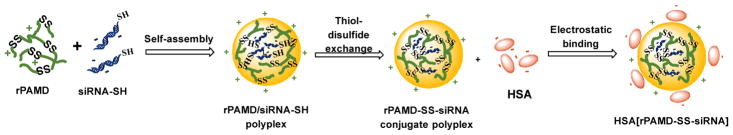
Abstract
This study reports on a simple method to prepare siRNA-polycation conjugate polyplexes by in situ thiol-disulfide exchange reaction. The conjugate polyplexes are prepared using thiol-terminated siRNA and a bioreducible branched polycationic inhibitor of the CXCR4 chemokine receptor (rPAMD). The rPAMD-SS-siRNA conjugate polyplexes exhibit improved colloidal stability and resistance against disassembly with heparin, serum, and physiological salt concentrations when compared with control conventional rPAMD/siRNA polyplexes. Coating the polyplexes with human serum albumin masks the positive surface charge and contributes to the enhanced in vitro gene silencing and improved safety in vivo. The conjugate polyplexes display improved in vivo reporter gene silencing following intravenous injection in tumor-bearing mice. Because the conjugate polyplexes retained the ability of rPAMD to inhibit CXCR4 and restrict cancer cell invasion, the developed systems show promise for future combination antimetastatic siRNA therapies of cancer.
Graphical Abstract
INTRODUCTION
Small interfering RNA (siRNA) has received increasing attention as therapeutic approach for treating various diseases through silencing specific genes.1–3 The use of naked unformulated siRNA, however, is associated with several inherent problems due to the poor cellular permeability, easy degradation by endogenous enzymes and off-target effects.4 To efficiently deliver siRNAs to the target tissues in vivo, a wide variety of non-viral carrier systems, such as cationic lipids and cationic polymers, have been developed to form polyelectrolyte complexes with the oppositely charged siRNA.5, 6 These cationic carriers provide high transfection efficiency in vitro but suffer from low activity in vivo. The cationic carriers may not form sufficiently strong interactions with anionic siRNA simply through electrostatic interactions because of the relatively small size of siRNA.7, 8 Polycations have a tendency to interact with multiple proteins in serum and cause aggregation which can lead to fatal disseminated intravascular coagulation-like condition.9–11 In addition, because of their dynamic nature, polyplexes are susceptible to disassembly by various competing biomacromolecules in the systemic circulation or at a glomerular basement membrane in the kidneys by the action of heparan sulfate.12–18 Many solutions have been proposed to the premature systemic disassembly, including modifications with hydrophobic residues and covalent crosslinking of the particles.12, 19–22 To overcome the limitations of siRNA in effective electrostatic condensation with polycations, polymerized siRNA was used and shown to form stable polyplexes with efficient intracellular translocation and targeted gene silencing in vitro.23, 24 Finally, direct covalent linkage of siRNA to synthetic polymers can significantly improve the performance during systemic siRNA delivery. 25–27
Human serum albumin (HSA) is the most abundant protein in human body. HSA is involved in many important biological functions, such as delivery of nutrients from circulating system to cells and maintenance of osmotic pressure and vascular integrity. As a natural transport protein, HSA is able to bind with various endogenous and exogenous molecules.28, 29 In addition, its target specificity for multiple receptors such as the glycoprotein-60 and a secreted protein acidic and rich in cysteine (SPARC) receptor may allow transport across endothelial barriers by caveolae-medicated transcytosis and improved uptake in tumors.30, 31 Several studies also showed that albumin-based drug delivery systems can accumulate in solid tumors and facilitate tumor targeting due to passive targeting by enhanced permeability and retention (EPR) effect.32, 33 Even though EPR effect remains controversial as a universal rationale for drug delivery in human tumors, it has been successfully observed and been taken advantage clinically in several types of tumors. Liposomal doxorubicin formulation was used for radio-scintigraphy and clear tumor accumulation was seen in the wholebody imaging.34 Another case of EPR effect observed in human tumor is the selective accumulation of Lipiodol in the tumor after intra-arterial infusion. Lipiodol staining has become a routine tumor detection method before hepatic tumor resection.35, 36 Albumin has been also explored in improving the safety and efficacy of polyplexes. Highly positively charged polyplexes are rapidly cleared from the circulation by the mononuclear phagocyte system due to increased protein binding or high affinity interactions with phagocytic cells.37, 38 In addition, polyplexes can be effectively taken up by Kupffer cells as foreign particles. Modifying the surface of the polyplexes with negatively charged albumin may contribute to decreasing the recognition and removal by reticuloendothelial organs, reduce toxicity and improve delivery of therapeutic nucleic acids.29, 39 PEI/DNA polyplexes coated with albumin showed improved gene transfer efficiency with negligible toxicity.40–42 Stable ternary polyplexes were developed by modification of PEI with ligands for albumin binding.43 Interestingly, even unmodified HSA improved the internalization and silencing efficiency of PEI/siRNA polyplexes in cancer cells.44 Unfortunately, very few studies report on the use of albumin-coated polyplexes in vivo.
CXCR4 is one of the most commonly expressed chemokine receptors, which plays a vital role in the process of metastasis in multiple types of cancer.45, 46 Binding with its ligand, stromal cell-derived factor-1 (SDF-1), divergent intracellular signaling transduction pathways are initiated, which can result in a variety of responses such as chemotaxis, cancer cell proliferation, migration and invasion.47 Driven by CXCR4 expressed in the primary tumor cells, the cells metastasize to secondary sites where SDF-1 is highly expressed, such as lung, liver, bone marrow and brain.48 Phosphatidylinositol-3-kinase (PI3K) pathway is activated by CXCR4/SDF-1 axis, leading to activation of protein kinase AKT, which is the key mediator in cancer cell migration and survival.49 Moreover, the secretion of matrix metalloproteinases (MMPs) is increased and then extracellular matrix further degrades after CXCR4 activation, which facilitates the invasion process.50 Mitogenactivated protein kinase (MAPK) pathways is stimulated by CXCR4/SDF-1 axis as well, resulting in increase in cancer cell proliferation and survival.51 Blocking the CXCR4/SDF-1 interaction by antagonizing CXCR4 can inhibit macrophage infiltration, induce tumor growth arrest and prevent metastatic spread.52, 53
We previously reported the synthesis of hyperbranched bioreducible polycationic CXCR4 inhibitors named reducible polymeric AMD3100 (rPAMD) (Scheme 1).54–57 The rPAMD had the ability to inhibit cancer cell invasion as a result of their CXCR4 inhibitory activity, while mediating efficient transfection in vitro. To further advance development of rPAMD for systemic delivery of siRNA, we hypothesized that conjugating thiol-modified siRNA to rPAMD will enhance stability against disassembly and that coating of such conjugates with HSA will decrease the positive surface charge and enhance safety of the formulations (Scheme 2). Here, we have tested a simple thiol-disulfide exchange conjugation strategy to prepare rPAMD-SS-siRNA polyplex conjugates and tested their stability in serum and against heparin exchange. Reporter gene silencing activity of the polyplex conjugates was tested in B16F10-luc mouse melanoma cells to determine how the covalent conjugation and HSA coating affects the silencing efficacy. Finally, the systemic siRNA delivery efficacy of the developed polyplex conjugates was tested in vivo in a syngeneic tumor model.
Scheme 1.
