ABSTRACT
Anabaena sp. strain PCC 7120 is a model strain for molecular studies of cell differentiation and patterning in heterocyst-forming cyanobacteria. Subtle differences in heterocyst development have been noticed in different laboratories working on the same organism. In this study, 360 mutations, including single nucleotide polymorphisms (SNPs), small insertion/deletions (indels; 1 to 3 bp), fragment deletions, and transpositions, were identified in the genomes of three substrains. Heterogeneous/heterozygous bases were also identified due to the polyploidy nature of the genome and the multicellular morphology but could be completely segregated when plated after filament fragmentation by sonication. hetC is a gene upregulated in developing cells during heterocyst formation in Anabaena sp. strain PCC 7120 and found in approximately half of other heterocyst-forming cyanobacteria. Inactivation of hetC in 3 substrains of Anabaena sp. PCC 7120 led to different phenotypes: the formation of heterocysts, differentiating cells that keep dividing, or the presence of both heterocysts and dividing differentiating cells. The expression of PhetZ-gfp in these hetC mutants also showed different patterns of green fluorescent protein (GFP) fluorescence. Thus, the function of hetC is influenced by the genomic background and epistasis and constitutes an example of evolution under way.
IMPORTANCE Our knowledge about the molecular genetics of heterocyst formation, an important cell differentiation process for global N2 fixation, is mostly based on studies with Anabaena sp. strain PCC 7120. Here, we show that rapid microevolution is under way in this strain, leading to phenotypic variations for certain genes related to heterocyst development, such as hetC. This study provides an example for ongoing microevolution, marked by multiple heterogeneous/heterozygous single nucleotide polymorphisms (SNPs), in a multicellular multicopy-genome microorganism.
KEYWORDS: Anabaena sp. strain PCC 7120, microevolution, heterocyst differentiation
INTRODUCTION
Cyanobacteria are prokaryotes that perform oxygenic photosynthesis. Some filamentous cyanobacteria can form heterocysts that fix N2 aerobically, contributing significantly to the global nitrogen cycle (1, 2). Anabaena (Nostoc) sp. strain PCC 7120 is the model strain used for molecular studies of heterocyst differentiation/patterning and multicellularity (3, 4). Upon N stepdown, some vegetative cells (usually ca. 10%) are induced to differentiate into heterocysts, and the number of vegetative cells between heterocysts roughly follows a Gaussian-type distribution (5), which is an example of a reaction-diffusion pattern (6, 7). Although the exact origin of this freshwater strain is not clear, it was first recorded as Nostoc muscorum and obtained from the algal collection of the Iowa State University in 1971 (8) and then deposited by R. Haselkorn in the Pasteur Culture Collection (PCC). From R. Haselkorn of Chicago University, C. Peter Wolk of the Michigan State University obtained this strain and developed the first conjugative gene transfer system (9). Most laboratories now working on Anabaena PCC 7120 obtained the strain directly or indirectly from either PCC, Chicago University, or Michigan State University. With the genetic tools available for Anabaena PCC 7120, many genes involved in heterocyst differentiation have been identified (10, 11).
Different laboratories working on the same genes sometimes find different phenotypes or spatial regulation. One example is nrrA, the expression of which was shown to be either upregulated in proheterocysts/heterocysts (12) or nondiscernible between vegetative cells and heterocysts (13). Another example is hetC, initially identified after the characterization of a transposon-generated mutant that formed no heterocysts but had increased expression of hetR, the master regulator gene of heterocyst differentiation (14). hetC mutants formed semiregularly spaced low autofluorescence cells, mostly doublets, at 48 h after N stepdown, while the wild type produced mature heterocysts before 24 h. The lowered autofluorescence in differentiating cells was probably due to the degradation of phycobilisomes, often used as a marker of heterocyst differentiation (14, 15). Whereas mature heterocysts are terminally differentiated cells, unable to divide, the low autofluorescence cells formed in hetC mutants were further shown to keep dividing, retaining upregulated expression of hetR (16) and some other genes involved in heterocyst differentiation (17). Unlike these reports, hetC mutants generated in the other two labs showed no or delayed heterocyst formation but no dividing differentiating cells (18, 19). In addition, heterocyst formation in a hetC background was fully or partially restored by the inactivation of patS or hetN, the two heterocyst inhibitor genes, even though the double mutants generated in the two labs differed in the frequency and function of heterocysts (18, 19). No adequate explanation is thus far available to explain the phenotypic differences observed in different laboratories while working on the same genes.
Genome microevolution events have been well studied in Escherichia coli (20, 21), Bacillus subtilis (22), Pseudomonas aeruginosa strain PAO1 (23), and some other bacterial pathogens (24–26). Microevolution and associated phenotypic changes or epistatic effects have also been reported in prokaryotes with multicellular behaviors, such as Caulobacter (27) and Myxococcus (28), or the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 (29–31). During a >40-year history, genetic events such as substitutions, deletions/insertions, and transpositions could accumulate in the genomes of Anabaena PCC 7120 substrains maintained in different labs, and such genomic variations may exert epistatic effects on the phenotypes of mutants of certain genes. Unlike most prokaryotes, cyanobacteria have one chromosome but in multicopies (32); in addition, Anabaena PCC 7120 has long multicellular filaments, with communicating channels connecting the cells along the filaments (4). These two characteristics can enable spontaneous mutations to accumulate more easily in an individual chromosome or individual cell in Anabaena PCC 7120 than those in unicellular single-copy-genome bacteria. Phenotypically discernible mutations (for example, altered heterocyst formation, shortened filaments, etc.) would have been discarded, but “cryptic” mutations could well be retained in “Anabaena PCC 7120” in different laboratories. In this study, we used 3 substrains as examples to compare their genome sequences/structures and to show the epistatic effects of genomic variations on the hetC phenotype. An evaluation of the differences between substrains should help us to better understand the function of certain genes in the background of microevolution and to design genetic experiments by taking into account such variations in Anabaena PCC 7120.
RESULTS
SNPs and indels (1 to 3 bp) in substrains of Anabaena sp. PCC 7120.
