Abstract
Mycobacterium sp. ELW1 co-metabolically degraded up to 1.8 µmol phenanthrene (PHE) in ~48 hours (hr) and the formation of hydroxyphenanthrene (OHPHE) metabolites, including 1-hydroxyphenanthrene (1-OHPHE), 3-hydroxyphenanthrene (3-OHPHE), 4-hydroxyphenanthrene (4-OHPHE), 9-hydroxyphenanthrene (9-OHPHE), 9,10-dihydroxyphenanthrene (1,9-OHPHE), and trans-9,10-dihydroxy-9,10-dihydrophenanthrene (trans-9,10-OHPHE), were identified and quantified over time. The monooxygenase responsible for co-metabolic transformation of PHE was inhibited by 1-octyne. First-order PHE transformation rates, kPHE, and half-lives, t1/2, for PHE-exposed cells ranged 0.16 – 0.51 hr−1 and 1.4 – 4.3 hr, respectively, and the 1-octyne controls ranged 0.015 – 0.10 hr−1 and 7.0 – 47 hr, respectively. While single compound standards of PHE and trans-9,10-OHPHE, the major OHPHE metabolite formed by ELW1, were not toxic to embryonic zebrafish (Danio rerio), single compound standards of minor OHPHE metabolites, 1-OHPHE, 3-OHPHE, 4-OHPHE, 9-OHPHE, and 1,9-OHPHE, were toxic, with effective concentrations (EC50s) ranging from 0.5– 5.5 µM. The metabolite mixtures formed by ELW1, and the reconstructed standard mixtures of the identified OHPHE metabolites, elicited a toxic response in zebrafish for the same 3 time points. EC50s for the metabolite mixtures formed by ELW1 were lower (more toxic) than those for the reconstructed standard mixtures of the identified OHPHE metabolites. Ten unidentified hydroxy PHE metabolites were measured in the derivatized mixtures formed by ELW1 and may explain the increased toxicity of the ELW1 metabolites mixture, relative to the reconstructed standard mixtures of the identified OHPHE metabolites.
TOC Art
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are compounds comprised of two or more fused benzene rings that form during the incomplete combustion of organic substances from both natural and anthropogenic sources.1 PAHs are also found in asphalt,2 crude oil,3 coal tar,4–6 and creosote.7,8 Many PAHs are considered to be probable or possible human and animal carcinogens by the International Agency for Research on Cancer (IARC), as well as probable human carcinogens by the U.S. EPA Integrated Risk Information System.9,10 Due to their ubiquity and known toxicity, remediation of PAH-contaminated soils and sediments is ongoing.
Many remediation technologies, including bioremediation, chemical oxidation, and thermal treatments, have been used for PAH removal.11 Bioremediation is a cost effective strategy that can be used in situ, ex situ, aerobically, anaerobically, and further enhanced through biostimulation and/or bioaugmentation.11–14 Two possible processes that occur during bioremediation are 1) degradation (the process of microorganisms utilizing PAHs as an energy source and to form new biomass) and/or 2) co-metabolism (the process of microorganisms transforming PAHs without metabolic benefits).15 However, bioremediation of PAHs is limited due to their low aqueous solubilities (0.003 to 234 µmol L−1 at 25°C) and high affinity for organic matter.16 These properties result in low bioavailability of PAHs to microorganisms. When microorganisms come in contact with PAHs, essential enzymes (oxygenases) need to be present in the cell for biodegradation and/or co-metabolism to occur.17
Microorganisms contain highly diverse and selective oxygenases and/or hydroxylases that are necessary for aerobic degradation and/or co-metabolism. The initial step in the transformation of PAHs requires oxidation of the rings by ring-hydroxylating oxygenases (RHOs), specifically either monooxygenases (RHMs) or dioxygenases (RHDs). Oxidation by RHOs occurs through the addition of one or two hydroxy groups, to regiospecific and stereoselective carbon atoms, to form dihydrodiols.18 The selection of the activation site is determined by the shape of the enzyme and determines the metabolite structures.19,20 Dihydrodiols undergo dehydrogenation to form PAH-diols using dihydrodiols dehydrogenases (DHDGs), followed by a ring-opening step performed by ring cleavage dioxygenases (RCD).18 The process is repeated as long as the microbe has structurally specific RHOs, DHDGs, and RCDs for the metabolites that are formed at each step. Co-metabolism of PAHs occurs through the broad specificity of RHOs. Studies have shown naphthalene dioxygenase, present in Pseudomonas sp. NCIB 9816 and P. aeruginosa strain PAO1, was able to oxidize 50 aromatic compounds and three-ringed PAHs, respectively.21,22 Dioxygenase complexes in Mycobacterium sp. strain AP1, Sphingomonas sp. strain LH128 and CHY-1 were able to oxidize PAHs with up to 5 rings.23–25 Microorganisms have different co-metabolism capabilities based on their RHOs.
