ABSTRACT
Sinorhizobium meliloti enters into beneficial symbiotic interactions with Medicago species of legumes. Bacterial exopolysaccharides play critical signaling roles in infection thread initiation and growth during the early stages of root nodule formation. After endocytosis of S. meliloti by plant cells in the developing nodule, plant-derived nodule-specific cysteine-rich (NCR) peptides mediate terminal differentiation of the bacteria into nitrogen-fixing bacteroids. Previous transcriptional studies showed that the intensively studied cationic peptide NCR247 induces expression of the exo genes that encode the proteins required for succinoglycan biosynthesis. In addition, genetic studies have shown that some exo mutants exhibit increased sensitivity to the antimicrobial action of NCR247. Therefore, we investigated whether the symbiotically active S. meliloti exopolysaccharide succinoglycan can protect S. meliloti against the antimicrobial activity of NCR247. We discovered that high-molecular-weight forms of succinoglycan have the ability to protect S. meliloti from the antimicrobial action of the NCR247 peptide but low-molecular-weight forms of wild-type succinoglycan do not. The protective function of high-molecular-weight succinoglycan occurs via direct molecular interactions between anionic succinoglycan and the cationic NCR247 peptide, but this interaction is not chiral. Taken together, our observations suggest that S. meliloti exopolysaccharides not only may be critical during early stages of nodule invasion but also are upregulated at a late stage of symbiosis to protect bacteria against the bactericidal action of cationic NCR peptides. Our findings represent an important step forward in fully understanding the complete set of exopolysaccharide functions during legume symbiosis.
IMPORTANCE Symbiotic interactions between rhizobia and legumes are economically important for global food production. The legume symbiosis also is a major part of the global nitrogen cycle and is an ideal model system to study host-microbe interactions. Signaling between legumes and rhizobia is essential to establish symbiosis, and understanding these signals is a major goal in the field. Exopolysaccharides are important in the symbiotic context because they are essential signaling molecules during early-stage symbiosis. In this study, we provide evidence suggesting that the Sinorhizobium meliloti exopolysaccharide succinoglycan also protects the bacteria against the antimicrobial action of essential late-stage symbiosis plant peptides.
KEYWORDS: antimicrobial activity, exopolysaccharide, symbiosis
INTRODUCTION
The nitrogen-fixing bacterium Sinorhizobium meliloti can enter into mutually beneficial symbiosis with Medicago plant species. Under nitrogen-limiting environmental conditions, Medicago plants secrete aromatic flavonoid compounds into the surrounding soil, signaling S. meliloti bacteria to synthesize and to secrete the lipochitooligosaccharide Nod factor. Nod factor induces plant root hair curling to trap surrounding bacteria. Subsequent signaling between the plant and the bacteria leads to the initiation of tubular structures called infection threads, which elongate in the root hair toward cortical root cell layers. These infection threads support bacterial propagation and invasion into the developing nodule, eventually leading bacteria to specialized host cells that use an endocytosis-like process to internalize bacteria. Intracellular S. meliloti bacteria are contained within a host-derived membrane compartment called a symbiosome, and host signals subsequently cause these internalized bacteria to differentiate into nitrogen-fixing bacteroids. S. meliloti bacteroids supply fixed nitrogen, in the form of ammonia (NH3), to the plant host in exchange for fixed carbon, in the form of dicarboxylic acids (1).
Much of our understanding of the signaling molecules and pathways required for nitrogen-fixing symbiosis stem from genetic studies of S. meliloti and the host plants Medicago truncatula and Medicago sativa. Research by our laboratory and others has shown that S. meliloti exopolysaccharides are key signaling molecules for infection thread initiation and growth (2–5). S. meliloti is capable of synthesizing the exopolysaccharides succinoglycan and galactoglucan, and both polysaccharides can independently support symbiosis with host plants. Succinoglycan is a polymer of octasaccharide subunits, each of which is composed of one galactose residue and seven glucose residues, modified with acetyl, succinyl, and pyruvyl groups (6). Galactoglucan is a polymer of disaccharide subunits, each of which is composed of a glucose residue and a galactose residue, modified with an acetyl group and a pyruvyl group, respectively (7). S. meliloti produces and secretes two forms of succinoglycan, i.e., a high-molecular-weight (HMW) form composed of hundreds of octasaccharide subunits (8) and a low-molecular-weight (LMW) form composed of one to three octasaccharide subunits (9). Of these S. meliloti polysaccharides, succinoglycan is the best understood in terms of its structure and functions during legume symbiosis. S. meliloti bacteria that harbor mutations abolishing succinoglycan biosynthesis, modification, or polymerization initiate legume symbiosis and promote root hair curling but either fail to induce the host to form an infection thread through which plant tissue invasion is achieved or fail to support continued growth of an infection thread, if one is initiated (3). Results of previous work suggest that S. meliloti bacteria that lack functional ExoY, the phosphoglycosyltransferase that initiates succinoglycan biosynthesis, elicit stronger expression of plant defense genes than do wild-type bacteria, suggesting that one potential mode of action of succinoglycan is to dampen the response of the plant immune system (10). This phenotype led to the hypothesis that one function of succinoglycan is to serve as a signaling molecule to the host plant. Recent research by Kawaharada et al. (11, 12) confirmed this hypothesis by showing that a host plant receptor selectively binds to bacterial exopolysaccharides and this recognition by the bacterial exopolysaccharide receptor is necessary for infection thread growth and bacterial invasion across the epidermal cell layers.
Work in the host plant M. truncatula identified signaling factors that S. meliloti encounters after entering host cells, inside the symbiosomes. These signals represent a family of more than 600 nodule-specific cysteine-rich (NCR) peptides that are essential for the differentiation of bacteria into nitrogen-fixing bacteroids (13–16). NCR peptides show high levels of diversity in amino acid sequence, length, and charge but have conserved patterns of either four or six cysteines (17). Recently, two NCR peptides, namely, NCR211 (18) and NCR169 (19), have been shown to be essential for Sinorhizobium-Medicago symbiosis. One NCR peptide that has been extensively studied for its physiological activities regarding S. meliloti is the cationic peptide NCR247. NCR247 possesses antimicrobial activity at relatively high concentrations (20 μM) (13, 20). In contrast, at a sublethal dose (4 μM), NCR247 induces massive transcriptional changes (about 20% of annotated genes) in cell cycle-synchronized bacterial cultures, inhibits cell division, and inhibits translation (15, 21). This group of genes includes all genes involved in the synthesis of succinoglycan. NCR247 has the sequence RNGCIVDPRCPYQQCRRPLYCRRR, which includes four cysteines, six arginines, and some hydrophobic residues. When NCR247 is oxidized, the presence of four cysteines allows the formation of three distinct oxidized regioisomers with different disulfide connectivities. Each regioisomer has unique effects on the action of NCR247, affecting its various physiological actions (21). The disulfide connectivity inside Medicago root nodules has not yet been established for any NCR peptide.
The exopolysaccharide succinoglycan is known to be important for early symbiosis. However, recently published studies suggest late-stage symbiotic functions of succinoglycan. Interestingly, the many genes whose transcription is modulated by NCR247 treatment include the ExoS-ChvI regulon, which includes the genes involved in exopolysaccharide synthesis, export, and polymerization (15, 22). Furthermore, a recent genomic, sequencing-based screen for transposon mutations that sensitize or protect S. meliloti versus NCR247 antimicrobial activity found that some of the genes induced by NCR247 treatment were among the 78 genes affecting NCR247 sensitivity (15, 23). Specifically, transposon insertions in genes exoW, exoV, exoT, and exoQ, which are part of the final stages of succinoglycan biosynthesis, resulted in significant loss of competitiveness during NCR247 treatment (23). Therefore, NCR247 induced expression of these genes, and increased exopolysaccharide expression may be required for resistance to the bactericidal effects of NCR247 during bacteroid differentiation (23). Additional evidence for late-stage functions of succinoglycan can be hypothesized based on published transcriptome data derived from laser-dissected nodule sections, which show that a number of genes encoding succinoglycan biosynthesis proteins are upregulated later during symbiosis (see Fig. S1A in the supplemental material) (24). It remains to be determined why genes that encode exopolysaccharide biosynthesis proteins are upregulated past the initial stage of symbiosis, for which they have been shown to be essential.
In this study, we investigated whether a second mode of action by which S. meliloti exopolysaccharides might exert their biological effects is by modulating the antimicrobial activity of NCR peptides later during symbiosis. To investigate this issue, we treated S. meliloti succinoglycan biosynthesis gain-of-function and loss-of-function mutants with a lethal dose of the model peptide NCR247 and demonstrated that the production of succinoglycan resulted in greater resistance to the antimicrobial action of NCR247. Using two independent approaches for peptide-polysaccharide interactions, we obtained evidence that succinoglycan and NCR247 interact directly and this direct interaction is affected by the presence of both the anionic pyruvyl and succinyl modifications and the noncharged acetyl modification. We provide evidence that this interaction is, at least partly, ionic in nature. Thus, our results suggest that, in addition to serving as a signaling molecule in incipient infection threads, the exopolysaccharide succinoglycan may provide a level of protection to S. meliloti cells when they are exposed to NCR247 and similar NCR peptides in the symbiosomes of host cells.
