Abstract
An intimate link between centrosome function and neurogenesis is revealed by the identification of many genes with centrosome-associated functions that are mutated in microcephaly disorders. Consistent with the major role of the centrosome in mitosis, mutations in these centrosome-related microcephaly (CRM) genes are thought to affect neurogenesis by depleting the pool of neural progenitor cells, primarily through apoptosis as a consequence of mitotic failure or premature differentiation as a consequence of cell cycle delay and randomization of spindle orientation. However, as suggested by the wide range of microcephaly phenotypes and the multifunctional nature of many CRM proteins, this picture of CRM gene function is incomplete. Here, we explore several examples of CRM genes pointing to additional functions that contribute to microcephaly, including regulation of cell cycle signaling, actin cytoskeleton, and Hippo pathway proteins, as well as functions in postmitotic neurons and glia. As these examples are likely just the tip of the iceberg, further exploration of the roles of microcephaly-related genes are certain to reveal additional unforeseen functions important for neurodevelopment.
INTRODUCTION
Centrosomes are supramolecular protein complexes critical for animal development, including formation and maturation of the most complex organ of all—the brain. Compelling evidence for a role in brain development stems from analysis of human patients that links mutations in at least 15 centrosome-related genes with a spectrum of microcephaly disorders (Table 1), including primary microcephaly (MCPH) and Seckel syndrome (SCKL), which have the common feature of reduced head and brain size reflecting fewer neurons (Duerinckx and Abramowicz, 2017; Nano and Basto, 2017). Centrosomes are multifunctional organelles, composed of pairs of centrioles surrounded by a dynamic pericentriolar matrix (PCM) of proteins, famous for their cell biological role as microtubule-organizing centers (MTOCs). In this capacity, the centrosome facilitates mitotic spindle formation, cell motility, intracellular trafficking, and immune synapse response, among other processes. Centrosomes also donate their core centriole structures to be repurposed as the basal bodies necessary for building motile and nonmotile cilia (Arquint et al., 2014; Woodruff et al., 2014; Lerit and Poulton, 2016; Vertii et al., 2016).
TABLE 1:
Centrosome-related microcephaly (CRM) genes.
| Gene | OMIM | Functions | Common phenotypes | Variable phenotypes |
|---|---|---|---|---|
| WDR62 | MCPH2 | PCM, spindle integrity and orientation, Aurora A activation | Microcephaly, cortical malformations | Cortical malformations including pachygyria, cortical thickening, lissencephaly, subcortical band heterotopia, polymicrogyria, corpus callosum defects |
| CDK5RAP2 | MCPH3 | PCM, spindle orientation, centriole duplication, Hippo pathway regulation? | Microcephaly | Short stature, simplified gyral patterning, corpus callosum defects, hearing loss |
| ASPM | MCPH5 | PCM, spindle integrity and orientation, regulation of actin cytoskeleton? | Microcephaly | Short stature, seizures, simplified gyral patterning |
| CPAP | MCPH6 SCKL4 | PCM, centriole duplication, centriole growth, ciliary disassembly | Microcephaly, short stature (SCKL) | Seizures |
| STIL | MCPH7 | Centriole duplication | Microcephaly | Holoprosencephaly |
| CEP135 | MCPH8 | PCM | Microcephaly | |
| CEP152 | MCPH9 SCKL5 | PCM, centriole duplication | Microcephaly, short stature (SCKL) | Simplified gyral patterning |
| CDK6 | MCPH12 | MTOC activity, cell cycle length | Microcephaly, simplified gyral patterning | |
| SAS6 | MCPH14 | Centriole duplication | Microcephaly | Seizures, abnormal ventricles, cerebellar hypoplasia |
| CEP63 | SCKL6 | PCM, centriole duplication, CDK1 recruitment | Microcephaly, short stature | |
| NIN | SCKL7 | MTOC activity | Microcephaly, short stature | Immature sulcus patterning |
| TUBGCP4 | MCCRP1 | MTOC activity | Microcephaly, short stature | Eye defects, simplified gyral patterning |
| PLK4 | MCCRP2 | Centriole duplication | Microcephaly, short stature | Eye defects, simplified gyral patterning, small cerebellum and brainstem |
| TUBGCP6 | MCCRP3 | MTOC activity | Microcephaly, eye defects | Corpus callosum defects |
| PCNT | MOPD2 | PCM, MTOC activity | Microcephaly, severe short stature |
Here lies the exciting mystery to be solved—linking the cell-biological roles of the centrosome with its roles in brain development. What precise neurogenic mechanisms are disrupted in centrosome-related microcephaly (CRM) mutants? Do different mutations in centrosome genes affect the same or different pathways? We highlight the complexity of the microcephaly disorder by showcasing commonalities and differences between phenotypes of centrosome MCPH and SCKL genes. Untangling CRM mutant contributions to the microcephaly phenotype requires expanding our current models.