Accumulation of PAH metabolites occurs when microbes stop degradation or co-metabolism at certain metabolites. These metabolites may be more toxic than the parent PAHs.26–30 Coal tar-contaminated soil, collected from a manufactured gas plant site, was bioremediated using an unknown microbial consortium in an aerobic bioreactor and showed statistically significant increases in developmental toxicity in embryonic zebrafish (Danio rerio), as well as genotoxicity using the DT40 bioassay.27 In addition, the toxicological response of PAH mixtures have been inconsistent, with some studies showing an additive effect (the sum of the toxicity of the individual compounds) or antagonist effect (less than the expected toxicity if the effects were additive).31–38
Because it is so ubiquitous in the environment and is often the most abundant PAH, phenanthrene (PHE) has been used as a model compound to study the ability for microbes to degrade PAHs.39–43 In addition, PHE is commonly described as a prototype PAH due to the replication of its 3-ring structure throughout higher-ringed, more carcinogenic PAHs, such as benzo[a]pyrene.40–42 PHE has also been used as a model PAH for human metabolism studies because its bay-region and K-region lead to the possible formation of more carcinogenic PAHs.39,43 PHE also has a moderate aqueous solubility of 6.73 µmol L−1, compared to the range of aqueous solubilities for other parent PAHs (0.003 to 234 µmol L−1), and this is important for bioremediation studies in aqueous systems.16,17
For this study, the ability of a novel microorganism, Mycobacterium sp. strain ELW1, to transform PHE in an aqueous system was evaluated. ELW1 was isolated from stream sediment using isobutene (2-methylpropene) as the single source of carbon for growth and energy.44 PCR sequencing of the 16S rRNA gene of ELW1 indicates that it is a Mycobacterium strain.44 The ability of ELW1 to utilize isobutene as a growth substrate has potential for the in situ stimulation of endogenous microbial populations through isobutene and oxygen addition, and may lead to a novel method to promote the initial oxidation of PAHs for subsurface bioremediation, through co-metabolic transformations.
The purpose of this study was to determine the rate of PHE transformation by ELW1, identify and quantify OHPHE metabolites formed by ELW1, and to characterize the toxicity of PHE metabolite mixtures formed by ELW1, using the embryonic zebrafish model. To our knowledge, we are the first to identify and quantify a wide range of phenanthrene microbial metabolites, as well as to evaluate their development toxicity in mixtures formed by a microorganism. In addition, we are the first to evaluate ELW1 for the transformation of PAHs. This study has broad implications for the use of bioremediation to clean up PAH contaminated sites, because it suggests that the toxicity of PAH metabolite mixtures formed should be considered as part of the initial assessment for site remediation.
MATERIALS AND METHODS
Chemicals
Standards for PHE, 16 OHPHEs, and isotopically-labeled PAHs were purchased from various vendors, listed in the Supporting Information Table S1. Dichloromethane (DCM), methanol (MeOH), acetone, ethyl acetate, and acetonitrile were purchased from EMD Millipore (Gibbstown, NJ). Toluene (≥ 99%), dimethyl sulfoxide (DMSO) (≥ 99%), the derivatizing agent, N,O-bis-(trimethylsilyl) trifluoroacetamide (BSTFA), and liquid 1-octyne were purchased from Sigma Aldrich (St. Louis, MO). Isobutene (C.P. grade) was acquired from Gas Innovations.
