ABSTRACT
The RNA polymerase (RNAP) binding protein A (RbpA) contributes to the formation of stable RNAP-promoter open complexes (RPo) and is essential for viability in mycobacteria. Four domains have been identified in the RbpA protein, i.e., an N-terminal tail (NTT) that interacts with RNAP β′ and σ subunits, a core domain (CD) that contacts the RNAP β′ subunit, a basic linker (BL) that binds DNA, and a σ-interaction domain (SID) that binds group I and group II σ factors. Limited in vivo studies have been performed in mycobacteria, however, and how individual structural domains of RbpA contribute to RbpA function and mycobacterial gene expression remains mostly unknown. We investigated the roles of the RbpA structural domains in mycobacteria using a panel of rbpA mutants that target individual RbpA domains. The function of each RbpA domain was required for Mycobacterium tuberculosis viability and optimal growth in Mycobacterium smegmatis. We determined that the RbpA SID is both necessary and sufficient for RbpA interaction with the RNAP, indicating that the primary functions of the NTT and CD are not solely association with the RNAP. We show that the RbpA BL and SID are required for RPo stabilization in vitro, while the NTT and CD antagonize this activity. Finally, RNA-sequencing analyses suggest that the NTT and CD broadly activate gene expression, whereas the BL and SID activate or repress gene expression in a gene-dependent manner for a subset of mycobacterial genes. Our findings highlight specific outcomes for the activities of the individual functional domains in RbpA.
IMPORTANCE Mycobacterium tuberculosis is the causative agent of tuberculosis and continues to be the most lethal infectious disease worldwide. Improved molecular understanding of the essential proteins involved in M. tuberculosis transcription, such as RbpA, could provide targets for much needed future therapeutic agents aimed at combatting this pathogen. In this study, we expand our understanding of RbpA by identifying the RbpA structural domains responsible for the interaction of RbpA with the RNAP and the effects of RbpA on transcription initiation and gene expression. These experiments expand our knowledge of RbpA while also broadening our understanding of bacterial transcription in general.
KEYWORDS: Mycobacterium, RNA polymerases, RbpA, eubacteria, transcription, transcriptional regulation
INTRODUCTION
Progress toward the World Health Organization (WHO) goal of eradicating Mycobacterium tuberculosis continues to be hampered by the estimated 480,000 new cases of multidrug-resistant M. tuberculosis infections among the overall 10.1 million new cases of tuberculosis worldwide in 2016 (1). Strategies for M. tuberculosis eradication include the development of novel therapies, which is aided by the identification of druggable targets. Bacterial transcription is carried out by the RNA polymerase (RNAP) and has been successfully targeted using rifampin, which remains a cornerstone of therapy for M. tuberculosis patients (2, 3). In addition to the subunits that constitute the RNAP holoenzyme in all bacteria (two α subunits and β, β′, ω, and σ subunits), mycobacteria also require two additional essential proteins, RNAP binding protein A (RbpA) and CarD, to form stable transcription initiation complexes (4–7). Unlike Escherichia coli, which has been used to define the events of bacterial transcription initiation, mycobacteria are unable to irreversibly form stable RNAP-promoter open complexes (RPo) and require both RbpA and CarD to reach RPo stability comparable to that of E. coli (4, 7, 8), which could explain the essentiality of these proteins (9–11). Both CarD and RbpA have also been shown to affect the sensitivity of mycobacteria to rifampin (12, 13). Therefore, improving our understanding of these transcription factors could provide an avenue to future therapies targeting CarD or RbpA while improving the efficacy of currently approved drugs.
RbpA was discovered in Streptomyces coelicolor as a protein that coimmunoprecipitates with the RNAP and is unique to the Actinobacteria phylum (14). RbpA consists of a central core domain (CD) flanked by an unstructured 26-amino-acid N-terminal tail (NTT) and a C-terminal σ-interaction domain (SID) linked to the CD by a 15-amino-acid basic linker (BL) (6, 11, 15, 16). The RbpA SID forms a stable binary complex with group I (σA in M. tuberculosis) and certain group II (σB in M. tuberculosis) σ factors (6, 14–16). Bacterial two-hybrid experiments in S. coelicolor showed that mutating the R88 residue within the RbpA SID to an alanine significantly weakened the interaction between S. coelicolor RbpA and the housekeeping σHrdB (15), highlighting the importance of this residue in the interaction. Based on structural studies, the BL makes electrostatic contacts with the DNA phosphate backbone of the nontemplate strand upstream of the −10 promoter element in the RPo conformation, the CD is positioned near the RNAP β′ zinc binding domain, and the NTT threads into the RNAP active site cleft between the β′ zinc binding domain and the σA4 domain (6, 16, 17). In support of a functional role for the BL, fluorescence anisotropy experiments showed that addition of M. tuberculosis RbpA to Mycobacterium bovis RNAP-σA holoenzyme in the presence of M. tuberculosis CarD decreased the dissociation constant (Kd) of RNAP binding to a vapB10 promoter template and an R79A mutation in the M. tuberculosis RbpA BL abolished the RbpA-mediated increases in RNAP affinity for the vapB10 promoter (16).
Most characterization of RbpA has been performed in vitro, and there have been only limited studies of how the domains of RbpA contribute to gene regulation in mycobacteria. In a recent study using Mycobacterium smegmatis, an R79A mutation in the RbpA BL and deletion of the NTT and CD resulted in slower growth of the bacteria (6). Herein, we expand on that work and compare the roles of each RbpA domain, both in vitro and in vivo, to show that only the SID is required for association with the RNAP and the activities of different domains affect the expression of distinct gene sets in the bacteria.
RESULTS
Individual RbpA structural domains are important for mycobacterial growth and viability.
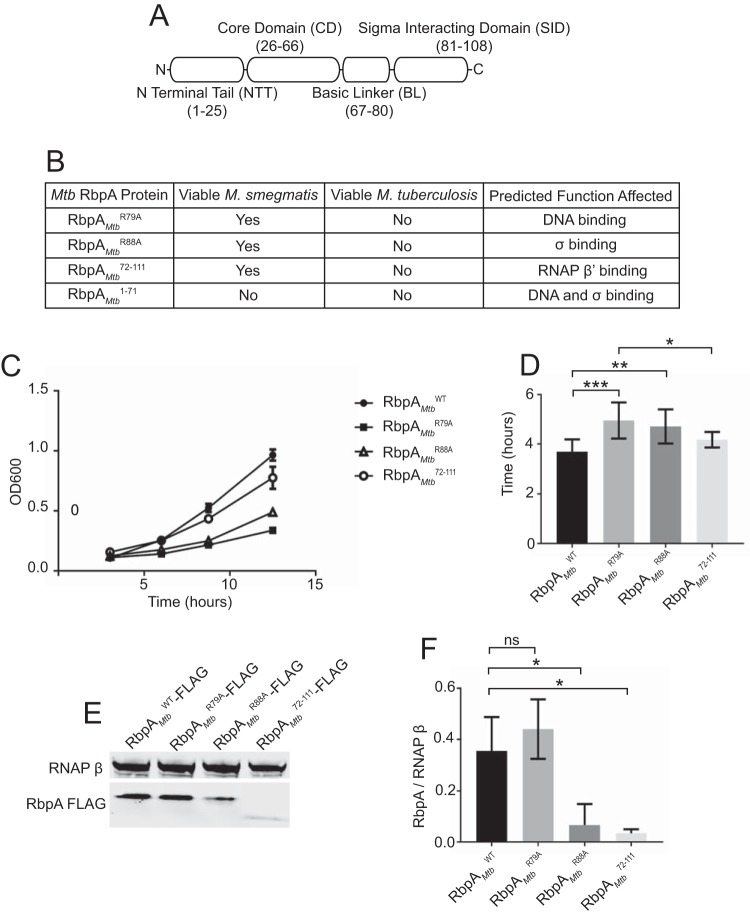
To distinguish the roles of the RbpA structural domains in mycobacteria (Fig. 1A), we first engineered merodiploid strains of M. tuberculosis and M. smegmatis that expressed rbpAMtb at the chromosomal attB site. The M. smegmatis and M. tuberculosis RbpA proteins are 92% identical. Expression of rbpAMtb at the attB site allowed deletion of the endogenous rbpA gene in both M. tuberculosis and M. smegmatis, demonstrating that the RbpA protein from M. tuberculosis can substitute for the M. smegmatis RbpA protein to support viability. We then attempted to replace the rbpAMtb gene at the attB site in M. tuberculosis and M. smegmatis with alleles encoding RbpAMtbR79A, RbpAMtbR88A, RbpAMtb1–71, or RbpAMtb72–111, using a previously described gene-swapping method (13, 18). The R79A mutation is within the BL and should disrupt DNA binding, the R88A mutation in the SID has been shown to weaken the affinity of RbpA for σ, the position 1 to 71 RbpA fragment is deleted for the BL and SID, and the position 72 to 111 RbpA fragment is deleted for the NTT and CD (Fig. 1) (11, 15, 16). Using the gene-swapping approach, we found that none of the RbpA mutants could support viability in M. tuberculosis (Fig. 1B), demonstrating that M. tuberculosis is highly sensitive to any kind of disruption in RbpA function. In contrast, all of the mutant rbpA alleles except that encoding RbpAMtb1–71 supported viability of M. smegmatis, thus providing us with a genetic system to study M. tuberculosis RbpA in vivo by using M. smegmatis strains expressing RbpAMtbWT, RbpAMtbR79A, RbpAMtbR88A, and RbpAMtb72–111. The inability to obtain strains expressing the RbpAMtb1–71 allele as the only rbpA allele demonstrated that the RbpA BL and SID are required for viability in mycobacteria.
