Abstract
Background
Previous research found that ALG3 is associated with cervical cancer, but the role of ALG3 in breast cancer was still unknown.
Material/Methods
The expression of ALG3 in breast carcinoma tissues was determined by immunochemistry. The ability of cellular proliferation, migration, and invasion was determined by CCK-8 assay, wound healing migration assay, and cell invasion assays, respectively. The binding between HSF2 and promoter of ALG3 was determined by ChIP assay.
Results
There was an increased expression of ALG3 in breast cancer tissues compared to normal breast tissues (p<0.05). High expression of ALG3 was significantly correlated with poor OS (p<0.05). ALG3 expression was significantly increased in cancer samples with advanced stages (stage III/IV) compared with those in the early stages of disease (stage I/II) (p<0.05). The staining intensity of ALG3 was significantly correlated to the tumor grade (grades 2–3 versus 1, p<0.05). Silencing ALG3 or HSF2 inhibited the proliferation, migration, and invasion abilities of MCF-7 cells. Silencing ALG3 retarded the growth of MCF-7 cells in vivo.
Conclusions
Silencing ALG3 inhibited MCF-7 cells growth in vitro and in vivo. HSF2 activated ALG3 and promoted the growth of breast carcinoma.
MeSH Keywords: Cell Proliferation, Early Detection of Cancer, Neoplasm Invasiveness
Background
Breast carcinoma is the second death cause of cancer and the most frequent cancer among females[1,2]. In recent years, surgery and postoperative chemotherapy have made significant advances in the treatment of breast cancer. However, the 5-year survival rate of breast cancer is still not high. In particular, treatment of triple-negative breast cancer is less effective and severely threatens the survival of patients. Triple-negative breast cancer is highly malignant, aggressive, resistant to chemotherapy, and has a poor prognosis[3–5]. However, the molecular mechanisms of breast cancer remain unclear[6–8].
ALG3 is the novel target of 3q27.1 amplification in esophageal squamous cell carcinoma. ALG3 is significantly increased and associated with lymph node metastasis [9]. In cervical cancer, ALG3 has increased expression. However, the role of ALG3 in the tumorigenesis still needs to be clarified [10].
Therefore, in this research, we firstly investigated the expression pattern of ALG3 in breast cancer, then we determined the function and the mechanism of ALG3 in breast cancer cells.
Material and Methods
Tissues
We enrolled breast cancer patients admitted to our hospital from January 2016 to January 2010 who were pathologically diagnosed with breast cancer and received a breast resection. The study was approved by the Ethics Commission of Three Gorges Central Hospital and informed consents from the patients were obtained. Ninety cases of breast cancer and normal paracancerous tissue were obtained.
Evaluation of immunohistochemical staining
The expression of ALG3 in breast cancer was detected by immunohistochemical staining in accordance with our previous studies [11,12]. All the sections were observed by 3 pathologists using a light microscope [9]. The intensity of ALG3-positive cells was rated as follows: 0, 1, 2, or 3, for negative, weak, moderate, and strong intensity, respectively. The percentage of ALG3-positive cells was scored as follows: 0 for no cytoplasm expression, 1 for 1–25% positive tumor cytoplasm, 2 for 26–50% positive tumor cytoplasm, 3 for 51–75% positive tumor cytoplasm, and 4 for 76–100% positive tumor cytoplasm [13]. We multiplied the intensity and percentage scores to obtain the final ALG3 staining score, defined as follows: a staining score less than 6 was considered low expression, while staining score of 7 or more was considered high expression. All values are showed as the mean ± standard error (mean ±SEM).
Cell culture and transfection procedure
MCF-7 cell lines were cultured in RPM1 1640 medium. The medium was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in an atmosphere with 5% carbon dioxide at 37°C. The siRNA sequences were obtained from Guangzhou RiboBio Co., LTD (Guangzhou, China). The siRNA sequences were as following: ALG3-1: 5′-GGUUUCGUGUACAUCUUUAUG-3′; ALG3-2: 5′-GGACCUGAGUCUACCCUCAGG-3′; and NC (negative control) siRNA: 5′-UUCUUCGAAGGUGUCACGUTT-3′. The lentiviral vectors expressing shRNA targeting ALG3 (called LV3-shALG3-1 and LV3-shALG3-2) were purchased from Guangzhou RiboBio Co., LTD (Guangzhou, China).
