Abstract
Hydroxyethyl starch (HES) is a clinically used polysaccharide colloidal plasma volume expander. The goal of this study was to synthesize HES modified with hydroxychloroquine (HCQ) as a novel polymeric drug with the ability to inhibit the invasive character of pancreatic cancer (PC) cells. HES was conjugated with HCQ using a simple carbonyldiimidazole coupling to prepare Chloroquine-modified HES (CQ-HES). CQ-HES with various degrees of HCQ substitution were synthesized and characterized. Atomic force microscopy was used to demonstrate a pH-dependent assembly of CQ-HES into well-defined nanoparticles. In vitro studies in multiple PC cell lines showed CQ-HES to have a similar toxicity profile as HCQ. Confocal microscopy revealed the propensity of CQ-HES to localize to lysosomes and mechanistic studies confirmed the ability of CQ-HES to inhibit autophagy in PC cells. Further studies demonstrated a greatly enhanced ability of CQ-HES to inhibit the migration and invasion of PC cells when compared with HCQ. The enhanced inhibitory actions of CQ-HES compared to HCQ appeared to arise in part from the increased inhibition of ERK and Akt phosphorylation. We found no significant HCQ release from CQ-HES, which confirmed that the observed activity was due to the action of CQ-HES as a polymeric drug. Due to its promising ability to block cancer cell invasion and the ability to form nanoparticles, CQ-HES has the potential as a drug delivery platform suitable for future development with chemotherapeutics to establish novel antimetastatic treatments.
Keywords: Hydroxychloroquine, hydroxyethyl starch, polymeric drug, pancreatic cancer, invasion, metastasis
Graphical Abstract
INTRODUCTION
Modified polysaccharides have shown great promise in the biomedical field. 1–8 A few non-synthetic and semi-synthetic formulations of glucans such as dextran and hydroxyethyl starch (HES) have been used clinically for the past few decades,9, 10 and their biocompatibility, biodegradability, low immunogenicity, and ability to be readily modified by various functional moieties make them attractive starting materials in the field of drug delivery.11–19 Modification with hydrophobic moieties such as cholesterol or hydrophobic drugs allows for assembly of the polysaccharides into nanoparticles that may improve the efficacy and safety of drug delivery.20, 21 For example, modifying HES with fatty acids allowed for assembly of HES micelles,22 and subsequently it was shown that propylating starch facilitates the assembly of large nanoparticles for encapsulation of doxetaxel.23 Using pentadecylphenol-modified dextran, Pramod et al. synthesized a dual delivery nanovesicle of camptothecin and doxorubicin, which displayed synergistic improvement in anticancer activity.24 Similarly, Wasiak et al. used dodecylamine to drive assembly of a dextran-based polymeric doxorubicin-conjugate.25 Senanayake et al. showed that modifying dextrin with both cholesterol and fluorouracil produced a polymer-drug nanogel conjugate that demonstrated sustained drug release and improved anticancer activity.26
In tumor delivery applications, these nanoparticle systems take advantage of the unique tumor biology through the enhanced permeability and retention (EPR) effect and allow for improved tumor accumulation.13–15 For most types of malignancies, poor tumor accumulation presents one of the biggest hurdles to developing new therapeutics. This is especially true for pancreatic cancer (PC), where the unique tumor microenvironment prevents drug penetration and attainment of therapeutic levels within pancreatic tumors.27, 28 In this sense, readily modifiable and biocompatible, polysaccharide-based drug delivery systems can improve tumor accumulation of chemotherapeutics without the concerns of eliciting immunogenic responses.29–31 Thus, there is an ongoing need to develop polysaccharide-based drug delivery systems that can provide meaningful improvements in survival and/or reductions in systemic toxicities for patients with difficult to treat malignancies where drug delivery or tumor accumulation is an issue.
HES is a modified polysaccharide consisting of approximately 20% amylose, a linear α-1,4-glucose polymer, and 80% amylopectin, a branched glucose polymer with both α-1,4- and α-1,6-glycosidic linkages. HES is prepared from natural sources like waxy maize starch by chemical modification with ethylene oxide. The hydroxyethylation increases the solubility and reduces enzymatic degradation of starch by serum amylases, thereby extending its plasma half-life.32 This increase in biostability is dependent on the average substitution per anhydroglucose unit (AGU) as well as the ratio between C2 and C6 substitution.33 Optimization of these substitutions has resulted in sufficiently prolonged plasma half-lives that have allowed for translation of HES into the clinical setting as a colloidal blood volume expander.11 Multiple HES formulations are available commercially that differ in their molecular weight and degree of hydroxyethylation.
HES lacks any known drug interactions, shows very low rates of hypersensitivity, and has very few known side effects.11, 12 These favorable properties make HES an attractive alternative to non-degradable polymers like polyethylene glycol (PEG) in the design of drug delivery systems. Modifying HES with hydrophobic moieties allows for assembly into nanoparticles that are capable of drug encapsulation and delivery.22, 34, 35 Using a hydrophobic drug to drive the assembly of polysaccharides and to enable encapsulation of another drug for combination therapy is an underappreciated approach to designing drug delivery systems. Herein, we explore this concept by proposing to use hydroxychloroquine (HCQ) as an HES modifier to create a polysaccharide delivery system with therapeutic benefits imparted by the known biological activity of HCQ. Chloroquine (CQ) and HCQ are 4-aminoquinoline derivatives that were synthesized nearly 80 years ago as antimalarial agents. Only a few drugs have remained in clinical use as long as these two compounds, which is due to their favorable safety profiles, efficacy, and powerful immune-modulating properties.36 Both CQ and HCQ are weakly basic compounds, which are readily protonated in acidic vacuoles, leading to their accumulation in lysosomes and neutralization of lysosomal pH.36 The buffering effect of CQ and HCQ leads to disruption of endolysosomal trafficking and autophagy, which have been one of the main reasons for renewed interest in these compounds as adjuncts in chemotherapy regimens.37 The weak electrolyte nature of HCQ may also allow for pH-controlled assembly of polymers modified with HCQ, while also providing the therapeutic benefits associated with autophagy inhibition. Multiple cancers, including PC, exhibit a strong dependence upon autophagy for growth and proliferation.38, 39 As an added potential benefit, HCQ has been shown to possess weak antagonistic properties towards the C-X-C chemokine receptor type 4 (CXCR4).40, 41 CXCR4 is commonly overexpressed in many types of cancer and promotes growth, survival, chemoresistance, and metastasis.42–44 CXCR4 inhibition decreases metastatic spread of CXCR4-expressing tumor cells and sensitizes the cells to chemotherapy.45, 46 In addition to modulating CXCR4 signaling, HCQ-modified drug delivery systems may increase tumor delivery through active targeting of the CXCR4 receptor. More recently, HCQ has been shown to possess other mechanisms that may impart anticancer effects.47–49 Regardless of the underlying mechanism, HCQ shows the ability to sensitize cancer cells to chemotherapy and radiotherapy in various cancers.50 Thus, HCQ may serve as a novel multi-functional moiety in drug delivery systems especially in PC where chemoresistance and poor tumor accumulation have made it very difficult to improve treatment options.51, 52
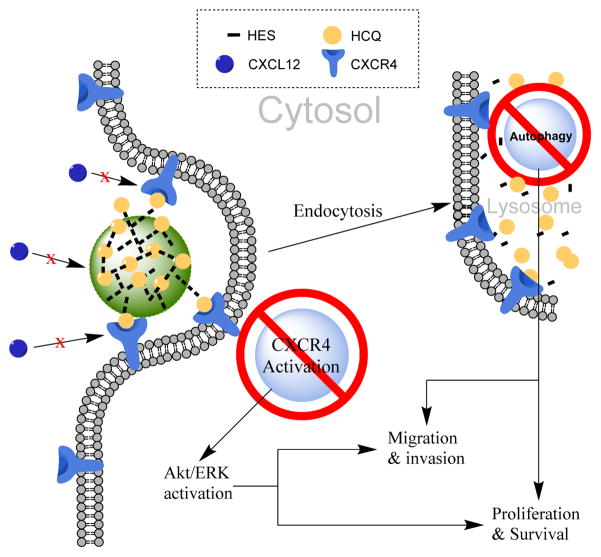
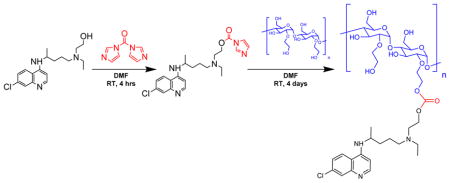
We recently reported on a chloroquine-containing HPMA copolymer that blocks CXCR4 signaling and shows the ability to reduce invasion in vitro and metastasis in vivo in an experimental lung metastasis model.53, 54 However, the HPMA-based polymer demonstrated only limited ability to inhibit autophagy, one of the main mechanisms by which HCQ exerts its therapeutic actions. This led us to investigate other HCQ-modified polymers that better retained the autophagy inhibitory properties of HCQ for improved anticancer activity. Thus, we extended the use of HCQ to polysaccharide modification by presenting the synthesis of HCQ-modified HES (CQ-HES), which display significant inhibition of autophagy in PC cells and demonstrate a new use for the well-established and safe drug HCQ as an assembling moiety in drug delivery systems (Scheme 1).
Scheme 1.
Proposed mechanism of action of CQ-HES.