Phenanthrene Transformation and Metabolite Formation Experiments
Pure Mycobacterium sp. strain ELW1 was originally isolated by, and acquired from, Michael Hyman at North Carolina State University.44 Methods used to grow the culture are described in detail in the Supporting Information. Briefly, pure cultures were grown in batch using 500 mL glass media bottles and mineral salt medium (MSM), with ~10% (v/v) isobutene in the headspace, until the cell density was OD600 > 0.7 (usually 6–8 days) measured using UV-VIS spectrophotometer analysis (Orion Aquamate 8000, ThermoFisher Scientific). Cellular protein was determined by lysing the cells in NaOH and calorimetrically determining the concentration using UV-VIS at 540 nm and a calibration curve (Detailed Methods, Protein Assay in Supporting Information). The cells were harvested, centrifuged, and re-suspended in fresh MSM. Cellular activity of the concentrated cells was determined by measuring the rate of isobutene consumption (Figure S6 and Table S10).
PHE transformation studies were conducted as resting cell tests, using ~25–40 mg of cells, in the absence of isobutene. PHE standards were prepared in MeOH and spiked in 303 mL MSM in 500 mL media bottles at a maximum of 0.0089% MeOH (v/v) in each reactor, which did not negatively impact the activity of ELW1 (Figure S8). Control reactors included 1-octyne controls (MSM, cells, PHE, and 1-octyne), PHE-only controls (MSM and PHE) and cell-only controls (MSM and cells). 1-Octyne (~7 µmol in the aqueous media) was used as a control to inhibit the alkene monooxygenase involved in isobutene oxidation to reduce PHE transformation and better understand the mechanism of transformation.44 Preparation of stock gas-phase 1-octyne and determination of optimal 1-octyne aqueous concentration is explained in detail in the Supporting Information. 1-Octyne controls and PHE-only controls were prepared with 0.033, 0.068, 0.14, and 0.84 µmol PHE. Reactors containing PHE-exposed cells (MSM, PHE, and cells) were prepared with 0.033, 0.068, 0.42, 0.84, 1.2, and 1.8 µmol PHE. All reactors were prepared in triplicate and equilibrated with isobutene and 1-ocytne (for 1-octyne controls) in a temperature-controlled 30°C room (on a rotary shaker table operating at 200 rpm) for ~1 hr prior to the addition of PHE and re-suspended cells (~25 mg per reactor).
Liquid samples were collected immediately after the addition of cells, followed by collections at approximately 1, 3, 5, 10, 22, 48, and 72 h, as needed, until PHE was no longer measured in the samples (see Chemical Analysis).
Sample Extraction and Preparation
Samples (5 – 15 mL) used to determine PHE transformation kinetics were collected from each of the triplicate reactors and spiked with labeled surrogates (listed in Table S3) immediately after collection to account for analyte loss during sample extraction. All samples were extracted using solid phase extraction (SPE), with a modified version of a previously published method.45 Bond Elut Plexa (30 mg, 3 mL) cartridges (Agilent Technologies, New Castle, DE) were preconditioned with 5 mL MeOH, followed by 5 mL deionized water. The liquid samples were added to the cartridges, analytes were retained on the sorbent, and the eluent was discarded. Analytes were eluted with 5 mL acetone and 16 mL DCM, dried with sodium sulfate, and concentrated using a TurboVapR evaporator (nitrogen gas, 30 °C water bath).
After SPE, the PHE transformation kinetics sample extracts were solvent exchanged to ethyl acetate, spiked with d10-Acenaphthene (internal standard) for a final volume of 300 µL, and analyzed for PHE using GC/MS (described below). The mean recovery of PHE across the entire analytical method was 54 ± 2.7% (Table S3).
