Aggregated polypeptides accumulating inside mitochondria are sequestered in a single cellular quality compartment, called IMiQ. Its formation retains proteotoxic aggregates in a distinct cellular localization, increasing mitochondrial fitness by relieving the protein quality control system of misfolded polypeptides.
Abstract
Aggregation processes can cause severe perturbations of cellular homeostasis and are frequently associated with diseases. We performed a comprehensive analysis of mitochondrial quality and function in the presence of aggregation-prone polypeptides. Despite a significant aggregate formation inside mitochondria, we observed only a minor impairment of mitochondrial function. Detoxification of aggregated reporter polypeptides as well as misfolded endogenous proteins inside mitochondria takes place via their sequestration into a specific organellar deposit site we termed intramitochondrial protein quality control compartment (IMiQ). Only minor amounts of endogenous proteins coaggregated with IMiQ deposits and neither resolubilization nor degradation by the mitochondrial protein quality control system were observed. The single IMiQ aggregate deposit was not transferred to daughter cells during cell division. Detoxification of aggregates via IMiQ formation was highly dependent on a functional mitochondrial fission machinery. We conclude that the formation of an aggregate deposit is an important mechanism to maintain full functionality of mitochondria under proteotoxic stress conditions.
INTRODUCTION
Cellular survival depends on the maintenance of protein function under normal and stress conditions, a process summarized as protein homeostasis. Recent evidence indicated that formation of large aggregate deposits might represent a cell-protective mechanism to counteract proteotoxic stress conditions (Tyedmers et al., 2010; Sontag et al., 2014). An organized formation of these deposits seems to be a regulated process that sequesters potential toxic molecules from the rest of the cellular environment and also may allow a specific removal (Sontag et al., 2017). In bacteria, misfolded proteins, in particular upon overexpression, accumulate in the form of one or two relatively large inclusion bodies that are typically localized at the cell poles (Winkler et al., 2010). In the yeast Saccharomyces cerevisiae, three different classes of aggregate deposits have been identified (Miller et al., 2015b). Cytosolic accumulations of misfolded proteins are either transported into the nucleus, forming INQ (intranuclear quality control compartment; Kaganovich et al., 2008; Miller et al., 2015a), or remain in the cytosol as CytoQ compartments. CytoQ represent peripheral stress-induced aggregate sites (Specht et al., 2011), also termed “stress foci” (Spokoini et al., 2012) or “Q-bodies” (Escusa-Toret et al., 2013). Aggregated proteins located in CytoQ or INQ may be resolubilized by the cytosolic disaggregase machinery consisting of members of the Hsp70 and Hsp100 chaperone families. In contrast, amyloid aggregates, formed by β-sheet–rich proteins, are stored separately in a compartment localized next to the vacuole, called IPOD (insoluble protein deposit; Kaganovich et al., 2008). Deposition sites for protein aggregates have also been described in mammalian cells. Overexpressed, folding-deficient proteins accumulate in the aggresome located in proximity of the microtubule-organizing center (Johnston et al., 1998). The deposition of aggregated polypeptides at specific sites inside a cell may fulfill at least three different purposes. 1) The misfolded polypeptides are neutralized by a sequestration from the cellular environment. 2) The localization of the deposits facilitates an eventual removal of the aggregates. 3) An asymmetric distribution of damaged polypeptides at few sites might prevent the transfer of the toxic molecules to the daughter cell during cell division. Generally, an asymmetric distribution of aging factors like protein aggregates seems to be an important aspect of cellular rejuvenation and determination of life expectancy (Nyström and Liu, 2014).
Owing to the endosymbiotic origin of mitochondria, protein homeostasis on the polypeptide level is mainly maintained by an endogenous protein quality control (PQC) system, comprising chaperones and ATP-dependent proteases (Voos, 2013). Additional mechanisms act on the organellar level to maintain overall mitochondrial quality. An accumulation of misfolded polypeptides in mitochondria elicits an unfolded proteins response (UPRmt) increasing the expression of mitochondrial PQC components (Haynes and Ron, 2010). Cells can also specifically remove dysfunctional mitochondria by a variation of autophagy, termed mitophagy (Wang and Klionsky, 2011). Mitophagy reactions are closely connected with the potential of mitochondria to undergo fission and fusion reactions. Mitochondrial dynamics contributes to quality control by content mixing and by complementation of mtDNA mutations (Westermann, 2010).
In this work, we expressed a folding-deficient, aggregation-prone reporter protein in yeast mitochondria to characterize mitochondria-specific proteotoxic effects. We analyzed the aggregation behavior of the fluorescent reporter by biochemical assays as well as by a microscopic analysis of mitochondrial morphology and distribution. We show that aggregation-prone mitochondrial polypeptides are sequestered into a large aggregate deposit site, thereby preventing overall proteotoxic damage to mitochondrial functions. We characterized the effects of mitochondrial fusion and fission components on aggregate deposit formation and analyzed transfer of aggregates to daughter cells during cell division. We conclude that aggregate deposition represents a protection mechanism under proteotoxic stress, when the reactivity of the mitochondrial PQC system is overwhelmed.
RESULTS
Aggregation-prone mitochondrial proteins accumulate at distinct cellular sites
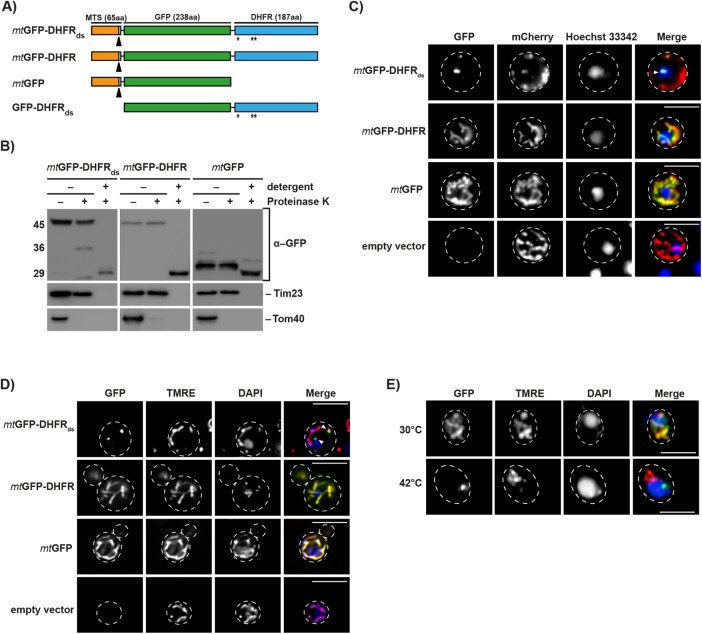
To test the sensitivity of mitochondria to the expression of aggregation-prone proteins, we designed a fusion protein, termed mtGFP-DHFRds, which consisted of green fluorescent protein (GFP) and dihydrofolate reductase (DHFR) from mouse, containing three specific mutations (Cys7Ser, Ser42Cys, and Arg49Cys) at sites required for protein stability. The fusion protein was targeted to the mitochondrial matrix due to an N-terminal mitochondrial targeting sequence (MTS) derived from the yeast protein cytochrome b2 (Figure 1A). As controls, we used two mitochondrially targeted fusion proteins containing either GFP fused to nonmutated DHFR or GFP alone (mtGFP-DHFR and mtGFP). The expression of all proteins was under the control of a galactose-inducible promoter (Figure 1A) to avoid indirect defects caused by a long-term accumulation of potentially toxic polypeptides. Proteins were typically expressed for only several hours after which expression was turned off by removal of galactose from the medium or addition of glucose to repress the promoter. After expression, all reporter proteins were processed and resistant against externally added proteases, similar to the endogenous mitochondrial protein Tim23, indicating their localization inside mitochondria (Figure 1B). As a control, the mitochondrial outer membrane protein Tom40 was digested and after detergent lysis of the mitochondria, both the reporter constructs as well as Tim23 became sensitive to protease digestion.
FIGURE 1:
Expression and localization of aggregation-prone mitochondrial reporter proteins. (A) Schematic depiction of the destabilized reporter construct mtGFP-DHFRds and the corresponding control constructs mtGFP-DHFR, mtGFP, and GFP-DHFRds. Indicated are the MTS (orange), the GFP domain of A. victoria (green), and the DHFR domain of Mus musculus. Destabilizing mutations in the DHFR domain are indicated (*). Cleavage site for mitochondrial processing peptidase is indicated by a black arrow. (B) Treatment of intact and lysed mitochondria containing destabilized reporter or control proteins with 2.5 µM proteinase K. Mitochondria were lysed by treatment with 0.5% Triton X-100. Proteinase K digestion was performed for 20 min at 4°C. Reporter proteins and control proteins of the outer (Tom40) and inner mitochondrial membrane (Tim23) were detected by Western blot. (C) Analysis of aggregate-containing live cells by fluorescence microscopy. Mitochondria-targeted destabilized GFP fusion proteins or controls (green) were expressed in yeast cells for 5 h. Cells also constitutively expressed the fusion protein b2(167)∆-mCherry (red) as a mitochondrial marker protein. Nuclei were visualized by staining with Hoechst-33342 (blue). IMiQ structures are indicated with a white arrow. Scale bar: 10 µm. (D) Cells expressing destabilized GFP-reporter or control proteins for 5 h were analyzed as above. Mitochondrial inner membrane potential (Δψ) was visualized by staining with TMRE (red) and nuclei stained with DAPI (blue). Scale bar: 10 µm. (E) Immunofluorescence of cells after expression of the folded control protein mtGFP-DHFR (green) for 5 h and subsequent incubation at the indicated temperatures for 4 h. Mitochondria were stained with TMRE and nuclei were visualized by staining with DAPI (blue). Scale bar: 10 µm.
