype of the ΔddrI mutant.
DdrI is involved in completion of cell division, genome segregation, and stability of the MP1 megaplasmid in D. radiodurans.
D. radiodurans cells depleted for DdrI were analyzed by fluorescence microscopy at three different growth states, exponential (A650, 0.3), transitional (A650, 1.5), and stationary (A650, 3.5) phases. The absence of the DdrI protein in exponentially growing D. radiodurans cells led to 55.2% (544/986) of the cells belonging to tetrads versus 34.5% (252/730) for the R1 strain (Fig. 6 and S5). Surprisingly, in the absence of DdrI, structures containing 8 cells (two-tetrads), representing 2.4% of the exponentially growing cells (24/986), were observed, and their proportion increased dramatically at transitional- and stationary-growth phases to reach 36.7% (480/1,308) and 78.0% (1,336/1,712) of the cells, respectively. In contrast, the proportion of wild-type cells belonging to the two-tetrads was very low, these structures being undetectable during exponential growth, and only 2.6% (32/1,246) of them were present at stationary phase. Moreover, the proportion of ΔddrI mutant cells belonging to dyads decreased rapidly during the cell cycle and dropped to 1.6% (28/1,712) at stationary phase (Fig. 6 and S5). These results suggest a putative role of the DdrI protein in the completion of D. radiodurans cell division.
FIG 6.
DdrI is required for the regulation of cell division and/or genomic DNA segregation. (A) Fluorescence microscopy analyses of the cell shape of exponential (A650, 0.3), transitional (A650, 1.5) and stationary (A650, 3.5) phases cells from GY9613 (wild type) and GY13630 (ΔddrI mutant) strains. The percentage of cells found in dyads (2 cells, blue), tetrads (4 cells, red) and two-tetrads (8 cells, green) are illustrated in the pie charts. Two-tetrads are indicated by green arrows. (B) Membranes were stained with FM4-64 (red) and nucleoids with DAPI (green). Anucleate cells are indicated by yellow arrows. n represents the total number of cells examined.
Interestingly, during exponential growth, 7.6% (15/197) of ΔddrI mutant cells were found to be anucleated, compared to <0.5% (<1/202) for the R1 wild-type cells. This phenotype was even more pronounced during the transitional- and stationary-growth phases, where 15.4% (32/208) and 25% (53/212) of the ΔddrI anucleate cells were observed, compared to <0.5% (<1/210) and 4% (8/201) for the R1 wild-type cells, respectively. These results suggest that DdrI may also be implicated in the regulation of genes involved in chromosome segregation. The drB0145 gene encoding a putative plasmid replication initiator protein and the divergently transcribed operon encompassing the drB0001 and drB0002 genes annotated as encoding a ParA ATPase involved in chromosome partitioning and a protein of the ParB family, respectively, share two putative overlapping DdrI binding motifs (Fig. 7A and Tables S1 and S2). To compare the maintenance of the MP1 megaplasmid harboring these genes in wild-type and ΔddrI mutant cells, we introduced a cassette expressing resistance to chloramphenicol into the drB0121 gene located on the MP1 replicon and previously shown to be inactivated without any effect on growth (13). We found that the proportion of chloramphenicol-resistant (Cmr) cells containing the MP1 megaplasmid diminished to 24% after 10 generations in the ΔddrI mutant when cells are grown without selection pressure, whereas the megaplasmid was stably maintained in wild-type cells (Fig. 7B). In contrast, when the Cmr cassette was introduced into the drC0017 gene of the CP1 plasmid, the proportion of Cmr cells after 10 generations did not decrease (Fig. 7B). These results indicate that DdrI participates in the maintenance of the MP1 megaplasmid.
FIG 7.
The absence of the DdrI protein reduces the maintenance of the MP1 megaplasmid in D. radiodurans. (A) Putative DdrI binding sites located upstream of genes related to DNA replication and segregation of the MP1 megaplasmid are indicated. The DdrI binding sites potentially involved in transcriptional regulation of drB0145 gene and of the divergently transcribed drB0001-drB0002 operon are indicated by brackets. (B) Aliquots of GY13673 (ddrI+ ΔdrC0017Ωcat), GY13674 (ΔddrI ΔdrC0017Ωcat), GY13696 (ddrI+ ΔdrB0121Ωcat), and GY13697 (ΔddrI ΔdrB0121Ωcat) cells containing either CP1Ωcat (left) or MP1Ωcat (right) were grown for 10 generations in TGY2X liquid medium without selection pressure (see Materials and Methods). Appropriate dilutions were then plated on both TGY agar and TGY agar plates supplemented with chloramphenicol to determine the proportions of cells harboring either MP1Ωcat or CP1Ωcat. The proportions of chloramphenicol-resistant cells are the averages of the results from at least three independent experiments.
All the morphological defects observed in cells devoid of DdrI were increased in stationary phase. Thus, we analyzed by Western blotting the expression of the DdrI-FLAG protein in exponential, transitional, and stationary phases of growth, and we showed that the expression of DdrI was highly induced at stationary phase in a DrRRA-dependent manner (Fig. S6), suggesting an important role of DdrI at this growth phase.
DdrI is required for plasmid p11559 maintenance in D. radiodurans.
During our strain constructions, we observed that transformation was less efficient in ΔddrI mutant bacteria. Thus, we analyzed the effect of the absence of DdrI protein on DNA transformation in D. radiodurans. As shown in Fig. 8A, the deletion of ddrI moderately decreased chromosomal DNA transformation frequency (approximately 5-fold) but dramatically affected the plasmid DNA transformation efficiency. Indeed, the transformation frequency of the p11559 plasmid (41), conferring spectinomycin resistance, dropped 20,000-fold in cells devoid of DdrI compared to wild-type cells (Fig. 8B). These observations suggest the requirement of DdrI for efficient overall DNA transformation or for replication and/or maintenance of plasmid p11559 in D. radiodurans.
FIG 8.
Deletion of ddrI dramatically reduces p11559 plasmid transformation frequency in D. radiodurans. (A and B) GY9613 (wild type) and GY13630 (ΔddrI mutant) competent cells were transformed with 300 ng of genomic DNA from the GY11733 strain (conferring rifampin resistance) (A) or 300 ng of p11559 plasmid DNA (conferring spectinomycin resistance) (B). Appropriate dilutions of the transformation mixtures were plated on TGY agar plates to determine the number of viable cells, and on TGY agar plates supplemented with rifampin or spectinomycin to determine the number of transformants. The transformation frequencies were calculated as the number of transformants divided by the number of viable cells in the transformation mixture. The values are the averages of the results from at least three independent experiments. (C) Aliquots of GY14142 (ddrI+), GY13670 (ΔddrI) and GY13644 (ΔddrI crp+) cells harboring the p11559 plasmid were grown for 10 generations in TGY2X liquid medium without selection pressure. Then, appropriate dilutions were spread on both TGY agar and TGY agar plates supplemented with spectinomycin to calculate the proportions of spectinomycin-resistant cells harboring p11559. The values are the averages of the results from at least three independent experiments.