FIG 1.
Multiple RbpA structural domains are important for mycobacterial growth and viability. (A) Diagram showing that M. tuberculosis RbpA is composed of an N-terminal tail (NTT) (amino acids 1 to 25), a core domain (CD) (amino acids 26 to 66), a basic linker (BL) (amino acids 67 to 80), and a σ-interaction domain (SID) (amino acids 81 to 111). (B) Table of M. tuberculosis and M. smegmatis strains engineered or determined to be nonviable with replacement of the RbpAMtbWT expression cassette with a cassette expressing RbpAMtbR79A, RbpAMtbR88A, RbpAMtb1–71, or RbpAMtb72–111. An empty expression cassette was transformed as a negative control, while replacement of RbpAMtbWT with RbpAMtbWT was used as a positive control. (C) Growth curves of M. smegmatis expressing RbpAMtbWT, RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, with nine replicates for each strain. (D) M. smegmatis doubling times calculated from the growth curves in panel C. Results are plotted as means ± standard deviations. Statistical significance was analyzed by analysis of variance (ANOVA) and Tukey's multiple-comparison test. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. All comparisons were included in the analysis, but only statistically significant differences are indicated in the figure. (E) Lysates from M. smegmatis ΔrbpA attB::tet-rbpA expressing RbpAMtbWT-FLAG (lane 1), RbpAMtbR79A-FLAG (lane 2), RbpAMtbR88A-FLAG (lane 3), or RbpAMtb72–111-FLAG (lane 4), analyzed with monoclonal antibodies specific for either RNAP β or FLAG. (F) Graphical representation of RbpA stability as the ratio of RbpA molecules per RNAP β, showing the means ± standard errors of the means of three replicates. Statistical significance was analyzed by ANOVA and Tukey's multiple-comparison test. *, P ≤ 0.05; ns, not significant.
To determine how each of these mutations in RbpA affected mycobacterial growth, the doubling times of M. smegmatis strains expressing the wild type (WT), RbpAMtbWT, or mutant RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111 were measured (Fig. 1C and D). The doubling times of the RbpAMtbR79A (4.3 h) and RbpAMtbR88A (4.4 h) strains were significantly longer than that of the RbpAMtbWT strain (3.2 h), indicating that the functions performed by the RbpA BL and SID are required for optimal M. smegmatis growth (Fig. 1D). Although the growth rate of the RbpAMtb72–111 (3.9 h) strain trended lower than that of the RbpAMtbWT strain, this difference was not statistically significant, indicating that loss of the RbpA NTT and CD has only a mild effect on M. smegmatis growth.
To determine whether the mutations in RbpA affected the RbpA protein levels in M. smegmatis, we engineered M. smegmatis strains that expressed the C-terminally FLAG-tagged RbpA proteins RbpAMtbWT-FLAG, RbpAMtbR79A-FLAG, RbpAMtbR88A-FLAG, and RbpAMtb72–111-FLAG as the only copy of the rbpA product and we measured the levels of RbpAMtbWT-FLAG, RbpAMtbR79-FLAG, RbpAMtbR88A-FLAG, and RbpAMtb72–111-FLAG proteins in cell lysates by Western blot analysis (Fig. 1E). The levels of RbpAMtbR88A-FLAG protein were significantly lower than the levels of RbpAMtbWT-FLAG. Therefore, the slower growth of the M. smegmatis strain expressing RbpAMtbR88A could in part be a result of lower levels of RbpA protein. The levels of RbpAMtb72–111-FLAG protein were also significantly lower in cell lysates, compared to the levels of RbpAMtbWT-FLAG. However, we found that this decrease in band intensity was due to issues with the detection of RbpAMtb72–111-FLAG with the anti-FLAG antibody. Therefore, it is unclear whether deletion of the RbpA NTT and CD decreases the levels of RbpAMtb72–111-FLAG in cell lysates.
The RbpA SID is necessary and sufficient for association with RNAP.