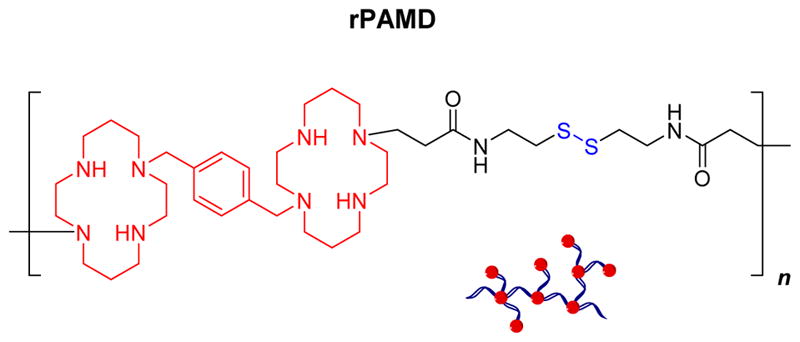
Structure of reducible polymeric AMD3100 (rPAMD).
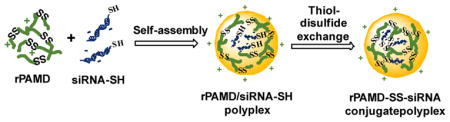
Scheme 2.
Preparation of HSA[rPAMD-SS-siRNA] conjugate polyplexes
RESULTS AND DISCUSSION
Preparation of albumin-coated polyplexes and conjugate polyplexes
Conventional siRNA polyplexes are formed by electrostatic interactions between polycations and the nucleic acid. One of the major challenges of these polyplexes when applied in vivo is their stability against disassembly with competing polyelectrolytes and other charged molecules in serum. Several viable solutions have been developed, including covalent conjugation of siRNA to polymer carriers.26 As part of our long-term goal to develop CXCR4-inhibiting polycations (rPAMD) for siRNA delivery, we have adopted siRNA conjugation as a strategy to enable systemic administration of rPAMD/siRNA polyplexes. We proposed that assembling the hyperbranched bioreducible rPAMD with 5′-thiol functionalized siRNA will lead to facile in situ stabilization of the formed conjugate polyplexes by thiol-disulfide exchange reaction between siRNA-SH and the abundant disulfides in rPAMD (Scheme 2). We proposed that electrostatic interactions would dominate the initial formation of the polyplexes and that the thiol-disulfide exchange will then proceed rapidly within the formed polyplexes because of the local high concentration of the disulfides and thiols within the polyplexes (Scheme 2).
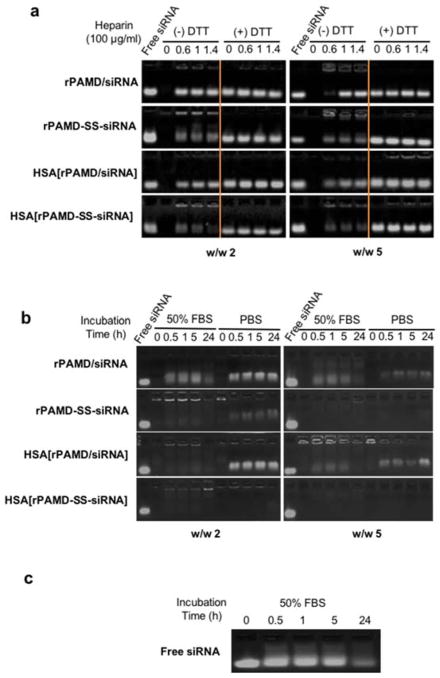
We have prepared both conventional polyplexes (rPAMD/siRNA) and conjugate polyplexes (rPAMD-SS-siRNA) at two different polymer/siRNA w/w ratios (2 and 5). The formation of the conjugate polyplexes was confirmed by assessing stability against polyelectrolyte exchange with heparin (Figure 1a). While no free siRNA was observed at any samples in the absence of heparin, the presence of as little as 60 μg/mL heparin caused complete dissociation of the conventional polyplexes prepared at w/w 2 and in the presence of 100 μg/mL heparin in case of polyplexes prepared at w/w 5. In contrast, the conjugate polyplexes showed strong resistance to heparin-induced disassembly at both w/w ratios, with only a small amount of free siRNA released by the action of heparin. The heparin exchange assay distinguishes between electrostatically and covalently bound siRNA. The small amount of free siRNA released from the conjugate polyplexes suggested that not all siRNA-SH participated in the thiol-disulfide exchange, most likely because of partial siRNA-SH oxidation. It is worth noting that the thiol-disulfide exchange may lead to a degradation of rPAMD. The extent of the disulfide cleavage, however, depends on the ratio of the siRNA-SH thiols to rPAMD disulfides present in the formulation. For example, formulations prepared at w/w ratio of 5 contain ~89 disulfides for each siRNA-SH. We assumed that the molecular weight of siRNA-SH is about 13,600 g/mol and that the molecular weight of the repeating unit per 1 disulfide in rPAMD is 764 g/mol. For a typical rPAMD/siRNA-SH formulation prepared at w/w ratio of 5, that translates to about 89 disulfides per each thiol (5/764) ÷ (1/13,600). In such case, the extent of rPAMD cleavage is negligible since only about 1% of the disulfides can be cleaved by the siRNA-SH.
Figure 1.
Stability of polyplexes and conjugate polyplexes against disassembly. (a) Heparin-induced siRNA release from the polyplexes and conjugate polyplexes prepared at w/w ratio = 2 and 5 in 5 mM HEPES buffer (pH 7.4) and incubated with increasing concentrations of heparin with (+) or without (−) 20 mM DTT. (b) Stability of the polyplexes and conjugate polyplexes in 50% FBS and PBS following incubation for 0–24 h. The release of free siRNA in both (a) and (b) was visualized by agarose gel electrophoresis. (c) Free siRNA degradation profile in 50% FBS.
Successful use of any polyplex stabilization strategy requires that the stabilization is reversible, and that siRNA is released in the cytoplasm. We have incubated the polyplexes with 10 mM DTT to simulate intracellular reducing environment and to confirm reversibility of the conjugate polyplexes. As shown in Figure 1a, disulfide reduction with DTT destabilized both the polyplexes and conjugate polyplexes and resulted in a complete release of siRNA. No differences were observed between polyplexes prepared at the different w/w ratios.
