ABSTRACT
SpoVD and PBP4b are structurally very similar high-molecular-weight, class B penicillin-binding proteins produced early during sporulation in Bacillus subtilis. SpoVD is known to be essential for endospore cortex synthesis and thereby the production of heat-resistant spores. The role of PBP4b is still enigmatic. Both proteins are synthesized in the cytoplasm of the mother cell. PBP4b remains in the cytoplasmic membrane of the mother cell, whereas SpoVD accumulates in the forespore outer membrane. By the use of SpoVD/PBP4b chimeras with swapped protein domains, we show that the N-terminal part of SpoVD, containing the single transmembrane region, determines the forespore targeting of the protein.
IMPORTANCE Beta-lactam-type antibiotics target penicillin-binding proteins (PBPs), which function in cell wall peptidoglycan synthesis. Bacteria of a subset of genera, including Bacillus and Clostridium species, can form endospores. The extreme resistance of endospores against harsh physicochemical conditions is of concern in clinical microbiology and the food industry. Endospore cortex layer biogenesis constitutes an experimental model system for research on peptidoglycan synthesis. The differentiation of a vegetative bacterial cell into an endospore involves the formation of a forespore within the cytoplasm of the sporulating cell. A number of proteins, including some PBPs, accumulate in the forespore. An understanding of the molecular mechanisms behind such subcellular targeting of proteins in bacterial cells can, for example, lead to a means of blocking the process of sporulation.
KEYWORDS: Bacillus subtilis, endospores, penicillin-binding proteins, protein structure-function, protein targeting, sporulation
INTRODUCTION
Bacterial cells use different survival strategies to deal with a variable environment and different habitats. The vegetative cells of Bacillus and Clostridium species can differentiate to form endospores resistant to physicochemical conditions that normally kill organisms. Sporulation is initiated by nutrient depletion and takes many hours for the cell to complete (1–4). Early in sporulation, an asymmetrically located septum is formed in the cell, resulting in a mother cell and a forespore. In subsequent developmental steps, the forespore is engulfed by the mother cell in a process suggested to be driven by cell wall remodeling (5). The endospore protective cortex (composed of modified peptidoglycan [6]) and coat (composed of multiple proteins [7]) layers are deposited in the engulfed forespore. When the endospore is mature, the mother cell lyses to release the spore into the surrounding environment. Sporulation is regulated by a cascade of sigma factors. First, σF is activated in the forespore, and this leads to the activation of σE in the mother cell, which triggers the engulfment of the forespore and initiates spore cortex assembly and many other processes. Some 150 transcriptional units in Bacillus subtilis depend on σE (8). Proteins encoded by σE-transcribed genes are synthesized in the mother cell cytoplasm, and many have the forespore as their final destination. The signals and mechanisms for bringing these proteins to the forespore during sporulation are poorly understood (9).
In this work, we have studied the different subcellular localizations of two very similar sporulation-specific membrane proteins in Bacillus subtilis, SpoVD and PBP4b (also known as PBPI or YrrR). They are class B, high-molecular-weight penicillin-binding proteins (PBPs) anchored to the membrane by a single transmembrane (TM) domain (Fig. 1; also see Fig. S1 in the supplemental material). The main part of such PBPs is exposed on the extracytoplasmic side of the membrane where peptidoglycan is assembled. This part has two large protein domains joined by a β-rich linker (10). The most C-terminal domain (TP) has d,d-transpeptidase activity and binds penicillin covalently to an active site serine residue. Between the TM and the TP, there is a domain of unknown function, here designated UF.
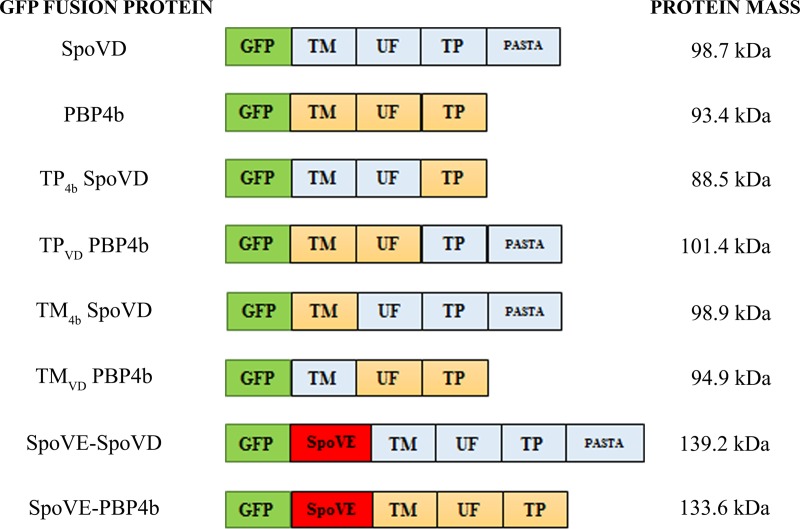
FIG 1.
Schematic illustration of GFP fusion proteins. The transmembrane (TM), unknown function (UF), transpeptidase (TP), and PASTA domains and SpoVE in fusion protein variants are schematically indicated (the relative sizes of the domains are not to scale). SpoVD and PBP4b domains and SpoVE are colored light blue, yellow, and red, respectively, whereas GFP is in green. The TM, UF, and TP domains of SpoVD comprise residues 1 to 40, 41 to 214, and 215 to 646, respectively. For PBP4b, they comprise residues 1 to 40, 41 to 235, and 236 to 584, respectively. The predicted molecular mass of each fusion protein is indicated.
B. subtilis cells have six different class B PBPs, which are involved in vegetative growth, sporulation, germination, or a not yet identified function (11). SpoVD is produced in the mother cell, under the control of σE, and is required for the production of heat-resistant endospores (12). PBP4b is encoded by the pbpI (yrrR) gene, which is under the control of both σE and σF (8, 13). The protein was named (14) following the convention of naming PBPs on the basis of their relative migration during denaturing polyacrylamide gel electrophoresis. The amino acid sequences of SpoVD and PBP4b are 27% identical and 42% similar (14) (Fig. S1). The presence of a penicillin-binding protein and serine/threonine kinase-associated (PASTA) domain at the very C-terminal end of SpoVD is unique among the sporulation-specific PBPs in B. subtilis (15). Strains lacking the transpeptidase activity of SpoVD, due to a mutation of the active site serine or the absence of the entire protein, produce spores without the cortex protective layer and accumulate peptidoglycan biosynthetic precursors in the mother cell cytoplasm (16, 17). PBP4b does not seem important for cortex synthesis, since the inactivation of pbpI alone or in combination with genes for other PBPs has no effect on the production of normal spores (14).
PBP4b and SpoVD are special among the high-molecular-weight PBPs in B. subtilis in that the TP domain contains two conserved cysteine residues: Cys328 and Cys353 in PBP4b and Cys332 and Cys351 in SpoVD (Fig. S1). One of these cysteine residues is located in the middle of the Ser-Xaa-Asp active-site motif (i.e., Xaa is Cys). The two cysteine residues in SpoVD can form a disulfide bond which, when present, blocks the enzyme activity of the protein (18). The close structural relatedness and common σE-dependent production in the mother cell during sporulation suggest that PBP4b and SpoVD have similar and perhaps overlapping activities. We have compared the temporal production and subcellular localization of PBP4b and SpoVD in sporulating cells and have used PBP4b/SpoVD chimeric proteins with swapped domains to identify determinants for subcellular protein sorting during sporulation.
RESULTS
SpoVD and PBP4b show similar expression patterns during sporulation.
Wei et al. demonstrated that the pbpI gene, encoding PBP4b, is expressed exclusively during sporulation but found no phenotype associated with the lack of the gene (14). Strains deleted for pbpI produced heat-resistant endospores with a normal cortex peptidoglycan composition and did not accumulate peptidoglycan precursors. The pbpI gene is cotranscribed with the downstream yrrS gene, which encodes a protein of unknown function. For our studies, we constructed an in-frame markerless deletion of pbpI to avoid a polar effect on the expression of yrrS (Table 1). This deletion of pbpI did not affect the ability of cells to produce heat-resistant spores (strain LMD158) (Table 2).
TABLE 1.