In addition to PHE, all extracts were analyzed for 1-hydroxynaphthalene (1-OHNAP), 2-hydroxynaphthalene (2-OHNAP), 1,5-dihydroxynaphthalene (1,5-OHNAP), 1,6-dihydroxynaphthalene (1,6-OHNAP), 2,3-dihydroxynaphthalene (2,3-OHNAP), 2,6-dihydroxynaphthalene (2,6-OHNAP), 2,7-dihydroxynaphthalene (2,7-OHNAP), 1-hydroxy-2-naphthoic acid (1-OH-2-NAP), 1-hydroxyphenanthrene (1-OHPHE), 3-hydroxyphenanthrene (3-OHPHE), 4-hydroxyphenanthrene (4-OHPHE), 9-hydroxyphenanthrene (9-OHPHE), 1,9-dihydroxyphenanthrene (1,9-OHPHE), trans-9,10-dihydroxy-9,10-dihydrophenanthrene (trans-9,10-OHPHE), and cis-9,10-dihydroxy-9,10-dihydrophenanthrene (cis-9,10-OHPHE) (Table S1, Figure S1). A 50 µL aliquote of the 300 µL sample extract was transferred into a 300 µL spring insert containing 100 µL acetonitrile and 20 µL toluene and concentrated to 20 µL using a fine stream of nitrogen.45,46 BSTFA (30 µL) was added to the extract and the mixture was incubated at 70 °C for 40 min.46 Storage stability of OHPHEs, as well as intra- and inter-day variability, has been previously assessed.45 The mean recovery of the OHPHEs across the entire analytical method ranged from 19 ± 1.5 to 83 ± 3.8% (Table S3).
Chemical Analysis
PHE and OHPHEs were analyzed on an Agilent 6890 gas chromatograph (GC) coupled to a 5973N mass spectrometer (MS), with electron ionization (EI) in selective ion monitoring (SIM) mode, using a DB-5MS (Agilent, 30 m × 0.25 mm I.D., 0.25 µm film thickness) column. OHPHEs were also analyzed in full scan mode. The instrument methods are described in Supporting Information. Commercially available standards were used to quantify OHPHEs that were able to be derivatized by BSTFA. SIM method parameters for BSTFA-derivatized OHPHEs, including windows, fragment ions, and retention times, are listed in Table S4. The estimated detection limits (EDLs) were calculated following EPA Method 8280 and was 0.51 pg µL−1 for PHE (Table S3) and ranged 0.86 – 5.0 pg µL−1 for OHPHEs (Table S3).47
Embryonic Zebrafish Bioassay
PHE and OHPHE mixtures formed by ELW1 were generated by exposing ELW1 to 1.8 µmol PHE in the same reactor bottles described above for the PHE transformation kinetic studies. At 5, 28, 51, 76, and 122 hr, 50 mL was collected from each reactor (n = 4) of the PHE-exposed cells, 1-octyne control, PHE-only control, and cells-only controls and combined. Samples were extracted as described above (Sample Extraction and Preparation) but were not spiked with surrogates to prevent toxicity caused by the addition of the surrogates. After SPE, the extracts were split gravimetrically, with 80% of the extract being used for toxicity testing (toxicity fractions) and the remaining 20% being used for chemical analysis (chemical fractions). Chemical fractions were solvent exchanged to ethyl acetate, spiked with d10-Acenaphthene (internal standard) for a final volume of 300 µL, and analyzed for PHE and OHPHEs as described above. These results were used to calculate the concentrations of PHE and OHPHEs in the toxicity fractions used for toxicity testing. The toxicity fractions were blown to dryness and reconstituted with 100 µL DMSO to a concentration of ~8 mM total PHE and OHPHE concentration.