To understand the genetic basis in the variation of Anabaena PCC 7120 when maintained separately in different laboratories, we employed second-generation sequencing to analyze the genomes of substrains HAU and IHB. Around 48 million paired-end reads were generated for the HAU substrain and 17 million reads for the IHB substrain (read length, 150 bp). Genomic changes, such as single nucleotide polymorphisms (SNPs) and small indels (insertions/deletions), in Anabaena PCC 7120 were then analyzed. SNPs were identified using three software programs, and consensus sites were retrieved. Compared to the reference genome sequence of Anabaena PCC 7120 (maintained in C. P. Wolk's lab until 2001 when it was sequenced) in the Cyanobase (33), HAU showed 161 SNPs and 48 indels in the chromosome (see Table S1 in the supplemental material) and 21 SNPs and 11 indels in plasmids (Table 1); IHB showed 149 SNPs and 38 indels in the chromosome (Table S1) and 21 SNPs and 9 indels in plasmids (Table 1). The two substrains shared 80 SNPs and 21 indels in the chromosome, and 9 SNPs and 4 indels in plasmids. Randomly chosen SNPs and indels (except multiple sites in the same gene), including 29 in HAU, 34 in IHB, and 45 in MSU (corresponding to 29 in HAU and 30 in IHB, 14 of them shared by HAU and IHB), were confirmed by PCR and traditional sequencing.
TABLE 1.
SNPs in plasmids of Anabaena sp. PCC 7120 substrains IHB and HAUa
| Plasmid | Position | Reference base | IHB |
HAU |
Gene(s) | Region | ||
|---|---|---|---|---|---|---|---|---|
| Base | Depthc | Base | Depth | |||||
| Alpha | 3289 | C | T | 139 | C | all7005, asr7006 | Intergenic | |
| Alpha | 8242 | T | T | C | 125 | all7011 | ORFb | |
| Alpha | 25546 | T | T | G | 159 | all7027 | ORF | |
| Alpha | 49297 | A | A | G | 353 | alr7063 | ORF | |
| Alpha | 72155 | T | T | C | 310 | all7084 | ORF | |
| Alpha | 77566 | T | G | 1,822 | T | alr7089 | ORF | |
| Alpha | 85981 | A | G | 2,144 | G | 225 | all7098 | ORF |
| Alpha | 92804 | G | A/G | 1,554/438 | G | all7106, alr7107 | Intergenic | |
| Alpha | 125193 | T | T | C | 132 | alr7129 | ORF | |
| Alpha | 147600 | G | G | A | 141 | asr7143 | ORF | |
| Alpha | 157737 | G | A | 2,081 | G | alr7157 | ORF | |
| Alpha | 161185 | C | T | 1,205 | C | all7160, all7161 | Intergenic | |
| Alpha | 183584 | C | T | 2,016 | T | 140 | all7185 | ORF |
| Alpha | 197224 | T | T | C | 188 | all7191 | ORF | |
| Alpha | 216050 | G | C | 1,931 | G | alr7206 | ORF | |
| Alpha | 216051 | T | C | 1,914 | T | alr7206 | ORF | |
| Alpha | 216052 | T | C | 1,895 | T | alr7206 | ORF | |
| Alpha | 231547 | A | G | 2,049 | G | 155 | all7218, alr7219 | Intergenic |
| Alpha | 268895 | T | T | C | 323 | alr7249 | ORF | |
| Alpha | 289163 | A | G | 1,827 | G | 139 | all7275 | ORF |
| Alpha | 300552 | A | G | 1,909 | G | 227 | alr7294, alr7295 | Intergenic |
| Alpha | 300902 | A | G | 1,879 | G | 199 | alr7295 | ORF |
| Alpha | 310851 | G | G | A | 175 | alr7299 | ORF | |
| Alpha | 326058 | T | T | C | 210 | alr7304 | ORF | |
| Alpha | 364530 | G | G | A | 224 | asr7330 | ORF | |
| Alpha | 407466 | G | A | 1,983 | G | asr7385, alr7386 | Intergenic | |
| Beta | 51863 | G | A | 483 | G | alr7555 | ORF | |
| Gamma | 23623 | T | C | 1,400 | C | 310 | all8023 | ORF |
| Gamma | 56266 | C | T | 2,065 | C | asl8049, asl8050 | Intergenic | |
| Gamma | 56708 | C | T | 2,062 | C | asl8049, asl8050 | Intergenic | |
| Gamma | 96179 | A | A | G | 439 | all8083, asl8084 | Intergenic | |
| Delta | 32318 | A | G | 446 | G | 493 | alr8542, alr8543 | Intergenic |
| Zeta | 4713 | A | C | 1,101 | C | 364 | alr9504, alr9505 | Intergenic |
Bases in boldface font were confirmed with PCR and traditional sequencing.
ORF, open reading frame.
Number of sequencing reads.
Approximately 76.7% of the SNPs and indels were located within open-reading frames, which is consistent with the percentage of coding sequences in the whole genome. In several genes, more than one SNP was found. For example, GTT was replaced with CCC at position 216050 to 216052 on the alpha plasmid in IHB and T and A were replaced with C and G at chromosomal positions 6386500 and 6387506 within alr5351 in HAU.
Nonsynonymous substitutions were found in at least 4 genes involved in heterocyst formation or patterning. In HAU, such substitutions were found in two genes required for formation of the polysaccharide layer (glycosyltransferase), alr2832 (chromosome 3449008, C→T [Pro→Ser]) and alr2840 (chromosome 3461181, G→A [Gly→Asp]), and in a gene for the formation of the glycolipid layer (glycolipid synthase), alr5351 (chromosome 6387506, A→G [Asn→Asp]). In IHB, the same substitution in alr5351 was found; in addition, a C→T (chromosome 5730046) substitution was found in the intergenic region upstream of patN (alr4812), a gene involved in pattern formation (34). Even so, heterocyst envelope formation and heterocyst patterning remained normal in the two substrains.
Heterogeneous/heterozygous SNPs.