The effect of physiological salt concentration on the integrity of the polyplexes was investigated by incubation in PBS. High salt concentrations can weaken the electrostatic interactions between siRNA and polycation and cause polyplex disassembly. The destabilizing effect of PBS was clearly demonstrated in rPAMD/siRNA polyplexes by the significant amount of released siRNA over the 24 h incubation period (Figure 1b). Polyplexes prepared at w/w 5 were more resistant to salt dissociation than those prepared at w/w 2. As above, the conjugate polyplexes displayed greatly enhanced stability in physiological saline as documented by no significant siRNA release. Combination of high ionic strength with the complex mix of proteins and other charged molecules found in serum can further destabilize the polyplexes and so the stability was further tested in 50% serum (FBS) (Figure 1b). Free siRNA was used as a reference (Figure 1c) for comparison. As expected, conventional polyplexes disassembled rapidly in the presence of serum and significant amount of the siRNA was degraded within 24 h as suggested by the fading band of the free siRNA in the gel. Conjugate polyplexes showed high stability against disassembly, although the appearance of fluorescence in the start well of the conjugate polyplexes at w/w 2 suggested that serum at least partially disrupted the polyplex structure and allowed binding of ethidium bromide to siRNA. We note that serum and tumor interstitial fluid may have some reductive properties that could potentially lead to partial extracellular polyplex destabilization. While the environment in serum in vivo is mostly oxidizing, the presence of low concentrations of reduced cysteine and GSH may partially degrade rPAMD.58 However, multiple published studies by us and other groups with polyplexes based on bioreducible polycations support sufficient stability of the disulfides in plasma circulation.21, 59, 60 Further evidence for serum stability of disulfides comes from extensive experience with drug-antibody conjugates such as Mylotarg in which the drug is attached to the antibody using a linker with sterically hindered disulfide.61
Colloidal characterization
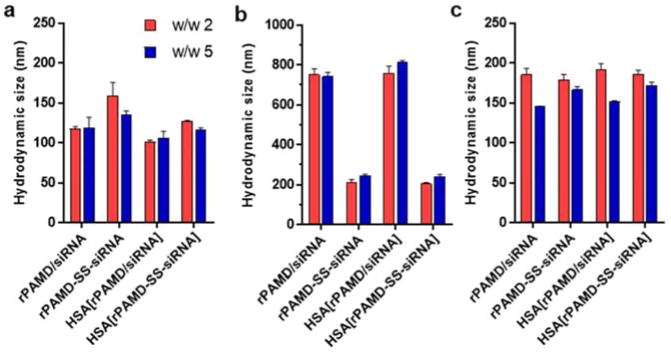
Hydrodynamic size and ζ potential are important parameters that determine the cellular uptake, pharmacokinetics and biodistribution of polyplexes. We have measured the size and zeta potential of polyplexes and conjugate polyplexes prepared in 5 mM HEPES (pH 7.4) at both w/w ratios by dynamic light scattering (Figure 2 and 3). Polyplexes and conjugate polyplexes with both tested w/w ratios showed similar sizes in a range from 120–160 nm (Figure 2a) and positive zeta potentials ranging from 15 to 22 mV (Figure 3). The size of the conjugate polyplexes was slightly larger than the sizes of the conventional polyplexes. We have then assessed colloidal stability of both formulations by measuring the changes in size following a 4 h incubation in PBS (Figure 2b). As expected, the rPAMD/siRNA polyplexes showed poor colloidal stability indicated by rapid aggregation with sizes reaching nearly 800 nm within 4 h. In contrast, the conjugate polyplexes remained relatively stable even in PBS as their sizes increased only to 210–246 nm. When combined with the data in Figure 1b, the improvement in colloidal stability may suggest intra-particle crosslinking of the conjugate polyplexes by the catalytic action of the free thiols in siRNA-SH, similar to previously reported increase in colloidal stability of crosslinked DNA polyplexes.62
Figure 2.
Hydrodynamic size. Polyplexes and conjugate polyplexes were prepared at w/w ratio = 2 and 5 in 5 mM HEPES buffer (pH 7.4) and coated with HSA. Hydrodynamic size was measured by dynamic light scattering. (a) Hydrodynamic size in 5 mM HEPES buffer (pH 7.4); (b) Hydrodynamic size after 4 h incubation in PBS, and (c) Hydrodynamic size after adjustment of pH from 7.4 to 5. All data shown as mean ± SD (n = 3).
Figure 3.
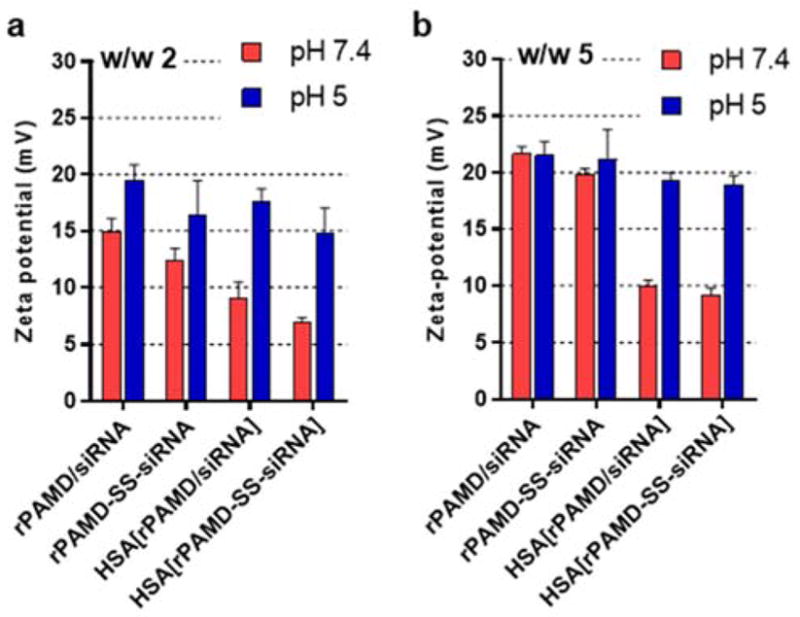
Zeta potential. Polyplexes and conjugate polyplexes were prepared at w/w ratio (a) 2 and (b) 5 in 5 mM HEPES buffer (pH 7.4), coated with HSA, and pH was adjusted to 5. All data shown as mean ± SD (n = 3).
Effect of albumin coating on polyplex properties
Both polyplex and conjugate polyplex showed highly positive surface charge, which made them ill-suited for in vivo use. Covalent conjugation of poly(ethylene glycol) (PEG) is the most common method to shield the positive surface charge of polyplexes. Here, we explored alternative approach to shielding the positive surface charge by adsorption of HSA. Polyplexes and conjugate polyplexes were prepared as described above and then, HSA was added and allowed to adsorb to the surface of the polyplexes (Scheme 2). We first confirmed that HSA has no effect on polyplex stability against disassembly with heparin, serum, or PBS (Figure 1). The polyplexes coated with HSA showed slightly smaller hydrodynamic size in 10 mM HEPES (pH 7.4) (Figure 2a) but no effect of HSA on colloidal stability in PBS was observed (Figure 2b). Importantly, adsorption of the negatively charged HSA significantly decreased the zeta potential of both polyplexes and conjugate polyplexes in solution with pH 7.4 (Figure 3), thus making them better suited for the subsequent in vivo testing.
The HSA coating is strongly dependent on pH and because of the isoelectric point of albumin (4.9), we predicted that the coating would be reversed during endo/lysosomal trafficking of the polyplexes. We have thus prepared the coated polyplexes at pH 7.4 and then decreased the pH to 5 and observed the effect of the pH change on particle size (Figure 2c) and zeta potential (Figure 3). While the pH change had no significant effect of the particle size, we have observed significant increase in the zeta potential for both polyplexes and conjugate polyplexes. The zeta potential increased to the levels comparable to those found for the non-coated polyplexes, thus strongly suggesting that HSA dissociated from the polyplexes. Such behavior may be beneficial for the intracellular trafficking of the polyplexes as it may lead to exposure of unmodified rPAMD/siRNA in the endo/lysosomes and ultimately improve cytoplasmic transport.
Cytotoxicity and transfection activity in vitro
Another important consideration for systemic siRNA delivery is to minimize toxicity of the polyplexes. The use of bioreducible polycations such as rPAMD substantially decreases cytotoxicity when compared with non-reducible polycations.63–65 To ensure that the formation of conjugate polyplexes and HSA coating had no reverse effects on cytotoxicity, we evaluated the effect of polyplexes and conjugate polyplexes on cell viability of a mouse melanoma B16F10-luc cell line (Figure 4). We have determined cell viability at three different rPAMD concentrations: 2, 8 and 15 μg/mL and found no significant adverse effect with any of the formulations as the cell viability remained above 90%.
Figure 4.
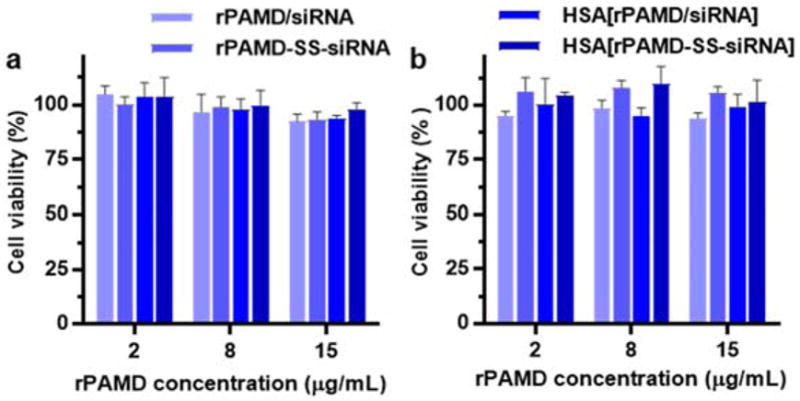
Cytotoxicity of polyplexes and conjugate polyplexes in B16F10-luc. Cells were treated with the formulations prepared at (a) w/w 2 and (b) w/w 5. Data are shown as mean viability ± SD (n = 3).