MATERIALS AND METHODS
Materials
HES (200 kDa, degree of substation 0.5) was purchased from Serumwerk Bernburg, Germany. Fetal bovine serum (FBS) was from Atlanta Biologicals. Mia Paca-1, Mia Paca-2, and AsPC-1 cell lines were kind gifts from Dr. Surinder Batra. CellTiter Blue was from Promega. Laemmli Sample Buffer, nitrocellulose membranes, 1× TBS with 1% casein, and Precision Plus Protein WesternC Standard were from Bio-Rad. 10% Tris-glycine Midi Protein gels were from Invitrogen. The following antibodies were used to manufacturer’s specifications: LC3B (Cell Signaling Technology, 2775S), Akt/ERK (Cell Signaling Technology, 12651s), GAPDH (Cell Signaling Technology, 2118S), and anti-rabbit (Cell Signaling Technology 7074S). Immun-StarTM AP Chemiluminescence Kit was from Bio-Rad. All other reagents, chemicals, and cell culture materials were obtained from Fisher Scientific.
Synthesis of CQ-HES
HCQ sulfate was first converted to its free base form. The HCQ sulfate salt (6 g, 13.8 mmol) was dissolved in 10 mL of deionized water. While stirring, a 30% NH4OH aqueous solution (5 mL, 38.2 mmol) was added dropwise to precipitate the free base HCQ, which was then isolated by extraction with 20 mL dichloromethane (DCM). The organic layer was dried with anhydrous Na2SO4 and the solvent was evaporated to produce HCQ as a colorless viscous solid. The synthesis of CQ-HES was performed using CDI coupling with slight alterations to a previously described method.55 Under nitrogen atmosphere, 389 mg (2.4 mmol) of carbonyl diimidazole (CDI) was added to a dry Schlenk tube, and 3 mL of dry dimethyl-formamide (DMF) was then transferred to the tube via syringe to dissolve the CDI. In a separate dried vessel, HCQ free base (200 mg, 0.6 mmol) was dissolved in 1 mL of dry DMF and transferred via syringe to the stirring CDI solution. The reaction mixture was stirred for 4 h at room temperature, and HCQ activation was monitored by thin layer chromatography (TLC) (1:1 DCM: ethyl acetate). Once complete, the solution was diluted with 5 mL DCM, and 2 mL of water was added to quench any unreacted CDI. The activated HCQ was extracted to DCM (20 mL), washed with water (10 mL × 2) then brine, and dried over Na2SO4. DCM removal in a rotary evaporator produced 220 mg (75%) of activated HCQ (HCQ-CI) as a colorless solid. 1H NMR (499 MHz, Chloroform-d) δ 8.43 (d, J = 5.8 Hz, 1H), 8.14 (d, J = 1.1 Hz, 1H), 8.08 – 7.94 (m, 3H), 7.41 (t, J = 1.4 Hz, 1H), 7.35 (dd, J = 9.0, 2.2 Hz, 1H), 7.09 – 7.02 (m, 1H), 6.45 (d, J = 5.9 Hz, 1H), 4.47 (t, J = 6.0 Hz, 2H), 3.76 (p, J = 6.6 Hz, 1H), 2.85 (t, J = 6.0 Hz, 2H), 2.64 – 2.51 (m, 4H), 1.80 (ddd, J = 16.2, 9.8, 6.6 Hz, 1H), 1.72 – 1.54 (m, 4H), 1.36 (d, J = 6.4 Hz, 3H), 1.04 (t, J = 7.1 Hz, 3H).
HES (300 mg, 1.8 mmol glucose units) was dried for 2 h at 105°C in a dried Schlenk tube. Then, 3 mL of DMF was added to the Schlenk tube under nitrogen atmosphere, and the mixture was heated to 60°C while stirring to dissolve the HES, followed by addition of various amount of HCQ-CI intermediate (see Table 1) in 2 mL of DMF. The resulting solution was stirred at room temperature for 4 days. The CQ-HES product was collected by precipitation with 500mL of acetone and further purified by dissolving in DMF and precipitated two more times to give the final product. NMR and elemental analysis were used to calculate degree of HES substitution with HCQ. 1H NMR (499 MHz, D2O) δ 8.68 (d, J = 6.2 Hz, 1H), 8.40 (d, J = 9.1 Hz, 1H), 8.16 (d, J = 2.2 Hz, 1H), 7.02 (d, J = 6.3 Hz, 1H), 5.83 (d, J = 123.5 Hz, 5H), 4.51 – 3.71 (m, 35H), 3.55 – 3.37 (m, 5H), 2.59 – 2.48 (m, 1H), 2.10 (d, J = 6.9 Hz, 4H), 1.68 (d, J = 6.5 Hz, 3H), 1.51 (td, J = 7.4, 2.4 Hz, 3H). All NMR spectra can be found in the supporting information (Figures S1–S3).
Table 1.
Characterization of CQ-HES
| HCQ-CI:AGUa | MWb | %Cl (w/w%)c | %HCQ (w/w%)d | HCQ:AGUe | yield, % | |
|---|---|---|---|---|---|---|
| HES | - | 1.480 ×105 | - | - | - | - |
| CQ-HES2.5 | 1:23 | 1.614 ×105 | 0.25 | 2.4 | 1:69/ND | 61.3 |
| CQ-HES8.5 | 1:15 | 2.079 ×105 | 0.94 | 8.8 | 1:17/1:15 | 72.9 |
| CQ-HES12.5 | 1:9 | 1.627 ×106 | 1.35 | 12.6 | 1:11/1:17 | 76.6 |
| CQ-HES17.5 | 1:8 | 1.707 ×106 | 1.88 | 17.6 | 1:8/1:11 | 79.5 |
| CQ-HES20 | 1:3.3 | 2.935 ×106 | 2.25 | 20.9 | 1:6/1:4.7 | 68.4 |
| CQ-HES30 | 1:1.7 | 2.862 ×105 | 3.39 | 31.2 | 1:3.5/1:4.1 | 81.7 |
CQ-HES# represents a formulation of CQ-HES with approximate HCQ w% = #.
ratio of HCQ-CI:AGU used in step 2 of synthesis (scheme 2), HCQ-CI = hydroxychloroquine-carbonylimidazole intermediate, AGU = anhydrous glucose unit.
MW = molecular weight of CQ-HES as determined by gel permeation chromatography.
%Cl (w/w%) = weight percent of chlorine in CQ-HES determined by elemental analysis.
%HCQ (w/w%) = weight percent hydroxychloroquine based on elemental analysis data.
HCQ:AGU = molecular ratio of hydroxychloroquine to anhydrous glucose unit based on elemental analysis/NMR.
Fluorescently labelled CQ-HES (fCQ-HES) was synthesized by using CQ-HES20 produced by the above procedure. Fluorescein (665 mg, 2 mmol) and CDI (389 mg, 2.4 mmol) were dissolved in DMF (4 mL) and stirred for 4 h at room temperature. TLC showed the formation of the intermediate and the unreacted fluorescein. Water (10 mL) was added to terminate the reaction after the mixture was stirred for 4 h. The fluorescein intermediate was extracted by DCM (50 mL) and washed by brine (10 mL). After evaporation, the intermediate (560 mg, 66 %) was obtained and used for the next step without further purification. To conjugate fluorescein to CQ-HES20, conditions were maintained in a similar fashion as described above with a few exceptions. CQ-HES20 was used instead of HES and was dried under vacuum for 3 days at room temperature. A ratio of 1:100 fluorescein-CI:AGU was used for the second step of the reaction, and the final fCQ-HES product was precipitate with methanol instead of acetone.
Polymer analysis
1H NMR spectroscopy was performed using the Bruker Avance-III HD 500 MHz NMR; 13C NMR spectroscopy was performed using the Bruker Avance-III HD 600 MHz NMR unless otherwise specified. Data was analyzed with Bruker TopSpin software (v3.5pl7). Approximately 10 mg samples were dissolved into 700 μL of either CDCl3 for HCQ and HCQ-CI or d6-DMSO for HCQ, HES, and CQ-HES unless otherwise specified. All CQ-HES were analyzed for carbon, nitrogen, hydrogen, and chlorine content using elemental analysis (Atlantic Micro Labs, Atlanta, Georgia). The molecular weight of CQ-HES was determined by gel permeation chromatography (GPC). The Agilent 1260 Infinity LC system was operated at a flow rate of 0.5 mL/min using a 0.1 M sodium acetate buffer (pH 5.0) with a TSKgel G3000PWXL-CP column (Part No. 0021873, Tosoh Bioscience LLC, King of Prussia, PA), miniDAWN TREOS multi-angle light scattering (MALS) detector, and an Optilab T-rEX refractive index detector from Wyatt Technology (Santa Barbara, CA). Data was analyzed using Astra 6.1 software. HPLC was used to assess the presence of unreacted HCQ or HCQ-CI in CQ-HES. The polymers were dissolved in acetate buffer (pH 5.0) at 5 mg/mL, the solutions were then filtered with 0.2 μm filter and analyzed on an Eclipse Plus C18 column (5 μm, 4.6×150mm) at a flow rate of 1 mL/min using a gradient mobile phase composed of 5–95% gradient of acetonitrile (0.01% trifluoroacetic acid) and HPLC grade water with 0.01% trifluoroacetic acid.