Fluorescence microscopy analysis of the control proteins mtGFP and mtGFP-DHFR revealed that they evenly distributed within the mitochondrial network and completely colocalized with another mitochondria-targeted fluorescent protein construct b2(167)∆-mCherry, containing a matrix-targeting presequence derived from the yeast protein cytochrome b2. This observation indicated their correct intracellular localization and a normal solubility. In contrast, in cells expressing the destabilized mtGFP-DHFRds, the reporter protein was typically concentrated into a dot-like structure. These agglomerations were also positive for b2-mCherry, showing that the dot structures were derived from mitochondria; however, while all GFP flourescence was restricted to the dot structures, the mCherry fusion proteins were predominately located in mitochondria-like structures (Figure 1C). The microscopic analysis showed that mtGFP-DHFRds–expressing cells under these conditions typically exhibited two dot structures: a smaller dot that was typically located within the mitochondrial network and a larger dot containing less mitochondrial material that was separated from the mitochondrial network and oriented more to the cell center. These dot structures represent a very prominent feature as essentially all mtGFP-DHFRds–expressing cells exhibited this phenomenon. As the live-cell imaging experiments do not detect nonfolded—and therefore nonfluorescent—GFP domains, we also analyzed yeast cells expressing the normal and the destabilized fusion proteins by indirect immunofluorescence using polyclonal GFP-reactive antibodies that detect both folded and unfolded polypeptides. We obtained images that were essentially identical to the live-cell microscopy (Supplemental Figure S1A), indicating that all expressed mtGFP-DHFRds polypeptides were located in the dot structures and no misfolded GFP domains accumulate elsewhere in the cell.
By staining with the membrane potential–dependent dye tetramethylrhodamine ethyl ester (TMRE) we tested whether the separated agglomerates retained some mitochondrial functionality. TMRE staining in cells expressing control proteins reflected the complete mitochondrial network, demonstrating an intact membrane potential (Figure 1D). The separate mitochondrial agglomerate containing mtGFP-DHFRds aggregates showed no TMRE staining, while the remaining mitochondrial network in these cells exhibited a normal membrane potential. This indicated that the majority of protein aggregates were separated within a specialized nonfunctional mitochondrial compartment. To investigate whether a sequestration of misfolded polypeptides also occurs under other stress conditions, we subjected cells expressing the normal mtGFP-DHFR fusion protein to a short sublethal heat shock at 42°C. In contrast to control conditions at 30°C, which showed a normal TMRE-positive distribution of the GFP fluorescence, the GFP signal in heat-stressed cells was focused in a dot structure separated from the normal, TMRE-stained mitochondria (Figure 1E). Hence, under heat stress, the behavior of mtGFP-DHFR became essentially identical to the aggregation-prone fusion protein. We also tested whether endogenous mitochondrial proteins were sequestrated from the mitochondrial network under stress conditions to determine whether this phenomenon is independent of the use of the GFP-DHFR reporter proteins. We followed the localization of an endogenous mitochondrial protein, the F1-ATPase subunit beta (Atp2), during incubation of yeast cells at elevated temperatures by immunofluorescence (Supplemental Figure S1B). We found that incubation at 42°C led to a fragmentation of mitochondria after 1 and 2 h incubation. At longer time points, mitochondria seem to generally adapt to the stress conditions as indicated by a recovery of the mitochondrial network at the cell periphery. However, after 4 h, bright Atp2-positive dots were observed located in the cell center near the nucleus, potentially indicating a separation of aggregated Atp2 polypeptides. Indeed, it could be shown by a Western blot analysis of total cell lysates that a small fraction of endogenous Atp2 polypeptides aggregated under these conditions (Supplemental Figure S1C). Taking the results together, we conclude that aggregated mitochondrial polypeptides accumulate in specific sites that are derived from mitochondria but are separated from the normal tubular network. We termed this novel mitochondrial aggregate site the intramitochondrial PQC compartment (IMiQ).
Mitochondrial protein homeostasis is maintained in the presence of destabilized proteins
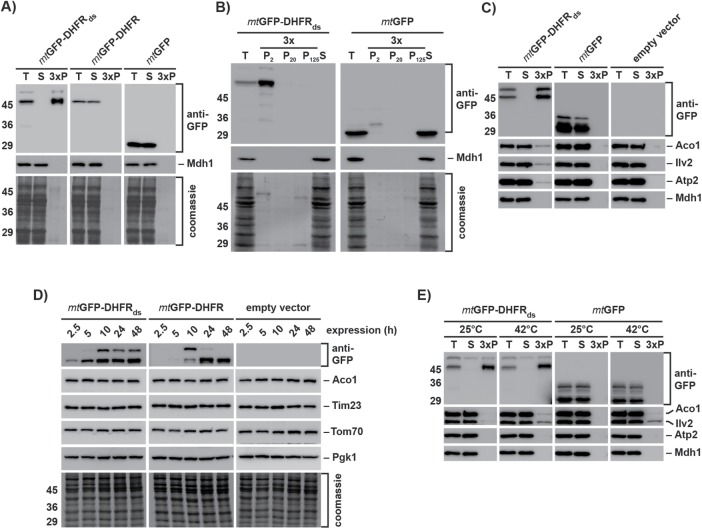
We analyzed the destabilized reporter protein by sedimentation assays to investigate the biochemical properties of the formed aggregates. Under native conditions and high-speed centrifugation, both mtGFP-DHFR and mtGFP were detected in the soluble fraction, while mtGFP-DHFRds was found exclusively in the pellet fraction, demonstrating complete aggregation under normal growth conditions (Figure 2A). Analysis of mtGFP-DHFRds expressing cells at different time points of expression (after 2.5 and 10 h) showed that the destabilized protein exclusively formed large inclusions, pelleting already at 2000 × g (Figure 2B and Supplemental Figure S2, A and B).
FIGURE 2:
Protein homeostasis is maintained in the presence of aggregated polypeptides. (A) The aggregation behavior of the destabilized reporter and control proteins was analyzed in WT yeast cell extracts after 10 h protein expression. Total cell extracts (T), supernatants (S), and aggregate pellet fractions after 20,000 × g centrifugation (P, 3 × amounts) were applied to SDS–PAGE and analyzed by Western blot using antibodies against GFP. Mdh1 was used as soluble control protein of the mitochondrial matrix. Total protein content is indicated by the respective Coomassie-stained blot membrane sections. (B) Differential centrifugation of WT yeast cell extracts containing destabilized reporter proteins. Cells expressing the indicated proteins for 2.5 h were lysed and pellet fractions (3 × amounts) after centrifugation at 2000 × g (P2), 20,000 × g (P20), and 125,000 × g (P125) were analyzed by SDS–PAGE and Western blot as above. (T, total cell extract; S, supernatant after 125,000 × g centrifugation). (C) Coaggregation of endogenous matrix proteins. Cells expressing the indicated proteins for 24 h were lysed and centrifuged at 20,000 × g. Supernatants (S) and pellet fractions (P; 3 × amounts) were analyzed by SDS–PAGE and Western blot as above using antibodies against the indicated proteins. (D) Long-term stability of mitochondrial proteins. Cells containing the indicated reporter plasmids were collected after the indicated expression times. Protein levels of the GFP-reporter proteins as well as an outer membrane protein (Tom70), an inner membrane protein (Tim23), and a mitochondrial matrix protein (Aco1) were analyzed by Western blot. The cytosolic protein Pgk1 was used as loading control. (E) Heat-dependent aggregation. Mitochondria isolated from cells expressing the indicated proteins for 5 h were incubated either at 25°C or at 42°C for 20 min. Aggregation of the reporter proteins and indicated endogenous matrix proteins was analyzed by lysis and centrifugation as described above.