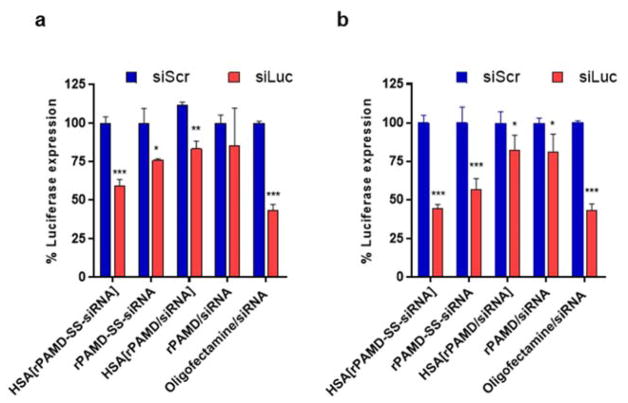
The ability of the polyplexes and conjugate polyplexes to silence expression of the luciferase reporter gene was studied in B16F10-luc cells (Figure 5). Oligofectamine/siRNA complexes were used as a control in all luciferase silencing studies. Treatment with formulations prepared with negative control (siScr) showed no effect on luciferase expression, confirming safety of the studied systems. Overall, the conjugate polyplexes rPAMD-SS-siRNA exhibited better luciferase silencing activity than rPAMD/siRNA polyplexes at both w/w ratios. As expected polyplexes prepared at w/w 5 showed better silencing activity than those at w/w 2. HSA coating had a small, but significant, positive effect on the silencing activity of the conjugate polyplexes but no effect on the activity of the conventional polyplexes. We found that HSA[rPAMD-SS-siRNA] prepared at w/w 5 showed the best luciferase silencing as they decreased the expression to ~45%. The best performing conjugate polyplexes showed activity fully comparable to the Oligofectamine control.
Figure 5.
Transfection activity of polyplexes and conjugate polyplexes. Formulations were prepared either with siLuc or control siScr at (a) w/w ratio 2 or (b) w/w ratio 5. Luciferase silencing was determined by in B16F10-luc cells. Data are shown as mean % luciferase expression ± SD (n = 3). * p < 0.05, ** p < 0.01, *** p < 0.001.
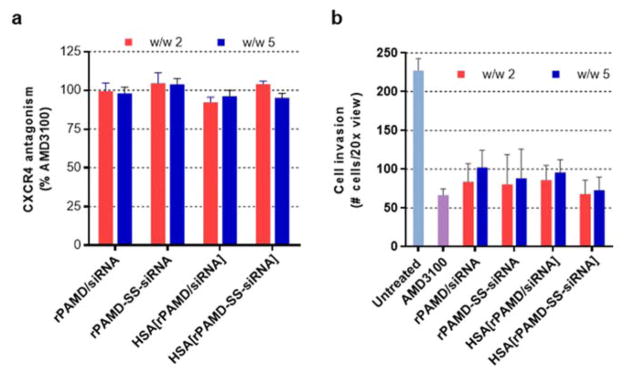
CXCR4 antagonism and inhibition of cancer cell invasion
We have developed rPAMD as antimetastatic inhibitors of the chemokine receptor CXCR4 and it was important to validate that the conjugate polyplexes retained the original CXCR4 inhibitory activity. First, we measured CXCR4 inhibition using CXCR4 redistribution assay by a high-content fluorescence microscopy analysis as described previously.66, 67 The assay is based on the inhibition of SDF-1 triggered endocytosis of EGFP-CXCR4 receptors. This is a phenotypic assay that uses automatic image analysis to quantify the extent of EGFP-CXCR4 internalization into the cells. We have used a small molecule CXCR4 inhibitor AMD3100 as a positive control and expressed the results as % of the AMD3100 activity. As shown in Figure 6a, we have found that all tested formulations used at concentrations used in the siRNA silencing studies showed CXCR4 inhibition indistinguishable from the activity of 300 nM AMD3100. We then investigated if the demonstrated CXCR4 inhibition resulted in the ability of the polyplexes and conjugate polyplexes to inhibit invasion of cancer cells. The important role of CXCR4 in the migration and invasion of multiple types of cancer cells and the ability of CXCR4 inhibitors to prevent the migration is well known.68 We have previously shown that both rPAMD and rPAMD/DNA polyplexes effectively inhibit CXCR4-mediated invasion of multiple cancer cells.57, 69 As shown in Figure 6b, all the polyplexes and conjugate polyplexes showed effective inhibition of cell invasion at w/w ratio either 2 or 5. Based on the quantification of the average numbers of invaded cells, 71% cancer cells were prevented from invading and migrating through the layer of Matrigel by control AMD3100. The polyplexes and conjugate polyplexes achieved similar activity as they inhibited invasion of 60–70% cancer cells. Both of the above experiments together demonstrated that the conjugate polyplexes retain the CXCR4 activity of rPAMD. While the mechanism of the CXCR4 inhibition by the conjugate polyplexes is not fully understood, we believe that excess rPAMD used in the formulations is most likely responsible for the observed activity. Additional CXCR4 inhibition is likely contributed as rPAMD is released and degraded in the cells following intracellular delivery and disassembly of the conjugate polyplexes.
Figure 6.
Inhibition of CXCR4 and cancer cell invasion. (a) CXCR4 inhibition of polyplexes and conjugate polyplexes in U2OS cells. AMD3100 (300nM) was used as a positive control. Data are shown as mean % CXCR4 antagonism relative to AMD3100 ± SD (n = 3). (b) Inhibition of cancer cell invasion. U2OS cells were treated with polyplexes or conjugate polyplexes (rPAMD 2.0 μg/mL) and allowed to invade through Matrigel upon stimulation with SDF-1 for 16 h. AMD3100 (300 nM) was used as positive control. Data are shown as mean number of invaded cells ± SD (n = 3).
Transfection activity in vivo
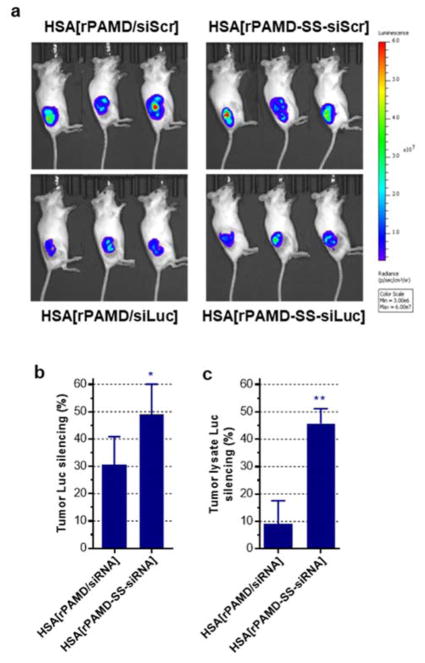
The ability of the conjugate polyplexes to deliver siRNA systemically after intravenous injection was investigated in mice bearing B16F10-luc tumors. We first conducted a dose-finding and preliminary toxicity evaluation of both conventional polyplexes and conjugate polyplexes both with and without HSA coating. We found that coating with HSA improves safety of both formulations and increases the estimated maximum tolerated dose about 2.5-fold (equiv. rPAMD dose 2.5 mg/kg vs. 1 mg/kg). We thus focused our attention on the HSA-coated formulations. The B16F10-luc cells were injected subcutaneously and the tumor growth was followed by IVIS bioluminescence imaging. We have started the treatments with HSA[rPAMD-SS-siRNA] and HSA[rPAMD/siRNA] when the tumors reached 50 mm3. Both conjugate polyplexes and conventional polyplexes were prepared with siLuc and siScr. The control siScr formulations were injected to establish 100% luciferase expression levels. Following the treatments and shortly before animal sacrifice, whole-body luminescence images were taken (Figure 7a) and luciferase expression in the tumor regions quantified. As shown in Figure 7b, the HSA-coated conjugate polyplexes showed increased luciferase silencing (~50%) when compared with the HSA-coated conventional polyplexes (~30%). The tumors were resected, mechanically homogenized, lysed and the luciferase silencing was validated in the tumor lysates as above (Figure 7c). The tumor lysate results confirmed the superior silencing activity of the conjugate polyplexes. The observed improved in vivo activity of the conjugate complexes confirmed the in vitro findings and showed the importance of the polyplex stabilization for systemic delivery. The primary benefit of coating with albumin appeared to be decrease in systemic toxicity most likely due to reduction in the surface charge. Potential effects of the HSA coating on pharmacokinetics and renal clearance remain to be investigated.