Total RNA isolation and RT-qPCR
Total RNAs were extracted as previously described [14]. The RT and qPCR reactions were conducted as previously described [9]. Relative mRNA expression was calculated using the comparative cycle threshold (CT) method (2−ΔΔCT).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was carried out using the ChIP Assay Kit (Cell Signaling Technology, 9003) according to the manufacturer’s instructions. Briefly, cross-linked chromatin was sonicated into 200- to 1000-bp fragments. Then, the chromatin was immunoprecipitated using anti-HSF2 antibody. Quantitative PCR was conducted according to the method described above.
CCK-8 assay
The cellular proliferation was determined by the CCK-8 assay according to the manufacture’s instruction [15,16].
Cell migration and invasion assays
The cell migration and invasion assays were performed as described in our previous studies[17, 18]. Briefly, MCF-7 cells were cultured in 6-well plates to about 80% confluence. The medium was replaced with serum-free medium. After wounding, the distance between 2 wounds was measured at 0 and 72 h. Invasion assays were performed as follows: the upper side was coated using matrigel basement membrane matrix for 2 h at 37°C; MCF-7 cells were added into the top chamber, and then incubated for 48 h; 6% paraformaldehyde was used to fix the invasive cells; then cells were stained in 0.5% crystal violet (Beyotime) and counted.
Western blot
Western Blot analysis was performed as previously described [19]. Three independent experiments were performed using Western blot analysis. The ALG3 (ab151211), HSF2 (ab52758), and GAPDH (ab8245) antibodies were purchased from Abcam, Inc. The antibody was diluted to 1: 1000.
Animal experiment
All animal experimental protocols were approved by the Committee on the Use and Care of Animals (Three Gorges Central Hospital). MCF-7 cells were infected with LV3-NC or LV3–1. The 6-week-old BALB/c nude mice were grouped into either the LV3-NC-infected group (n=5) or the LV3-1-infected group (n=5). The infected MCF-7 cells were injected subcutaneously into the left armpit. Twenty-eight days later, the weight and volume of tumors were calculated.
Statistical analysis
All statistical analyses were performed using SPSS 19.0 software. Each experiment was performed in triplicate. Student’s t-test or analysis of variance (ANOVA) was used for statistical analysis. The differences were considered significant when P<0.05.
Results
High expression of ALG3 in breast cancer was associated with poor outcome
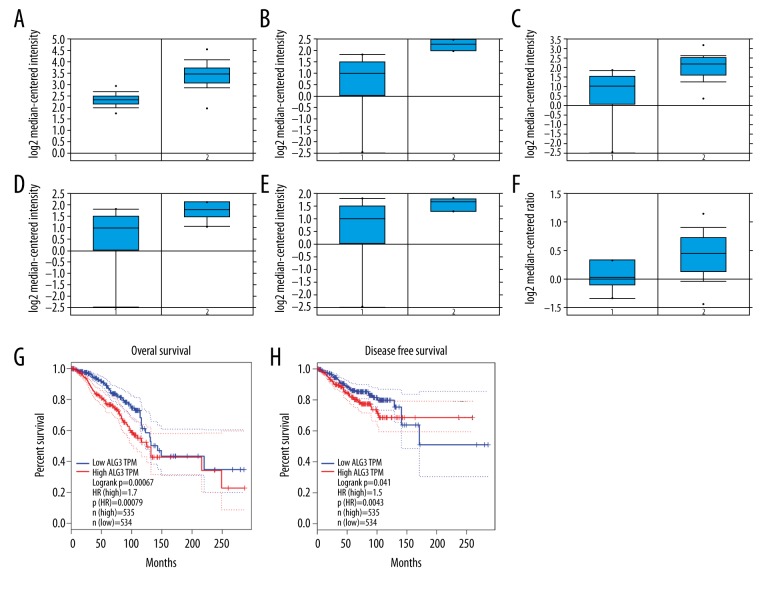
The use of the Oncomine database and GEPIA database was according to methods described in previous studies [20–22]. We first investigated the expression of ALG3 in breast cancer using the Oncomine database. We found that the mRNA expression of ALG3 was significantly up-regulated in breast cancer patients in 14 datasets. In the Curtis Breast dataset, the expression of ALG3 was increased in breast cancer tissues compared with normal tissues, with a fold change=2.135 (Figure 1A). In the Radvanyi Breast dataset, overexpression of ALG3 was found in all types of breast cancer samples compared with normal tissues: ductal breast carcinoma in situ with fold change=3.413 (Figure 1B), invasive ductal breast carcinoma with fold change=2.980 (Figure 1C), invasive lobular breast carcinoma with fold change=2.408 (Figure 1D), and invasive mixed breast carcinoma with fold change=2.184 (Figure 1E). In the Finak Breast dataset, the expression of ALG3 in breast cancer also increased compared with normal tissues (Figure 1F). Evaluation of the ALG3 expression with OS and RFS was performed using the GEPIA Online Tool (http://gepia.cancer-pku.cn/detail.php). ALG3 high expression was significantly correlated with poor OS and RFS for all invasive breast cancer patients (Figure 1G, 1H).