RESULTS
Strains that overproduce succinoglycan have enhanced resistance to NCR247 antimicrobial activity.
Previously, we showed that mutations that eliminate the final processing steps of succinoglycan production sensitize S. meliloti to the antimicrobial activity of NCR247, and we hypothesized that succinoglycan production may provide protection against NCR247 (23). An alternative possibility is that mutations in genes that affect the final processing steps of succinoglycan stress and weaken cellular structures, increasing the sensitivity of the cells to antimicrobial peptides, such as NCR247, that target the cell membrane. To begin testing the hypothesis that succinoglycan production is protective against NCR247, we treated strains that overproduce succinoglycan and parental wild-type strains with a dose of NCR247 that is lethal to the wild-type S. meliloti strain Sm1021, and we monitored bacterial survival hourly over a 5-h period during treatment. In this experiment, we used Sm1021 derivatives carrying either the chvID52E (25) or exoS96 (26) mutation, both of which result in hyperactivation of the ExoS-ChvI two-component system. The ExoS-ChvI two-component system regulates a suite of genes, including those for succinoglycan biosynthesis (25), and expression of the ExoS-ChvI regulon is stimulated by bacteriostatic levels of NCR247 (15). We found that strains bearing either the chvID52E (Fig. 1A) or exoS96 (Fig. 1B) alleles exhibited significant increases in resistance to NCR247, relative to their parental wild-type or exoY mutant control strains, which possess similar antimicrobial sensitivities to NCR247 under our experimental conditions (23). The protective effect of the chvID52E and exoS96 alleles was dependent on succinoglycan production, as loss-of-function mutations in either exoY (Fig. 1A) or exoA (Fig. 1B), which encodes the UDP-galactose-undecaprenyl-phosphate galactose-I-phosphate transferase or a glycosyltransferase, respectively, that initiate succinoglycan production and therefore are essential for succinoglycan production (22), significantly increased NCR247 sensitivity, compared to the S. meliloti Sm1021 derivatives with the chvID52E or exoS96 alleles (Fig. 1A and B). These observations show that constitutive activation of the ExoS-ChvI regulon enhances the resistance of S. meliloti to NCR247 and this increased resistance is dependent on a functional succinoglycan biosynthesis pathway.
FIG 1.
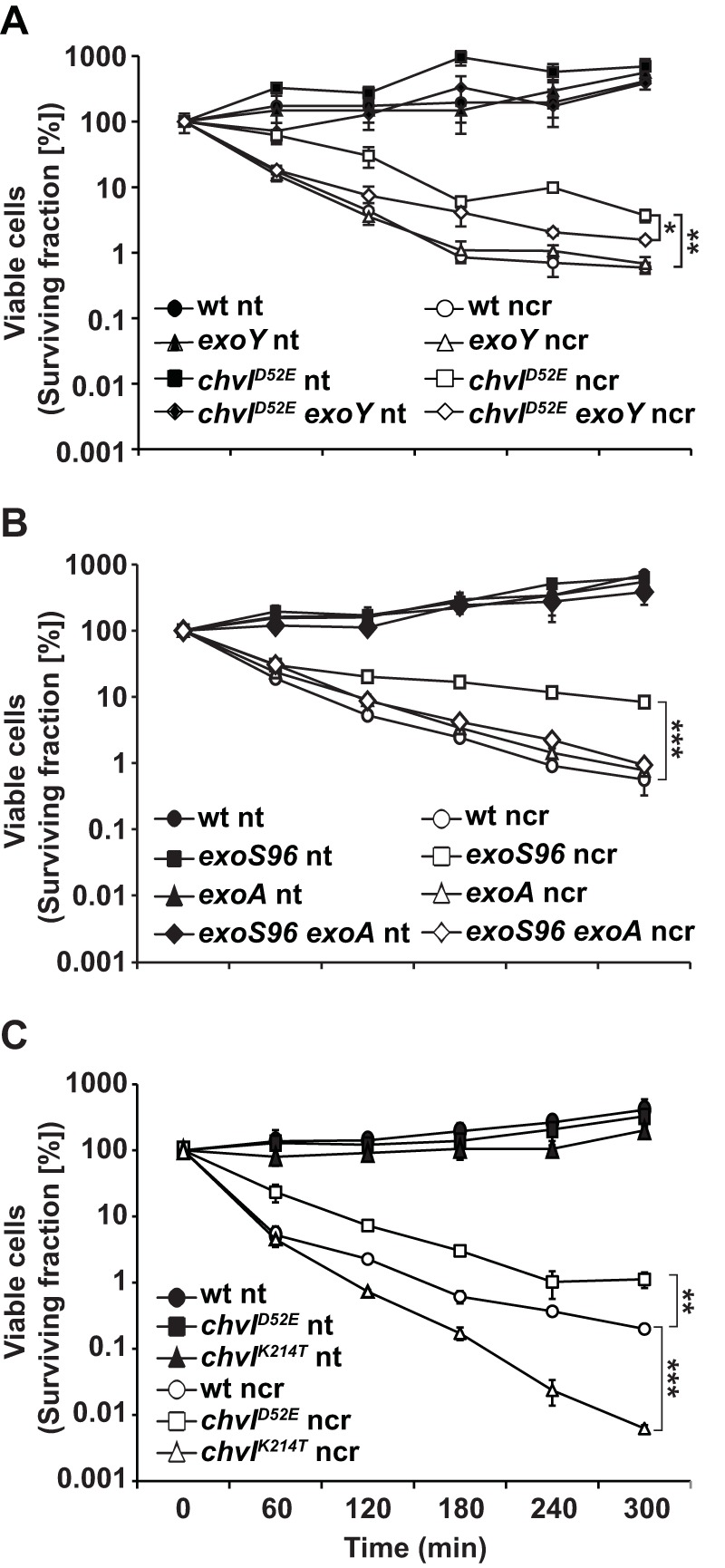
Protection against the antimicrobial activity of NCR247 with hyperactivation of exo gene regulators. Early-exponential-phase cells of the defined strains were treated for 5 h with 20 μM NCR247 in MOPS-GS buffer with CAs, and the viable cells were enumerated at the defined time points. Data points and error bars indicate the means ± standard deviations. The results shown are representative of trends observed in at least two independent experiments. For space reasons, significant differences are indicated only for the 5-h time points. wt, wild-type; nt, not treated; ncr, NCR247. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
We also assayed the sensitivity of bacteria carrying the partial loss-of-function allele ChvIK214T, which significantly depresses expression of the ExoS-ChvI regulon, including succinoglycan biosynthesis genes (27). Consistent with the aforementioned results, we found that the ChvIK214T strain exhibited enhanced sensitivity to NCR247 antimicrobial activity, relative to wild-type cells (Fig. 1C).
Exogenous addition of succinoglycan protects S. meliloti against the antimicrobial activity of NCR247.
We investigated whether exogenous addition of succinoglycan could protect S. meliloti that was simultaneously treated with a bactericidal dose of NCR247. We lyophilized the supernatant of cultures of exoS96 mutant cells, redissolved the supernatant in water, determined the polysaccharide concentration using anthrone-sulfuric acid assays, and then added defined concentrations of polysaccharide to S. meliloti wild-type cells, along with NCR247. We found that 10 μg ml−1 of exogenous succinoglycan increased the survival rates of NCR247-treated cells by about 10-fold, relative to cells treated with NCR247 alone (Fig. 2A). Interestingly, we observed a similar level of protection afforded to NCR247-treated S. meliloti cells with succinoglycan at 50 μg ml−1 (Fig. 2A). Thus, these findings indicate that the presence of succinoglycan can provide protection from the antimicrobial effects of NCR247 for S. meliloti.
FIG 2.
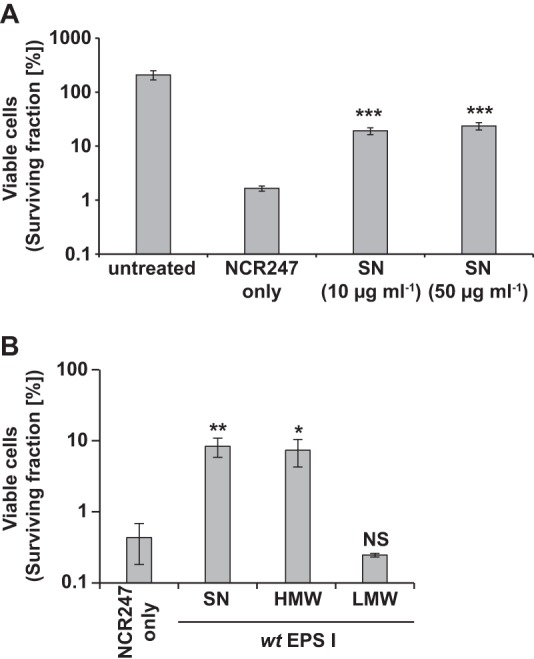
Protection against the antimicrobial activity of cationic NCR247 with extracellular addition of succinoglycan. Early-exponential-phase S. meliloti cells were treated for 5 h with 20 μM NCR247 and 10 μg ml−1 of the defined succinoglycan preparations in MOPS-GS buffer with CAs, and then the number of viable cells was determined. Bars and error bars indicate the means ± standard deviations. The results shown are representative of trends observed in at least two independent experiments. SN, supernatant; wt, wild-type; EPS, exopolysaccharide (succinoglycan); NS, not significant. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05.