Figure 7.
In vivo luciferase gene silencing by HSA-coated conjugate polyplexes. Mice with subcutaneous B16F10-luc tumors were given daily intravenous injections for 4 days of the HSA-coated polyplexes or conjugate polyplexes prepared with either siLuc or control siScr. (a) Bioluminescence images of the tumor-bearing mice before sacrifice. (b) Luciferase silencing determined from the region of interest analysis of the tumor-associated signal in live animals. Luciferase expression in animals treated with siScr was set as 100%. (c) Luciferase silencing in homogenized tumor tissues. Data are expressed as mean ± SEM (n = 7). * p < 0.05, ** p < 0.01.
CONCLUSION
In conclusion, we described a simple approach to prepare polyplexes stabilized against dissociation using in situ thiol-disulfide exchange between siRNA-SH and bioreducible branched polycations. Our results demonstrated not only increased overall stability of the conjugate polyplexes but also enhanced gene silencing activity in vitro and in vivo. Further stabilization of the polyplexes by surface coating with albumin provided additional enhancement of gene silencing in vitro and decrease of toxicity in vivo. Using polycations that inhibit CXCR4, we confirmed that even when conjugated to the siRNA, these polycations retain their inhibitory activity as suggested by strong ability to prevent invasion of cancer cells. Overall, these initial findings provide impetus for further evaluation of the conjugate polyplexes in systemic anticancer and antimetastatic siRNA therapies.
MATERIALS AND METHODS
Materials
AMD3100 base form was purchased from Biochempartner (China). N, N′-cystamine-bisacrylamide (CBA) was obtained from Polysciences, Inc. (Warrington, PA). Ethidium bromide (EtBr) was from Fisher Bioreagents (Fair Lawn, N.J.). Dithiothreitol (DTT) was obtained from Alfa Aesar (Heysham, LA3 2XY, England). Scrambled siRNA (siScr, sense: 5′-AUG AAC GUG AAU UGC UCA AUU-3′), luciferase siRNA (siLuc, sense: 5′-GGA CGA GGA CGA GCA CUU CUU-3′), thiol-modified scrambled siRNA (siScr-SH, sense: 5′-S-S-AUG AAC GUG AAU UGC UCA AUU-3′), thiol-modified luciferase siRNA (siLuc-SH, sense: 5′-S-S-GGA CGA GGA CGA GCA CUU CUU-3′) were custom-synthesized by Dharmacon, GE, and deprotected by tris(2-carboxyethyl) phosphine hydrochloride (TCEP) solution following the manufacturer’s instructions. Dulbecco’s Modified Eagle Medium (DMEM), RPMI-1640 Medium, Dulbecco’s Phosphate Buffered Saline (PBS), and Fetal Bovine Serum (FBS) were from Thermo Scientific (Waltham, MA). Cell culture inserts for 24-well plates with 8.0 μm pores (Translucent PET Membrane, cat# 353097) and BD Matrigel™ Basement Membrane Matrix (cat# 354234) were obtain from BD Biosciences (Billerica, MA). Human SDF-1 was from Shenandoah Biotechnology, Inc. (Warwick, PA). All other reagents were from Fisher Scientific and used as received unless otherwise noted.
Synthesis and characterization of rPAMD
rPAMD was synthesized through Michael-type polyaddition with equal molar ratio of AMD3100 and a reducible bisacrylamide CBA.56 Briefly, AMD3100 (100.4 mg, 0.4 mmol) and CBA (52 mg, 0.4 mmol) were dissolved in methanol/water mixture (2 mL, 7/3 v/v). The reaction was stirred under nitrogen at room temperature for 48 h. Excess AMD3100 (10 mg) was added to react with any residual CBA acrylamide groups. After 6 h of continuous stirring, 1.25 M HCl was added dropwise to the mixture until the pH reached 4. The resulting HCl salt of rPAMD was collected and dialyzed (molecular weight cut-off 3.5 kDa) against acidified water (pH 4) for 3 days before final lyophilization. 1H-NMR was used to confirm the chemical composition of the synthesized rPAMD and AMD3100 content in the polymers. The completion of the reaction was validated by the disappearance of the acrylamide peak of CBA. The weight- and number-average molecular weights, as well as the polydispersity index (PDI, Mw/Mn) was characterized by gel permeation chromatography (GPC) operated in 0.1 M sodium acetate buffer (pH 5.0) using Agilent 1260 Infinity LC system equipped with a miniDAWN TREOS multi-angle light scattering (MALS) detector and a Optilab T-rEX refractive index detector (Wyatt Technology, Santa Barbara, CA). The column used was Tosoh Bioscience TSKgel G3000PWXL-CP eluted at a flow rate of 0.5 mL/min. Results were analyzed using Astra 6.1 software from Wyatt Technology. The molecular weight (Mn) of rPAMD was 12.6 kDa and the polydispersity index was 1.4.
Preparation and characterization of HSA-coated polyplexes
rPAMD/siRNA polyplexes were prepared in 5 mM HEPES buffer (pH 7.4) at predetermined w/w ratios by mixing equal volume of rPAMD and siRNA solutions. The polyplexes were allowed to form at room temperature for 20 min before use. Similarly, rPAMD-SS-siRNA conjugate polyplexes were prepared by mixing of rPAMD and siRNA-SH in 5 mM HEPES buffer (pH 7.4) and kept at room temperature for 2 h before use. To prepare HSA-coated polyplexes, which are named as HSA[rPAMD/siRNA] and HSA[rPAMD-SS-siRNA], HSA solution in 5 mM HEPES buffer (pH 7.4) was added to the prepared polyplexes or conjugate polyplexes at a HSA/rPAMD w/w ratio of 5. The mixture was allowed to stand at room temperature for 20 min before use. Hydrodynamic diameter and ζl-potential were determined by dynamic light scattering using a ZEN3600 Zsizer Nano-ZS (Malvern Instruments Ltd., Massachusetts, United States). Results were expressed as mean ± standard deviation (SD) of three experimental runs for each sample.
Agarose gel electrophoresis
Polyplexes were prepared as above and incubated with increasing concentrations of heparin with or without 10 mM DTT. 18 μL of each sample (siRNA concentration: 20 μg/mL) were loaded onto a 2.5% agarose gel containing 0.5 μg/mL EtBr and run for 30 min under electrophoresis at 75 V in 0.5X Tris/Borate/EDTA (TBE) running buffer. Free siRNA of the same concentration was used as the control. The gel was then visualized under UV.