During the natural DNA transformation process in D. radiodurans, only one strand of incoming DNA is transported into the cytoplasm. Bouthier de la Tour et al. (7) previously demonstrated that the reconstitution of an intact plasmid by single-strand annealing (SSA) requires the DdrB protein, likely through its single-strand annealing activity. To test whether DdrI regulates ddrB gene expression, we analyzed, by Western blotting, the cellular level of DdrB-FLAG-tagged protein expressed in R1 strain and in the ΔddrI mutant. As shown in Fig. S7A, the absence of DdrI protein results in a significant increase in the DdrB-FLAG protein level. This result suggests that DdrI negatively regulates the expression of ddrB, in accordance with the location of its putative binding site downstream of the ddrB promoter and overlapping the RDRM recognized by the DdrO repressor (Fig. S7B) (18). Therefore, the strong reduction in plasmid DNA transformation efficiency in the ΔddrI mutant may be related to a defect in stable plasmid maintenance rather than impairment of the SSA process. To test this hypothesis, we analyzed the maintenance of the p11559 plasmid in the ΔddrI mutant (p11559) and R1 (p11559) strains grown for 10 generations without selection pressure. As shown in Fig. 8C, the spectinomycin-resistant (Spcr) population of cells harboring plasmid p11559 diminished from 89.7% in the wild-type strain to approximately 10% in the ΔddrI mutant after 10 generations. Interestingly, the induced plasmid-based production of E. coli CRP in the ΔddrI mutant bacteria resulted in an intermediate 57% proportion of Spcr cells after growth without any selection pressure for 10 generations. These results suggest that some putative DdrI target sites, located on plasmid p11559 or on the whole genome of D. radiodurans, may be involved in stable maintenance of plasmid p11559 and are shared with the E. coli CRP.
DISCUSSION
To date, approximately 100 genes have been annotated as encoding transcriptional regulators and transcription factors in the D. radiodurans genome, but only a few of them have been functionally studied. Cells devoid of the DdrI protein, a transcription regulator of the CRP family, display a pleotropic phenotype including growth defects under laboratory standard conditions, sensitivity to genotoxic agents, oxidative stress (13, 26; this work), heat shock treatment, as well as defects in cell division and replicon stability.
We showed here that the expression of E. coli CRP (CRPEc) in a D. radiodurans ΔddrI mutant partially restores growth and resistance to UVC, MMC, Paraquat, and heat shock, strongly suggesting that DdrI is a protein belonging to the CRP family, and that both the CRPEc and DdrI regulators can share the regulation of several common target genes. In agreement with this, the alignment of DNA-binding sites previously identified for several members of the CRP family revealed a consensus sequence among diverse bacterial species (see Fig. S2 in the supplemental material). These data led us to perform an in silico prediction of the potential DdrI targets in the D. radiodurans genome, using the FITBAR Web tool (31). A pseudopalindromic 5′-TGTGA(N6)TCACA-3′ consensus sequence was extrapolated from the 115 DdrI candidate target genes identified (Table S2 and Fig. 3A). This motif is in agreement with binding sites of the HTH regulator family members, which bind to DNA as dimers. EMSAs showed the important role played by cAMP for DNA binding on a DNA sequence containing this site (Fig. 3). These results suggest that DdrI likely undergoes a conformational change upon cAMP binding necessary for its DNA binding, as previously shown for E. coli CRP (42–44) and other CRP family members (45, 46), contrary to the SdrP factor of Thermus thermophilus, which acts independently of any added effector molecule (47). Although adenylate cyclases lack a specific signature, making them difficult to predict (48), DRA0006 was proposed to contain domains that are similar to those of adenylate cyclases (26). Moreover, D. radiodurans encodes several putative cAMP-binding proteins, and its cAMP intracellular concentration was shown to increase following gamma irradiation (49), suggesting a potential function of cAMP as a second messenger in D. radiodurans.
The 115 in silico-predicted DdrI target genes are involved in various cellular processes, including replication and DNA repair, response to oxidative stress and heat shock treatment, and signal transduction (Table 1), and several genes are similar to known CRPEc targets (particularly those involved in central carbon metabolism, transport of various metabolites, cell wall biogenesis, and lipid metabolism) (Table S2). Surprisingly, several putative DdrI binding sites are distant from the beginning of the coding sequences (>250 bp). However, we showed that dr2381 is strongly upregulated (56-fold) after heat shock treatment in a DdrI-dependent manner (Fig. 5), in spite of the distance of the putative DdrI binding site (604 bp) (Tables S1 and S2).
To better understand the sensitivity of the ΔddrI mutant to heat shock treatment, we tested, by RT-qPCR, the expression levels of heat shock responsive genes. Here, we showed that ddrI was induced by heat shock treatment and that the expression of the DdrI protein was under the control of DrRRA, even under normal growth conditions. This control is direct since the DrRRA regulator binds to the promoter region of the ddrI gene (25). We thus propose that the heat shock stimulus activates a two-component system consisting of a not-yet-identified histidine kinase and its cognate response regulator DrRRA (25), which in turn induces the transcription of ddrI. After 5 min at 48°C, all the genes tested were upregulated in a DdrI-dependent manner except the drA0199 gene, encoding a nodulation-related protein, and drA0143, encoding a 3-hydroxyacyl-coenzyme A (3-hydroxyacyl-CoA) dehydrogenase (Fig. 5). A direct DdrI control is expected for the genes exhibiting a predicted DdrI binding site in their upstream regulatory region (Tables S1 and S2), in contrast to the other genes tested, for which DdrI may act indirectly. However, the in silico prediction may not identify targets with less homology to the consensus sequence, and additional targets may exist in vivo, as suggested by the binding of DdrI shown by EMSAs, to sequences located upstream of different genes not found in Tables S1 and S2 (26). Among these sequences, some are located upstream of dr0997 (ddrI), dr1689 (glgC), dr1819 (uvsE), dr1921 (sbcD), and dr2220 (terB) but contain 7-bp spacers and not 6 bp for CRPEc and the CRP from other bacterial species (Fig. S2). However, it is important to note that CRP and related regulators are able to recognize and bind to relatively divergent sites, which are reflected in the very low informational content of their consensus sequence, and in E. coli, genomic SELEX experiments also identified new CRP binding sites not predicted in silico (27).