Hydrolytic stability of CQ-HES
CQ-HES solutions were prepared by slowly adding 200 μL of stock CQ-HES (50 mg/mL in DMSO) to a 2 mL stirring solution of 0.01 N HCl, water, PBS, or acetate buffer (pH 5.0) producing a final concentration of 5 mg/mL. A 200 μL aliquot was sampled at the following time points: 0, 24, 48, 72, and 168 hours. CQ-HES of similar concentrations were subjected to acid hydrolysis, 1 N HCl at 100ºC for 4 hours, and subsequently neutralized with equal parts 1 N NaOH.56 Samples were filtered with 0.2 micron filters and analyzed for free HCQ content using HPLC and polymer size using GPC.
Dynamic Light Scattering (DLS)
CQ-HES solutions were prepared by slowly adding 200 μL of stock CQ-HES (50 mg/mL in DMSO) to a 2 mL stirring solution of water, PBS, or acetate buffer (pH 5.0) producing a final concentration of 5 mg/mL. After preparation, solutions were then subjected to dialysis (Thermo Scientific, 3,500 MWCO snakeskin dialysis tubing) for 24 hours using the same solutions they were initially diluted in as dialysis media (water, PBS, or acetate buffer respectively). The solutions were analyzed on a Zetasizer Nano ZS (Malvern Instruments Ltd., UK) using a 633 nm laser at 25ºC. All samples were analyzed using the backscattering angle of 173º. Overall average particle size, distribution, and polydispersity indexes were obtained using the manufacturer provided DTS software.
Atomic force microscopy (AFM)
AFM samples were prepared as described in the previous sections and diluted 500-fold in the respective media (10 μg/mL). A droplet of each sample (5–10 μL) was immediately deposited on freshly cut mica for total incubation time of 2 min. Samples were rinsed carefully with water and dried under a flow of argon. AFM images in air were acquired using MultiMode AFM NanoScope IV system (Bruker Instruments, Santa Barbara, CA) operating in tapping mode with 1.5 Hz scanning rate. TESPA probes from Bruker were used for tapping mode imaging. The probes had a spring constant of about 40 N/m and a resonance frequency between 300 and 320 kHz. Particle sizes were analyzed using ImageJ and plotted with GraphPad Prism 5.
Cell Culture
AsPC-1, MiaPaca-1, and MiaPaca-2 cells were cultured in DMEM high glucose medium supplemented with 10% FBS. U2OS cells stably expressing functional EGFP-CXCR4 fusion protein (Fisher Scientific) were cultured in DMEM high glucose supplemented with 10% FBS, 2 mM L-glutamine, 1% Pen-Strep, and 0.5 mg/mL G418. All cell lines were maintained at 37°C with 5% CO2. All cell lines were grown to 70–80% confluency before subculturing using trypsin, 0.05% for U2OS and 0.25% for PC cell lines.
Cytotoxicity
Cytotoxicity studies were performed using the CellTiter-Blue viability assay. Cells were seeded at 2,000–3,000 cells/well in a 96 well plate. The medium was replaced after 24 h with medium containing 10% FBS and different concentration of CQ-HES or HCQ. The cells were incubated for 72 hours, and afterwards, the medium was aspirated and replaced with 100 μL of the CellTiter-Blue solution. The cells were left to incubate for one hour, and the fluorescence was measured using a SpectraMax M5. The cell viability was calculated as a relative percentage, 57sample/[A]untreated ×100%. Dose response curves were constructed and analyzed using GraphPad Prism 5. IC50 values were calculated as the CQ-HES or HCQ concentration required to achieve 50% growth inhibition relative to untreated cells.
Western Blot
Cells were treated in a 6-well plate and harvested with ice cold RIPA lysis buffer containing protease and phosphatase inhibitors. Samples were centrifuged for 10 minutes at 14,000 RPM, cellular supernatant was collected, and the amount of protein in each sample was analyzed using BCA protein assay kit. Sample protein levels were adjusted with RIPA buffer, diluted with equal parts of 2× Laemmli Sample Buffer containing 5% 2-mercaptoethanol, and heated for 10 minutes at 90°C. Samples (20 μg) were electrophoresed (10% Tris-Glycine Midi Protein Gels) and transferred to nitrocellulose membranes. Membranes were blocked for 1 hour at room temperature using 1× TBS (1% casein). Primary antibodies were run overnight at 4°C in 1× TBS (1% casein) to manufacturer’s specifications. Afterwards, membranes were washed thrice with TBS and incubated for 1 hour at room temperature with secondary antibody at manufacturer’s specifications. Chemiluminescence was initiated using the Immun-StarTM AP Chemiluminescence Kitand subjected to LucentBlue X-ray films. Western blots were analyzed using Image Studio Lite (LI-COR).
Confocal microscopy
The LSM 510 Zeiss confocal microscope was used to image intracellular trafficking of fCQ-HES (5 μM HCQ, 490/520 exc/em) in Mia Paca-1 cells. LysoTracker Red DND-99 (577/590 exc/em) was used to fluorescently label lysosomes.
Cell migration
Cell migration was assessed using a wound healing assay as previously described.58 Briefly, the cells were seeded in 12-well plates. Upon growing to near confluence, a sterile 200 μL pipette tip was used to create a wound. The cells were then washed twice with PBS, and media containing 10% FBS and respective HCQ, CQ-HES, or control was added to the wells. Using an AMG EVOS XL Core Imaging System, the cells were then imaged at regular time intervals: 0, 6, 12, and 24 hours. Wound healing was assessed as the percent of wound closure (± standard deviation) compared to time zero measured as the distance between wound borders with 10 measurements being taken for each biological replicate at each time point.
Cell invasion
Cell invasion was performed as previously described.59 Briefly, Matrigel was thawed on ice and then diluted 1:4 with serum free medium. A final volume of 40 uL of the Matrigel solution was placed into cell culture inserts and incubated for 2 hours at 37°C in 24 well plates. Cells were harvested using AccutaseTM cell detachment solution and re-suspended in serum-free medium. Cells were diluted with serum-free medium to their respective drug (HCQ or CQ-HES) or vehicle concentration and a final concentration of 200,000 cells per insert. Medium with 10% FBS was then added to the lower chamber of the wells, and the cells were allowed to incubate overnight at 37°C. Cells were fixed in 100% methanol, stained with 0.2% Crystal Violet, and imaged using AMG EVOS XL Core Imaging System. Invasion was assessed as the average cells/area imaged ± standard deviation. Invasion was also assessed in an SDF-1 dependent manner using U2OS cells constitutively expressing CXCR4. The method was similar to the above except for following changes: 1) cell density – 100,000 cells/insert; 2) DMEM supplemented with 2 mM L-glutamine; and 3) SDF-1 (100 ng/mL) in serum free media in the lower well instead of serum-containing media.
RESULTS
Synthesis and characterization of CQ-HES
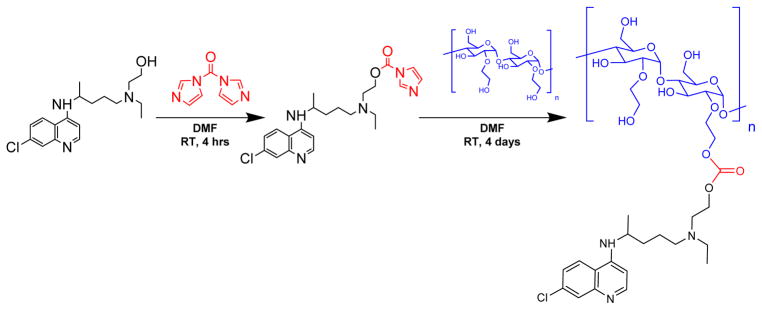
Because of the abundant hydroxyl groups, many linker chemistries can be implemented to conjugate a drug or hydrophobic group to HES.60, 61 HCQ contains a primary hydroxyl group that is a prime target for conjugation. The secondary alkyl-aryl amine also present in the HCQ structure is not a suitable site for conjugation due to its low reactivity and its importance for binding to the CXCR4 receptor.40 Although hydroxyethylation increases the stability of HES, the polysaccharide remains susceptible to degradation in extreme pH conditions. Multiple previous reports have used mild carbodiimide and CDI chemistries for polysaccharide modifications.34, 55 Conjugation of HCQ to HES required a linker chemistry suitable for conjugating two hydroxyls. Thus, we selected the two-step CDI approach, which not only allowed mild conditions for conjugation of HCQ to HES, but it also resulted in the formation of a potentially biodegradable carbonate bond. In the first step of this procedure, the hydroxyl of HCQ was activated with CDI to produce an HCQ-CI intermediate, which was then used in the subsequent step to react with HES to afford CQ-HES (Scheme 2). Adjusting the ratio of HCQ-CI with HES allowed us to control the degree of HES substitution with HCQ (Table 1).
Scheme 2.
Synthesis of CQ-HES.
NMR analysis of CQ-HES demonstrated the presence of the characteristic aromatic signal of HCQ at 7.5–8.5 ppm, as well as the presence of complex signal signature from the HES at 2–6 ppm (Figure 1). Furthermore, CQ-HES displayed the unique presence of a carbonyl group at 155 ppm with long-range coupling solely to the protons adjacent to the former hydroxyl groups, which were found to have shifted from 3.5 to 4.1 ppm. The remaining coupling relationships of HCQ in CQ-HES were preserved as seen from the 1D and 2D NMR spectra and validated the proposed linkage between the hydroxyls and no side-reactions occurring through the amine groups (see supporting information – Figures S2 and S3). Lastly, no starting materials or intermediate species, i.e. CDI or imidazole, were apparent in the NMR spectra of CQ-HES. HPLC further confirmed the absence of any small molecule impurities in the CQ-HES samples. The degree of substitution of HES and content of HCQ was first calculated from the NMR spectra using the ratio of the anomeric C1 proton (* at 5–6 ppm) in HES to one of the aromatic protons (** at 8.43 ppm) in HCQ (Figure 1). The content of HCQ was then validated using elemental analysis (Table 1).