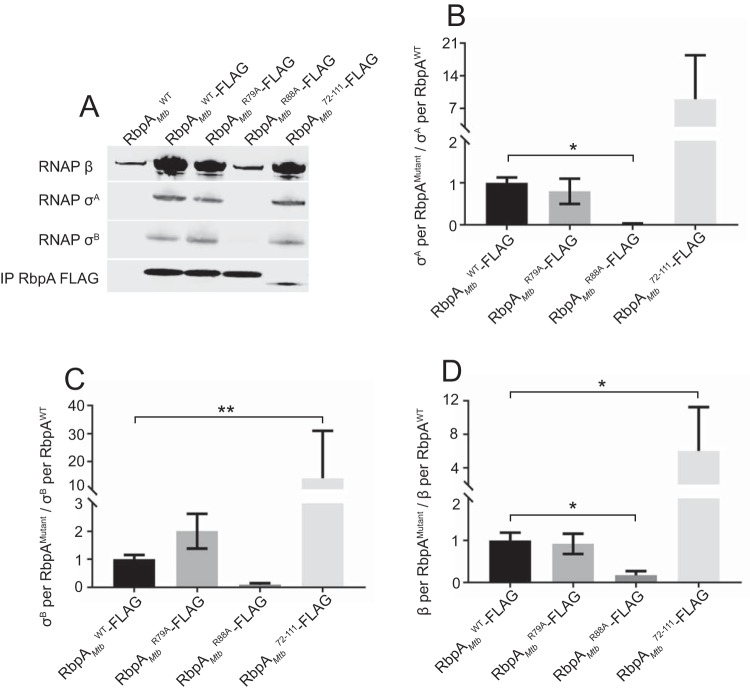
Structural studies indicate that RbpA engages in four different macromolecular interactions in mycobacterial RNAP-promoter initiation complexes, i.e., (i) the RbpA NTT binding to RNAP β′ and σ, (ii) the RbpA CD binding to RNAP β′, (iii) the RbpA BL binding to DNA, and (iv) the RbpA SID binding to σ (6, 11, 16); however, it is not known which of these interactions are required for the association of RbpA with the RNAP. To address this gap in knowledge, we performed coimmunoprecipitation experiments analyzing the amounts of σA, σB, and RNAP β subunit that coimmunoprecipitated with the RbpA-FLAG-tagged proteins (Fig. 2). The levels of σA and σB coimmunoprecipitated with RbpAMtbR88A-FLAG were dramatically reduced, compared to those coimmunoprecipitated with RbpAMtbWT-FLAG (Fig. 2A, B, and C), as expected based on the importance of R88 for σ binding (15). In addition to the decreases in σA and σB levels, the levels of RNAP β coimmunoprecipitated with RbpAMtbR88A-FLAG were significantly reduced (Fig. 2A and D). In crystallographic studies, the R88 in the RbpA SID is not positioned to bind directly to the core RNAP subunits; therefore, we conclude that the reduced β coimmunoprecipitated with RbpAMtbR88A-FLAG is due to the reduced RbpA-σ interaction. This indicates that the interaction between the RbpA SID and the σ subunit is the primary determinant of the association of RbpA with the RNAP. In contrast, deletion of the NTT and CD (RbpAMtb72–111) did not decrease the amounts of RNAP β, σA, or σB associated with RbpA. Therefore, despite the observations that the CD was positioned to interact with RNAP β′ and the NTT was positioned to interact with RNAP β′ and σ (6, 17), these interactions are not necessary for association with RNAP. Notably, although the levels of RNAP β, σA, and σB coimmunoprecipitated per molecule of RbpAMtb72–111 appear to be increased in Fig. 2, we found that the differences were due to lower levels of RbpAMtb72–111 detection by Western blot analysis (data not shown). RbpAMtbR79A-FLAG coimmunoprecipitated similar levels of β and σA, compared to RbpAMtbWT-FLAG. Coimmunoprecipitated levels of σB trended higher with RbpAMtbR79A-FLAG but were not statistically significantly different from those observed with RbpAMtbWT-FLAG (Fig. 2). Collectively, our data show that the RbpA SID is both necessary and sufficient for interaction with RNAP.
FIG 2.
The RbpA SID is necessary and sufficient for the association of RbpA with the RNAP. (A) Western blot analysis of lysates immunoprecipitated for FLAG-tagged RbpA. Monoclonal antibodies specific for FLAG were used to detect RbpAMtb-FLAG protein variants (bottom row). RNAP β coimmunoprecipitated by the FLAG-tagged RbpA constructs was detected with a monoclonal antibody specific for RNAP β, and both σA and σB were detected using a monoclonal antibody specific for a shared epitope in E. coli σ70. (B to D) Amounts of σA (B), σB (C), and RNAP β (D) coimmunoprecipitated by RbpA, based on band intensity, and expressed as the ratio of σA, σB, or RNAP β to RbpA, with eight replicates for each strain. Results are shown as means ± standard deviations. Statistical significance was determined by one-way ANOVA and Kruskal-Wallis multiple-comparison test. *, P ≤ 0.05; **, P ≤ 0.01.
RbpA mutants exhibit distinct kinetic phenotypes on the pathway to RPo formation.
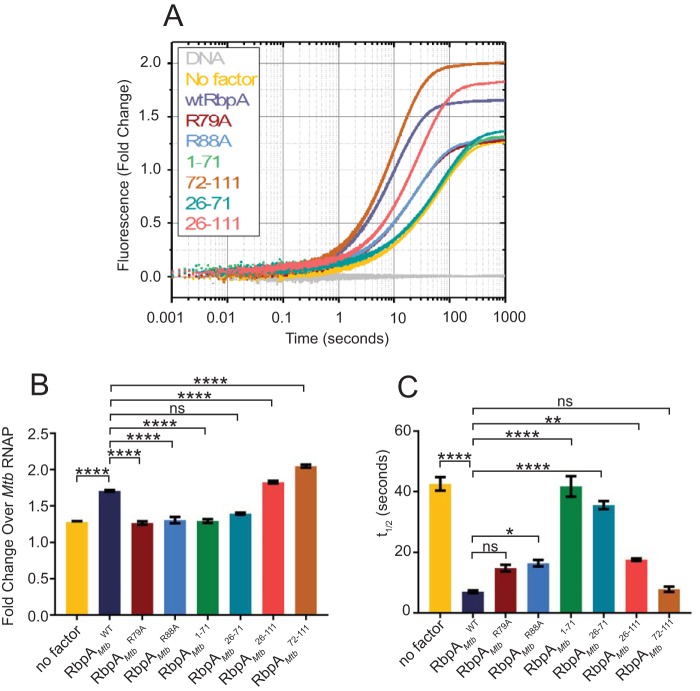
RbpA has been proposed to accelerate a forward kinetic step in the formation of RPo, resulting in more stable RPo at equilibrium (6, 7). A real-time fluorescence assay (7) was used to determine the effects of RbpA mutants on RPo formation by the M. tuberculosis RNAP. Briefly, a Cy3 label was incorporated onto the +2 dT nucleotide, with respect to the +1 transcription start site, of the nontemplate strand of the M. tuberculosis rRNA rrnAP3 promoter (19). The Cy3 label is positioned within the transcription bubble such that, upon opening of the promoter DNA, a 2-fold fluorescence enhancement is observed (20); this allows quantitation of the kinetics of RPo equilibration, by monitoring the change in fluorescence as a function of time, and the stability of RPo, by using the equilibrium fluorescence value (4, 7, 21). Incubating RbpAMtbWT at a saturating concentration (2 μM) with 35 nM M. tuberculosis RNAP-σA holoenzyme and the Cy3-labeled rrnAP3 promoter resulted in a greater amount of RPo at equilibrium than observed with RNAP-σA holoenzyme and the rrnAP3 promoter alone (Fig. 3A), consistent with the known role of RbpA in stabilizing the otherwise unstable mycobacterial RNAP open complex (6, 7). When the same concentrations of RbpAMtbR79A and RbpAMtbR88A were added, no enhancement of the amount of RPo at equilibrium over RNAP-σA holoenzyme and the rrnAP3 promoter alone was observed (Fig. 3A and B), demonstrating the importance of these residues. For a qualitative description of the kinetics, we calculated t1/2 values (the time required to reach the midpoint of the final equilibrium fluorescence). Interestingly, these mutants exhibited approximately 3-fold faster kinetics (RbpAMtbR79A t1/2 of 14.8 ± 1.1 s and RbpAMtbR88A t1/2 of 16.4 ± 1.1 s), compared with the RNAP-σA holoenzyme and the rrnAP3 promoter alone (t1/2 of 43 ± 2 s) (Fig. 3C). The finding of faster kinetics accompanied by no change in the equilibrium fluorescence value suggests that these mutants retain the ability to stabilize the transition state on the pathway to RPo but have lost the ability to stabilize RPo itself. This behavior is analogous to the classic model for enzyme activity (22), in which the transformation of substrate to product is accelerated without changes in the final equilibrium between the two states. In this scenario, the mutant RbpA proteins may increase the rate of opening and the rate of closing equally, such that the ratio of rates remains constant. These results suggest that the interactions between RbpA and both the promoter DNA (R79) and σ factor (R88) are essential for RPo stabilization and that RbpA is still capable of catalyzing promoter opening even in the presence of these mutations.
FIG 3.
