Abstract
Dynapenia is defined as the age-related loss of muscle strength, and plays a significant role in the loss of physical function and increased risk of disability among older individuals. The need for an early diagnosis supports the search for a biomarker that reflects muscle ‘weakening’. This has previously proven difficult due to patient heterogeneity at presentation and lack of understanding of the underlying molecular mechanisms. The aim of the present study was to identify potential urinary biomarkers of dynapenia in patients undergoing potentially curative surgery for upper gastrointestinal cancer. Maximum isometric knee extensor strength (strain gauge) and maximum leg extensor power (Nottingham power rig) measurements were taken. Cut-off values for dynapenia were based on the Allied Dunbar national fitness survey. Values below the 5th percentile for the population matched for age and sex on the Allied Dunbar national fitness survey were used to stratify the cohort into dynapenic or normal. Urine samples taken at induction of anaesthesia were analysed by SELDI-TOF mass spectrometry using CM10 and IMAC30 chip-types to establish statistically significant m/z peak fingerprint patterns, followed by in-gel LC-MS/MS to identify molecular constituents. Statistical analysis of decision-tree calculations using Biomarker Pattern software resulted in models with sensitivities of 86 and 96%, specificities of 81 and 89%, and overall correctness of 84 and 93%, when applied to the entire cohort for power and strength measurement-based stratifications using the IMAC30 chip-type and the CM10 chip-type, respectively. The molecular identities of 10 peaks of interest were further investigated. After subtraction of potentially unrelated proteins, they were identified as fragments of Annexin A1, collagen α-1 (XV), perlecan and myotrophin. These results demonstrate that urinary screening can be used to define cancer-associated muscle weakness, and the identification of potential biomarkers could be invaluable in establishing a rapid test to measure and assess dynapenia in the clinical setting.
Keywords: SELDI-TOF, dynapenia, muscle wasting, urine biomarker
Introduction
Cancer cachexia is a syndrome of muscle and fat loss leading to progressive functional impairment (1,2). It is thought to be due to complex and variable host-tumour interactions (3,4). Currently, cachexia is defined by the presence of weight loss greater than 5% or greater than 2% in the presence of low muscle mass or low BMI (5). This definition also introduces the concept of ‘pre-cachexia’, a phase of minimal weight loss that may be a stage at which intervention is best targeted. By relying on weight loss, the current cancer cachexia definition is dependent on gross assessment of the patient phenotype rather than identification of the underlying pathological process. Weight loss (often self-reported) may be difficult to determine accurately and may be complicated by factors which act to increase body mass such as fluid accumulation or fat gain because of chemotherapeutic or hormonal treatment. The radiological detection of low muscle mass is known to be associated with adverse patient outcome, particularly in the presence of obesity (5). However, the use of low muscularity in the diagnosis of cachexia is hampered by the fact that measures of muscle mass are often only available as a single measure without dynamic reference to loss or gain. As a result, slim individuals with a low pre-morbid muscle mass risk are grouped along with those with severe cachexia. One alternative to measuring the quantity of fat and muscle tissue in body composition is to measure the tissue quality or function.
Dynapenia is defined as ‘the age associated loss of muscle strength that is not caused by neurologic or muscular diseases’ (6). It predisposes patients to an increased risk of functional limitations and mortality. Development of dynapenia is variable according to pre-existing host characteristics such as initial BMI, body composition, physical activity, food intake and pre-existing co-morbidities. These factors are often concurrently present and interact to lead to muscle wasting (6).
Early recognition of dynapenia and assessment of its progression or regression may be difficult with the common measures used to diagnose muscle wasting, e.g., CT scan. Furthermore, repeated measures of muscle strength and power may be hampered in routine clinical practice by patient frailty and the specialised equipment required. However, safe, accessible and non-invasive tools to detect dynapenia biochemically are currently lacking because the precise molecular mechanisms that define it are currently poorly characterised (7).
Assessment of the urinary protein profile to determine metabolites which may act as biomarkers is one potential method for diagnosing or monitoring disease. Urine is an ideal sample source for the clinical setting, as it can be obtained easily, is relatively stable and the collection of samples is non-invasive. In addition, urine contains a relatively small number of proteins which are present at lower concentrations compared with serum or plasma therefore making them easier to detect (8,9). Despite the relative simplicity of urine this method is complicated by the difficulty in standardising urine samples across individuals to determine the clinically relevant concentration of any particular urinary biomarker. One solution to this is to examine the whole urine protein and peptide profile with SELDI-TOF-MS mass spectrometry (MS) and identify potential biomarkers from a urinary proteomic ‘fingerprint’ of multiple peaks corresponding to the urinary metabolites of a potential marker. Previous studies have found the technique of SELDI-TOF-MS ideally suited for urine analysis, with a combination of high throughput, speed and relatively low cost (10). The main drawback of SELDI-TOF-MS is the comparatively medium resolution of the spectra obtained. However, this is adequate to resolve peaks in the 1,000 to 25,000 Da range from spectra with <500 peaks. The presence or absence of dynapenic muscle is predicted by a decision tree model consisting of the presence or absence of multiple urinary protein/peptide peaks.
SELDI-TOF MS is an established technique for peptide/protein biomarker discovery (11). This method has been successfully performed in a recent study where SELDI-TOF MS was capable of identifying urinary diagnostic markers of upper gastrointestinal (uGI) cancer (12). As excess or abnormal skeletal muscle protein breakdown or synthesis is likely to result in the presence of metabolites in the urine just as cancer-associated markers appear in urine, an identical methodology should be suitable for the detection of protein/peptide markers for the presence of dynapenia.
With this in mind, we aimed to identify potential urinary biomarkers for dynapenia in patients with uGI cancer.
Materials and methods
Materials
Buffers, gels and SELDI chips were purchased from Bio-Rad (Hemel Hempstead, UK), and all other chemicals were obtained from Sigma-Aldrich (Gillingham, UK), unless stated otherwise in the text.
Sample collection
Patients were recruited from the regional uGI or hepatopancreaticobiliary mutli-disciplinary team meetings. Patients had to be over the age of 18, be able to give informed consent and be undergoing potentially curative surgery. There were otherwise no exclusion criteria. Ethical approval was provided by the Lothian region ethics board. Urine was sampled into a sterile container at the time of anaesthesia following an overnight fast. Samples were then snap frozen in liquid nitrogen and stored at −40°C until use. Samples requiring to be stored for over a month were stored at −80°C. The study conformed to the standards set by the Declaration of Helsinki.