Figure 1.
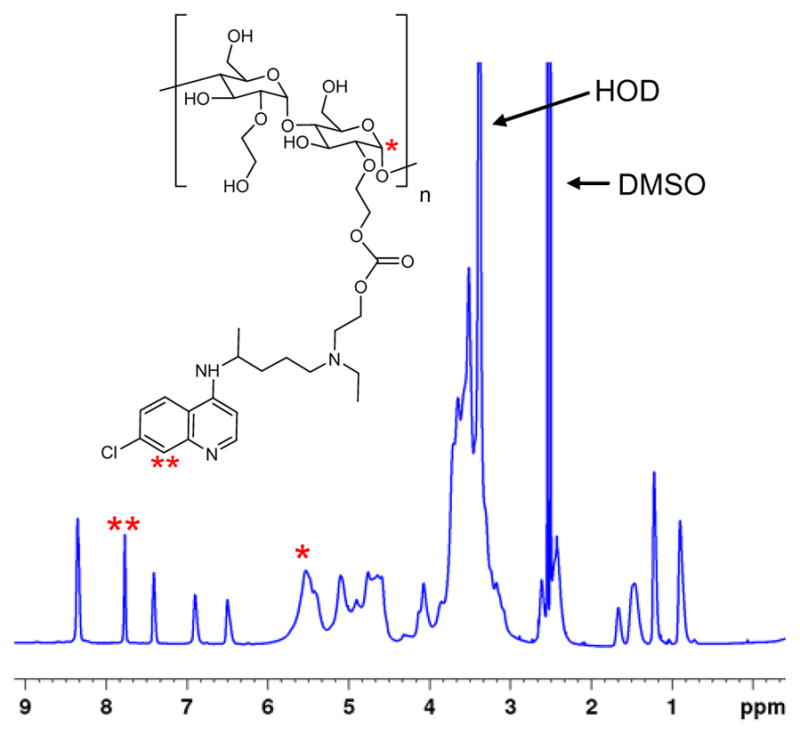
1H-NMR spectrum of CQ-HES. Inset shows the structure of CQ-HES. For complete peak assignments, see supporting information (Figures S1–S2).
As shown in Table 1, by varying the HCQ:HES ratio (HCQ-CI:AGU) from ~1:20 to 1:2, we were able to synthesize CQ-HES with HCQ content ranging from 2.4 to 32 w%. The corresponding degree of substitution of the glucose units in HES with HCQ ranged from 1:69 to 1:3.5. Because of the HCQ substitution, the molecular weight of CQ-HES increased with increasing degree of HCQ substitution in the HES chain (Table 1).
Solubility and particle formation
All CQ-HES readily dissolved in DMF and DMSO up to 200 mg/mL. However, dissolving CQ-HES directly in water or aqueous buffers proved difficult even with extensive sonication and the solubility was found to be dependent upon the HCQ content and pH of the solvent. In PBS, only CQ-HES2.5 and CQ-HES8.5 (HCQ w% ~ 2.5% and 8.5%, respectively) were soluble. In water, CQ-HES17.5 was soluble up to 5 mg/mL. In the acidic pH of the acetate buffer, the amines in HCQ become protonated affording an increase the solubility, and as a result all CQ-HES formulations were soluble at 5 mg/mL (see supporting information – Table S1). Solubility of CQ-HES was further evaluated in acetate buffer, water, or PBS. Solutions prepared using CQ-HES20 and CQ-HES30 were initially soluble in water or PBS at 1.5 mg/mL, but aggregated and precipitated within one hour of preparation. All other lower HCQ-containing CQ-HES remained soluble up to 20 mg/mL with no signs of aggregation or precipitation.
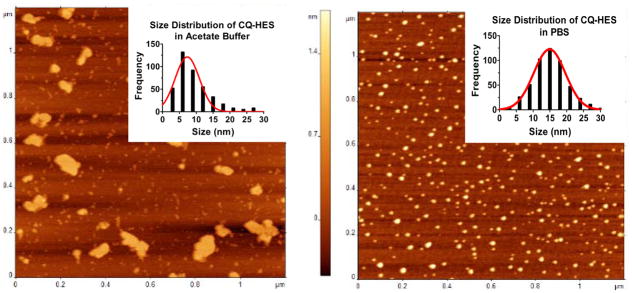
CQ-HES20 had high HCQ content and substantial aqueous solubility and was thus selected for further study of its physical and biological properties. We first analyzed the hydrodynamic size of CQ-HES by DLS. Control HES dissolved in acetate buffer, water, or PBS revealed hydrodynamic sizes of ~20 nm. CQ-HES20 showed a slight increase in hydrodynamic size to ~30 nm in all three solutions (see supporting information – Table S2). Using the empirical calculator in the Zetasizer software, HES (200 kDa) showed a predicted size of 16–26 nm, while conservative estimates for CQ-HES (250–300 kDa) were 17–33 nm. AFM was then used to directly visualize possible formation of nanoparticles. In water and acetate buffer, CQ-HES20 displayed irregularly shaped nanostructures indicative of single molecule arrangements with a few interspersed thin-film aggregates (Figure 2). However, images of CQ-HES20 formulated in PBS revealed the formation of well-defined nanoparticles. Analysis of the formed particles described a normally distributed population with an average diameter of 15 nm (range 2–30 nm) (Figure 2).
Figure 2.
AFM and corresponding size distribution of CQ-HES20 in acetate buffer solution (left) and PBS (right).
Hydrolytic stability of CQ-HES
Hydrolytic stability of the HES backbone and of the carbonate linker between HCQ and HES were assessed. CQ-HES20 was incubated in 0.01 N HCl, acetate buffer, water, or PBS (pH 2, 5.0, 7.0, or 7.4 respectively) for 72 h at 37ºC. No release of free HCQ or any changes in the molecular weight of CQ-HES were observed during the experiment using HPLC and GPC, suggesting relative stability of the carbonate linker. However, hydrolytic cleavage of CQ-HES20 and release of HCQ was observed after incubation in 1 M HCl for 4 h. These pieces of evidence validate the success of the conjugation, efficiency of the purification procedure, and the stability of CQ-HES.
Cytotoxicity
The presence of HCQ provides CQ-HES with a weak polycationic character. It was important to first assess whether this results in any unexpected cytotoxicity. To address this issue, we tested the cytotoxic effects of CQ-HES20 in three different PC cell lines, AsPC-1, MiaPaca-1, and MiaPaca-2. We found no difference in the cytotoxicity of CQ-HES and parent HCQ (see supporting information – Figure S4). Repeating these cytotoxicity studies at densities (6×105 cells/cm2) used in the migration, invasion, and cellular signaling studies described below, showed no discernable toxicity of either CQ-HES or HCQ until 300 μM (Supporting Figure S4). Thus, we selected 100 μM concentrations of HCQ and CQ-HES to be used in the subsequent studies as the cytotoxic effects should be of little consequence in the modifications of cellular behaviors addressed below.
Inhibition of autophagy and intracellular trafficking of CQ-HES
Because the amines in HCQ act as proton acceptors, sufficiently high concentrations of HCQ can neutralize the acidic lysosome and inhibit protease activity and autophagosomal fusion, effectively inhibiting autophagy.62 Using CDI chemistry, we aimed to conjugate HCQ to HES through the hydroxyl group of HCQ so that the amines remain unaltered and retain the biological activity of HCQ. However, the conjugation of HCQ with HES could have unanticipated effects on the ability of CQ-HES to inhibit autophagy. To investigate this assertion, we determined the buffering ability of CQ-HES (supporting information – Figure S5). CQ-HES displayed a comparable buffering capacity to HCQ within physiological ranges of pH 3.6–7.9. This buffering capacity was solely the result of the HCQ in CQ-HES as HES lacked any buffering ability by itself. We further sought to determine any difference in biological activity for autophagy inhibition between CQ-HES20 and HCQ by measuring intracellular levels of the autophagosome marker LC3B in three PC cells. All three PC cell lines showed an increase in both LC3B-I and II after treatment with CQ-HES20 that was comparable to free HCQ (Figure 3) indicating a preservation of the autophagy inhibitory activity of HCQ in CQ-HES.
Figure 3.
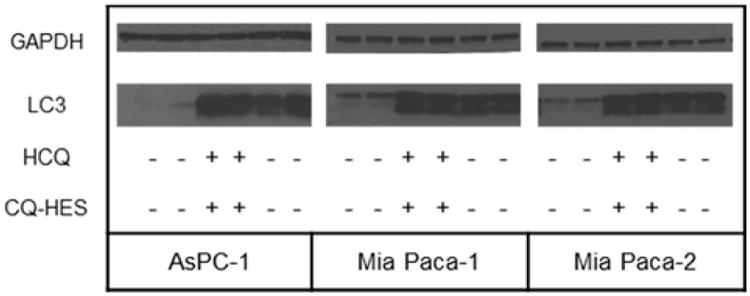
Autophagy analyzed by western blot of LC3.