RbpA mutants exhibit distinct effects on RPo formation. (A) Fluorescence fold changes, compared to DNA alone, which were was used to monitor RPo formation and stability in real time, using fixed amounts of M. tuberculosis RNAP (35 nM), Cy3-labeled (+2 thymine nontemplate strand) M. tuberculosis rrnAP3 promoter DNA (1 nM), and RbpA (2 μM). Time courses are shown as an average of at least 5 replicates. (B) Total fluorescent fold changes, normalized to RNAP-σA-rrnAP3 alone, for all RbpA constructs. (C) t1/2 values, calculated as the time required to reach one-half of the final fluorescence intensity, for each sample. For panels B and C, means ± standard errors of the means are plotted. Statistical significance was analyzed by ANOVA and Tukey's multiple-comparison test. *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant. Only comparisons between RbpAWT and each of the RbpA mutant constructs are shown in the figure.
To further investigate the domain requirements for RPo stabilization, we repeated the experiments described above with RbpAMtb1–71 (containing the NTT and CD) and RbpAMtb26–71 (containing the CD only), and we observed minimal enhancement in RPo stability (Fig. 3A and B) and an identical rate of RPo equilibration (t1/2 of 42 ± 3 s), relative to the RNAP-σA holoenzyme and the rrnAP3 promoter alone (Fig. 3A and C), indicating that the NTT and CD are unable to affect RPo stability on their own. Conversely, RbpAMtb72–111 showed the greatest amount of RPo at equilibrium, even higher than that of RbpAMtbWT (Fig. 3A and B), with kinetics (t1/2 of 7.8 ± 0.9 s) similar to those of RbpAMtbWT (t1/2 of 6.9 ± 0.5 s) (Fig. 3C). The finding that RbpAMtb72–111 exhibits similar kinetics but a greater amount of RPo at equilibrium, compared with RbpAMtbWT, raises the possibility that the NTT and CD negatively affect RbpA activity under these conditions. To determine whether it was the NTT and/or the CD that antagonized RbpA-mediated RPo stabilization, we assayed an RbpA protein with deletion of just the NTT (RbpAMtb26–111). RbpAMtb26–111 yielded a greater fold change in fluorescence than did RbpAMtbWT but smaller change than did RbpAMtb72–111 (Fig. 3A and B), suggesting that both the CD and NTT are responsible for the antagonistic effect on RbpA-dependent RPo stability. RbpAMtb26–111 exhibited approximately 2-fold slower kinetics of RPo equilibration (t1/2 of 17.6 ± 1.2 s) than did RbpAMtb72–111 (t1/2 of 7.8 ± 0.9 s) and RbpAMtbWT (t1/2 of 6.9 ± 0.5 s) (Fig. 3A and C). One possibility consistent with this observation is that, in the presence of the rest of the domains, the NTT decreases the amount of RPo at equilibrium by increasing a reverse rate leading toward the RNAP-promoter closed complex (RPc). Importantly, performing these experiments with multiple RNAP concentrations suggests that the effect of each RbpA construct is limited by DNA-bound kinetic intermediates and not the rates of association and dissociation of RNAP to and from promoter DNA (see Fig. S1 in the supplemental material). Taken together, these results suggest that residues R79 and R88 are essential for RPo stabilization and that the NTT and CD can inhibit RPo formation.
Truncation of the RbpA NTT/CD and mutations in the RbpA BL and SID result in distinct gene expression changes in M. smegmatis.
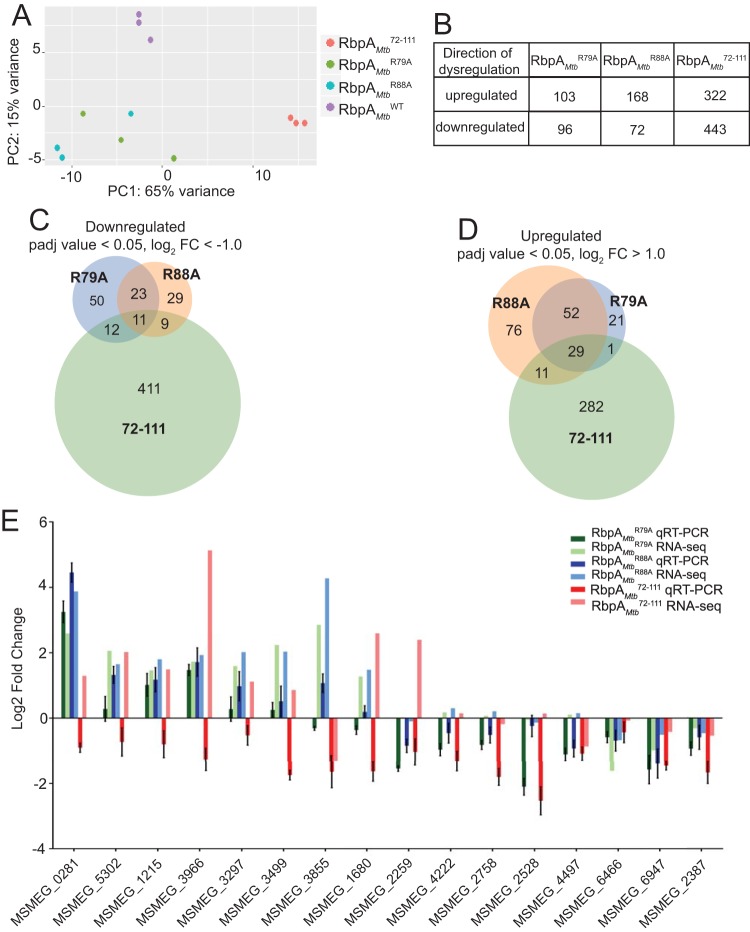
To determine how the individual RbpA domains contribute to gene expression, we performed RNA-sequencing (RNA-seq) experiments with cultures of M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, RbpAMtb72–111, or RbpAMtbWT (Table S1). The only previous analysis of this type focused on the gene expression profiles that resulted from deletion of the RbpA NTT and CD in M. smegmatis, but it did not investigate the roles of the other RbpA domains (6). Principal-component analysis (PCA) of the RNA-seq data was performed and provided a general overview of how gene expression patterns among the RbpA mutants clustered in relationship to each other. Three distinct sample clusters were apparent from the PCA results (Fig. 4A), indicating three different gene expression patterns. The first cluster included the three RbpAMtbWT replicates, the second cluster included the three RbpAMtb72–111 replicates, and the third cluster included the replicates from both RbpAMtbR79A and RbpAMtbR88A. The PCA results indicate that loss of the RbpA NTT/CD affects a gene subset that is different from the genes affected by mutations in the RbpA BL and SID. The number of genes significantly (adjusted P values of <0.05) upregulated or downregulated 2-fold in the RbpA mutants varied, with 766 genes being differentially expressed in RbpAMtb72–111, compared to 199 genes in RbpAMtbR79A and 244 genes in RbpAMtbR88A (Fig. 4B; also see Table S2).
FIG 4.
Truncation of the RbpA NTT/CD and mutations in the RbpA BL and SID result in distinct gene expression changes in M. smegmatis. (A) PCA results showing sample distances across two principal components (PC), generated using read counts of RNA collected from M. smegmatis expressing RbpAMtbWT, RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, mapped to the M. smegmatis mc2155 genome and normalized with regularized logarithmic transformation. Each point represents one of three replicates for RbpAMtbWT, RbpAMtbR79A, RbpAMtbR88A, and RbpAMtb72–111. (B) Numbers of genes significantly (adjusted P values of ≤0.05) upregulated or downregulated 2-fold in M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, relative to M. smegmatis expressing RbpAMtbWT. FC, fold change. (C) Venn diagram showing overlap of the genes downregulated 2-fold (adjusted P values of ≤0.05) in M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, relative to M. smegmatis expressing RbpAMtbWT. (D) Venn diagram showing overlap of the genes upregulated 2-fold (adjusted P values of ≤0.05) in M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, relative to M. smegmatis expressing RbpAMtbWT. (E) qRT-PCR and RNA-seq log2 fold changes for 16 genes in M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111, relative to M. smegmatis expressing RbpAMtbWT. Transcript levels were normalized to an MS2 RNA spike-in control that was added at a constant level of 1 ng/1 billion cells. Means ± standard errors of the means of three replicates are shown for each M. smegmatis strain.