To test for potential proteotoxic effects of mitochondrial protein aggregation, we analyzed a cosedimentation of the proteins Aco1, Ilv2, and Atp2, which are already known to be thermo-unstable in yeast mitochondria (Bender et al., 2011). Only very minor amounts (∼1%) of the tested proteins cosedimented with mtGFP-DHFRds aggregates after 10 h expression, increasing only slightly after long-term expression (Figure 2C and Supplemental Figure S2C). The major fraction of the three proteins remained soluble. As a control, the thermo-stable protein Mdh1 did not coaggregate with mtGFP-DHFRds aggregates. To test for coaggregation in a more comprehensive manner, we analyzed the subset of proteins cosedimenting with mtGFP-DHFRds by a 2D–PAGE approach. A general evaluation of the 2D-spot pattern again revealed only minor amounts of cosedimenting proteins compared with the amount of the destabilized reporter protein (Supplemental Figure S3). We identified 16 coaggregating proteins, mostly abundant metabolic enzymes, like Ilv1 and Ilv3 (involved in amino acid biosynthesis), Cit1 (trichloroacetic acid [TCA] cycle), or Atp1 and Atp2 (subunits of the ATP synthase F1 part). To assess the overall cellular reactivity to the accumulation of mitochondrial aggregates, we followed the protein levels of mitochondrial proteins of the matrix, inner mitochondrial membrane, as well as outer mitochondrial membrane (Figure 2D). The amounts of all tested proteins were stable during the experimental time frame, excluding changes in mitochondrial protein biogenesis or turnover due to proteotoxic stress.
We incubated isolated mitochondria at 42°C for 40 min to analyze the coaggregation of endogenous mitochondrial proteins with mtGFP-DHFRds under heat stress conditions. Although Aco1 and Ilv2 started to aggregate at higher temperatures as expected, the amount of aggregated proteins was similar in mitochondria containing the misfolded mtGFP-DHFRds (Figure 2E). Hence, even under heat stress conditions, the expression of an aggregation-prone polypeptide did not result in additional coaggregation of other endogenous mitochondrial proteins, most likely due to the sequestration of the destabilized fusion proteins in the IMiQ compartment.
Mitochondrial PQC system is relieved by sequestration of destabilized proteins
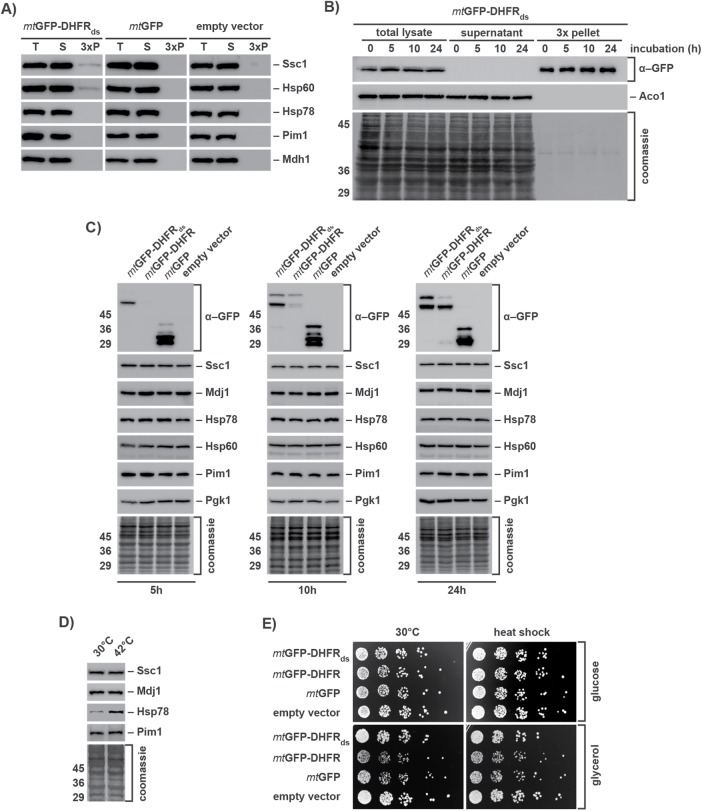
We assayed aggregate contents after long-term expression of mtGFP-DHFRds in order to analyze recruitment of mitochondrial PQC components to IMiQ deposits. Hsp78, the mitochondrial ClpB-type chaperone, as well as the matrix AAA+ protease Pim1 did not cosediment with the protein aggregates. In contrast, as we already observed by 2D–PAGE analysis, a small amount of the mitochondrial Hsp70-type chaperone Ssc1 could be found in the pellet fraction, its amounts slightly increasing from 5 to 24 h expression. The chaperonin Hsp60 was also found in the pellet fraction after 10 and 24 h (Figure 3A and Supplemental Figure S2D). Although Ssc1 as well as Hsp60 interacted with mitochondrial aggregates, we estimated that only ∼1% of the total protein amount of both proteins was found associated with the destabilized reporter proteins.
FIGURE 3:
Components of the mitochondrial PQC system are not affected by the accumulation of aggregated proteins. (A) Coaggregation of mitochondrial PQC system components. WT yeast cells expressing reporter proteins for 24 h were lysed and centrifuged at 20,000 × g. Total cell extracts (T), supernatants (S), and pellet fractions (P; 3 × amounts) were analyzed by Western blot using antibodies against GFP and the indicated mitochondrial PQC components. (B) Long-term stability of IMiQ aggregates. Expression of mtGFP-DHFRds was blocked after 2.5 h by addition of 2% glucose. Cells were further incubated at 30°C for the indicated times. Aggregates were analyzed by Western blot as described. Aco1 was used as a control protein. (C) Protein levels of mitochondrial PQC components. Cell extracts containing destabilized or control proteins expressed for 5, 10, or 24 h were analyzed by Western blot using the indicated antibodies. The cytosolic protein Pgk1 was used as loading control. (D) Protein levels of mitochondrial PQC components in cells incubated at 30 or 42°C for 4 h were analyzed by Western blot as above. (E) Growth phenotype of mtGFP-DHFRds expressing cells. After 10 h expression of indicated reporter proteins, cells were incubated at 30 or 42°C (heat shock) for 1 h and grown on fermentable (2% glucose) or nonfermentable (3% glycerol) medium at 30°C.
We also tested the long-term stability of the mitochondrial aggregates. We stopped the expression of mtGFP-DHFRds by the addition of glucose after 2.5 h and incubated the cells additionally for up to 24 h (Figure 3B). At all time points, mtGFP-DHFRds remained in the aggregate pellet. The protein amounts were very similar to the beginning of the experiment, indicating that the IMiQ inclusion was neither resolubilized nor degraded in the tested time frame. We tested whether the accumulation of misfolded polypeptides would be also able to elicit an UPRmt reaction. We determined the protein levels of several mitochondrial PQC components after different time points of mtGFP-DHFRds expression and found that even after 24 h none of the mitochondrial members of the PQC system were up-regulated, demonstrating that the presence of mitochondrial aggregates was not able to trigger a stress response (Figure 3C). As a control, we performed a heat stress experiment by incubating cells for 4 h at 42°C. As expected, ClpB-type chaperone Hsp78 showed a significant up-regulation. We conclude that effective sequestration of misfolded proteins by IMiQ formation prevented an occupation and subsequent overload of the mtPQC system.
Mitochondrial health and functionality are maintained by IMiQ formation
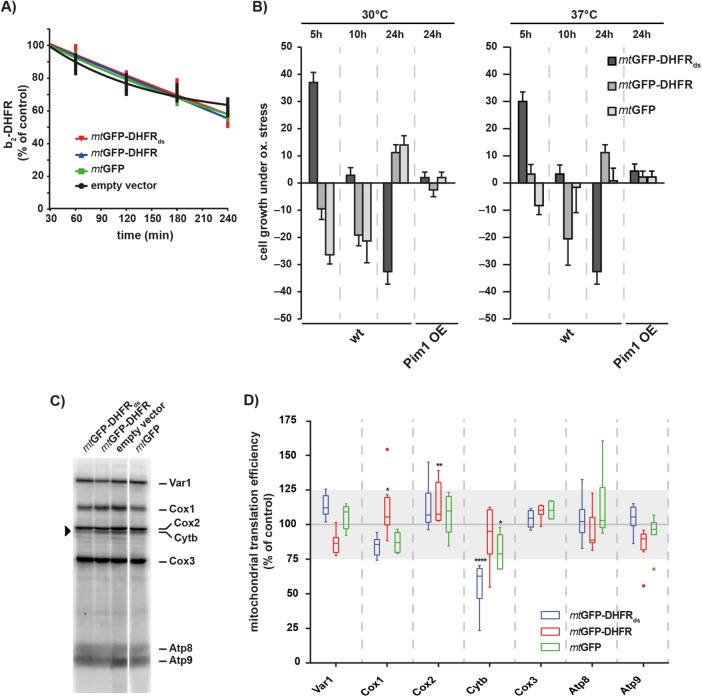
As an expression of the aggregation-prone mtGFP-DHFRds had neither a significant effect on general protein solubility nor on the function of the mitochondrial PQC system, we performed a comprehensive analysis of mitochondrial processes to assess the functional impact of mitochondrial aggregation reactions. To investigate growth phenotype, cells were grown either on fermentable glucose medium or on nonfermentable glycerol medium. In both cases, cells expressing mtGFP-DHFRds grew like control cells, indicating that destabilized proteins within mitochondria had no general toxic effect (Figure 3D). We measured the inner membrane potential (Δψ) of these mitochondria using the fluorescent dye DiSC3, which is quenched in a Δψ-dependent manner (Gärtner et al., 1995). Recovery of fluorescence upon addition of sodium azide was similar in mitochondria containing mtGFP-DHFRds compared with control mitochondria (Supplemental Figure S4A), indicating normal membrane integrity and respiratory efficiency. We also examined the ability of mitochondria to import precursor proteins from the cytosol, a process that requires a coordinated function of import machinery and molecular chaperones. The in vitro import of the radiolabeled preprotein Su9(70)-DHFR into mitochondria isolated from yeast strains expressing the destabilized mtGFP-DHFRds or control proteins exhibited no differences, indicating full activity of the import machinery (Supplemental Figure S4B). A structural analysis of mitochondrial protein complexes of the oxidative phosphorylation system, TCA cycle, as well as the preprotein translocase of the outer membrane (TOM) by BN–PAGE did not show any alterations caused by a presence of protein aggregates (Supplemental Figure S4C). Taken together, the experiments indicate that mitochondrial protein homeostasis was not affected by the accumulation of misfolded proteins.
