Abstract
Liquid chromatography-selected reaction monitoring mass spectrometry (LC-SRM) is not only a proven tool for clinical chemistry, but also a versatile method to enhance the capability to quantify biomarkers for tumor biology research. As the treatment of cancer continues to evolve, the ability to assess multiple biomarkers to assign cancer phenotypes based on the genetic background and the signaling of the individual tumor becomes paramount to our ability to treat the patient. In breast cancer, the American Society of Clinical Oncology (ASCO) has defined biomarkers for patient assessment to guide selection of therapy: estrogen receptor, progesterone receptor, and the HER2/Neu receptor tyrosine kinase; therefore, these proteins were selected for LC-SRM assay development. Detailed molecular characterization of these proteins is necessary for patient treatment, so expression and phosphorylation assays have been developed and applied. In addition, other LC-SRM assays were developed to further evaluate tumor biology (e.g. Ki-67 for proliferation and vimentin for tumor aggressiveness related to the epithelial-to-mesenchymal transition). These measurements combined with biomarkers for tissue quality and histological content are implemented in a three tier multiplexed assay platform, which is translated from cell line models into frozen tumor tissues banked from breast cancer patients.
Keywords: Breast Cancer Biomarkers, Liquid chromatography-selected reaction monitoring mass spectrometry, Quantitative Proteomics, Estrogen Receptor, HER2
Introduction
Three tumor markers are classified as level of evidence I in breast cancer by the American Society of Clinical Oncology (ASCO). Estrogen receptor alpha (ERα)1,2 and progesterone receptors alpha and beta (PRα and PRβ)3,4 are nuclear hormone receptors; human epidermal growth factor receptor 2 (HER2)5,6 is a protein tyrosine kinase. Because of their significance in regulating growth and survival of breast tumor cells, patients are assigned molecular classifications and prescribed targeted therapies based on biomarker measurements of ERα, PR, and HER2. Estrogen receptor modulators (e.g. tamoxifen), aromatase inhibitors (e.g. anastrozole), estrogen receptor antagonists (e.g. fulvestrant), and mTOR inhibitors (e.g. everolimus) are used in treatment for hormone receptor-positive breast cancers; while monoclonal antibody therapeutics (e.g. trastuzumab and pertuzumab) or small molecule tyrosine kinase inhibitors (e.g. Lapatinib), are used to treat HER2-positive breast cancers.7 The measurement of these biomarkers drives the selection of treatment regimens in breast cancer patients; improvements in the ability to evaluate these molecular endpoints and gather additional information may enable physicians to enhance the efficacy of therapy and continue to refine personalized treatment strategies for breast cancer patients.8
In particular, the signaling and oncogene addiction in each tumor will define the best opportunities for treatment with targeted therapies. Therefore, the quantification of post-translational modification levels in addition to expression may prove to be valuable, because ERα,9,10 PR, and HER2 are all regulated by phosphorylation. The combination of expression and phosphorylation levels may provide additional diagnostic and prognostic information and elucidate mechanisms of drug resistance.11 Serine 167 (S167) is an estrogen-independent phosphorylation site of ERα, as a result of mTOR/S6K1 and MAPK/RSK pathway activation.12 S294 is a ligand-dependent phosphorylation site, which is modified by CDK7 when ERα is bound to ligands like estradiol and tamoxifen.13 S162 PRβ increases in response to progesterone and can be induced by the cyclin A-Cdk2 complex.14–16 Tyrosine 877 (Y877) is a HER2 phosphorylation site located in the activation loop of the kinase domain regulating kinase activity17 and mediated by TNFα through c-Src activation.18 Y735 and Y1248 are another two phosphosites of HER2, which mediate protein-protein interactions with Src homology 2 domain-containing-transforming protein C1 (SHC).19 These phosphorylation sites and others could be important to evaluate in breast cancer patients to define the most appropriate modes of treatment. Immunohistochemical assessments of Ki-67 20–21 and vimentin 22–23 are also widely used for monitoring proliferation and aggressiveness via the epithelial-to-mesenchymal transition in breast cancer, because they serve as prognostic indicators. Vimentin overexpression contributes to the aggressive phenotype of tumors and poor prognosis especially in triple-negative breast cancer (TNBC), a highly heterogeneous disease, which doesn’t express ERα and PR and has no overexpression of HER2.24,25 In clinical studies, techniques such as enzyme-linked immunosorbent assays (ELISA) and immunohistochemical analysis (IHC) are used extensively to evaluate protein expression levels for ER, PR, and HER2 to classify breast cancer subtypes.26–29 However, all these techniques need specific antibodies for each target and assay multiplexing is limited. In addition, complex characterization of multiple phosphorylations can be challenging if antibody reagents for each analyte are not of sufficient quality. Liquid chromatography-selected reaction monitoring mass spectrometry (LC-SRM)30–33 complements antibody-based measurement techniques and has emerged as an important technique for clinical biomarker measurements. The technique is very versatile due to its capacity for multiplexing, which enables LC-SRM to be applied to study tumor biology in situ.34,35
For biomarker quantification in breast cancer, Narumi et al.36 have performed large-scale phosphoproteome quantification by using IMAC coupled with iTRAQ and verified 15 phosphopeptides by selected reaction monitoring mass spectrometry (SRM) using stable isotope-labeled standard peptides in breast cancer tissue samples. Song et al.37 have presented a gel-assisted digestion method to identify breast cancer biomarker candidates by using label-free quantification. Collodoro et al.38 have identified six biomarkers in MCF-7/BOS cells exposed to 17β-estradiol and found three related in a network centered on estrogen receptor and AKT1. Schoenherr et al.39 have implemented a peptide-based immuno-SRM assay for quantification of ER and HER2 expression in both cell lysates and human tissues with good agreement with the ER/HER2 status measured by clinical assays. Sprung et al.40 compared the reproducibility of SRM assay-based peptide quantification between formalin-fixed, paraffin-embedded (FFPE) samples and frozen carcinoma tissues and evaluated the precision of HER2 quantification as a feasibility study of SRM application in FFPE tissues. Additional publications from Catenacci et al. illustrated the utility of HER2 quantification in FFPE specimens and correlated well with clinical assays.41,42 Domanski et al.43 developed a phosphatase-based phosphopeptide quantitation (PPQ)-SRM method for determination of phosphorylation stoichiometry of HER2, ERα, RAF, and ERK1 in peptides spiked into E. coli digest. However, these previous applications usually focus on a single peptide assay for determining protein expression levels. Britton et al.9,10 and Held et al.14 have mapped ERα phosphosites by LC-MS/MS and set up selected reaction monitoring methods for quantification of ERα expression and phosphorylation in estradiol and/or EGF stimulated breast cancer cell lines.