HMW succinoglycan is essential for protection against the antimicrobial action of NCR247.
S. meliloti secretes two forms of succinoglycan, a HMW form and a LMW form (9). We tested whether both the HMW and LMW forms of succinoglycan could protect S. meliloti from the antimicrobial effects of NCR247. Using the supernatant of the succinoglycan-overexpressing exoS96 mutant strain, we separately precipitated the HMW and LMW fractions of succinoglycan, as described previously (9), and then tested the effects of each fraction on the bactericidal action of NCR247. We found that addition of HMW succinoglycan (10 μg ml−1) enhanced the survival of S. meliloti by 1 order of magnitude, relative to cultures treated with NCR247 alone (Fig. 2B). In contrast, addition of the LMW form (10 μg ml−1) had no effect on the survival of S. meliloti treated with NCR247 (Fig. 2B). Thus, the HMW form of succinoglycan is necessary to enhance the survival of S. meliloti treated with NCR247.
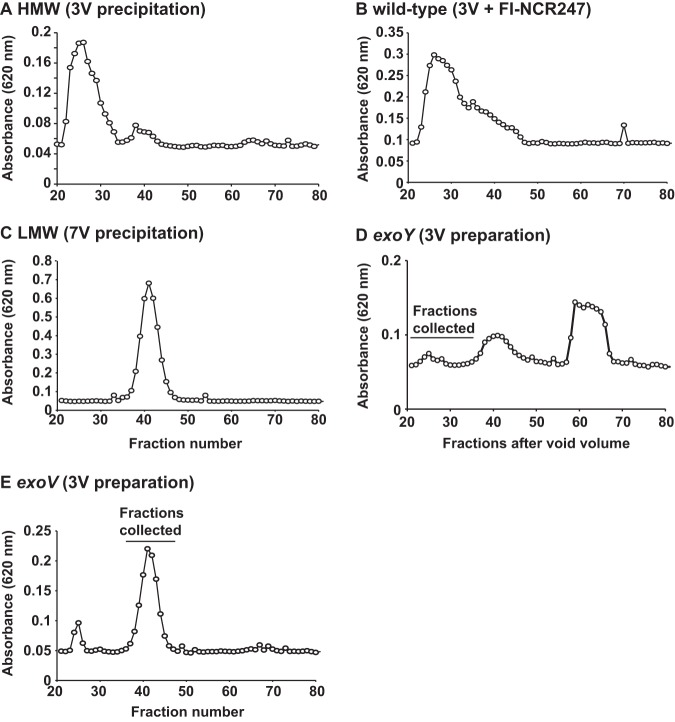
As an additional control, we analyzed the supernatant of an exoY mutant after precipitation with 3 volumes of ethanol. As reported recently (23), an exoY mutant that lacks the ability to synthesize succinoglycan does not show increased sensitivity to NCR247 (see Fig. S2 in the supplemental material). We collected fractions 20 to 35 after separation of wild-type and exoY supernatants on a Bio-Gel P-6 column, after ethanol precipitation, and assessed whether the preparations were able to provide protection against the antimicrobial action of NCR247 (see below for size exclusion chromatography data). We found that the supernatant collected from the exoY mutant did not contain any significant amounts of polysaccharide corresponding to HMW succinoglycan (see Fig. 5A and D). As expected, we found that the polysaccharide material collected from fractions 20 to 35 from wild-type supernatant did protect against NCR247 (Fig. S2) and the material recovered from the corresponding fractions of the exoY mutant did not have the same effect on NCR247 sensitivity (Fig. S2). This result confirms our findings that HMW succinoglycan protects against the antimicrobial action of NCR247.
FIG 5.
Graphical representation of the polysaccharide contents of fractions collected from size exclusion chromatography. A defined amount of succinoglycan (500 μg) was loaded onto a Bio-Rad Bio-Gel P-6 column, 1.6-ml fractions were collected, and the polysaccharide content of each fraction was determined using the anthrone-sulfuric acid assay. 3V, supernatant precipitated with 3 volumes of ethanol; 7V, 3-volume fraction precipitated with an additional 7 volumes of ethanol.
Bacterial protection by HMW succinoglycan is not universal for cationic peptides.
We found that HMW succinoglycan has protective properties during NCR247 treatment of S. meliloti. Therefore, we tested whether this protective function is a general property during challenge with cationic peptides or whether our findings are specific for NCR247. To test our hypothesis, we treated an Sm1021 wild-type strain with the antibiotic polymyxin B (28), the bee venom component melittin (29), human α-defensin 5 (HD5) (30), the human antimicrobial peptide LL-37 (31), and the bovine antimicrobial peptide fragment Bac71–35 (32). Our results indicate that the protective effects of succinoglycan against cationic peptides are not universal. Succinoglycan protects against the antimicrobial effects of only Bac7 to a similar degree as for NCR247 (Fig. 3A). These results suggest that there is something special about the effects of succinoglycan on the antimicrobial effects of NCR247.
FIG 3.
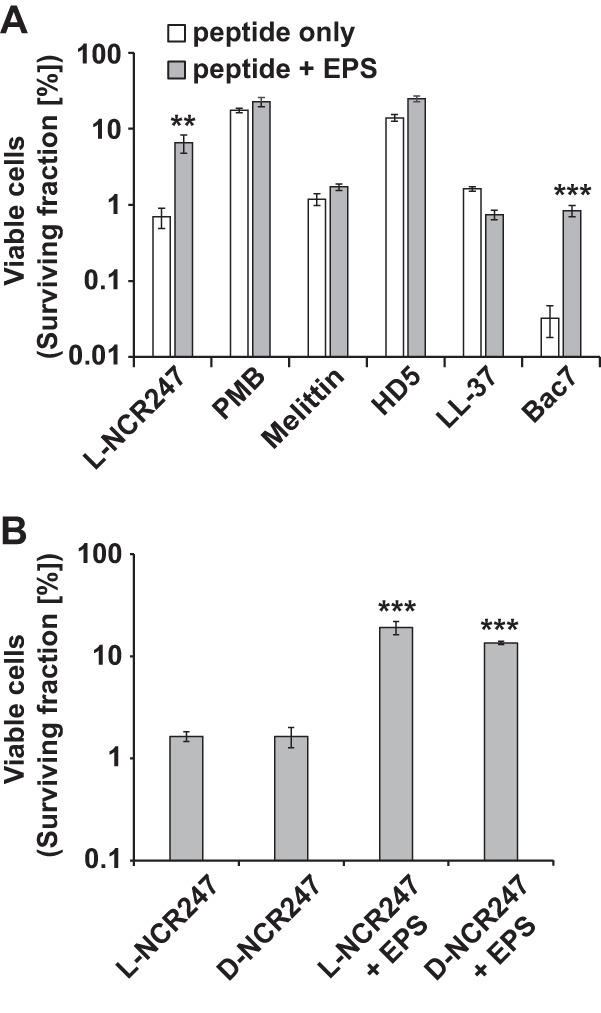
S. meliloti succinoglycan protection against NCR247 and Bac7. (A) S. meliloti Sm1021 cells were treated for 5 h with l-NCR247 (20 μM), polymyxin B (PMB) (1.75 μg ml−1), melittin (1 μM), human defensin 5 (HD5) (20 μM), the human antimicrobial peptide LL-37 (0.5 μM), or Bac71–35 (0.5 μM) and then recovered on LBMC agar plates supplemented with 200 μg ml−1 streptomycin. Results shown are representative of trends observed in at least two independent experiments. (B) Early-exponential-phase S. meliloti wild-type cells were treated with 20 μM l- or d-NCR247 and supplemented with 10 μg ml−1 wild-type HMW succinoglycan as indicated. Bars and error bars indicate means ± standard deviations. The results shown are representative of trends observed in at least two independent experiments. EPS, exopolysaccharide (succinoglycan). **, P ≤ 0.01; ***, P ≤ 0.001.
Bacterial protection by HMW succinoglycan is not dependent on chiral interactions.