B. subtilis strains used in this work
| Strain | Genotype and phenotypea | Reference and/or originb |
|---|---|---|
| 1A1 | trpC2 | BGSCc |
| LMD6 | trpC2 spoVEΩpLSSD6 Eryr | 18 |
| LMD12 | trpC2 spoVD::spc Spcr | 18 |
| LMD15 | trpC2 pheA1 amyE::ery spoVD::spc Eryr Spcr | 18 |
| LMD101 | trpC2 ΔspoVD | 16 |
| LMD120 | trpC2 spoVEΩpLSSD6 qcr::neo Eryr Neor | This work; LUH180→LMD6 |
| LMD121 | trpC2 ΔspoVE qcr::neo Neor | This work; pLEB27+LMD120→LMD6 |
| LMD156 | trpC2 (greA-pbpI-yrrS)ΩpLMS2 Cmlr | This work; pLMS2→1A1 |
| LMD158 | trpC2 ΔpbpI | This work; pBKJ223→LMD156 |
| LMD159 | trpC2 spoVD::spc ΔpbpI Spcr | This work; LMD12→LMD158 |
| LMD162 | trpC2 ΔspoVD ΔpbpI amyE::ery Eryr | This work; pLEB2+LMD15→LMD159 |
| LMD163 | trpC2 ΔspoVD amyE::Pxyl-gfp-spoVD Spcr | This work; pLMS3→LMD101 |
| LMD164 | trpC2 ΔspoVD amyE::Pxyl-gfp-spoVD1–214-pbpI236–584 Spcr | This work; pLMS4→LMD101 |
| LMD165 | trpC2 ΔspoVD amyE::Pxyl-gfp-pbpI1–235-spoVD215–646 Spcr | This work; pLMS5→LMD101 |
| LMD166 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD Spcr | This work; pLMS3→LMD162 |
| LMD167 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD2–214-pbpI236–584 Spcr | This work; pLMS4→LMD162 |
| LMD168 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI1–235-spoVD215–646 Spcr | This work; pLMS5→LMD162 |
| LMD169 | trpC2 ΔpbpI amyE::Pxyl-gfp-pbpI Spcr | This work; pLMS6→LMD158 |
| LMD170 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI Spcr | This work; pLMS6→LMD162 |
| LMD172 | trpC2 ΔspoVD amyE::Pxyl-gfp-pbpI1–40-spoVD41–646 Spcr | This work; pLMS7→LMD101 |
| LMD173 | trpC2 ΔspoVD amyE::Pxyl-gfp-spoVD2–40-pbpI41–584 Spcr | This work; pLMS8→LMD101 |
| LMD174 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI1–40-spoVD41–646 Spcr | This work; pLMS7→LMD162 |
| LMD175 | trpC2 ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD1–40-pbpI41–584 Spcr | This work; pLMS8→LMD162 |
| LMD179 | trpC2 ΔspoVD amyE::PσE-gfp-spoVD Spcr | This work; pLMS9→LMD101 |
| LMD180 | trpC2 ΔspoVD amyE::PσE-gfp-pbpI1–40-spoVD41–646 Spcr | This work; pLMS11→LMD101 |
| LMD181 | trpC2 ΔspoVD amyE::PσE-gfp-spoVD2–40-pbpI41–584 Spcr | This work; pLMS12→LMD101 |
| LMD182 | trpC2 ΔpbpI amyE::PσE-gfp-pbpI Spcr | This work; pLMS10→LMD158 |
| LMD185 | trpC2 ΔspoVD ΔpbpI amyE::PσE-gfp-spoVD Spcr | This work; pLMS9→LMD162 |
| LMD186 | trpC2 ΔspoVD ΔpbpI amyE::PσE-gfp-pbpI Spcr | This work; pLMS10→LMD162 |
| LMD187 | trpC2 ΔspoVD ΔpbpI amyE::PσE-gfp-pbpI1–40-spoVD41–646 Spcr | This work; pLMS11→LMD162 |
| LMD188 | trpC2 ΔspoVD ΔpbpI amyE::PσE-gfp-spoVD2–40-pbpI41–584 Spcr | This work; pLMS12→LMD162 |
| LMD189 | trpC2 ΔspoVD amyE::Pxyl-gfp-spoVE-spoVD Spcr | This work; pLEB48→LMD101 |
| LMD190 | trpC2 ΔspoVD amyE::Pxyl-gfp-spoVE-pbpI Spcr | This work; pLEB47→LMD101 |
| LMD193 | trpC2 ΔspoVE qcr::neo amyE::Pxyl-gfp-spoVE-pbpI Spcr Neor | This work; pLEB47→LMD121 |
| LMD194 | trpC2 ΔspoVE qcr::neo amyE::Pxyl-gfp-spoVE-spoVD Spcr Neor | This work; pLEB48→LMD121 |
| LUH180 | trpC2 qcr::neo Neor | 51 |
Cmlr, Spcr, Eryr, and Neor indicate resistance to chloramphenicol, spectinomycin, erythromycin, and neomycin, respectively. The superscript numbers for genes encoding chimeric proteins indicate amino acid positions in the respective wild-type protein. Pxyl and PσE indicate xylose-inducible promoter and the promoter of the spoVD gene, respectively.
An arrow indicates transformation of the indicated strain with plasmid or chromosomal DNA and selection for antibiotic resistance on TBAB plates.
BGSC, Bacillus Genetic Stock Center, Columbus, OH.
TABLE 2.
Sporulation efficiencies of B. subtilis strains
| Strain | Relevant genotype | Yield (%) of heat-resistant sporesa |
|---|---|---|
| 1A1 | Wild type | 67 ± 4b |
| LMD101 | ΔspoVD | 0c |
| LMD158 | ΔpbpI | 55 ± 9 |
| LMD162 | ΔspoVD ΔpbpI amyE::ery | 0c |
| LMD163 | ΔspoVD amyE::Pxyl-gfp-spoVD | 83 ± 5 |
| LMD166 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD | 57 ± 2 |
| LMD167 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD2–214-pbpI236–584 | 0c |
| LMD168 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI1–235-spoVD215–646 | 0c |
| LMD170 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI | 0c |
| LMD174 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-pbpI1–40-spoVD41–646 | 0c |
| LMD175 | ΔspoVD ΔpbpI amyE::Pxyl-gfp-spoVD2–40-pbpI41–584 | 0c |
| LMD185 | ΔspoVD ΔpbpI amyE::PσE-gfp-spoVD | 68 ± 9d |
| LMD186 | ΔspoVD ΔpbpI amyE::PσE-gfp-pbpI | 0c,d |
| LMD187 | ΔspoVD ΔpbpI amyE::PσE-gfp-pbpI1–40-spoVD41–646 | 0c,d |
| LMD188 | ΔspoVD ΔpbpI amyE::PσE-gfp-spoVD2–40-pbpI41–584 | 0c,d |
Cells were sporulated for 2 days at 37°C in NSMP supplemented with 0.2% (wt/vol) xylose. The heat-resistant spore yield was assayed by incubation at 80°C for 10 min and calculated as CFU after heating divided by CFU of nonheated culture. The results are presented as the averages from three independent experiments ± SEMs.
Data from reference 15. The cells were grown in the absence of xylose.
No colonies were obtained when 100 μl heated culture was plated on TBAB. Three independent experiments were done, including one biological replicate using another clone.
The cells were grown in the absence of xylose.
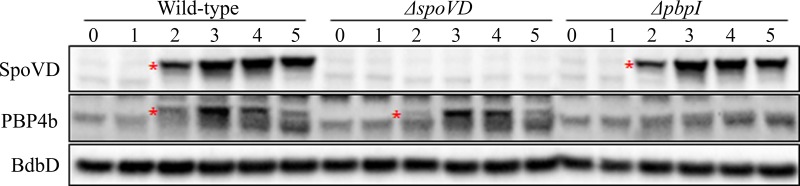
To study PBP4b, we first produced in Escherichia coli a water-soluble variant (sPBP4b) of the protein lacking the TM domain (see Fig. S2A in the supplemental material). Transpeptidase activity of the isolated sPBP4b was confirmed by the binding of Bocillin FL, penicillin G, and ampicillin (Fig. S2B). Using an antiserum generated against the purified sPBP4b and an available SpoVD antiserum, we probed the lysates of sporulating B. subtilis cells for the two proteins. As an internal control, we also probed for BdbD, which is a membrane-bound disulfide oxidoreductase acting on newly synthesized SpoVD (18, 19) and is produced constitutively (20). PBP4b (∼65 kDa) appeared at the second hour after the onset of sporulation, very similar to the SpoVD protein (Fig. 2). This observed temporal expression pattern is in agreement with transcription of the pbpI gene, which begins at the 1st to 2nd hour after the initiation of sporulation and peaks at the 3rd hour (14, 21). Furthermore, the cellular level of PBP4b protein was not affected by the deletion of spoVD, and the level of SpoVD was unaffected by deletion of pbpI (Fig. 2).