FEWER NEURONS, SMALLER BRAIN
Microcephaly is defined by a reduction in brain size reflecting a reduction in the number of neurons. What then is the link between CRM mutations and loss of neurons? Is it simply that centrosomes are required for mitosis and thus disrupting centrosome function reduces the efficacy of cell division, resulting in fewer cells? To put this hypothesis in perspective, we briefly overview mammalian brain development.
The brain develops from a neuroepithelial tube of polarized neural progenitor cells (NPCs) with apical cilia extending into the ventricle (Figure 1; Dwyer et al., 2016). NPCs progress through phases of cell division beginning with expansion of their numbers via symmetrical proliferative divisions. NPCs then begin to divide asymmetrically, generating one daughter that remains an NPC and one daughter that differentiates into an intermediate neural progenitor or a neuron that migrates basally. NPCs can also undergo a final symmetrical division to generate two neurons. The balance between proliferative and differentiative divisions is a key determinant of the final number of neurons in the brain. Current models of microcephaly mainly attribute the disorder to a reduction of the NPC pool, either through increased apoptosis or through premature differentiation. Therefore, understanding how centrosome function is linked to differentiation and apoptosis is key to understanding the roles of CRM genes in brain development.
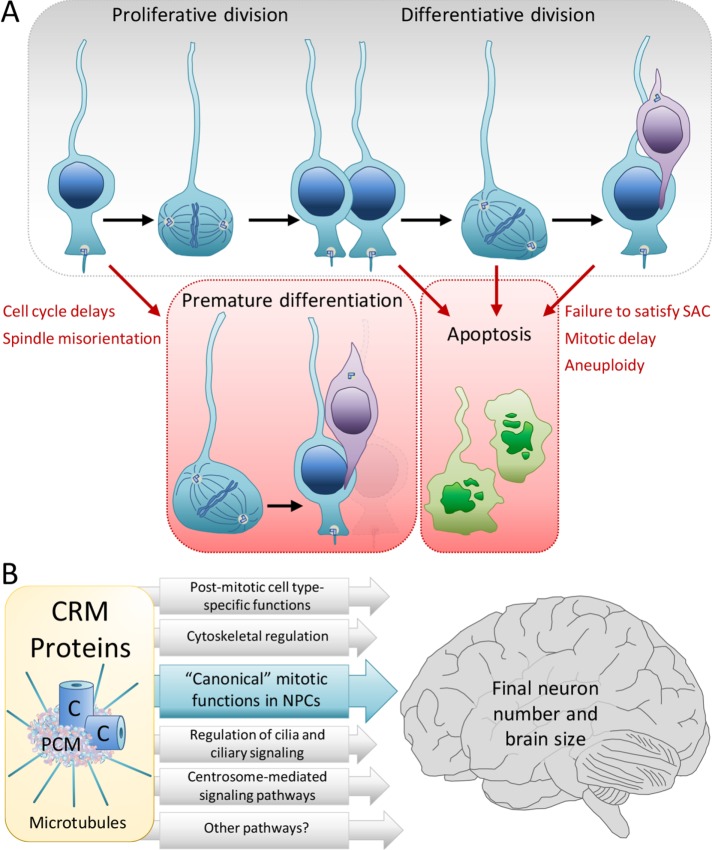
FIGURE 1:
Canonical and noncanonical roles for centrosome-related microcephaly (CRM) genes in neurogenesis and brain size. (A) Neural progenitor cells (NPCs) undergo a series of symmetric proliferative divisions during early neurogenesis to expand the NPC pool. These cells then switch to an asymmetric mode of division that generates neurons and maintains the NPC pool throughout the later stages of neurogenesis (top). Defects in CRM genes can disrupt neurogenic divisions, resulting in loss of NPCs through premature differentiation due to spindle misorientation and cell cycle delays, or activation of apoptotic pathways due to failure to satisfy the SAC, mitotic delays, or aneuploidy (bottom). The end result of the depleted NPC pool is a reduction in final neuron number and ultimately brain size. (B) Schematic showing canonical mitotic functions for CRM genes (blue) and additional noncanonical roles (gray) that collectively contribute to proper neurogenesis and brain size.