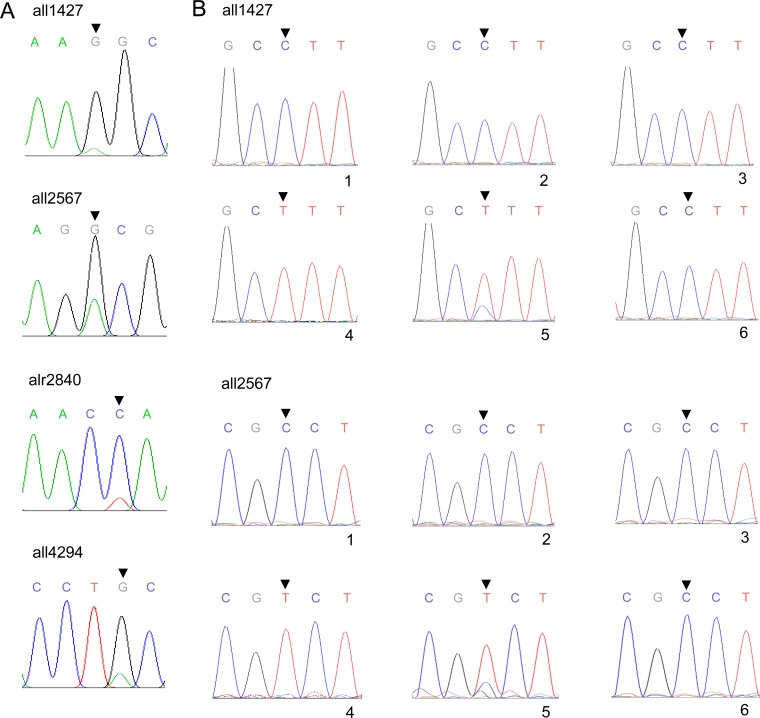
From the second-generation sequencing data, we identified some heterogeneous/heterozygous SNPs in the IHB sample (Table 1; Table S1), and one of the two bases at such genomic positions was always identical to the reference. This indicated that some bases were being substituted for in the culture, generating SNPs in some copies of the genome (heterozygous), some cells of filaments, or some filaments of the population (heterogeneous). In Fig. 1A, 4 such genomic positions were directly shown with traditional sequencing. Furthermore, we randomly picked 6 colonies derived from sonicated filaments of substrain IHB on plates and cultured them in liquid medium. PCR products were generated with DNA templates from the six colonies for regions with the heterogeneous/heterozygous SNPs. In Fig. 1B and in Fig. S2, the sequences of 6 regions are shown for the 6 colonies, indicating that SNPs at these genomic positions could be completely segregated.
FIG 1.
Traditional sequencing showing ongoing substitutions in Anabaena PCC 7120 substrain IHB. Sequencing was performed with PCR products generated using DNA extracted from substrain IHB or single colonies of IHB. (A) Sequencing before colony isolation. (B) Sequencing after colony isolation (the complementary strand compared to that in panel A). 1 to 6, DNA extracted with cells derived from 6 colonies.
Transpositions and fragment deletions in genomes of substrains.
In addition to SNPs and small indels, genome structural variations were also identified. To perform such analyses, it is necessary to assemble sequencing reads of the genome into the circular chromosome and plasmids. However, the assembly of short reads from second-generation sequencing could not meet this requirement. Thus, the genomes of Anabaena PCC 7120 substrains IHB, HAU, and MSU were also resequenced using third-generation long-read sequencing technology. For each substrain, a 10-kb library was constructed and sequenced. In a total, 61,322 reads (501 Mb) with an average read length of 8,165 bp obtained for substrain HAU, 86,704 reads (624 Mb) with an average read length of 7,198 bp were obtained for IHB, and 781,164 reads (4.9 Gb) with an average read length of 6,276 bp were obtained for MSU. IHB and HAU genome sequences were each assembled into a complete circular chromosome and five plasmids (assembled as circular plasmid alpha, circular plasmid beta, linear plasmid gamma, linear plasmid delta, and linear plasmid epsilon). Plasmid zeta was missing in the data. This was probably due to the smaller size of the plasmid (5,584 bp) compared to the insert length (around 10-kb DNA fragments for library construction) of the sequencing library. However, with a much larger number of reads, MSU genome sequences were assembled into a complete circular chromosome and all six circular plasmids (a low ratio of smaller size DNA might have been included in the “10-kb” DNA preparation).
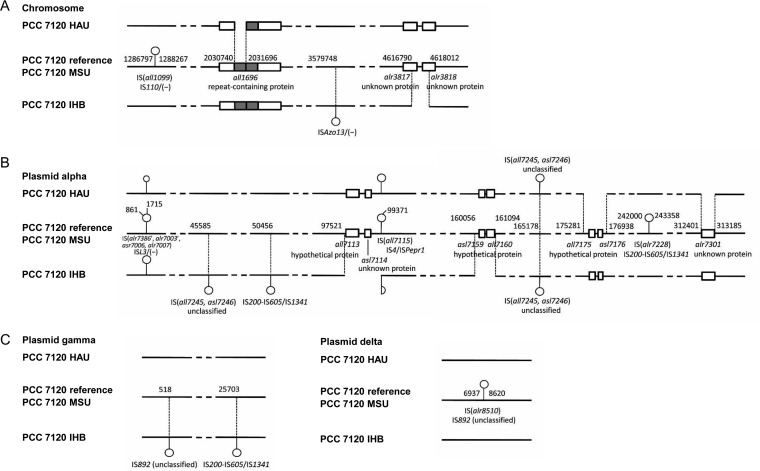
The genome structural variations are summarized in Fig. 2. In comparison to the reference genome, that for substrain HAU had fragment deletions at the chromosomal position 2030740 to 2031696 and alpha plasmid positions 861 to 1715 [within the complex IS(alr7386′, alr7003′, asr7006, alr7007)], 175281 to 176938, and 312401 to 313185; it also lacked insertion sequences (ISs) at the chromosomal position 1286797 to 1288267, the alpha plasmid position 242000 to 243358, and the delta plasmid position 6937 to 8620. Substrain IHB had fragment deletions at the chromosomal position 4616790 to 4618012 and alpha plasmid positions 97521 to 99371 [including a part of IS(all7115)] and 160056 to 161094; it lacked ISs at the chromosomal position 1286797 to 1288267, the alpha plasmid position 242000 to 243358, and the delta plasmid position 6937 to 8620 but instead acquired ISs at the chromosomal position 3579748, alpha plasmid positions 45585, 50456, and 165178, and gamma plasmid positions 518 and 25703 (according to the positions in the reference). In addition, three short sequence stretches (in short tandem repeat [STR] regions) were found in all three substrains but not in the reference genome (see Fig. S3); these differences should be due to erroneous sequence assembly of the Cyanobase reference genome. All these structural variations were confirmed with PCR.
FIG 2.
Differences in genome structure between Anabaena PCC 7120 substrains and the reference genome. (A) Structural differences in the chromosome. (B) Structural differences in the alpha plasmid. (C) Structural differences in gamma and delta plasmids.
Phenotypes of hetC mutants generated from substrains.
The relatively high number of genome variations showed no significant effects on heterocyst differentiation in the “wild-type” substrains, but certain variations may exert epistatic effects on the phenotypes of mutants generated with these substrains.