Cell culture
Human epithelial osteosarcoma U2OS cells that stably express functional EGFP-CXCR4 fusion protein were obtained from Fisher Scientific and cultured in DMEM supplemented with 2 mM L-glutamine, 1% Pen-Strep, 0.5 mg/mL G418 and 10% FBS. Mouse melanoma cell line B16F10 expressing luciferase (B16F10-luc) was purchased from PerkinElmer and maintained in RPMI with 10% FBS.
Cytotoxicity
Cytotoxicity of the polyplexes and conjugate polyplexes was determined by Cell Titer Blue assay. The cells were plated in 96-well plates at a density of 10,000 cells/well (B16F10-luc) and 8,000 cells/well (U2OS). The next day, the culture medium was removed and replaced by 150 μL of serum-free medium containing polyplexes or conjugate polyplexes prepared at w/w 2 and 5 (rPAMD concentration was 2, 8 and 15 μg/mL for B16F10-luc cells and 2 μg/mL for U2OS cells). For B16F10-luc, the medium was replaced with fresh growth medium after 4 h incubation and the cells were cultured for another 48 h. For U2OS, the cells were incubated with polyplexes or conjugate polyplexes for 16 h. Then, the medium in each well was removed and a mixture of 100 μL serum-free media and 20 μL of CellTiter-Blue reagent (CellTiter 96®Aqueous Non-Radioactive Cell Proliferation Assay, Promega) was added. After 1 h incubation, the fluorescence (F) was measured using Synergy 2 Microplate Reader (BioTek, VT) at an excitation wavelength of 560 nm and an emission wavelength of 590 nm. The relative cell viability (%) was calculated as [F]sample/[F]untreated × 100%.
CXCR4 redistribution assay
U2OS cells overexpressing EGFP-CXCR4 were plated in black 96-well plates with optical bottom 24 h before experiment at a density of 8000 cells per well. On the day of the assay, cells were washed twice with 100 μL assay buffer (DMEM supplemented with 2 mM L-Glutamine, 1% FBS, 1% Pen-Strep and 10 mM HEPES). Then different formulations (2.0 μg/mL of polymer) or AMD3100 (300 nM) were added in the assay buffer containing 0.25% DMSO and incubated with cells for 30 min at 37°C. SDF-1 was then added to each well to make the final concentration 10 nM and cells were incubated at 37°C for another 1 h. After incubation, 4% formaldehyde at room temperature for 20 min, followed by 4 times of PBS washing and staining with 1 μM Hoechst 33258 for 30 min. Fluorescent microscopy was conducted using EVOS fluorescence microscope (20x). High content analysis was used to quantify the internalization of CXCR4 receptors using Cellomics ArrayScan VT1 Reader and SpotDetector V3 BioApplication software. CXCR4 inhibitory activity of 300 nM AMD3100 was considered as positive control and set as 100% and cells treated with SDF-1 only was set as 0%. CXCR4 activity of all the tested particles was expressed as mean % inhibition ± SD (n = 3).
Cell invasion assay
Each transwell insert was coated with 40 μL diluted ice-cold Matrigel (1:3 v/v with serum-free medium) and placed in 37°C incubator for 2 h prior to experiment. CXCR4+ U2OS cells were trypsinized and resuspended in serum-free medium containing AMD3100 (300 nM), rPAMD/siRNA, HSA[rPAMD/siRNA], rPAMD-SS-siRNA or HSA[rPAMD-SS-siRNA] (rPAMD 2.0 μg/mL). 100,000 treated cells in 300 μL medium were added to each insert, and 20 nM SDF-1 in serum-free medium was added to the corresponding wells in the companion plate as the chemoattractant. Cells were allowed to invade through the Matrigel layer towards SDF-1 at 37°C for about 18 h. The non-invaded cells were then removed using a cotton swab, and the invaded cells at the bottom of the insert were fixed with 100% methanol and stained with 0.2% Crystal Violet for 10 min. The images were taken by EVOS xl microscope (20x) and the number of invaded cells counted in triplicate.
siRNA transfection in vitro
All transfection experiments were conducted in 48-well plates using cells at their logarithmic growth phase. B16F10-luc cells were seeded at a density of 8,000 cells/well 24 h prior to transfection. Different formulations with 100 nM siRNA were prepared as mentioned above in 250 μL serum-free medium and then added onto cells. After 4 h incubation, the medium was removed and replaced with fresh culture medium. Cells were cultured for another 48 h before measuring luciferase expression. For harvesting cells, 100 μL of 0.5x cell culture lysis buffer (Promega, Madison, WI) was added to the cells after discarding the culture medium and then lysed for at least 20 min at room temperature. The cell lysate was then transferred to 1.5 mL tubes and all the samples were centrifuged at 10,000 g for 10 min at 4°C. To measure the luciferase expression, 100 μL of 0.5 mM luciferin solution was automatically injected into each well containing 20 μL of cell lysate supernatant and the luminescence was integrated over 10 s using GloMax 96 Microplate Luminometer (Promega). Total cellular protein in the cell lysate was measured by the bicinchoninic acid protein assay using calibration curve constructed with standard bovine serum albumin solutions (Pierce, Rockford, IL). Transfection activity was expressed as % luciferase activity of siScr treated groups ± SD of triplicate samples.
siRNA transfection in vivo
All animal experiments were conducted in compliance with the guidelines of the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center. Six-week-old female C57BL/6 albino mice were subcutaneously injected with one million B16F10-luc cells and randomized to different treatment groups to avoid cage effects. When a tumor reached a volume of 50 mm3, the mice (in groups of 7) were injected intravenously (i.v.) with different polyplexes or conjugate polyplexes at siRNA dose of 1 mg/kg body weight daily for 4 days. Dextrose was added into each formulation to make a concentration of 5% before injection. The luminescence signal was detected in tumor before and after treatment using the Perkin Elmer IVIS bioluminescence imaging system. 24 h after the last injection, all mice were sacrificed, and the tumors were harvested and homogenized in 1x Cell Culture Lysis Reagent (Promega, Madison, WI) followed by centrifugation at 10,000 g for 5 min. The luciferase expression was measured as described in 2.7. Transfection activity was expressed as % luciferase expression relative to scrambled siRNA (siScr) control groups ± SD.
Statistical analysis
All the statistical analysis was conducted using GraphPad Prism software. An unpaired t test was used to compare two different groups. When comparing more than two groups, one-way ANOVA with multiple comparison was applied. A P value < 0.05 is considered statistically significant. The n value refers to the number of biological replicates except for hydrodynamic size and zeta-potential measurements (technical replicates).