Consistent with the PCA results, there was significant overlap in upregulated and downregulated genes between the RbpAMtbR79A and RbpAMtbR88A strains (Fig. 4C and D and Table 1), indicating that the SID and BL perform functions that contribute to the expression of a common subset of M. smegmatis genes. Also consistent with the PCA results, the upregulated and downregulated genes in RbpAMtb72–111 had little overlap with those in either RbpAMtbR79A or RbpAMtbR88A (Fig. 4C and D). Therefore, the number of shared downregulated or upregulated genes between RbpAMtb72–111 and either RbpAMtbR79A or RbpAMtbR88A was underenriched (Table 1).
TABLE 1.
Overlap enrichment of differentially expressed genes in M. smegmatis strains expressing RbpAMtbR79A, RbpAMtbR88A, or RbpAMtb72–111a
| Overlap comparison | Hypergeometric P value | Fold enrichment |
|---|---|---|
| RbpAMtbR79A vs RbpAMtbR88A, upregulated | 1.81E−11 | +1.52 |
| RbpAMtbR79A vs RbpAMtbR88A, downregulated | 0.0003 | +1.59 |
| RbpAMtbR79A vs RbpAMtb72–111, upregulated | 0.094 | −1.22 |
| RbpAMtbR79A vs RbpAMtb72–111, downregulated | 1.63E−07 | −2.03 |
| RbpAMtbR88A vs RbpAMtb72–111, upregulated | 0.001 | −1.43 |
| RbpAMtbR88A vs RbpAMtb72–111, downregulated | 0.0005 | −1.69 |
Overlap enrichment of differentially upregulated (log2 fold change of >1.0; adjusted P value of <0.05) and differentially downregulated (log2 fold change of <−1.0; adjusted P value of <0.05) genes for each possible comparison between RbpAMtbR79A, RbpAMtbR88A, and RbpAMtb72–111 was evaluated by calculating a hypergeometric P value for fold overenrichment (positive values in the fold enrichment column) or underenrichment (negative values in the fold enrichment column). Fold enrichment values were calculated by dividing the number of overlapping genes observed for each comparison by the number of overlapping genes expected when the null hypothesis of no enrichment was accepted for the same comparison. P values indicate the probability that the underenrichment or overenrichment in the overlap of differentially expressed genes of the two strains being compared would occur randomly. P values of <0.05 are statistically significant.
Given that RbpA stabilizes RNAP-σA-rrnAP3 RPo in vitro and the R79 and R88 residues are essential for this activity, it might be expected that the RbpA BL and SID cooperate to activate transcription from all promoters that RbpA regulates. Similarly, the ability of the NTT and CD to antagonize RbpA-mediated RPo stabilization would lead to the hypothesis that expression from RbpA-regulated genes would be increased in their absence. However, this was not supported by the RNA-seq data, in which similar numbers of transcripts were upregulated and downregulated in each RbpA mutant (Fig. 4B). These data could mean that domains within RbpA can promote both activation and repression of gene expression. However, it is also possible that there was general downregulation or upregulation of gene expression in the RbpA mutants that we were unable to detect due to the addition of equal amounts of RNA from each strain into the sequencing reaction. To explore this possibility, we performed spike-in experiments (23) in which we isolated RNA from cultures of M. smegmatis expressing RbpAMtbR79A, RbpAMtbR88A, RbpAMtb72–111, or RbpAMtbWT and added 1 ng of MS2 bacteriophage RNA (Roche) per 1 billion bacterial cells to the RNA samples. cDNA was generated for each sample, and quantitative reverse transcription-PCR (qRT-PCR) was performed to determine transcript levels for 16 M. smegmatis genes relative to MS2 RNA, which was used as a proxy to represent cell number. The 16 M. smegmatis genes analyzed included genes that were significantly upregulated or downregulated in RbpA mutants during RNA-seq experiments. When results were normalized to MS2 RNA levels, all 16 genes, including the genes considered highly upregulated in the RNA-seq analysis, were downregulated in RbpAMtb72–111 compared to RbpAMtbWT, suggesting that overall transcript levels in RbpAMtb72–111 are decreased (Fig. 4E). Therefore, despite the findings that deletion of the NTT and CD had only a mild effect on the growth rate (Fig. 1C and D) and enhanced RPo stabilization activity in vitro (Fig. 3A and B), the NTT and CD are required for WT levels of gene expression in M. smegmatis. In contrast, qRT-PCR results for the RbpAMtbR79A and RbpAMtbR88A mutants were similar to the RNA-seq results, indicating that RbpAMtbR79A and RbpAMtbR88A mutants do indeed lead to both upregulation and downregulation of gene expression. When we analyzed the genes that were most upregulated or downregulated with the RbpAMtbR79A and RbpAMtbR88A mutants, they fell into multiple diverse functional classes (Table S2), indicating that RbpA activity likely affects multiple cellular processes.
DISCUSSION
In this study, we investigated the functions of the individual RbpA structural domains to gain insight into the complex in vivo roles of RbpA. To study the roles of the RbpA NTT and CD, we truncated the N-terminal 71 amino acids of RbpA. The role of the RbpA BL was probed using a point mutation at R79, which has been implicated in the interaction between RbpA and DNA (16). Finally, we investigated the RbpA SID by using a point mutation at R88, which is one of the key residues needed for the interaction between RbpA and σ (15) but had yet to be studied in mycobacteria in vivo. We found that the function of each RbpA structural domain is required for M. tuberculosis viability and wild-type growth rates in M. smegmatis and disruption of the RbpA BL and SID functions causes a more severe growth defect than loss of the NTT and CD. Our data indicate that M. tuberculosis has a more stringent requirement for RbpA activity, similar to what we observed for CarD (5, 13).
We determined that the RbpA SID interaction with σ is the only interaction required for the association of RbpA with the RNAP; the RbpA R88A substitution resulted in not only loss of the interactions with σA and σB but also almost complete loss of association with the core RNAP β subunit. In contrast, deletion of the NTT and CD did not negatively affect the association of RbpA with RNAP, suggesting that the RbpA NTT and CD serve functions distinct from interaction with RNAP. The RbpA R88A substitution also resulted in decreased RbpA protein levels. Previous studies investigating CarD mutants with altered affinities for the RNAP found that CarD protein levels correlated with CarD affinity for the RNAP (21). Our data showing that RbpAMtbR88A has a lower affinity for the RNAP and is present in lower abundance in the cell supports a model in which RbpA protein levels are also affected by its ability to interact with the RNAP. CarD was shown to be a target of the Clp protease in M. tuberculosis and, similarly, RbpA levels were >2-fold higher in a M. tuberculosis strain lacking Clp protease subunits, suggesting that RbpA protein levels may also be regulated by the Clp protease (24).
Previous studies investigating the effect of RbpA on RPo stability reported that R79 is required for RPo stabilization, whereas both the NTT and CD are dispensable (6, 7). We have expanded on these findings by determining that the RbpA-σ interaction is required for enhanced RPo stability and the NTT and CD antagonize this activity (Fig. 3). Furthermore, our results suggest that the effect of RbpA on the kinetics of RPo equilibration can be differentiated from its effect on equilibrium levels of RPo and that RbpA can affect both forward and reverse rates on the pathway to RPo, at the concentrations tested. The similar effects of RbpA BL and SID mutations on RPo stabilization (Fig. 3) mirror the significant overlap in the expression profiles of RbpAMtbR79A and RbpAMtbR88A (Fig. 4). In contrast, the truncation of NTT and CD, which affects RPo stability differently than mutations in RbpA BL and CD, results in an expression profile significantly different from that of RbpAMtbR79A and RbpAMtbR88A.