The literature suggests a clear link. In NPCs, defects in spindle stability can cause prolonged mitosis and a delay in satisfying the spindle assembly checkpoint (SAC), leading to apoptosis (Chen et al., 2014; Sgourdou et al., 2017). Defects in cell fate and differentiation are also controlled, in part, by centrosomes through mitotic spindle misorientation (Li et al., 2017), and through mother and daughter centriole inheritance (Wang et al., 2009). Additionally, premature differentiation of NPCs can be triggered by improper centrosome-mediated cell cycle regulation (Capecchi and Pozner, 2015) or delayed ciliary disassembly (Gabriel et al., 2016). In Drosophila, loss of both centrosomes and SAC causes increased cell death, premature differentiation, and a decreased proliferation rate of neural stem cells (Poulton et al., 2017), pointing to the critical importance of mitotic functions in brain growth.
Thus, defects in centrosomes can increase both apoptosis and differentiation. This big-picture view is well substantiated, but many critical details remain unclear. It is also puzzling why the list of CRM mutations is not more expansive, including all genes critical for mitosis, differentiation, and apoptosis. As one investigates each CRM mutant in more detail, it becomes clear that the seemingly linear pathway to a smaller brain is much more complex.
CRM PROTEINS: BOUND TOGETHER, BUT FUNCTIONING INDEPENDENTLY
Centrosome proteins form a highly interconnected and dynamic network, allowing centrosomes to play many roles (Galletta et al., 2016). This does not mean, however, that all centrosome proteins are required for all centrosome functions. In fact, many centrosome proteins have multiple cell type–specific and cell cycle–dependent roles, controlled by specific biochemical modifications and binding partners. For example, CPAP plays critical roles in centriole duplication (Tang et al., 2011), spindle pole integrity (Chou et al., 2016), and ciliary disassembly (Gabriel et al., 2016). There are also several moonlighting roles for CRM proteins away from the centrosome. For example, Drosophila Ana2 (STIL) functions both at the centriole (in procentriole formation) and away from the centriole at the cell cortex (in spindle pole orientation; Wang et al., 2011). Therefore, while CRM proteins have clear overlapping functions, they are likely to participate in unique mechanisms or pathways that contribute to the control of brain size.
Independent roles for CRM genes are further supported by the observation that CRM mutations, in different genes or the same gene, cause MCPH with variant additional phenotypes. Some examples include WDR62 mutations, which show several additional structural defects in the brain cortex (Bilgüvar et al., 2010); CPAP mutations, which are associated with either MCPH or SCKL (Bond et al., 2005; Al-Dosari et al., 2010); and CDK5RAP2 mutations, which are linked to MCPH, a more severe SCKL-like phenotype with deafness (Lancaster et al., 2013), or a more minor defect affecting only the corpus callosum (Jouan et al., 2016).