CRP measurement
Serum CRP concentration was measured using an automated immunoturbidimetric assay. Using this assay, a concentration of 10 mg/l represents the upper limit of normal range with most healthy individuals having a serum concentration <2 mg/l.
Dynapenia stratification
Maximum isometric knee extensor strength (strain gauge) (IKES). Maximum IKES (13) was measured with the participant seated in an adjustable straight-backed chair with the pelvis secured and the knee flexed at strain gauge and data acquisition system (PowerLab; ADInstruments, Ltd., Oxford, UK). Following instruction, the participant made a maximum voluntary contraction which was held for 5 sec. Three separate measurements (Newtons) were obtained for each limb and the highest value from the dominant limb used in subsequent analysis. The coefficient of variation for IKES is 6.9% for a single session and 10% across sessions occurring over several days (14).
Maximum leg extensor power (LEP) (Nottingham Power Rig)
Maximum LEG (Nottingham Power Rig) was measured using the Nottingham Power Rig. The participant was seated on the rig with the seat position adjusted so that in full extension the footplate was fully depressed. The participant pushed as hard and as fast as possible against the footplate to accelerate the fly wheel. The final velocity of the flywheel was used to calculate the average power output (Watts) during a single maximal thrust of the lower limb. The process was repeated five times with each limb and the highest value from the dominant limb used in subsequent analysis. The coefficient of variation for repeated tests of leg extensor power measured using the Nottingham power rig in healthy individuals was 8.7% (15).
Definition of dynapenia
Dynapenia was defined as a strength and/or power below the 5th percentile for the population matched for age and sex based on the Allied Dunbar national fitness survey (16).
SELDI-TOF-MS
SELDI-chips (CM10 and IMAC30) were prepared for sample application according to the protocol and as previously reported (13). Briefly, IMAC30 chips were loaded with 0.1 M CuSO4, washed with water, neutralised with 0.1 M NaAc (pH 4.0) and washed with water, followed by two washes with 0.1 M NaHPO4. NaCl (0.5 M), and CM10 chips were washed twice with 0.1 M NaHPO4 (pH 4.0). All the chips were processed in a bioprocessor-assembly by incubating 0.1 ml urine and 0.1 ml binding buffer [CM10: 0.1 M NaHPO4 (pH 4.0); IMAC30: 0.1 M NaHPO4, 0.5 M NaCl] for 1 h at room temperature with vigorous shaking. This was followed by three washes with 0.2 ml binding buffer for 5 min each at room temperature with vigorous shaking and two washes with 0.2 ml water at room temperature with vigorous shaking. They were then air-dried and 2 times 1 ml energy absorbing matrix [sinapinic acid (SPA), in 50% ACN, 0.5% TFA)] was added. Air-dried chips were analysed in a PCS4000 SELDI-TOF instrument (Bio-Rad) by measuring the 1,000 to 25,000 Da range with a laser setting of 2.5 mJ and spectra were exported as ‘.xml’ files. The SELDI instrument was calibrated using the ProteinChip All-In-One peptide standard (Bio-Rad). Source voltage was 25,000 V, and detector voltage was 2,946 V. Quality control and consistency was ensured by using one random pool of urines on one spot per chip each. Spectral alignments of quality controls ensured consistency of all spectra.
Data processing
ProteinChip Data Manager software (PCDMS) version 4.1 with integrated Biomarker Wizard cluster analysis (Bio-Rad) was used for analysis. SELDI-TOF-MS traces were split into two groups: Control and dynapenia. The baseline was subtracted from individual m/z traces and profiles were normalised using total ion current, followed by identification of peak clusters using the cluster analysis tool. Peaks were selected in the first pass where the signal to noise (S/N) ratio was >5, had a valley depth of at least 3, and in the second pass S/N ratio was 2 and the valley depth 2. The cluster mass window was set to 0.2% of mass. Clustered peaks were only included if they occurred in at least 10% of all spectra. The resulting P-values, receiver-operating characteristic (ROC) areas, average and median m/z values, and intensities of the clustered peaks were exported and saved as ‘.csv’ files and used for model building. Heat-maps using Pearson's correlation and principle component analysis plots were calculated to assess global group divisions (i.e., dynapenia and control). A two-sample t-test was used to compare mean normalized intensities between the case and control groups. P<0.05 was considered statistically significant.
Model building and validation
Clustered peak lists were analysed with the Biomarker Pattern software (BPS; Bio-Rad). The m/z versus intensity matrices were analysed using decision tree-analysis, selecting the standard error rule of minimum cost-tree regardless of size, and the method used was Gini. V-fold testing was set to 1,000. A total of 15 dynapenic samples and 15 control samples were randomly chosen and used as the learning and testing datasets for both the CM10- and IMAC30-based datasets. The remainder of 19 (IMAC30) or 12 (CM10) samples were used as the validation dataset for blind-testing. Sensitivity was defined as the probability of predicting dynapenia cases, and the specificity was defined as the probability of predicting control samples.
Peak isolation and identification
Peaks observed in the CM10 and IMAC30 chip-types which showed marked expressional differences between control and dynapenia samples, or branching points in the models were further investigated. Urine (0.5 ml) from positive or negative samples in relation to specific peaks was added to 30 ml CM10 or IMAC30 spin column resins (Bio-Rad) and 0.75 ml binding buffer [0.1 M NaHPO4 (pH 4.0) for CM10 resins, and or 0.1 M NaHPO4 (pH 7.0) including 0.5 M NaCl for IMAC30 resins] and incubated for 1 h at room temperature under constant agitation. Unbound material was removed and the resin was washed 4 times with 0.3 ml binding buffer. Bound material was separated by electrophoresis on a 16.5% Tris-Tricine gel (Bio-Rad), and gel bands in the region of 2 to 10 kDa were excised after Coomassie staining (BioSafe Coomassie; Bio-Rad). Positive and negative samples were both chosen on the presence and absence of a specific m/z peak to be identified based on SELDI-TOF-MS analysis. Proteins and peptides from gel bands were digested in situ with trypsin, the resulting peptides eluted with ACN, and analysed by LC-MS/MS as described (12). Fragmentation spectra were then processed by Xcalibur and BioWorks software (Thermo Fisher Scientific, Loughborough, UK) and submitted to the Mascot search engine (Matrix Science, London, UK) using UniProt/SwissProt (release July 2010, Homo sapiens, 18,055 sequences) as the reference database. Mascot search parameters were: Enzyme specificity trypsin, maximum missed cleavage 1, fixed modifications cysteine carbamidomethylation, variable modification methionine oxidation, precursor mass tolerance +/-3 Da, fragment ion mass tolerance +/-0.4 Da. Only Mascot hits with a false discovery rate of <0.05 were taken into consideration. Proteins with at least two peptide matches were then analysed by pattern matching based on SELDI-TOF-MS measured expression levels of peaks of interest (expected abundance in selected samples) and observed presence of proteins. Peptide distribution of identified peptides within a protein as well as calculated molecular mass of identified proteins was also used to assess whether breakdown products were likely to account for mass variances between the expected mass and the molecular weight of the full-length protein.