Acknowledgments
This work was supported in part by the University of Nebraska Medical Center and in part by a grant from the National Institutes of Health (R01 EB015216)
ABBREVIATIONS
- rPAMD
reducible polymeric plerixafor
- HSA
human serum albumin
- siRNA-SH
siRNA modified with thiol
- siScr
scrambled siRNA
- siLuc
luciferase siRNA
Footnotes
NOTES
The authors declare no competing financial interest.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129:107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Li X, Huang L. Non-viral nanocarriers for siRNA delivery in breast cancer. J Control Release. 2014;190:440–450. doi: 10.1016/j.jconrel.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Rev Genetics. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Sen J, Bathula SR, Yang Q, Fittipaldi R, Huang L. Novel cationic lipid that delivers siRNA and enhances therapeutic effect in lung cancer cells. Mol Pharm. 2009;6:696–705. doi: 10.1021/mp800136v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HJ, Ishii A, Miyata K, Lee Y, Wu S, Oba M, Nishiyama N, Kataoka K. Introduction of stearoyl moieties into a biocompatible cationic polyaspartamide derivative, PAsp(DET), with endosomal escaping function for enhanced siRNA-mediated gene knockdown. J Control Release. 2010;145:141–148. doi: 10.1016/j.jconrel.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Kebbekus P, Draper DE, Hagerman P. Persistence length of RNA. Biochemistry. 1995;34:4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- 9.Dobrovolskaia MA, Patri AK, Simak J, Hall JB, Semberova J, De Paoli Lacerda SH, McNeil SE. Nanoparticle size and surface charge determine effects of PAMAM dendrimers on human platelets in vitro. Mol Pharm. 2011;9:382–393. doi: 10.1021/mp200463e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verbaan FJ, Oussoren C, van Dam IM, Takakura Y, Hashida M, Crommelin DJA, Hennink WE, Storm G. The fate of poly(2-dimethyl amino ethyl)methacrylate-based polyplexes after intravenous administration. Int J Pharm. 2001;214:99–101. doi: 10.1016/s0378-5173(00)00642-6. [DOI] [PubMed] [Google Scholar]
- 11.Jones CF, Campbell RA, Brooks AE, Assemi S, Tadjiki S, Thiagarajan G, Mulcock C, Weyrich AS, Brooks BD, Ghandehari H. Cationic PAMAM Dendrimers Aggressively Initiate Blood Clot Formation. ACS nano. 2012;6:9900–9910. doi: 10.1021/nn303472r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. Balancing cationic and hydrophobic content of PEGylated siRNA polyplexes enhances endosome escape, stability, blood circulation time, and bioactivity in vivo. ACS nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naeye B, Deschout H, Caveliers V, Descamps B, Braeckmans K, Vanhove C, Demeester J, Lahoutte T, De Smedt SC, Raemdonck K. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials. 2013;34:2350–2358. doi: 10.1016/j.biomaterials.2012.11.058. [DOI] [PubMed] [Google Scholar]
- 14.Zuckerman JE, Choi CH, Han H, Davis ME. Polycation-siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc Natl Acad Sci USA. 2012;109:3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng M, Librizzi D, Kilic A, Liu Y, Renz H, Merkel OM, Kissel T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly (ethylene glycol) copolymers. Biomaterials. 2012;33:6551–6558. doi: 10.1016/j.biomaterials.2012.05.055. [DOI] [PubMed] [Google Scholar]
- 16.Park J, Park J, Pei Y, Xu J, Yeo Y. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Adv Drug Del Rev. 2016;104:93–109. doi: 10.1016/j.addr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S, Dagnaes-Hansen F, Nielsen EJB, Wengel J, Besenbacher F, Howard KA, Kjems J. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther. 2009;17:1225–1233. doi: 10.1038/mt.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Béhé M, Kissel T. Stability of siRNA polyplexes from poly(ethylenimine) and poly(ethylenimine)-g-poly(ethylene glycol) under in vivo conditions: Effects on pharmacokinetics and biodistribution measured by Fluorescence Fluctuation Spectroscopy and Single Photon Emission Computed Tomography (SPECT) imaging. J Control Release. 2009;138:148–159. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Oupicky D, Ogris M, Howard KA, Dash PR, Ulbrich K, Seymour LW. Importance of lateral and steric stabilization of polyelectrolyte gene delivery vectors for extended systemic circulation. Mol Ther. 2002;5:463–472. doi: 10.1006/mthe.2002.0568. [DOI] [PubMed] [Google Scholar]
- 20.Oupicky D, Parker AL, Seymour LW. Laterally stabilized complexes of DNA with linear reducible polycations: Strategy for triggered intracellular activation of DNA delivery vectors. J Am Chem Soc. 2002;124:8–9. doi: 10.1021/ja016440n. [DOI] [PubMed] [Google Scholar]
- 21.Oupicky D, Carlisle RC, Seymour LW. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001;8:713–724. doi: 10.1038/sj.gt.3301446. [DOI] [PubMed] [Google Scholar]
- 22.Chen G, Wang K, Hu Q, Ding L, Yu F, Zhou Z, Zhou Y, Li J, Sun M, Oupický D. Combining Fluorination and Bioreducibility for Improved siRNA Polyplex Delivery. ACS Appl Mater Interfaces. 2017;9:4457–4466. doi: 10.1021/acsami.6b14184. [DOI] [PubMed] [Google Scholar]
- 23.Lee SY, Huh MS, Lee S, Lee SJ, Chung H, Park JH, Oh YK, Choi K, Kim K, Kwon IC. J Control Release. 2010;141:339–346. doi: 10.1016/j.jconrel.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nature Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 25.Kim JS, Oh MH, Park JY, Park TG, Nam YS. Protein-resistant, reductively dissociable polyplexes for in vivo systemic delivery and tumor-targeting of siRNA. Biomaterials. 2013;34:2370–2379. doi: 10.1016/j.biomaterials.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Lee SH, Kang YY, Jang HE, Mok H. Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics. Adv Drug Del Rev. 2016;104:78–92. doi: 10.1016/j.addr.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Oishi M, Hayama T, Akiyama Y, Takae S, Harada A, Yamasaki Y, Nagatsugi F, Sasaki S, Nagasaki Y, Kataoka K. Supramolecular assemblies for the cytoplasmic delivery of antisense oligodeoxynucleotide: polyion complex (PIC) micelles based on poly(ethylene glycol)-SS-oligodeoxynucleotide conjugate. Biomacromolecules. 2005;6:2449–2454. doi: 10.1021/bm050370l. [DOI] [PubMed] [Google Scholar]
- 28.Paál K, Müller J, Hegedûs L. High affinity binding of paclitaxel to human serum albumin. Eur J Biochem. 2001;268:2187–2191. doi: 10.1046/j.1432-1327.2001.02107.x. [DOI] [PubMed] [Google Scholar]
- 29.Wilson B, Lavanya Y, Priyadarshini S, Ramasamy M, Jenita JL. Albumin nanoparticles for the delivery of gabapentin: preparation, characterization and pharmacodynamic studies. Int J Pharm. 2014;473:73–79. doi: 10.1016/j.ijpharm.2014.05.056. [DOI] [PubMed] [Google Scholar]
- 30.Larsen MT, Kuhlmann M, Hvam ML, Howard KA. Albumin-based drug delivery: harnessing nature to cure disease. Mol Cell Ther. 2016;4:3. doi: 10.1186/s40591-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desai N, Trieu V, Damascelli B, Soon-Shiong P. SPARC Expression Correlates with Tumor Response to Albumin-Bound Paclitaxel in Head and Neck Cancer Patients. Transl Oncol. 2009;2:59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piao L, Li H, Teng L, Yung BC, Sugimoto Y, Brueggemeier RW, Lee RJ. Human serum albumin-coated lipid nanoparticles for delivery of siRNA to breast cancer. Nanomedicine. 2013;9:122–129. doi: 10.1016/j.nano.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller BG, Leuenberger H, Kissel T. Albumin nanospheres as carriers for passive drug targeting: an optimized manufacturing technique. Pharm Res. 1996;13:32–37. doi: 10.1023/a:1016064930502. [DOI] [PubMed] [Google Scholar]
- 34.Harrington KJ, Mohammadtaghi S, Uster PS, Glass D, Peters AM, Vile RG, Stewart JSW. Effective targeting of solid tumors in patients with locally advanced cancers by radiolabeled pegylated liposomes. Clin Cancer Res. 2001;7:243–254. [PubMed] [Google Scholar]