We also showed that DdrI is involved in the upregulation of tnpA and tnpB, the two genes of the ISDra2F transposon, in response to heat shock treatment, presumably indirectly, since no predicted DdrI target sequence was found in ISDra2. The subterminal stem-loop necessary for ISDra2 transposition (40) has been shown to act as a transcription terminator, avoiding any transcription readthrough in the IS. We previously showed that ISDra2F transposition is increased after exposure to ionizing radiation (37, 40) but did not result from an increase in TnpA expression during the 180-min postirradiation incubation (40). The consequences of the increased expression of the TnpA transposase and the TnpB negative regulator of transposition upon heat shock treatment, in the presence of DdrI, and to a lower extent in the absence of DdrI (Fig. 5), require further investigations.
We further showed that DdrI was induced in the stationary phase of growth in a DrRRA-dependent manner and that DdrI deficiency leads to a striking increase in two-tetrad-forming cell subpopulations during the cell cycle, compared to the wild-type strain, indicating that DdrI is important for the accurate completion of cell division, particularly during the stationary phase. Chou and Tan (50) previously showed that the addition of NaCl could induce 8-, 16-, and 32-cell unit formation and attributed this phenomenon to the failure of cells to septate after cell wall formation. Once a tetrad forms during active growth, adjacent pairs seem to separate by delamination. Thus, the DdrI protein might regulate some genes involved in this delamination process. DdrI deficiency also leads to a striking increase in anucleate cells during the cell cycle, compared to the wild-type strain, indicating that DdrI is important for genome segregation. The Par system has been shown in Caulobacter crescentus and Bacillus subtilis to be required for active chromosome segregation by assisting segregation of the replication origin (for a review, see references 51 and 52). In D. radiodurans, each genome replicon, except the CP1 plasmid, has its own Par proteins encoded by parAB operons. The deletion of parB1 from chromosome I resulted in a significantly higher level of anucleation than that measured in the wild type (53), a phenotype similar to that observed for the ΔddrI mutant. In addition, functional characterization of the partitioning proteins of D. radiodurans suggested the possibility of a role of ParA2, encoded on chromosome II, in the regulation of cell division by nucleoid compaction at the vicinity of septum growth (54), and it was proposed that interaction of Par proteins with divisome proteins may play important roles in genome segregation (55). A predicted DdrI target sequence was found 545 bp upstream of the promoter region of the drA0001-drA0002 operon encoding the ParA2 and ParB2 chromosome-partitioning proteins (Tables S1 and S2), suggesting that the high anucleation levels observed in the ΔddrI mutant may be partly related to a defect in completion of chromosome segregation due to downregulation of the parA2B2 operon. In D. radiodurans lacking topoisomerase IV (Topo IV), its DNA gyrase (encoded by gyrA and gyrB) is expected to ensure the Topo IV-mediated functions in chromosome segregation, namely, the decatenase function required for partitioning the daughter chromosomes before cell division (56). The gyrA gene was shown to contain a putative DdrI binding site 112 bp upstream of its coding region (Tables S1 and S2) and to be upregulated in a DdrI-dependent manner upon heat shock treatment (Fig. 5). Therefore, the lack of DdrI may also impair GyrA expression and thus accurate chromosome segregation.
We demonstrated that DdrI is required for stable maintenance of the natural MP1 megaplasmid. Two putative DdrI binding sites were identified upstream of the promoter region of drB0145 (encoding a plasmid replication initiator protein) and the divergently transcribed operon encompassing the drB0001 and drB0002 genes encoding ParA3 and ParB3, respectively (Fig. 7). Therefore, the involvement of DdrI in the stable maintenance of MP1 may be mediated through its positive control of both drB0145 and parA3B3 operon expression, as suggested above for the parA2B2 operon expressed by chromosome II. Unfortunately, as chromosome II expresses essential genes, it was not possible to analyze the effect of DdrI deficiency on its maintenance. In addition, DdrI was shown to be required for stable maintenance of the p11559 plasmid (Fig. 8) but not for those of the natural CP1 plasmid. The reduction in p11559 stability may explain the partial complementation of the ΔddrI mutant by CRPEc expressed from this plasmid, even if the experiments were performed under constant selection pressure. The p11559 plasmid (41) contains the basic replicon of the deinococcal SARK plasmid pUE10 (57), which is different from those of CP1, and both plasmids likely employ different mechanisms for their own replication, explaining their different stabilities in the absence of DdrI.
In conclusion, we propose that DdrI may regulate, directly or indirectly, a large variety of genes involved in DNA metabolism, DNA repair, response to oxidative stress or to heat shock, regulation of transcription, and signal transduction (Table 1), as well as many genes involved in ribosome structure and translation and in various metabolic pathways (Tables S1 and S2). We also showed that this regulator may be involved in cell division, chromosome segregation, and plasmid stability. The ddrI gene transcription levels are under the control of the DrRRA response regulator, but DdrI itself putatively regulates the expression of other regulators (Tables 1, S1, and S2). Our results indicate that DdrI is a major regulator in D. radiodurans, and further studies will better explain the cascade of regulatory pathways associated with the various phenotypes observed for a ΔddrI mutant.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used are listed in Table 2. E. coli strain DH5α was the general cloning host, and strain SCS110 was used to propagate plasmids prior to their introduction into D. radiodurans via transformation (58). All D. radiodurans strains were derivatives of strain R1 ATCC 13939 and were grown at 30°C in TGY2X liquid medium or tryptone-glucose-yeast extract (TGY) plates (15). E. coli strains were grown at 37°C in lysogeny broth (LB). Media were supplemented with the appropriate antibiotics used at the following concentrations: 75 μg/ml spectinomycin, 100 μg/ml hygromycin, 6 μg/ml kanamycin, 3.5 μg/ml chloramphenicol, and 25 μg/ml rifampin for D. radiodurans and 50 μg/ml spectinomycin, 25 μg/ml kanamycin, and 35 μg/ml chloramphenicol for E. coli. The expression of E. coli CRP in D. radiodurans or His6::DdrI in Rosetta (DE3) was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the medium. The transformation of D. radiodurans with genomic DNA, PCR products, or plasmid DNA was performed as previously described (15).