Mascot-SELDI matrix matching
Observed proteins with at least two peptide matches from the LC-MS/MS analysis were then further analysed by pattern matching based on SELDI-TOF-MS measured expression levels of peaks of interest (expected abundance in selected samples). This was carried out using software written in-house, which compares observed protein expression patterns in a pre-defined set of samples (LC-MS/MS results) against a matrix of peak patterns (SELDI-TOF clustered peak intensities, where estimated peaks are set to null) in the same set of samples. The scoring was based on sensitivity (percent observed over expected) and specificity (percent not observed over not expected), and results are presented in descending order of cumulative scores.
Results
Patients
A total of 51 patients were recruited, of whom 44% had oesophagogastric cancer and the remainder had a mixture of pancreatic and duodenal cancer. A total of 49 patients underwent strength testing of which 22 (44.9%) were classified as dynapenic; 42 of these patients also underwent power testing of which 23 (54.8%) were dynapenic; 13 patients had both low strength and power. The mean age of participants was 65 years with ages ranging from 43 to 82. Patients on average had lost 5.6% of their body weight, mean BMI was 24.10 and CRP was 3. Demographic data are shown in Table I.
Table I.
Expression profiles of potential biomarker peaks identified in cluster and decision tree-analysis models.
| Chip-type | P-value | ROC area | m/z average | m/z STD | Intensity average | Intensity STD | Average fold change in dynapenia | Median fold change in dynapenia | Frequency % found in control | Frequency % found in dynapenia | Sensitivity | Specificity | SELDI normalized intensity cut-off value | Model score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMC30 | 0.213 | 0.623 | 2,199.692 | 1.191 | 35.053 | 35.303 | 0.87 | 1.92 | 85 | 91 | 77 | 59 | 19.565 | 55 |
| IMC30 | 0.748 | 0.539 | 5,880.582 | 0.452 | 3.724 | 6.828 | 0.75 | 1.01 | 15 | 9 | 86 | 33 | 0.836 | 39 |
| IMC30 | 0.015 | 0.311 | 6,362.808 | 7.783 | 11.323 | 13.123 | 0.72 | 0.36 | 96 | 59 | 77 | 74 | 6.033 | 75 |
| IMC30 | 0.044 | 0.331 | 6,460.539 | 7.200 | 7.782 | 8.992 | 0.71 | 0.43 | 81 | 55 | 82 | 59 | 5.652 | 38 |
| IMC30 | 0.016 | 0.270 | 7,478.491 | 8.908 | 8.185 | 8.419 | 0.53 | 0.37 | 93 | 68 | 82 | 59 | 7.202 | 0 |
| IMC30 | 0.018 | 0.290 | 7,683.885 | 11.419 | 17.201 | 17.923 | 0.49 | 0.52 | 96 | 95 | 50 | 85 | 7.396 | 0 |
| IMC30 | 0.042 | 0.342 | 11,756.315 | 5.845 | 13.023 | 34.690 | 0.49 | 0.49 | 96 | 86 | 73 | 67 | 3.408 | 100 |
| IMC30 | 0.049 | 0.357 | 11,967.707 | 5.838 | 3.082 | 8.058 | 0.52 | 0.39 | 63 | 36 | 64 | 74 | 1.041 | 87 |
| CM10 | 0.003 | 0.228 | 10,867.511 | 4.409 | 6.917 | 6.609 | 0.43 | 0.30 | 68 | 43 | 61 | 89 | 3.041 | 76 |
| CM10 | 0.017 | 0.690 | 2,445.428 | 1.569 | 38.118 | 47.170 | 2.79 | 2.57 | 68 | 91 | 74 | 68 | 15.310 | 100 |
| CM10 | 0.018 | 0.301 | 12,726.534 | 4.194 | 1.392 | 1.669 | 0.40 | 0.42 | 47 | 9 | 74 | 74 | 0.987 | 73 |
| CM10 | 0.031 | 0.690 | 2,583.045 | 0.712 | 6.470 | 7.567 | 1.93 | 2.12 | 21 | 43 | 83 | 53 | 2.393 | 79 |
Peaks of interest were analysed by m/z clustering with a window of 0.2% of mass in 49 (IMAC30 dataset) or 42 (CM10 dataset) samples. Both average m/z and intensities with standard errors are included. P-values were calculated using the Mann-Whitney nonparametric test, and the ROC area with cancer as the positive group. The average and median fold change was calculated using intensity values of individual peak clusters from the control and cancer groups. Frequency % describes the likelihood of a particular peak to be found in a specific group by peak-clustering using a S/N of 5. Sensitivity and specificity values were calculated by computational decision-tree modelling using the peak in question as the sole predictor, and the SELDI normalised cut-off values are the associated mass spectrometric intensity values where the reported sensitivity and specificity levels are met. The model score is derived from the tree-analysis modelling and describes the importance of a peak within the model. ROC, receiver-operating characteristic.
Data collection
Mass spectra data were collected in the m/z range of 1,000 to 25,000 for the 49 strength-measurement-based and 42 power-measurement- based patient urine samples using the IMAC30 (Cu2+-complexed) chip-type and the CM10 chip-type respectively. A total of 99 (IMAC30) and 106 (CM10) cluster peaks across the entire datasets were identified. Fifteen of these peak clusters in the IMAC30, and 6 in the CM10 datasets had P-values <0.05. Analysis using peak clustering and group distribution demonstrated that both control and dynapenia groups shared a general overlap in PCA for both chip-types but were distinct enough to allow a degree of separation in heat-map plotting using Pearsons correlation.