Mitochondrial aggregation influences functional aspects of the protease Pim1
Proteolytic processes have been previously shown to play a major role during mitochondrial aggregation reactions by a prevention of the accumulation of misfolded polypeptides (Bender et al., 2011). To determine whether mitochondrial aggregates interfered with proteolytic processes, we performed degradation assays of an in vitro imported radiolabeled reporter protein b2(167)Δ-DHFR. The degradation efficiency in mitochondria isolated from aggregate-containing cells was similar to control mitochondria, indicating that the overall degradation processes in the mitochondrial matrix remained intact (Figure 4A). Previous experiments in our group showed an involvement of the matrix AAA+ protease Pim1 in the removal of polypeptides damaged by reactive oxygen species (ROS; Bender et al., 2010). When we performed cellular growth analysis in the presence of the ROS-inducing chemical menadione, cells expressing mtGFP-DHFRds showed an increased ROS resistance after 5 h expression. However, cells became increasingly sensitive to ROS after longer expression times, resulting in a significantly decreased ROS resistance after 24 h expression (Figure 4B). Cells expressing the control proteins mtGFP-DHFR and mtGFP showed the opposite pattern. We confirmed these observations by performing the ROS resistance analysis at an elevated culture temperature of 37°C and found a similar behavior for cells containing mtGFP-DFHRds aggregates. The increased ROS sensitivity suggested that mitochondrial aggregate formation influenced Pim1-related functions. We performed the ROS resistance assay in Pim1 overexpressing cells. Indeed, elevated levels of the protease Pim1 completely rescued the ROS sensitivity phenotype of cells expressing mtGFP-DHFRds even at elevated temperatures.
FIGURE 4:
Mitochondrial aggregates interfere with Pim1 function. (A) Protein degradation in the mitochondrial matrix. Radiolabeled cytb2(167)Δ-DHFR was imported for 20 min into the matrix of isolated mitochondria expressing the indicated proteins for 10 h. After the import reaction, samples were withdrawn at the indicated time points and analyzed by SDS–PAGE and autoradiography. Residual protein levels were quantified using MultiGauge (Fujifilm). Mean values and SEM bars (n = 5) are shown. (B) ROS resistance of IMiQ-containing cells. WT or Pim1-overexpression (OE) cells expressing destabilized reporter or control proteins for the indicated times were grown on fermentable minimal medium (2% glucose) in the presence of 20 mM menadione at 30 or 37°C. Growth inhibition zones were measured and quantified as resistance to menadione relative to control cells. 0% represents cells containing empty vector; 100% represents complete resistance to menadione. Shown are mean values and SEM (n = 5). (C, D) In organello translation rates. Isolated mitochondria containing destabilized reporter or control proteins (10 h expression; indicated in colors) were incubated with [35S]methionine/cysteine for 45 min at 30°C (4.4 µCi/ reaction). Translation products were analyzed by 15% urea-SDS–PAGE and detected by digital autoradiography. (D) Quantification of three independent experiments (representative autoradiogram shown in C). The values were normalized to protein levels of translation products in mitochondria isolated from cells containing empty vector. Quantifications are shown as a box–whisker diagram. Significance was tested by two-way analysis of variance test (*: p < 0.05; **: p < 0.01; ****: p < 0.0001). Gray area indicates a typical 25% fluctuation range between individual translation experiments.
To analyze whether mitochondrial gene expression was affected by the presence of mitochondrial aggregates, we performed an in organello translation assay, in which newly synthesized proteins were labeled by the incorporation of [35S]methionine/cysteine in isolated mitochondria. After separation of the translation products, we detected seven of eight mitochondria-encoded proteins (Figure 4, C and D). Interestingly, only the translation of cytochrome b was significantly reduced in mtGFP-DHFRds containing mitochondria. None of the other mitochondria-encoded proteins showed significant alterations. As Pim1 was shown to be specifically required for the maturation of cytochrome b mRNA in yeast mitochondria (van Dyck et al., 1998), this observation confirms a negative influence of IMiQ accumulation on Pim1 functions.
IMiQ is stored at the nucleus to allow asymmetric distribution during budding
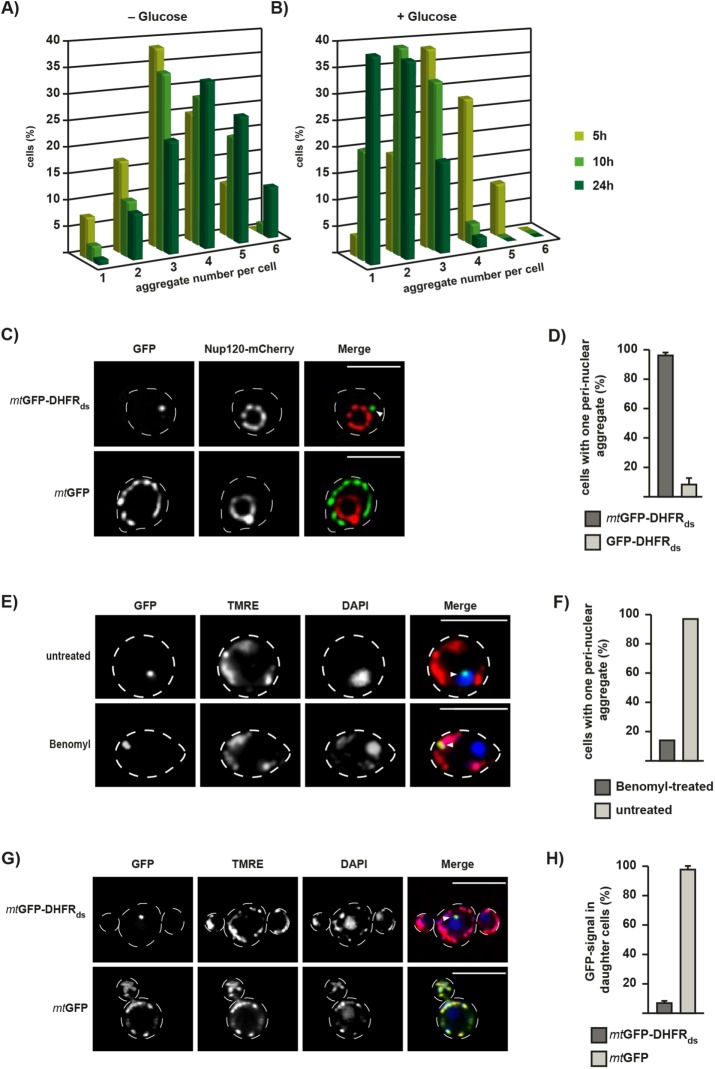
We analyzed the dynamics of mitochondrial aggregate structures during mtGFP-DHFRds expression over time. Yeast cells were incubated in the presence of galactose to obtain a continuous expression or alternatively, glucose was added after 2.5 h to repress further expression. After a subsequent incubation for up to 24 h, the GFP-positive dot structures per cell were counted (Figure 5, A and B). Here, the focal plane was moved through each individual cell to find all dot-like aggregate structures. After 5 h expression, each cell contained on average three inclusions. During constitutive expression of mtGFP-DHFRds, the number of aggregates increased over time, leading to four to six dots per cell after 24 h. Upon inhibition of expression, the number of aggregates decreased over time, resulting in only one or two dots per cell after 24 h, while total protein levels of the expressed constructs remained similar over 24 h (Figure 3B). These results suggest that misfolded proteins first form several smaller inclusions within mitochondria, which fuse at longer incubation times to one or two larger aggregate compartments.
