Abstract
The aim of the present study was to examine the expression and role of vascular endothelial growth factor A (VEGFA) and microRNA (miRNA or miR)-381 in tumor thrombi from patients with hepatocellular carcinoma and portal vein tumor thrombus (PVTT). Tumor thrombi and adjacent paired tissues were collected from 39 patients with hepatocellular carcinoma with PVTT. VEGFA expression levels were assessed using reverse transcription-quantitative polymerase chain reaction and western blotting. miRNAs that may regulate VEGFA expression were predicted using bioinformatics analysis and confirmed via a dual luciferase reporter assay. The effects of VEGFA and its upstream miRNA on proliferation of the proliferation of EAhy926 human venous endothelial cells were analyzed using an MTT assay. Compared with the paired adjacent tissues, VEGFA was significantly upregulated at both the mRNA and protein level in tumor thrombi (P<0.05). VEGFA was predicted to be a target of miR-381 and this was confirmed experimentally. miR-381 expression was significantly downregulated in tumor thrombi from patients with PVTT compared with paired adjacent tissues (P<0.05). In addition, transfection with antagomirs against miR-381 or short interfering RNA against VEGFA significantly inhibited EAhy926 cell proliferation (P<0.05). In conclusion, the results of the present study indicate that VEGFA is upregulated in tumor thrombi whereas miR-381 is downregulated. VEGFA is regulated by miR-381 and both may be associated with the development of PVTT.
Keywords: microRNA-381, vascular endothelial growth factor, hepatocellular carcinoma, tumor thrombus
Introduction
Primary hepatocellular carcinoma (HCC) is one of the most common malignant tumors of the digestive system, with high intrahepatic recurrence and metastasis rates leading to poor overall prognosis (1–3). Trousseau (4) reported that patients with cancer suffered complications due to venous thrombosis; the comorbidity of venous thrombosis and cancer was thus named Trousseau syndrome. Venous thrombosis is a common complication that occurs during the development of malignant tumors and is the second leading cause of death in patients with Trosseau syndrome after the cancer itself (5). A previous study suggested that malignant tumor is a high-risk factor for venous thrombosis, with ~12.3% of patients with cancer developing thrombi within 6 months of diagnosis (6). However, the mechanisms underlying the development of tumor thrombosis secondary to malignant tumors is yet to be elucidated.
The development of HCC is dependent on angiogenesis (7). Vascular endothelial growth factor A (VEGFA) serves an important role in the normal physiology and angiogenesis of various abnormal pathologies (8). The expression of VEGF in HCC cells is high due to local hypoxia stimulation, particularly near tumor necrotic areas (9,10). Inhibiting the expression of VEGF or blocking the corresponding signal transduction pathway and other pathways has an anti-angiogenic effect (11). Humanized monoclonal antibodies that bind to VEGFA and block its activity have been approved for the treatment of neoplastic diseases (12).
Several mRNA and microRNA (miR or miRNA) molecules are associated with Trousseau syndrome (13), including integrin β3 and the polyphosphate-factor XII signaling pathway (14). The occurrence of tumor thrombi is associated with neovascularization (15). VEGF, which is also secreted by tumor cells, and its signal transduction pathway serve important roles in the development of tumor thrombosis (16–18). In tumor angiogenesis, there are interactions between the coagulation process, inflammatory cytokines and malignant tumor growth and metastasis (19). The high permeability of tumor neovascular tissues induces plasma leakage resulting in increased specific volume, increased blood viscosity and blood stasis, all risk factors for thrombosis (16).
In the present study, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting were used to analyze the expression of VEGFA in the tumor thrombi of patients with HCC with portal vein tumor thrombus (PVTT). miRNAs that regulate the expression of VEGFA were predicted by bioinformatics methods and validated using a dual luciferase reporter assay; it was demonstrated that VEGFA is targeted by miR-381. The association between miR-381 and VEGFA was investigated and their roles in venous endothelial cell proliferation were examined using an MTT assay. The results of the present study may provide a novel insight into the regulatory mechanisms of PVTT development in HCC.
Materials and methods
Patients
A total of 39 patients with HCC and PVTT were enrolled in the present study. All patients were diagnosed and underwent tumor thrombus resection between August 2013 and June 2016 at the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China). PVTT samples and paired adjacent tissues were collected from all patients. The cohort comprised 18 male and 21 female patients with a median age of 46.6 years (range, 26–65 years). All patients were diagnosed by a clinician and diagnoses were confirmed by histopathology. Prior to surgical resection, patients did not receive hormone, herbal, radiotherapy or chemotherapy treatment. PVTT was confirmed by Doppler ultrasound or based on symptoms and patient history. Written informed consent was obtained from every patient and the study was approved by the Ethics Review Board of Guangxi Medical University.