To understand the cellular localization of CQ-HES, we performed live cell imaging studies and observed the cellular trafficking of CQ-HES. Lysosomes were stained with LysoTracker Red, and the cells were incubated with fluorescein-labeled CQ-HES (fCQ-HES). FCQ-HES displayed a rapid internalization with ultimate localization and retention in lysosomes as evident by co-localization with the LysoTracker signal (Figure 4). After substantial accumulation of CQ-HES in the lysosomes, the fluorescence of the LysoTracker Red decreased suggesting a neutralization of the acidic pH in the lysosomes. This evidence taken together with the acid-base titration data indicates that the functionality of HCQ in CQ-HES is preserved and the localization of HCQ to the lysosomes is maintained in CQ-HES although the mechanism by which this occurs may be different.
Figure 4.
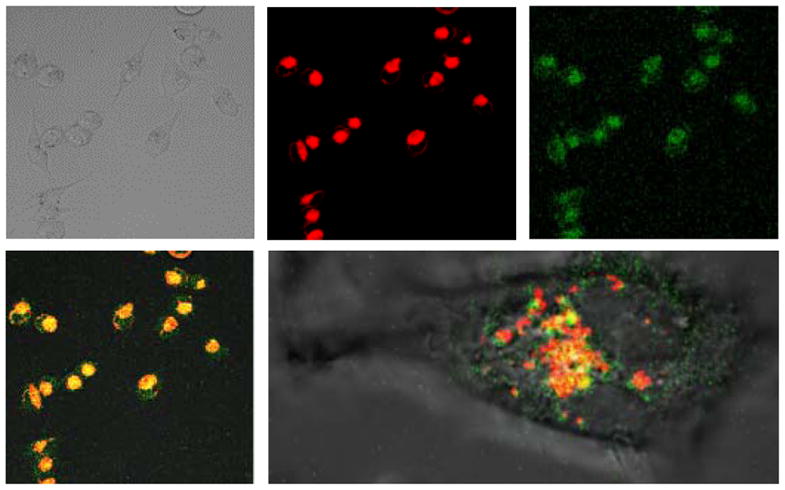
Cellular localization of CQ-HES using live cell imaging of Mia Paca-1 cells. Transmitted light (top left), LysoTracker Red (top middle), fluorescein-labeled CQ-HES (top right), colocalization (bottom left), and colocalization of an individual cell (bottom right).
Effect of CQ-HES on the migration and invasion of PC cells
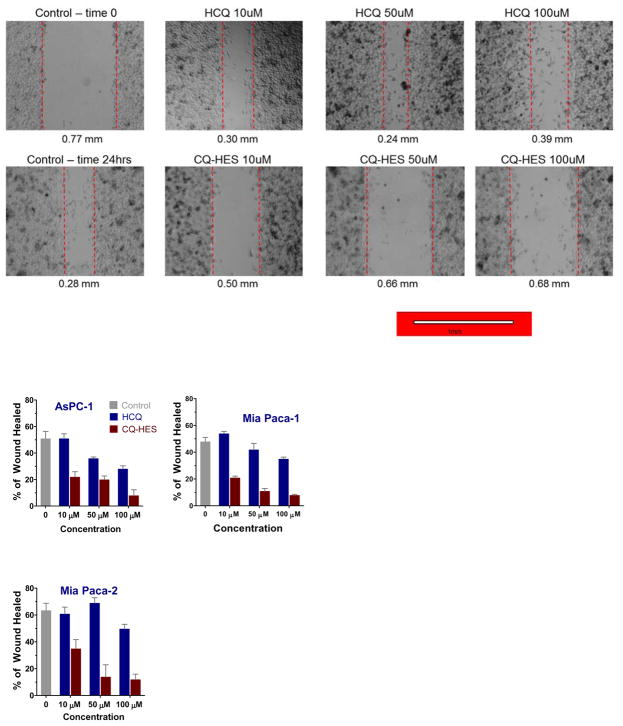
Following the successful demonstration of the strong autophagy inhibition of CQ-HES, we explored the other potential HCQ target in cancer, the CXCR4 chemokine receptor, by evaluating the related effects of CQ-HES on the motility of cancer cells assessed first by a wound healing assay (Figure 5). Twenty-four hours after creating the wound, the untreated control group showed ~50–60% wound closure in all three PC cell lines. In contrast, a considerable inhibition of the wound closure was observed in cells treated with CQ-HES. The inhibitory effect of CQ-HES was observed in all three PC cell lines even at the lowest concentration used (10 μM). The highest CQ-HES concentration (100 μM) showed ~80% inhibition over the control group, while the 10 μM concentration still displayed significant migration inhibition of ~40–60% over control. In contrast, the effects of CQ-HES on wound healing substantially exceeded the effects of HCQ alone, as HCQ did not reach significant inhibition until doses of approximately 50 μM.
Figure 5.
Effect of CQ-HES20 on PC cell migration determined by wound healing assay at 24 hours with representative figures from Mia Paca-2 cells displayed above.
Next, we examined the ability of CQ-HES to inhibit cancer cell invasion using a Boyden chamber Matrigel invasion assay with 10% FBS media as the source of chemotactic signals (Figure 6). Negative controls lacking FBS demonstrated a minimal degree of random invasion, while the positive controls containing FBS showed a strong ability to drive invasion in all three cell lines through the Matrigel. As in the wound healing assay, CQ-HES demonstrated a greatly enhanced ability to inhibit PC cell invasion when compared with equivalent concentrations of HCQ. At 1 μM, CQ-HES was able to reduce invasion by ~80% in AsPC-1 cells and ~70% in MiaPaca-2 cells, but only a modest reduction of ~10% was observed in MiaPaca-1 cells. Increasing the concentration to 50 μM achieved a nearly complete inhibition of cell invasion in all three cell lines. In contrast, there was no significant effect on invasion with 1 μM HCQ in any of the cell lines tested. Regardless of cell type or concentration, the effects of CQ-HES were substantially greater than that of comparable levels of HCQ.
Figure 6.
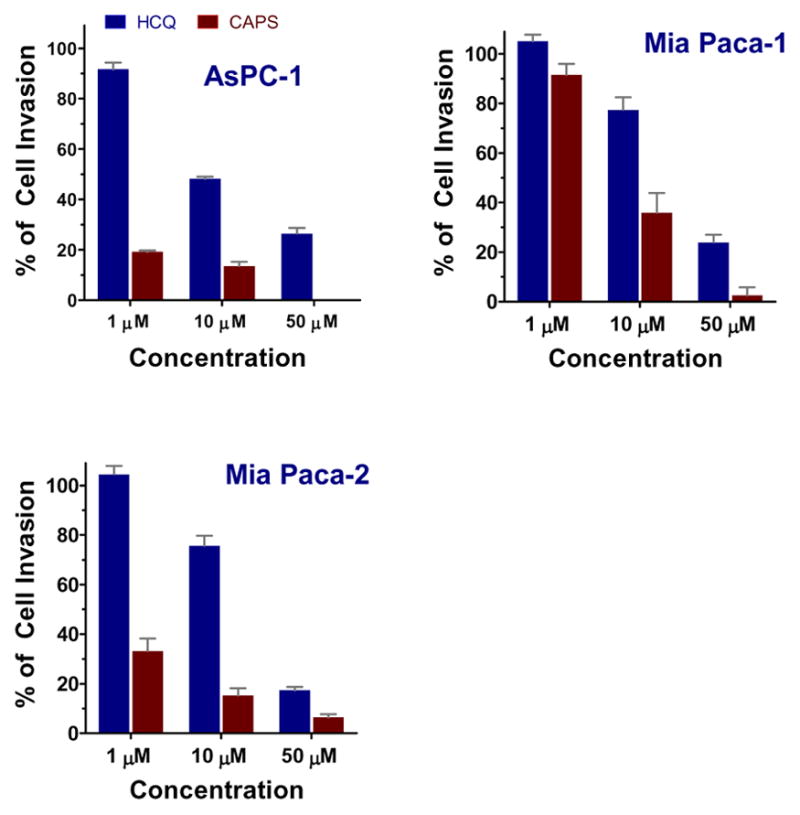
Effect of CQ-HES20 on FBS-driven PC cell invasion at 24 hours.
Effect of CQ-HES on CXCR4 signaling
Through QSAR modeling, direct competition binding assays, and through effects on downstream activation components, HCQ has been identified as an antagonist of the CXCR4 receptor.40, 45 We have previously shown that chloroquine-containing HPMA copolymers inhibited CXCR4-mediated breast cancer cell invasion and decreased lung metastasis in vivo. Thus, we sought to confirm the involvement of CXCR4 antagonism in the ability of CQ-HES to inhibit PC cell migration and invasion. First, using U2OS cells overexpressing CXCR4-EGFP fusion protein, we found that CQ-HES inhibited CXCR4-mediated cell invasion to a similar extent as HCQ at equivalent concentrations that was similar to AMD3100 (300 nM), a potent CXCR4 antagonist (Figure 7). We then determined cellular localization of the CXCR4 receptor (see supporting information – Figure S8) after 1 h activation with its ligand SDF-1 (100 ng/mL). CXCR4 readily translocated from the cell surface to focalized points within the cytosol after SDF-1 activation, indicating rapid receptor activation and internalization. Treatment with both HCQ and CQ-HES displayed modest decreases in CXCR4 internalization following SDF-1 stimulation, indicating partial antagonism.