A multiplexed LC-SRM assay for quantifying expression and phosphorylation of ERα, PR, and HER2 is reported here using protein enrichment to enable measurement of multiple molecular endpoints. In addition to quantification of protein expression, nine phosphorylation sites (pS154, pS167, pS294, pY537, pS554, and pS559 from ERα; pS162 from PRβ; pY735 and pY877 from HER2) have been included in the assay platform. These assays have been applied to study the changes associated with drug treatment in breast cancer cell lines and implemented on the pre-treatment frozen tissue specimens from breast cancer patients that are either HER2+/− or ER/PR+/− (n = 20 total with 5 per group). Expression levels from a group of high abundance proteins including ALBU, ACTB, HSP7C, ATPB, LDHA, LDHB, G3P, LMNA, ENOA, HBA, and HBB are monitored as tissue quality control biomarkers and compared with tissue evaluation by a pathologist. Based on different expression levels of the targets, either SDS-PAGE or immunoprecipitation (IP) is used for enrichment.
The ability to examine the protein expression as well as the phosphorylation status in these biomarkers with quantitative mass spectrometry in a single sample could improve our understanding of their role in the development and progression of breast cancer. The quantitative evaluation of protein expression and phosphorylation status of these targets in tumor tissues could further refine the classification of molecular subtypes and support clinical decision-making including selection of targeted therapeutics. The integration of tissue quality control measurements, quantification of proliferation biomarkers, and ER/PR/HER2 evaluation could improve patient assessment and open novel avenues for patient classification, prognosis, and selection of therapy in breast cancer.
Materials and Methods
Cell Lines and Reagents
The human MCF7 (ER+),44 T47D (PR+),45 BT474 (HER2+),46 and HCC38 (triple negative)47 breast cancer cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA) for assay development against the proteins listed in parentheses and grown under the recommended conditions: 37°C, 5% CO2, Dulbecco’s modified Eagle’s (D5796, Sigma, St. Louis, MO) or RPMI-1640 culture media (R8758, Sigma, St. Louis, MO), supplemented with 10% fetal bovine serum (SH30910.03, Thermo Scientific) and 1% penicillin/streptomycin (P0781 Sigma, St. Louis, MO). β-Estradiol (E2758), NaF (S7920), Na2MoO4 (243655), acetic acid (A6283), Coomassie Brilliant Blue G (B5133), ammonium bicarbonate (A6141), and tris(2-carboxyethyl)phosphine hydrochloride (C4706) were purchased from Sigma-Aldrich (St. Louis, MO). Iodoacetamide (RPN6302V) was purchased from GE Healthcare (Chicago, IL). Human EGF (100–15) was purchased from PeproTech (Rocky Hill, NJ). Lapatinib ditosylate (S1028) was purchased from Selleck (Houston, TX). Promegestone or R5020 (NLP004005MG) was purchased from PerkinElmer (Boston, MA). HPLC grade solvents (water and acetonitrile) were supplied by Burdick and Jackson (Honeywell, Muskegon, MI). Antibodies were purchased from Santa Cruz Biotechnology (ERα antibody F-10), Cell Signaling Technologies (PR antibody 6A1, 3172), and Calbiochem (HER2 antibody OP15). Pierce Protein A/G UltraLink Resin (53132) was purchased from Thermo Scientific. Sequencing grade modified trypsin (V5111) was purchased from Promega (Madison, WI). ERα (DYC5715-2), PR (DYC5415-2) and HER2 (DYC1129-2) DuoSet IC ELISA kits and other related reagents were purchased from R&D Systems (Minneapolis, MN).
Assay Development
Peptides were selected for each of the three biomarker proteins based on known biology as well as publically available MS data. Peptides were infused to optimize transitions, and methods were developed using Skyline.48 Peptide sequences, transitions, and spiked amounts of stable isotope-labeled standard for each experiment have been listed in Supplementary Table 1. For other proteins, existing LC-MS/MS data and LC-SRM screening were used for peptide selection.35 For tissue QC proteins, the following peptides were selected VDPVNFK (HBA), LLVVYPWTQR (HBB), LVNEVTEFAK (ALBU), SADTLWDIQK (LDH-B), AGFAGDDAPR (ACTB), GTLDPVEK (HSP7C), IGLFGGAGVGK (ATPB), VTLTSEEEAR (LDHA), SGAQASSTPLSPTR (LMNA), VGVNGFGR (G3P), and YISPDQLADLYK (ENOA). In addition, SLYASSPGGVYATR and QDVDNASLAR were used to monitor vimentin; TSDTETEPSK and SPPPESVDTPTSTK were chosen for Ki-67 quantification. Peptide standards were synthesized using standard FMOC chemistry at the 25 μmol scale, purified, and characterized as previously described.35 Amino acid analysis was performed by the Protein Chemistry Laboratory at Texas A & M University in order to quantify the amount of each peptide in stock solutions (1 mM), which were then stored at −80ºC until use.
Evaluation of ER/PR/HER2 Assays
Assays for the peptides from ERα, PR, and HER2 were combined and scheduled into one LC-SRM method, which included 27 peptides and 9 phosphopeptides. Assays were evaluated with calibration curves in both buffer and matrix to determine characteristics, including linear range, lower limit of quantification (LOQ), accuracy, and precision. Additional details are included in Supplementary Methods with example data in Supplementary Figure 1 and Supplementary Tables S2–S5.
Cell Treatment and Cell Lysis
Cells were grown to 80% confluence and then plated in 15 cm tissue culture dishes at a density of 9.5 x 106 cells each, and allowed to attach overnight. On the following day, after removing the media, the cells were washed three times with PBS. Phenol red-free Dulbecco’s modified Eagle’s medium (17-205-CV, Cellgro, Corning Life Sciences) supplemented with 10% charcoal stripped FCS (12676-029, Gibco, ThermoFisher) was used to “serum-starve” the cells for 24 hours prior to treatment by removing lipids, hormones, and selected growth factors. MCF7 and BT474 cell lines were treated with Estradiol (10 nM for 30 min.), EGF (50 ng/mL for 10 min.), Estradiol plus EGF (10 nM and 50 ng/mL for 20 min), and Promegestone (100 nM for 1 hr.). Without starvation, both cell lines were treated with Lapatinib (1 μM for 1 hr.) and either harvested or rescued by treatment by EGF (50 ng/mL for 10 min.) and then harvested.
After treatment, the cells were washed twice with room temperature PBS and harvested on ice using a cell scraper and collected into 1.0 mL of cold cell lysis buffer [100 mM NaCl, 20 mM Tris (pH 7.5), 0.5% NP40, 100 mM NaF, 10 mM Na3VO4, 50 mM Na2MoO4, 10 μL/mL phosphatase inhibitor cocktail 3 (P0044, Sigma-Aldrich, St. Louis, MO), 320 nM okadaic acid (Cat# O7885, Sigma-Aldrich), 100 U/mL deoxyribonuclease I (Cat# D2821, Sigma-Aldrich), and 1 tablet/10 mL Roche mini-complete protease inhibitor cocktail (Cat# 11836153001, Roche Applied Science, Indianapolis, IN). The cell lysate was kept on ice and sonicated twice using 10 second pulses. Debris was pelleted by centrifugation at 18,000 × g for 15 minutes at 4°C. Supernatants were decanted and an aliquot (5 μL) was retained for protein quantification using Bradford Assay (23236, Thermo Scientific). A second aliquot (20 μL or 2% of the total volume) is set aside for western blotting to monitor the pre-IP protein expression levels and enable qualitative assessment of immunoprecipitation (IP) efficiency (see example data in Supplementary Figure S2); Western blots were performed for speed and to provide a method orthogonal to the LC-SRM analysis.