We have found that RbpAMtbR79A and RbpAMtbR88A mutants can result in both upregulation and downregulation of transcript levels in M. smegmatis, depending on the gene. Upregulation of gene expression in RbpA mutants could be due to direct effects with RbpA acting as a repressor in some promoter contexts, due to differences in basal initiation kinetics. However, this observation could also be explained by indirect effects with RbpA enhancing the expression of a transcription factor that represses the expression of a set of genes. Future studies that expand analysis of RbpA past the limited promoters that have been explored in vitro will be necessary to address these possibilities.
Our data from spike-in control qRT-PCR experiments suggest that gene expression is globally downregulated in the M. smegmatis RbpAMtb72–111 mutant. This suggests that the NTT and CD are required for efficient gene expression, and it complicates interpretations of the RbpAMtb72–111 RNA-seq data in this study. This finding may also have an impact on a previously published RNA-seq data set for the M. smegmatis RbpAMtb72–111 strain (6). When we compared our RNA-seq data set for RbpAMtb72–111 with the previously reported data, we found that there was no significant overlap in genes that registered as upregulated or downregulated. This could be due to a difference in the culturing methods used in the two studies and/or it could be related to the finding that gene expression in general is less robust. How the NTT and CD mechanistically promote efficient gene expression while antagonizing RPo stability on the rrnAP3 promoter in vitro remains an open question for future studies.
MATERIALS AND METHODS
Media and bacterial strains. (i) Mycobacterium tuberculosis.
The Erdman strain was grown at 37°C in 7H9 (broth) or 7H10 (agar) medium supplemented with 60 μl/liter oleic acid, 5 g/liter bovine serum albumin (BSA), 2 g/liter dextrose, and 0.003 g/liter catalase (oleic acid-albumin-dextrose-catalase [OADC]), 0.5% glycerol, and 0.05% Tween 80 (broth). The M. tuberculosis merodiploid strain was constructed by integrating pMSG430-rbpAMtbWT (expressing RbpAMtbWT from a constitutive Pmyc1-tetO promoter; kanamycin resistant) into the attB site of the Erdman strain. A specialized transducing phage with homology to M. tuberculosis H37Rv nucleotides 2307223 to 2307826 and 2303122 to 2308681 was used to replace all except the start and stop codons of the endogenous rbpA gene with a hygromycin resistance cassette in the merodiploid strain, thus generating ΔrbpA attB::tet-rbpAMtbWT. Gene swapping was used to construct strains of mycobacteria expressing different rbpA alleles and to test their viability, as described previously (13, 18). The M. tuberculosis ΔrbpA attB::tet-rbpAMtbWT strain was transformed with pDB19-rbpAMtbWT (expressing RbpAMtbWT from a constitutive Pmyc1-tetO promoter; zeocin resistant) to replace the pMSG430-rbpAMtbWT construct at the attB site of the M. tuberculosis ΔrbpA attB::tet-rbpAMtbWT strain. The transformants were selected with zeocin, and loss of the pMSG430-rbpAMtbWT construct was confirmed by verifying their inability to grow in the presence of kanamycin. The M. tuberculosis ΔrbpA::tet-rbpAMtbWT strain transformed with pDB19-rbpAMtbWT was named csm323. Csm323 was transformed with pMSG430-rbpAMtbR79A, pMSG430-rbpAMtbR88A, pMSG430-rbpAMtb1–71, or pMSG430-rbpAMtb72–111 (expressing RbpAMtbR79A, RbpAMtbR88A, RbpAMtb1–71, or RbpAMtb72–111, respectively, from a constitutive Pmyc-tetO promoter; kanamycin resistant) to replace the pDB19-rbpAMtbWT construct at the attB site of csm323. The transformants were selected with kanamycin; when positive transformants in M. tuberculosis csm323 could not be obtained (as was the case for pMSG430-rbpAMtbR79A, pMSG430-rbpAMtbR88A, pMSG430-rbpAMtb1–71, and pMSG430-rbpAMtb72–111 transformations), the mutations were deemed nonviable.
(ii) Mycobacterium smegmatis.
All M. smegmatis strains were derived from mc2155 and grown at 37°C in LB medium supplemented with 0.5% dextrose, 0.5% glycerol, and 0.05% Tween 80 (broth). The M. smegmatis merodiploid strain was constructed by integrating pMSG430-rbpAMtbWT into the attB site of mc2155. The M. smegmatis merodiploid strain was transformed with pDB88, with homology to mc2155 nucleotides 3928650 to 3929246 and 3929589 to 3930405, to replace the endogenous rbpA, using two-step allelic exchange as described previously (25), thus generating ΔrbpA::tet-rbpAMtbWT, which was named csm275. Csm275 was transformed with pDB19-rbpAMtbWT to replace the pMSG430-rbpAMtbWT construct at the attB site of the M. smegmatis ΔrbpA attB::tet-rbpAMtbWT strain. The transformants were selected with zeocin, and loss of the pMSG430-rbpAMtbWT construct was confirmed by verifying their inability to grow in the presence of kanamycin. The M. smegmatis ΔrbpA::tet-rbpAMtbWT strain transformed with pDB19-rbpAMtbWT was named csm291. Csm291 was transformed with pMSG430-rbpAMtbWT, pMSG430-rbpAMtbR79A, pMSG430-rbpAMtbR88A, pMSG430-rbpAMtb1–71, pMSG430-rbpAMtb72–111, pMSG430-rbpAMtbWT-FLAG, pMSG430-rbpAMtbR79A-FLAG, pMSG430-rbpAMtbR88A-FLAG, pMSG430-rbpAMtb1–71-FLAG, or pMSG430-rbpAMtb72–111-FLAG to replace the pDB19-rbpAMtbWT construct at the attB site of csm291. Each FLAG tag repeated the sequence for FLAG twice (2×FLAG). The transformants were selected with kanamycin, and loss of the pDB19-rbpAMtbWT construct was confirmed by verifying their inability to grow in the presence of zeocin. When positive transformants in csm291 could not be obtained (as was the case for pMSG430-rbpAMtb1–71 and pMSG430-rbpAMtb1–71-FLAG transformations), the mutations were deemed nonviable. Csm291 strains transformed with pMSG430-rbpAMtbR79A, pMSG430-rbpAMtbR88A, pMSG430-rbpAMtb72–111, pMSG430-rbpAMtbWT-FLAG, pMSG430-rbpAMtbR79A-FLAG, pMSG430-rbpAMtbR88A-FLAG, and pMSG430-rbpAMtb72–111-FLAG were named csm322, csm314, csm328, csm313, csm329, csm327, and csm347, respectively.
Antibiotics and chemicals.
In mycobacterial cultures, 20 μg/ml kanamycin and 12.5 μg/ml zeocin were used. In E. coli cultures, 40 μg/ml kanamycin, 50 μg/ml chloramphenicol, 50 μg/ml streptomycin, and 100 μg/ml ampicillin were used.
Western blotting and immunoprecipitation.