Figure 1.
ALG3 expression was elevated and associated with poor outcome in breast cancer. The mRNA expression of ALG3 in breast cancer was analyzed using data derived from the Oncomine database. The expression of ALG3 in breast cancer was increased compared to normal breast tissues (A–F). Evaluation of the ALG3 genes with OS and RFS was performed using the GEPIA Online Tool (http://gepia.cancer-pku.cn/detail.php) (G, H).
The increased expression of ALG3 was associated with cancer progression
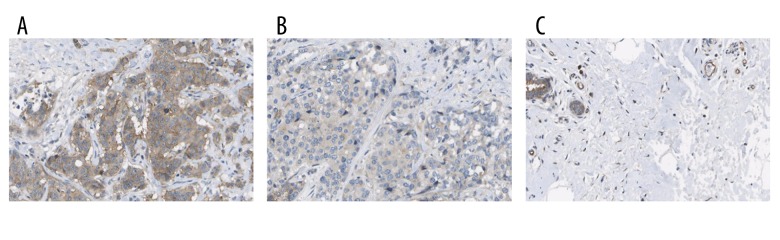
ALG3 was mostly localized in the cytoplasm and cell membrane (Figure 2A, 2B). The ALG3 expression was highly stained in breast cancer (Figure 2A, 2B). ALG3 expression in glandular cells showed medium staining. However, ALG3 showed medium staining in glandular cells and was not detected in adipocytes (Figure 2C). ALG3 expression was significantly increased in breast carcinoma samples compared with normal breast tissue (p<0.05; Figure 2A–2C and Table 1). The Mann-Whitney U test was used to determine the associations between ALG3 expression and clinicopathologic variables of 90 breast cancer tissues (Table 1). The relationship between ALG3 expression and age was not significant (p>0.05; Table 1). ALG3 expression was significantly increased in cancer samples with advanced stages (stage III/IV) as compared with those in the early stages of disease (stage I/II) (p<0.05). The staining intensity of ALG3 was significantly related to the tumor grade (grades 2–3 versus 1, p<0.05).
Figure 2.
ALG3 expression was detected by immunohistochemistry. ALG3 was located in cytoplasm and membranes (A, B). The expression of ALG3 in breast cancer tissues (A, B) was increased compared to the normal breast tissues (C). Original magnification, 200×.
Table 1.