Breast Cancer Tissue Sample Processing and Evaluation
Pre-treatment frozen tissue specimens from breast cancer patients (18< age < 90) were obtained from the Tissue Core at H. Lee Moffitt Cancer Center; additional details are provided in Supplementary Table S6. Protocols have been approved by as non-human subjects research (MCC 17239 and MCC 17241) prior to tissue release. One aliquot of 30 mg tissue was requested from patients classified in other of the following four groups ER+/PR+, ER−/PR−, HER2+, or HER2− (n = 5 per group); each group should be mutually exclusive (e.g. no ER+/HER2+ patients, etc.). Hematoxylin and eosin (H&E) stained slides were obtained for each tissue aliquot for review by a board-certified pathologist (SB, AM) and digitized with a high-throughput slide scanning instrument (ScanScope XT, Aperio); example images are appended to the Supplementary Figures (additional views available upon request). The frozen tumor tissues were powdered (BioPulverizer, BioSpec, Bartlesville, OK) and transferred into a 2 ml microcentrifuge tube using liquid N2. After evaporation of the nitrogen, 1.5 ml of cold lysis buffer was added. After sonication and centrifugation, 10 μg of tissue homogenate was fractionated by SDS-PAGE to enable quantification of the tissue QC biomarkers. A second aliquot (50 μg) of the tissue homogenate was used for SDS-PAGE to isolate bands for HER2 and Ki-67. The remaining bulk of the tissue homogenate (~1 mg) was used for multiplexed IP-LC-SRM.
Immunoprecipitation and SDS-PAGE Fractionation
The overall workflow has been shown in Supplementary Figure S3. For tissue QC, 10 μg of total protein were prepared by rapid gel electrophoresis to enable detergent removal and buffer exchange. For HER2 and Ki-67 quantification, 50 μg of total protein was separated using 4–12% Criterion XT gels (Bio-Rad) and gel bands were selected for in-gel digestion. For IP experiments in cell lines, 2 mg total protein was used. ERα antibody (10 μL with approximately 2 μg of antibody) was added to the cell lysate or tissue homogenate on ice. After 5 minutes, PR antibody (15 μL containing 20 μg) was added. Then, immune complexes were formed at 4°C for 1.5 hours. Protein A/G UltraLink Resin (20 μL of 50% slurry) was washed three times with 1 mL 0.5 % NP-40 and resuspended in 0.5 % NP-40. The washed resin was added to the lysate and incubated overnight at 4°C. The beads were then pelleted at 1,000 × g for 30 seconds. The supernatant was removed, and an aliquot (2% or 20 μL) is retained for western blotting (to compare pre-IP and post-IP levels to assess capture efficiency). Proteins eluted from the beads are separated by SDS-PAGE prior to in-gel digestion, as previously published; 34,35 additional details are provided in the Supplementary Methods.
Nano-LC-SRM Analysis
LC-SRM data were acquired using a nanoLC system (U3000, Dionex, Sunnyvale, CA) interface with a triple quadrupole mass spectrometer (Vantage, Thermo, San Jose, CA), equipped with a nanoelectrospray ion source operated in positive ion mode. The methods have been previously described,34,35 and additional details are available in the Supplementary Methods. Data were acquired by XCalibur 2.1 TSQ Vantage software versions and analyzed with Skyline.48 Raw data was imported to Skyline for automated peak selection and manual verification. Fragmentation patterns (transition ranking) and retention time of proteolytic and synthetic peptides were also compared. In order to eliminate potential interferences, the allowable variation in transition ratios was set to 5%; transitions for the proteolytic peptides from each biological sample observed with higher levels of variation than the SIS peptide were removed prior to quantification. If the signal of the proteolytic peptides was poor, ion signals were manually evaluation and only transitions with sufficient peak intensity and quality were selected for quantification. Peak areas were exported to text files (csv) for analysis (Microsoft Excel).
ELISA
Cell lysates and tissue homogenates were diluted in order to the working range of each ELISA kit. Values included 1:50 for the samples used for ERα and PR ELISA measurements and 1:100, 1:500, and 1:1,500 for HER2 measurements in cell lines and patient samples HER2 (to accommodate the wide range of expression levels observed for HER2). Standards and cell line samples were measured in duplicate and patient specimens were measured in triplicate using a microplate reader (Versa Max, Molecular Devices, Sunnyvale, CA, USA). Based on the manufacturer’s recommendations, the standard curves were generated for ERα (15.6 – 500 pg/ml), PR (12.5 – 800 pg/ml), and HER2 (62.5 – 4000 pg/ml) and used to quantify those biomarkers in each sample.
Results and Discussion
LC-SRM Assay Development and Validation for ERα, PR, and HER2
Breast cancer cell lines MCF7 (ERα+), T47D (PR+), and BT474 (HER2+) have been selected specifically for LC-SRM assay development for ERα, PR, and HER2. To provide additional detail, MCF7 44 and T47D 45 are luminal A subtype cell lines and considered positive for ER and PR, but negative for HER2. BT474 46 is a luminal B subtype cell line, which is considered positive for ER and HER2. Based on the protein expression level, co-immunoprecipitation was used to enrich ERα and PR by adding the two antibodies sequentially with a 5 minutes’ waiting period. SDS-PAGE separation is used for further purification of both HER2 from the total protein and ERα/PR from the immunoprecipitation. General workflow with the region of the excised gel bands is illustrated in Supplementary Figure S3. Supplementary Table S1 shows all the peptide sequences monitored for ERα, PR, and HER2 expression and phosphorylation with the loading amounts of stable isotope-labeled standard peptides on the column.