Collectively, these data suggest that many pathways are likely in play, and that a single model of neurogenic defects cannot explain all cases of CRM. To further probe this idea, we will next examine specific CRM genes to identify whether microcephaly is due to a role in differentiation, mitosis, apoptosis, or yet another unforeseen role.
ASPM: REGULATING THE ACTIN CYTOSKELETON TO CONTROL TISSUE ARCHITECTURE?
ASPM is the most commonly mutated CRM gene, accounting for 25–50% of all MCPH cases (Thornton and Woods, 2009). Mouse models of ASPM microcephaly have reduced cortical layers exhibiting premature differentiation of NPCs (Fish et al., 2006; Capecchi and Pozner, 2015). Early studies in ASPM-depleted mice point to a defect in NPC spindle orientation with increased asymmetric divisions and a subsequent decrease in the progenitor pool as the primary mechanism underlying microcephaly (Fish et al., 2006); subsequent work indicates that this model is incomplete.
More recently, ASPM was shown to regulate time spent in G1 by protecting Cyclin E from ubiquitin-mediated degradation, so that loss of ASPM can cause premature differentiation via cell cycle lengthening (Capecchi and Pozner, 2015). Drosophila mutants of the ASPM orthologue asp also have a smaller brain with spindle and cell division defects, suggesting a conserved function (Rujano et al., 2013; Schoborg et al., 2015). Interestingly, separation of function mutations show that reduced brain size is at least partially independent of spindle defects (Schoborg et al., 2015). Instead, the reduced brain size in asp mutant flies is related to its role in regulating the actin cytoskeleton to control neuroepithelial architecture (Rujano et al., 2013). These results are consistent with experiments in mice showing that randomization of spindle orientation is associated with premature differentiation, but insufficient to cause reduction in cortical layers (Li et al., 2017). Furthermore, ASPM mutant mice also exhibit disrupted apical epithelial architecture in the ventricular zone (Jayaraman et al., 2016), suggesting that regulation of the actin cytoskeleton is a conserved mechanism contributing to proper brain size by Asp/ASPM. Thus, the prominent role of ASPM in spindle organization appears to play a relatively minor role in microcephaly. Exploring other roles for ASPM in more depth is a critical future research focus.
WDR62: A GLIAL-SPECIFIC FUNCTION IN MAMMALS?
WDR62, the second most commonly mutated gene in human MCPH patients, also appears to have unexpected additional roles beyond its function in NPC division, which might underlie microcephaly. WDR62 is best known for its functions in maintaining mitotic centrosome and spindle integrity by recruiting CPAP, both through a complex with CEP63 and ASPM, and through activation of Aurora A kinase (Chen et al., 2014; Chou et al., 2016; Jayaraman et al., 2016). WDR62 mutants have defective attachment of centrosomes to mitotic spindles, disorganized PCM, abnormal microtubule nucleation, and improper spindle orientation (Bogoyevitch et al., 2012; Chen et al., 2014; Ramdas Nair et al., 2016; Sgourdou et al., 2017). Furthermore, centrosome and spindle defects in WDR62 mutant mouse NPCs prevent satisfaction of SAC and cause mitotic delay and apoptosis, leading to a reduction in cortical layers (Chen et al., 2014; Sgourdou et al., 2017).
In Drosophila, Wdr62 mutants also have reduced PCM recruitment and show reduced brain size (Ramdas Nair et al., 2016; Lim et al., 2017), indicating conserved function. This work, however, shows a surprising deviation from the canonical WDR62 function, as small brains in Wdr62 mutant flies are linked to a deficit in postmitotic glial cells rather than neural stem cells. Wdr62 depletion in neural stem cells is not sufficient to reduce brain size, whereas Wdr62 depletion in glial cells causes loss of both glia and stem cells and reduced brain size, suggesting that glial signaling is necessary to maintain neural stem cell identity (Lim et al., 2017). This glia-specific function depends on the interaction between Wdr62 and Aurora A, indicating further conservation between mammals and flies. Although glial cells have been shown to regulate mammalian NPC numbers (Cunningham et al., 2013), it is unclear whether WDR62 is involved in such processes; further studies are warranted.