Association of ALG3 expression with clinicopathological characteristics in 90 patients of breast cancer.
| No. of patients (n=90) | ALG3 expression | P value | ||
|---|---|---|---|---|
| Low no. (%) | High no. (%) | |||
| Characteristics | ||||
| Age (years) | >0.05 | |||
| <50 | 48 | 22 (45.80%) | 26 (54.20%) | |
| ≥50 | 42 | 20 (47.62%) | 22 (52.38%) | |
| Normal breast | 90 | 76 (84.44%) | 14 (15.56%) | <0.05 |
| Cancer tissues | 90 | 16 (51.11%) | 74 (48.89%) | |
| FIGO stage | <0.05 | |||
| I/II | 52 | 12 (23.08%) | 40 (76.92%) | |
| III/IV | 38 | 4 (10.53%) | 34 (89.47%) | |
| Grade | ||||
| 1 | 42 | 10 (23.80%) | 32 (76.20%) | |
| 2 | 28 | 4 (14.29%) | 24 (85.71%) | |
| 3 | 20 | 2 (10.00%) | 18 (90.00%) | |
| Grade 2–3 versus 1 | <0.05 | |||
Silencing ALG3 inhibited the proliferation, migration, and invasion ability of breast cancer cells
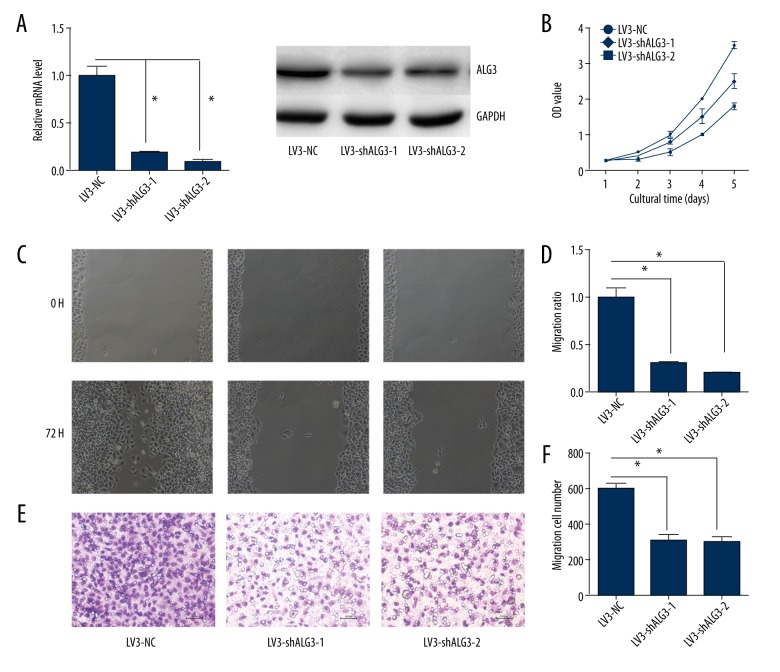
The expression of ALG3 was decreased in LV3-shALG3-1- and LV3-shALG3-2-infected MCF-7 cells compared with LV3-NC-infected MCF-7 cells (Figure 3A). The cell proliferation, migration, and invasion in MCF-7 cells infected with LV3-shALG3-1 and LV3-shALG3-2 were reduced compared to MCF-7 cells infected with LV3-NC (P<0.05; Figure 3B–3F).
Figure 3.
ALG3 regulates cellular proliferation, migration, and invasion. (A) ALG3 was down-regulated by infection with LV3-shALG3-1 or LV3-shALG3-2. (B) Breast cancer MCF-7 cells were infected with LV3-NC, LV3-shALG3-1, or LV3-shALG3-2. Cell proliferation was assessed by CCK-8. (C, D) Breast cancer MCF-7 cell migration ability was detected by the wound healing assay. The migration of LV3-shALG3-1- or LV3-shALG3-2-infected MCF-7 cells was lower compared to LV3-NC-infected cells. (E, F) Breast cancer MCF-7 cell invasion ability was detected by Matrigel invasion assays. The invasion ability of LV3-shALG3-1- or LV3-shALG3-2-infected MCF-7 cells was decreased compared with LV3-NC-infected cells. Original magnification, 200×. Error bars represent standard error. * p<0.05, and **p<0.001.