In order to determine whether matrix effects have a significant adverse effect on the ionization efficiency and intensity of the peptides in the assay, response curves in the presence of background peptides expected to be present during analysis of endogenous ER, PR and HER2 from breast cancer cells have been built. Triple negative HCC38 cell lysates have been used for assay validation including dynamic range, LOD and LOQ determination in calibration curves in matrix to give a background. As an example, Supplementary Figure S1 shows the LC-SRM calibration curves in buffer (A) and matrix (B) for the ERα unmodified peptide, LASTNDKGSMAMESAK, and calibration curves in buffer (C) and matrix (D) for the corresponding phosphopeptide, LApSTNDKGSMAMESAK. Twelve data points are used for calculation in each calibration curve from 0 (buffer or matrix blank) to 1 pmol. The equations of the calibration curves are also provided in Supplementary Figure S1. The slopes of the equations are similar between the calibration curves in buffer and matrix in the same peptide, while the intercepts indicate the difference between the calibration curves from background interference. All the calibration curves have a linear correlation within the ranges from LLOQ to ~1 pmol. Detailed results with CV and accuracy from normal calibration curve in buffer and matrix for the ER unmodified peptide, LASTNDKGSMAMESAK, and phosphopeptide, LApSTNDKGSMAMESAK, have been shown in Supplementary Table S2. LLOQ values were determined by the lowest amount injected into the LC-MS system to give a CV ≤ 20%. For the unmodified peptide, LASTNDKGSMAMESAK, even though it was detectable from 14 amol in buffer, the CV (44.6%) exceeds the quality criterion (15%). LLOQ was determined to be 276 amol based on the CV (1.8%). In matrix, the LLOQ was determined to be 138 amol with CV 8.7%. For the phosphopeptide, LApSTNDKGSMAMESAK, LLOQ values were determined to be 16 amol (CV 14.8%) in buffer and 164 fmol (CV 4.4%) in matrix. A summary of LLOQ values in fmol for ER, PR, and HER2 peptides in buffer and matrix is shown in Supplementary Table S3.
Mixed cell lysates from MCF7, T47D, and BT474 with EGF and estradiol stimulation to increase phosphorylation levels have been used for assay evaluation, including accuracy and precision. Accuracy was measured by spike and recovery experiments for each target peptide in the assay platform. Supplementary Table S4 shows the results of accuracy for each target peptides in three spiking levels (100 fmol, 500 fmol, and 1000 fmol). Most peptides were quantified in the acceptable accuracy range from 80% to 120%.
The reproducibility of LC-SRM performance alone as well as the reproducibility of the entire sample preparation (co-immunoprecipitation of ERα and PR, SDS-PAGE separation, in-gel tryptic digestion, and LC-SRM) was evaluated. Supplementary Table S5 shows the CV values for the precision from these tests.
LC-SRM Assay Application in Drug Treated Cell Lines
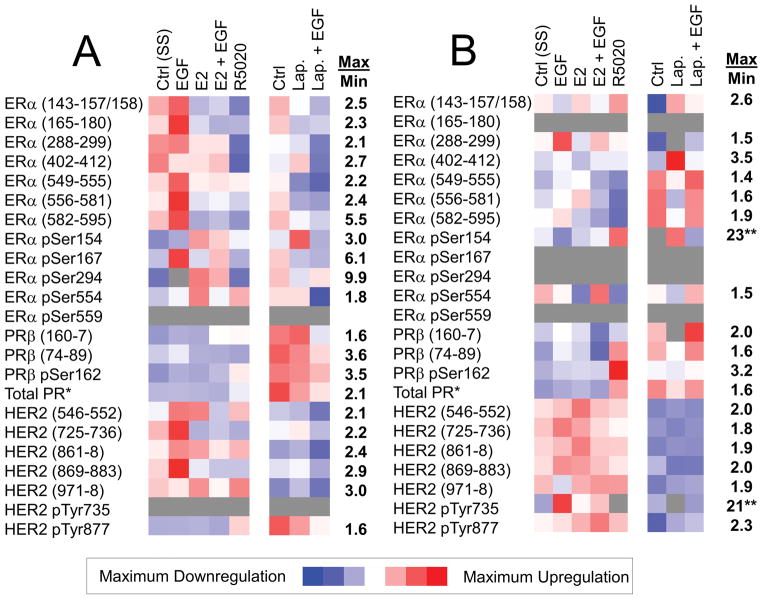
The breast cancer cell lines, MCF7 and BT474, have been used to study the different levels of expression and phosphorylation of ER, PR, and HER2 under a series of drug treatments, including EGF or/and ligand (estradiol) stimulation, promegestone (R5020) stimulation, tyrosine kinase inhibitor (Lapatinib) inhibition, and Lapatinib inhibition followed by EGF stimulation (Figure 1). Minimum quantitative estimates (fmol/mg total protein) of each target peptide from these three proteins have been calculated using SIS peptides, as shown in Supplementary Table S7. The phosphorylation stoichiometry (%) has also been calculated for each treatment and shown in Supplementary Table S8. Phosphorylation of ERα pSer167 in MCF7 cells increased with estradiol (1.6-fold increase) and EGF (2.8-fold increase), when compared to control, which is consistent with expectation. Ligand dependent phosphorylation of ERα pSer294 in MCF7 was clearly increased by estradiol (4.0-fold increase), which is consistent with known biology. Similarly, phosphorylation of PR pSer162 increased in both MCF7 and BT474 cells after stimulation with promegestone (R5020). Phosphorylation of HER2 pTyr735 in BT474 cells is increases with EGF treatment, is not observed after treatment with Lapatinib, but can be rescued by co-treatment with EGF and Lapatinib. Therefore, this ER/PR/HER2 assay can not only measure the expression and phosphorylation levels, but also confirm these aspects of known biology, which have been selected as positive controls in these cell line models. With quantification of both unmodified and phosphopeptides, the phosphorylation stoichiometry can be determined to further assess patient specimens. This additional level of detail could ultimately assist in selection of the targeted therapy appropriate for each patient.
Figure 1. LC-SRM Analysis of Expression and Phosphorylation during Ligand Stimulation and Drug Treatment Experiments in Cell Lines.
Measurements in ER+ MCF7 cells (A) and in HER2+ BT474 cells (B) are shown. The heat maps show relative quantification from lowest (blue) to highest (red); grey indicates missing data. A general scale bar for the level of change is presented; the numerical values (right-hand columns) indicate the maximum to minimum ratio for the relative quantification data presented in the heat maps. *In both panels, total PR is quantified using a peptide common to both isoforms (PRα 725–31, PRβ 561–7). **In the BT474 drug treatment experiments, LLOQ was used for missing minimum values to estimate the maximum-to-minimum ratio.