TABLE 2.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or other relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MG1655 | K-12 F− λ− ilvG rfb-50 rph-1 | Laboratory stock |
| DH5α | supE44 ΔlacU(ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| SCS110 | endA dam dcm supE44 Δ(lac-proAB) (F′traD36 proAB lacIqZΔM15) | Laboratory stock |
| Rosetta (DE3) | F− ompT hsdS20 gal dcm lacY1 (DE3)/pRARE6 (Cmr) | |
| D. radiodurans | ||
| GY9613 | ATCC 13939, R1 | 64 |
| GY11733 | Rifr (rpoBΔ9 Rifr)a | 41 |
| GY12830 | ddrB::spa-cat | 7 |
| GY13109 | Δ(ISDra2F)ΩtetA | 40 |
| GY13630 | ΔddrIΩhph | This work |
| GY13641 | ΔddrIΩhph Δ(ISDra2F)ΩtetA | This work |
| GY13643 | ΔddrIΩhph Δ(ISDra2F)ΩtetAΩPddrI::ddrI-cat | This work |
| GY13644 | ΔddrIΩhph/p14750 | This work |
| GY13645 | ΔddrIΩPddrI::his6::ddrI-kan | This work |
| GY13653 | ΔddrIΩhph ddrB::spa-cat | This work |
| GY13663 | ddrI::flag-cat | This work |
| GY13670 | ΔddrIΩhph/p11559 | This work |
| GY13673 | ΔdrC0017Ωcat | This work |
| GY13674 | ΔddrIΩhph ΔdrC0017Ωcat | This work |
| GY13679 | ΔdrRRAΩhph ddrI::flag-cat | This work |
| GY13681 | ΔdrRRAΩhph | This work |
| GY13696 | ΔdrB0121Ωcat | This work |
| GY13697 | ΔddrIΩhph ΔdrB0121Ωcat | This work |
| GY14142 | R1/p11559 | This work |
| Plasmids | ||
| p11086 | Source of Pout::kan cassette | Laboratory stock |
| p11559 | Expression vector; Pspac-term116, PtufA::lacI, Spcr in E. coli and D. radiodurans | 41 |
| p12625 | Source of Pkat::hph cassette | Laboratory stock |
| p12723 | Source of flag-cat cassette | This work |
| p14738 | pET28a; PT7lac::His6::ddrI-termT7 | This work |
| p14750 | p11559; Pspac::crp-term116 | This work |
| pPS6 | Source of Ptuf1::cat cassette | 65 |
| pET28a | pET expression system, PT7lac, N-terminal His6 tag | Novagen |
Rifr, rifampin resistant.
Construction of D. radiodurans mutant and tester strains.
GY13630 ΔddrI, GY13681 ΔdrRRA, GY13673 ΔdrC0017, and GY13696 ΔdrB0121 mutant strains were constructed by replacement of the corresponding locus with the appropriate antibiotic resistance cassette using the tripartite ligation method (41). The GY13674 ΔdrC0017 ΔddrI and GY13697 ΔdrB0121 ΔddrI double-mutant strains were constructed by transforming the ΔdrC0017Ωcat and ΔdrB0121Ωcat single mutants, respectively, with genomic DNA from the GY13630 ΔddrIΩhph strain. The ΔddrI mutant, expressing an ectopic ddrI gene under the control of its own promoter, was constructed by replacement of the tetA gene in the GY13641 ΔddrIΩhph ΔISDra2FΩtetA strain, with a fragment encompassing the PddrI-ddrI and the Cmr cassette. The D. radiodurans strain GY13663, expressing the DdrI protein fused to a FLAG tag at the C terminus (DdrI-FLAG), was constructed using the tripartite ligation method (41). The ΔddrI mutant strain GY13653 expressing the DdrB-SPA protein and the ΔdrRRA mutant strain GY13679 expressing the DdrI-FLAG protein were obtained by transforming the GY13630 ΔddrI or the GY13681 ΔdrRRA strain with genomic DNA from GY12830 and GY13663 expressing the appropriate tagged proteins, respectively. To assess the functionality of the His6::DdrI-tagged protein, GY13645 expressing the His6::DdrI-tagged protein was constructed by replacement of the ddrI locus with a fragment encompassing the ddrI coding sequence fused to a His6 tag at the N terminus and the kanamycin resistance (Kanr) cassette. The purity of the strains was systematically confirmed by diagnostic PCR and sequencing (see Fig. S8A and B for the genetic structure and the purity of the ΔddrI mutant and Fig. S8C and D for those of ΔdrC0017Ωcat, ΔdrC0017Ωcat ΔddrI, ΔdrB0121Ωcat, and ΔdrB0121Ωcat ΔddrI mutants). The oligonucleotides used for strain construction and diagnostic PCR will be provided on request.
DNA manipulations.
Plasmid DNA was extracted from E. coli using the NucleoSpin plasmid miniprep kit (Macherey-Nagel). D. radiodurans chromosomal DNA was isolated as described previously (59). PCRs were carried out with Phusion DNA polymerase (Thermo Scientific) to amplify fragments subsequently used for cloning, or with GoTaq Flexi DNA polymerase (Promega) for diagnostic PCR. PCR products were purified using the PCR cleanup kit (Macherey-Nagel).
Plasmid construction.
The crp coding sequence was amplified by PCR using genomic DNA from E. coli strain MG1655 as the template. The PCR fragment was digested by NdeI and XhoI and ligated into the expression vector p11559, resulting in plasmid p14750 expressing crp under the control of the Pspac promoter inducible by IPTG.
The ddrI coding sequence was amplified by PCR using genomic DNA of the wild-type GY9613 strain as the template. The PCR fragment was digested by NdeI and XhoI and ligated into the expression vector pET-28a (Novagen). The resulting p14738 plasmid expresses the DdrI protein fused to a His6 tag at the N terminus under the control of the T7 lac promoter inducible by IPTG.
His6::DdrI-tagged protein purification.
The p14738 plasmid, expressing the His6::DdrI-tagged protein under the control of the T7 lac promoter, was used to transform E. coli Rosetta (DE3) cells. Transformants were grown in LB medium supplemented with the appropriate antibiotics at 37°C to an A600 of 0.45, and the expression of the His6::DdrI protein was induced by IPTG at a final concentration of 1 mM for 3 h. Cells were harvested (3,500 × g, 10 min, 4°C), resuspended in lysis buffer (20 mM Tris-HCl [pH 7.8], 800 mM NaCl, 5% glycerol, 10 mM imidazole, 0.03% Triton X-100) at a rate of 0.1 ml of lysis buffer for A600 of 1, and disrupted using an ultrasonic cell disrupter (350 W, 3 × 2 min). The cleared lysate was recovered by centrifugation (13,000 × g, 1 h, 4°C) and mixed gently overnight at 4°C with 1 ml nickel-nitrilotriacetic acid (Ni-NTA) slurry equilibrated with lysis buffer (0.5 ml bed volume; Qiagen). The lysate–Ni-NTA mixture was then washed by 5 elution steps with 5 bed volumes of elution buffer (20 mM Tris-HCl [pH 7.8], 200 mM NaCl, 5% glycerol, and 0.03% Triton X-100) containing increasing concentrations of imidazole (40 mM, 60 mM, 100 mM, 150 mM, and 200 mM). The purity of the DdrI protein was verified by SDS-gel electrophoresis, and the protein fraction, eluted in the presence of 150 mM imidazole, was dialyzed on a PD10 column (GE Healthcare) with equilibration buffer (50 mM Tris-HCl [pH 7.8], 100 mM NaCl, and 30% glycerol), according to the manufacturer's protocol. Dithiothreitol (DTT) and EDTA were added at a final concentration of 1 mM, and the His6::DdrI protein was stored at −80°C.