FIG 2.
SpoVD and PBP4b in B. subtilis sporulating cells. Shown are immunoblots of cell lysates of the B. subtilis wild-type (1A1), ΔspoVD (LMD101), and ΔpbpI (LMD158) strains (Table 1). Samples were taken from the cultures at 0, 1, 2, 3, 4, and 5 h after the induction of sporulation by resuspension. Fifteen micrograms of protein was loaded in each lane. The top and middle panels show blots probed with anti-SpoVD and anti-PBP4b serum, respectively. SpoVD (71 kDa) and PBP4b (65.5 kDa) are indicated by asterisks. The bottom panel shows the blot probed with anti-BdbD serum, serving as an internal control for equal loading of cell material.
Functional properties of GFP fusion proteins.
To investigate the subcellular localization of PBP4b compared to that of SpoVD in sporulating B. subtilis cells, we constructed N-terminal fusions of the two proteins to green fluorescent protein (GFP) (Fig. 1). The gene encoding the respective fusion protein, and under the control of a xylose-inducible promoter, was inserted as a single copy at the amyE locus in strains deleted for pbpI or spoVD or deleted for both these genes (Table 1). The deletion of spoVD (strain LMD101) resulted in phase-gray heat-sensitive spores lacking the cortex layer (16). The defect was complemented by the gene encoding GFP-SpoVD (strain LMD163) (Table 2), confirming that the fusion protein is functional. The ΔspoVD ΔpbpI double mutant was also complemented by GFP-SpoVD (strain LMD166) (Table 2). GFP-PBP4b did not complement the lack of SpoVD (strain LMD170), as expected. An immunoblot analysis demonstrated that the GFP-PBP4b fusion protein was produced and membrane bound in cells grown for sporulation (Fig. 3). The blot also showed that the cellular concentration of the PBP4b fusion protein was higher than that for SpoVD.
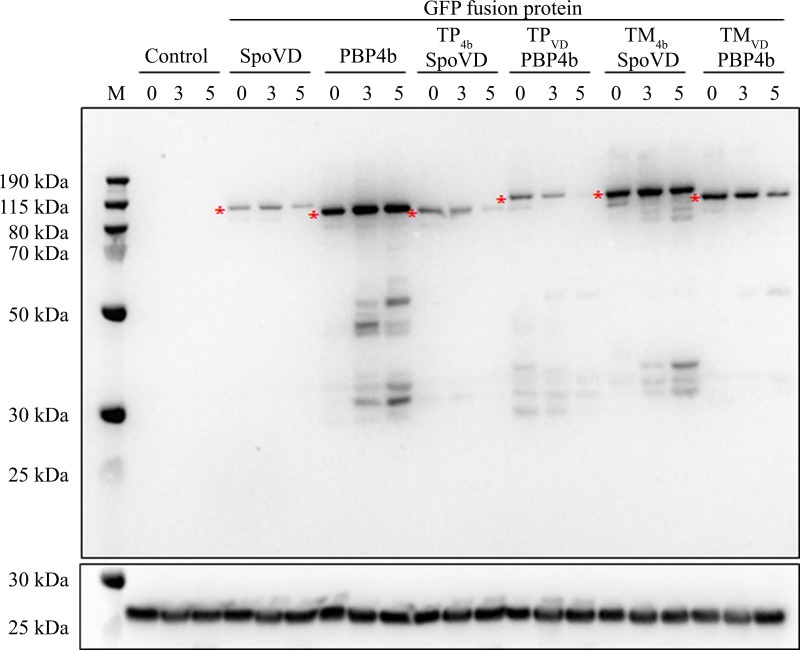
FIG 3.
GFP fusion proteins in sporulating cells. Shown is an immunoblot for GFP-SpoVD, GFP-PBP4b, and four GFP-SpoVD/PBP4b chimeras. Proteins in the membrane fraction from ΔspoVD ΔpbpI strains containing the gene encoding the respective GFP fusion protein, expressed under the control of a xylose-inducible promoter, were separated by SDS-PAGE. The B. subtilis strains were LMD162 (lacks fusion protein), LMD166, LMD170, LMD167, LMD168, LMD174, and LMD175 (Table 1). Samples for analysis were taken at 0, 3, and 5 h after the induction of sporulation by resuspension. Equal amounts of protein (11 μg) were loaded in each lane. The upper panel is a blot probed with anti-GFP serum. Each GFP fusion protein variant is indicated by an asterisk. The lower panel is a blot probed with anti-BdbD serum. Molecular mass markers (lane M) are indicated.
PBP4b accumulates in the mother cell cytoplasmic membrane.
We then determined the subcellular distribution of GFP-PBP4b compared to that of GFP-SpoVD in sporulating cells using fluorescence microscopy with strains LMD166 and LMD170. One hour after the induction of sporulation by resuspension, the membrane dye FM4-64 was added to the cultures, and each hour during the time period from 1 to 5 h, a sample was taken for microscopy. Beginning at the start of the experiment, xylose was included in the growth medium. The GFP fusion proteins were therefore produced constitutively and also present before the asymmetric septum formed in the sporulating cells (Fig. 3). The sporulation stage of individual cells was classified on the basis of the FM4-64 membrane staining profile and the phase-contrast image. We could not differentiate cells at a late stage of engulfment from those having a fully engulfed spore. Therefore, in this work, the term “forespore in cell” refers both to a late stage in engulfment and to completed forespore engulfment.
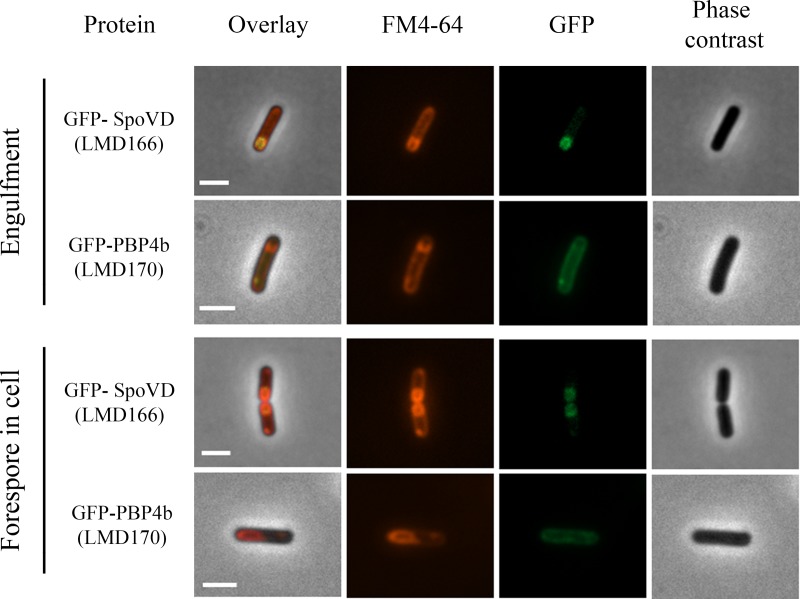
At 1 h into sporulation, when most LMD166 cells were in the preseptation stage, GFP-SpoVD was distributed evenly in the cytoplasmic membrane. At 2 to 3 h, some cells had formed asymmetric septa and initiated the engulfment process as observed by curved septa. At this stage, GFP-SpoVD was enriched at the asymmetric septum. At 4 h, most cells showed fluorescent forespores and only a weak fluorescence intensity from the mother cell membranes. The same distribution of fluorescence was seen in cells with a developed phase-bright spore. GFP-PBP4b showed a different subcellular membrane distribution after the asymmetric septum had formed compared to that of GFP-SpoVD. Fluorescence from GFP-PBP4b in strain LMD170 was observed in the mother cell membrane, independent of sporulation stage, and no signal was found associated with the forespore (Fig. 4; see Fig. S3A and B).
FIG 4.