Tissue Evaluation by H&E and LC-SRM Quantification of Tissue Quality Control Biomarkers
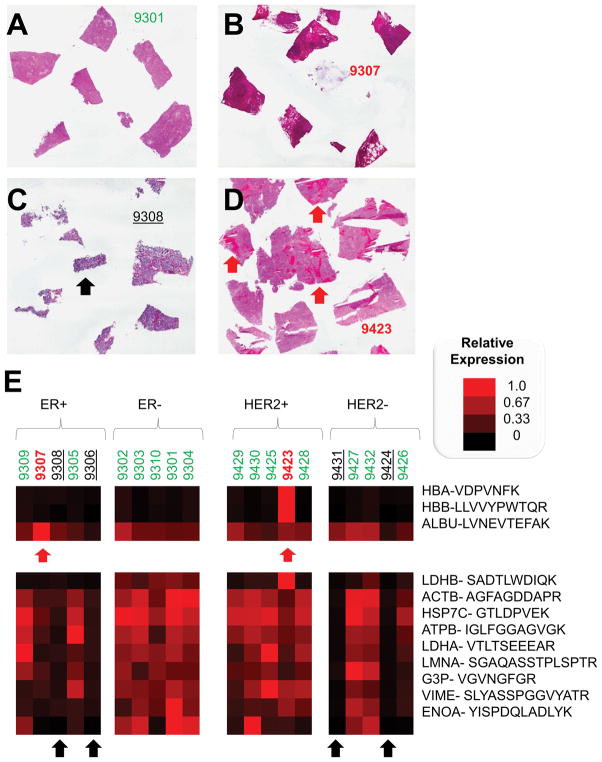
Pre-treatment frozen tissues have been used for LC-SRM assays in 20 patients (5 and 5 for ER+/−, as well as 5 and 5 for HER2+/−) using 30 mg wet weight of frozen tumor tissue per patient. Images of H&E stained slides for each tissue have been obtained for tissue evaluation (see Supplementary Figures). Figure 2(A–D) shows examples of the H&E stained images of tumor tissues to illustrate an example of a highly cellular tumor as well as some of the challenges with biomarker assessment in bulk tumors: ascites, necrosis, and blood.
Figure 2. Representative Hematoxylin and Eosin Stained Tissue Sections for Good Quality and Challenging Tissue Specimens and Relative Quantification of Tissue Quality Control Proteins.
Frozen sections from tumors have been selected to illustrate high cellularity tumor tissue (A) as well as samples with ascites (B), necrosis (C), and blood (D). In panel E, albumin and hemoglobin are measured with LC-SRM to define tissues that may include ascites or whole blood, which contributes significantly to the total protein amount derived from the tumor tissue. Samples with high levels of these proteins, which could disrupt the biomarker measurements, are marked with red arrows. Cellular markers include lactate dehydrogenases, actin, heat shock cognate 71, nuclear lamin, GAPDH, vimentin, and enolase. Samples with low amounts of these cellular proteins are marked with black arrows to indicate samples that may lack sufficient cellularity for IP-LC-SRM of bulk tumor tissue. The scale bar for relative expression is shown (right).
Total protein recovery from these tissues based on Bradford assays averaged 6% of the total wet weight of tumor. High abundance proteins, including the cellularity biomarkers and blood components, have also been selected and quantified to evaluate the tissue quality by LC-SRM. Normalized expression levels of selected cellularity biomarkers, such as ACTB, G3P and LMNA, were used to examine the 20 patient samples and biological replicates of 2 QC cell lines. As shown in Figure 2E and Supplementary Figure S4, the expression of cellularity biomarkers is lower in patients 9308 and 9306 (ER+/PR+) and patients 9431 and 9424 (HER2−) especially for LMNA, which is a structural component of cell nuclei, which may serve a similar function to counting nuclei in evaluation of H&E stained sections. Normalized expression of blood cell components such as ALBU, HBA, HBB have been shown in Figure 2E and Supplementary Figure S4. Patient 9423 had a much higher amount of HBA and HBB than others which indicates that this sample contained less total intracellular protein, which impacted quantification of HER2 (in fmol/mg total protein). Therefore, the evaluation of tissue QC using high abundance proteins helped determine potential confounding factors for tissue analysis and complemented the pathologist’s (SB) review of the H&E stained slides.
LC-SRM Assay Implementation in Breast Cancer Patient tissues
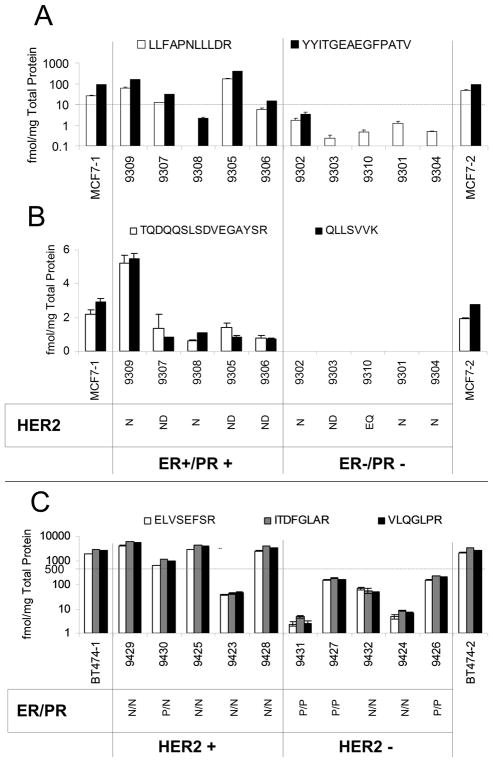
ERα/PR/HER2 assays have been applied to the pre-treatment frozen tissue specimens from breast cancer patients that are either ER/PR+/− or HER2+/− (n = 5 for each of the four groups). At the beginning and end of each batch, biological replicates of the cell lines, MCF7 (ER+) and BT474 (HER2+), are used for quality control of sample processing and mass spectrometry performance. For molecular subtype classification, 10 fmol ER per mg total protein ER and 500 fmol HER2 per mg total protein in tissue homogenate were used as cut-offs for determination of positive or negative biomarker status.27,49 As shown in Figure 3A and 3C as well as Supplementary Table S9, ERα and HER2 expression quantified by LC-SRM did match well with clinical categories except patient 9308 (ER+) and 9423 (HER2+), which have been indicated to be of poor quality upon pathology review and tissue QC by LC-SRM analysis. PR expression and phosphorylation has also been determined, as shown in Figure 3B and Supplementary Table S9. Patient 9309 has a higher expression level of PR than others (~5 fmol/mg total protein). In addition, ER phosphorylation levels have been measured as shown in Table 8 and Supplementary Figure S5. Patients 9309 and 9305 from the ER+ group had higher levels of ERα pSer294. As ERα Ser294 phosphorylation depends upon ligand (estradiol) stimulation, these two patients could potentially have better response to anti-hormone therapy.
Figure 3. LC-SRM Assay Implementation in Breast Tumors.
IP-LC-SRM quantification of ER expression (A) and PR expression (B) in tumor tissues and the positive control, MCF7 cells, indicates separation between the ER+ and ER− patients with the exception of 9306, which had low cellularity (see Figure 2E). The dotted line indicates a cutoff value of 10 fmol/mg, which can be used for molecular classification. SDS-PAGE-LC-SRM HER2 expression measurements (C) in another set of tumor tissues and the BT474 positive control also indicate the ability to differentiate between groups using a cutoff value of 500 fmol/mg total protein for molecular classification, with the exception of sample 9423, which had a high degree of blood infiltration (see Figure 2E). Both ER and HER2 cutoffs are based on existing clinical values for intact protein measurements. Clinical calls based on IHC and FISH have been added to the bottom of panels B and C to indicate the status for the biomarkers not assessed by LC-SRM in these samples using the following designations: positive (P), negative (N), no data (ND), and equivocal (EQ).