For immunoprecipitation, 1-liter cultures were pelleted by centrifugation, resuspended in 20 ml of 1× phosphate-buffered saline (PBS) with complete protease inhibitor cocktail (Roche), and lysed with high-pressure (30 lb/in2) cell disruption (CF model; Constant Systems, Daventry, UK). The lysate was treated with DNase I (New England BioLabs), added to anti-FLAG affinity gel (clone M2; Sigma-Aldrich, St. Louis, MO), and rotated overnight at 4°C. The protein-agarose matrix was washed three times with NP-40 buffer (10 mM sodium phosphate [pH 8.0], 150 mM NaCl, 1% Nonidet-40, 1× complete protease inhibitor cocktail). The immunoprecipitated protein complexes were eluted with 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 150 μg/ml FLAG peptide (Sigma-Aldrich), 1× complete protease inhibitor cocktail. Protein samples were mixed with SDS-PAGE loading buffer and run on a 4 to 12% Bis-Tris protein gel (Invitrogen). For the Western blot analysis, σA and σB were detected using a mouse monoclonal antibody against E. coli σ70 (clone 2G10; Neoclone, Madison, WI), RNAP β was detected using a mouse monoclonal antibody against E. coli RNAP β (clone 8RB13; Neoclone), and FLAG-tagged RbpA was detected using an anti-FLAG mouse monoclonal antibody (Sigma-Aldrich). Secondary LiCor IRDye 800CW goat anti-mouse IgG polyclonal antibodies were used to detect the primary antibodies. Secondary antibody near-infrared fluorescence was detected with the LiCore Odyssey version 3.0 imaging system, and band intensity was analyzed with Image Studio Lite version 4.0.
Protein purification for biochemical assays.
Plasmids containing the Mycobacterium tuberculosis H37Rv genomic DNA encoding the different M. tuberculosis RNAP holoenzyme subunits were a gift from Jayanta Mukhopadhyay (Bose Institute, Kolkata, India) (26, 27). Expression was carried out in accordance with the method described by Banerjee et al. (26), with minor exceptions. Briefly, E. coli BL21(DE3) cells were transformed with plasmids pET-Duet-rpoB-rpoC (encoding the β and β′ subunits), pAcYc-Duet-sigA-rpoA (encoding an N-terminal 10×His-tagged σA subunit and α subunit), and pCDF-rpoZ (encoding the ω subunit) and were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.8. The culture was then treated with 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown overnight at 16°C. Cells were harvested via centrifugation (4,070 × g for 15 min at 4°C), and the resultant pellets were stored at −80°C. M. tuberculosis RNAP-σA holoenzyme was purified according to methods used previously for the M. bovis RNAP core complex (4). M. tuberculosis RbpA constructs were cloned into pET-SUMO (Thermo Fisher Scientific) and transformed into E. coli BL21(DE3). Cultures were grown at 37°C to an OD600 of 0.8, and protein overexpression was induced with the addition of 0.5 mM IPTG overnight at 16°C. Cells were harvested by centrifugation (4,070 × g for 15 min at 4°C), and the cell pellets were stored at −80°C. The cells were resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 5 mM imidazole, 300 mM NaCl, 5 mM β-mercaptoethanol, protease inhibitor [Sigma-Aldrich]) and lysed by sonication at 4°C. Soluble lysate was separated from insoluble lysate by centrifugation (2,700 × g for 20 min at 4°C). RbpA was purified from the soluble lysate by Ni2+ affinity chromatography (Gold Biotechnology). Ni2+ columns were washed with wash buffer (50 mM Na2HPO4 [pH 8.0], 20 mM imidazole, 300 mM NaCl) until no protein was detected with NanoDrop spectrophotometer OD280 readings. RbpA was eluted from the Ni2+ affinity columns with elution buffer (50 mM Na2HPO4 [pH 8.0], 250 mM imidazole, 300 mM NaCl, 5 mM β-mercaptoethanol). The His-SUMO tag was cleaved from the RbpA constructs with His-Ulp1 protease during overnight dialysis at 4°C (20 mM Tris-HCl [pH 8.0], 250 mM NaCl, 20 mM imidazole, 1 mM β-mercaptoethanol). The His-SUMO tag and His-Ulp1 were separated from RbpA by a second round of Ni2+ affinity chromatography, and the cleaved RbpA was collected as the flowthrough fraction. Cleaved RbpA was dialyzed overnight at 4°C in storage buffer (20 mM Tris-HCl [pH 8.0], 250 mM NaCl, 1 mM β-mercaptoethanol), concentrated to approximately 200 μM (Vivaspin 20, molecular weight cutoff of 3,000; GE Healthcare), and stored at −80°C.
Preparation of fluorescent promoter DNA template.
A Cy3-labeled promoter template of 150 bp +2 nontemplate dT, containing nucleotides 1470151 to 1470300 of the M. tuberculosis Erdman genomic DNA, including the rrnAP3 promoter, was prepared as described previously (4, 7, 21).
Stopped-flow fluorescence assay.
Stopped-flow experiments were performed as described previously (4, 7, 21), with notable exceptions. Prior to data acquisition, M. tuberculosis RNAP-σA holoenzyme, with or without RbpA protein, was incubated at 37°C for 10 min. All experiments were conducted with equal-volume mixing of 2 nM Cy3-labeled rrnAP3 promoter DNA with 70 or 200 nM M. tuberculosis RNAP-σA holoenzyme, with or without 4 μM RbpA protein. Thus, the final concentrations upon mixing were 1 nM DNA and 35 or 100 nM RNAP-σA holoenzyme, with or without 2 μM RbpA protein. Accounting for all contributions from protein storage buffers, the final reaction buffer conditions upon equal-volume mixing were as follows: 20 mM Tris (pH 8.0), 77.5 mM NaCl, 10 mM MgCl2, 5 μM ZnCl2, 20 μM EDTA, 5% (vol/vol) glycerol, 1 mM dithiothreitol, and 0.1 mg/ml BSA. Experiments were performed with an SX-20 stopped-flow spectrophotometer (Applied Photophysics, Leatherhead, UK) with a dead time of ∼1 ms and a total shot volume of ∼100 μl. Samples were excited using a 535-nm fixed-wavelength light-emitting diode (LED) light source with a 550-nm shortpass filter, and emission was monitored using a 570-nm longpass filter. Data were collected at 37°C for 1,000 s by sampling 5,000 points over a logarithmic decay. Each protein condition is represented by the average of at least 5 shots obtained using multiple RNAP preparations, plotted as the fold change over DNA alone according to the formula (F − Fo)/Fo, where Fo is the buffer-subtracted reading for DNA alone and F is the buffer-subtracted reading for DNA mixed with protein.
RNA-seq analysis.
M. smegmatis strains csm275, csm322, csm314, and csm328 were cultured to an OD600 of 0.4 to 0.6, pelleted, resuspended in TRIzol (Thermo Fisher Scientific), and lysed by bead beating (FastPrep; MP Bio, Santa Ana, CA). RNA was extracted with chloroform, precipitated with isopropanol, and resuspended in water. RNA was treated with DNase I (Thermo Fisher Scientific), and RNA integrity and quality were analyzed with an Agilent bioanalyzer. rRNA was removed from samples using the Illumina Ribo-Zero rRNA removal kit. cDNA libraries were generated using an adapted Illumina TruSeq library preparation kit and were quality controlled by analysis of the cDNA size distribution with the Agilent TapeStation. cDNA libraries were pooled and sequenced in a single lane of an Illumina HiSeq 2000 Rapid Run flow cell with a 50-bp single-end read format. Sequencing reads were demultiplexed and converted to a FASTQ format using Illumina bcl2fastq script. Adapter sequences were trimmed from the raw reads, which were then aligned with the M. smegmatis mc2155 reference genome (GenBank accession number NC_008596) using the STAR aligner (28). Sequence alignment map (SAM) files generated from alignments were converted to BAM files using SAMTools (29), and aligned reads were counted per genome feature using the BioConductor package Subread featureCounts function (30). Differential expression analysis and subsequent PCA were performed with BioConductor DESeq2 (31). Venn diagrams were made with an online tool (https://www.stefanjol.nl/venny). Hypergeometric P values and enrichment values were calculated using an online calculator (http://systems.crump.ucla.edu/hypergeometric). The hypergeometric distribution describes the probability of k successes in s draws, without replacement, from a population of size N that contains exactly M successes. N was defined at the total number of differentially expressed genes in the two RbpA mutant constructs being compared, s was defined as the number of differentially upregulated or downregulated genes in one RbpA mutant included in the comparison, M was defined as the number of differentially upregulated or downregulated genes in the second RbpA mutant included in the comparison, and k was defined as the number of differentially upregulated or downregulated genes shared by the two RbpA mutants in the comparison.
qRT-PCR analysis.