Fluorescence and phase-contrast microscopy images of sporulating B. subtilis LMD166 (GFP-SpoVD) and LMD170 (GFP-PBP4b) cells at the forespore engulfment and forespore in the cell stages. The fusion proteins were expressed from genes dependent on the xylose promoter. Cells were taken for microscopy at hourly intervals after sporulation induction by resuspension. The sporulation stage was determined on the basis of the FM4-64 fluorescence and phase-contrast images. Engulfment occurred 2 to 3 h after induction and the forespore in the cell stage was observed about 4 h after induction. For forespore-containing cells, strain LMD166 typically (86%, n = 139 cells) showed GFP fluorescence mainly from the forespore membrane, whereas strain LMD170 typically (90%, n = 29 cells) showed GFP fluorescence mainly from the mother cell membrane. Bars, 2 μm.
To exclude the possibility that the xylose promoter or the presence of xylose in the growth medium during sporulation causes an aberrant distribution of the GFP fusion proteins, for example, due to unbalanced protein production, we made use of the spoVD promoter, which is σE dependent and repressed by SpoIIID. The constructed gfp fusion genes under this promoter were, as before, inserted at the amyE locus in the ΔspoVD ΔpbpI genomic background. Strain LMD185 (containing GFP-SpoVD) showed a normal production of heat-resistant spores, whereas strain LMD186 (containing GFP-PBP4b) produced only heat-sensitive spores, as expected (Table 2). The presence of GFP fusion proteins in both strains, and the production of these proteins only during sporulation, was verified by immunoblot analysis (see Fig. S4). The different subcellular localizations of SpoVD and PBP4b in sporulating cells were confirmed and appeared more clearly than in the experiments based on the xylose promoter, because the respective proteins were not present at the initiation of sporulation (see Fig. S5). The finding of PBP4b in the mother cell membranes in sporulating cells is consistent with results by Ojkic et al. obtained using the spoIID promoter for gene expression of fusion proteins (5).
To analyze whether the SpoVD protein (and cortex synthesis) affects the subcellular distribution of GFP-PBP4b in sporulating cells, we also analyzed strain LMD169, deleted for only pbpI (Fig. S3C). Up to the stage of asymmetric septation, there was no difference compared to LMD170 cells (GFP-PBP4b in ΔspoVD ΔpbpI double-deletion background). However, starting at engulfment and proceeding until the forespore in cell stage, we observed two different patterns for LMD169. GFP fluorescence was found either only in the mother cell membrane (as seen for LMD170) or in both the mother cell membrane and the forespore. Late in sporulation, at the phase-gray state, no fluorescence was associated with the forespore. Thus, the only notable difference between strains LMD170 (ΔspoVD ΔpbpI) and LMD169 (ΔpbpI) was that a minority fraction of LMD169 cells (23%, n = 141) at approximately 5 h into sporulation showed fluorescence from the forespores in addition to the mother cell cytoplasmic membranes. Similar results were obtained with the corresponding strains LMD186 (ΔspoVD ΔpbpI) compared to LMD182 (ΔpbpI), in which the GFP-PBP4b fusion gene is expressed from the spoVD promoter (data not shown).
We conclude from the fluorescence microscopy that PBP4b during sporulation is retained in the mother cell membrane compared to SpoVD, which accumulates in the forespore outer membrane. In concert with the engulfment of the forespore, SpoVD disappears from the mother cell cytoplasmic membrane.
Forced targeting of PBP4b to the forespore.
The heat sensitivity of spores resulting from the lack of SpoVD is not complemented by PBP4b as shown here (Table 2) and previously by Wei et al. (14). The mother cell cytoplasmic membrane is an inappropriate localization for PBP4b to functionally substitute for a lack of SpoVD, and we hypothesized that this explains the failure of complementation. In the forespore outer membrane, SpoVD physically interacts with SpoVE (22), which is a membrane integral protein of 39 kDa belonging to the SEDS (shape, elongation, division, and sporulation) family of proteins (23). The expression of SpoVE is dependent on σE, and the protein is targeted to the forespore outer membrane. SpoVE is required for endospore cortex synthesis and thus the formation of heat-resistant spores. The accumulation of SpoVD in the forespore outer membrane depends on SpoVE (22). A tandem fusion protein of SpoVE and SpoVD is active as demonstrated before by complementation of ΔspoVD and ΔspoVE mutants (22).
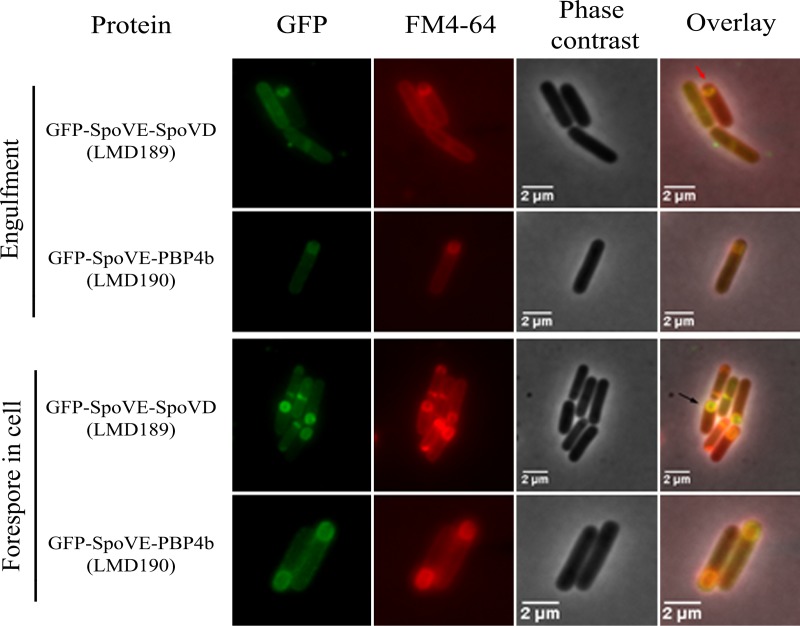
To bring PBP4b to the forespore outer membrane, we constructed a gene encoding a GFP-SpoVE-PBP4b triple-fusion protein and, as a control, a corresponding gene encoding GFP-SpoVE-SpoVD (Fig. 1). By immunoblotting, it was confirmed that the fusion protein variants were present in membranes during sporulation (see Fig. S6). As determined by fluorescence microscopy, GFP-SpoVE-SpoVD (strain LMD189) localized to the forespore membrane, while GFP-SpoVE-PBP4b (strain LMD190) was found in both the mother cell and forespore membranes (Fig. 5; see Fig. S7). The GFP-SpoVE-SpoVD protein in strains LMD194 (ΔspoVE) and LMD189 (ΔspoVD) enabled the formation of heat-resistant spores (Table 3), confirming that the triple-fusion protein is localized to the forespore. Also, the GFP-SpoVE-PBP4b variant complemented SpoVE deficiency (strain LMD193), demonstrating by function that this fusion protein is incorporated into the forespore. GFP-SpoVE-PBP4b did not complement the lack of SpoVD (strain LMD190) (Table 3). Our results indicate that despite forespore membrane localization, PBP4b as a fusion with SpoVE is for some other reason unable to perform the essential task in cortex peptidoglycan synthesis executed by SpoVD.
FIG 5.
Fluorescence and phase-contrast microscopy images of sporulating B. subtilis LMD189 (GFP-SpoVE-SpoVD) and LMD190 (GFP-SpoVE-PBP4b) cells at the forespore engulfment and forespore in the cell stages. The fusion proteins were expressed from genes dependent on the xylose promoter. Cells were taken for microscopy at hourly intervals after sporulation was induced by resuspension. The sporulation stage of cells was determined on the basis of the FM4-64 fluorescence and phase-contrast images. The red arrow indicates a cell in the forespore engulfment stage and the black arrow indicates a cell with a forespore. For forespore-containing cells, LMD189 typically (93%, n = 329) showed GFP fluorescence mainly from the forespore membrane, whereas LMD190 (96%, n = 491 cells) showed fluorescence from both the mother cell and the forespore membranes.
TABLE 3.