Decision-tree analysis
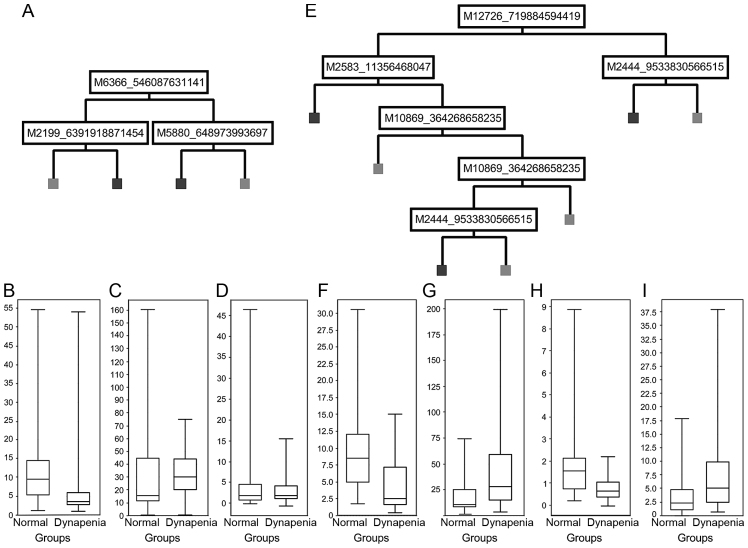
Decision-tree modelling using the BPS of peak clusters of 15 random samples from each cohort was validated by the remainder of the entire cohort (12 control and 7 dynapenic samples for strength, and 4 control and 8 dynapenic samples for power). Decision tree models are shown in Fig. 1A and E. The IMAC30 chipset-based model (Fig. 1A) showed a sensitivity of 57% and a specificity of 58% with an overall correctness of 58% for the validation data-set. Application of the derived model to the entire cohort showed a sensitivity of 86%, specificity of 81% and an overall correctness of 84%. Application of the CM10 chipset-based model (Fig. 1E) to the validation dataset gave a sensitivity of 88%, specificity of 50% and an overall correctness of 75%. For the entire cohort, the values are 96% sensitivity, 89% specificity and 93% overall correctness.
Figure 1.
Decision tree analysis. Cluster peaks involved in the tree-analysis model using (A) the IMAC30 chip-type or (E) the CM10 chip-type stratifying dynapenia (on the right) and control (on the left) were plotted according to their normalised intensity values (B, C, D, F, G, H and I, y-axis) for peak clusters of (B) m/z 6,366, (C) m/z 2,199, (D) m/z 5,880, (F) m/z 10,869, (G) m/z 2,445, (H) m/z 12,726, and (I) m/z 2,583.
Potential biomarkers
Statistical analysis of the various m/z cluster peaks involved in the models and potential marker peaks showing low P-values are shown in Table II. We could identify 8 peak clusters in the IMAC30-based dataset which had either high model score values, or showed P-values <0.05, of which 6 were downregulated and two unchanged in dynapenia (e.g., m/z 6,366 in Fig. 1B; m/z 2,199 in Fig. 1C; and m/z 5880 in Fig. 1D). The CM10-based chipset showed four peak clusters of interest, of which two were upregulated, and two downregulated in dynapenia (e.g., m/z 10,869 in Fig. 1F; m/z 2,445 in Fig. 1G; m/z 12,726 in Fig. 1H; and m/z 2,583 in Fig. 1I).
Table II.
List of potential biomarkers identified.
| m/z peak | Basis | Regulation | Protein ID | Protein name | Mass (Da) | Mascot score | Average number of peptides | Average emPAI | Percentage of sequence coverage | Expected percentage of sequence coverage | Pattern matching | Mascot-SELDI matrix matching scores |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6,366 | Strength, | Down | ACTA_HUMAN | Actin, aortic smooth muscle | 42,009 | 97 | 8 | 0.27 | 11.2 | 15.2 | A | 50//100 |
| IMAC30 | G3P_HUMAN | GAPDH | 36,053 | 66 | 4.6 | 0.16 | 10.8 | 17.7 | A | 50//100 | ||
| HORN_HUMAN | Hornerin | 282,390 | 66 | 11.3 | 0.04 | 4.4 | 2.3 | A | 50//100 | |||
| 6,461 | Strength, | Down | ALBU_HUMAN | Serum albumin precursor | 69,367 | 190 | 18.1 | 0.4 | 12.7 | 9.3 | A | 71//66 |
| IMAC30 | ||||||||||||
| 7,474 | Strength, | Down | G3P_HUMAN | GAPDH | 36,053 | 66 | 4.6 | 0.16 | 10.8 | 20.7 | A | 50//100 |
| IMAC30 | MASP2_HUMAN | Mannan-binding lectin | 75,702 | 65 | 9.3 | 0.13 | 6.3 | 9.9 | A | 50//100 | ||
| serine protease 2 precursor | ||||||||||||
| MMRN2_HUMAN | Multimerin-2 precursor | 104,409 | 32 | 3.7 | 0.04 | 4.2 | 7.2 | A | 50//100 | |||
| MTPN_HUMAN | Myotrophin | 12,895 | 64 | 5.5 | 0.5 | 26.8 | 58.0 | A | 50//100 | |||
| PEBP1_HUMAN | Phosphatidylethanolamine- | 21,057 | 109 | 3.5 | 0.41 | 20.9 | 35.5 | A | 50//100 | |||
| binding protein 1 | ||||||||||||
| 7,679 | Strength, | Down | HBB_HUMAN | Hemoglobin subunit β | 15,998 | 108 | 5.3 | 0.91 | 32.3 | 48.0 | A | 66//100 |
| IMAC30 | ||||||||||||
| 11,756 | Strength, | Down | ALBU_HUMAN | Serum albumin precursor | 69,367 | 190 | 18.1 | 0.4 | 12.7 | 17.0 | A | 66//100 |
| IMAC30 | GFAP_HUMAN | Glial fibrillary acidic protein | 49,937 | 61 | 4 | 0.07 | 11.2 | 23.5 | A | 66//100 | ||
| 11,966 | Strength, | Down | G3P_HUMAN | GAPDH | 36,053 | 66 | 4.6 | 0.16 | 10.8 | 33.2 | A | 60//100 |
| IMAC30 | ||||||||||||
| 10,869 | Power, | Down | ANXA1_HUMAN | Annexin A1 | 38,714 | 126 | 6.5 | 0.3 | 16.3 | 28.1 | A, M | 100//100 |
| CM10 | CAP7_HUMAN | Azurocidin precursor | 27,342 | 43 | 2.7 | 0.14 | 10 | 39.8 | A, M | 100//100 | ||
| CEL_HUMAN | Bile salt-activated | 79,322 | 37 | 3.5 | 0.08 | 3.4 | 13.7 | A | 100//100 | |||
| lipase precursor | ||||||||||||
| COFA1_HUMAN | Collagen α-1(XV) | 141,720 | 34 | 7.5 | 0.04 | 4.7 | 7.7 | A | 100//100 | |||
| chain precursor | ||||||||||||
| HV303_HUMAN | Ig heavy chain V–III | 12,582 | 45 | 3.7 | 0.31 | 9.4 | 86.4 | A, M | 100//100 | |||
| region VH26 precursor | ||||||||||||
| HV307_HUMAN | Ig heavy chain V–III | 13,668 | 45 | 3.8 | 0.29 | 9.2 | 79.5 | A, M | 100//100 | |||
| region CAM | ||||||||||||
| 2,445 | Power, | Up | MASP2_HUMAN | Mannan-binding lectin | 75,702 | 65 | 9.3 | 0.13 | 6.3 | 3.2 | A | 100//100 |
| CM10 | serine protease 2 precursor | |||||||||||
| PGBM_HUMAN | Basement membrane-specific | 468,830 | 198 | 16.8 | 0.05 | 3.2 | 0.5 | A | 100//100 | |||
| heparan sulfate proteoglycan | ||||||||||||
| core protein precursor | ||||||||||||
| 2,583 | Power, | Up | CATB_HUMAN | Cathepsin B precursor | 38,791 | 44 | 3.7 | 0.1 | 6.1 | 6.7 | A | 66//100 |
| CM10 | KV113_HUMAN | Ig κ chain V-I region Lay | 11,834 | 46 | 2.8 | 0.33 | 17.6 | 21.8 | A | 66//100 | ||