One way to probe the nature of a molecular interaction is to change the chirality of one of the interacting biomolecules (32, 33). Therefore, we obtained and purified an all-d-enantiomer of NCR247, made up of only d-amino acids, and found its lethality to be indistinguishable from that of the l-enantiomer (Fig. 3B). To test whether succinoglycan also protects against the NCR247 d-enantiomer, we assayed and compared the degree to which exogenously added HMW succinoglycan antagonized the l- and d-enantiomers of NCR247, and we found that HMW succinoglycan antagonized both NCR247 enantiomers, to similar extents (Fig. 3B). These observations suggest that succinoglycan antagonizes NCR247 largely through mechanisms that do not involve chiral interactions.
HMW succinoglycan directly interacts with NCR247.
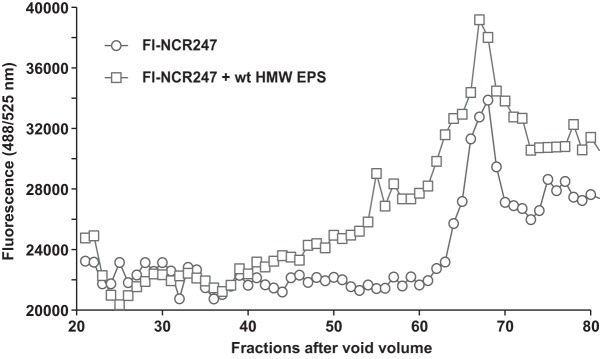
Since NCR247 carries six positive charges, it seemed possible that the mechanism underlying succinoglycan attenuation of NCR247 bactericidal activity involved a direct interaction between the anionic polysaccharide and the cationic peptide. To test this, we used two approaches with two different modified NCR247 peptides. First, we utilized size exclusion chromatography to investigate changes in the retention times of a fluorescein isothiocyanate (FITC)-labeled derivative of the reduced NCR247 peptide (FI-NCR247) (21) and the HMW fractions of wild-type succinoglycan. The bactericidal activity of FI-NCR247 was the same as that of the unmodified reduced NCR247 peptide (21). Initially, we used CoCl2 to establish the column volume of a Bio-Gel P-6 column (fractions 70 to 80) (Fig. S3). Subsequently, we established that FI-NCR247 eluted slightly earlier, in fractions 63 to 70 (Fig. 4), by measuring the FITC fluorescence intensity for each elution fraction. S. meliloti HMW succinoglycan was shown previously to run in the early-eluting fractions that correspond to the void volume of a comparable column setup (9). We found that wild-type HMW succinoglycan eluted just after fraction 20 (Fig. 5A); therefore, we analyzed only elution fractions between fractions 20 and 80 for the following experiments. When FI-NCR247 was coloaded with HMW succinoglycan, much of the FITC fluorescence of FI-NCR247 eluted much earlier, in a broad range from fraction 40 to fraction 62, compared to the curve representing only FI-NCR247 (Fig. 4). This shift in FI-NCR247 fluorescence is consistent with FI-NCR247 binding directly to HMW succinoglycan but slowly coming off as the HMW succinoglycan progresses down the column. Analysis of the polysaccharide content in each fraction using the anthrone-sulfuric acid assay showed that most of the wild-type HMW succinoglycan still eluted in fractions 20 to 30 when coloaded with FI-NCR247 (Fig. 5B). We also determined that LMW succinoglycan eluted between HMW succinoglycan and FI-NCR247, in fractions 37 and 44 (Fig. 5C).
FIG 4.
Effects of purified succinoglycan on NCR247 mobility on a size exclusion column. FI-NCR247 was mixed with 500 μg of the HMW succinoglycan fraction from the wild-type (wt) strain, in a final volume of 2 ml, and loaded onto a Bio-Rad Bio-Gel P-6 (fine mesh) size exclusion column. The fluorescence of each fraction, not including the void volume, was measured (excitation wavelength, 488 nm; emission wavelength, 525 nm) and plotted. All data are the averages of at least three independent experiments. EPS, exopolysaccharide (succinoglycan).
As a second approach to investigate the direct interactions between NCR247 and succinoglycan, we used Octet (ForteBio) biolayer interferometry. This method differs from our size exclusion column experiments in multiple ways. This methodology is used to determine direct molecular interactions based on light interference patterns changed by molecular binding of an analyte molecule in solution to an immobilized, biosensor-tip-bound, ligand molecule. An increase in binding signal is the net difference of binding and dissociation processes. Our experimental setup used streptavidin-coated biosensor tips loaded with biotinylated NCR247 peptide (see Materials and Methods for peptide details). An experiment using the ForteBio Octet platform for biolayer interferometry involves multiple stages. After incubation of the biosensor tips in assay buffer, the first stage is ligand (biotin-NCR247) binding to the streptavidin-coated biosensor tips (Fig. 6A). Ligand binding is followed by analyte (HMW succinoglycan) association in assay buffer (Fig. 6A). Finally, the association step is stopped by placing the biosensor tips into assay buffer without solubilized succinoglycan. The results shown in Fig. 6A represent two representative concentrations of NCR247 and 10 μg ml−1 of wild-type HMW succinoglycan. Based on preliminary experiments, we decided to use NCR247 at a final concentration of 0.7 μM to load the biosensor tips for all subsequent experiments. All results shown for the following experiments focus on the association and dissociation stages of our binding studies, with the data being normalized on the y axis to the beginning of the association stage.
FIG 6.
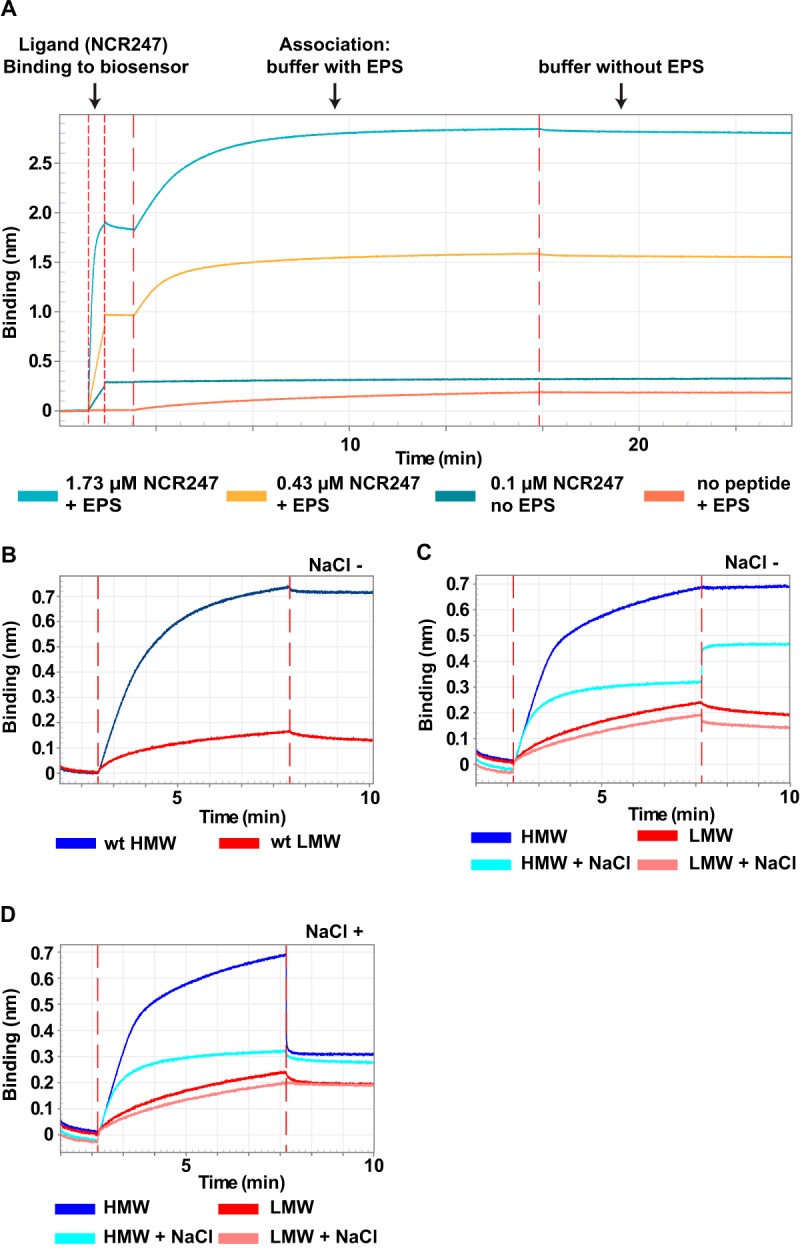
Direct interaction of NCR247 with succinoglycan. Octet biolayer interferometry assays were performed as described in Materials and Methods. (A) Overview of all stages of an Octet biolayer interferometry assay. The steps include NCR247-biotin binding to the biosensor tip and the association of solubilized succinoglycan with immobilized NCR247-biotin. In a final step, the loaded biosensor tips are immersed in assay buffer without succinoglycan. (B to D) Octet biolayer interferometry assay results. The concentration of free succinoglycan was kept constant at 10 μg ml−1 for the HMW and LMW forms. Where indicated, the ammonium acetate assay buffer was supplemented with 0.1 M NaCl. Times of 1.7 to 2.7 min, buffer; 2.7 to 8.2 min, buffer plus HMW or LMW succinoglycan; 8.2 to 10 min, buffer without salt (NaCl −) (B and C) or with 0.1 M salt (NaCl +) (D) (times varied minimally between experiments). Results shown are representative of trends observed in at least two independent experiments. wt, wild-type; EPS, exopolysaccharide (succinoglycan).