FIGURE 5:
Formation of IMiQ leads to aggregate sequestration at the nucleus. (A, B) Quantification of aggregate numbers in cells expressing mtGFP-DHFRds. Expression was either continuous up to 24 h (A) or stopped after 2.5 h by the addition of 2% glucose (B). Live cells were analyzed by fluorescence microscopy and GFP-positive dots were counted in 200 cells each after 5 h (light green), 10 h (green), and 24 h (dark green). IMiQ structures in all focal planes throughout a cell were counted. The entirety of all expressing cells was set to 100%. (C) Perinuclear localization of IMiQ. Yeast cells expressing the nuclear pore protein Nup120-mCherry (red), mtGFP-DHFRds, or the control protein mtGFP (green) for 5 h were analyzed by fluorescence microscopy. Arrow indicates IMiQ structure. Scale bar: 5 µm. (D) Quantification of aggregate localization. Cells expressing mtGFP-DHFRds (dark-gray) or cytosolic GFP-DHFRds (light-gray) for 5 h that contained mitochondrial aggregates located in the nuclear periphery were counted (100 cells each, set to 100%). Mean values and SEM bars are shown (n = 3). (E) Perinuclear localization of IMiQ is dependent on microtubule function. Cells expressing mtGFP-DHFRds were treated with 100 µg/ml benomyl (Sigma Aldrich, Germany) for 5 h and analyzed by fluorescence microscopy. Mitochondria were visualized by TMRE staining (red) and nuclei by DAPI staining (blue). Scale bar: 10 µm. (F) Quantification of benomyl-treated cells. Cells were analyzed as described in D. (G) Distribution of IMiQ structures during cell division. Fluorescence microscopy of live yeast cells expressing the indicated proteins for 5 h. Expression was stopped by the addition of 2% glucose and cells were incubated at 30°C for an additional 3 h. Mitochondria were visualized by TMRE staining (red) and nuclei by DAPI staining (blue). Scale bar: 10 µm. (H) Quantification of yeast cell buds containing aggregate structures. Cells expressing indicated reporter proteins were analyzed by fluorescence microscopy as above. 100 daughter cells during the budding process (as assessed by phase-contrast imaging) were analyzed for the presence of GFP-positive dots in three independent experiments. Shown are mean values and SEM (n = 3).
To investigate the cellular localization of the sequestered aggregate structures, we coexpressed mtGFP-DHFRds with mCherry-tagged Nup120, a protein of the nuclear pore complex (Skruzny et al., 2009). Although mtGFP localization reflected the mitochondrial network and was not located at the nucleus, the IMiQ structure formed by mtGFP-DHFRds was found in a perinuclear localization in ∼96% of all expressing cells, close to but not inside the nucleus (Figure 5, C and D). To determine whether this localization of the IMiQ is a specific feature, we expressed a destabilized GFP-DHFRds fusion protein, which did not contain a MTS, in the cytosol. Although these cells also showed prominent cytosolic aggregate structures visible as single dots, a perinuclear localization was only observed in ∼8% of all cells analyzed (Figure 5D). As IMiQs represent structures that seem to be separated from the typical mitochondrial network, we tested if the perinuclear association of IMiQs was an active, microtubule-dependent process. Indeed, yeast cells expressing mtGFP-DHFRds that were treated with the microtubule toxin benomyl still formed a single, fluorescence-positive aggregate dot. However, in this case the aggregate structure remained within normal mitochondrial structures as were visualized by staining with the mitochondrial dye TMRE (Figure 5E). Only ∼15% of the benomyl-treated cells contained the perinuclear dot structure typical for IMiQ formation (Figure 5F). These observations indicated that the perinuclear localization of IMiQ deposits was a specific and active process.
To test whether the IMiQ deposit is distributed to daughter cells during cell division, we analyzed cells expressing either mtGFP or mtGFP-DHFRds for 5 h, stopped the expression by the addition of glucose, and then incubated the cells for an additional 3 h. Nonaggregating mtGFP was distributed to the daughter cells in 98% of all expressing cells, correlating with a normal distribution of mitochondria from the mother cell to the bud (Figure 5, G and H). In contrast, while TMRE staining of mitochondria revealed a normal propagation of mitochondria per se, only 7% of cells expressing mtGFP-DHFRds exhibited aggregate structures in the daughter cells. This observation argues that the IMiQ compartment was not distributed to daughter cells, resulting in an eventual elimination of protein aggregates during multiple rounds of cell division.
Detoxification of mitochondrial aggregates by IMiQ formation requires Hsp78 and a functional fission and fusion machinery
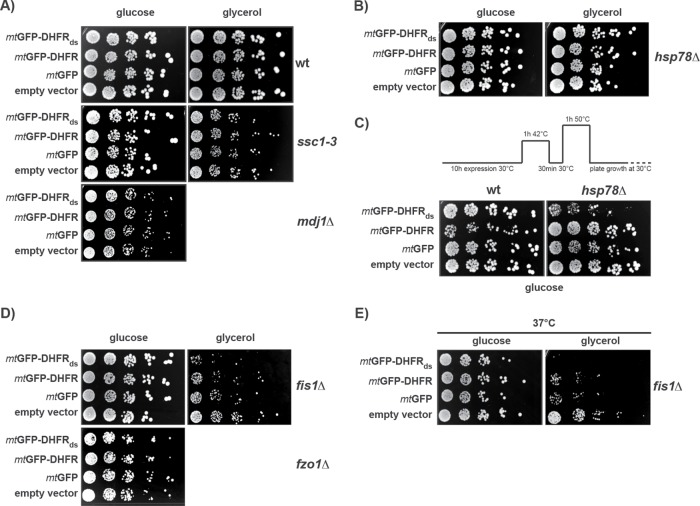
To characterize which cellular factors are involved in IMiQ formation, we analyzed mutant yeast strains lacking different mitochondrial proteins involved in protein and organellar quality control. We compared the growth rates of the respective mutant strains expressing the destabilized mtGFP-DHFRds or its control proteins on fermentable and nonfermentable carbon sources. We found that in mutant cells, where members of the mitochondrial Hsp70 system were deactivated (ssc1–3 and mdj1∆) cellular growth rates were not affected by aggregate formation (Figure 6A). Off note, mdj1 deletions strains are respiratory deficient, preventing an analysis on nonfermentable carbon sources. Indeed, immunofluorescence images of cells expressing mtGFP-DHFRds showed no Ssc1-reactive staining of the IMiQ structure (Supplemental Figure S1A), indicating that the mitochondrial Hsp70 Ssc1 does not colocalize with the aggregates to the IMiQ deposit. Also, in hsp78∆ cells lacking the mitochondrial disaggregase, no significant growth differences between cells containing mitochondria with and without aggregated polypeptides were observed under normal conditions (Figure 6B). However, when we tested the thermotolerance of hsp78∆ cells in the presence and absence of aggregate accumulation (Figure 6C), the survival of wild-type (WT) cells was not influenced by the presence of aggregate proteins in mitochondria, whereas hsp78∆ cells expressing mtGFP-DHFRds showed a significantly reduced survival rate under these heat stress conditions. As control, expression of the wt HSP78 gene complemented the growth defect of the hsp78Δ cell (Supplemental Figure S5A). These results support our conclusion that IMiQ formation allows a functional detoxification of damaged proteins independently of the mitochondrial PQC system, at least under normal conditions. In addition, we conclude that the ClpB-type chaperone Hsp78 plays a role in the detoxification process.
FIGURE 6:
Analysis of aggregate proteotoxicity in different mutant cells. Cellular factors involved in IMiQ formation. (A–E) Growth rates of mutant yeast cells expressing mtGFP-DHFRds or control proteins. After galactose induction for 10 h, cells from the indicated mutant strains were spotted on selective plates containing fermentable (2% glucose) or nonfermentable carbon source (3% glycerol) and incubated at 30°C to assess mitochondrial function. (A) Hsp70 chaperone system. Temperature-sensitive mutant ssc1–3 cells (Gambill et al., 1993) were pretreated for 1 h at 37°C to induce the nonpermissive phenotype. The mdj1∆ deletion strain was respiration deficient and did not grow on nonfermentable carbon sources. (B) Growth analysis of hsp78∆ deletion cells. (C) Analysis of acquired thermotolerance in WT and hsp78∆ deletion cells expressing mtGFP-DHFRds or control proteins for 10 h. After protein expression at 30°C, cells were subjected to a short heat shock and recovery as indicated. Cells were then incubated at a lethal temperature of 50°C for 1 h and spotted on selective plates containing 2% glucose. (D) Growth analysis of fis1∆ and fzo1Δ deletion cells. (E) Growth of fis1Δ cells expressing the destabilized reporter protein for 10 h under mild thermal stress. Cells were spotted as described above and incubated at 37°C.