Figure 7.
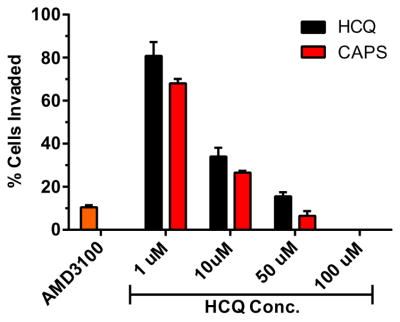
The effects of CQ-HES on SDF-1-mediated invasion in CXCR4-expressing U2OS cells.
To understand the effect of CQ-HES on CXCR4 signaling at the molecular level, we examined two downstream pathways (Akt, ERK) affected by CXCR4 activation with SDF-1. The ability of CQ-HES to inhibit phosphorylation of Akt and ERK was assessed using Western blot to measure the ratio of phosphorylated Akt and ERK to the total cellular content of the corresponding protein. Treatment of U2OS cells with 100 ng/mL SDF-1 for 1 h induced phosphorylation of both Akt and ERK (Figure 8). The phosphorylation of both proteins was inhibited significantly by CQ-HES treatment. HCQ treatment showed substantially lower inhibitory activity than CQ-HES as did treatment with the commercial CXCR4 antagonist, AMD3100. Thus, the ability of CQ-HES to inhibit downstream signaling pathways of CXCR4 involved in cell motility may explain its enhanced effects on migration and invasion.
Figure 8.
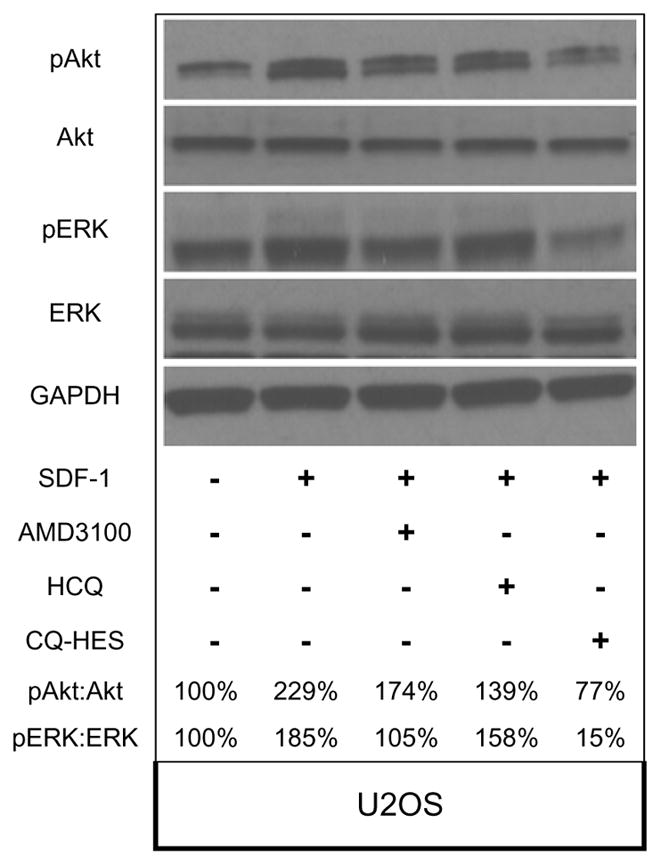
Antagonistic effects of AMD3100, HCQ, and CQ-HES on CXCR4 downstream signaling after SDF-1 stimulation. Levels of pAkt:Akt and pERK:ERK are presented as percentage ratios to control group that did not receive SDF-1.
DISCUSSION
In our previous reports, we have synthesized an antimetastatic polymeric drug (pCQ) based on HPMA copolymers containing HCQ.53, 54 The previous polymers showed the ability to reduce invasion of cancer cells in vitro and lung metastasis in vivo. We have demonstrated the observed effects were at least in part a result of blocking CXCR4 signaling; however, the pCQ polymers abrogated the ability of HCQ to inhibit autophagy, a potential anticancer mechanism. Herein, we improved upon the original concept of a polymeric HCQ-based drug by presenting a simple synthesis of a polysaccharide HCQ drug (CQ-HES) with preserved autophagy inhibition.
Examining the DLS data, we observed a corresponding increase in hydrodynamic size of HES from ~20 nm to CQ-HES with a size of ~30 nm. These sizes are in accordance with previous reports of HES-modified compounds22, 34 and with estimates from the empirical size calculator of the Zetasizer software, which predict 200 kDa polysaccharides to be 16–26 nm depending on branching status, while conservative calculations for CQ-HES20 of 250–300 kDa to be 17–33 nm. However, hydrodynamic sizes obtained from DLS may be misleading, and a complementary method to validate nanoparticle formation and assess morphology should always be included. Thus to further characterize any particle formation, we used AFM, which showed that HES, although displaying a hydrodynamic sizes of ~20 nm via DLS, failed to demonstrate any nanoparticle formation irrespective of pH conditions (supporting information Figure S3). On the other hand, CQ-HES assembled into nanoparticles in a pH-dependent manner. In acidic aqueous conditions, only amorphous and linear structures were observed, while 15 nm diameter particles with well-defined spherical shapes and a narrow size distribution were observed when CQ-HES was formulated in PBS. The well-defined nanoparticle distribution and sizes of CQ-HES defined by AFM is in contrast to the sizes and PDI obtained from DLS. The discrepancies between DLS and AFM data could be derived from a number of factors including the difference in hydration status for aqueous samples used in DLS and lack thereof after drying for visualization with AFM. Irrespective, these findings suggested CQ-HES possesses pH-dependent assembling properties, although we are uncertain if these nanoparticles may be stable enough to survive in vivo applications.
By conjugating HCQ to HES through the drug hydroxyl group, we were able to preserve the biological activity of HCQ. Like pCQ previously studied, CQ-HES displayed the ability to partially inhibit CXCR4 signaling in cancer cells, and these effects were more pronounced than comparable levels of HCQ. Unlike pCQ, CQ-HES strongly inhibited autophagy to a similar degree as free HCQ. Because of its large size, CQ-HES was endocytosed and transported to lysosomes where it could disrupt autophagy by neutralizing the pH in the acidic organelle. We believe the difference in functionality between CQ-HES and pCQ arises from the nature of the chemical backbone, size, and related effects on their intracellular trafficking. CQ-HES has a molecular weight greater than 200 kDa and a highly hydrophilic glucose backbone. These properties greatly favor trafficking and retention into endosomes and lysosomes. On the other hand, pCQ with its hydrophobic methacrylate backbone and smaller molecular weight showed the ability to avoid lysosomal trafficking and disrupt autophagy. The autophagy inhibition by CQ-HES also explains why the cytotoxicity of CQ-HES was similar to HCQ. Furthermore, the relatively stable conjugation of HCQ to HES may have prolonged the pharmacologic effects of CQ-HES. However, more studies are needed to fully understand the mechanism of action and intracellular stability of CQ-HES. Future studies will determine how HCQ is released from CQ-HES both in vitro and in vivo and evaluate the drug loading potential and anticancer activity of CQ-HES particularly in the context of PC.
CQ-HES inhibited CXCR4 signaling to a greater extent than HCQ. The explanation for the improved CQ-HES activity is likely through increased antagonism of the receptor at the cellular surface, retention of the receptor in the endosome, or a combination of both. Once the CXCR4 receptor is stimulated by SDF-1, it becomes internalized at which point it is either recycled or degraded.63, 64 HCQ interferes with the receptor trafficking by preventing recycling and decreasing the surface receptor concentration, thus minimizing long-term activation of the pathway.65, 66 In the case of CQ-HES, where multiple HCQ molecules reside on a single chain of HES, the overall avidity is substantially enhanced in comparison to the affinity of HCQ due to the multivalency effect.67 Thus, CQ-HES may preferentially serve a dual function of not only antagonizing CXCR4, but also may act to prevent receptor recycling following internalization through a receptor mediated process further diminishing CXCR4 signaling. Our data suggest, however, that these effects are not limited to CXCR4 and that other receptors could also be sequestered, leading to a general reduction in receptor presentation on the cell surface. The fact that CQ-HES decreased invasion when either SDF-1 or FBS was used as chemoattractants, informed us that it is most likely a combination of both receptor antagonism and intracellular sequestration, in addition to autophagy inhibition, that leads to decreased invasion and motility seen in cells treated with CQ-HES.
CONCLUSIONS
The goal of this study was to synthesize a polysaccharide-based polymeric drug of HCQ, which retained the biological activity of HCQ and secondarily demonstrated nanoparticle assembling characteristics. We have chosen to utilize a system composed of HES modified with CDI linker chemistry because of its biocompatibility and biodegradability, which represents a novel class of multifunctional, biocompatible drug delivery systems. The methods presented herein provide a means to conjugate HCQ to HES while also controlling the degree of substitution and preserving the biological activity of HCQ, autophagy inhibition and CXCR4 antagonism. Furthermore, CQ-HES displayed the propensity to assemble into nanoparticles and a superior ability to reduce migration and invasion over comparable levels of HCQ. Future studies will investigate various formulation methods for controlling the nanoparticle formation properties of CQ-HES, determine the release profile of HCQ from CQ-HES in vitro and in vivo, and assess the effects on the pharmacokinetics of HCQ. Due to its propensity to readily form nanoparticles in a pH-dependent manner, we will also explore the use of CQ-HES as a multifunctional drug delivery vehicle for targeting and reducing local invasion and metastasis of aggressive malignancies like PC.