We found that wild-type HMW succinoglycan bound rapidly to the immobilized biotinylated NCR247 peptide, as indicated by the rapidly increasing binding signal (Fig. 6B). We did not observe significant reduction in the binding signal when the biosensors were placed back into assay buffer (Fig. 6B). These findings confirmed our results obtained with the size exclusion column (Fig. 4). Because biolayer interferometry requires only very small amounts of ligand and analyte, we were also able to test the interaction of NCR247 with LMW succinoglycan. We observed significantly less binding, as indicated by the nearly flat binding curve, compared to the HMW succinoglycan curve (Fig. 6B).
The interaction between NCR247 and succinoglycan is salt dependent.
To assess whether the observed interactions between HMW succinoglycan and NCR247 are ionic in nature, we conducted further experiments with a modified association buffer. To increase the ionic strength, we added NaCl to a final concentration of 0.1 M. We found that 0.1 M NaCl in the association buffer drastically reduced the binding of HMW succinoglycan to biosensor-bound NCR247, compared to HMW succinoglycan without additional NaCl (Fig. 6C). This observation indicates that NaCl ions compete with succinoglycan molecules for binding to cationic NCR247. The small increase in binding signal after the biosensor was moved from the NaCl-containing buffer back into assay buffer without NaCl might be a consequence of different optical properties of the buffers that affect sensor readings in the presence of HMW succinoglycan (Fig. 6C). For LMW succinoglycan, NaCl in the association buffer had very little effect on the low level of binding observed (Fig. 6C). Next, we tested whether we could use NaCl to force dissociation of succinoglycan from the NCR247-loaded biosensor tips. When NCR247-loaded biosensors with bound HMW succinoglycan were placed in assay buffer containing 0.1 M NaCl, the binding signal rapidly decreased to the level observed when succinoglycan was allowed to associate with NCR247 in assay buffer supplemented with 0.1 M NaCl (Fig. 6D). Taken together, these observations (Fig. 6C and D) suggest that the molecular basis of the interaction between NCR247 and succinoglycan is at least partially ionic.
Succinyl, pyruvyl, and acetyl modifications of succinoglycan affect the interaction between NCR247 and succinoglycan.
Succinoglycan is decorated with three well-characterized side modifications, namely, succinyl, acetyl, and pyruvyl residues; S. meliloti mutants that lack intact exoH, exoZ, and exoV genes lack these respective modifications. To further investigate the molecular characteristics of the interaction between succinoglycan and NCR247, we decided to purify succinoglycan from these three S. meliloti mutants. We collected the supernatants from these mutants and precipitated succinoglycan with 3 volumes of ethanol.
Succinoglycan produced by an exoH mutant lacks the anionic succinyl modification (34). An exoH mutant that has been well characterized predominantly produces HMW succinoglycan (8). Just like wild-type HMW succinoglycan, HMW succinoglycan from an exoH mutant was able to protect against the antimicrobial action of NCR247 (Fig. 7). Biolayer interferometry assays showed that exoH mutant succinoglycan displayed a substantially increased binding rate and a greater degree of binding, compared to wild-type HMW succinoglycan (Fig. 8A; also see Fig. S4A in the supplemental material). We also observed that the addition of NaCl to the assay buffer after the NCR247-exoH succinoglycan association resulted in a slower reduction in the binding signal from the NCR247-coated biosensor tip, compared to wild-type succinoglycan (Fig. 8A). This finding indicates that NCR247 forms a stronger association with exoH succinoglycan than with wild-type succinoglycan (Fig. 8A) and that the loss of this particular anionic succinoglycan modification results in increased NCR247-succinoglycan binding. It is interesting that, despite its negative charge, the succinyl group seems to impair the ability of succinoglycan to interact with NCR247, and the improved binding observed in the absence of the succinyl modification might involve hydrophobic interactions rather than ionic interactions.
FIG 7.
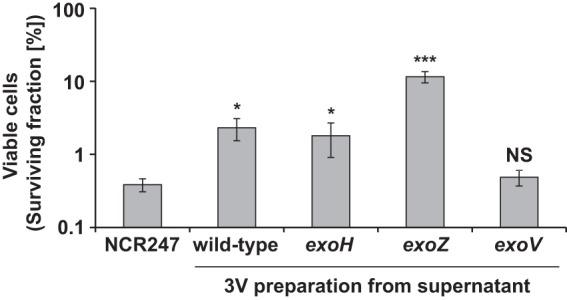
Protection by HMW succinoglycan from exoH and exoZ mutants against the antimicrobial action of NCR247. Early-exponential-phase cells of S. meliloti were treated for 5 h with 20 μM NCR247 and 10 μg ml−1 of the defined succinoglycan preparations in MOPS-GS buffer with CAs, and then the number of viable cells was determined. Bars and error bars indicate the means ± standard deviations. The results are representative of at least two independent experiments. NS, not significant; 3V, supernatant precipitated with 3 volumes of ethanol. ***, P ≤ 0.001; *, P ≤ 0.05.
FIG 8.
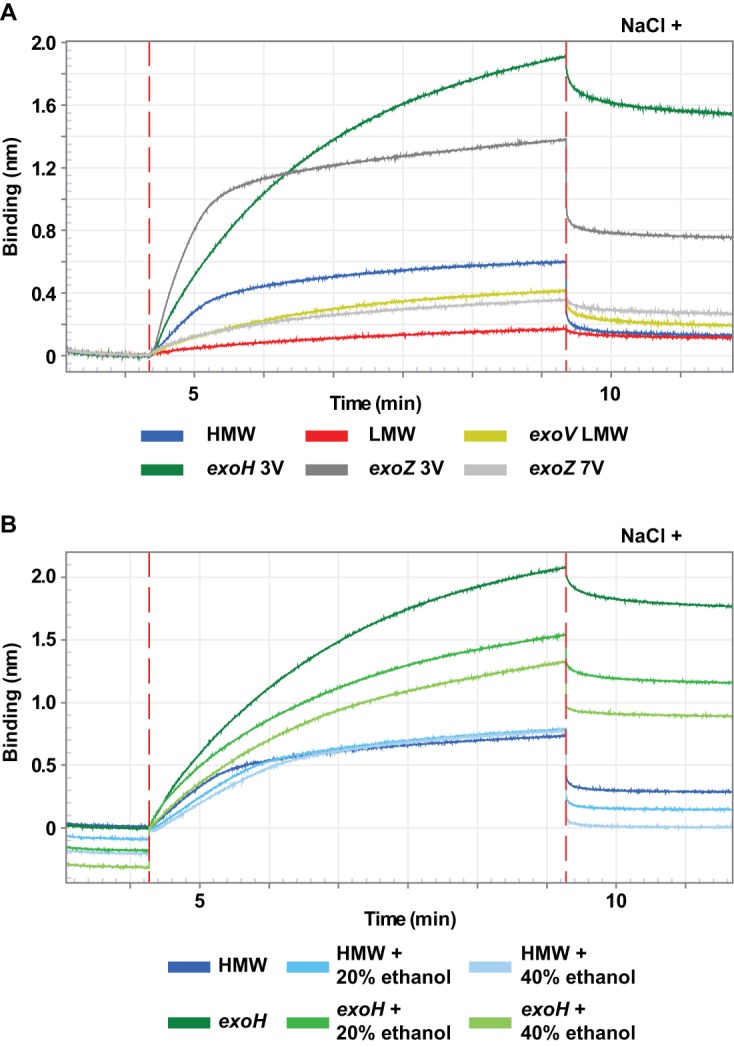
Succinoglycan-modifying residues affecting polysaccharide binding to NCR247. Summary plots of biolayer interferometry experiments with succinoglycan preparations from S. meliloti mutants that are unable to modify succinoglycan with one of three modifications, i.e., succinyl (exoH), pyruvyl (exoV), or acetyl (exoZ). Octet biolayer interferometry assays were performed as described in Materials and Methods. The concentration of free succinoglycan was kept constant at 10 μg ml−1 for all derivatives. Where indicated, the ammonium acetate assay buffer was supplemented with 0.1 M NaCl or the indicated amounts of ethanol. Results shown are representative of trends observed in at least two independent experiments. 3V, supernatant precipitated with 3 volumes of ethanol; 7V, 3-volume fraction precipitated with an additional 7 volumes of ethanol.