RNA extraction and RT-qPCR
Total RNAs were isolated from thrombi and adjacent tissues using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Following quantification using a spectrophotometer at 490 nm, RNA (1 µg) was reverse transcribed into cDNA using a TIANScript II cDNA 1st strand cDNA Synthesis kit (Tiangen Biotech Co., Ltd., Beijing, China). miRNA was reverse transcribed from total RNA using an miRcute miRNA cDNA Synthesis kit (Tiangen Biotech Co., Ltd.). qPCR was performed using the SuperReal SYBR Green PreMix (Tiangen Biotech Co., Ltd.) for mRNA, or the miRcute miRNA qPCR Detection kit (Tiangen Biotech Co., Ltd.) for miRNA using a Bio-Rad IQ5 thermocycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). For mRNA, the reaction mixture was incubated at 94°C for 2 min followed by 35 cycles at 94°C for 30 sec, 55°C for 30 sec and 71°C for 1 min. For miRNA, the reaction mixture was incubated at 95°C for 5 min followed by 40 cycles at 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec. Primers used were as follows: VEGFA. forward 5′-TTGCCTTGCTGCTCTACCTC-3′ and reverse 5′-AAATGCTTTCTCCGCTCTGA-3′; β-actin, forward 5′-TGACGTGGACATCCGCAAAG-3′ and reverse 5′-CTGGAAGGTGGACAGCGAGG-3′; miR-381, forward 5′-ACACTCCAGCTGGGTATACAAGGGCAAGCT-3′ and reverse 5′-TGGTGTCGTGGAGTCG-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′. The relative expression levels were evaluated using the 2−ΔΔCq method (20). β-actin and small nuclear U6 were used as internal controls for mRNA and miRNA, respectively.
Western blotting
Proteins were extracted using radioimmunoprecipitation assay buffer and protease inhibitor phenylmethane sulfonyl fluoride. The protein concentration was determined by using a bicinchoninic acid assay kit [RTP7102; Real-Times (Beijing) Biotechnology Co., Ltd., Beijing, China]. A total of 20 µg/lane protein was separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. After blocking with 5% skimmed milk at room temperature for 1 h, the membranes were probed with the rabbit anti-VEGFA (1:1,000; ab46154) or rabbit anti-β-actin (1:5,000; ab129348; both Abcam, Cambridge, MA, USA) at 4°C overnight. After washing five times with Tris-buffered saline and Tween-20 (5 min each time), the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:3,000; ab6721; Abcam) at room temperature for 1 h. Signal detection was performed using an enhanced chemiluminescence reaction kit (ab65623; Abcam) on a Gel Doc XR+ system (Bio-Rad Laboratories, Inc.). The acquired images were analyzed using Image lab 3.0 (Bio-Rad Laboratories, Inc.) and the relative protein expression was expressed as the densitometric value ratio of VEGFA to the β-actin band.
Bioinformatics analysis
To further identify the miRNAs that may regulate the expression of VEGFA the following bioinformatics databases were used: miRanda (34.236.212.39/microrna/home.do), TargetScan (targetscan.org), PiTa (genie.weizmann.ac.il/pubs/mir07/mir07_data.html), RNAhybrid (bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and PICTA (pictar.mdc-berlin.de/) were used.
Dual-luciferase reporter gene assay
Based on the results of bioinformatics prediction, the conservative miR-381 binding sequence on 3′-untranslated region (UTR) of wild type (WT) or the mutant VEGFA mRNA was cloned. The primers to clone the WT miR-381 binding sequence were designed by CE Design V1.04 (Vazyme, Nanjing, China) ad follows: WT, forward 5′-tga tga aag ctg cgc act agt AAAGAGTAGGGTTTTTTTTCAGTATTCTT-3′ and reverse 5′-aaaagatcctttattaagcttTGCTGGGGAGCCAGGGGA-3′. Primers for the mutated sequence were constructed using a Mut Express II Fast Mutagenesis kit V2 C214-01/02 (Vazyme). Mutant, forward 5′-GTACCGGTTTaacaataATAAAATTCATGTTTCCAATCTCTCTCT-3′ and reverse 5′-tattgttAAACCGGTACAAATAAGAGAGCAAG-3′. The mutant binding sequence is underlined below:
2821 tgtatcttttgctctctcttgctctcttatttgtaccggtttTTGTATataaaattcatg
2881 tttccaatctctctctccctgatcggtgacagtcactagcttatcttgaacagatattta
Luciferase reporter plasmids were generated by inserting WT or mutant sequences of VEGFA into the multiple cloning site (Spe-1 and HindIII) downstream of the luciferase reporter gene in the pMIR-REPORT™ Luciferase (Thermo Fisher Scientific, Inc.). The 293T cells (American Type Culture Collection, Manassas, VA, USA) were transfected with 0.8 µg constructed luciferase reporters and 100 nM antagomir (agomiR)-381 (Sangon Biotech, Shanghai, China) or negative control RNA (NC) using ExFect Transfection reagent (cat. no. T101-02; Vazyme, Piscataway, NJ, USA). A total of 10 ng pMIR-REPORT™ β-gal Control Plasmid was transfected as an internal control for transfection efficiency. Luminescence was measured at 24 h after transfection using a Dual-Luciferase® Reporter Assay System (Promega Corp., Madison, WI, USA) according to the manufacturer's protocol Measurements of luminescence were performed using a Glomax 20/20 (Promega Corp.).