| KV305_HUMAN | Ig κ chain V–III region WOL | 11,746 | 117 | 6.8 | 1.58 | 40.4 | 22.0 | A | 66//100 | |||
| KV310_HUMAN | Ig κ chain V–III region VH | 12,757 | 47 | 3.2 | 0.51 | 18.9 | 20.2 | A | 66//100 |
The basis relates to either the leg strength-measurement based model using the IMAC30 dataset, or the leg power-measurement based CM10 dataset. Pattern matching was done using either an automated approach (A) using in-house written software, or by manual comparison (M) of the observed presence of proteins in 4 positive and 4 negative urine samples based on the measured normalised intensities of m/z peak clusters. The automated identification involved Mascot-identified protein expressions in 10 samples for the IMAC30 chipset, and 17 samples in the CM10 chip-type in the 2 to 10 kDa range, which were compared to the expected expression pattern found by SELDI in the same samples. The Mascot-SELDI matrix matching scores were calculated as percentages for sensitivity and specificity. Protein ID is the Swiss-Prot identifier. emPAI, exponentially modified protein abundance index; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Six of these clustered peaks, which showed statistical significance from the IMAC30 dataset, and four from the CM10 chipset were further investigated by enrichment of urinary proteins from 10 (IMAC30) and 17 (CM10) samples each. Gel bands were excised after electrophoresis and LC-MS/MS downstream processing, followed by Mascot searching and identification of proteins contained within the samples, as described earlier (10). All the lead m/z clusters had P-values <0.05.
The downregulated m/z 10,869 peak cluster observed in the CM10 dataset was enriched in four positive samples and four negative samples, which did not contain this peak based on SELDI-MS and served as controls. This peak cluster is also the most significant based on ROC area values, which signifies a combination of false-positive and false-negative discovery rates. Mascot searching matched this peak to either Annexin A1, azurocidin or IgG (Table II). All other potential markers were identified by Mascot pattern matching. This was performed using a dataset of 600 proteins identified for the IMAC chip-type from 10 urine samples, identical to samples used in this study, and 950 proteins for CM10 chips from 17 urine samples, also identical to samples used in this study, in the region of 2–10 kDa. Each individual identification was based on Mascot scores >16 and consisting of ≥2 peptides each (17). The scores were calculated as a percentage of the expected pattern in the Mascot-identified protein list compared to the measured pattern of peaks found by SELDI-TOF above the baseline (sensitivity), thereby setting all estimated peaks as null values, which were used to calculate the specificity. Table II lists all molecules identified using this approach.
We found several fragments of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (m/z 6,366, 7,474 and 11,966), all of which were downregulated in dynapenia using strength as the stratifier. Other potential components of the downregulated m/z 6,366 cluster are fragments of aortic smooth muscle actin (ACTA2) and hornerin. Both the downregulated m/z 6,461 and 11,756 clusters match the expression pattern of albumin. Mannan-binding lectin serine protease 2 precursor (MASP2) is associated both with the downregulated m/z 7,474 and upregulated m/z 2,445. Other potential candidates for the downregulated m/z 7,474 peak cluster are Multimerin-2 (MMRN2), also known as EMILIN-3 or elastin microfibril interface located protein 3, myotrophin (MTPN) and phosphatidylethanolamine-binding protein 1 (PEBP1). Hemoglobin subunit β was identified as the constituent of the m/z 7,679 peak cluster. Glial fibrillary acidic protein (GFAP) was matched to the m/z 11,756 peak cluster. The m/z 10,869 cluster, observed in the CM10 chip based dataset which used power measurements for stratification, also matched the observed expression patterns for Bile salt-activated lipase precursor (CEL) and collagen α-1 (XV) chain precursor (COL15A1). The m/z 2,445 cluster matches basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG2), and the m/z 2,583 peak cluster matched the expression profiles of cathepsin B and several regions of the immunoglobulin κ light chain. The m/z 12,726 peak cluster found in the CM10-based screen could not be identified through pattern matching as its molecular mass is too big compared to the gel band-cutting range.
Additionally, we tested whether any of our proposed m/z peaks correlated with age or gender and found that none of the peak clusters showed any significant age- or sex-related expression differences. We also observed that the m/z distribution of individual peaks within a peak cluster shows the expected pattern of a heterogeneous population of urinary molecular species. However, this does not exclude that some peak clusters may consist of more than one unique protein or peptide. The high degree of overlap of m/z peaks in some of the potential markers in due to the way the DataManager software interprets ‘missing’ peaks (e.g., non-existent or below the S/N ratio) and attempts to estimate a value centred at the m/z cluster value.