As mitochondrial fusion and fission represent a major aspect of organellar quality control, we also tested the effect of factors required for the regulation of mitochondrial dynamics. A knockout strain lacking the mitochondrial fission factor Fis1 exhibited a growth defect on a nonfermentable carbon source when cells expressed the aggregation reporter (Figure 6D). In contrast, growth on a fermentable carbon source was similar to control cells, indicating some mitochondrial proteotoxicity already under normal conditions. When we increased the growth temperature of fis1∆ cells to 37°C, representing mild heat stress conditions, the expression of the destabilized polypeptides caused an essentially lethal phenotype on nonfermentable medium (Figure 6E). Expression of the wt FIS1 gene complemented the severe growth defect of the fis1Δ cells (Supplemental Figure S5B). Deletion cells of the fusion factor Fzo1 expressing mtGFP-DHFRds grew like control cells and showed no aggregate toxicity (Figure 6D). Unfortunately, fzo1∆ cells were respiratory deficient in general and could not be analyzed under nonfermentable conditions. These results indicate that the absence of a functional fission process leads to a severe perturbation of mitochondrial activity in the presence of aggregated polypeptides and to a complete abrogation of the protective effect of IMiQ formation, in particular under cellular stress conditions.
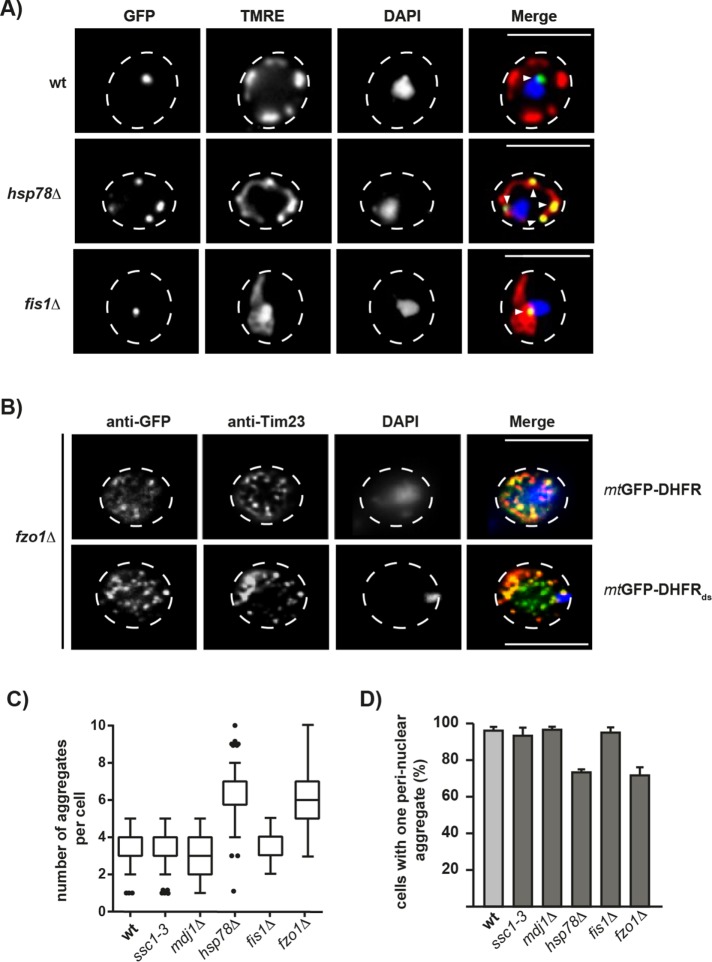
When we characterized cellular aggregate distribution in the mutant strains hsp78∆ and fis1∆, both exhibiting aggregate proteotoxicity, by fluorescence microscopy, we observed significant differences from WT cells. Instead of a single IMiQ deposit, hsp78∆ cells showed multiple aggregate structures that were clearly colocalized with normal mitochondrial structures. The aggregate structures in hsp78∆ cells also did not accumulate near the nucleus (Figure 7A). In contrast, fis1∆ cells showed a single aggregate structure located closely to the nucleus, reminiscent of the IMiQ structure. However, due to the inability of mitochondrial fission in fis1∆ cells, all mitochondrial material was clumped together in a single large structure also containing the fluorescent aggregate (Figure 7A). In contrast, the fusion-deficient fzo1∆ cells, exhibit a large number of small GFP-positive structures, as was shown by immunofluorescence characterization (Figure 7B). In the case of cells expressing the stable mtGFP-DHFR, these structures largely overlap with Tim23-positive signals and represent the mitochondrial fragment in the fusion mutant. The inner membrane preproteins translocase component was used as a mitochondrial marker protein. In the case of mtGFP-DHFRds most of the dot structures did not overlap with Tim23, most likely indicating aggregate structures that have separated from the residual mitochondria. We conclude that although many smaller aggregates form initially, in the fusion-defective cells they accumulate further but are unable to coalesce to a single IMiQ deposit. These observations were corroborated by a quantitative microscopic analysis of aggregate number and localization in the different mutant strains. Both hsp78∆ and fzo1∆ cells showed a significant increase in aggregate dot number (Figure 7C). Despite the increased number of cellular aggregate structures in hsp78∆ and fzo1∆ cells, fewer cells contained dots in the direct vicinity of the nucleus (Figure 7D). This indicates that both internal mitochondrial components localized in the matrix compartment (Hsp78 chaperone activity) as well as external processes affecting mitochondria as a whole (mitochondrial fusion) contribute to IMiQ formation. Interestingly, fis1∆ cells did not exhibit any differences in aggregate number and localization despite exhibiting a pronounced proteotoxicity. We conclude that a physical separation of the aggregates from the mitochondrial network together with a fusion of aggregate-containing mitochondrial particles are both necessary to reach the full mito-protective effects of IMiQ formation.
FIGURE 7:
Cellular factors involved in IMiQ formation. (A) Live-cell fluorescence microscopy of hsp78∆ and fis1Δ cells expressing the destabilized reporter protein mtGFP-DHFRds. Mitochondria were visualized by TMRE staining (red) and nuclei by DAPI staining (blue). Arrows indicate aggregate structures. Scale bar: 10 µm. (B) Immunofluorescence of fzo1∆ yeast cells expressing the aggregation reporter proteins mtGFP-DHFR and mtGFP-DHFRds as indicated after 5 h of induction with galactose. Cells were fixed and incubated with antisera against GFP (green) and Tim23 (red) as described in the Supplemental Experimental Procedures. Nuclei were visualized by staining with DAPI (blue). Scale bar: 10 µm. (C) Aggregate number in yeast mutant cells. The indicated mutant cells expressing mtGFP-DHFRds were analyzed by fluorescence microscopy. GFP-positive aggregate dot numbers were evaluated in 200 cells each and shown as a box–whisker diagram. (D) Localization of aggregates in mutant cells. A perinuclear localization of aggregate structures was analyzed in the indicated mutant strains as described in Figure 5D. For each strain, 100 cells were counted and set to 100%. Shown are mean values and SEM (n = 3).
DISCUSSION
Sequestration of cytosolic protein aggregates into distinct subcellular compartments has been recently described as a PQC pathway that reduces the proteotoxic potential of an accumulation of misfolded polypeptides (Kaganovich et al., 2008; Miller et al., 2015a). Here we describe a novel quality control compartment for destabilized proteins in mitochondria of S. cerevisiae, IMiQ. Aggregation-prone mitochondrial proteins are accumulated and sequestered in a specific compartment distinct from the cellular mitochondrial network. The IMiQ finally accumulates in a microtubule-dependent reaction at a single site in the periphery of the nucleus. IMiQ formation essentially maintains mitochondrial homeostasis and prevents the proteotoxic impact of aggregates on mitochondrial functions. Thus, in addition to INQ, CytoQ, and IPOD, IMiQ represents a fourth PQC compartment identified in yeast. Deposition of aggregates in distinct cellular locations (sequestration) is a secondary defense response to proteotoxic stress and takes place when the primary defense system, consisting of molecular chaperones and proteases, is not functional or overwhelmed. Notably, we observed IMiQ formation not only in the presence of destabilized reporter proteins, but also with endogenous mitochondrial proteins in WT cells under severe stress conditions. In our system, IMiQ formation occurred in the presence of a fully functional mitochondrial PQC system, indicated by normal mitochondrial protein biogenesis and degradation rates.
As indicated by our phenotypic analysis of cells expressing destabilized proteins, aggregate accumulation in the form of IMiQ deposits relieves the mitochondrial PQC system from potential proteotoxic polypeptides to maintain mitochondrial fitness. A similar cytoprotective role of aggregate sequestration has been already observed in the case of cytosolic Q-body formation (Escusa-Toret et al., 2013). IMiQ formation protected the metabolic activity of the majority of mitochondria in the affected cells thereby enabling normal cellular growth rates, even in the presence of additional stress. The membrane potential of the part of the mitochondrial network not containing aggregates remained largely intact, while the membranes surrounding the IMiQ did not exhibit a potential. Taking the results together, mitochondria remained surprisingly resilient to the effects of aggregate accumulation. We conclude from our observations that aggregate sequestration in the form of IMiQ is a major factor in the resistance to proteotoxic impacts.