Supplementary Material
Acknowledgments
This work was supported in part by the University of Nebraska Medical Center and in part by the National Institutes of Health (EB015216, EB020308, EB019175).
Footnotes
References
- 1.Besheer A, Caysa H, Metz H, Mueller T, Kressler J, Mäder K. Benchtop-MRI for in vivo imaging using a macromolecular contrast agent based on hydroxyethyl starch (HES) International Journal of Pharmaceutics. 2011;417(1–2):196–203. doi: 10.1016/j.ijpharm.2010.10.051. [DOI] [PubMed] [Google Scholar]
- 2.Mizrahy S, Peer D. Polysaccharides as building blocks for nanotherapeutics. Chemical Society Reviews. 2012;41(7):2623–2640. doi: 10.1039/c1cs15239d. [DOI] [PubMed] [Google Scholar]
- 3.Noga M, Edinger D, Kläger R, Wegner SV, Spatz JP, Wagner E, Winter G, Besheer A. The effect of molar mass and degree of hydroxyethylation on the controlled shielding and deshielding of hydroxyethyl starch-coated polyplexes. Biomaterials. 2013;34(10):2530–2538. doi: 10.1016/j.biomaterials.2012.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Freichels H, Wagner M, Okwieka P, Meyer RG, Mailander V, Landfester K, Musyanovych A. (Oligo)mannose functionalized hydroxyethyl starch nanocapsules: en route to drug delivery systems with targeting properties. Journal of Materials Chemistry B. 2013;1(34):4338–4348. doi: 10.1039/c3tb20138d. [DOI] [PubMed] [Google Scholar]
- 5.Tang S, Zhao Z, Chen G, Su Y, Lu L, Li B, Liang D, Jin R. Fabrication of ampicillin/starch/polymer composite nanofibers with controlled drug release properties by electrospinning. Journal of Sol-Gel Science and Technology. 2016;77(3):594–603. [Google Scholar]
- 6.Harling S, Schwoerer A, Scheibe K, Daniels R, Menzel H. A new hydrogel drug delivery system based on hydroxyethylstarch derivatives. Journal of microencapsulation. 2010;27(5):400–8. doi: 10.3109/02652040903367301. [DOI] [PubMed] [Google Scholar]
- 7.Bachelder EM, Beaudette TT, Broaders KE, Dashe J, Fréchet JMJ. Acetal-Derivatized Dextran: An Acid-Responsive Biodegradable Material for Therapeutic Applications. Journal of the American Chemical Society. 2008;130(32):10494–10495. doi: 10.1021/ja803947s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meenach SA, Kim YJ, Kauffman KJ, Kanthamneni N, Bachelder EM, Ainslie KM. Synthesis, Optimization, and Characterization of Camptothecin-Loaded Acetalated Dextran Porous Microparticles for Pulmonary Delivery. Molecular pharmaceutics. 2011;9(2):290–298. doi: 10.1021/mp2003785. [DOI] [PubMed] [Google Scholar]
- 9.Foster JM, Sleightholm R, Watley D, Wahlmeier S, Patel A. The Efficacy of Dextran-40 as a Venous Thromboembolism Prophylaxis Strategy in Cytoreductive Surgery and Hyperthermic Intraperitoneal Chemotherapy. The American surgeon. 2017;83(2):134–140. [PubMed] [Google Scholar]
- 10.Jones CI, Payne DA, Hayes PD, Naylor AR, Bell PR, Thompson MM, Goodall AH. The antithrombotic effect of dextran-40 in man is due to enhanced fibrinolysis in vivo. J Vasc Surg. 2008;48(3):715–22. doi: 10.1016/j.jvs.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Treib J, Baron JF, Grauer MT, Strauss RG. An international view of hydroxyethyl starches. Intensive care medicine. 1999;25(3):258–68. doi: 10.1007/s001340050833. [DOI] [PubMed] [Google Scholar]
- 12.Brecher ME, Owen HG, Bandarenko N. Alternatives to albumin: starch replacement for plasma exchange. Journal of clinical apheresis. 1997;12(3):146–53. doi: 10.1002/(sici)1098-1101(1997)12:3<146::aid-jca8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 13.Debele TA, Mekuria SL, Tsai HC. Polysaccharide based nanogels in the drug delivery system: Application as the carrier of pharmaceutical agents. Materials Science and Engineering: C. 2016;68:964–981. doi: 10.1016/j.msec.2016.05.121. [DOI] [PubMed] [Google Scholar]
- 14.Wei X, Senanayake TH, Warren G, Vinogradov SV. Hyaluronic Acid-Based Nanogel–Drug Conjugates with Enhanced Anticancer Activity Designed for the Targeting of CD44-Positive and Drug-Resistant Tumors. Bioconjugate Chemistry. 2013;24(4):658–668. doi: 10.1021/bc300632w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L, Liu M, Wang J, Zhai G. Chondroitin sulfate-based nanocarriers for drug/gene delivery. Carbohydr Polym. 2015;133:391–9. doi: 10.1016/j.carbpol.2015.07.063. [DOI] [PubMed] [Google Scholar]
- 16.Masina N, Choonara YE, Kumar P, du Toit LC, Govender M, Indermun S, Pillay V. A review of the chemical modification techniques of starch. Carbohydrate Polymers. 2017;157:1226–1236. doi: 10.1016/j.carbpol.2016.09.094. [DOI] [PubMed] [Google Scholar]
- 17.Domnina YA, Yeo Y, Tse JY, Bellas E, Kohane DS. Spray-dried lipid-hyaluronan-polymethacrylate microparticles for drug delivery in the peritoneum. Journal of biomedical materials research. Part A. 2008;87(3):825–31. doi: 10.1002/jbm.a.31741. [DOI] [PubMed] [Google Scholar]
- 18.Abouelmagd SA, Ku YJ, Yeo Y. Low molecular weight chitosan-coated polymeric nanoparticles for sustained and pH-sensitive delivery of paclitaxel. Journal of drug targeting. 2015;23(7–8):725–35. doi: 10.3109/1061186X.2015.1054829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo Y, Ito T, Bellas E, Highley CB, Marini R, Kohane DS. In situ cross-linkable hyaluronan hydrogels containing polymeric nanoparticles for preventing postsurgical adhesions. Annals of surgery. 2007;245(5):819–24. doi: 10.1097/01.sla.0000251519.49405.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ercole F, Whittaker MR, Quinn JF, Davis TP. Cholesterol Modified Self-Assemblies and Their Application to Nanomedicine. Biomacromolecules. 2015;16(7):1886–1914. doi: 10.1021/acs.biomac.5b00550. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Zhao M, Zhao L. Well-defined hydroxyethyl starch-10-hydroxy camptothecin super macromolecule conjugate: cytotoxicity, pharmacodynamics research, tissue distribution test and intravenous injection safety assessment. Drug delivery. 2016:1–9. doi: 10.3109/10717544.2015.1110844. [DOI] [PubMed] [Google Scholar]
- 22.Besheer A, Hause G, Kressler J, Mäder K. Hydrophobically Modified Hydroxyethyl Starch: Synthesis, Characterization, and Aqueous Self-Assembly into Nano-Sized Polymeric Micelles and Vesicles. Biomacromolecules. 2007;8(2):359–367. doi: 10.1021/bm0609487. [DOI] [PubMed] [Google Scholar]
- 23.Dandekar P, Jain R, Stauner T, Loretz B, Koch M, Wenz G, Lehr CM. A hydrophobic starch polymer for nanoparticle-mediated delivery of docetaxel. Macromolecular bioscience. 2012;12(2):184–94. doi: 10.1002/mabi.201100244. [DOI] [PubMed] [Google Scholar]
- 24.Pramod PS, Shah R, Chaphekar S, Balasubramanian N, Jayakannan M. Polysaccharide nano-vesicular multidrug carriers for synergistic killing of cancer cells. Nanoscale. 2014;6(20):11841–55. doi: 10.1039/c4nr03514c. [DOI] [PubMed] [Google Scholar]
- 25.Wasiak I, Kulikowska A, Janczewska M, Michalak M, Cymerman IA, Nagalski A, Kallinger P, Szymanski WW, Ciach T. Dextran Nanoparticle Synthesis and Properties. PLoS ONE. 2016;11(1):e0146237. doi: 10.1371/journal.pone.0146237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senanayake TH, Warren G, Vinogradov SV. Novel Anticancer Polymeric Conjugates of Activated Nucleoside Analogues. Bioconjugate Chemistry. 2011;22(10):1983–1993. doi: 10.1021/bc200173e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, Lolkema MP, Buchholz M, Olive KP, Gress TM, Tuveson DA. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60(6):861–8. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 28.Komar G, Kauhanen S, Liukko K, Seppanen M, Kajander S, Ovaska J, Nuutila P, Minn H. Decreased blood flow with increased metabolic activity: a novel sign of pancreatic tumor aggressiveness. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(17):5511–7. doi: 10.1158/1078-0432.CCR-09-0414. [DOI] [PubMed] [Google Scholar]
- 29.Li M, Tang Z, Zhang D, Sun H, Liu H, Zhang Y, Zhang Y, Chen X. Doxorubicin-loaded polysaccharide nanoparticles suppress the growth of murine colorectal carcinoma inhibit the metastasis of murine mammary carcinoma in rodent models. Biomaterials. 2015;51:161–72. doi: 10.1016/j.biomaterials.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Liu K, Jiang X, Hunziker P. Carbohydrate-based amphiphilic nano delivery systems for cancer therapy. Nanoscale. 2016;8(36):16091–16156. doi: 10.1039/c6nr04489a. [DOI] [PubMed] [Google Scholar]