Cell culture and transfection
Human venous endothelial cells EAhy926 (American Type Culture Collection) were cultured in Dulbecco's Modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) with 10% heat-inactivated fetal bovine serum (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 1% penicillin and streptomycin at 37°C in a humidified atmosphere containing 5% CO2. A total of 3×105 cells were seeded in a 24-well plate and transfected with 25 pmol of agomiR-381 (Sangon Biotech) using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. As a control, EAhy926 cells were transfected with NC RNA without specifically targeting any human gene products. To silence the expression of VEGFA, cells were transfected with 50 nM VEGFA siRNA (siVEGFA) or NC (Sangon Biotech, Shanghai, China). Cells were collected at 48 h post-transfection for further experiments.
MTT assay
Cells were seeded into 96-well plates at a density of 2,000 cell/well. Cells were cultured at in a humidified atmosphere containing 5% CO2 at 37°C. At 24, 48, and 72 h, 20 µl of MTT reagent (5 mg/ml; JRDUN biotechnology, Shanghai, China) were added and cells were incubated at 37°C for 4 h until purple precipitate was visible. On the last day the culture supernatant was removed and 150 µl dimethyl sulfoxide per well was added. The 96-well plate was oscillated for 10 min until purple precipitate was dissolved. The absorbance was measured at 490 nm on a microplate reader. A cell growth curve was generated based on these absorbance values.
Statistical analysis
Data analysis was performed using SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA) and expressed as the mean standard deviation. Differences between groups were evaluated for significance using one-way analysis of variance with Student Newman Keuls, Tamhane's T2 or Dunnett's T3 post hoc tests. P<0.05 was considered to indicate a statistically significant difference.
Results
VEGFA is upregulated in tumor thrombi
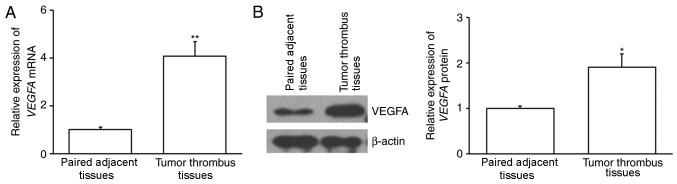
The expression of VEGFA mRNA was assessed using RT-qPCR. The results demonstrated that VEGFA mRNA expression was significantly increased in tumor thrombus samples compared with paired adjacent tissues (P<0.01; Fig. 1A). The expression of VEGFA protein was also assessed using western blotting and the results revealed that it was significantly upregulated in tumor thrombi compared with paired adjacent tissues (P<0.05; Fig. 1B). This suggests that VEGFA may be associated with the formation of tumor thrombi.
Figure 1.
VEGFA expression in patients with PVTT. Tumor thrombi and their paired adjacent tissues were collected from patients with HCC and PVTT. The expression of VEGFA (A) mRNA and (B) protein was analyzed using quantitative-polymerase chain reaction and western blotting, respectively. *P<0.05 and **P<0.01 vs. paired adjacent tissues. VEGFA, vascular endothelial growth factor A; PVTT, portal vein tumor thrombus; HCC, hepatocellular carcinoma.
VEGFA is a direct target of miR-381
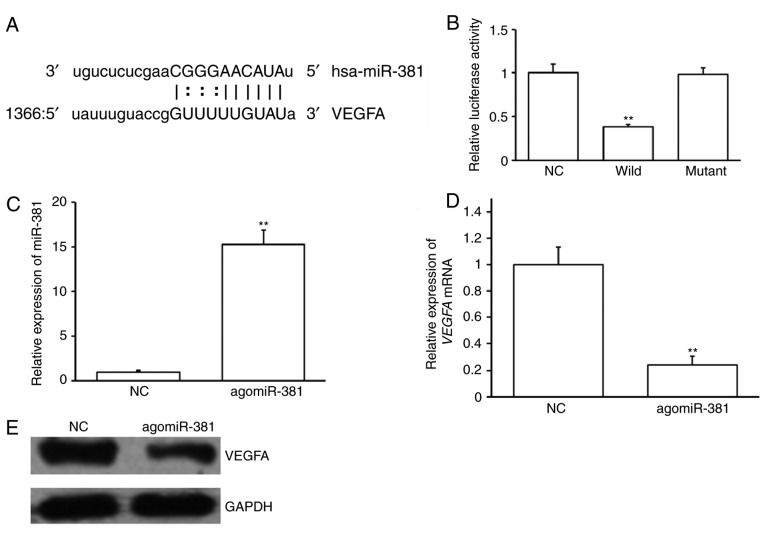
Potential upstream miRNA regulators of VEGFA were investigated. miRNAs that may bind to the 3′-UTR of VEGFA were predicted using bioinformatics. An miR-381 binding site was identified at the 3′-UTR of VEGFA mRNA (Fig. 2A). A dual luciferase reporter assay was performed to confirm this prediction. Cotransfection of agomiR-381 and pMIR-REPORT-VEGFA WT construct significantly decreased luciferase activity compared with NC and pMIR-REPORT-VEGFA WT construct (P<0.05; Fig. 2B). However, cotransfection with agomiR-381 and pMIR-REPORT-VEGFA mutant construct had no significant effect on luciferase activity (Fig. 2B). These results suggest that VEGFA is a possible target of miR-381, with miR-381 binding to its 3′-UTR sequence.