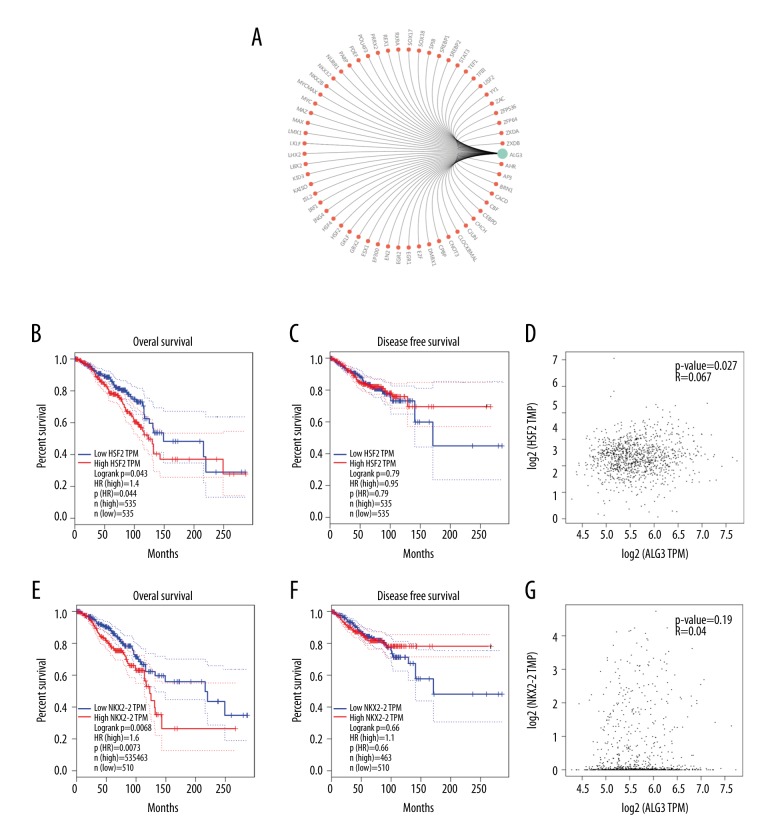
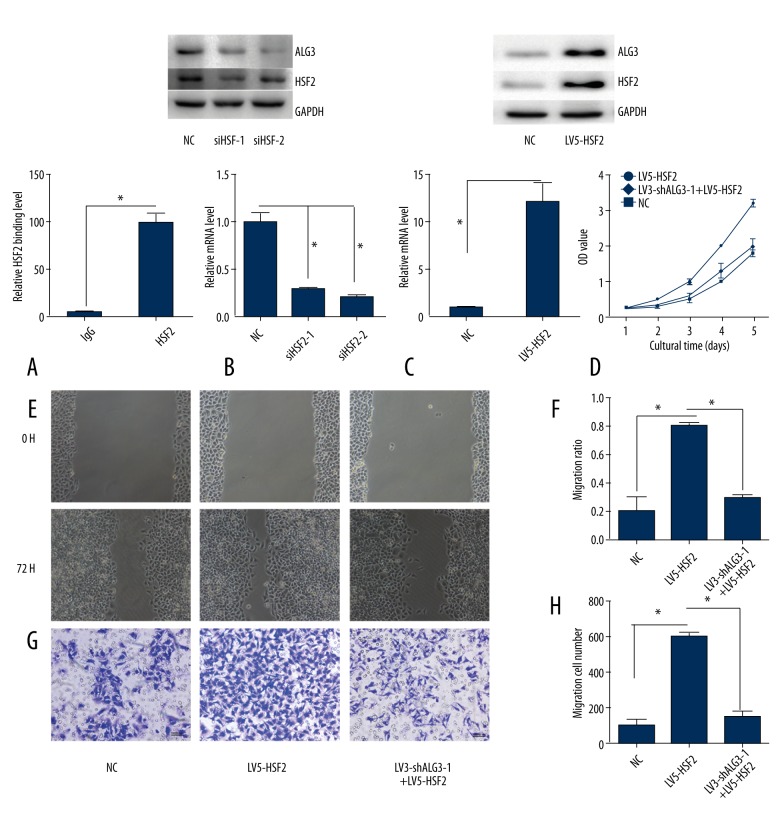
ALG3 is the potential target gene of HSF2 and NKX2-2
In order to investigate the potential mechanism of ALG3 regulation malignant behaviors in breast cancer, we predicted the transcriptional factor related to ALG3 using GCBI (https://www.gcbi.com.cn/gclib/html/index). We found that ALG3 is the potential target gene of HSF2 and NKX2-2(Figure 4A). We also found that HSF2 or NKX2-2 high expression was significantly correlated with poor OS for all invasive breast cancer patients (Figure 4B, 4E). However, there were no significant differences in RFS for all invasive breast cancer patients (p values 0.79 and 0.66, respectively) (Figure 4C, 4F). The expression of HSF2 was well correlated with ALG3 (Figure 4D). However, the expression of NKX2-2 was not correlated with ALG3 (Figure 4G).