Discussion
In the present study, approximately 45% of patients with uGI cancer were classified as being dynapenic based on strength and power measurements. One potential limitation of the study is that there are currently no defined cut points for dynapenia and we therefore stratified patients based on the Allied Dunbar national fitness survey using a strength and/or power measurement below the 5th percentile for the population matched for age and sex. The populations described in other similar studies of skeletal muscle attenuation in healthy individuals are also small however, and different (although similar) values could be obtained depending on the series chosen (18).
In this cohort SELDI-TOF-MS could analyse and screen urine from uGI cancer patients for prospective dynapenia biomarkers to establish a proteomic fingerprint pattern which potentially can be used in clinical diagnostics. The main drawback of SELDI-TOF-MS is the comparatively medium resolution of the spectra obtained, however this is adequate to resolve peaks in the 1,000 to 25,000 Da range from spectra with less than 500 peaks. We found that both IMAC30 and CM10 are useful chip-types for the analysis of human urine (10), and we could generate models based on the full analysis of 51 physical-activity measurement-based dynapenic and non-dynapenic samples from cancer patients. The samples were measured on CM10 and IMAC30 chips, and using the tree-analysis method, we established statistical models with overall sensitivities of 86% (IMAC30) and 96% (CM10) and specificities of 81% (IMAC30) and 89% (CM10) across the entire datasets. Using expression pattern matching, we could assign several proteins identified in urine to our proposed biomarkers.
Cathepsin B and IgGs are upregulated in uGI cancer, and therefore the presence of both molecules at m/z 2,583 in our panel of potential candidates to identify muscle-loss markers is not surprising (19). However, those molecules are very unlikely to be viable potential biomarkers in dynapenia. Nevertheless, it cannot be excluded that modulated levels of specific immunoglobulins are associated with dynapenia per se.
Annexin A1 was reported to have an anti-inflammatory role and is downregulated in nasal epithelial cells from cystic fibrosis patients (20), but was shown to be upregulated in muscular dystrophy (21). Azurocidin was described as a neutrophil-derived protein, and is mainly expressed in proliferating smooth muscle cells. It was identified for its antimicrobial activity and was reported to show strong immunoregulatory effects on host cells (22). The downregulation of fragments of IgG heavy chain, whose expression pattern matches the expected pattern from the SELDI-MS scans can be an artefact or may be due to an as of yet unidentified mechanism. GAPDH has been shown to interact with several muscular gene products such as ataxin-1 and androgen receptor (23), and its role in glycolysis is well documented. A downregulation of GAPDH could therefore be an indication of starvation.
We also associated actin (ACTA2) to one of our proposed m/z cluster peaks. This molecule is expressed in muscle tissues and is a major constituent of the contractile apparatus. Mutations of this gene can cause coronary artery disease and stroke (24). The downregulated expression of this molecule associated with dynapenia could be due to a failing of regeneration of muscle cells. The same m/z peak cluster may also contain fragments of hornerin, which is described in the literature as a component of epithelial cells, and is downregulated in skin-associated diseases such as psoriasis (25). This molecule is potentially a contaminant in our dataset.
The downregulated m/z 6,461 and 11,756 clusters match the expression pattern of albumin. However, albumin itself is a common molecule found in all samples in various levels, and specific breakdown fragments can be detected in many samples. The downregulation of albumin associated with muscle loss was also observed in a sarcopenia stratification model (unpublished results), and its association with muscle dysfunction remains unclear.
Various fragments of MASP2 were found to be associated both with a downregulation of m/z 7,474 and an upregulation of m/z 2,445. However, the upregulation of this molecule appears to be more significant based on the Mascot-SELDI-matrix matching scores. Levels of MASP2 were reported to be elevated in neonatal infections (26), and mouse models lacking this molecule showed protection of myocardial and gastrointestinal ischemia/reperfusion injury (27). The possible role of this molecule in dynapenia remains unclear.
MMRN2 is a secreted protein described to be associated with tumour progression (28) and part of the autophagy system (29). It is a potential candidate for the downregulated m/z 7,474 peak cluster, however, its involvement in muscle loss is unknown, and due to the potential involvement with cancer is not a suitable dynapenia marker.
MTPN was shown to have a strong growth promoting activity on cultured primary skeletal muscle cells (30). The observed m/z of 7,474 would also fit the published molecular weight of the mature and deglycosylated form of 7 kDa. Elevated plasma levels of MTPN have been described in the literature to be associated with heart failure (31). However, our observed downregulation would fit with a potential hypothesis that one of the effects of dynapenia is a reduced re-growth of muscle cells.
PEBP1 is known to be downregulated in various cancers, such as prostate, breast, gastrointestinal stromal tumours, melanoma, and epithelial ovarian cancer (31). This fits our observed downregulation of this marker, and is therefore not considered to be of relevance in dynapenia. Hemoglobin subunit β was identified as the constituent of the m/z 7,679 peak cluster, which is downregulated in dynapenia. This might be a potential misidentification, or might be indicative of kidney dysfunction. GFAP was shown to be a marker for axonal damage in chronic neuropathies (32), and has been reported to be associated with trauma and was found frequently in various gliomas (33). Its modulation is therefore potentially due to cancer and cancer conditions rather than dynapenia. Bile salt-activated lipase precursor (CEL) is mainly expressed and secreted in the pancreas and mammary glands, and has a role in vitamin and fat absorption. Defects in this gene cause diabetes and pancreatic exocrine dysfunction (34). Its role in muscle ablation and degeneration is not apparent.
COL15A1, which also matches the expression pattern of the downregulated peak cluster at m/z 10,869, is mainly expressed in muscle cells, and in moderate levels in the kidney and pancreas (35), and is part of the basement membrane zone. Mouse studies have shown that a lack of this molecule does not cause developmental defects, however, progressive histological changes characteristic for muscular diseases were visible a few months after birth, and they were more vulnerable than controls to exercise-induced muscle injury (36). COL15A1 therefore appears to function as a structural component needed to stabilize skeletal muscle cells and microvessels, which could explain our observed apparent downregulation of this protein in dynapenia.