M. smegmatis strains csm275, csm322, csm314, and csm328 were cultured to an OD600 of 0.5 to 0.7, pelleted, resuspended in TRIzol (Thermo Fisher Scientific), and lysed by bead beating (FastPrep; MP Bio). RNA was extracted with chloroform, precipitated with isopropanol, and resuspended in water. MS2 bacteriophage RNA (Roche) was added to the bacterial RNA at a ratio of 1 ng of MS2 RNA per 1 billion bacteria, RNA was treated with DNase I (Thermo Fisher Scientific), and cDNA was synthesized with the Superscript III first-strand synthesis system (Invitrogen). qRT-PCR was performed with a SYBR green qPCR kit (Bio-Rad), and MSMEG_0281, MSMEG_5302, MSMEG_1215, MSMEG_3966, MSMEG_3297, MSMEG_3499, MSMEG_3855, MSMEG_1680, MSMEG_2259, MSMEG_4222, MSMEG_2758, MSMEG_2528, MSMEG_4497, MSMEG_6466, MSMEG_6947, and MSMEG_2387 transcript levels were measured and normalized to spike-in MS2 RNA transcript levels. Primers are listed in Table S3 in the supplemental material.
Accession number(s).
The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (32) and are accessible through GEO Series accession number GSE107123.
Supplementary Material
ACKNOWLEDGMENTS
C.L.S. and E.A.G. were supported by grant GM107544 from the NIH. C.L.S. was also supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award. J.P. and A.L.G. were supported by NIGMS grant GM007067. A.L.G. was also supported by a Stephen I. Morse Graduate Fellowship. G.S.-C. was supported by NIH grant R25HG006687. D.J. was supported by a Gary K. Ackers Fellowship and an Elliot L. Elson Education and Training Fellowship. J.J.M. was supported by NIH training grant AI007172-36A1. Purchase of the stopped-flow fluorescence equipment was made possible by equipment supplement 3R01GM107544-04S1 from the NIH. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis; the center is partially supported by NCI Cancer Center Support grant P30CA91842 to Siteman Cancer Center and by ICTS/CTSA grant UL1TR000448 from the National Center for Research Resources, a component of the NIH, and the NIH Roadmap for Medical Research.
This publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the NIH.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00690-17.
REFERENCES
- 1.World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/250441/9789241565394-eng.pdf;jsessionid=428E77D20AE697AC55FF9BAE89B48DC5?sequence=1. [Google Scholar]
- 2.Calvori C, Frontali L, Leoni LTG. 1965. Effect of rifamycin on protein synthesis. Nature 207:417–418. doi: 10.1038/207417a0. [DOI] [PubMed] [Google Scholar]
- 3.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 4.Rammohan J, Manzano AR, Garner AL, Stallings CL, Galburt EA. 2015. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res 43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garner AL, Weiss LA, Manzano AR, Galburt EA, Stallings CL. 2014. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol Microbiol 93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubin EA, Fay A, Xu C, Bean JM, Glickman MS, Darst SA, Campbell EA. 2017. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife 6:e22520. doi: 10.7554/eLife.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rammohan J, Ruiz Manzano A, Garner AL, Prusa J, Stallings CL, Galburt EA. 2016. Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD. Nucleic Acids Res 44:7304–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis E, Chen J, Leon K, Darst SA, Campbell EA. 2015. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res 43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma AK, Chatterji D. 2014. Dual role of MsRbpA: transcription activation and rescue of transcription from the inhibitory effect of rifampicin. Microbiology 160:2018–2029. doi: 10.1099/mic.0.079186-0. [DOI] [PubMed] [Google Scholar]
- 11.Bortoluzzi A, Muskett FW, Waters LC, Addis PW, Rieck B, Munder T, Schleier S, Forti F, Ghisotti D, Carr MD, O'Hare HM. 2013. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J Biol Chem 288:14438–14450. doi: 10.1074/jbc.M113.459883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey A, Verma AK, Chatterji D. 2011. Molecular insights into the mechanism of phenotypic tolerance to rifampicin conferred on mycobacterial RNA polymerase by MsRbpA. Microbiology 157:2056–2071. doi: 10.1099/mic.0.047480-0. [DOI] [PubMed] [Google Scholar]
- 13.Weiss LA, Harrison PG, Nickels BE, Glickman MS, Campbell EA, Darst SA, Stallings CL. 2012. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J Bacteriol 194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paget MSB, Molle V, Cohen G, Aharonowitz Y, Buttner MJ. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the σR regulon. Mol Microbiol 42:1007–1020. doi: 10.1046/j.1365-2958.2001.02675.x. [DOI] [PubMed] [Google Scholar]
- 15.Tabib-Salazar A, Liu B, Doughty P, Lewis RA, Ghosh S, Parsy ML, Simpson PJ, O'Dwyer K, Matthews SJ, Paget MS. 2013. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res 41:5679–5691. doi: 10.1093/nar/gkt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubin EA, Tabib-Salazar A, Humphrey LJ, Flack JE, Olinares PDB, Darst SA, Campbell EA, Paget MS. 2015. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc Natl Acad Sci U S A 112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyaci H, Chen J, Lilic M, Palka M, Mooney RA, Landick R, Darst SA, Campbell EA. 2018. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts. Elife 7:e34823. doi: 10.7554/eLife.34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pashley CA, Parish T. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol Lett 229:211–215. doi: 10.1016/S0378-1097(03)00823-1. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-y-Merchand JA, Colstonl MJ, Cox RA. 1996. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology 142:667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 20.Ko J, Heyduk T. 2014. Kinetics of promoter escape by bacterial RNA polymerase: effects of promoter contacts and transcription bubble collapse. Biochem J 463:135–144. doi: 10.1042/BJ20140179. [DOI] [PubMed] [Google Scholar]
- 21.Garner AL, Rammohan J, Huynh JP, Onder LM, Chen J, Bae B, Jensen D, Weiss LA, Ruiz Manzano A, Darst SA, Campbell EA, Nickels BE, Galburt EA, Stallings CL. 2017. Effects of increasing the affinity of CarD for RNA polymerase on Mycobacterium tuberculosis growth, rRNA transcription, and virulence. J Bacteriol 199:e00698-16. doi: 10.1128/JB.00698-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albery WJ, Knowles JR. 1976. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry 15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 23.Chen K, Hu Z, Xia Z, Zhao D, Li W, Tyler JK. 2016. The overlooked fact: fundamental need for spike-in control for virtually all genome-wide analyses. Mol Cell Biol 36:662–667. doi: 10.1128/MCB.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raju RM, Unnikrishnan M, Rubin DHF, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. 2012. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog 8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkan D, Stallings CL, Glickman MS. 2011. An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene 470:31–36. doi: 10.1016/j.gene.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee R, Rudra P, Prajapati RK, Sengupta S, Mukhopadhyay J. 2014. Optimization of recombinant Mycobacterium tuberculosis RNA polymerase expression and purification. Tuberculosis 94:397–404. doi: 10.1016/j.tube.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee R, Rudra P, Saha A, Mukhopadhyay J. 2015. Recombinant reporter assay using transcriptional machinery of Mycobacterium tuberculosis. J Bacteriol 197:646–653. doi: 10.1128/JB.02445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao Y, Smyth GK, Shi W. 2014. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.