Figure 4.
(A) Bioinformatic prediction of transcription factors binding to ALG3 promoter. (B, C) Evaluation of the HSF2 genes with OS and RFS was performed using the GEPIA Online Tool (http://gepia.cancer-pku.cn/detail.php). (D) The correlation between HSF2 and ALG3 was analyzed (http://gepia.cancer-pku.cn/detail.php). (E, F) Evaluation of the NKX2-2 genes with OS and RFS was performed using the GEPIA Online Tool (http://gepia.cancer-pku.cn/detail.php). (G) The correlation between NKX2-2 and ALG3 was analyzed (http://gepia.cancer-pku.cn/detail.php).
HSF2 promoted the expression of ALG3 and increased the proliferation, migration, and invasion ability of breast cancer cells
The ChIP assay confirmed that HSF2 directly bound to the promoter of ALG3 (Figure 5A). However, we did not detect binding between NKX2-2 and the promoter of ALG3. Silencing HSF2 reduced the expression of ALG3 (Figure 5B). Overexpression of HSF2 promoted the expression of ALG3 (Figure 5C). We also observed that overexpression of HSF2 increased the proliferation, migration, and invasion ability of breast cancer cells. This phenomenon was partly abolished by silencing ALG3 (Figure 5D–5H).
Figure 5.
HSF2 promoted the expression of ALG3 and increased the proliferation, migration, and invasion ability of breast cancer cells. (A) The ChIP assay confirmed that HSF2 was bound to the ALG3 promoter. (B) Silencing HSF2 inhibited the expression of ALG3. (C) Overexpression of HSF2 increased the expression of ALG3. (D) Cellar proliferation was detected by CCK-8. (E, F) Breast cancer MCF-7 cell migration ability was detected by the wound healing assay. (G, H) Breast cancer MCF-7 cell invasion ability was detected by Matrigel invasion assays.
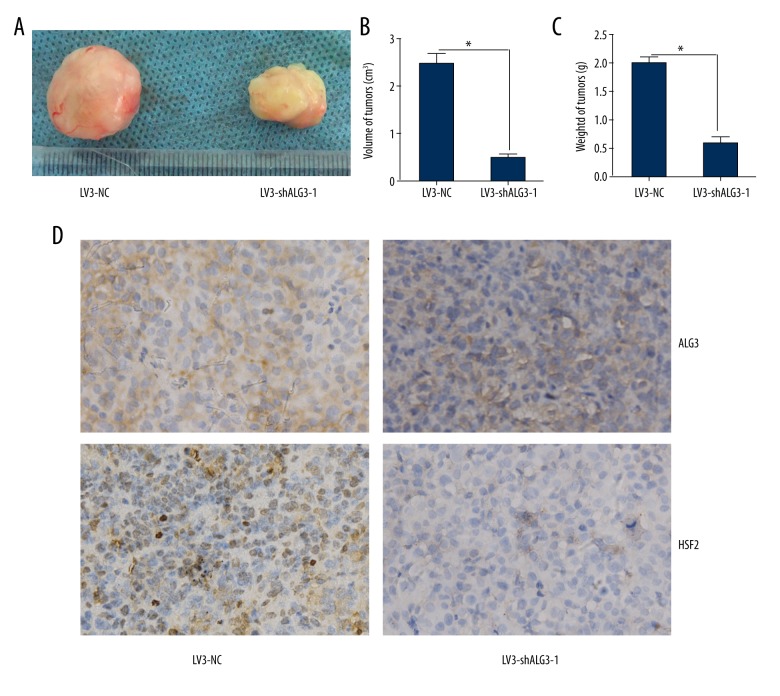
Silencing HSF2 inhibited the growth of MCF-7 cells in vivo
To determine the effect of HSF2 on the growth of MCF-7 cells in vivo, an animal model was constructed. MCF-7 cells infected with LV3-NC and LV3-shALG3-1 formed tumors in all nude mice. The average volume of tumors was decreased in the LV3-shALG3-1 group compared to that in the LV3-NC group (p<0.05) (Figure 6A, 6B). The average weight of tumors in the LV3-shALG3-1 group was decreased compared to that of the LV3-NC group (Figure 6C). The expression of ALG3 and HSF2 in tumors derived from the LV3-shALG3-1-infected group was decreased compared to that in the LV3-NC group (Figure 6D).