Basement membrane-specific HSPG2, also known as Perlecan, which matched the upregulated m/z 2,445 cluster, is a secreted inter-cellular protein, which serves as an attachment site for cells, and its roles are in vascularisation, development and vascular response to injury (37). Deficiencies in this molecule have been shown to cause hypertrophy. It also appears to be important in maintaining fast muscle mass, fiber composition and in regulating myostatin signaling (38). The observed upregulation of a fragment of this molecule is likely due to atrophy and might be directly linked to dynapenia.
In conclusion, stratification of our cancer cohort based on power and strength measurements using dynapenia as a decider yielded a set of potential urinary biomarkers by SELDI-TOF MS. The most likely lead candidates in this study are Annexin A1 and COL15A1 chain, which were downregulated, in leg-power measurement-based dynapenia. The dynapenia model based on leg-strength measurement includes downregulated MTPN as well as upregulated Perlecan as the most promising potential biomarker candidates. Further studies including an extended cohort will help determine the validity of our findings in discriminating between strength and power, and specific assays monitoring expression levels of our proposed biomarkers will help to translate our findings into the clinical setting.
Acknowledgements
We thank N.A. Stephens for patient recruitment.
Glossary
Abbreviations
- BPS
Biomarker Pattern software
- emPAI
exponentially modified protein abundance index
- uGI
upper gastrointestinal
- OGJ
oesophagogastric junction
- PCDMS
ProteinChip Data Manager software
- SPA
sinapinic acid
Funding
The funding of this study was provided by the University of Edinburgh.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
HH carried out the SELDI TOF-MS and contributed to the manuscript. AM and CG undertook the patient assessments and AM helped with the manuscript. RJES and JM wrote and critically revised the manuscript. AC performed the confirmatory mass spectrometry analysis. KCHF and JAR designed the project and JAR gave final approval for the version to be published. All authors approved of the study.
Ethics approval and consent to participate
Ethics approval was obtained from the Lothian Research Ethics Board and all the patients provided written, informed consent.
Consent for publication
Not applicable.
Competing interests
The authors have declared no conflict of interest.
References
- 1.Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, Jatoi A, Loprinzi C, MacDonald N, Mantovani G, et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011;12:489–495. doi: 10.1016/S1470-2045(10)70218-7. [DOI] [PubMed] [Google Scholar]
- 2.Fearon KC. Cancer cachexia and fat-muscle physiology. N Engl J Med. 2011;365:565–567. doi: 10.1056/NEJMcibr1106880. [DOI] [PubMed] [Google Scholar]
- 3.Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: Mediators, signaling, and metabolic pathways. Cell Metab. 2012;16:153–166. doi: 10.1016/j.cmet.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Skipworth RJ, Stewart GD, Dejong CH, Preston T, Fearon KC. Pathophysiology of cancer cachexia: Much more than host-tumour interaction? Clin Nutr. 2007;26:667–676. doi: 10.1016/j.clnu.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Tan BH, Birdsell LA, Martin L, Baracos VE, Fearon KC. Sarcopenia in an overweight or obese patient is an adverse prognostic factor in pancreatic cancer. Clin Cancer Res. 2009;15:6973–6979. doi: 10.1158/1078-0432.CCR-09-1525. [DOI] [PubMed] [Google Scholar]
- 6.Clark BC, Manini TM. What is dynapenia? Nutrition. 2012;28:495–503. doi: 10.1016/j.nut.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kushnir MM, Mrozinski P, Rockwood AL, Crockett DK. A depletion strategy for improved detection of human proteins from urine. J Biomol Tech. 2009;20:101–108. [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalantari S, Jafari A, Moradpoor R, Ghasemi E, Khalkhal E. Human Urine Proteomics: Analytical techniques and clinical applications in renal diseases. Int J Proteomics. 2015;782798 doi: 10.1155/2015/782798. doi: 10.1155/2015/782798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffrey RE. A review of experimental design best practices for proteomics based biomarker discovery: Focus on SELDI-TOF. Methods Mol Biol. 2010;641:167–183. doi: 10.1007/978-1-60761-711-2_10. [DOI] [PubMed] [Google Scholar]
- 11.Wright GL, Jr, Cazares LH, Leung SM, Nasim S, Adam BL, Yip TT, Schellhammer PF, Gong L, Vlahou A. Proteinchip(R) surface enhanced laser desorption/ionization (SELDI) mass spectrometry: A novel protein biochip technology for detection of prostate cancer biomarkers in complex protein mixtures. Prostate Cancer Prostatic Dis. 1999;2:264–276. doi: 10.1038/sj.pcan.4500384. [DOI] [PubMed] [Google Scholar]
- 12.Husi H, Stephens N, Cronshaw A, MacDonald A, Gallagher I, Greig C, Fearon KC, Ross JA. Proteomic analysis of urinary upper gastrointestinal cancer markers. Proteomics Clin Appl. 2011;5:289–299. doi: 10.1002/prca.201000107. [DOI] [PubMed] [Google Scholar]
- 13.Harridge SD, Kryger A, Stensgaard A. Knee extensor strength, activation, and size in very elderly people following strength training. Muscle Nerve. 1999;22:831–839. doi: 10.1002/(SICI)1097-4598(199907)22:7<831::AID-MUS4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Todd G, Gorman RB, Gandevia SC. Measurement and reproducibility of strength and voluntary activation of lower-limb muscles. Muscle Nerve. 2004;29:834–842. doi: 10.1002/mus.20027. [DOI] [PubMed] [Google Scholar]
- 15.Arden NK, Spector TD. Genetic influences on muscle strength, lean body mass, and bone mineral density: A twin study. J Bone Miner Res. 1997;12:2076–2081. doi: 10.1359/jbmr.1997.12.12.2076. [DOI] [PubMed] [Google Scholar]
- 16.Summary. Health Education Authority; London: 1992. Activity and Health Research: Allied Dunbar National Fitness Survey: Main Findings. [Google Scholar]
- 17.Husi H, Skipworth RJE, Cronshaw A, Fearon KCH, Ross JA. Proteomic identification of potential cancer markers in human urine using subtractive analysis. Int J Oncol. 2016;48:1921–1932. doi: 10.3892/ijo.2016.3424. [DOI] [PubMed] [Google Scholar]