CPAP: POSTER BOY FOR MULTIPLE PATHWAYS TO MICROCEPHALY?
CPAP is a multifunctional CRM gene, with roles in centriole duplication and elongation, PCM organization, and ciliary disassembly (Tang et al., 2011; Zheng et al., 2014; Gabriel et al., 2016; Sharma et al., 2016). Humans with CPAP mutations present with a range of phenotypic severity, and studies of various CPAP microcephaly models suggest distinct underlying mechanisms. For example, an MCPH CPAP variant with a single amino acid substitution in the TCP domain fails to localize efficiently to the centriole, fails to support centriole duplication, and is defective in recruiting several PCM components in cultured NPCs (Tang et al., 2011; Zheng et al., 2014). In contrast, NPCs in SCKL patient-derived organoids with a mutation deleting CPAP’s CC5 domain have proper centriole duplication, spindle morphology, and recruitment of key PCM components. However, their NPCs have defects in ciliary disassembly, and the increased time required to resorb the cilium causes a corresponding delay in the G1-S transition, leading to a loss of NPCs through premature differentiation (Gabriel et al., 2016). CPAP null mutant mice have NPCs with normal spindle orientation, chromosome segregation, and interphase cell cycle progression; however, NPCs undergo increased apoptosis due to both prometaphase delay and premature differentiation (Bazzi and Anderson, 2014; Insolera et al., 2014). Further, CPAP depletion in neurons impairs neuronal migration, revealing an additional postmitotic function for CPAP (Garcez et al., 2015). Nonetheless, that such distinct mechanisms stemming from CPAP have all been implicated in microcephaly suggests that additional phenotypic complexity is masked by broad clinical definitions.
Given that ASPM, WDR62, and CPAP utilize novel cellular mechanisms in both mitotic and postmitotic cells to control brain size, a key question emerges—is it possible that other CRMs control brain size by mechanisms unrelated to their canonical cell division functions?
CDK5RAP2: A KEY REGULATOR OF THE HIPPO SIGNALING PATHWAY?
Disruption of PCM organizing and spindle pole focusing functions of CDK5RAP2 play a major role in CDK5RAP2 mutant microcephaly (Fong et al., 2008; Kodani et al., 2015; Chavali et al., 2016); however, recent work suggests the possibility of additional defects in centrosome-mediated signaling pathways, such as Hippo (Sukumaran et al., 2017). Mutant CDK5RAP2 patient–derived cells and CDK5RAP2 mouse models link premature differentiation and apoptosis with a number of mitosis-related phenotypes, including defective centriole duplication, mitotic PCM disorganization, spindle misorientation, and aneuploidy (Buchman et al., 2010; Lizarraga et al., 2010; Lancaster et al., 2013; Yigit et al., 2015). Interestingly, however, CDK5RAP2 was recently shown to interact with Hippo pathway proteins, and CDK5RAP2 MCPH patient-derived cells show altered Hippo pathway protein levels, indicating abnormal Hippo signaling (Sukumaran et al., 2017). The Hippo pathway is a key regulator of cell proliferation, apoptosis, and organ size (Yu et al., 2015). Further, many Hippo pathway components are apically localized (Yu and Guan, 2013), and in Drosophila neural stem cells, phosphorylation by the Hippo pathway kinase Warts is required for the localization of some apical complex proteins (Keder et al., 2015). Disruption of Hippo signaling could potentially affect cell polarity and prevent proper localization of apical cell fate determinants, thereby altering cell fate decisions. Thus, Hippo signaling is well situated to play additional roles in determining brain size. Given the proposed roles of centrosomes and cilia as major centers of signal transduction (Arquint et al., 2014), signaling pathways converging on the centrosome are likely to contribute to CRM in some mutants as well.