- 31.Hu H, Li Y, Zhou Q, Ao Y, Yu C, Wan Y, Xu H, Li Z, Yang X. Redox-Sensitive Hydroxyethyl Starch-Doxorubicin Conjugate for Tumor Targeted Drug Delivery. ACS applied materials & interfaces. 2016;8(45):30833–30844. doi: 10.1021/acsami.6b11932. [DOI] [PubMed] [Google Scholar]
- 32.Jungheinrich C, Neff TA. Pharmacokinetics of hydroxyethyl starch. Clinical pharmacokinetics. 2005;44(7):681–699. doi: 10.2165/00003088-200544070-00002. [DOI] [PubMed] [Google Scholar]
- 33.Treib J, Haass A, Pindur G, Seyfert UT, Treib W, Grauer MT, Jung F, Wenzel E, Schimrigk K. HES 200/0.5 is not HES 200/0.5. Influence of the C2/C6 hydroxyethylation ratio of hydroxyethyl starch (HES) on hemorheology, coagulation and elimination kinetics. Thrombosis and haemostasis. 1995;74(6):1452–6. [PubMed] [Google Scholar]
- 34.Goszczyński TM, Filip-Psurska B, Kempińska K, Wietrzyk J, Boratyński J. Hydroxyethyl starch as an effective methotrexate carrier in anticancer therapy. Pharmacology Research & Perspectives. 2014;2(3):e00047. doi: 10.1002/prp2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao K, Li D, Xu W, Ding J, Jiang W, Li M, Wang C, Chen X. Targeted hydroxyethyl starch prodrug for inhibiting the growth and metastasis of prostate cancer. Biomaterials. 2017;116:82–94. doi: 10.1016/j.biomaterials.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Browning DJ. Hydroxychloroquine and Chloroquine Retinopathy. Springer New York; New York, NY: 2014. Pharmacology of Chloroquine and Hydroxychloroquine; pp. 35–63. [Google Scholar]
- 37.Kimura T, Takabatake Y, Takahashi A, Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer research. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 38.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’Antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes & development. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M-C, Wang H-C, Hou Y-C, Tung H-L, Chiu T-J, Shan Y-S. Blockade of autophagy reduces pancreatic cancer stem cell activity and potentiates the tumoricidal effect of gemcitabine. Molecular cancer. 2015;14(1):1–17. doi: 10.1186/s12943-015-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Yip MLR, Shen X, Li H, Hsin L-YC, Labarge S, Heinrich EL, Lee W, Lu J, Vaidehi N. Identification of Anti-Malarial Compounds as Novel Antagonists to Chemokine Receptor CXCR4 in Pancreatic Cancer Cells. PLoS ONE. 2012;7(2):e31004. doi: 10.1371/journal.pone.0031004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debnath BXS, Grande F, Garofalo A, Neamati N. Small Molecule Inhibitors of CXCR4. Theranostics. 2013;3(1):47–75. doi: 10.7150/thno.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Guo L, Zhao H, Zhao J, Weng H, Zhao B. CXCR4 over-expression and survival in cancer: a system review and meta-analysis. Oncotarget. 2015;6(7):5022–40. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krieg A, Riemer JC, Telan LA, Gabbert HE, Knoefel WT. CXCR4-A Prognostic and Clinicopathological Biomarker for Pancreatic Ductal Adenocarcinoma: A Meta-Analysis. PLoS One. 2015;10(6):e0130192. doi: 10.1371/journal.pone.0130192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balkwill FR. The chemokine system and cancer. The Journal of pathology. 2012;226(2):148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 45.Balic A, Sorensen MD, Trabulo SM, Sainz B, Jr, Cioffi M, Vieira CR, Miranda-Lorenzo I, Hidalgo M, Kleeff J, Erkan M, Heeschen C. Chloroquine targets pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling. Mol Cancer Ther. 2014;13(7):1758–71. doi: 10.1158/1535-7163.MCT-13-0948. [DOI] [PubMed] [Google Scholar]
- 46.Arora S, Bhardwaj A, Singh S, Srivastava SK, McClellan S, Nirodi CS, Piazza GA, Grizzle WE, Owen LB, Singh AP. An undesired effect of chemotherapy: gemcitabine promotes pancreatic cancer cell invasiveness through reactive oxygen species-dependent, nuclear factor kappaB- and hypoxia-inducible factor 1alpha-mediated up-regulation of CXCR4. J Biol Chem. 2013;288(29):21197–207. doi: 10.1074/jbc.M113.484576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, De Jesus R, McAllister G, Hoffman GR, Bray K, Lemon L, Lucas J, Fantin VR, Abraham RT, Murphy LO, Nyfeler B. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proceedings of the National Academy of Sciences. 2016;113(1):182–187. doi: 10.1073/pnas.1515617113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen HW, Leonard DA. Chloroquine inhibits cyclization of squalene oxide to lanosterol in mammalian cells. The Journal of biological chemistry. 1984;259(13):8156–62. [PubMed] [Google Scholar]
- 49.King MA, Ganley IG, Flemington V. Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. 2016;35(34):4518–4528. doi: 10.1038/onc.2015.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell death & disease. 2013;4(10):e838. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long J, Zhang Y, Yu X, Yang J, LeBrun DG, Chen C, Yao Q, Li M. Overcoming drug resistance in pancreatic cancer. Expert opinion on therapeutic targets. 2011;15(7):817–28. doi: 10.1517/14728222.2011.566216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, Sarkar FH. Pancreatic cancer: understanding and overcoming chemoresistance. Nature reviews. Gastroenterology & hepatology. 2011;8(1):27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 53.Yu F, Xie Y, Sleightholm RL, Li J, Oupicky D. Polymeric chloroquine as an inhibitor of cancer cell migration and experimental lung metastasis. J Control Release. 2016 doi: 10.1016/j.jconrel.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu F, Xie Y, Wang Y, Peng Z-H, Li J, Oupický D. Chloroquine-Containing HPMA Copolymers as Polymeric Inhibitors of Cancer Cell Migration Mediated by the CXCR4/SDF-1 Chemokine Axis. ACS Macro Lett. 2016:342–345. doi: 10.1021/acsmacrolett.5b00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morimoto N, Obeid R, Yamane S, Winnik FM, Akiyoshi K. Composite nanomaterials by self-assembly and controlled crystallization of poly(2-isopropyl-2-oxazoline)-grafted polysaccharides. Soft Matter. 2009;5(8):1597–1600. [Google Scholar]
- 56.Haebel S, Hejazi M, Frohberg C, Heydenreich M, Ritte G. Mass spectrometric quantification of the relative amounts of C6 and C3 position phosphorylated glucosyl residues in starch. Analytical Biochemistry. 2008;379(1):73–79. doi: 10.1016/j.ab.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Cancer Facts & Figures 2003. American Cancer Society; 2003. [Google Scholar]
- 58.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods in molecular biology (Clifton, NJ) 2005;294:23–9. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 59.Li J, Oupicky D. Effect of biodegradability on CXCR4 antagonism, transfection efficacy and antimetastatic activity of polymeric Plerixafor. Biomaterials. 2014;35(21):5572–5579. doi: 10.1016/j.biomaterials.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adak S, Banerjee R. A green approach for starch modification: Esterification by lipase and novel imidazolium surfactant. Carbohydrate Polymers. 2016;150:359–368. doi: 10.1016/j.carbpol.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 61.Nevozhay D, Budzynska R, Kanska U, Jagiello M, Omar MS, Boratynski J, Opolski A. Antitumor properties and toxicity of dextran-methotrexate conjugates are dependent on the molecular weight of the carrier. Anticancer research. 2006;26(2a):1135–43. [PubMed] [Google Scholar]
- 62.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neel NF, Schutyser E, Sai J, Fan G-H, Richmond A. Chemokine receptor internalization and intracellular trafficking. Cytokine & growth factor reviews. 2005;16(6):637–658. doi: 10.1016/j.cytogfr.2005.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Busillo JM, Benovic JL. Regulation of CXCR4 Signaling. Biochimica et biophysica acta. 2007;1768(4):952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. The Journal of cell biology. 1980;85(3):839–852. doi: 10.1083/jcb.85.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jopling HM, Odell AF, Pellet-Many C, Latham AM, Frankel P, Sivaprasadarao A, Walker JH, Zachary IC, Ponnambalam S. Endosome-to-Plasma Membrane Recycling of VEGFR2 Receptor Tyrosine Kinase Regulates Endothelial Function and Blood Vessel Formation. Cells. 2014;3(2):363–385. doi: 10.3390/cells3020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitov PI, Bundle DR. On the Nature of the Multivalency Effect: A Thermodynamic Model. Journal of the American Chemical Society. 2003;125(52):16271–16284. doi: 10.1021/ja038223n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.