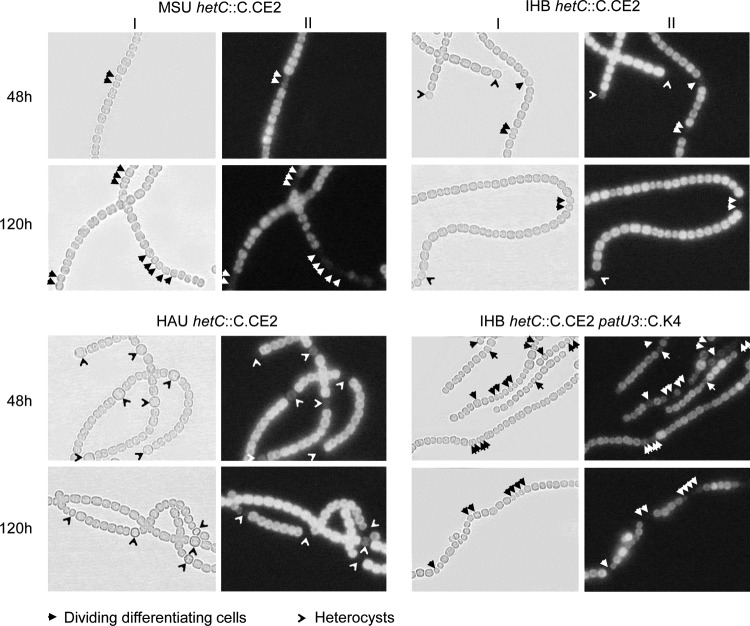
hetC is a gene upregulated in differentiating heterocysts, and different phenotypes of hetC-null mutants have been reported: differentiating cells formed heterocysts with impaired functions (18, 19) or continued dividing with partial differentiation (16, 17). The hetC sequences, including its promoter region, were unchanged in all the three substrains compared to that in the reference strain. In this study, we generated hetC::C.CE2 mutants from substrains HAU, IHB, and MSU by double crossover with the same plasmid, pHB1281. As shown in Fig. 3, HAU hetC::C.CE2 formed heterocysts, IHB hetC::C.CE2 formed dividing differentiating cells (DDC; around 75% of differentiating sites) and heterocysts, and MSU hetC::C.CE2 formed DDC only. The interruption of hetC could be complemented by hetC carried on a pDU1-based plasmid (see Fig. S4), indicating that the mutant phenotype was not due to a polar effect on the downstream gene (complementation also reported in reference 14). In addition, we also inactivated patU3, a pattern formation gene (35), in IHB hetC::C.CE2 to see the effect on DDC and heterocysts. An IHB patU3-null mutant formed multiple contiguous heterocysts (35), whereas the IHB hetC::C.CE2 patU3::C.K4 double mutant formed no heterocysts but DDC in longer chains (up to 5 cells at 48 h) compared to that in IHB hetC::C.CE2 (up to 2 cells at 48 h). This further supported the observation that the inactivation of hetC in certain genetic backgrounds indeed eliminates heterocyst formation, resulting in the DDC phenotype.
FIG 3.
Light (I) and phycobilisome fluorescence (II) micrographs of hetC mutants generated from substrains of Anabaena PCC 7120 and a hetC patU3 double mutant of substrain IHB. Micrographs were taken at indicated hours after N stepdown.
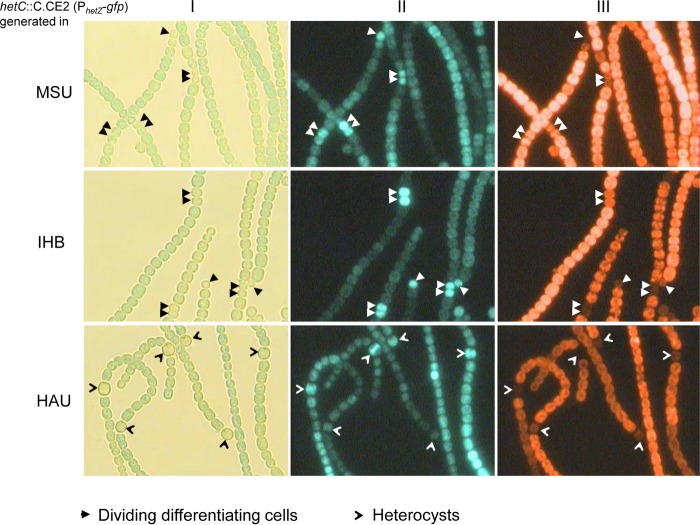
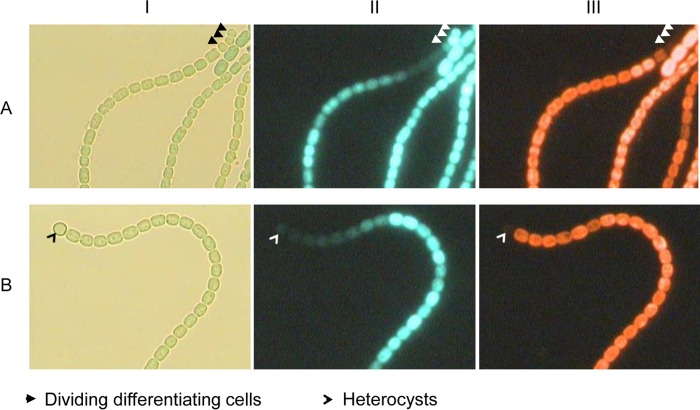
It has been proposed that at a late stage of heterocyst differentiation, HetC may play a role in the transport of patS–hetN-dependent inhibitors (18, 19). The transport of inhibitors from heterocysts to vegetative cells was probed with a HetR-CFP fusion expressed from a pDU1-based plasmid (19). We wondered if HetC had the same function in the three substrains we used. Because the promoter of hetZ is directly regulated by HetR and sensitive to the PatS-derived inhibitor (36), we introduced a similar plasmid carrying PhetZ-gfp (pHB1465, as described in reference 35) into the hetC::C.CE2 mutants (Fig. 4). In the hetC mutants of substrains IHB and MSU, PhetZ-gfp was upregulated in all or most differentiating cells at 48 h after N stepdown relative to the expression of PhetZ-gfp in vegetative cells on the same filaments. In the HAU hetC mutant with PhetZ-gfp, however, the difference between the GFP fluorescence of heterocysts and that of vegetative cells was much less discernible. We also generated an IHB hetC patA double mutant carrying PhetZ-gfp. The IHB hetC patA double mutant formed terminal DDC or heterocysts due to a lack of patA function (37). At 48 h after N stepdown, a gradient of GFP fluorescence was formed in cells adjacent to the terminal DDC or heterocysts in the double mutant with PhetZ-gfp (Fig. 5). Apparently, at least in certain substrains of Anabaena PCC 7120, the inhibition of PhetZ-gfp in vegetative cells by differentiating cells is not dependent on the function of hetC.
FIG 4.
Light (I), GFP fluorescence (II), and phycobilisome fluorescence (III) micrographs of hetC::C.CE2 (PhetZ-gfp) mutants generated from Anabaena PCC 7120 substrains.