CONCLUDING REMARKS
Microcephaly is an extremely complex disorder, with nearly 30 genes linked to it to date. Many of these genes encode proteins, which can be classified into several broad functional groups, including DNA damage response, centromere organization, cell cycle control, chromatin regulation, and centrosome-related proteins. At first glance, CRM proteins seem to be the easiest class to investigate, given their role in mitotic spindle formation and its link to premature NPC differentiation and apoptosis. However, this model has fallen out of favor in light of studies showing that cell fate determination can be altered without causing microcephaly (Li et al., 2017). The likely explanation is a complex blend of mitotic and nonmitotic function for CRM genes in both progenitors and postmitotic cells during brain development.
Characterization of microcephalic mutants and identification of novel neurogenic mechanisms underlying the phenotype require research models with complex neurodevelopment, and thus animal models and cultured brain organoids are well suited to the task. Considering the substantial similarities between Drosophila and mammalian neurogenesis (Homem and Knoblich, 2012) and the apparently well-conserved roles of microcephaly-associated genes between these species, we anticipate that studies in simple model organisms will reveal gene functions important for microcephaly, especially given the wide range of genetic manipulations allowing interrogation of mitotic and postmitotic roles. Similarly, we anticipate that as cerebral organoid culture becomes increasingly standardized, reproducible, and accessible, it will become an immensely powerful system for elucidating mechanisms of neurogenesis. Such model systems are particularly useful because they allow testing of different mutant isoforms with a common genetic background. While patient-derived cells are certainly informative, genetic background effects are expected to be significant, especially since many patients are consanguineous. Recapitulating human mutations allows characterization of mutation-specific defects in neurogenesis; however, to identify and tease apart specific mechanisms that contribute to microcephaly phenotypes, experiments using separation of function mutations are required. Thus, the study of microcephaly-associated genes is an exciting field of research that is well suited to a combination of basic cell and developmental biological analysis, which promises to reveal a more complete picture of how complex pathways cooperate to give rise to our most complex organ.
Acknowledgments
N.M.R. is supported by the Division of Intramural Research at the National Institutes of Health/National Heart, Lung, and Blood Institute (1ZIAHL006126).
Abbreviations used:
- CRM
centrosome-related microcephaly
- MCPH
microcephaly primary hereditary
- MTOC
microtubule-organizing center
- NPC
neural progenitor cell
- PCM
pericentriolar matrix
- SAC
spindle assembly checkpoint
- SCKL
Seckel syndrome.