EMSA.
A single-stranded Cy5-labeled oligonucleotide (10 μM), 5′-Cy5-CATCAGAAAATGTGTGTCTGGTGAAGCAGTTTTGTGAAAAAACTCACAGCCTGGGATAGAAAACAGCACTATCACAGCAT-3′ (80 nt), containing a putative DdrI binding site (bold letters) at position 33, was incubated with the complementary single-stranded 80-nt oligonucleotide (10 μM) in 10 μl of annealing buffer (40 mM Tris-HCl [pH 7.8], 50 mM NaCl, 20 mM MgCl2). The mixture was heated at 90°C for 10 min and cooled for 3 h at room temperature. Thirty femtomoles from the resulting double-stranded substrate was incubated for 20 min at 30°C with recombinant protein His6::DdrI (0.5 or 1 μM) in 15 μl of binding buffer (10 mM Tris HCl [pH 7.8], 20 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1 mg/ml bovine serum albumin [BSA], 10% glycerol) in the presence or absence of 200 μM cAMP and poly(dI-dC) (13 or 33 μg/ml). The samples were loaded onto an 8% polyacrylamide (37.5/1 [wt/wt] acrylamide-bisacrylamide) gel in 0.25× Tris-borate-EDTA. After migration (20 V/cm, 3 h, 4°C), bands were visualized using a phosphorimager (Typhoon Trio imager; GE Healthcare).
Cross-linking of DdrI.
Three micrograms of the DdrI protein was preincubated with 0 or 200 μM cAMP in 10 mM sodium phosphate buffer (pH 7) at 30°C for 20 min. Then, 0.05% glutaraldehyde was added or not added and the samples were incubated at 30°C for 30 min in a final volume of 20 μl. After incubation, 5 μl of 5× Laemmli buffer (312 mM Tris-HCl [pH 6.8], 50% glycerol, 250 mM DTT, 0.1% bromophenol blue) was added, and the samples were loaded onto a 15% SDS-polyacrylamide gel. After migration (14 V/cm, 2 h), the gel was stained using Coomassie blue.
Purification of total RNA and cDNA synthesis.
Bacteria were grown to an A650 of 0.5 at 30°C and transferred or not transferred to 48°C for 5 min. Then, bacteria (10 ml) were harvested and centrifuged (3,500 × g, 10 min at 4°C). The pellets were stored at −80°C for 2 h and total RNA were isolated using the FastRNA Pro Blue kit (MP Biomedicals, Irvine, CA) and the FastPrep-24 instrument, according to the manufacturer's protocols. The RNA concentration was measured using the DeNovix DS-11 spectrophotometer (SelectScience). One microgram of total RNA was then treated with DNase I amplification grade (Invitrogen), according to the manufacturer's instructions, except that 1 μl of DNase I was added every 5 min during the 15-min incubation. RNA purity and integrity were analyzed by 1.4% agarose gel electrophoresis. Subsequently, 250 ng of RNA was then transcribed into cDNA by reverse transcriptase using the Maxima first-strand cDNA synthesis kit (Thermo Scientific), according to the manufacturer's instructions, in a final volume of 200 μl. The reverse transcriptase negative control, containing all reagents for the reverse transcription reaction except the Maxima enzyme mix, was carried out to assess, by quantitative PCR (qPCR) experiments, genomic DNA contamination of the RNA samples.
qPCR analyses.
qPCR assays were performed using 2 μl of cDNA as the template and the iTaq Universal SYBR Green Supermix (Bio-Rad), according to the manufacturer's instructions. For all genes analyzed, the primer pairs were designed using the IDT PrimerQuest tool (Integrated DNA Technologies) and will be provided upon request. The thermal cycling protocol used on the Bio-Rad CFX 96 real-time PCR detection system was 1st step, 95°C for 2 min; 2nd step, 39 cycles of 95°C for 5 s and 60°C for 30 s; and 3rd step, melt curve from 60°C to 95°C, with an increment of 0.5°C for 5 s. The amplification specificities were verified by examining the melt curve profile, and the amplification efficiencies were assayed by generating standard curves (using serial dilutions of the cDNA). All genes tested exhibited the recommended amplification efficiency of 90 to 110% and R2 values of >0.98. qPCR assays performed using the reverse transcriptase negative control as the template confirmed the absence of genomic DNA contamination (data not shown). Four candidate reference genes, tuf1 (dr0309), recN (dr1477), amyE (dr1472), and hpi (dr2508), were evaluated for their expression stability on all samples by geNorm analysis (60) (qbase+). The tuf1 (dr0309) and recN (dr1477) genes were validated as reference genes. Expression values in wild-type and ΔddrI mutant (GY13630) bacteria after 5 min at 48°C relative to 30°C were determined for candidate genes and were normalized to the expression values of tuf1 and recN using the REST 2009 software (http://www.gene-quantification.de/rest-2009.html). RT-qPCRs were performed in triplicate from three or four biological replicates.
Western blot analysis of tagged proteins DdrI-FLAG or DdrB-SPA.
Exponential-growth cultures (A650, 0.45) of bacteria expressing the DdrI-FLAG protein were grown at 30°C and then transferred to 48°C, 20-ml aliquots taken at different times (0, 0.5, or 1 h) were centrifuged (3,500 × g, 10 min, 4°C). The pellets were resuspended in 150 μl of 1× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and protein extracts were prepared as previously described (61). Protein concentrations were determined by Bradford assay (Bio-Rad protein assay dye reagent), and 10 μg of proteins was loaded on 12% SDS-PAGE gels (stain-free precast gels; Bio-Rad) and transferred onto a polyvinylidene difluoride (PVDF) membrane (GE Healthcare). The Western blot analyses were performed as previously described (18) with a 1:5,000 dilution of anti-FLAG rabbit primary antibodies (Thermo Scientific). For an analysis of expression of DdrB-SPA protein, 20 ml of exponential-growth cultures (A650, 0.5) of GY12830 and GY13653 expressing the DdrB-SPA protein was prepared, and 5 μg of proteins was analyzed by Western blot experiments with a 1:5,000 dilution of anti-FLAG rabbit primary antibodies (Thermo Scientific), as described above.