To assess whether the improved binding of the HMW succinoglycan obtained from the exoH mutant might be due to hydrophobic interactions, potentially involving the methyl groups on the acetyl and pyruvyl groups, we performed Octet binding assays in the presence of ethanol. We found that the ability of wild-type HMW succinoglycan to bind NCR247 was not affected by ethanol (Fig. 8B, association stage). The wild-type HMW samples incubated in the presence of ethanol showed higher rates of dissociation when the sensor tips were placed in ammonium acetate buffer supplemented with NaCl (Fig. 8B, dissociation stage). In contrast, NCR247 binding to succinoglycan precipitated from the exoH mutant supernatant was drastically reduced in the presence of ethanol and was dependent on the ethanol concentration (Fig. 8B). These findings indicate that hydrophobic interactions between NCR247 and succinoglycan play a greater role when the succinyl modification is missing from succinoglycan.
Mutation of the exoZ gene results in the secretion of succinoglycan lacking the noncharged acetyl modification (35). Functional ExoZ is not essential for successful symbiosis (36). We precipitated HMW succinoglycan and then LMW succinoglycan, with an additional 7 volumes of ethanol, from the supernatant of this mutant. We found that exoZ mutant HMW succinoglycan was able to protect S. meliloti against the antimicrobial action of NCR247 (Fig. 7). Our results using biolayer interferometry showed that HMW and LMW succinoglycan derived from the exoZ mutant bound NCR247 more efficiently than did the corresponding samples prepared from wild-type supernatant (Fig. 8A; also see Fig. S4B). Interestingly, as was the case for the succinyl modification, the effect of the acetyl modification appeared to be to attenuate the interaction between NCR247 and succinoglycan.
Finally, we examined whether succinoglycan from an exoV mutant, which lacked succinoglycan with the pyruvyl modification, was able to bind to NCR247. The pyruvyl modification of succinoglycan was hypothesized to be important for the formation of HMW succinoglycan, suggesting that an exoV mutant would be unable to produce significant amounts of HMW succinoglycan (37). We observed that the succinoglycan in the supernatant of the exoV mutant that precipitated after the addition of 3 volumes of ethanol contained a small amount of HMW succinoglycan and a correspondingly larger amount of LMW molecules, compared to the wild-type sample prepared with 3 volumes of ethanol (compare Fig. 5A and E). This preparation did not provide protection from the antimicrobial activity of NCR247 (Fig. 7).
We purified fractions 37 to 47, which presumably contained LMW succinoglycan, from the exoV mutant (Fig. 5E). We found that the succinoglycan from those fractions bound less well than HMW succinoglycan from wild-type S. meliloti but showed the same NCR247-binding profile as the exoV fraction precipitated with 3 volumes of ethanol (Fig. S4C) and the LMW succinoglycan from an exoZ mutant (Fig. 8A). We were unable to purify enough material from the HMW peak for our experiments (Fig. 5E). These findings confirm that the lack of the anionic pyruvyl modification interferes with the polymerization of succinoglycan subunits in HMW material (37) but permits the LMW material to interact with NCR247 to some degree. However, this effect does not seem to be great enough to result in protection from the antimicrobial action of NCR247 (Fig. 7).
DISCUSSION
Previously, our laboratory reported that the intensively studied NCR247 peptide induced global transcriptional changes in S. meliloti at a sublethal concentration (15). The genes whose expression was increased included those involved in the biosynthesis of succinoglycan. Furthermore, our research also identified several examples of genes required for succinoglycan synthesis that were important for protecting cells from the antimicrobial effects of NCR247 (23). These observations suggest that NCR peptides upregulate expression of succinoglycan biosynthesis genes and succinoglycan then exerts a protective role that is potentially important for symbiosis. Therefore, we asked whether S. meliloti succinoglycan is able to protect the bacteria from the antimicrobial activities of the cationic model peptide NCR247.
We provide data suggesting that succinoglycan can protect S. meliloti against the antimicrobial activities of NCR247. We also show that succinoglycan must be present in its HMW form to exert this protective function. Furthermore, we used two independent methodologies with two distinct modified NCR247 peptides to provide evidence that the protective function of NCR247 is relayed by direct molecular interactions between the cationic NCR247 peptide and the anionic HMW succinoglycan. Therefore, in addition to its role in promoting and maintaining directed infection thread growth, our data suggest that S. meliloti exopolysaccharides may have an additional symbiotic function, namely, interacting with cationic NCR peptides to reduce their potential bactericidal effects.
Succinoglycan is essential for S. meliloti to establish successful symbiosis with Medicago plant species, by mediating directed and efficient infection thread growth inside root hairs (3). This conclusion is consistent with a previously published, comprehensive, nodule laser-dissection gene expression study (24) that showed that all exo genes that are involved in succinoglycan biosynthesis are expressed at relatively high levels during nodule infection in early symbiotic stages and their expression then decreases (see Fig. S1A in the supplemental material) (24). However, eight of the succinoglycan biosynthesis genes are then reexpressed at significant levels in the nitrogen fixation zone (ZIII), where some of the commonly studied NCR peptides also display elevated gene expression (Fig. S1B). These observations from nodule laser-dissection studies are consistent with the evidence presented here showing that succinoglycan may have a second, late-stage role during symbiosis, in which it helps modulate the potential toxicity of NCR peptides.
Our results suggest that it is the HMW form of succinoglycan that is required for protection against the antimicrobial action of NCR247, with the anionic charge of succinoglycan allowing it to function as a chelator to mask the cationic charges of NCR247. The observation that increasing the amount of HMW succinoglycan does not result in complete protection (Fig. 2A) raises the possibility that succinoglycan forms a proximal protective layer around the bacteria that limits the antimicrobial effect of NCR247 on the bacteria. Our results also indicate that this effect is not stereospecific and thus is most likely due to electrostatic interactions, with possible hydrophobic contributions as well. Another nonexclusionary possibility is that succinoglycan binds to a currently unidentified bacterial receptor that is important for NCR247 activity, which results in buffering of NCR247 antimicrobial effects. The magnitude of this protective effect was not seen equally with all cationic antimicrobial peptides we investigated (Fig. 3). Among the antimicrobial peptides we investigated, the protective effect of succinoglycan was strongest for NCR247 and the bovine proline-rich Bac7 peptides. These peptides have been studied in the context of the symbiotically essential S. meliloti BacA protein (20, 38, 39); however, currently it is not known whether there is a functional connection between BacA and succinoglycan. To assess how broad this exopolysaccharide-mediated protection is against NCR peptides, future work should examine NCR peptides of different charges and other antimicrobial peptides. LMW succinoglycan may not be able to form a charged mesh that is dense enough to chelate the attacking NCR247. However, a recent study found LMW succinoglycan to be important for bacterial protection against acidic pH levels, similar to those encountered by bacteria inside symbiosomes (40).
It is interesting that loss of either the succinyl modification or the acetyl modification increased the interaction between HMW succinoglycan and NCR247. These observations raise the possibility that at least one biological role for these poorly understood exopolysaccharide modifications is to tune their interactions with cationic NCR peptides so that lethality can be minimized while still allowing the peptides to exert their important effects during differentiation into bacteroids and during nitrogen fixation. Because loss of the pyruvyl modification also affects the efficiency of polymerization of succinoglycan octasaccharide subunits into polymers, it is more difficult to assess the role of the pyruvyl modification. Nevertheless, our data are consistent with the anionic pyruvyl modification playing an important role in the interactions with the cationic NCR peptides.
It was reported recently that NCR peptides, especially cationic NCR peptides, interact with the membranes of differentiating bacteria and mediate the formation of outer membrane vesicles or other changes, such as pore formation (41). By being secreted and forming a proximal protective layer around the bacteria, succinoglycan, with its anionic and noncharged modifications, may be acting as a regulator of NCR peptide biological activity on the bacterial membranes. This effect might be accomplished by direct binding to a select group of NCR peptides, possibly cationic ones. A delicate balance between hydrophilic and hydrophobic interactions with NCR peptides could be a key element for this interaction.
In conclusion, we propose that, in addition to its well-studied role in promoting infection thread initiation and growth early in symbiosis, succinoglycan plays a second independent role later in symbiosis. Our data suggest that HMW succinoglycan modulates the exposure of the internalized rhizobia to the cationic NCR peptides produced within the nodule, to achieve the levels and balance of NCR peptides necessary for differentiation into bacteroids and bacteroid maintenance. An additional possible late-stage role could be that LMW succinoglycan protects bacteria inside symbiosomes against pH stress (40).
MATERIALS AND METHODS
Bacterial strains and reagents.
All bacterial strains used in this study are listed in Table 1. S. meliloti strains were grown in lysogeny broth supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4 (LBMC) and containing the desired antibiotics. Antibiotics were used at the following concentrations: 200 μg ml−1 streptomycin, 100 μg ml−1 neomycin, 25 μg ml−1 gentamicin, and 50 μg ml−1 hygromycin.
TABLE 1.