FIG 5.
Light (I), GFP fluorescence (II), and phycobilisome fluorescence (III) micrographs of hetC::C.CE2-patA::C.K2d (PhetZ-gfp) mutants generated from Anabaena PCC 7120 substrain IHB. (A) GFP fluorescence gradient in cells adjacent to terminal DDC. (B) GFP fluorescence gradient in cells adjacent to a terminal heterocyst.
DISCUSSION
In the past decade, with the evolution of sequencing techniques, genome variations between substrains have been investigated in more and more bacterial strains. In this study, we analyzed the differences between 3 substrains of Anabaena PCC 7120, a research model for cell differentiation in filamentous cyanobacteria, and compared the phenotypes of hetC-null mutants generated from these substrains with the same plasmid.
Employing second-generation sequencing technology, we screened genome variations in 2 substrains, HAU and IHB, and identified 182 and 170 SNPs, respectively, in comparison to the reference genome sequence published in 2001 (33). Because randomly selected SNPs, 21 to 33 for each of the three substrains, were 100% confirmed with PCR and traditional sequencing, the long lists of SNPs and small indels in Table 1 and Table S1 in the supplemental material are considered to be highly reliable. Compared to Caulobacter crescentus (27) and Myxococcus xanthus (28), the number of genome variations between substrains of Anabaena PCC 7120 is much higher. This could be explained by the combined effect of multicopy genome and long multicellular filaments of Anabaena: upon purifying a strain by streaking on plates, spontaneous mutations have much higher chances to be retained in such cyanobacterial species than those in unicellular single-copy-genome bacteria. The cyanobacterium Synechocystis PCC 6803, unicellular but with a multicopy genome, also accumulated a higher number of genome variations (31) than other bacteria in the laboratory.
Of the substrains of Anabaena PCC 7120 used in the literature after 1984, most were derived from those of the Haselkorn laboratory and the Wolk laboratory, and a small number were derived from the PCC. In this study, we compared one obtained from the PCC in 1984 and one obtained from the Wolk laboratory in 1992 with the reference genome generated with the Wolk laboratory substrain (MSU) in 2001. According to the history of the three substrains, it is not surprising to have specific genome variants for each of them; however, it is surprising that substrains HAU and IHB shared 89 SNPs and 25 indels relative to the reference genome sequence, because IHB is not in the same lineage as HAU/MSU (see Fig. S1). We hypothesize that this could be due to the different growth media for Anabaena PCC 7120: the medium used in the Wolk laboratory was AA/8 (38), while that in the Zhang laboratory (HAU) (39) and in the Xu laboratory (IHB) (40) was BG11. The 89 SNPs and 25 indels might have been present in the culture of AA/8 at very low percentages but became absolutely dominant after serial transfer in BG11 medium in the Zhang laboratory. Further traced back to the 1970s, these SNPs should have been present in the source strain in the Haselkorn laboratory and that of the PCC; therefore, they were also found in the Xu laboratory strain (IHB) grown in BG11. With a multicopy genome, filamentous cyanobacteria may keep some SNPs and indels at very low percentages in the population under one condition and boost up these SNPs and indels under an alternative condition. Other culture conditions that may differ between laboratories, such as light intensity and shaking, may also contribute to the microevolution observed among substrains.
In addition to the 89 SNPs and 25 indels shared by substrains HAU and IHB (the corresponding reference bases were therefore specifically found in MSU), 93 SNPs, 34 indels, and 4 fragment deletions were specifically found in HAU, and 81 SNPs, 22 indels, 3 fragment deletions, and 5 extra copies of different ISs were specifically found in IHB. In comparison to HAU and IHB genomes, three extra copies of ISs were found in the reference genome, confirmed by third-generation sequencing and PCR with MSU genomic DNA. As with the presence of many identical SNPs and indels in HAU and IHB, the presence of IS(all7245, asl7246) at the alpha plasmid position 165178 (according to the reference position) in HAU and IHB but not in MSU could also be attributed to the selection pressure in different growth media. The insertion sequence that harbors transposase-encoding genes all7245 and asl7246 could not be directly defined computationally, according to Wolk et al. (41). In our study, however, we found the presence of extra copies of the IS at alpha plasmid position 45585 in IHB and at alpha plasmid position 165178 in HAU and IHB; therefore, we were able to extract the whole sequence of IS(all7245, asl7246) (see Fig. S5). Unlike typical ISs, IS(all7245, asl7246) has no inverted repeats at the two ends.
In the substrain IHB, some specific heterogeneous/heterozygous SNPs were identified. Such SNPs represent ongoing microevolution in the population and could be completely segregated in cell lineages derived from sonicated filaments. The presence of heterogeneous/heterozygous SNPs in substrain IHB was probably due to many years of serial transfer in culture collections PCC and FACHB before being stored in a −70°C freezer in the Xu laboratory. The slow growth mode in culture collections might have greatly delayed the process of genetic drift or selection and therefore retained the heterogeneous/heterozygous SNPs in the population. Such SNPs were not found in HAU (the substrain MSU was not analyzed with second-generation sequencing and therefore has no information regarding heterogeneous/heterozygous SNPs).
It has been shown in Myxococcus that changes in genetic background may alter the phenotype of the same mutation (28). For most genes involved in heterocyst differentiation and patterning, such as ntcA (42, 43), hetR (36, 44, 45), hetP (46, 47), hetZ (hetZ mutants generated with substrains IHB and MSU in reference 35), patS (35, 48, 49), and patA (37, 50), insertion/deletion in different substrains of Anabaena PCC 7120 resulted in similar phenotypes; however, hetC is an exception (14, 16–19) (Fig. 3 and 4). hetC (insertion/deletion) mutants generated in different labs with different plasmids resulted in different phenotypes, producing dividing differentiating cells (14, 16, 17) or differentiation-delayed nonfunctional heterocysts (18, 19). In this study, we used the same plasmid to generate hetC insertion mutants from 3 substrains of Anabaena PCC 7120, and the mutants produced either dividing differentiating cells, heterocysts, or both. HetC had been proposed to export PatS- or HetN-derived peptide inhibitors from heterocysts to vegetative cells (18, 19); however, using PhetZ-gfp as the indicator, we also showed the difference in the inhibition of GFP fluorescence in vegetative cells by heterocysts/dividing differentiating cells (Fig. 4). In a hetC-patA double mutant generated with substrain IHB, a gradient of PhetZ-gfp expression was clearly shown in vegetative cells in the neighbor of terminal heterocysts/dividing differentiating cells (Fig. 5). At the least, HetC is not involved in the transport of the heterocyst differentiation inhibitor in this substrain. For these reasons, the different phenotypes of hetC mutants reported could not be explained by different ways of generating the mutants but could be associated with the genome variations in substrains. In addition, the different phenotypes of hetC mutants implied that the role of hetC in heterocyst differentiation can be compromised by other mutations. Consistently, orthologs of hetC are found only in approximately half of heterocyst-forming species (Table 2). As the different substrains have been separated for a relatively short period, the different hetC phenotypes depending on the genomic backgrounds represent an ongoing rapid microevolution event. In the future, it might be possible to find out the causal mutation by screening each of the SNPs, indels, deletions, or transpositions.