Sensitivity assays to DNA-damaging agents UVC and mitomycin C.
Bacteria were grown in TGY2X liquid medium to an A650 of 1 and then serially diluted in TGY2X and spotted on TGY agar plates supplemented or not supplemented with mitomycin C (40 ng/ml), exposed or not exposed, to UVC rays (600 J/m2) at a dose rate of 3.5 J/m2/s. TGY agar plates were supplemented with spectinomycin and 1 mM IPTG for strains harboring the p14750 plasmid to induce E. coli crp gene expression.
Sensitivity assay to oxidative stress generated by Paraquat.
D. radiodurans strains were analyzed for their sensitivity to Paraquat-methyl viologen (Sigma-Aldrich) by using the disc inhibition assay, as follows. Exponential-phase cultures (1.5 ml; A650, 0.5) were spread onto TGY plates (supplemented with spectinomycin and 1 mM IPTG for strain GY13644). Sterilized 6-mm-diameter filter paper discs (Dominique Dutscher) were placed on the agar surface. Then, 10 μl of various concentrations (4, 6, 8, and 10 mM) of Paraquat (freshly diluted in sterile H2O) was spotted onto each disc. After incubation at 30°C for 2 days, the diameters of the growth inhibition zones were measured.
Sensitivity to heat shock.
Overnight cultures of bacteria were grown at 30°C, serially diluted in TGY2X, spotted on TGY agar plates, and then incubated at 30°C, 37°C, or 39°C for 3 to 5 days.
Fluorescence microscopy.
Cells in exponential-growth (A650, 0.3), transitional-growth (A650, 1.5), and stationary-growth (A650, 3.5) phases were fixed by adding toluene (3% final concentration) and kept at 4°C overnight. DNA and membranes were stained by incubation with 2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and 6 μg/ml FM4-64 (Life Technologies) for 5 min at room temperature. One microliter of the cell suspension (in 10 mM MgSO4) was then immobilized onto 1% agarose-coated slides and observed by fluorescence microscopy on a wide-field Leica DM RXA microscope. Images were captured with a 5-MHz Micromax 1300Y charge-coupled-device (CCD) camera (Roper Instruments) with the appropriate filters and analyzed with the MetaMorph and ImageJ softwares.
In silico prediction of putative DdrI targets.
The in silico prediction of potential DdrI targets in the D. radiodurans genome was performed using the Fast Investigation Tool for Bacterial and Archaeal Regulons (FITBAR) Web tool dedicated to statistically robust predictions of prokaryotic regulons (31). This analysis was performed on the sequenced D. radiodurans genome (33) by using the 166 binding sites recognized by the CRP of E. coli (27) as the matrix (see Table S1 in the supplemental material). A total of 478 putative DdrI binding sites were found in the D. radiodurans genome and sites located in intergenic regions at distances of <650 bp were listed (Table S1). The genome of D. radiodurans has a GC content of 67%, while that of E. coli is 50.8%. Thus, a matrix based on the set of 176 sequences among the potential DdrI intergenic targets was used in a second prediction to take this GC content discrepancy between D. radiodurans and E. coli into account and to increase the informational content of the consensus sequence; 269 sites were found in the D. radiodurans genome, with a majority (198 sites) located in intergenic regions, and sites located in intergenic regions at distances <650 bp were listed (Table S2). Position-specific scoring matrices (PSSM) were generated from the 166 CRP E. coli or the 176 putative DdrI binding sites (listed in Table S1) using log-odds PSSM (62). Chromosome scanning was concentrated on the intergenic regions, on both DNA strands, and false positives in the prediction were discerned by using compound importance sampling (CIS; described in reference 63) as a P value calculation algorithm. The list of putative DdrI binding sites (Tables S1 and S2) was sorted by position, and only predicted sites located from bp −650 to +3 of the downstream coding sequence were conserved.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adriana Bailone for valuable discussions and for her critical reading of the manuscript, Yvan Zivanovic for his constant help with D. radiodurans genome analysis, and Michael DuBow for polishing our English.
L.M. gratefully acknowledges the Ministère de l'Enseignement Supérieur, de la Recherche et de l'Innovation (MESRI) for her doctoral training grant. This work was supported by the Centre National de la Recherche Scientifique, the University Paris-Sud, Electricité de France (RB2017-02 to F.C.) and the Agence Nationale de la Recherche (ANR “Radioresistance”-11-BSV3-01701 to S.S.).
This work was carried out in compliance with the current laws governing genetic experimentation in France.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00129-18.