Figure 2.
VEGFA is a target of miR-381. (A) The results of bioinformatics software revealed the binding sequence of miR-381 on the 3′UTR of VEGFA mRNA. (B) Wild and mutant constructs were cotransfected with NC. Luciferase activities were assayed 24 h post transfection. (C) EAhy926 cells were transfected with agomiR-381 or NC. The expression of miR-381 was analyzed. (D) Following transfection, VEGFA mRNA expression was assessed. (E) VEGFA protein expression was analyzed by immunoblotting following agomiR-381 transfection. **P<0.01 vs. NC. VEGFA, vascular endothelial growth factor A; miR, microRNA; 3′UTR, 3′-untranslated region; Wild, wild type VEGFA 3′UTR luciferase reporter construct; mutant, mutant VEGFA 3′UTR luciferase reporter construct; NC, negative control; agomiR, antagomir.
To confirm the results of the dual luciferase reporter assay, EAhy926 cells were transfected with agomiR-381 or NC. AgomiR-381 transfection significantly increased miR-381 expression (P<0.01; Fig. 2C) and significantly decreased VEGFA mRNA expression in EAhy926 cells (P<0.01; Fig. 2D). AgomiR-381 transfection also markedly reduced VEGFA protein levels (Fig. 2E). These results suggest that the expression of VEGFA is regulated by miR-381.
miR-381 is downregulated in tumor thrombi
The expression of miR-381 was assessed using RT-qPCR. miR-30b expression was significantly downregulated in tumor thrombi compared with adjacent paired tissues (P<0.01; Fig. 3). This indicates that miR-381 may contribute to the development of PVTT.
Figure 3.
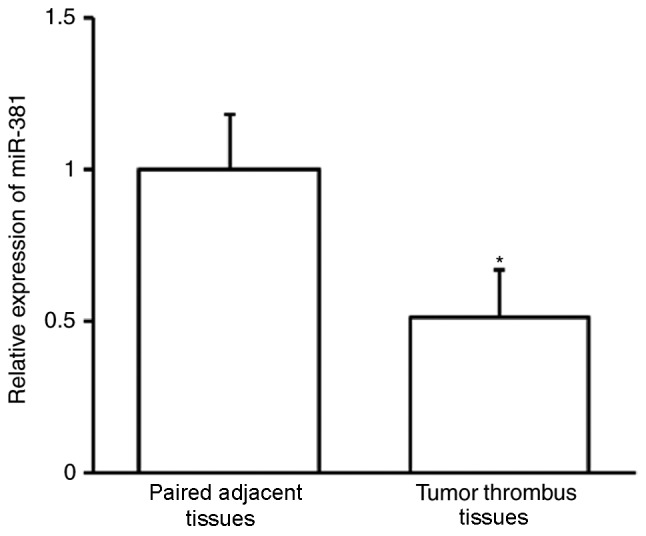
Reverse transcription-quantitative polymerase chain reaction was used to assess miR-381 expression in tumor thrombi and their paired adjacent tissues. *P<0.05 vs. paired adjacent tissues.
Overexpression of miR-381 inhibits EAhy926 cell proliferation
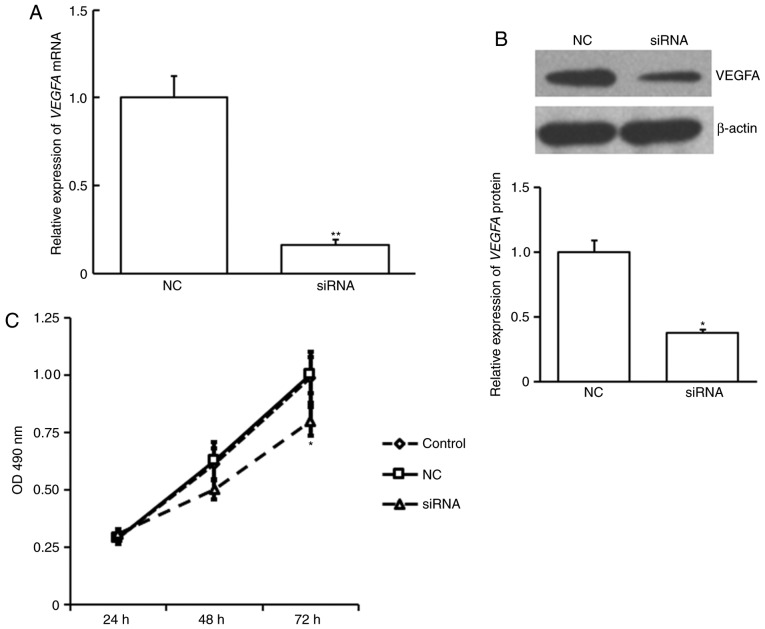
To investigate whether miR-381 regulates cell proliferation, EAhy926 cells were transiently transfected with agomiR-381 or NC. An MTT proliferation assay revealed that EAhy926 cells transfected with miR-381 exhibited significantly decreased cell proliferation compared with untreated cells or those transfected with NC (P<0.05; Fig. 4). This result suggests that miR-381 is able to inhibit the proliferation of venous endothelial cells.