Single OHPHE standards, mixture extracts formed by ELW1 for various time points, and reconstructed OHPHE standard mixtures (prepared in the same OHPHE ratios as the mixture extracts formed by ELW1 using single OHPHE standards, Table S8) were assessed in embryonic zebrafish according to Truong et al.48 At 6 hours post-fertilization (hpf), dechorinated zebrafish embryos (n = 32) were exposed to different concentrations (ranging from 0.01 – 60 µM) of single OHPHE standards, mixture extracts formed by ELW1 at various time points, and reconstructed OHPHE standard mixtures. Briefly, embryos were dechorionated and exposed to 0 – 50 µM OHPHE standards (32 animals per concentration, 6 concentrations) from 6–120 hours post fertilization (hpf). For the mixtures, embryos were exposed to 6 nominal water (zebrafish embryo media) concentrations, from 0.01 to 60 µM, depending on the concentration of the stock produced. Although the high-end test concentration (60 µM) was above the water solubility of PHE (~ 7 µM), the solubility of PHE was likely enhanced slightly by the DMSO present and zebrafish embryo media. In addition, there was no evidence of PHE being insoluble at this high-end dose. Embryos were statically exposed and evaluated at 24 and 120 hpf for 22 morbidity and mortality endpoints. A Fisher’s Exact test was performed for each of the 22 endpoints and a cumulative “any” effect to allow for detection for any aberrant animals.49
In addition to the bioassay, the toxicity of the reconstructed OHPHE standard mixtures (EC50,mix) was predicted using the concentration addition approach.50,51 This method combines the proportion of each of the toxic OHPHEs in the mixture, the toxic contribution of each OHPHE to the mixture, and assumes that each toxic compound has the same pathway of action in an organism using this equation:51
| (1) |
where pi is the fraction of compound i in the mixture and EC50,i is the EC50 values for the individual OHPHE standards.
Statistical Analyses
Statistical analyses were performed using JMP 12 software (by SAS). Differences between triplicate means for both zero-order and first-order rate constants were evaluated using Student t-tests with statistical significance resulting in a p-value ≤ 0.05.
RESULTS AND DISCUSSION
Phenanthrene Transformation and Metabolite Formation by ELW1
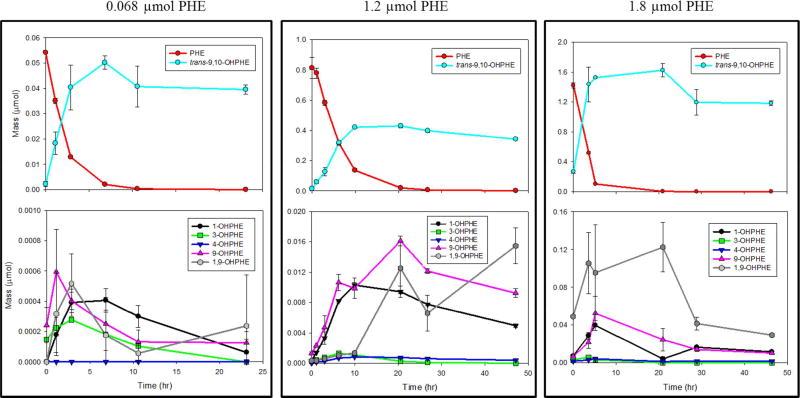
PHE was 100% co-metabolically transformed by ELW1 within 29 hr, at initial PHE masses of 0.033, 0.068, 0.42, 0.84, and 1.8 µmol, and within 47 hr at 1.2 µmol (Figure 1A). In the 1-octyne controls, 26–49% of PHE was transformed (Figure 1B), while in the PHE-only controls, 6–31% of PHE was lost, likely due to adsorption to the reactor walls (Figure 1C). Our measured concentrations of PHE and OHPHE in the PHE-exposed cells and 1-octyne controls account for transformation and adsorption losses combined. 1-Octyne effectively, but not completely, inhibited PHE degradation (Figure 1B).44 The enzyme responsible for PHE oxidation is most likely an alkene monooxygenase found at high levels in isobutene-grown cells of ELW1.44 The activity of the pre-exposed ELW1 cells, determined by the rate of isobutene consumption, was similar for all experiments (Figure S6 and Table S10).
Figure 1.
Transformation of initial PHE masses by ELW1 (with standard error bars, n = 3). (A) PHE-exposed cells (MSM, cells, and PHE) (solid lines), (B) 1-octyne, alkene monooxygenase inhibitor controls (MSM, cells, and PHE) (dashed lines), and (C) PHE-only, no-cells control (MSM and PHE) (dotted lines).