Sporulation efficiencies of B. subtilis strains containing GFP-SpoVE-PBP4b or GFP-SpoVE-SpoVD triple-fusion protein
| Strain | Relevant genotype | Yield (%) of heat-resistant sporesa |
|---|---|---|
| LMD101 | ΔspoVD | 0b |
| LMD121 | ΔspoVE | 0b |
| LMD189 | ΔspoVD amyE::Pxyl-gfp-spoVE-spoVD | 87 ± 1 |
| LMD190 | ΔspoVD amyE::Pxyl-gfp-spoVE-pbpI | 0b |
| LMD193 | ΔspoVE amyE::Pxyl-gfp-spoVE-pbpI | 77 ± 4 |
| LMD194 | ΔspoVE amyE::Pxyl-gfp-spoVE-spoVD | 73 ± 1 |
Cells were sporulated for 2 days at 37°C in NSMP supplemented with 0.2% (wt/vol) xylose. The results are presented as the averages from three independent experiments ± SEMs.
No colonies were obtained when 100 μl heated culture was plated on TBAB. Three independent experiments were done, including one biological replicate using another clone.
N-terminal domain of SpoVD mediates forespore localization.
To study if an individual domain of SpoVD functions as the determinant for forespore targeting, we constructed a series of genes encoding various SpoVD/PBP4b chimeras. The protein sequences were divided into three parts: the TM (comprising the N-terminal end and the transmembrane region), the UF (with the domain of unknown function), and the TP (the C-terminal part with transpeptidase activity and, in the case of SpoVD, containing one PASTA sequence). Borders for the TM and TP domains were chosen on the basis of an amino acid sequence comparison (Fig. S1) and the predicted secondary structure of SpoVD (18). The four constructed chimeric proteins, all with GFP at the N terminus, are schematically presented in Fig. 1. The prefixes in the names of the chimeras indicate the domain that has been swapped.
The chimeric proteins were all present in sporulating cells, although the relative amounts varied as determined by an immunoblot (Fig. 3). None of the chimeric proteins produced in ΔspoVD ΔpbpI cells complemented the sporulation defect, i.e., strains LMD167, LMD168, LMD174, and LMD175 produced phase-gray heat-sensitive spores (Table 2). The same results were obtained with ΔspoVD single-mutant strains containing the different chimeras (strains LMD164, LMD165, LMD172, and LMD173) (data not shown). This excluded the remote possibility that a deletion of pbpI in the chromosome somehow interferes with the function of chimeric proteins.
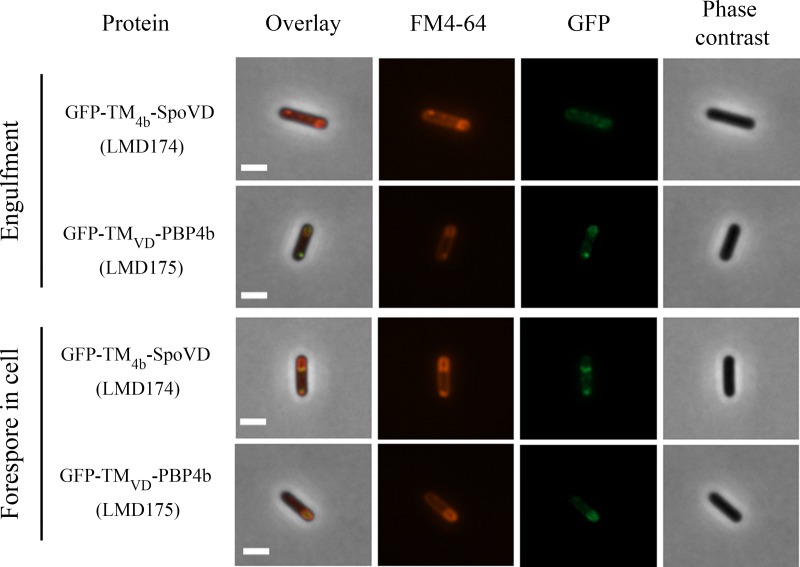
Fluorescence microscopy showed that chimeras with the TM domain of PBP4b (strains LMD168 and LMD174) had the same subcellular distribution as PBP4b. In contrast, those containing the TM domain of SpoVD (strains LMD167 and LMD175) accumulated in the forespore (Fig. 6; see also Fig. S8 and S9). Generally, the chimeric proteins resulted in a less-distinct distribution of GFP fluorescence in the cells compared to that of GFP fusions with the wild-type proteins. For example, strains LMD167 and LMD175 showed GFP signal accumulation at the forespore membranes but also weak GFP signals from the mother cell membranes. LMD174 cells (containing GFP-TM4b-SpoVD) at the stage of engulfed forespore showed fluorescence at one of the poles of the forespores. This pattern was not observed for GFP-PBP4b.
FIG 6.
Fluorescence and phase-contrast microscopy images of sporulating B. subtilis LMD174 (GFP-TM4b-SpoVD) and LMD175 (GFP-TMVD-PBP4b) cells at the forespore engulfment and forespore in the cell stages. The proteins were expressed from genes dependent on the xylose promoter. Cells were taken for microscopy at hourly intervals after sporulation induction by resuspension. The sporulation stage of cells was determined on the basis of the FM4-64 fluorescence and phase-contrast images. For forespore-containing cells, strain LMD175 typically (80%, n = 131 cells) showed fluorescence from the entire forespore membrane, whereas LMD174 (100%, n = 48 cells) showed a different pattern, with fluorescence from only one pole of the forespore and from the entire mother cell membrane. Bars, 2 μm.
To confirm the localization patterns for the TM domain-swapped chimeric proteins fused to GFP, we also analyzed strains in which the genes for the proteins were expressed from the spoVD promoter (instead of the xylose-inducible promoter) and in both ΔspoVD ΔpbpI double and ΔspoVD single-deletion genomic backgrounds. The amounts of chimeric proteins were relatively small in the cells (see Fig. S4). Strains LMD181 and LMD188, containing chimeric PBP4b with the TM of SpoVD, contained little fusion protein; therefore, the intensity of GFP fluorescence did not enable us to determine the subcellular localization of this fusion protein in these strains. The results with strains LMD180 and LMD187 confirmed the localization pattern for GFP-TM4b-SpoVD seen in experiments based on the xylose promoter.
Our findings with the SpoVD/PBP4b chimeric proteins suggest that the TM domain of SpoVD comprises a signal for forespore targeting in sporulating B. subtilis cells.
DISCUSSION
During sporulation in B. subtilis, many membrane proteins produced in the mother cell localize selectively to the forespore and play a role in endospore morphogenesis. Three mechanistic models for how proteins are targeted to the forespore have been proposed (24). In the first model, called targeted insertion, the protein is directly and selectively inserted into the outer membrane that surrounds the forespore. The second model, selective degradation, assumes a random insertion of the protein into all membranes available in the mother cell and subsequent proteolytic elimination of the protein only in the cytoplasmic membrane. The third model involves a random insertion of the protein into membranes followed by lateral diffusion and capture in the forespore outer membrane before engulfment is completed.
SpoVD and PBP4b in B. subtilis are paralogous (see Fig. S1 in the supplemental material), high-molecular-weight class B PBPs synthesized early during sporulation in the cytoplasm of the mother cell. Later in sporulation, SpoVD but not PBP4b accumulates in the forespore. Using SpoVD/PBP4b chimeras with swapped protein domains, we demonstrate that the N-terminal 40 residues of SpoVD comprise a forespore targeting signal. This segment comprises the N-terminal 11 residues exposed to the cytoplasm of the mother cell, a transmembrane region of approximately 20 residues, and a short extracytoplasmic sequence (Fig. 1 and Fig. S1). Also, for the related B. subtilis essential high-molecular-weight class B enzymes PBP2a and PBP2b, the TM domains play roles in the subcellular localization of the proteins (25).
PBPs generally constitute components of dynamic multienzyme complexes. They are, for example, present in the so-called elongasome and divisome for lateral cell wall and division septum peptidoglycan synthesis, respectively. The localization of B. subtilis PBP1 (class A enzyme) to the septa of vegetative cells is dependent on several cell division proteins, including FtsZ, PBP2b, DivIB, and DivIC (26). Similarly, in E. coli, the positioning of PBP3 (FtsI, a class B enzyme) at the cell division septum depends on the prior localization of FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsBL, and FtsW (27). FtsW and PBP3 directly interact and can be isolated as a subcomplex (28). The N-terminal 65 residues of PBP3, and in particular, residues Arg23, Leu39, and Gln46, are important for binding to FtsW (29–32). The periplasmic loops between transmembrane segments 7 and 8 as wells as segments 9 and 10 in FtsW seem important for the interaction with PBP3 (33, 34).