Bacterial strains used in this study
| Sinorhizobium meliloti strain | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Sm1021 | Wild-type derivative of SU47; Strepr | 45 |
| chvID52E strain (EC220) | Succinoglycan gain-of-function allele in Sm1021 wild-type background, chvlD52E-chvI+; Strepr Hygr | 25 |
| chvIK214T strain (DW570) | Succinoglycan partial-loss-of-function allele, chvIK214T; Strepr Neor | 46 |
| exoS96 strain | Sm1021 exoS96::Tn5 Strepr Neor | 26 |
| exoY strain | Sm1021 exoY210::Tn5 Strepr Neor | 47 |
| exoA strain | Sm1021 exoA32::Tn233 Strepr Gentr | 7 |
| exoS96-exoA strain | exoS96 exoA double mutant; Strepr Gentr Neor | This study |
| chvID52E-exoY strain | chvlD52E exoY double mutant; Strepr Hygr | This study |
| exoV strain | Sm1021 exoV::Tn5 (exoV2) Strepr Neor | 37 |
| exoH strain | Sm1021 exoH::Tn5 Strepr Neor | 34, 48 |
| exoZ strain | Sm1021 exoZ::Tn5 Strepr Neor Rm8431 | 48 |
Strepr, streptomycin resistant; Neor, neomycin resistant; Gentr, gentamycin resistant; Hygr, hygromycin resistant.
Generation of double mutants.
The exoS96 exoA and chvID52E exoY double mutants were constructed using ϕM12 phage transduction, by transducing the exoY::Tn5 and exoA::Tn233 mutations into the chvID52E and exoS96 mutants, respectively, using established methodology (42).
Synthesis of NCR247 peptide derivatives.
The oxidized NCR247 peptide (mixture of all regioisomers) was prepared as described previously (21). The reduced l- and d-forms of NCR247 were commercially synthesized by GenScript (USA) and then purified by our laboratory to a purity of 99%.
The synthesis of biotin-NCR247-Bpared was performed using standard 9-fluorenylmethoxy carbonyl (Fmoc) solid-phase peptide synthesis techniques, in a custom-made 25-ml glass reaction vessel outfitted with a medium-porosity frit and a T-bore for N2 gas bubbling. The synthesis of biotin-NCR247-Bpared was performed on a 0.01-mmol scale with Fmoc-Arg(pentamethyldihydrobenzofuran)-Novasyn TGA resin; at amino acid position 3, Gly was replaced with Lys (NCR247[G3K-biotin]) and, at position 20, Tyr was replaced with 4-benzoyl-l-phenylalanine (Bpa) (NCR247[G3K-biotin, Y20Bpa]). Fmoc-Lys-biotin-OH and Nα-Fmoc-4-benzoyl-l-phenylalanine were coupled three times using Nα-Fmoc-4-benzoyl-l-phenylalanine (10 equivalents) and Fmoc-Lys-biotin-OH and were dissolved in dimethylformamide (DMF) (15 ml) containing hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU) (10 equivalents), 1-hydroxy-7-azabenzotriazole (HOAt) (10 equivalents), and N,N-diisopropylethylamine (DIPEA) (20 equivalents). After coupling of the N-terminal Arg residue, the resin was thoroughly washed with methylene chloride and the Fmoc was removed at 60°C for 10 min using 20 ml of a solution containing trifluoroacetic acid (TFA), 1,2-ethanedithiol (EDT), water, and triisopropylsilane (TIS) (94:2.5:2.5:1). The mixture was concentrated to a volume of ∼5 ml by using a gentle stream of nitrogen. Ice-cold diethyl ether was added to the resulting concentrate, which resulted in precipitation of the crude peptides, and the mixture was centrifuged in a Beckman Coulter GS-6R rotor (3,500 rpm for 15 min at 4°C). The organic supernatant was decanted, and the pelleted precipitate was redissolved in 5 ml of 5% TFA in water-acetonitrile (3:1). Preparative reverse-phase high-performance liquid chromatography (RP-HPLC) purification (10 to 60% acetonitrile in TFA over 30 min, at 10 ml/min) afforded reduced biotin-NCR247-Bpared (2.5 mg; overall yield of 10%). The crude product was reduced by addition of 1 mM tris(2-carboxy)ethyl-1-phosphine (TCEP) in Tris-HCl (pH 8.0) and was purified to yield biotin-NCR247-Bpared.
Peptide killing assays.
The defined strains were grown to the early exponential phase (optical density at 600 nm [OD600] of 0.1 to 0.3), washed three times with 0.85% saline (0.85 g NaCl in 100 ml), and resuspended to a final OD600 of 0.1 in 3-(N-morpholino)propanesulfonic acid (MOPS)-buffered minimal medium (50 mM MOPS, 1 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, 0.004 mM biotin [pH 7.0]) (MOPS-GS buffer) supplemented with 1% Casamino Acids (CAs). The cells were then treated with 20 μM NCR247 peptide, and the surviving cells were recovered by spotting 10-μl aliquots, in triplicate, on LBMC-streptomycin plates.
Purification of succinoglycan.
Succinoglycan was purified and isolated using an exopolysaccharide-overproducing S. meliloti exoS96 mutant strain, as described previously (9, 43). Cultures were grown at 30°C in glutamate-d-mannitol-salts (GMS) medium (1× M9 medium with 12 mM d-mannitol, 5 mM glutamic acid, 86 mM NaCl, 0.008 mM thiamine, and 0.004 mM biotin [pH 7.0]) supplemented with biotin and thiamine, in a final volume of 1 liter containing streptomycin and neomycin, with shaking at 160 rpm. The supernatant containing all exopolysaccharides was then collected (20,000 × g for 30 min), and secreted HMW succinoglycan and LMW succinoglycan were purified using a two-step ethanol precipitation process, as described previously (9). First, the supernatant was precipitated with the addition of 3 volumes of ethanol (6,000 × g for 30 min). Second, the supernatant of the first precipitation reaction was precipitated again with the addition of 7 volumes of ethanol (6,000 × g for 30 min). After precipitation, HMW succinoglycan and LMW succinoglycan pellets were resuspended in up to 5 ml of deionized water, lyophilized, and resuspended in deionized water until they were used in the defined experiments at working concentrations based their polysaccharide contents, as determined with the anthrone-sulfuric acid assay (44).
Size exclusion chromatography.
Size exclusion chromatography of purified total succinoglycan was performed as described previously, with modifications, on a column (1.5 by 90 cm) of Bio-Gel P-6 resin (fine mesh; Bio-Rad) (9). The column was preequilibrated with 0.1 M ammonium acetate (pH 5.0). For purification of single succinoglycan fractions, approximately 500 μg of purified succinoglycan, suspended in a final volume of 2 ml of 0.1 M ammonium acetate buffer (pH 5.0), was loaded onto the column. Whenever mentioned, FI-NCR247 was loaded at a final concentration of 50 μg ml−1. Fractions (1.6 ml) were collected, and the carbohydrate content of each fraction was determined by using the anthrone-sulfuric acid assay (44). In brief, in a 96-well plate, 50 μl of the size exclusion chromatography fraction was mixed with 150 μl of glacial sulfuric acid containing anthrone (2 g liter−1). The samples were left at 4°C for 20 min and then kept at 70°C for 1 h, followed by incubation at room temperature for 20 min. Then the absorbance at 620 nm was measured for each sample using a Tecan Spark 10M plate reader.
Biolayer interferometry.
Biolayer interferometry was carried out using a ForteBio Octet RED96 biolayer interferometer, following the manufacturer's instructions for a standard kinetic assay. Streptavidin-coated biosensor tips were incubated in 200 μl of assay buffer (0.1 M ammonium acetate [pH 5.0]), each for 60 s. Then biotinylated NCR247 was loaded onto each biosensor tip at the defined concentration until the binding signal reached a value of >1.4. Biosensor tip loading was followed by incubation in assay buffer for 60 s. Association between the ligand NCR247 and the analyte HMW succinoglycan (10 μg ml−1 in assay buffer) was observed over a time frame of 300 s, in assay buffer supplemented as indicated in the corresponding figures. To stop binding kinetics for dissociation, the biosensor tips were placed back into assay buffer not containing any succinoglycan, with or without supplementation with 0.1 M NaCl (as indicated in the corresponding figures), for 120 s. Data analysis was performed using Octet data analyses 8.2 software.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants GM31030 (to G.C.W.) and P30 ES002109 (to the Massachusetts Institute of Technology Center for Environmental Health Sciences). G.C.W. is an American Cancer Society Professor. E.J.C. received funding from the California State University Special Fund for Research, Scholarship, and Creative Activity.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Marco Scocchi for providing the Bac71–35 peptide, and we thank Elizabeth Nolan for providing the HD5 peptide. The Octet biolayer interferometry system (NIH S10 OD016326) is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00665-17.