- 35.Konno T, Maeda H, Iwai K, Tashiro S, Maki S, Morinaga T, Mochinaga M, Hiraoka T, Yokoyama I. Effect of arterial administration of high-molecular-weight anticancer agent SMANCS with lipid lymphographic agent on hepatoma: a preliminary report. Eur J Cancer Clin Oncol. 1983;19:1053–1065. doi: 10.1016/0277-5379(83)90028-7. [DOI] [PubMed] [Google Scholar]
- 36.Maki S, Konno T, Maeda H. Image enhancement in computerized tomography for sensitive diagnosis of liver cancer and semiquantitation of tumor selective drug targeting with oily contrast medium. Cancer. 1985;56:751–757. doi: 10.1002/1097-0142(19850815)56:4<751::aid-cncr2820560409>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Byrne JD, Napier ME, DeSimone JM. More effective nanomedicines through particle design. Small. 2011;7:1919–1931. doi: 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gustafson HH, Holt-Casper D, Grainger DW, Ghandehari H. Nanoparticle uptake: The phagocyte problem. Nano Today. 2015;10:487–510. doi: 10.1016/j.nantod.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bienk K, Hvam ML, Pakula MM, Dagnæs-Hansen F, Wengel J, Malle BM, Kragh-Hansen U, Cameron J, Bukrinski JT, Howard KA. An albumin-mediated cholesterol design-based strategy for tuning siRNA pharmacokinetics and gene silencing. J Control Release. 2016;232:143–151. doi: 10.1016/j.jconrel.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Syga MI, Nicolì E, Kohler E, Shastri VP. Albumin Incorporation in Polyethylenimine–DNA Polyplexes Influences Transfection Efficiency. Biomacromolecules. 2016;17:200–207. doi: 10.1021/acs.biomac.5b01308. [DOI] [PubMed] [Google Scholar]
- 41.Carrabino S, Di Gioia S, Copreni E, Conese M. Serum albumin enhances polyethylenimine-mediated gene delivery to human respiratory epithelial cells. J Gene Med. 2005;7:1555–1564. doi: 10.1002/jgm.799. [DOI] [PubMed] [Google Scholar]
- 42.Rhaese S, von Briesen H, Rübsamen-Waigmann H, Kreuter J, Langer K. Human serum albumin–polyethylenimine nanoparticles for gene delivery. J Control Release. 2003;92:199–208. doi: 10.1016/s0168-3659(03)00302-x. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura Y, Sato H, Nobori T, Matsumoto H, Toyama S, Shuno T, Kishimura A, Mori T, Katayama Y. J Biomat Sci, Pol Ed. 2017. Modification of ligands for serum albumin on polyethyleneimine to stabilize polyplexes in gene delivery; pp. 1–23. [DOI] [PubMed] [Google Scholar]
- 44.Nicoli E, Syga MI, Bosetti M, Shastri VP. Enhanced Gene Silencing through Human Serum Albumin-Mediated Delivery of Polyethylenimine-siRNA Polyplexes. PLoS One. 2015;10:e0122581. doi: 10.1371/journal.pone.0122581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, Guo L, Zhao H, Zhao J, Weng H, Zhao B. CXCR4 over-expression and survival in cancer: A system review and meta-analysis. Oncotarget. 2015;6:5022–5040. doi: 10.18632/oncotarget.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleightholm RL, Neilsen BK, Li J, Steele MM, Singh RK, Hollingsworth MA, Oupicky D. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol Ther. 2017;179:158–170. doi: 10.1016/j.pharmthera.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 47.Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nature Rev Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 48.Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 49.Luker KE, Luker GD. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 50.Fernandis AZ, Prasad A, Band H, Klosel R, Ganju RK. Regulation of CXCR4-mediated chemotaxis and chemoinvasion of breast cancer cells. Oncogene. 2004;23:157–167. doi: 10.1038/sj.onc.1206910. [DOI] [PubMed] [Google Scholar]
- 51.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 52.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Xie Y, Oupický D. Potential of CXCR4/CXCL12 chemokine axis in cancer drug delivery. Curr Pharmacol Rep. 2016;2:1–10. doi: 10.1007/s40495-015-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Li J, Oupický D. Polymeric plerixafor: effect of PEGylation on CXCR4 antagonism, cancer cell invasion, and DNA transfection. Pharm Res. 2014;31:3538–3548. doi: 10.1007/s11095-014-1440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Kumar S, Rachagani S, Sajja BR, Xie Y, Hang Y, Jain M, Li J, Boska MD, Batra SK, Oupicky D. Polyplex-mediated inhibition of chemokine receptor CXCR4 and chromatin-remodeling enzyme NCOA3 impedes pancreatic cancer progression and metastasis. Biomaterials. 2016;101:108–120. doi: 10.1016/j.biomaterials.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J, Oupicky D. Effect of biodegradability on CXCR4 antagonism, transfection efficacy and antimetastatic activity of polymeric Plerixafor. Biomaterials. 2014;35:5572–5579. doi: 10.1016/j.biomaterials.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Zhu Y, Hazeldine ST, Li C, Oupicky D. Dual-function CXCR4 antagonist polyplexes to deliver gene therapy and inhibit cancer cell invasion. Angew Chem Int Ed Engl. 2012;51:8740–8743. doi: 10.1002/anie.201203463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oupický D, Li J. Bioreducible Polycations in Nucleic Acid Delivery: Past, Present, and Future Trends. Macromol Biosci. 2014;14:908–922. doi: 10.1002/mabi.201400061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou QH, Miller DL, Carlisle RC, Seymour LW, Oupicky D. Ultrasound-enhanced transfection activity of HPMA-stabilized DNA polyplexes with prolonged plasma circulation. J Control Release. 2005;106:416–427. doi: 10.1016/j.jconrel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Neu M, Germershaus O, Mao S, Voigt KH, Behe M, Kissel T. Crosslinked nanocarriers based upon poly(ethylene imine) for systemic plasmid delivery: In vitro characterization and in vivo studies in mice. J Control Release. 2007;118:370–380. doi: 10.1016/j.jconrel.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Wu AM, Senter PD. Arming antibodies: prospects and challenges for immunoconjugates. Nat Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 62.Trubetskoy VS, Loomis A, Slattum PM, Hagstrom JE, Budker VG, Wolff JA. Caged DNA does not aggregate in high ionic strength solutions. Bioconjug Chem. 1999;10:624–628. doi: 10.1021/bc9801530. [DOI] [PubMed] [Google Scholar]
- 63.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JF. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjug Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 64.Li J, Manickam DS, Chen J, Oupicky D. Effect of cell membrane thiols and reduction-triggered disassembly on transfection activity of bioreducible polyplexes. Eur J Pharm Sci. 2012;46:173–180. doi: 10.1016/j.ejps.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu C, Li J, Zhu Y, Chen J, Oupický D. Opposing influence of intracellular and membrane thiols on the toxicity of reducible polycations. Biomaterials. 2013;34:8843–8850. doi: 10.1016/j.biomaterials.2013.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie Y, Wehrkamp CJ, Li J, Wang Y, Wang Y, Mott JL, Oupicky D. Delivery of miR-200c Mimic with Poly(amido amine) CXCR4 Antagonists for Combined Inhibition of Cholangiocarcinoma Cell Invasiveness. Mol Pharm. 2016;13:1073–1080. doi: 10.1021/acs.molpharmaceut.5b00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, Lepadatu AM, Zhu Y, Ciobanu M, Wang Y, Asaftei SC, Oupický D. Examination of Structure–Activity Relationship of Viologen-Based Dendrimers as CXCR4 Antagonists and Gene Carriers. Bioconjug Chem. 2014;25:907–917. doi: 10.1021/bc500191q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balkwill FR. The chemokine system and cancer. J Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Li J, Chen Y, Oupicky D. Balancing polymer hydrophobicity for ligand presentation and siRNA delivery in dual function CXCR4 inhibiting polyplexes. Biomater Sci. 2015;3:1114–1123. doi: 10.1039/C5BM00003C. [DOI] [PMC free article] [PubMed] [Google Scholar]