Figure 4.
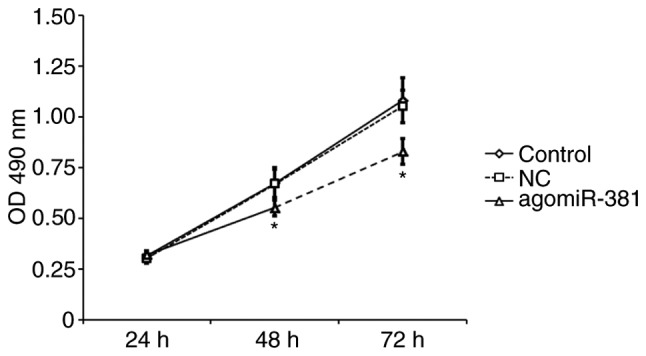
Overexpression of miR-381 inhibits EAhy926 cell proliferation. EAhy926 cells were transfected with agomiR-381 or NC and cell proliferation was analyzed by MTT assay. *P<0.05 vs. NC. miR, microRNA; agomiR, antagomir; NC, negative control; OD, optical density.
Effect of VEGFA on cell proliferation
Eahy926 cell proliferation was examined following siVEGFA transfection. siVEGFA was demonstrated to significantly decrease VEGFA expression at the mRNA (P<0.01; Fig. 5A) and protein level (P<0.05; Fig. 5B). siVEGFA significantly inhibited cell proliferation at 72 h post transfection compared with NC or untransfected control cells (P<0.05; Fig. 5C). This suggests that VEGFA also serves a role in venous endothelial cell proliferation.
Figure 5.
Suppression of VEGFA inhibits EAhy926 cell proliferation. EAhy926 cells were transfected with VEGFA siRNA or NC. (A) VEGFA mRNA expression was analyzed using reverse transcription-quantitative polymerase chain reaction. (B) VEGFA protein expression was analyzed using western blotting. (C) Cell proliferation was analyzed using an MTT assay. *P<0.05, **P<0.01 by one-way ANOVA, compared with NC transfected cells. VEGFA, vascular endothelial growth factor A; si, small interfering; NC, negative control.
Discussion
In the present study, the expression of VEGFA and its upstream regulator, miR-381, was assessed in tumor thrombus and paired adjacent tissues from patients with HCC with PVTT. The association between miR-381 and VEGFA was also analyzed. The effects of miR-381 and VEGFA on venous endothelial cell proliferation were investigated and the mechanism of secondary development of PVTT in cases of HCC was discussed. To the best of our knowledge, the present study is the first to investigate the role of miR-381 and VEGF in tumor thrombosis in patients with HCC.
The 5-year survival rate of patients with HCC is <12% in the USA, ~32.5% overall and 59.1% in patients in the early stages in China (21,22). A number of studies have investigated the association between venous thromboembolism and malignant tumors; for example, it has been reported that almost 10% of patients with renal cell carcinoma develop venous thrombosis (23,24). In addition, 60% of patients with tumor thrombus have tumor metastasis (25). For patients with tumors and venous thrombosis, complete resection of the tumor tissues is currently the only effective treatment option (24). Venous thrombosis causes complications in patients with tumors; tumor cells are able to flow through the blood vessels to the portal vein and gradually grow to form a tumor thrombus, while the tumor cells can also spread along the vein in the liver (26,27).