B. subtilis mutants deficient in SpoVD or SpoVE show very similar phenotypes by producing heat-sensitive endospores without a detectable cortex layer and by accumulating peptidoglycan synthesis precursors (17). Dworkin and coworkers (22) have demonstrated that SpoVE is required for forespore localization of SpoVD, that SpoVD and SpoVE physically interact, and that SpoVD protects the SpoVE polypeptide from degradation. B. subtilis has two SpoVE paralogues, FtsW and RodA. These three SEDS family proteins have been proposed to be lipid II flippases, but it was recently suggested that they act as transglycosylases (23, 35, 36). They are known to interact with a cognate class B PBP, i.e., in B. subtilis, FtsW interacts with PBP2b, RodA with several cell elongation PBPs (PBP2a and PBPH), and SpoVE with SpoVD (22, 37). The SpoVE forespore localization depends, directly or indirectly, on SpoIIQ, SpoIIIAH, and SpoIVFA (9). SpoIVFB is one of the best-studied proteins that localize specifically to the outer forespore membrane. Th functional localization of SpoIVFB relies on a second membrane integral protein called SpoIVFA. The collective data presented by Rudner et al. (24) are compatible with a model in which SpoIVFB is inserted into the cytoplasmic membrane followed by diffusion to, and capture in, the outer forespore membrane. SpoIIIAH (important for the localization of SpoVE and thereby also SpoVD to the forespore) reaches its final subcellular destination through an interaction with the forespore membrane protein SpoIIQ. The extracellular domains of these two proteins interact and become trapped in the space between the inner and outer forespore membranes (38).
For SpoVD, we have previously shown that neither the PASTA domain nor the transpeptidase activity of the TP domain are important for the accumulation of SpoVD in the forespore outer membrane (15, 16). Our findings in this work suggest that the N-terminal domain of SpoVD determines the forespore localization of the protein. Early in sporulation, the SpoVD polypeptide is synthesized on ribosomes in the mother cell cytoplasm, and the membrane insertion of the polypeptide is presumably mediated by the Sec system with the N-terminal segment of SpoVD acting as an uncleaved signal sequence. The maturation of SpoVD by the folding of the UF and TP domains involves StoA, CcdA, and probably other proteins (19). It remains to be discovered how SpoVD subsequently becomes concentrated in the forespore, but it apparently occurs by lateral diffusion in the membrane and capture by SpoVE before the engulfment of the forespore is completed, as described for other proteins (24). Alternate mechanisms, i.e., selective degradation of SpoVD in the mother cell membrane or selective insertion of the protein into the asymmetric septum, are not compatible with the available experimental data. There are, for example, no results indicating selective degradation of SpoVD but not PBP4b in sporulating cells (Fig. 2).
The function of PBP4b in B. subtilis remains enigmatic, because no phenotype has been associated with a lack of this protein alone or in combination with other deficiencies. The protein shows transpeptidase activity, i.e., it covalently binds penicillin (14) (this work). Most Bacillus species contain genes for two sporulation-specific, high-molecular-weight class B PBPs corresponding to SpoVD and PBP4b in B. subtilis, with the characteristic pair of cysteine residues in a conserved sequence in the transpeptidase domain (19) as a signature for identification. It appears unlikely that PBP4b, produced during sporulation, is without function under all possible conditions. The laboratory lineage of B. subtilis we have used for our studies might lack the component for some nonessential feature that depends on PBP4b and which is present in wild-type isolates. In this perspective, as an example, PBP4b is expected to interact with one hitherto unidentified SEDS family protein. The observed inability of PBP4b to replace SpoVD in endospore cortex synthesis is apparently not explained by only an improper subcellular localization of the protein during sporulation, i.e., in the mother cell cytoplasmic membrane rather than the forespore membrane. We show in this work that a SpoVE-PBP4b fusion protein in the forespore outer membrane complements SpoVE deficiency but not SpoVD deficiency (Table 3 and Fig. 5). Notably, a similar B. subtilis SpoVE-PBP2b fusion protein was found unable to complement a spoVE deletion mutant despite correct forespore targeting (22). This suggests that PBP2b in contrast to PBP4b somehow interferes with the function of SpoVE in the fusion protein. From these findings and the results with SpoVD/PBP4b chimeric protein variants with swapped domains, we conclude that PBP4b cannot functionally interact with SpoVE for cortex peptidoglycan synthesis. Multiple interactions within enzyme complexes (divisome, elongasome, etc.) are required for cell wall synthesis and presumably also for endospore cortex peptidoglycan synthesis. The situation may be further complicated by the fact that the functions of the UF domains in PBP4b and SpoVD might be different. Diverse functions of the UF domain among class B PBPs are illustrated by the interaction of PBP2 with MreC in cell wall synthesis in Helicobacter pylori (39), the recruitment of FtsN to PBP3 in E. coli (29), and the intramolecular domain interaction in PBP2a of methicillin-resistant Staphylococcus aureus (40).
MATERIALS AND METHODS
Bacterial strains and growth media.
The B. subtilis strains used in the work are presented in Table 1. Escherichia coli TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 λ−] was used for cloning plasmid DNA and E. coli Tuner [F− ompT hsdSB(rB− mB−) gal dcm lacY1 (DE3)] was used for the production of glutathione S-transferase (GST)–sPBP4b fusion protein. E. coli cells were grown at 37°C in 2× yeast-tryptone agar (YTA) medium, LB medium, or on LB agar plates (41). B. subtilis cells were grown at 37°C in minimal medium (42), nutrient sporulation medium with phosphate (NSMP) (43), or resuspension sporulation medium (44) or on tryptose blood agar base (TBAB) plates (Difco). Antibiotics were used when appropriate as follows: ampicillin (100 μg/ml) and kanamycin (40 μg/ml) for E. coli and chloramphenicol (4 μg/ml), tetracycline (15 μg/ml), spectinomycin (100 or 150 μg/ml), erythromycin (1 μg/ml), and neomycin (4 μg/ml) for B. subtilis. TBAB medium supplemented with 1% (wt/vol) soluble starch was used to test for amylase activity in B. subtilis colonies.
DNA techniques.
DNA manipulation was performed using standard methods (41). Plasmid DNA from E. coli was isolated using the Quantum Prep plasmid miniprep kit (Bio-Rad) or QIAfilter plasmid Midi kit (Qiagen). Chromosomal DNA of B. subtilis was isolated by the procedure described by Marmur (45) or the cetyl trimethylammonium bromide preparation procedure adapted from Wilson (46), where the cells were suspended in 75 mM NaCl containing 25 mM Tris-HCl (pH 7.5), 25 mM EDTA, 2 mg/ml lysozyme, and 50 μg/ml RNase A.
PCR was carried out using Phusion high-fidelity DNA polymerase (Finnzymes). Table S1 in the supplemental material shows the sequences of the oligonucleotides (primers) used in this work. PCR products were purified using Amicon Ultra 0.5-ml 30K centrifugal filters (Merck Millipore) or the QIAquick PCR purification kit (Qiagen). Dephosphorylation of DNA was performed using Antarctic alkaline phosphatase (New England BioLabs) at 37°C for 15 min, followed by heat inactivation at 65°C for 5 min. DNA was eluted from agarose gels using the Jetsorb gel extraction kit (Genomed GmbH) or GeneJET gel extraction kit (Thermo Scientific). DNA was ligated using T4 DNA ligase (New England BioLabs) at 16°C overnight.
Unless stated otherwise, E. coli cells were transformed using chemically competent cells (47). B. subtilis cells were grown to natural competence (48), and approximately 0.5 μg of DNA was added to 0.5 ml competent cells, unless another amount of DNA is stated. All DNA fragments cloned in plasmids were verified by Sanger sequencing at Eurofins MWG, Germany.
Construction of plasmids.
The plasmids used in the work are presented in Table S2. The construction of plasmids is described in the supplemental material.
Construction of B. subtilis strains.
The origin of strains used in this work is presented in Table 1. The insertion of genes encoding fusion proteins at the amyE locus was confirmed by a lack of amylase production or by the loss of erythromycin resistance (for LMD162 derivatives) and by PCR using primers Ewa54 and Ewa56 (Table S1).