Footnotes
REFERENCES
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS. Novel CENPJ mutation causes Seckel syndrome. J Med Genet. 2010;47:411–414. doi: 10.1136/jmg.2009.076646. [DOI] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk A-M, Nigg EA. Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130464. doi: 10.1098/rstb.2013.0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci USA. 2014;111:E1491–E1500. doi: 10.1073/pnas.1400568111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgüvar K, Oztürk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizog˘lu D, Tüysüz B, Cag˘layan AO, Gökben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Yeap YYC, Qu Z, Ngoei KR, Yip YY, Zhao TT, Heng JI, Ng DCH. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J Cell Sci. 2012;125:5096–5109. doi: 10.1242/jcs.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Buchman JJ, Tseng H-C, Zhou Y, Frank CL, Xie Z, Tsai L-H. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron. 2010;66:386–402. doi: 10.1016/j.neuron.2010.03.036. [DOI] [PubMed] [Google Scholar]
- Capecchi MR, Pozner A. ASPM regulates symmetric stem cell division by tuning cyclin E ubiquitination. Nat Commun. 2015;6:8763. doi: 10.1038/ncomms9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali PL, Chandrasekaran G, Barr AR, Tátrai P, Taylor C, Papachristou EK, Woods CG, Chavali S, Gergely F. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat Commun. 2016;7:11005. doi: 10.1038/ncomms11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-F, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat Commun. 2014;5:3885. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou E-J, Hung L-Y, Tang C-JC, Hsu W-B, Wu H-Y, Liao P-C, Tang TK. Phosphorylation of CPAP by aurora-A maintains spindle pole integrity during mitosis. Cell Rep. 2016;14:2975–2987. doi: 10.1016/j.celrep.2016.02.085. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martínez-Cerdeño V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci. 2013;33:4216–4233. doi: 10.1523/JNEUROSCI.3441-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerinckx S, Abramowicz M. The genetics of congenitally small brains. Semin Cell Dev Biol. 2017 doi: 10.1016/j.semcdb.2017.09.015. [DOI] [PubMed] [Google Scholar]
- Dwyer ND, Chen B, Chou S-J, Hippenmeyer S, Nguyen L, Ghashghaei HT. Neural stem cells to cerebral cortex: emerging mechanisms regulating progenitor behavior and productivity. J Neurosci. 2016;36:11394–11401. doi: 10.1523/JNEUROSCI.2359-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. ASPM specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K-W, Choi Y-K, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the -tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, Pozniakovsky A, Poser I, Noack F, Telugu NS, Calegari F, et al. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J. 2016;35:803–819. doi: 10.15252/embj.201593679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Fagerstrom CJ, Schoborg TA, McLamarrah TA, Ryniawec JM, Buster DW, Slep KC, Rogers GC, Rusan NM. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat Commun. 2016;7:12476. doi: 10.1038/ncomms12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Diaz-Alonso J, Crespo-Enriquez I, Castro D, Bell D, Guillemot F. Cenpj/CPAP regulates progenitor divisions and neuronal migration in the cerebral cortex downstream of Ascl1. Nat Commun. 2015;6:6474. doi: 10.1038/ncomms7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CCF, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi S-H. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Kodani A, Gonzalez DM, Mancias JD, Mochida GH, Vagnoni C, Johnson J, Krogan N, Harper JW, Reiter JF, et al. Microcephaly proteins WDR62 and ASPM define a mother centriole complex regulating centriole biogenesis, apical complex, and cell fate. Neuron. 2016;92:813–828. doi: 10.1016/j.neuron.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouan L, Ouled Amar Bencheikh B, Daoud H, Dionne-Laporte A, Dobrzeniecka S, Spiegelman D, Rochefort D, Hince P, Szuto A, Lassonde M, et al. Exome sequencing identifies recessive CDK5RAP2 variants in patients with isolated agenesis of corpus callosum. Eur J Hum Genet. 2016;24:607–610. doi: 10.1038/ejhg.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keder A, Rives-Quinto N, Aerne BL, Franco M, Tapon N, Carmena A. The Hippo pathway core cassette regulates asymmetric cell division. Curr Biol. 2015;25:2739–2750. doi: 10.1016/j.cub.2015.08.064. [DOI] [PubMed] [Google Scholar]