TABLE 2.
Predicted proteins similar to HetC and two paralogues in heterocyst-forming cyanobacteria
| Strain | Protein (no. of aaa; % identity) |
||
|---|---|---|---|
| Alr2817 (1,044 [HetC]) | Alr1927 (1,011) | Alr1614 (893) | |
| Nostoc azolla 0708 | Aazo_0728 (1,013; 72) | Aazo_1455 (892; 78) | |
| Anabaena cylindrica PCC 7122 | Anacy_0157 (1,013; 72) | ||
| Anabaena sp. strain 90 | ANA_C10349 (902; 68) | ANA_C11095 (867; 77) | |
| Anabaena sp. strain wa102 | AA650_20450 (906; 77) | ||
| Anabaena variabilis ATCC 29413 | Ava_1097 (1,042; 97) | Ava_4383 (1,011; 97) | Ava_4225 (865; 98) |
| Calothrix sp. strain 336/3 | IJ00_13565 (1,013; 53) | IJ00_03740 (1,010; 74) | IJ00_14935 (889; 79) |
| Calothrix sp. strain PCC 6303 | Cal6303_5242 (933; 73) | ||
| Calothrix sp. strain PCC 7507 | Cal7507_1646 (1,058; 65) | Cal7507_1651 (1,016; 78) | Cal7507_2015 (885; 84) |
| Nostoc punctiforme PCC 73102 | Npun_F3519 (1,034; 75) | Npun_R0865 (871; 83) | |
| Nostoc sp. strain PCC 7107 | Nos7107_2162 (1,019; 59) | Nos7107_5410 (901; 79) | |
| Nostoc sp. strain PCC 7524 | Nos7524_3642 (1,054; 70) | Nos7524_3636 (1,041; 80) | Nos7524_3051 (893; 88) |
| Rivularia sp. strain PCC 7116 | Riv7116_0387 (1,032; 45) | Riv7116_0383 (1,029; 62) | Riv7116_5020 (857; 65) |
aa, amino acids.
Our studies with Anabaena PCC 7120 showed that a filamentous cyanobacterial strain may accumulate a high number of genome variations in substrains relative to that in research model strains of unicellular cyanobacteria or other bacteria. Accordingly, the functions of some heterocyst-related genes, especially those not conserved in different heterocyst-forming species, may change with the genetic background. A conclusion regarding the function of such a gene should be considered more cautiously than before.
MATERIALS AND METHODS
General.
The history of the Anabaena PCC 7120 substrains used in this study is summarized in Fig. S1 in the supplemental material. Anabaena sp. PCC 7120 substrain IHB was obtained from the freshwater algal culture collection at the Institute of Hydrobiology, Chinese Academy of Sciences, where it has been maintained since a strain exchange with the Pasteur Culture Collection in 1984. The substrain MSU was obtained in 2010 from Ruanbao Zhou, who had brought it from the Wolk lab at Michigan State University to South Dakota State University in 2008. The substrain HAU was obtained in 2008 from C.-C. Zhang's lab at Huazhong Agricultural University, derived from the substrain of C.-C. Zhang's lab at the Aix-Marseille Université, and further traced back to a transfer from the Wolk lab in 1991. Substrains HAU and MSU were frozen at −70°C at the Xu lab until recovery for use. The substrains and derivatives were cultured in BG11 (51) on a rotary shaker under a light of 30 μE · m−2 · s−1, and antibiotics were added as required (40). To induce heterocyst differentiation, Anabaena filaments collected by centrifugation were washed 3 times with BG110 (without nitrate) and resuspended in the same medium. Microscopy was performed as previously described (17). Photomicrographs were taken through an Olympus BX41 microscope equipped with a JVC3 charge-coupled device color video camera. GFP fluorescence was visualized using a Chroma Sapphire GFP filter set. Phycobiliprotein fluorescence was observed using the red long-pass WG fluorescence cube (BP510-550, BA590) from Olympus.
Construction of hetC mutants and a plasmid for complementation.
hetC::C.CE2 mutants were constructed with Anabaena sp. PCC 7120 substrains using pHB1281 as described before (17). The plasmid was transferred into the PCC 7120 substrains by conjugation (52). Double-crossover mutants were generated by positive selection using sacB (38). A pDU1-based plasmid with the wild type hetC, called pHB3589, was introduced into the MSU hetC::C.CE2 mutant for complementation. A hetC::C.CE2-patU3::C.K4 double mutant was constructed by inactivating patU3 in IHB hetC::C.CE2 by using pHB1156 (35). A hetC::C.CE2-patA::C.K2d double mutant was constructed by inactivating patA in IHB hetC::C.CE2 by using pHB213 (35).
hetC with its promoter was generated by PCR using PhetC-1 (5′-ACCTATCTCCGCCCTATGG-3′)/hetCcom-2 (5′-TACCTTGATAGGGGCTTACAAATG-3′) as primers and PCC 7120 IHB DNA as the template, was cloned into pMD18-T (T-vector, TaKaRa), and was confirmed by sequencing. The resulting plasmid pHB3570 was cut with XbaI and blunted with T4 DNA polymerase. From pRL57 (53), the omega cassette (Smr, Spr) was excised with DraI and cloned into the blunted XbaI site of pHB3570, producing pHB3577. The omega-hetC structure was then excised with PstI-BamHI from pHB3577, blunted with T4 DNA polymerase, cloned into EcoRI-cut and T4 DNA polymerase-blunted pRL25C (54), producing pHB3589.
Second-generation sequencing and identification of SNPs/indels.
Anabaena PCC 7120 substrains IHB and HAU were sequenced using second-generation short-read sequencing technology on an Illumina platform. One paired-end library was constructed for each substrain and was sequenced on the Illumina HiSeq 2500 for 150 cycles on each end according to the manufacturer's protocol (Illumina). Around 17 million and 48 million paired-end reads were generated for PCC 7120 substrains IHB and HAU, respectively. The raw reads were filtered by removing reads containing an adaptor or were of low quality. The resulting high-quality reads were used for following analyses.