(i) In-frame deletion of pbpI.
Strain LMD158 deleted for pbpI was constructed using the markerless gene replacement method described by Janes and Stibitz (49), with some modifications. B. subtilis 1A1 was transformed with pLMS2 containing an I-SceI restriction site, resulting in integration of the plasmid at the region flanking the pbpI locus by a single-crossover event (Campbell-type recombination). The obtained strain, LMD156, was transformed with pBKJ223, which encodes the I-SceI endonuclease. Cleavage by I-SceI introduces one double-strand break in the chromosomal DNA, and this triggers double-crossover (homologous recombination) repair, resulting in a markerless deletion of pbpI or retained pbpI. Tetracycline-resistant transformants were scored for loss of the allelic exchange plasmid by patching single colonies onto TBAB-chloramphenicol plates. Chloramphenicol-sensitive clones were then passed at least two times on TBAB plates to rid the cells of pBKJ223. The pbpI in-frame deletion in the obtained strain LMD158 was confirmed by PCR (primers MSI011/MSI012) and DNA sequencing.
(ii) Construction of spoVD pbpI double-deletion strain LMD162.
According to the protocol described by Liu et al. (18), strain LMD158 was transformed with chromosomal DNA from strain LMD12. Through a double-crossover event, spoVD was changed to spoVD::spc resulting in strain LMD159 that has spc inserted into spoVD and pbpI deleted. The deletion of spoVD in LMD159 was obtained using transformation (congression) with plasmid pLEB2 (that had been cleaved by ScaI) together with chromosomal DNA from strain LMD15 (spoVD::spc and containing an erythromycin resistance gene inserted into the amyE locus). Erythromycin-resistant transformants were selected on TBAB plates and tested for spectinomycin sensitivity. Chromosomal DNA was isolated from one erythromycin-resistant and spectinomycin-sensitive transformant named LMD162. The spoVD and pbpI double deletion was confirmed by PCR (primers Ewa1/Ewa4 and MSI011/MSI012, respectively).
(iii) In-frame deletion of spoVE.
Strain LMD121 carrying a markerless in-frame deletion of spoVE was constructed by congression. Briefly, LMD6 was transformed with a mixture of pLEB27 and LMD120 chromosomal DNA. The integration of the neomycin marker of LMD120 chromosomal DNA into the qcr locus of LMD6 enabled the selection of transformants. A double-crossover event between the insert in pLEB27 and regions flanking spoVE in the LMD6 chromosome led to an in-frame deletion of spoVE and loss of erythromycin resistance. Neomycin-resistant clones (850 colonies in total) were screened for a loss of erythromycin resistance by replica plating. The spoVE in-frame deletion in the chromosome of one clone, named LMD121, was confirmed by PCR (using primers Ewa32/Ewa35) and DNA sequence analysis.
Purification of sPBP4b and generation of antiserum.
sPBP4b was produced as a GST-sPBP4b fusion protein in E. coli Tuner(DE3)/pLEB28, and the thrombin-cleaved protein was isolated as described in the supplemental material. The yield of purified sPBP4b was 0.4 mg/liter culture. The identity and activity of the isolated protein were confirmed by mass spectrometry and the covalent binding of penicillins (Fig. S2). Antiserum against sPBP4b was obtained as described in the supplemental material.
Sporulation and heat resistance assay.
Sporulation was induced by nutrient exhaustion in NSMP as described in reference 50 or by the resuspension method (44). For strains that require xylose for the induction of gene expression, the medium was supplemented with 0.2% (wt/vol) xylose at the start of culture. Spore heat resistance assays were carried out on 2-day-old cultures in the case of nutrient exhaustion and on overnight cultures in the case of the resuspension method. The sporulation efficiency was analyzed by heating 5 ml of culture at 80°C for 10 min. Serial dilutions of heated and unheated samples were spread on TBAB plates. After overnight incubation at 37°C, the colonies were counted and the spore yield was calculated.
Light microscopy.
One hour after sporulation was induced by resuspension, the membrane stain dye FM4-64 (Invitrogen) was added to the culture at a final concentration of 0.5 μg/ml. Then, 500-μl samples were taken every hour from 1 to 5 h after resuspension. A total of 1 to 10 μl cell suspension was added to an agarose pad (1% agarose in phosphate-buffered saline [PBS]) on a microscopy glass slide and covered with a coverslip. Phase-contrast and fluorescence images were acquired using a Zeiss Axio Imager.Z2 microscope equipped with X-Cite 120 Illumination (EXFO Photonic Solutions Inc.), an ORCA-flash 4.0 C11440 camera (Hamamatsu Photonics), and Volocity software (PerkinElmer). For strains LMD189 and LMD190, the phase-contrast and fluorescence images were acquired using a Zeiss Observer.Z1 inverted microscope equipped with HXP 120V Illuminator (Leistungselektronik Jena GmbH), an ORCA-flash 4.0LT C11440 camera (Hamamatsu Photonics), and Zen 2.3 Pro (Zeiss). Typical exposure times used to obtain fluorescence images of cells were 600 ms for GFP and 200 ms for FM4-64. The images were processed and analyzed using the Volocity or FIJI (ImageJ) software. Background fluorescence was subtracted from each image using the autofluorescence of sporulating strain LMD162 (ΔspoVD ΔpbpI) or LMD101 (ΔspoVD). Images were saved in TIFF format.
Preparation of B. subtilis cell extracts.
Fifty milliliters cell culture was collected by centrifugation at 5,000 × g at 4°C for 15 min. The cell pellet was washed with 20 ml ice-cold 50 mM potassium phosphate buffer (pH 8.0) and centrifuged as before. The cells were resuspended in 1 ml ice-cold 20 mM Na-MOPS (morpholinepropanesulfonic acid) buffer (pH 7.4) containing 0.5 mM EDTA and 1× complete protease inhibitor cocktail (Roche) and transferred into a prechilled screw-cap microcentrifuge tube containing approximately 500 mg of 0.1-mm diameter zirconia glass beads. Phenylmethanesulfonyl fluoride was added from a 250 mM stock solution in 96% ethanol to a 1 mM final concentration. The cells were disrupted using a FastPrep-24 (MP Biomedicals) bead beating system (setting, 6.5 m/s, 3 cycles of 45 s). Samples were kept for 5 min on ice between cycles. The glass beads and unbroken cells were removed from lysates by two consecutive centrifugation steps, the first for 15 s and the second for 5 min (5,000 × g at 4°C). After each centrifugation step, the cleared cell lysate was transferred to a fresh tube. A 150-μl volume was taken out, and the remaining bulk lysate was centrifuged at 48,000 × g at 4°C for 1 h. The upper part of the supernatant (approximately 500 μl) was taken as the soluble cell fraction and the remaining supernatant was discarded. The pellet was suspended in 0.4 ml cold 20 mM Na-MOPS buffer (pH 7.4) and subjected to centrifugation as before. The washed pellet containing membranes was suspended in 120 μl cold 20 mM Na-MOPS buffer (pH 7.4). The samples were stored at −20°C. The protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) and with bovine serum albumin as the standard.
Immunoblot analysis.
Proteins in extracts were fractionated by SDS-PAGE (200 V, 150 mA) using precast bis-Tris 10% acrylamide gels (Invitrogen) and MES (morpholineethanesulfonic acid) running buffer. A prestained protein ladder, PageRuler Plus (Thermo Scientific), was used as a molecular mass marker. After SDS-PAGE, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore) using wet electrotransfer (30 V, 0.1 A, 4°C, overnight). The transfer buffer was Laemmli buffer (3.03 g Tris and 14.4 g glycine per liter) containing 20% (vol/vol) methanol. The antibodies used were anti-GFP (1:5,000; GenScript), anti-BdbD (20) (1:3,000), anti-SpoVD (18) (1:3,000), and anti-PBP4b (this work) (1:3,000) diluted as indicated in 1% fat-free skimmed milk in 0.1% Tween 20 in PBS (TBS). Immunodetection was carried out by chemiluminescence using donkey anti-rabbit antibodies conjugated to horseradish peroxidase (1:3,000 dilution; Amersham Biosciences) and Super Signal West Pico chemiluminescence substrate (Pierce Chemical). Luminescence was detected using a GelDoc imager (Bio-Rad).