Blocking cell signaling pathways or inhibiting tumor neovascularization to slow tumor growth is a major focus of tumor targeted therapy and the use of such neoadjuvant therapies have been described in several case studies (28–32). In humans, VEGFA is one of the most effective angiogenic factors as it is able to promote angiogenesis and increase blood supply (33). In the present study, it was demonstrated that VEGFA was significantly upregulated in tumor thrombi compared with normal tissues. Furthermore, silencing VEGFA inhibited EAhy926 cell proliferation. VEGF is also an important factor in HCC (34) and the data herein further suggest that VEGFA serves a crucial role in the development of PVTT.
miRNAs participate in the development of tumors by promoting the downregulation of mRNA expression and regulating protein-coding gene activity (35–37). In the present study, miR-381 was predicted to be an upstream regulator of VEGFA, which is consistent with a previous report in MG-63 cells (38). It has been reported that miR-381 is significantly downregulated in colon cancer tissues, and this induces the proliferation and invasion of colon cancer cells by increasing liver receptor homolog-1 upregulation (39). miRNA-381 in the p53/pituitary tumor-transforming 1 negative feedback loop inhibits the growth of pituitary tumors (40). miR-381 is also associated with multidrug resistance gene 1 and serves a role in multiple drug resistance (41). The combination of miR-381 and miR-424 inhibits the activity of cluster of differentiation 2 in renal cells by targeting WEE1 (42). Furthermore, it has also been demonstrated that miR-381 is associated with lung adenocarcinoma (43). Together, these studies demonstrate that miR-381 is a critical regulator in the development of various tumors. To further investigate the regulatory mechanism of miR-381 on VEGFA, miR-381 was overexpressed in EAhy926 cells via agomiR-381 transfection and cell proliferation was suppressed. Previous clinical studies and reports have indicated an association between tumor cell proliferation and PVTT (44–46). Additionally, miR-381 overexpression resulted in decreased VEGFA mRNA expression. The results of a dual-luciferase reporter assay demonstrated that miR-381 is able to directly target VEGFA. Collectively, the results of the present study indicate that miR-381 is able to directly regulate VEGFA expression.
In summary, VEGFA was upregulated in tumor thrombi from patients with HCC and PVTT, whereas miR-381 was downregulated. miR-381 is able to regulate VEGFA. Furthermore, miR-381 and VEGFA may regulate the proliferation of venous endothelial cells. Therefore, the findings of the present study suggest that miR-381 and VEGFA may be associated with the development of PVTT. However, a limitation of the present study is that the sample size was relatively small and primarily made up of individuals from the Chinese population. In order to further investigate the clinical applications of miR-381 and VEGF in tumor thrombosis, transgenic mice with miR-381 or VEGF knockout should be used in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JW and SZW performed the experiments, analyzed the data and drafted the manuscript. TRH conceived and designed the study and wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from every patient and the study was approved by the Ethics Review Board of Guangxi Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wedd JP, Nordstrom E, Nydam T, Durham J, Zimmerman M, Johnson T, Purcell Thomas W, Biggins SW. Hepatocellular carcinoma in patients listed for liver transplantation: Current and future allocation policy and management strategies for the individual patient. Liver Transpl. 2015;21:1543–1552. doi: 10.1002/lt.24356. [DOI] [PubMed] [Google Scholar]
- 2.Kuszyk BS, Beauchamp NJ, Jr, Fishman EK. Neurovascular applications of CT angiography. Semin Ultrasound CT MR. 1998;19:394–404. doi: 10.1016/S0887-2171(98)90016-6. [DOI] [PubMed] [Google Scholar]
- 3.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584–10597. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trousseau Armand. Lectures on clinical medicine, delivered at the Hotel-Dieu, Paris. Translated and edited with notes and appendices by P. Victor Bazire. Med Clin. 1872;1:1801–1867. [Google Scholar]
- 5.Thodiyil PA, Kakkar AK. Variation in relative risk of venous thromboembolism in different cancers. Thromb Haemost. 2002;87:1076–1077. doi: 10.1055/s-0037-1613136. [DOI] [PubMed] [Google Scholar]
- 6.Blom JW, Vanderschoot JP, Oostindier MJ, Osanto S, van der Meer FJ, Rosendaal FR. Incidence of venous thrombosis in a large cohort of 66,329 cancer patients: Results of a record linkage study. J Thromb Haemost. 2006;4:529–535. doi: 10.1111/j.1538-7836.2006.01804.x. [DOI] [PubMed] [Google Scholar]