REFERENCES
- 1.Confalonieri F, Sommer S. 2011. Bacterial and archael resistance to ionizing radiation. J Phys 261:012005. doi: 10.1088/1742-6596/261/1/012005. [DOI] [Google Scholar]
- 2.Slade D, Radman M. 2011. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev 75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krisko A, Radman M. 2013. Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Cold Spring Harb Perspect Biol 5:a012765. doi: 10.1101/cshperspect.a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmins J, Moe E. 2016. A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: major findings, functional and mechanistic insight and challenges. Comput Struct Biotechnol J 14:168–176. doi: 10.1016/j.csbj.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. 2006. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569–573. [DOI] [PubMed] [Google Scholar]
- 6.Bentchikou E, Servant P, Coste G, Sommer S. 2010. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet 6:e1000774. doi: 10.1371/journal.pgen.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouthier de la Tour C, Boisnard S, Norais C, Toueille M, Bentchikou E, Vannier F, Cox MM, Sommer S, Servant P. 2011. The deinococcal DdrB protein is involved in an early step of DNA double strand break repair and in plasmid transformation through its single-strand annealing activity. DNA Repair (Amst) 10:1223–1231. doi: 10.1016/j.dnarep.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin-Zaidman S, Englander J, Shimoni E, Sharma AK, Minton KW, Minsky A. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254–256. doi: 10.1126/science.1077865. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman JM, Battista JR. 2005. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol 5:17. doi: 10.1186/1471-2180-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly MJ. 2012. Death by protein damage in irradiated cells. DNA Repair (Amst) 11:12–21. doi: 10.1016/j.dnarep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 11.Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Venkateswaran A, Zhai M, Kostandarithes HM, Brim H, Makarova KS, Wackett LP, Fredrickson JK, Daly MJ. 2005. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev 29:361–375. doi: 10.1016/j.fmrre.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, Battista JR. 2004. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168:21–33. doi: 10.1534/genetics.104.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulermo R, Onodera T, Coste G, Passot F, Dutertre M, Porteron M, Confalonieri F, Sommer S, Pasternak C. 2015. Identification of new genes contributing to the extreme radioresistance of Deinococcus radiodurans using a Tn5-based transposon mutant library. PLoS One 10:e0124358. doi: 10.1371/journal.pone.0124358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narumi I, Satoh K, Kikuchi M, Funayama T, Yanagisawa T, Kobayashi Y, Watanabe H, Yamamoto K. 2001. The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J Bacteriol 183:6951–6956. doi: 10.1128/JB.183.23.6951-6956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonacossa de Almeida C, Coste G, Sommer S, Bailone A. 2002. Quantification of RecA protein in Deinococcus radiodurans reveals involvement of RecA, but not LexA, in its regulation. Mol Genet Genomics 268:28–41. doi: 10.1007/s00438-002-0718-x. [DOI] [PubMed] [Google Scholar]
- 16.Satoh K, Ohba H, Sghaier H, Narumi I. 2006. Down-regulation of radioresistance by LexA2 in Deinococcus radiodurans. Microbiology 152:3217–3226. doi: 10.1099/mic.0.29139-0. [DOI] [PubMed] [Google Scholar]
- 17.Ludanyi M, Blanchard L, Dulermo R, Brandelet G, Bellanger L, Pignol D, Lemaire D, de Groot A. 2014. Radiation response in Deinococcus deserti: IrrE is a metalloprotease that cleaves repressor protein DdrO. Mol Microbiol 94:434–449. doi: 10.1111/mmi.12774. [DOI] [PubMed] [Google Scholar]
- 18.Devigne A, Ithurbide S, Bouthier de la Tour C, Passot F, Mathieu M, Sommer S, Servant P. 2015. DdrO is an essential protein that regulates the radiation desiccation response and the apoptotic-like cell death in the radioresistant Deinococcus radiodurans bacterium. Mol Microbiol 96:1069–1084. doi: 10.1111/mmi.12991. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Xu Q, Lu H, Lin L, Wang L, Xu H, Cui X, Zhang H, Li T, Hua Y. 2015. Protease activity of PprI facilitates DNA damage response: Mn2+-dependence and substrate sequence-specificity of the proteolytic reaction. PLoS One 10:e0122071. doi: 10.1371/journal.pone.0122071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Lapidus A, Copeland A, Kim E, Land M, Mavrommatis K, Pitluck S, Richardson PM, Detter C, Brettin T, Saunders E, Lai B, Ravel B, Kemner KM, Wolf YI, Sorokin A, Gerasimova AV, Gelfand MS, Fredrickson JK, Koonin EV, Daly MJ. 2007. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One 2:e955. doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Groot A, Dulermo R, Ortet P, Blanchard L, Guerin P, Fernandez B, Vacherie B, Dossat C, Jolivet E, Siguier P, Chandler M, Barakat M, Dedieu A, Barbe V, Heulin T, Sommer S, Achouak W, Armengaud J. 2009. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 5:e1000434. doi: 10.1371/journal.pgen.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu G, Lu H, Wang L, Chen H, Xu Z, Hu Y, Tian B, Hua Y. 2010. DdrB stimulates single-stranded DNA annealing and facilitates RecA-independent DNA repair in Deinococcus radiodurans. DNA Repair (Amst) 9:805–812. doi: 10.1016/j.dnarep.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Bouthier de la Tour C, Mathieu M, Meyer L, Dupaigne P, Passot F, Servant P, Sommer S, Le Cam E, Confalonieri F. 2017. In vivo and in vitro characterization of DdrC, a DNA damage response protein in Deinococcus radiodurans bacterium. PLoS One 12:e0177751. doi: 10.1371/journal.pone.0177751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchard L, Guerin P, Roche D, Cruveiller S, Pignol D, Vallenet D, Armengaud J, de Groot A. 2017. Conservation and diversity of the IrrE/DdrO-controlled radiation response in radiation-resistant Deinococcus bacteria. Microbiologyopen 6:e00477. doi: 10.1002/mbo3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Xu G, Chen H, Zhao Y, Xu N, Tian B, Hua Y. 2008. DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol Microbiol 67:1211–1222. doi: 10.1111/j.1365-2958.2008.06113.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang S, Xu H, Wang J, Liu C, Lu H, Liu M, Zhao Y, Tian B, Wang L, Hua Y. 2016. Cyclic AMP receptor protein acts as a transcription regulator in response to stresses in Deinococcus radiodurans. PLoS One 11:e0155010. doi: 10.1371/journal.pone.0155010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada T, Fujita N, Yamamoto K, Ishihama A. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081. doi: 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl TA, Baumbach J, Jungwirth B, Puhler A, Tauch A. 2008. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J Biotechnol 135:340–350. doi: 10.1016/j.jbiotec.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Bai G, McCue LA, McDonough KA. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J Bacteriol 187:7795–7804. doi: 10.1128/JB.187.22.7795-7804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redfield RJ, Cameron AD, Qian Q, Hinds J, Ali TR, Kroll JS, Langford PR. 2005. A novel CRP-dependent regulon controls expression of competence genes in Haemophilus influenzae. J Mol Biol 347:735–747. doi: 10.1016/j.jmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Oberto J. 2010. FITBAR: a web tool for the robust prediction of prokaryotic regulons. BMC Bioinformatics 11:554. doi: 10.1186/1471-2105-11-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. 1986. Information content of binding sites on nucleotide sequences. J Mol Biol 188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 33.Hua X, Hua Y. 2016. Improved complete genome sequence of the extremely radioresistant bacterium Deinococcus radiodurans R1 obtained using PacBio single-molecule sequencing. Genome Announc 4:e00886-16. doi: 10.1128/genomeA.00886-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid AK, Howell HA, Battista JR, Peterson SN, Lidstrom ME. 2005. Global transcriptional and proteomic analysis of the Sig1 heat shock regulon of Deinococcus radiodurans. J Bacteriol 187:3339–3351. doi: 10.1128/JB.187.10.3339-3351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid AK, Howell HA, Battista JR, Peterson SN, Lidstrom ME. 2005. HspR is a global negative regulator of heat shock gene expression in Deinococcus radiodurans. Mol Microbiol 55:1579–1590. doi: 10.1111/j.1365-2958.2005.04494.x. [DOI] [PubMed] [Google Scholar]