Correlating with the absence of major functional defects, we observed only a minor extent of coaggregation between the destabilized proteins and other endogenous protein components in the mitochondria. Even under heat stress conditions, no increased coaggregation even of aggregation-prone proteins (Bender et al., 2011) was observed. A similar behavior was observed for typical members of the mitochondrial PQC machinery. Only minor amounts of matrix chaperones were found associated with the aggregates, indicating that the mitochondrial protein folding capacity remained essentially normal. This observation is in clear contrast to other systems, where significant amounts of molecular chaperones, in particular of the Hsp70 and Hsp100 families, were found associated with aggregate deposits in other systems (Kaganovich et al., 2008; Winkler et al., 2010). As we did not observe an obvious change in the amounts of Hsp60 or mtHsp70 chaperones in the cells expressing aggregation-prone mitochondrial polypeptides, we conclude that the protective effect of IMiQ formation also prevented the triggering of a mitochondrial stress response. The absence of a disaggregation reaction also indicated that the IMiQ is an essentially inert compartment. Previous experiments from our group have indicated that mtHsp70 is mainly involved in the prevention of aggregation at physiological conditions, but not at elevated temperatures (Bender et al., 2011). Hence, it is conceivable that under long-term stress, other chaperone-independent protective reactions, like IMiQ formation, become more relevant. The only chaperone system involved in IMiQ formation was Hsp78, the mitochondrial ClpB. Cells lacking Hsp78 contained a significantly larger number of smaller aggregates that were still part of the mitochondrial network and not able to merge to a single IMiQ. A similar effect was already observed during the formation of cytosolic protein deposits, in which deletion of Hsp104 led to a block in the progression of aggregate deposition, resulting in an accumulation of multiple peripheral protein aggregates (Escusa-Toret et al., 2013). Interestingly, this inability to sequester aggregate polypeptides resulted in a strong defect of acquired thermotolerance when the destabilized reporter protein was expressed in hsp78∆ cells. The ATP-dependent matrix protease Pim1 seemed to be the only mitochondrial PQC component that was negatively affected after expression of aggregation-prone polypeptides. Although its overall proteolytic activity did not seem to be altered by the presence of aggregates, a careful phenotypic analysis demonstrated a specific defect in mitochondrial translation of cytochrome b and an increased sensitivity of cells to ROS. Both observations directly indicate a functional defect of Pim1 as biogenesis of cytochrome b requires its activity (van Dyck et al., 1998) and pim1∆ cells were shown to be more sensitive to ROS treatments (Bender et al., 2010). Degradation of damaged proteins mediated by Pim1 becomes particularly important under oxidative stress conditions that do not allow refolding to the active conformation (Bayot et al., 2010; Bender et al., 2010).
Interestingly, the IMiQ deposit represented a very stable compartment. Even after longer incubation periods, we observed neither a resolubilization of the IMiQ-sequestered polypeptides nor a degradation by the mitochondrial PQC system. In contrast, polypeptides accumulated in amorphous aggregates like CytoQ and INQ can be resolubilized and degraded by the cellular PQC machinery (Kaganovich et al., 2008). In the case of aggregated polyQ-containing proteins, accumulated as IPOD, a removal by autophagy has been shown (Kaganovich et al., 2008; Lu et al., 2014). Similarly, in mammalian cells an autophagic degradation of proteins collected in aggresomes has been demonstrated (Gamerdinger et al., 2011; Ouyang et al., 2012). Our observation of unchanged aggregate amounts at least up to 48 h incubation would argue against a clearance process similar to stress-induced mitophagy as has been observed in mammalian cells (Wang and Klionsky, 2011).
A major advantage of the localization of aggregated polypeptides to distinct deposition sites is the possibility to get rid of the aggregates by an asymmetric distribution during cell division. All cellular aggregate compartments analyzed so far showed this behavior (Aguilaniu et al., 2003; Rujano et al., 2006; Erjavec et al., 2007; Lindner et al., 2008). Indeed, the IMiQ aggregates were also retained in the mother cells during the budding process, essentially producing aggregate-free daughter cells. Interestingly, asymmetric segregation of protein aggregates seems to be correlated in many cases with an association of the aggregates with different cellular organelles (Spokoini et al., 2012). It has been suggested that an attachment to nucleus or vacuole, like in the cases of INQ and IPOD, resulting in a low number of relatively immobile aggregates, reduces the probability of inheritance to the new daughter cells (Coelho et al., 2014). A recent report also described a prominent role of mitochondria in retention of cytosolic protein aggregates as tethering to mitochondria restricts their mobility and couples aggregate inheritance to the asymmetric distribution of damaged mitochondria (Higuchi et al., 2013; Zhou et al., 2014). The perinuclear association of the IMiQ deposit likely serves this purpose in the case of intramitochondrial aggregates.
Our experiments demonstrated that mitochondrial aggregate deposition and separation is highly dependent on fission and fusion processes inside the mitochondrial network (Westermann, 2010). A deletion of the fission factor Fis1 led to cytotoxicity in the presence of aggregation-prone polypeptides and a deletion of the fusion factor Fzo1 resulted in a failure to form IMiQs. These observations suggest that IMiQ formation requires a separation of the aggregates from the rest of the mitochondrial network combined with a gradual merger of the aggregate compartments into a single deposit site. However, it cannot be excluded that IMiQ formation involves the recently described generation of a specific form of mitochondrial vesicles (Sugiura et al., 2014), which may first collect the aggregated polypeptides before a deposition as an IMiQ takes place. An involvement of mitochondrial dynamics in protecting organelle quality has been already discussed in the context of neurodegenerative diseases that often comprise different instances of mitochondrial dysfunction (Lin and Beal, 2006). In mammalian cells, the cellular machinery controlling mitochondrial dynamics seems to allow a specific removal of damaged mitochondria while retaining intact organelles (Youle and van der Bliek, 2012). Our results show that IMiQ formation is an important variant of general cytoprotective reactions involved in quality control of mitochondria. After our identification of a specific mitochondrial aggregate sequestration mechanism that protects mitochondrial function and integrity under proteotoxic stress conditions, it will be of high interest to assess the relevance of IMiQ formation in a disease context.
MATERIALS AND METHODS
Plasmids and strains
In general, all constructions of plasmids were done by conventional cloning procedures. A summary of plasmids used in this study is given in Supplemental Table S1. All used yeast strains have been grown as described unless otherwise mentioned. Cells were inoculated in selective minimal medium containing 2% raffinose and incubated overnight at 30°C. Overnight cultures were diluted in selective minimal medium containing 3% glycerol to an OD600 of 0.2 and grown at 30°C to an OD600 of 1. Cells were harvested and resuspended in selective minimal medium containing 3% galactose and incubated at 30°C for indicated times to induce the expression of reporter proteins. A summary of yeast strains used in this study is given in Supplemental Table S2.
Plasmid pBG8099, encoding the enhanced GFP from Aequorea victoria with a mitochondrial matrix-targeting sequence derived from yeast cytochrome b2 (cytb2(84)∆-GFP) under control of the galactose-inducible promotor GAL10/1, was used to generate subsequent aggregation reporter proteins. In detail, the stop codon within the cytb2(84)Δ-GFP gene was removed by PCR by amplifying the coding sequence using primer without stop codon and reintroduced into pBG8099 via BamHI and EcoRI restriction sites (sites were introduced with primer sequence) resulting in pMB022. DNA fragments of encoding dihydrofolate reductase from mice were amplified by PCR from pSN2 (DHFR) and pUHE5377-8 (DHFRds) to yield the respective domains for construction of pMB028 and pMB032. The DHFR-encoding fragments were cloned into pMB022 via EcoRI (sites were introduced with primer sequence). To remove the MTS (for construction of pMB029) GFP was amplified without stop codon and cloned into pBG8099 via BamHI and EcoRI restriction sites. Subsequently, DHFRds was amplified and inserted as described above. pMB031 was constructed by removing cytb2(84)-GFP of pBG8099 by BamHI and EcoRI digestion. Subsequently, blunt ends were generated within the plasmid by the Klenow fragment. For construction of pMB034, the mCherry open reading frame (ORF) was amplified by PCR. Subsequently, the GFP ORF within pGB8118-1 (encoding cytb2(167)-GFP) was exchanged for the amplified mCherry ORF via ClaI and XhoI restriction sites (sites were introduced with primer sequence). For construction of pTB10 and pWJ09 the corresponding WT genes were amplified by PCR from yeast genomic DNA using primers that anneal ∼500 base pairs 5′ and 3′ of the ORF and cloned by standard procedures into the respective single-copy yeast expression plasmids.
Full sequence information of the expression plasmids encoding the used reporter protein constructs is provided as Supplemental Material.