As shown in Supplementary Figure S6, vimentin has been quantified in these tumors also using two peptides: SLYASSPGGVYATR and QDVDNASLAR. Because both cell lines are epithelial; little vimentin is observed, when compared to the tumor tissues. Across the sample group, vimentin expression levels differ by more than a factor of 20, but we have not explored a cutoff to establish epithelial tumors versus mesenchymal tumors. Vimentin expression levels and vimentin/actin ratios could not be correlated with stromal content from the H&E stained sections (see Supplementary Figure S6), and the tumor samples from triple negative breast cancer patients, which have a higher percentage of mesenchymal-type tumors, did show high levels of vimentin expression in certain cases (e.g. 9301, 9304, and 9432); some HER2+ tumors, but none of the ER+ tumors in this small sample set had vimentin expression levels as high.
Comparison of LC-SRM and ELISA Quantification of ERα, PR, and HER2 Expression
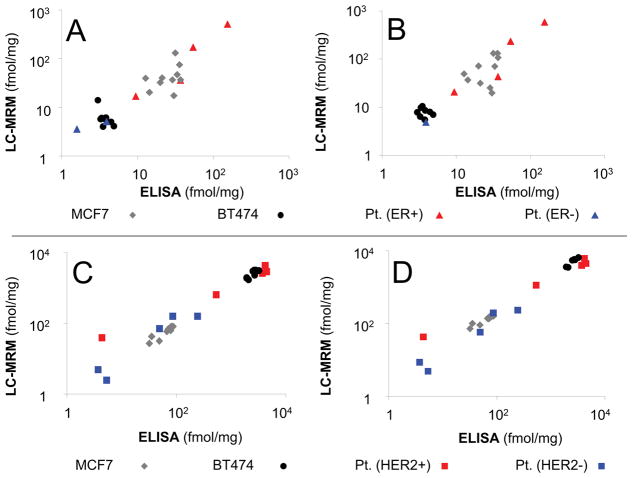
ELISA and LC-SRM quantification of ERα, PR, and HER2 correlate well (R2 > 0.9) in patient samples and cell line models, as shown in Figure 4 and Supplementary Figures S7–S9 and Supplementary Table S10. Immunogens for ELISA contain a.a. 1–116 for ERα, a.a. 1–189 for PR, and a.a. 23–652 for HER2; strong agreement is observed between methods, even though the only peptides selected for LC-SRM that share sequence with the immunogens are the PR peptide, VLSPLMSR, (a.a. 160–167, selected for monitoring phosphorylation), and the HER2 peptide, VLQGLPR (a.a. 546–552).
Figure 4. Comparison LC-SRM and ELISA for ER and HER2 Quantification in Cell Lines and Breast Tumor Tissues.
LC-SRM data for 2 peptides from ER, LLFAPNLLLDR (A) and YYITGEAEGFPATV (B), are compared against ELISA quantification for two control cell lines and 10 patient samples. The sample coding (upper right) applies to both panels. LC-SRM data for 2 peptides from HER2, ELVSEFSR (C) and ITDFGLAR (D), are compared against ELISA for the same 2 cell lines, but 10 different patient samples. The sample coding (lower right) applies to both panels. Additional data are shown in Supplementary Figures S7–9.
Concluding Remarks
Here, a multi-tier platform for quantification of breast cancer biomarkers has been established by using SDS-PAGE and IP for protein enrichment prior to LC-SRM analysis. High abundance proteins, which are used as tissue QC biomarkers (including ALBU, ACTB, HSP7C, ATPB, LDHA, LDHB, G3P, LMNA, ENOA, HBA, and HBB), have been quantified from 10 μg aliquots of cell lysate/tissue homogenate after SDS-PAGE for buffer exchange and in-gel digestion. THE EMT biomarker, vimentin, could also be quantified from this tissue QC sample. The proliferation biomarker, Ki-67, has very high molecular weight and is likely incompatible with SDS-PAGE protein fractionation; despite identification of 2 candidate peptides and subsequent assay development, this protein could not be quantitatively assessed in tumors in these experiments. Expression levels for highly amplified HER2 can be quantified from this same minimal amount of protein (10 μg), but to examine multiple peptides and phosphorylation sites a larger amount of total protein (50 μg) was fully separated in gel (or immunoprecipitated). IP-LC-SRM for HER2 would have less interference and lower amounts of background, but HER2 expression is approximately 1,000-fold higher than ER and PR leading to a significant challenge in multiplexing the IP step for all of these proteins. ERα and PR require IP enrichment from 1–2 mg total protein to generate a sufficient sample for LC-SRM analysis. The resulting LC-SRM assays have been applied to drug treatments in cell lines, compared with ELISA, and applied to frozen tumor specimens to illustrate the potential for their clinical translation. Continuing development and integration of specific biomarker measurements with background proteome characterization can further improve patient evaluation, similar to the use of the HER2/CEP17 ratio in FISH (see Supplementary Figure S10). Specifically, additional protein biomarkers of EMT (e.g. N-cadherin and E-cadherin) could provide additional utility for patient assessment.
In general, this multi-tier platform with integration of tissue quality control biomarkers’ measurements, quantification of proliferation and EMT biomarkers, and ER/PR/HER2 evaluation including protein expression and phosphorylation, could improve patient assessment and open novel avenues for patient classification, prognosis, and selection of targeted therapy in breast cancer. Similar approaches could be used to examine protein biomarkers relevant to other cancers.
Supplementary Material
Significance.
Different enrichment strategies have been combined with quantitative proteomics to evaluate protein biomarkers, including expression and phosphorylation levels for estrogen receptor, progesterone receptor, and the HER2 receptor tyrosine kinase, in breast tumor tissue.
Acknowledgments
This project was supported by a sponsored research agreement from Proteome Sciences plc. The Tissue Core, Collaborative Data Services Core, Analytic Microscopy, and Proteomics Core are supported by the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant and the Moffitt Foundation. LC-SRM instruments were purchased with grants from the Bankhead-Coley Cancer Research program of the Florida Dept. of Health (06BS-02-9614, 09BE-04). Figures were exported to TIFF using GNU Image Manipulation Program. Virginia Johnson and Larry Dangott at the Texas A&M Protein Chemistry Laboratory completed amino acid analysis. We would like to thank Michelle Fournier, Marek Wloch, and Vonetta Williams for assistance with tissue and data procurement and Mark Lloyd and Joseph Johnson for slide scanning. Jason Held, Gary Scott, and Chris Benz were exceptionally helpful in discussion of the project.
Footnotes
Conflict of Interest Statement
This work was conducted as an academic-industrial partnership funded by Proteome Sciences plc.