The genome sequence of strain PCC 7120 (33) downloaded from NCBI (accession no. GCA_000009705.1) was used as the reference for SNP identification. High-quality reads from each strain were mapped to the reference genome using BWA (55). The SAM files were converted to sorted BAM files using SAMtools version 1.3.1 (56). SNPs were called using both SAMtools and the Genome Analysis Toolkit (GATK) version 3.5 (57) and were filtered via variant filtration in GATK with the following parameters: QD, <10.0; FS, >10.0; MQ, <30.0; DP, <4; ReadPosRankSum, <−8.0. CLC Genomics Workbench version 10 was also used to call SNPs. The consensus sites between the results of three software programs were retrieved. The annotation for SNP sites was added using Annovar (58). Small insertions and deletions (indels) were identified using both GATK and CLC Genomics Workbench. The consensus indel sites identified by the two software programs were retrieved.
Third-generation sequencing and genome structure comparison.
Third-generation sequencing was performed on PacBio platforms (Pacific Biosciences, Menlo Park, CA). A 10-kb library was constructed for each substrain using the PacBio DNA template prep kit. For substrains HAU and IHB, each library was sequenced in one SMRT cell on PacBio RS II. The library for substrain MSU was sequenced in one SMRT cell on PacBio Sequel. The raw reads were filtered to retrieve high-quality subreads.
Substrain HAU and IHB genomes were assembled using HGAP3 (59), while the substrain MSU genome was assembled using Falcon, an assembler developed by PacBio. Each genome was aligned against the reference genome by BLAST, and unmapped regions were extracted and analyzed. The naming and classification of insertion sequences (ISs) in this study were according to Wolk et al. (41).
Accession number(s).
Genome sequences of substrains HAU and IHB were deposited in the NCBI SRA (Sequence Read Archive) under accession numbers SRR6049608 (IHB, Illumina), SRR6049609 (IHB, PacBio), SRR6049610 (HAU, Illumina), SRR6049611 (HAU, PacBio), and SRR6049612 (MSU, PacBio).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31270132 and 31770044) and the State Key Laboratory of Freshwater Ecology and Biotechnology at IHB, CAS (2016FBZ09).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00076-18.
REFERENCES
- 1.Zehr JP. 2011. Nitrogen fixation by marine cyanobacteria. Trends Microbiol 19:162–173. doi: 10.1016/j.tim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Issa AA, Abd-Alla MH, Ohyama T. 2014. Nitrogen fixing cyanobacteria: future prospect, p 23–48. In Ohyama T. (ed), Advances in biology and ecology of nitrogen fixation. InTechOpen, London, United Kingdom. doi: 10.5772/56990. [DOI] [Google Scholar]
- 3.Kumar K, Mella-Herrera RA, Golden JW. 2010. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol 2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrero A, Stavans J, Flores E. 2016. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol Rev 40:831–854. doi: 10.1093/femsre/fuw029. [DOI] [PubMed] [Google Scholar]
- 5.Wolk CP, Quine MP. 1975. Formation of one-dimensional patterns by stochastic processes and by filamentous blue-green algae. Dev Biol 46:370–382. doi: 10.1016/0012-1606(75)90113-X. [DOI] [PubMed] [Google Scholar]
- 6.Meinhardt H, Gierer A. 2000. Pattern formation by local self-activation and lateral inhibition. Bioessays 22:753–760. doi:. [DOI] [PubMed] [Google Scholar]
- 7.Kondo S, Miura T. 2010. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 8.Adolph KW, Haselkorn R. 1971. Isolation and characterization of a virus infecting the blue-green alga Nostoc muscorum. Virology 46:200–208. doi: 10.1016/0042-6822(71)90023-7. [DOI] [PubMed] [Google Scholar]
- 9.Wolk CP, Vonshak A, Kehoe P, Elhai J. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci U S A 81:1561–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Elhai J, Wolk CP. 2008. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p 383–422. In Herrero T, Flores E (ed), Cyanobacteria: molecular biology, genomics and evolution. Horizon Scientific Press, Norwich, UK. [Google Scholar]
- 11.Muro-Pastor AM, Hess WR. 2012. Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol 20:548–557. doi: 10.1016/j.tim.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Ehira S, Ohmori M. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol 59:1692–1703. doi: 10.1111/j.1365-2958.2006.05049.x. [DOI] [PubMed] [Google Scholar]
- 13.Muro-Pastor AM, Olmedo-Verd E, Flores E. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol Lett 256:171–177. doi: 10.1111/j.1574-6968.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 14.Khudyakov I, Wolk CP. 1997. hetC, a gene coding for a protein similar to bacterial ABC protein exporters, is involved in early regulation of heterocyst differentiation in Anabaena sp. strain PCC 7120. J Bacteriol 179:6971–6978. doi: 10.1128/jb.179.22.6971-6978.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baier K, Lehmann H, Stephan DP, Lockau W. 2004. NblA is essential for phycobilisome degradation in Anabaena sp. strain PCC 7120 but not for development of functional heterocysts. Microbiology 150(Pt 8):2739–2749. doi: 10.1099/mic.0.27153-0. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Wolk CP. 2001. Role for hetC in the transition to a nondividing state during heterocyst differentiation in Anabaena sp. J Bacteriol 183:393–396. doi: 10.1128/JB.183.1.393-396.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Xu X. 2005. Regulation by hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. J Bacteriol 187:8489–8493. doi: 10.1128/JB.187.24.8489-8493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corrales-Guerrero L, Flores E, Herrero A. 2014. Relationships between the ABC-exporter HetC and peptides that regulate the spatiotemporal pattern of heterocyst distribution in Anabaena. PLoS One 9:e104571. doi: 10.1371/journal.pone.0104571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Videau P, Rivers OS, Higa KC, Callahan SM. 2015. ABC transporter required for intercellular transfer of developmental signals in a heterocystous cyanobacterium. J Bacteriol 197:2685–2793. doi: 10.1128/JB.00304-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenaillon O, Barrick JE, Ribeck N, Deatherage DE, Blanchard JL, Dasgupta A, Wu GC, Wielgoss S, Cruveiller S, Médigue C, Schneider D, Lenski RE. 2016. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good BH, McDonald MJ, Barrick JE, Lenski RE, Desai MM. 2017. The dynamics of molecular evolution over 60,000 generations. Nature 551:45–50. doi: 10.1038/nature24287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeigler DR, Nicholson WL. 2017. Experimental evolution of Bacillus subtilis. Environ Microbiol 19:3415–3422. doi: 10.1111/1462-2920.13831. [DOI] [PubMed] [Google Scholar]