Aggregate sedimentation assay
The assay used in this study was adapted from Koplin et al. (2010). In brief, cell pellets were resuspended in zymolyase buffer (1.2 M sorbitol, 50 mM KPi, pH 7.2, 200 mM KCl2, 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 3 mg/ml zymolyase 20T (Amsbio, UK) and incubated for 30 min at 30°C. Washed cell pellets were resuspended in lysis buffer (0.5% Triton X-100, 50 mM KPi, pH 7.2, 10 mM DTT, 1 mM EDTA, 1× Protease inhibitor [Roth, Germany], 1 mM PMSF). After shaking for 20 min at 4°C, cleared supernatants were centrifuged at high speed (20,000 × g) for 20 min at 4°C. The pellet was washed once in wash buffer (0.5% Triton X-100, 20 mM KPi, pH 7.2, 1 mM PMSF) and subsequently washed in wash buffer without detergent. Aggregated proteins were analyzed by SDS–PAGE and Western blot. Aggregate pellet fractions were applied in threefold excess compared with total or supernatant fractions. Aggregation analysis in isolated mitochondria was performed as described above. For incubation of mitochondria at elevated temperatures, mitochondria were resuspended in buffer A (1.2 M sorbitol, 50 mM KPi, pH 7.2, 100 mM KCl, 10 mM MgCl2, 2 mM ATP, 2 mM NADH, 1 mM PMSF) before lysis.
Fluorescence microscopy
Live-cell microscopy was performed by transferring cells into selective medium with 3% glycerol at pH 7.4. For estimation of mitochondrial membrane potential, cells were stained with 0.5 µM TMRE for 15 min at 30°C and stained with 10 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) for 5 min at 30°C. Finally, cells were immobilized on a thin agarose pad (1% [wt/vol] agarose in selective medium with 3% glycerol at pH 7.4). Microscopic pictures were performed by an EVOSFl Cell Imaging System (AMG, Germany) with a 100× oil immersion objective lens (NA 1.28; AMG, Germany) or a TCS SP8 confocal microscope with a 63× oil immersion objective lens (NA 1.4; Leica, Germany) and processed using Fiji (Schindelin et al., 2012).
Yeast growth analysis
For the growth spot assay, 107 cells were used for serial dilutions up to 1:810. Diluted cells were spotted on selective medium plates containing either 2% glucose or 3% glycerol and incubated at the indicated temperatures. For the ROS growth inhibition assay, 107 cells were resuspended in 200 µl H2O and plated on selective medium plates containing 2% glucose. Small, round, sterile filter paper strips soaked with 2 µl 20 mM menadione were placed onto the agar, causing a circular growth inhibition zone. Plates were incubated at 30°C.
Degradation of radiolabeled preproteins in isolated mitochondria
Experiments assessing mitochondrial degradation of imported proteins were performed as described (Major et al., 2006). Radiolabeled cytb2(167)-DHFR was imported into 150 µg isolated mitochondria, which were then centrifuged at 14,000 × g for 10 min and resuspended in buffer B (250 mM sucrose, 10 mM MOPS/KOH, pH 7.2, 80 mM KCl, 5 mM MgCl2, 3% [wt/vol] bovine serum albumin [BSA], 10 mM KPi, pH 7.2, 3 mM ATP, 4 mM NADH, 10 mM creatine phosphate, 50 µg/ml creatine kinase). After an incubation at 25°C, samples of 15 µg mitochondria were taken, reisolated, and washed once with SEM buffer (250 mM sucrose, 10 mM MOPS/KOH, pH 7.2, 1 mM EDTA) containing 1 mM PMSF. Radiolabeled reporter proteins were detected by SDS–PAGE and digital autoradiography. Quantification of residual proteins was performed using ImageJ64 (Schneider et al., 2012).
In organello translation
Translation reactions were performed as described (Westermann et al., 2001). In brief, 30 µg isolated mitochondria per reaction was resuspended in translation buffer (643 mM sorbitol, 160 mM KCl, 16 mM KPi, 21.5 mM Tris/HCl, pH 7.4, 13.5 mM MgSO4, 3.21 mg/ml BSA, 21.5 mM ATP, 0.13 µM amino acid mix [–Met], 0.53 mM GTP, 1.42 mM creatine phosphate, 36 µg/ml creatine kinase, 1.21 mg/ml α-ketoglutarate) and incubated for 3 min at 30°C. Translation reaction was triggered by the addition of 4.4 µCi/ reaction [35S]methionine/cysteine (PerkinElmer, Germany) and incubated for 45 min at 30°C. By addition of 50 mM MOPS/KOH, pH 7.2, and 10 mM methionine, the translation reaction was stopped and further incubated for 20 min at 30°C. Mitochondria were washed twice in wash buffer (0.6 M sorbitol, 1 mM EDTA, 5 mM MOPS/KOH, pH 7.2, 1 mM methionine). Mitochondrial proteins were finally separated by urea-SDS–PAGE containing 15% acrylamide with 1.1 M urea and detected by digital autoradiography. Quantification of translation products was performed using ImageJ64 (Schneider et al., 2012).
Miscellaneous
All chemicals used in this study were analytical grade and purchased from Carl Roth (Karlstuhe, Germany) if not indicated otherwise. Enzymes and endonucleases were purchased from New England Biolabs (Frankfurt/Main, Germany). All shown single experimental figures represent typical results from at least three biological replicates. Specificity of all antisera used in this study was previously verified by comparisons of band patterns between WT and the respective knockout strains.
Supplementary Material
Acknowledgments
We thank E. Hurt for providing the plasmid pRS314-NUP120-mCherry. We are grateful to E. Deuerling for the opportunity to perform the microscopic analysis in her laboratory. Work in our laboratory was supported by the Deutsche Forschungsgemeinschaft (Grant No. VO 657/5-2 to W.V.).
Abbreviations used:
- Δψmt
electric potential across the inner mitochondrial membrane
- BN–PAGE
blue-native PAGE
- DHFR
dihydrofolate reductase
- IMM
inner mitochondrial membrane
- IMS
intermembrane space
- MPP
mitochondrial processing peptidase
- MTS
mitochondrial targeting sequence
- OMM
outer mitochondrial membrane
- TIM
translocase of the inner membrane
- TMRE
tetramethylrhodamine ethyl ester
- TOM
translocase of the outer membrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-01-0027) on December 6, 2017.
REFERENCES
- Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Bayot A, Gareil M, Rogowska-Wrzesinska A, Roepstorff P, Friguet B, Bulteau AL. Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1. J Biol Chem. 2010;285:11445–11457. doi: 10.1074/jbc.M109.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender T, Leidhold C, Ruppert T, Franken S, Voos W. The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics. 2010;10:1426–1443. doi: 10.1002/pmic.200800619. [DOI] [PubMed] [Google Scholar]
- Bender T, Lewrenz I, Franken S, Baitzel C, Voos W. Mitochondrial enzymes are protected from stress-induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol Biol Cell. 2011;22:541–554. doi: 10.1091/mbc.E10-08-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Lade SJ, Alberti S, Gross T, Tolic IM. Fusion of protein aggregates facilitates asymmetric damage segregation. PLoS Biol. 2014;12:e1001886. doi: 10.1371/journal.pbio.1001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner F, Voos W, Querol A, Miller BR, Craig EA, Cumsky MG, Pfanner N. Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants. J Biol Chem. 1995;270:3788–3795. doi: 10.1074/jbc.270.8.3788. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Ron D. The mitochondrial UPR—protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- Major T, von Janowsky B, Ruppert T, Mogk A, Voos W. Proteomic analysis of mitochondrial protein turnover: identification of novel substrate proteins of the matrix protease Pim1. Mol Cell Biol. 2006;26:762–776. doi: 10.1128/MCB.26.3.762-776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015a;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SB, Mogk A, Bukau B. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol. 2015b;427:1564–1574. doi: 10.1016/j.jmb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Nyström T, Liu B. The mystery of aging and rejuvenation—a budding topic. Curr Opin Microbiol. 2014;18:61–67. doi: 10.1016/j.mib.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Ouyang H, Ali YO, Ravichandran M, Dong A, Qiu W, MacKenzie F, Dhe-Paganon S, Arrowsmith CH, Zhai RG. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem. 2012;287:2317–2327. doi: 10.1074/jbc.M111.273730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Schneider C, Racz A, Weng J, Tollervey D, Hurt E. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 2009;7:e8. doi: 10.1371/journal.pbio.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Samant RS, Frydman J. Mechanisms and functions of spatial protein quality control. Annu Rev Biochem. 2017;86:97–122. doi: 10.1146/annurev-biochem-060815-014616. [DOI] [PubMed] [Google Scholar]
- Sontag EM, Vonk WI, Frydman J. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Curr Opin Cell Biol. 2014;26:139–146. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- van Dyck L, Neupert W, Langer T. The ATP-dependent PIM1 protease is required for the expression of intron-containing genes in mitochondria. Genes Dev. 1998;12:1515–1524. doi: 10.1101/gad.12.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim Biophys Acta. 2013;1833:388–399. doi: 10.1016/j.bbamcr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- Westermann B, Herrmann JM, Neupert W. Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol. 2001;65:429–438. doi: 10.1016/s0091-679x(01)65025-8. [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, Sourjik V, Weiss M, Frangakis AS, Mogk A, Bukau B. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–923. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.