- 7.Folkman J. What is the evidence that tumors are angiogenesis dependent? J National Cancer Institute. 1990;82:4–7. doi: 10.1093/jnci/82.1.4. [DOI] [PubMed] [Google Scholar]
- 8.Cosmai L, Gallieni M, Liguigli W, Porta C. Renal toxicity of anticancer agents targeting vascular endothelial growth factor (VEGF) and its receptors (VEGFRs) J Nephroll. 2017;30:171–180. doi: 10.1007/s40620-016-0311-8. [DOI] [PubMed] [Google Scholar]
- 9.Wu L, Zhang YS, Ye ML, Shen F, Liu W, Hu HS, Li SW, Wu HW, Chen QH, Zhou WB. Overexpression and correlation of HIF-2α, VEGFA and EphA2 in residual hepatocellular carcinoma following high-intensity focused ultrasound treatment: Implications for tumor recurrence and progression. Exp Ther Med. 2017;13:3529–3534. doi: 10.3892/etm.2017.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rong W, Yang L, Yin L, Gao Y, Xiao T, Cheng S. PSG9 promotes angiogenesis by stimulating VEGFA production and is associated with poor prognosis in hepatocellular carcinoma. Sci China Life Sci. 2017;60:528–535. doi: 10.1007/s11427-016-0226-7. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Kumar D, Gorain M, Butti R, Datta S, Agarwal S, Gupta S, et al. miRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8:e2706. doi: 10.1038/cddis.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu CH, Kang YK, Yang TS, Shun CT, Shao YY, Su WC, Su WC, Sandoval-Tan J, Chiou TJ, Jin K, et al. Bevacizumab with erlotinib as first-line therapy in Asian patients with advanced hepatocellular carcinoma: A multicenter phase II study. Oncology. 2013;85:44–52. doi: 10.1159/000350841. [DOI] [PubMed] [Google Scholar]
- 13.Meyer G. Venous thromboembolism and cancer. Rev Prat. 2015;65:216–219. (In French) [PubMed] [Google Scholar]
- 14.Bianconi D, Schuler A, Pausz C, Geroldinger A, Kaider A, Lenz HJ, Kornek G, Scheithauer W, Zielinski CC, Pabinger I, et al. Integrin beta-3 genetic variants and risk of venous thromboembolism in colorectal cancer patients. Thromb Res. 2015;136:865–869. doi: 10.1016/j.thromres.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kokudo T, Hasegawa K, Kokudo N. Liver, pancreas, biliary tract cancer. I. Surgical treatment of hepatocellular carcinoma associated with vascular tumor thrombosis. Gan To Kagaku Ryoho. 2014;41:1209–1211. (In Japanese) [PubMed] [Google Scholar]
- 16.Posch F, Thaler J, Zlabinger GJ, Königsbrügge O, Koder S, Zielinski C, Pabinger I, Ay C. Soluble vascular endothelial growth factor (sVEGF) and the risk of venous thromboembolism in patients with cancer: Results from the Vienna cancer and thrombosis study (CATS) Clin Cancer Res. 2016;22:200–206. doi: 10.1158/1078-0432.CCR-14-3358. [DOI] [PubMed] [Google Scholar]
- 17.Babu Govind K, Bhat GR. Cancer-associated thrombotic microangiopathy. Ecancermedicalscience. 2016;10:649. doi: 10.3332/ecancer.2016.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans CE, Grover SP, Saha P, Humphries J, Kim JW, Modarai B, Smith A. Suppression of angiogenic response in local vein wall is associated with reduced thrombus resolution. Thromb Res. 2014;134:682–685. doi: 10.1016/j.thromres.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Di Vito C, Navone SE, Marfia G, Hadi Abdel L, Mancuso ME, Pecci A, Crisà FM, Berno V, Rampini P, Campanella R, Riboni L. Platelets from glioblastoma patients promote angiogenesis of tumor endothelial cells and exhibit increased VEGF content and release. Platelets. 2017;28:585–594. doi: 10.1080/09537104.2016.1247208. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 22.Yang BH, Xia JL, Huang LW, Tang ZY, Chen MS, Li JQ, Liang AM, Mo QG, Lu HS, Dai CL, et al. Changes of clinical aspect of primary liver cancer in China during the past 30 years-control study for 3,250 cases with primary liver cancer. Zhonghua Yi Xue Za Zhi. 2003;83:1053–1057. (In Chinese) [PubMed] [Google Scholar]
- 23.Karnes RJ, Blute ML. Surgery insight: Management of renal cell carcinoma with associated inferior vena cava thrombus. Nat Clin Pract Urol. 2008;5:329–339. doi: 10.1038/ncpuro1122. [DOI] [PubMed] [Google Scholar]
- 24.Lambert EH, Pierorazio PM, Shabsigh A, Olsson CA, Benson MC, McKiernan JM. Prognostic risk stratification and clinical outcomes in patients undergoing surgical treatment for renal cell carcinoma with vascular tumor thrombus. Urology. 2007;69:1054–1058. doi: 10.1016/j.urology.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 25.Lam JS, Klatte T, Kim HL, Patard JJ, Breda A, Zisman A, Pantuck AJ, Figlin RA. Prognostic factors and selection for clinical studies of patients with kidney cancer. Crit Rev Oncol Hematol. 2008;65:235–262. doi: 10.1016/j.critrevonc.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Squillaci E, Fanucci E, Sciuto F, Masala S, Sodani G, Carlani M, Simonetti G. Vascular involvement in pancreatic neoplasm: A comparison between spiral CT and DSA. Dig Dis Sci. 2003;48:449–458. doi: 10.1023/A:1022568128376. [DOI] [PubMed] [Google Scholar]