- Kodani A, Yu TW, Johnson JR, Jayaraman D, Johnson TL, Al-Gazali L, Sztriha L, Partlow JN, Kim H, Krup AL, et al. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. Elife. 2015;4:e07519. doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit DA, Poulton JS. Centrosomes are multifunctional regulators of genome stability. Chromosome Res. 2016;24:5–17. doi: 10.1007/s10577-015-9506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Kroll T, Moll J, Frappart L, Herrlich P, Heuer H, Ploubidou A. Spindle misorientation of cerebral and cerebellar progenitors is a mechanistic cause of megalencephaly. Stem Cell Rep. 2017;9:1071–1080. doi: 10.1016/j.stemcr.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim NR, Shohayeb B, Zaytseva O, Mitchell N, Millard SS, Ng DCH, Quinn LM. Glial-specific functions of microcephaly protein WDR62 and interaction with the mitotic kinase AURKA are Essential for Drosophila brain growth. Stem Cell Rep. 2017;9:32–41. doi: 10.1016/j.stemcr.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han A-P, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano M, Basto R. Consequences of centrosome dysfunction during brain development. Adv Exp Med Biol. 2017;1002:19–45. doi: 10.1007/978-3-319-57127-0_2. [DOI] [PubMed] [Google Scholar]
- Poulton JS, Cuningham JC, Peifer M. Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J Cell Biol. 2017;216:1255–1265. doi: 10.1083/jcb.201607022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas Nair A, Singh P, Salvador Garcia D, Rodriguez-Crespo D, Egger B, Cabernard C. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Rep. 2016;14:1100–1113. doi: 10.1016/j.celrep.2015.12.097. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Sanchez-Pulido L, Pennetier C, le Dez G, Basto R. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat Cell Biol. 2013;15:1294–1306. doi: 10.1038/ncb2858. [DOI] [PubMed] [Google Scholar]
- Schoborg T, Zajac AL, Fagerstrom CJ, Guillen RX, Rusan NM. An Asp-CaM complex is required for centrosome-pole cohesion and centrosome inheritance in neural stem cells. J Cell Biol. 2015;211:987–998. doi: 10.1083/jcb.201509054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourdou P, Mishra-Gorur K, Saotome I, Henagariu O, Tuysuz B, Campos C, Ishigame K, Giannikou K, Quon JL, Sestan N, et al. Disruptions in asymmetric centrosome inheritance and WDR62-Aurora kinase B interactions in primary microcephaly. Sci Rep. 2017;7:43708. doi: 10.1038/srep43708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Aher A, Dynes NJ, Frey D, Katrukha EA, Jaussi R, Grigoriev I, Croisier M, Kammerer RA, Akhmanova A, et al. Centriolar CPAP/SAS-4 imparts slow processive microtubule growth. Dev Cell. 2016;37:362–376. doi: 10.1016/j.devcel.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran SK, Stumpf M, Salamon S, Ahmad I, Bhattacharya K, Fischer S, Müller R, Altmüller J, Budde B, Thiele H, et al. CDK5RAP2 interaction with components of the Hippo signaling pathway may play a role in primary microcephaly. Mol Genet Genom. 2017;292:365–383. doi: 10.1007/s00438-016-1277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C-JC, Lin S-Y, Hsu W-B, Lin Y-N, Wu C-T, Lin Y-C, Chang C-W, Wu K-S, Tang TK. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011;30:4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome. Trends Genet. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Hehnly H, Doxsey S. The centrosome, a multitalented Renaissance organelle. Cold Spring Harb Perspect Biol. 2016;8:a025049. doi: 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li S, Januschke J, Rossi F, Izumi Y, Garcia-Alvarez G, Gwee SS, Soon SB, Sidhu HK, Yu F, et al. An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev Cell. 2011;21:520–533. doi: 10.1016/j.devcel.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, Imai JH, Lian W-N, Vallee RB, Shi S-H. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit G, Brown KE, Kayserili H, Pohl E, Caliebe A, Zahnleiter D, Rosser E, Bögershausen N, Uyguner ZO, Altunoglu U, et al. Mutations in CDK5RAP2 cause Seckel syndrome. Mol Genet Genom Med. 2015;3:467–480. doi: 10.1002/mgg3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Guan K-L. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Zhao B, Guan K-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gooi LM, Wason A, Gabriel E, Mehrjardi NZ, Yang Q, Zhang X, Debec A, Basiri ML, Avidor-Reiss T, et al. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc Natl Acad Sci USA. 2014;111:E354–E363. doi: 10.1073/pnas.1317535111. [DOI] [PMC free article] [PubMed] [Google Scholar]