REFERENCES
- 1.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battisti L, Lara JC, Leigh JA. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc Natl Acad Sci U S A 89:5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellock BJ, Cheng HP, Walker GC. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J Bacteriol 182:4310–4318. doi: 10.1128/JB.182.15.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urzainqui A, Walker GC. 1992. Exogenous suppression of the symbiotic deficiencies of Rhizobium meliloti exo mutants. J Bacteriol 174:3403–3406. doi: 10.1128/jb.174.10.3403-3406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendis HC, Madzima TF, Queiroux C, Jones KM. 2016. Function of succinoglycan polysaccharide in Sinorhizobium meliloti host plant invasion depends on succinylation, not molecular weight. mBio 7:e00606-16. doi: 10.1128/mBio.00606-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A 82:6231–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 8.York GM, Walker GC. 1998. The succinyl and acetyl modifications of succinoglycan influence susceptibility of succinoglycan to cleavage by the Rhizobium meliloti glycanases ExoK and ExsH. J Bacteriol 180:4184–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang LX, Wang Y, Pellock B, Walker GC. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J Bacteriol 181:6788–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. 2008. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci U S A 105:704–709. doi: 10.1073/pnas.0709338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaharada Y, Nielsen MW, Kelly S, James EK, Andersen KR, Rasmussen SR, Fuchtbauer W, Madsen LH, Heckmann AB, Radutoiu S, Stougaard J. 2017. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun 8:14534. doi: 10.1038/ncomms14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszynski A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, Vinther M, Andersen SU, Krusell L, Thirup S, Jensen KJ, Ronson CW, Blaise M, Radutoiu S, Stougaard J. 2015. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 13.Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z, Farkas A, Mikulass K, Nagy A, Tiricz H, Satiat-Jeunemaître B, Alunni B, Bourge M, Kucho KI, Abe M, Kereszt A, Maroti G, Uchiumi T, Kondorosi E, Mergaert P. 2010. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 14.Farkas A, Maroti G, Durgo H, Gyorgypal Z, Lima RM, Medzihradszky KF, Kereszt A, Mergaert P, Kondorosi E. 2014. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc Natl Acad Sci U S A 111:5183–5188. doi: 10.1073/pnas.1404169111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penterman J, Abo RP, De Nisco NJ, Arnold MF, Longhi R, Zanda M, Walker GC. 2014. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci U S A 111:3561–3566. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maróti G, Downie JA, Kondorosi É. 2015. Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr Opin Plant Biol 26:57–63. doi: 10.1016/j.pbi.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132:161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D. 2015. An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci U S A 112:15238–15243. doi: 10.1073/pnas.1500123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath B, Domonkos A, Kereszt A, Szucs A, Abraham E, Ayaydin F, Boka K, Chen Y, Chen R, Murray JD, Udvardi MK, Kondorosi E, Kalo P. 2015. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci U S A 112:15232–15237. doi: 10.1073/pnas.1500777112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A, Longhi R, Boncompagni E, Hérouart D, Dall'Angelo S, Kondorosi E, Zanda M, Mergaert P, Ferguson GP. 2011. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9:e1001169. doi: 10.1371/journal.pbio.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shabab M, Arnold MF, Penterman J, Wommack AJ, Bocker HT, Price PA, Griffitts JS, Nolan EM, Walker GC. 2016. Disulfide cross-linking influences symbiotic activities of nodule peptide NCR247. Proc Natl Acad Sci U S A 113:10157–10162. doi: 10.1073/pnas.1610724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuber TL, Walker GC. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269–280. doi: 10.1016/0092-8674(93)90418-P. [DOI] [PubMed] [Google Scholar]
- 23.Arnold MFF, Shabab M, Penterman J, Boehme KL, Griffitts JS, Walker GC. 2017. Genome-wide sensitivity analysis of the microsymbiont Sinorhizobium meliloti to symbiotically important, defensin-like host peptides. mBio 8:e01060-17. doi: 10.1128/mBio.01060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrere S, Sallet E, Courcelle E, Moreau S, Debelle F, Capela D, de Carvalho-Niebel F, Gouzy J, Bruand C, Gamas P. 2014. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser capture microdissection coupled to RNA-seq. Plant J 77:817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 25.Chen EJ, Fisher RF, Perovich VM, Sabio EA, Long SR. 2009. Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti. J Bacteriol 191:6833–6842. doi: 10.1128/JB.00734-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty D, Leigh JA, Glazebrook J, Walker GC. 1988. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J Bacteriol 170:4249–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EJ, Sabio EA, Long SR. 2008. The periplasmic regulator ExoR inhibits ExoS/ChvI two-component signalling in Sinorhizobium meliloti. Mol Microbiol 69:1290–1303. doi: 10.1111/j.1365-2958.2008.06362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orwa JA, Govaerts C, Busson R, Roets E, Van Schepdael A, Hoogmartens J. 2001. Isolation and structural characterization of polymyxin B components. J Chromatogr A 912:369–373. doi: 10.1016/S0021-9673(01)00585-4. [DOI] [PubMed] [Google Scholar]
- 29.Tosteson MT, Tosteson DC. 1981. The sting: melittin forms channels in lipid bilayers. Biophys J 36:109–116. doi: 10.1016/S0006-3495(81)84719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ericksen B, Wu Z, Lu W, Lehrer RI. 2005. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother 49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gudmundsson GH, Agerberth B, Odeberg J, Bergman T, Olsson B, Salcedo R. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur J Biochem 238:325–332. doi: 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- 32.Podda E, Benincasa M, Pacor S, Micali F, Mattiuzzo M, Gennaro R, Scocchi M. 2006. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim Biophys Acta 1760:1732–1740. doi: 10.1016/j.bbagen.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Wei G, de Leeuw E, Pazgier M, Yuan W, Zou G, Wang J, Ericksen B, Lu WY, Lehrer RI, Lu W. 2009. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem 284:29180–29192. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leigh JA, Reed JW, Hanks JF, Hirsch AM, Walker GC. 1987. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell 51:579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- 35.Reuber TL, Walker GC. 1993. The acetyl substituent of succinoglycan is not necessary for alfalfa nodule invasion by Rhizobium meliloti Rm1021. J Bacteriol 175:3653–3655. doi: 10.1128/jb.175.11.3653-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buendia AM, Enenkel B, Köplin R, Niehaus K, Arnold W, Pünier A. 1991. The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol Microbiol 5:1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 37.Glucksmann MA, Reuber TL, Walker GC. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J Bacteriol 175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marlow VL, Haag AF, Kobayashi H, Fletcher V, Scocchi M, Walker GC, Ferguson GP. 2009. Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J Bacteriol 191:1519–1527. doi: 10.1128/JB.01661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wehmeier S, Arnold MF, Marlow VL, Aouida M, Myka KK, Fletcher V, Benincasa M, Scocchi M, Ramotar D, Ferguson GP. 2010. Internalization of a thiazole-modified peptide in Sinorhizobium meliloti occurs by BacA-dependent and -independent mechanisms. Microbiology 156:2702–2713. doi: 10.1099/mic.0.039909-0. [DOI] [PubMed] [Google Scholar]
- 40.Hawkins JP, Geddes BA, Oresnik IJ. 2017. Succinoglycan production contributes to acidic pH tolerance in Sinorhizobium meliloti Rm1021. Mol Plant Microbe Interact 30:1009–1019. doi: 10.1094/MPMI-07-17-0176-R. [DOI] [PubMed] [Google Scholar]
- 41.Montiel J, Downie JA, Farkas A, Bihari P, Herczeg R, Balint B, Mergaert P, Kereszt A, Kondorosi E. 2017. Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc Natl Acad Sci U S A 114:5041–5046. doi: 10.1073/pnas.1704217114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finan TM, Hartweig E, LeMieux K, Bergman K, Walker GC, Signer ER. 1984. General transduction in Rhizobium meliloti. J Bacteriol 159:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zevenhuizen LPTM, van Neerven ARW. 1983. (1→2)-β-d-Glucan and acidic oligosaccharides produced by Rhizobium meliloti. Carbohydr Res 118:127–134. doi: 10.1016/0008-6215(83)88041-0. [DOI] [Google Scholar]
- 44.Loewus FA. 1952. Improvement in anthrone method for determination of carbohydrates. Anal Chem 24:219. doi: 10.1021/ac60061a050. [DOI] [Google Scholar]
- 45.Meade HM, Long SR, Ruvkun GB. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wells DH, Chen EJ, Fisher RF, Long SR. 2007. ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti. Mol Microbiol 64:647–664. doi: 10.1111/j.1365-2958.2007.05680.x. [DOI] [PubMed] [Google Scholar]
- 47.York GM, Walker GC. 1997. The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low-molecular-weight succinoglycan. Mol Microbiol 25:117–134. doi: 10.1046/j.1365-2958.1997.4481804.x. [DOI] [PubMed] [Google Scholar]
- 48.Long S, Reed JW, Himawan J, Walker GC. 1988. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol 170:4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.