- 27.Chow LC, Rubin GD. CT angiography of the arterial system. Radiol Clin North Am. 2002;40:729–749. doi: 10.1016/S0033-8389(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 28.Hakenberg OW. Comment on Di Silverio et al: Neodajuvant therapy with sorafenib in advanced renal cell carcinoma with vena cava extension submitted to radical nephrectomy. Urol Int. 2008;80:454. doi: 10.1159/000136481. [DOI] [PubMed] [Google Scholar]
- 29.Shuch B, Riggs SB, LaRochelle JC, Kabbinavar FF, Avakian R, Pantuck AJ, Patard JJ, Belldegrun AS. Neoadjuvant targeted therapy and advanced kidney cancer: Observations and implications for a new treatment paradigm. BJU Int. 2008;102:692–696. doi: 10.1111/j.1464-410X.2008.07660.x. [DOI] [PubMed] [Google Scholar]
- 30.Karakiewicz PI, Suardi N, Jeldres C, Audet P, Ghosn P, Patard JJ, Perrotte P. Neoadjuvant sutent induction therapy may effectively down-stage renal cell carcinoma atrial thrombi. Eur Urol. 2008;53:845–848. doi: 10.1016/j.eururo.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Harshman LC, Srinivas S, Kamaya A, Chung BI. Laparoscopic radical nephrectomy after shrinkage of a caval tumor thrombus with sunitinib. Nat Rev Urol. 2009;6:338–343. doi: 10.1038/nrurol.2009.84. [DOI] [PubMed] [Google Scholar]
- 32.Bex A, Van der Veldt AA, Blank C, Meijerink MR, Boven E, Haanen JB. Progression of a caval vein thrombus in two patients with primary renal cell carcinoma on pretreatment with sunitinib. Acta Oncol. 2010;49:520–523. doi: 10.3109/02841860903521111. [DOI] [PubMed] [Google Scholar]
- 33.Roberts E, Cossigny DA, Quan GM. The role of vascular endothelial growth factor in metastatic prostate cancer to the skeleton. Prostate Cancer. 2013;2013:418340. doi: 10.1155/2013/418340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan JJ, Zhang YN, Liao JZ, Ke KP, Chang Y, Li PY, Wang M, Lin JS, He XX. miR-497 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting VEGFA and AEG-1. Oncotarget. 2015;6:29527–29542. doi: 10.18632/oncotarget.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Mohan R, Özcan S, Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic β-cells. J Biol Chem. 2012;287:31155–31164. doi: 10.1074/jbc.M112.362632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Tsai HC, Tzeng HE, Huang CY, Huang YL, Tsai CH, Wang SW, Wang PC, Chang AC, Fong YC, Tang CH. WISP-1 positively regulates angiogenesis by controlling VEGF-A expression in human osteosarcoma. Cell Death Dis. 2017;8:e2750. doi: 10.1038/cddis.2016.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Zhao Q, Fan L, Zhang Z, Tan B, Liu Y, Li Y. Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015;75:137–141. doi: 10.1016/j.biopha.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 40.Liang HQ, Wang RJ, Diao CF, Li JW, Su JL, Zhang S. The PTTG1-targeting miRNAs miR-329, miR-300, miR-381, and miR-655 inhibit pituitary tumor cell tumorigenesis and are involved in a p53/PTTG1 regulation feedback loop. Oncotarget. 2015;6:29413–29427. doi: 10.18632/oncotarget.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y, Ohms SJ, Li Z, Wang Q, Gong G, Hu Y, Mao Z, Shannon MF, Fan JY. Changes in the expression of miR-381 and miR-495 are inversely associated with the expression of the MDR1 gene and development of multi-drug resistance. PLoS One. 2013;8:e82062. doi: 10.1371/journal.pone.0082062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B, Duan L, Yin G, Tan J, Jiang X. Simultaneously expressed miR-424 and miR-381 synergistically suppress the proliferation and survival of renal cancer cells-Cdc2 activity is up-regulated by targeting WEE1. Clinics (Sao Paulo) 2013;68:825–833. doi: 10.6061/clinics/2013(06)17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O. MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thoracic Oncol. 2012;7:1069–1077. doi: 10.1097/JTO.0b013e31824fe976. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Zheng J, Chen P, Liu Q, Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;90:705–712. doi: 10.1016/j.biopha.2017.02.067. [DOI] [PubMed] [Google Scholar]
- 45.Hou G, Liu G, Yang Y, Li Y, Yuan S, Zhao L, Wu M, Liu L, Zhou W. Neuraminidase 1 (NEU1) promotes proliferation and migration as a diagnostic and prognostic biomarker of hepatocellular carcinoma. Oncotarget. 2016;7:64957–64966. doi: 10.18632/oncotarget.11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang Y, Yu H, Zhang L, Wang K, Guo W, Shi J, Liu S, Wu M, Wang H, Cheng S. Experimental study on enhancement of the metastatic potential of portal vein tumor thrombus-originated hepatocellular carcinoma cells using portal vein serum. Chin J Cancer Res. 2014;26:588–595. doi: 10.3978/j.issn.1000-9604.2014.10.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.