Figure 6.
ALG3 regulated tumorigenesis in the nude mouse model. (A–C) Mean tumor volume and weight on day 28 after tumor cell injection. LV3-shALG3-1- or LV3-NC-infected MCF-7 cells were implanted s.c. into the left armpit. (D) Immunohistochemical analysis of ALG3 or HSF2 expression were performed on tumor xenografts. Representative images are shown (original magnification ×200). * p<0.05.
Discussion
In this research, we found an increased expression of ALG3 in breast cancer tissues. We also observed that silencing ALG3 inhibited the malignant behavior of breast cancer cells. Mechanistic research revealed that HSF2 activated ALG3 and promoted malignant behavior in breast cancer. Based on the results above, ALG3 is a potential therapeutic target and biological marker of breast carcinoma.
In cervical cancer, ALG3 has increased expression. We did not investigate the function of ALG3 in cervical cancer. Our research revealed that the expression of ALG3 was elevated in breast cancer tissues, suggesting that ALG3 may play a role in breast cancer. The high expression of ALG3 was associated the cancer progression and poor outcome. Silencing ALG3 inhibited the malignant behavior of breast cancer. Our results indicate that ALG3 promotes the growth of breast cancer and that ALG3 is a potential target in breast cancer.
Increased expression of heat shock factor 2 has been reported in numerous types of cancers. HSF2 in lung cancer was up-regulated compared with the matched normal tissues [23]. The low expression of HSF2 was correlated with high Gleason score, metastasis, and poor survival of prostate cancer patients. This suggested that HSF2 is a suppressor of prostate cancer invasion [24]. The expression of HSF2 is significantly increased in lung cancer tissues. Overexpression of HSF2 promotes cell growth and cell migration [23]. The transcription of miR-183/-96/-182 is regulated by HSF2. It increased breast cancer cell growth by promoting more rapid completion of mitosis, promoted cell migration, and was essential for cell survival [25]. Our study found that the high expression of HSF2 was associated the poor outcome in breast carcinoma. Silencing HSF2 inhibited the malignant behavior of MCF-7 cells. HSF2 promoted cell growth, and cell migration was dependent on induction of up-regulation of ALG3. We also found that silencing ALG3 inhibited the expression of HSF2. It appears that ALG3 affects HSF2 through positive feedback regulation. Our data suggest that HSF2 is involved in breast cancer growth and metastasis.