- 18.Manini T, Clark B. Dynapenia and aging: An update. J Gerontol A Biol Sci Med Sci. 2012;67:28–40. doi: 10.1093/gerona/glr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Leary MF, Vainshtein A, Carter HN, Zhang Y, Hood DA. Denervation-induced mitochondrial dysfunction and autophagy in skeletal muscle of apoptosis-deficient animals. Am J Physiol Cell Physiol. 2012;303:C447–C454. doi: 10.1152/ajpcell.00451.2011. [DOI] [PubMed] [Google Scholar]
- 20.Bensalem N, Ventura AP, Vallée B, Lipecka J, Tondelier D, Davezac N, Dos Santos A, Perretti M, Fajac A, Sermet-Gaudelus I, et al. Down-regulation of the anti-inflammatory protein annexin A1 in cystic fibrosis knock-out mice and patients. Mol Cell Proteomics. 2005;4:1591–1601. doi: 10.1074/mcp.M500019-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Waddell LB, Lemckert FA, Zheng XF, Tran J, Evesson FJ, Hawkes JM, Lek A, Street NE, Lin P, Clarke NF, et al. Dysferlin, annexin A1, and mitsugumin 53 are upregulated in muscular dystrophy and localize to longitudinal tubules of the T-system with stretch. J Neuropathol Exp Neurol. 2011;70:302–313. doi: 10.1097/NEN.0b013e31821350b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez ML, Ruan X, Kumar P, Grammas P, Pereira HA. Functional modulation of smooth muscle cells by the inflammatory mediator CAP37. Microvasc Res. 2004;67:168–181. doi: 10.1016/j.mvr.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Koshy B, Matilla T, Burright EN, Merry DE, Fischbeck KH, Orr HT, Zoghbi HY. Spinocerebellar ataxia type-1 and spinobulbar muscular atrophy gene products interact with glyceraldehyde-3-phosphate dehydrogenase. Hum Mol Genet. 1996;5:1311–1318. doi: 10.1093/hmg/5.9.1311. [DOI] [PubMed] [Google Scholar]
- 24.Guo DC, Papke CL, Tran-Fadulu V, Regalado ES, Avidan N, Johnson RJ, Kim DH, Pannu H, Willing MC, Sparks E, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am J Hum Genet. 2009;84:617–627. doi: 10.1016/j.ajhg.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Meyer-Hoffert U, Reithmayer K, Paus R, Hansmann B, He Y, Bartels J, Gläser R, Harder J, Schröder JM. Highly complex peptide aggregates of the S100 fused-type protein hornerin are present in human skin. J Invest Dermatol. 2009;129:1446–1458. doi: 10.1038/jid.2008.370. [DOI] [PubMed] [Google Scholar]
- 26.St Swierzko A, Cedzynski M, Domzalska-Popadiuk I, MacDonald SL, Borkowska-Klos M, Atkinson AP, Szala A, Jopek A, Jensenius JC, Kawakami M, et al. Mannan-binding lectin-associated serine protease-2 (MASP-2) in a large cohort of neonates and its clinical associations. Mol Immunol. 2009;46:1696–1701. doi: 10.1016/j.molimm.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 27.Schwaeble WJ, Lynch NJ, Clark JE, Marber M, Samani NJ, Ali YM, Dudler T, Parent B, Lhotta K, Wallis R, et al. Targeting of mannan-binding lectin-associated serine protease-2 confers protection from myocardial and gastrointestinal ischemia/reperfusion injury. Proc Natl Acad Sci USA. 2011;108:7523–7528. doi: 10.1073/pnas.1101748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soltermann A, Ossola R, Kilgus-Hawelski S, von Eckardstein A, Suter T, Aebersold R, Moch H. N-glycoprotein profiling of lung adenocarcinoma pleural effusions by shotgun proteomics. Cancer. 2008;114:124–133. doi: 10.1002/cncr.23349. [DOI] [PubMed] [Google Scholar]
- 29.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi T, Takeshita K, Tsuchida N, Kitano K, Kawabata S, Iwanaga S, Ito T. Purification of a novel muscle cell growth factor S-myotrophin from porcine skeletal muscle. Int J Biochem Cell Biol. 1998;30:897–908. doi: 10.1016/S1357-2725(98)00052-1. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wang S, Tang X, Zhang A, Grabinski T, Guo Z, Hudson E, Berghuis B, Webb C, Zhao P, Cao B. Development and evaluation of monoclonal antibodies against phosphatidylethanolamine binding protein 1 in pancreatic cancer patients. J Immunol Methods. 2010;362:151–160. doi: 10.1016/j.jim.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 32.Notturno F, Capasso M, DeLauretis A, Carpo M, Uncini A. Glial fibrillary acidic protein as a marker of axonal damage in chronic neuropathies. Muscle Nerve. 2009;40:50–54. doi: 10.1002/mus.21323. [DOI] [PubMed] [Google Scholar]
- 33.Husain H, Savage W, Grossman SA, Ye X, Burger PC, Everett A, Bettegowda C, Diaz LA, Jr, Blair C, Romans KE, Holdhoff M. Pre- and post-operative plasma glial fibrillary acidic protein levels in patients with newly diagnosed gliomas. J Neurooncol. 2012;109:123–127. doi: 10.1007/s11060-012-0874-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hui DY, Hayakawa K, Oizumi J. Lipoamidase activity in normal and mutagenized pancreatic cholesterol esterase (bile salt-stimulated lipase) Biochem J. 1993;291:65–69. doi: 10.1042/bj2910065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kivirikko S, Saarela J, Myers JC, Autio-Harmainen H, Pihlajaniemi T. Distribution of type XV collagen transcripts in human tissue and their production by muscle cells and fibroblasts. Am J Pathol. 1995;147:1500–1509. [PMC free article] [PubMed] [Google Scholar]
- 36.Eklund L, Piuhola J, Komulainen J, Sormunen R, Ongvarrasopone C, Fássler R, Muona A, Ilves M, Ruskoaho H, Takala TE, Pihlajaniemi T. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci USA. 2001;98:1194–1199. doi: 10.1073/pnas.98.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mongiat M, Fu J, Oldershaw R, Greenhalgh R, Gown AM, Iozzo RV. Perlecan protein core interacts with extracellular matrix protein 1 (ECM1), a glycoprotein involved in bone formation and angiogenesis. J Biol Chem. 2003;278:17491–17499. doi: 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- 38.Xu Z, Ichikawa N, Kosaki K, Yamada Y, Sasaki T, Sakai LY, Kurosawa H, Hattori N, Arikawa-Hirasawa E. Perlecan deficiency causes muscle hypertrophy, a decrease in myostatin expression, and changes in muscle fiber composition. Matrix Biol. 2010;29:461–470. doi: 10.1016/j.matbio.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author upon reasonable request.