- 36.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, Moffat KS, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan JJ, Lam P, McDonald L, Utterback T, Zalewski C, Makarova KS, Aravind L, Daly MJ, Minton KW, Fleischmann RD, Ketchum KA, Nelson KE, Salzberg S, Smith HO, Venter JC, Fraser CM. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mennecier S, Servant P, Coste G, Bailone A, Sommer S. 2006. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol Microbiol 59:317–325. doi: 10.1111/j.1365-2958.2005.04936.x. [DOI] [PubMed] [Google Scholar]
- 38.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev 65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pasternak C, Dulermo R, Ton-Hoang B, Debuchy R, Siguier P, Coste G, Chandler M, Sommer S. 2013. ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol Microbiol 88:443–455. doi: 10.1111/mmi.12194. [DOI] [PubMed] [Google Scholar]
- 40.Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S. 2010. Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet 6:e1000799. doi: 10.1371/journal.pgen.1000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mennecier S, Coste G, Servant P, Bailone A, Sommer S. 2004. Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol Genet Genomics 272:460–469. doi: 10.1007/s00438-004-1077-6. [DOI] [PubMed] [Google Scholar]
- 42.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. 2004. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol 14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Passner JM, Schultz SC, Steitz TA. 2000. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 Å resolution. J Mol Biol 304:847–859. doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 44.Scott SP, Jarjous S. 2005. Proposed structural mechanism of Escherichia coli cAMP receptor protein cAMP-dependent proteolytic cleavage protection and selective and nonselective DNA binding. Biochemistry 44:8730–8748. doi: 10.1021/bi0479609. [DOI] [PubMed] [Google Scholar]
- 45.Stapleton M, Haq I, Hunt DM, Arnvig KB, Artymiuk PJ, Buxton RS, Green J. 2010. Mycobacterium tuberculosis cAMP receptor protein (Rv3676) differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J Biol Chem 285:7016–7027. doi: 10.1074/jbc.M109.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green J, Stapleton MR, Smith LJ, Artymiuk PJ, Kahramanoglou C, Hunt DM, Buxton RS. 2014. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr Opin Microbiol 18:1–7. doi: 10.1016/j.mib.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agari Y, Kashihara A, Yokoyama S, Kuramitsu S, Shinkai A. 2008. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol Microbiol 70:60–75. doi: 10.1111/j.1365-2958.2008.06388.x. [DOI] [PubMed] [Google Scholar]
- 48.Gancedo JM. 2013. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol Rev Camb Philos Soc 88:645–668. doi: 10.1111/brv.12020. [DOI] [PubMed] [Google Scholar]
- 49.Kamble VA, Rajpurohit YS, Srivastava AK, Misra HS. 2010. Increased synthesis of signaling molecules coincides with reversible inhibition of nucleolytic activity during postirradiation recovery of Deinococcus radiodurans. FEMS Microbiol Lett 303:18–25. doi: 10.1111/j.1574-6968.2009.01855.x. [DOI] [PubMed] [Google Scholar]
- 50.Chou FI, Tan ST. 1991. Salt-mediated multicell formation in Deinococcus radiodurans. J Bacteriol 173:3184–3190. doi: 10.1128/jb.173.10.3184-3190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Badrinarayanan A, Le TB, Laub MT. 2015. Bacterial chromosome organization and segregation. Annu Rev Cell Dev Biol 31:171–199. doi: 10.1146/annurev-cellbio-100814-125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Possoz C, Junier I, Espeli O. 2012. Bacterial chromosome segregation. Front Biosci 17:1020–1034. doi: 10.2741/3971. [DOI] [PubMed] [Google Scholar]
- 53.Charaka VK, Misra HS. 2012. Functional characterization of the role of the chromosome I partitioning system in genome segregation in Deinococcus radiodurans. J Bacteriol 194:5739–5748. doi: 10.1128/JB.00610-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charaka VK, Mehta KP, Misra HS. 2013. ParA encoded on chromosome II of Deinococcus radiodurans binds to nucleoid and inhibits cell division in Escherichia coli. J Biosci 38:487–497. doi: 10.1007/s12038-013-9352-5. [DOI] [PubMed] [Google Scholar]
- 55.Maurya GK, Modi K, Misra HS. 2016. Divisome and segrosome components of Deinococcus radiodurans interact through cell division regulatory proteins. Microbiology 162:1321–1334. doi: 10.1099/mic.0.000330. [DOI] [PubMed] [Google Scholar]
- 56.Devigne A, Bouthier de la Tour C, Guérin P, Lisboa J, Quevillon-Cheruel S, Armengaud J, Sommer S, Servant P. 2016. PprA protein is involved in chomosome segregation via its physical and functional interaction with DNA gyrase in irradiated Deinococcus radiodurans bacteria. MSphere 1:e00036-15. doi: 10.1128/mSphere.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meima R, Lidstrom ME. 2000. Characterization of the minimal replicon of a cryptic Deinococcus radiodurans SARK plasmid and development of versatile Escherichia coli-D. radiodurans shuttle vectors. Appl Environ Microbiol 66:3856–3867. doi: 10.1128/AEM.66.9.3856-3867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meima R, Rothfuss HM, Gewin L, Lidstrom ME. 2001. Promoter cloning in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol 183:3169–3175. doi: 10.1128/JB.183.10.3169-3175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Norais C, Servant P, Bouthier-de la-Tour C, Coureux PD, Ithurbide S, Vannier F, Guerin PP, Dulberger CL, Satyshur KA, Keck JL, Armengaud J, Cox MM, Sommer S. 2013. The Deinococcus radiodurans DR1245 protein, a DdrB partner homologous to YbjN proteins and reminiscent of type III secretion system chaperones. PLoS One 8:e56558. doi: 10.1371/journal.pone.0056558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:research0034.1–research0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouthier de la Tour C, Toueille M, Jolivet E, Nguyen HH, Servant P, Vannier F, Sommer S. 2009. The Deinococcus radiodurans SMC protein is dispensable for cell viability yet plays a role in DNA folding. Extremophiles 13:827–837. doi: 10.1007/s00792-009-0270-2. [DOI] [PubMed] [Google Scholar]
- 62.Durbin R, Eddy S, Krogh A, Mitchison G. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 63.Barash Y, Elidan G, Kaplan T, Friedman N. 2005. CIS: compound importance sampling method for protein-DNA binding site p-value estimation. Bioinformatics 21:596–600. doi: 10.1093/bioinformatics/bti041. [DOI] [PubMed] [Google Scholar]
- 64.Anderson AW, Nordon HC, Cain RF, Parrish G, Duggan G. 1956. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol 10:575–577. [Google Scholar]
- 65.Passot FM, Nguyen HH, Dard-Dascot C, Thermes C, Servant P, Espeli O, Sommer S. 2015. Nucleoid organization in the radioresistant bacterium Deinococcus radiodurans. Mol Microbiol 97:759–774. doi: 10.1111/mmi.13064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.