References
- 1.Le Romancer M, Poulard C, Cohen P, Sentis S, Renoir JM, Corbo Cracking the estrogen receptor’s posttranslational code in breast tumors. L Endocr Rev. 2011;32:597–622. doi: 10.1210/er.2010-0016. [DOI] [PubMed] [Google Scholar]
- 2.Anbalagan M, Huderson B, Murphy L, Rowan BG. Post-translational modifications of nuclear receptors and human disease. Nucl Recept Signal. 2012;10:e001. doi: 10.1621/nrs.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemm DL, Sherman L, Boonyaratanakornkit V, Schrader WT, Weigel NL, Edwards DP. Differential hormone-dependent phosphorylation of progesterone receptor A and B forms revealed by a phosphoserine site-specific monoclonal antibody. Mol Endocrinol. 2000;14:52–65. doi: 10.1210/mend.14.1.0413. [DOI] [PubMed] [Google Scholar]
- 4.Hopp TA, Weiss HL, Hilsenbeck SG, Cui Y, Allred DC, Horwitz KB, Fuqua SA. Breast cancer patients with progesterone receptor PR-A-rich tumors have poorer disease-free survival rates. Clin Cancer Res. 2004;10:2751–60. doi: 10.1158/1078-0432.ccr-03-0141. [DOI] [PubMed] [Google Scholar]
- 5.Colomer R, Montero S, Lluch A, Ojeda B, Barnadas A, Casado A, Massutí B, Cortés-Funes H, Lloveras B. Circulating HER2 extracellular domain and resistance to chemotherapy in advanced breast cancer. Clin Cancer Res. 2000;6:2356–62. [PubMed] [Google Scholar]
- 6.Asgeirsson KS, Agrawal A, Allen C, Hitch A, Ellis IO, Chapman C, Cheung KL, Robertson JF. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007;9:R75. doi: 10.1186/bcr1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SH, Jeon J, Kim SI. Personalized medicine in breast cancer: a systematic review. J Breast Cancer. 2012;15:265–72. doi: 10.4048/jbc.2012.15.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartlett JM, Rea D, Rimm DL. Quantification of hormone receptors to guide adjuvant therapy choice in early breast cancer: better methods required for improved utility. J Clin Oncol. 2011;29:3715–6. doi: 10.1200/JCO.2011.37.3704. [DOI] [PubMed] [Google Scholar]
- 9.Britton DJ, Scott GK, Schilling B, Atsriku C, Held JM, Gibson BW, Benz CC, Baldwin MA. A novel serine phosphorylation site detected in the N-terminal domain of estrogen receptor isolated from human breast cancer cells. J Am Soc Mass Spectrom. 2008;19:729–40. doi: 10.1016/j.jasms.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atsriku C, Britton DJ, Held JM, Schilling B, Scott GK, Gibson BW, Benz CC, Baldwin MA. Systematic mapping of posttranslational modifications in human estrogen receptor-alpha with emphasis on novel phosphorylation sites. Mol Cell Proteomics. 2009;8:467–80. doi: 10.1074/mcp.M800282-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi N, Iwamoto T, Gonzalez-Angulo AM, Ferrer-Lozano J, Lluch A, Niikura N, Bartholomeusz C, Nakamura S, Hortobagyi GN, Ueno NT. Prognostic impact of phosphorylated HER-2 in HER-2+ primary breast cancer. Oncologist. 2011;16:956–65. doi: 10.1634/theoncologist.2010-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamnik RL, Holz MK. mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor alpha serine 167 phosphorylation. FEBS Lett. 2010;584:124–8. doi: 10.1016/j.febslet.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Held JM, Britton DJ, Scott GK, Lee EL, Schilling B, Baldwin MA, Gibson BW, Benz CC. Ligand binding promotes CDK-dependent phosphorylation of ER-alpha on hinge serine 294 but inhibits ligand-independent phosphorylation of serine 305. Mol Cancer Res. 2012;10:1120–32. doi: 10.1158/1541-7786.MCR-12-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Beck CA, Poletti A, Edwards DP, Weigel NL. Identification of phosphorylation sites unique to the B form of human progesterone receptor. In vitro phosphorylation by casein kinase II. J Biol Chem. 1994;269:31034–40. [PubMed] [Google Scholar]
- 15.Knotts TA, Orkiszewski RS, Cook RG, Edwards DP, Weigel NL. Identification of a phosphorylation site in the hinge region of the human progesterone receptor and additional amino-terminal phosphorylation sites. J Biol Chem. 2001;276:8475–83. doi: 10.1074/jbc.M009805200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Beck CA, Poletti A, Clement JP, 4th, Prendergast P, Yip TT, Hutchens TW, Edwards DP, Weigel NL. Phosphorylation of human progesterone receptor by cyclin-dependent kinase 2 on three sites that are authentic basal phosphorylation sites in vivo. Mol Endocrinol. 1997;11:823–32. doi: 10.1210/mend.11.6.0006. [DOI] [PubMed] [Google Scholar]
- 17.Bose R, Molina H, Patterson AS, Bitok JK, Periaswamy B, Bader JS, Pandey A, Cole PA. Phosphoproteomic analysis of HER2/neu signaling and inhibition. Proc Natl Acad Sci U S A. 2006;103:9773–8. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivas MA, Tkach M, Beguelin W, Proietti CJ, Rosemblit C, Charreau EH, Elizalde PV, Schillaci R. Transactivation of ErbB-2 induced by tumor necrosis factor alpha promotes NF-kappaB activation and breast cancer cell proliferation. Breast Cancer Res Treat. 2010;122:111–24. doi: 10.1007/s10549-009-0546-3. [DOI] [PubMed] [Google Scholar]
- 19.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100012. 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11:174–83. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 21.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, McShane L, Paik S, Penault-Llorca F, Prudkin L, Regan M, Salter J, Sotiriou C, Smith IE, Viale G, Zujewski JA, Hayes DF International Ki-67 in Breast Cancer Working Group. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–64. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassanein M, Huiart L, Bourdon V, Rabayrol L, Geneix J, Nogues C, Peyrat JP, Gesta P, Meynard P, Dreyfus H, Petrot D, Lidereau R, Noguchi T, Eisinger F, Extra JM, Viens P, Jacquemier J, Sobol H. Prediction of BRCA1 Germ-Line Mutation Status in Patients with Breast Cancer Using Histoprognosis Grade, MS110, Lys27H3, Vimentin, and KI67. Pathobiology. 2013;80:219–227. doi: 10.1159/000339432. [DOI] [PubMed] [Google Scholar]
- 23.Karihtala P, Auvinen P, Kauppila S, Haapasaari KM, Jukkola-Vuorinen A, Soini Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138:81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita N, Tokunaga E, Kitao H, Hisamatsu Y, Taketani K, Akiyoshi S, Okada S, Aishima S, Morita M, Maehara Y. Vimentin as a poor prognostic factor for triple-negative breast cancer. J Cancer Res Clin Oncol. 2013;139:739–46. doi: 10.1007/s00432-013-1376-6. [DOI] [PubMed] [Google Scholar]