Conclusions
We found increased expression of ALG3 in breast cancer. High expression of ALG3 was related to cancer progression and poor outcome. We also found that HSF2 activated the transcription of ALG3 and promoted breast cancer growth and metastasis. ALG3 is a potential oncotarget of breast carcinoma.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.van de Ven S, Smit VT, Dekker TJ, et al. Discordances in ER, PR and HER2 receptors after neoadjuvant chemotherapy in breast cancer. Cancer Treat Rev. 2011;37(6):422–30. doi: 10.1016/j.ctrv.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Ladoire S, Enot D, Senovilla L, et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy. 2016;12(5):864–75. doi: 10.1080/15548627.2016.1154244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Ma Y, He HW, et al. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy. 2015;11(12):2323–34. doi: 10.1080/15548627.2015.1074372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basset C, Bonnet-Magnaval F, Navarro MG, et al. Api5 a new cofactor of estrogen receptor alpha involved in breast cancer outcome. Oncotarget. 2017;8(32):52511–26. doi: 10.18632/oncotarget.17281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Y, Chen T, Li G, et al. Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget. 2017;8(32):52156–77. doi: 10.18632/oncotarget.11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiping Z, Qingshan W, Shuai Z, et al. A summary of relationships between alternative splicing and breast cancer. Oncotarget. 2017;8(31):51986–93. doi: 10.18632/oncotarget.17727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, Meng Y, Yu T, et al. Ubiquitin E3 ligase MARCH7 promotes ovarian tumor growth. Oncotarget. 2015;6(14):12174–87. doi: 10.18632/oncotarget.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YW, Bae SM, Kim YW, et al. Gene expression profiles in squamous cell cervical carcinoma using array-based comparative genomic hybridization analysis. Int J Gynecol Cancer. 2007;17(3):687–96. doi: 10.1111/j.1525-1438.2007.00834.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu S, Jiang B, Li H, et al. Wip1 is associated with tumorigenity and metastasis through MMP-2 in human intrahepatic cholangiocarcinoma. Oncotarget. 2017;8(34):56672–83. doi: 10.18632/oncotarget.18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa F, Toscani D, Chillemi A, et al. Expression of CD38 in myeloma bone niche: A rational basis for the use of anti-CD38 immunotherapy to inhibit osteoclast formation. Oncotarget. 2017;8(34):56598–611. doi: 10.18632/oncotarget.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Q, Huang F, Wang X, et al. High Eg5 expression predicts poor prognosis in breast cancer. Oncotarget. 2017;8(37):62208–16. doi: 10.18632/oncotarget.19215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poillet-Perez L, Jacquet M, Hervouet E, et al. GABARAPL1 tumor suppressive function is independent of its conjugation to autophagosomes in MCF-7 breast cancer cells. Oncotarget. 2017;8(34):55998–6020. doi: 10.18632/oncotarget.19639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Fu S, Chen Q, et al. The function of oxytocin: A potential biomarker for prostate cancer diagnosis and promoter of prostate cancer. Oncotarget. 2017;8(19):31215–26. doi: 10.18632/oncotarget.16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.JingSong H, Hong G, Yang J, et al. siRNA-mediated suppression of collagen type iv alpha 2 (COL4A2) mRNA inhibits triple-negative breast cancer cell proliferation and migration. Oncotarget. 2017;8(2):2585–93. doi: 10.18632/oncotarget.13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Bi J, Peng Y, et al. Nuclear receptor NR4A1 is a tumor suppressor down-regulated in triple-negative breast cancer. Oncotarget. 2017;8(33):54364–77. doi: 10.18632/oncotarget.17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue MQ, Liu J, Sang JF, et al. Expression characteristic of CXCR1 in different breast tissues and the relevance between its expression and efficacy of neo-adjuvant chemotherapy in breast cancer. Oncotarget. 2017;8(30):48930–37. doi: 10.18632/oncotarget.16893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Jin L, Ye F, et al. Isoform expression patterns of EPHA10 protein mediate breast cancer progression by regulating the E-Cadherin and β-catenin complex. Oncotarget. 2017;8(18):30344–56. doi: 10.18632/oncotarget.15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagi T, Nagai K, Shimizu H, Matsuzawa SI. Melanoma antigen A12 regulates cell cycle via tumor suppressor p21 expression. Oncotarget. 2017;8(40):68448–59. doi: 10.18632/oncotarget.19497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HY, Zeng, Liang YK, et al. GATA3 and TRPS1 are distinct biomarkers and prognostic factors in breast cancer: Database mining for GATA family members in malignancies. Oncotarget. 2017;8(21):34750–61. doi: 10.18632/oncotarget.16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx247. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong YH, Cheng HZ, Peng H, et al. Heat shock factor 2 is associated with the occurrence of lung cancer by enhancing the expression of heat shock proteins. Oncol Lett. 2016;12(6):5106–12. doi: 10.3892/ol.2016.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Björk JK, Åkerfelt M, Joutsen J, et al. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene. 2016;35(14):1770–84. doi: 10.1038/onc.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li P, Sheng C, Huang L, et al. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014;16(6):473. doi: 10.1186/s13058-014-0473-z. [DOI] [PMC free article] [PubMed] [Google Scholar]