- 23.Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. 2015. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol 10:599–611. doi: 10.2217/fmb.15.3. [DOI] [PubMed] [Google Scholar]
- 24.Pinto M, Borges V, Antelo M, Pinheiro M, Nunes A, Azevedo J, Borrego MJ, Mendonça J, Carpinteiro D, Vieira L, Gomes JP. 2016. Genome-scale analysis of the non-cultivable Treponema pallidum reveals extensive within-patient genetic variation. Nat Microbiol 2:16190. doi: 10.1038/nmicrobiol.2016.190. [DOI] [PubMed] [Google Scholar]
- 25.Guerrini V, Subbian S, Santucci P, Canaan S, Gennaro ML, Pozzi G. 2016. Experimental evolution of Mycobacterium tuberculosis in human macrophages results in low-frequency mutations not associated with selective advantage. PLoS One 11:e0167989. doi: 10.1371/journal.pone.0167989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harkins CP, Pettigrew KA, Oravcová K, Gardner J, Hearn RMR, Rice D, Mather AE, Parkhill J, Brown SJ, Proby CM, Holden MTG. 2018. The microevolution and epidemiology of Staphylococcus aureus colonization during atopic eczema disease flare. J Investig Dermatol 138:336–343. doi: 10.1016/j.jid.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marks ME, Castro-Rojas CM, Teiling C, Du L, Kapatral V, Walunas TL, Crosson S. 2010. The genetic basis of laboratory adaptation in Caulobacter crescentus. J Bacteriol 192:3678–3788. doi: 10.1128/JB.00255-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradley MD, Neu D, Bahar F, Welch RD. 2016. Inter-laboratory evolution of a model organism and its epistatic effects on mutagenesis screens. Sci Rep 6:38001. doi: 10.1038/srep38001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tajima N, Sato S, Maruyama F, Kaneko T, Sasaki NV, Kurokawa K, Ohta H, Kanesaki Y, Yoshikawa H, Tabata S, Ikeuchi M, Sato N. 2011. Genomic structure of the cyanobacterium Synechocystis sp. PCC 6803 strain GT-S. DNA Res 18:393–399. doi: 10.1093/dnares/dsr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanesaki Y, Shiwa Y, Tajima N, Suzuki M, Watanabe S, Sato N, Ikeuchi M, Yoshikawa H. 2012. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res 19:67–79. doi: 10.1093/dnares/dsr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trautmann D, Voss B, Wilde A, Al-Babili S, Hess WR. 2012. Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803.DNA Res 19:435–448. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol Lett 323:124–131. doi: 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8:20–213; 227–253. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 34.Masukawa H, Sakurai H, Hausinger RP, Inoue K. 2017. Increased heterocyst frequency by patN disruption in Anabaena leads to enhanced photobiological hydrogen production at high light intensity and high cell density. Appl Microbiol Biotechnol 101:2177–2188. doi: 10.1007/s00253-016-8078-3. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Du Y, Khudyakov I, Fan Q, Gao H, Ning D, Wolk CP, Xu X. 2007. A gene cluster that regulates both heterocyst differentiation and pattern formation in Anabaena sp. strain PCC 7120. Mol Microbiol 66:1429–1443. doi: 10.1111/j.1365-2958.2007.05997.x. [DOI] [PubMed] [Google Scholar]
- 36.Du Y, Cai Y, Hou S, Xu X. 2012. Identification of the HetR-recognition sequence upstream of hetZ in Anabaena sp. strain PCC 7120. J Bacteriol 194:2297–2306. doi: 10.1128/JB.00119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J, Scappino L, Haselkorn R. 1992. The patA gene product, which contains a region similar to CheY of Escherichia coli, controls heterocyst pattern formation in the cyanobacterium Anabaena 7120. Proc Natl Acad Sci U S A 89:5655–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai YP, Wolk CP. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol 172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang J, Shi L, Tan H, Janicki A, Zhang CC. 2009. Mutual regulation of ntcA and hetR during heterocyst differentiation requires two similar PP2C-type protein phosphatases, PrpJ1 and PrpJ2, in Anabaena sp. strain PCC 7120. J Bacteriol 191:6059–6066. doi: 10.1128/JB.01271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ning D, Xu X. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447–453. doi: 10.1099/mic.0.26747-0. [DOI] [PubMed] [Google Scholar]
- 41.Wolk CP, Lechno-Yossef S, Jäger KM. 2010. The insertion sequences of Anabaena sp. strain PCC 7120 and their effects on its open reading frames. J Bacteriol 192:5289–5303. doi: 10.1128/JB.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei TF, Ramasubramanian TS, Golden JW. 1994. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol 176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frías JE, Flores E, Herrero A. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol 14:823–832. [DOI] [PubMed] [Google Scholar]
- 44.Buikema WJ, Haselkorn R. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev 5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 45.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol 9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 46.Fernández-Piñas F, Leganés F, Wolk CP. 1994. A third genetic locus required for the formation of heterocysts in Anabaena sp. strain PCC 7120. J Bacteriol 176:5277–5283. doi: 10.1128/jb.176.17.5277-5283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higa KC, Callahan SM. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol Microbiol 77:562–574. doi: 10.1111/j.1365-2958.2010.07257.x. [DOI] [PubMed] [Google Scholar]
- 48.Yoon HS, Golden JW. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 49.Corrales-Guerrero L, Mariscal V, Flores E, Herrero A. 2013. Functional dissection and evidence for intercellular transfer of the heterocyst-differentiation PatS morphogen. Mol Microbiol 88:1093–1105. doi: 10.1111/mmi.12244. [DOI] [PubMed] [Google Scholar]
- 50.Hou S, Zhou F, Peng S, Gao H, Xu X. 2015. The HetR-binding site that activates expression of patA in vegetative cells is required for normal heterocyst patterning in Anabaena sp. PCC 7120. Sci Bull (Beijing) 60:192–201. doi: 10.1007/s11434-014-0724-5. [DOI] [Google Scholar]
- 51.Castenholz RW. 2001. Oxygenic photosynthetic bacteria, p 474–487. In Boone DR, Castenholz RW, Garrity GM (ed), Bergey's manual of systematic bacteriology. Volume 1: the Archaea and the deeply branching and phototrophic Bacteria. Springer-Verlag, New York, NY. [Google Scholar]
- 52.Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 53.Elhai J, Wolk CP. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 54.Wolk CP, Cai Y, Cardemil L, Flores E, Hohn B, Murry M, Schmetterer G, Schrautemeier B, Wilson R. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J Bacteriol 170:1239–1244. doi: 10.1128/jb.170.3.1239-1244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.