- 25.Karihtala P, Auvinen P, Kauppila S, Haapasaari KM, Jukkola-Vuorinen A, Soini Y. Vimentin, zeb1 and Sip1 are up-regulated in triple-negative and basal-like breast cancers: association with an aggressive tumour phenotype. Breast Cancer Res Treat. 2013;138:81–90. doi: 10.1007/s10549-013-2442-0. [DOI] [PubMed] [Google Scholar]
- 26.Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, Tang P. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 27.Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguilar Z, Wilson C, Rong HM, Bauerfeind I, Felber M, Wang HJ, Beryt M, Seshadri R, Hepp H, Slamon DJ. Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst. 2003;95:142–53. doi: 10.1093/jnci/95.2.142. [DOI] [PubMed] [Google Scholar]
- 28.Colomer R, Montero S, Lluch A, Ojeda B, Barnadas A, Casado A, Massutí B, Cortés-Funes H, Lloveras B. Circulating HER2 extracellular domain and resistance to chemotherapy in advanced breast cancer. Clin Cancer Res. 2000;6:2356–62. [PubMed] [Google Scholar]
- 29.Sun CF, Wu TL, Tsao KC, Wu JT. Development of two ELISA for estrogen and progesterone receptor with sufficient sensitivity for fine needle aspirate and core biopsy. J Clin Lab Anal. 2001;15:138–43. doi: 10.1002/jcla.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoofnagle AN, Becker JO, Wener MH, Heinecke JW. Quantification of thyroglobulin, a low-abundance serum protein, by immunoaffinity peptide enrichment and tandem mass spectrometry. Clin Chem. 2008;54:1796–804. doi: 10.1373/clinchem.2008.109652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012;58:777–81. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–73. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 33.Gerber SA, Kettenbach AN, Rush J, Gygi SP. The absolute quantification strategy: application to phosphorylation profiling of human separase serine 1126. Methods Mol Biol. 2007;359:71–86. doi: 10.1007/978-1-59745-255-7_5. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Gruidl M, Remily-Wood E, Liu RZ, Eschrich S, Lloyd M, Nasir A, Bui MM, Huang E, Shibata D, Yeatman T, Koomen JM. Quantification of beta-catenin signaling components in colon cancer cell lines, tissue sections, and microdissected tumor cells using reaction monitoring mass spectrometry. J Proteome Res. 2010;9:4215–27. doi: 10.1021/pr1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remily-Wood ER, Liu RZ, Xiang Y, Chen Y, Thomas CE, Rajyaguru N, Kaufman LM, Ochoa JE, Hazlehurst L, Pinilla-Ibarz J, Lancet J, Zhang G, Haura E, Shibata D, Yeatman T, Smalley KS, Dalton WS, Huang E, Scott E, Bloom GC, Eschrich SA, Koomen JM. A database of reaction monitoring mass spectrometry assays for elucidating therapeutic response in cancer. Proteomics Clin Appl. 2011;5:383–96. doi: 10.1002/prca.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narumi R, Murakami T, Kuga T, Adachi J, Shiromizu T, Muraoka S, Kume H, Kodera Y, Matsumoto M, Nakayama K, Miyamoto Y, Ishitobi M, Inaji H, Kato K, Tomonaga T. A strategy for large-scale phosphoproteomics and SRM-based validation of human breast cancer tissue samples. J Proteome Res. 2012;11:5311–22. doi: 10.1021/pr3005474. [DOI] [PubMed] [Google Scholar]
- 37.Song MN, Moon PG, Lee JE, Na M, Kang W, Chae YS, Park JY, Park H, Baek MC. Proteomic analysis of breast cancer tissues to identify biomarker candidates by gel-assisted digestion and label-free quantification methods using LC-MS/MS. Arch Pharm Res. 2012;35:1839–47. doi: 10.1007/s12272-012-1018-6. [DOI] [PubMed] [Google Scholar]
- 38.Collodoro M, Lemaire P, Eppe G, Bertrand V, Dobson R, Mazzucchelli G, Widart J, De Pauw E, De Pauw-Gillet MC. Identification and quantification of concentration-dependent biomarkers in MCF-7/BOS cells exposed to 17β-estradiol by 2-D DIGE and label-free proteomics. J Proteomics. 2012;75:4555–69. doi: 10.1016/j.jprot.2012.04.032. [DOI] [PubMed] [Google Scholar]
- 39.Schoenherr RM, Whiteaker JR, Zhao L, Ivey RG, Trute M, Kennedy J, Voytovich UJ, Yan P, Lin C, Paulovich AG. Multiplexed quantification of estrogen receptor and HER2/Neu in tissue and cell lysates by peptide immunoaffinity enrichment mass spectrometry. Proteomics. 2012;12:1253–60. doi: 10.1002/pmic.201100587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sprung RW, Martinez MA, Carpenter KL, Ham AJ, Washington MK, Arteaga CL, Sanders ME, Liebler DC. Precision of Multiple Reaction Monitoring Mass Spectrometry Analysis of Formalin-Fixed, Paraffin-Embedded Tissue. J Proteome Res. 2012;11:3498–505. doi: 10.1021/pr300130t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellappan S, Blackler A, Liao WL, O’Day E, Xu P, Thyparambil S, Cecchi F, Hembrough T, Catenacci DV. Therapeutically Induced Changes in HER2, HER3, and EGFR Protein Expression for Treatment Guidance. J Natl Compr Canc Netw. 2016;14:503–7. doi: 10.6004/jnccn.2016.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nuciforo P, Thyparambil S, Aura C, Garrido-Castro A, Vilaro M, Peg V, Jimenez J, Vicario R, Cecchi F, Hoos W, Burrows J, Hembrough T, Ferreres JC, Perez-Garcia J, Arribas J, Cortes J, Scaltriti M. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Mol Oncol. 2016;10:138–47. doi: 10.1016/j.molonc.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Domanski D, Murphy LC, Borchers CH. Assay development for the determination of phosphorylation stoichiometry using multiple reaction monitoring methods with and without phosphatase treatment: application to breast cancer signaling pathways. Anal Chem. 2010;82:5610–20. doi: 10.1021/ac1005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–16. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 45.Keydar I, Chen L, Karby S, Weiss FR, Delarea J, Radu M, Chaitcik S, Brenner HJ. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer. 1979;15:659–70. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 46.Lasfargues EY, Coutinho WG, Redfield ES. Isolation of two human tumor epithelial cell lines from solid breast carcinomas. J Natl Cancer Inst. 1978;61:967–78. [PubMed] [Google Scholar]
- 47.Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, Tomlinson G, Tonk V, Ashfaq R, Leitch AM, Minna JD, Shay JW. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int J Cancer. 1998;78:766–74. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 48.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher B, Costantino J, Redmond C. A Randomized Clinical Trial Evaluating Tamoxifen in the Treatment of Patients with Node-Negative Breast Cancer Who Have Estrogen-Receptor–Positive Tumors. N Engl J Med. 1989;320:479–484. doi: 10.1056/NEJM198902233200802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.