Supplementary Material
ACKNOWLEDGMENTS
We thank Fanny Passot and Klas Flärdh, Lund University, for their expert instructions and assistance in operating the fluorescence microscopes and Claes von Wachenfeldt for advice on the design of protein chimeras.
Funding was provided by the Swedish Research Council (grant 2015-02547).
We have no conflict of interest to declare.
L.H. and E.B.-F. planned and supervised the research work. D.L. performed the initial experiments and purified and characterized sPBP4b. M.S. and E.B.-F. performed most of the experiments. M.S., L.H., and E.B.-F. analyzed data and wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00163-18.
REFERENCES
- 1.Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 2.Hilbert DW, Piggot PJ. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ojkic N, Lopez-Garrido J, Pogliano K, Endres R. 2016. Cell-wall remodelling drives engulfment during Bacillus subtilis sporulation. Elife 5:e18657. doi: 10.7554/eLife.18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popham D, Bernhards C. 2016. Spore peptidoglycan, p 157–177. In Driks A, Eichenberger P (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 7.Driks A, Eichenberger P. 2016. The spore coat, p 179–200. In Driks A, Eichenberger P (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 8.Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972. doi: 10.1016/S0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- 9.Dworkin J. 2016. Protein targeting during Bacillus subtilis sporulation, p 145–156. In Driks A, Eichenberger P (ed), The bacterial spore: from molecules to systems. ASM Press, Washington, DC. [Google Scholar]
- 10.Sauvage E, Kerff F, Terrak M, Ayala J, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 11.Scheffers DJ, Pinho MG. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel RA, Drake S, Buchanan CE, Scholle R, Errington J. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol 235:209–220. doi: 10.1016/S0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. 2006. The forespore line of gene expression in Bacillus subtilis. J Mol Biol 358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 14.Wei Y, McPherson DC, Popham DL. 2004. A mother cell-specific class B penicillin-binding protein, PBP4b, in Bacillus subtilis. J Bacteriol 186:258–261. doi: 10.1128/JB.186.1.258-261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukowska-Faniband E, Hederstedt L. 2015. The PASTA domain of penicillin-binding protein SpoVD is dispensable for endospore cortex peptidoglycan assembly in Bacillus subtilis. Microbiology 161:330–340. doi: 10.1099/mic.0.000011. [DOI] [PubMed] [Google Scholar]
- 16.Bukowska-Faniband E, Hederstedt L. 2013. Cortex synthesis during Bacillus subtilis sporulation depends on the transpeptidase activity of SpoVD. FEMS Microbiol Lett 346:65–72. doi: 10.1111/1574-6968.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasudevan P, Weaver A, Reichert E, Linnstaed S, Popham D. 2007. Spore cortex formation in Bacillus subtilis regulated by accumulation of peptidoglycan precurosors under the control of sigma K. Mol Microbiol 65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Carlsson Moller M, Petersen L, Soderberg CA, Hederstedt L. 2010. Penicillin-binding protein SpoVD disulphide is a target for StoA in Bacillus subtilis forespores. Mol Microbiol 75:46–60. doi: 10.1111/j.1365-2958.2009.06964.x. [DOI] [PubMed] [Google Scholar]
- 19.Bukowska-Faniband E, Hederstedt L. 2017. Transpeptidase activity of penicillin-binding protein SpoVD in peptidoglycan synthesis conditionally depends on the disulfide reductase StoA. Mol Microbiol 105:98–114. doi: 10.1111/mmi.13689. [DOI] [PubMed] [Google Scholar]
- 20.Crow A, Lewin A, Hecht O, Carlsson Moller M, Moore GR, Hederstedt L, Le Brun NE. 2009. Crystal structure and biophysical properties of Bacillus subtilis BdbD. An oxidizing thiol:disulfide oxidoreductase containing a novel metal site. J Biol Chem 284:23719–23733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eijlander R, de Jong A, Krawczyk A, Holsappel S, Kuipers O. 2014. SporeWeb: an interactive journey through the complete sporulation cycle of Bacillus subtilis. Nucleic Acids Res 42:D685–D691. doi: 10.1093/nar/gkt1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fay A, Meyer P, Dworkin J. 2010. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol 399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Real G, Fay A, Eldar A, Pinto S, Henriques A, Dworkin J. 2008. Determinants for the subcellular localization and function of a nonessential SEDS protein. J Bacteriol 190:363–376. doi: 10.1128/JB.01482-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudner DZ, Pan Q, Losick RM. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A 99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y. 2008. Effects of protein domains on localization of penicillin-binding proteins 2a and 2b in Bacillus subtilis. Masters thesis. Virgina Polytechnic Institute and State University, Blacksburg, VA. [Google Scholar]
- 26.Scheffers DJ, Errington J. 2004. PBP1 is a component of the Bacillus subtilis cell division machinery. J Bacteriol 186:5153–5156. doi: 10.1128/JB.186.15.5153-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buddelmeijer N, Beckwith J. 2002. Assembly of cell division proteins at the E. coli cell center. Curr Opin Microbiol 5:553–557. doi: 10.1016/S1369-5274(02)00374-0. [DOI] [PubMed] [Google Scholar]
- 28.Fraipont C, Alexeeva S, Wolf B, van der Ploeg R, Schloesser M, den Blaauwen T, Nguyen-Disteche M. 2011. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157:251–259. doi: 10.1099/mic.0.040071-0. [DOI] [PubMed] [Google Scholar]
- 29.Wissel MC, Weiss DS. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J Bacteriol 186:490–502. doi: 10.1128/JB.186.2.490-502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piette A, Fraipont C, Den Blaauwen T, Aarsman ME, Pastoret S, Nguyen-Disteche M. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J Bacteriol 186:6110–6117. doi: 10.1128/JB.186.18.6110-6117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marrec-Fairley M, Piette A, Gallet X, Brasseur R, Hara H, Fraipont C, Ghuysen JM, Nguyen-Disteche M. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol Microbiol 37:1019–1031. doi: 10.1046/j.1365-2958.2000.02054.x. [DOI] [PubMed] [Google Scholar]
- 32.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastoret S, Fraipont C, den Blaauwen T, Wolf B, Aarsman ME, Piette A, Thomas A, Brasseur R, Nguyen-Disteche M. 2004. Functional analysis of the cell division protein FtsW of Escherichia coli. J Bacteriol 186:8370–8379. doi: 10.1128/JB.186.24.8370-8379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leclercq S, Derouaux A, Olatunji S, Fraipont C, Egan AJ, Vollmer W, Breukink E, Terrak M. 2017. Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep 7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz N. 2015. Lipid flippases for bacterial peptidoglycan biosynthesis. Lipid Insights 8(Suppl 1):21–31. doi: 10.4137/LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gueiros-Filho F. 2007. Cell division. Caister Academic Press, Norfolk, UK. [Google Scholar]
- 38.Doan T, Marquis KA, Rudner DZ. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol 55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- 39.Contreras-Martel C, Martins A, Ecobichon C, Trindade DM, Mattei PJ, Hicham S, Hardouin P, Ghachi ME, Boneca IG, Dessen A. 2017. Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex. Nat Commun 8:776. doi: 10.1038/s41467-017-00783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peacock S. 2015. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci U S A 44:1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortnagel P, Freese E. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol 95:1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholson WL, Setlow P. 1990. Sporulation, germination and outgrowth, p 391–450. In Harwood CR, Cutting SM (ed), Molecular biology methods for Bacillus. John Wiley & Sons, Ltd, Chichester, United Kingdom. [Google Scholar]
- 45.Marmur J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218. doi: 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- 46.Wilson K. 1997. Preparation of genomic DNA from bacteria. In Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds), Current protocols in molecular biology. John Wiley & Sons, Inc, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D, Jessee J, Bloom FR. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol 204:63–113. doi: 10.1016/0076-6879(91)04006-A. [DOI] [PubMed] [Google Scholar]
- 48.Hoch JA. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol 204:305–320. doi: 10.1016/0076-6879(91)04015-G. [DOI] [PubMed] [Google Scholar]
- 49.Janes B, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953. doi: 10.1128/IAI.74.3.1949-1953.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erlendsson LS, Moller M, Hederstedt L. 2004. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J Bacteriol 186:6230–6238. doi: 10.1128/JB.186.18.6230-6238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, Le Brun NE. 1998. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J Biol Chem 273:8860–8866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.