All samples were analyzed for 16 hydroxy PAH metabolites with commercially available standards (Table S1, Figure S1), at three different initial PHE masses (Figure 2 and Figure S2). The metabolites detected in both the PHE-exposed cells (Figure 2) and 1-octyne controls (Figure S2), included only OHPHE compounds: 1-OHPHE, 3-OHPHE, 4-OHPHE, 9-OHPHE, 1,9-OHPHE, and trans-9,10-OHPHE. The OHPHE masses were significantly higher (p-value < 0.05) in the PHE-exposed cells than in the 1-octyne controls for the same mass of PHE tested. At all PHE masses tested, the primary OHPHE metabolite formed by both PHE-exposed cells and 1-octyne controls was trans-9,10-OHPHE, with its percent contribution to the total OHPHE metabolite mass ranging from 72–100%. In both cases, trans-9,10-OHPHE was formed as PHE was transformed (Figure 2 and Figure S2). In the PHE-exposed cells, the trans-9,10-OHPHE concentration remained constant for the duration of each experiment, with no further transformation during the time course of the experiment (Figure 2). Neither cis-9,10-OHPHE or 1-OH-2-NAP were detected in the samples. In addition, OHPHE metabolites were not detected in the PHE-only or cells-only controls.
Figure 2.
Mass of PHE transformed and OHPHE metabolites formed by ELW1 in PHE-exposed cells for initial PHE masses of 0.068 µmol, 1.2 µmol, and 1.8 µmol (with standard error bars, n = 3). Note that there are differences in the x- and y-axes between plots.
Several studies have identified PHE metabolites during degradation and co-metabolism of PHE by other microorganisms. According to the summary of PHE degradation pathways by microorganisms, published by Mallick et al, both trans-9,10-OHPHE and cis-9,10-OHPHE are possible products resulting from the initial oxidation of the 9 and 10 positions during PHE metabolism.18 The trans-9,10-OHPHE isomer is formed from an epoxide at the 9,10 position (9,10-epoxy-9,10-dihydroxyphenanthrene), or co-oxidaxation, by a monooxygenase and is a dead-end product.18,40,52 Cis-9,10-OHPHE, on the other hand, is formed by a dioxygenase and may be further oxidized to form 9,10-dihydroxyphenanthrene (9,10-OHPHE).18,40,53 Trans-9,10-OHPHE was the sole metabolite formed during co-metabolism of PHE by Mycobacterium strain S1 grown in the presence of anthracene.54 However, the formation of cis-dihydrodiols, including cis-3,4-dihydroxy-3,4-dihydrophenanthrene (cis-3,4-OHPHE) by Mycobacterium sp. strain 6PY1, M. vanbaalenii strain PYR-1, and Sphingomonas sp. strain A4,55–58 cis-9,10-OHPHE by M. aromativorans JS19b1,57–60 and cis-1,2-dihydroxy-1,2-dihydrophenanthrene (cis-1,2-OHPHE) by M. aromativorans JS19b1,57,59 have been measured in other studies. Kim et al also measured additional OHPHE metabolites including 2-and 3-OHPHE, 9,10-OHPHE, and one additional phenanthrenediol.57 Identification of other PHE metabolites structures further in the PHE degradation pathways presented in Mallick et al,18 including coumarins, benzocourmarins, and 1-OH-2-NAP, have also been made.53,60–62 In our study, 1-OH-2-NAP was not detected, suggesting PHE co-metabolism by ELW1 did not form cis-1,2-OHPHE or cis-3,4-OHPHE, or that ELW1 was not able to transform these products to 1-OH-2-NAP.18 Based on the consistency of trans-9,10-OHPHE concentrations over time and its high contribution to the total mass of the OHPHE metabolites formed by ELW1, trans-9,10-OHPHE did not appear to undergo further transformation by ELW1.
Kinetics and Mass Balance
Average biomass-normalized zero-order PHE transformation rates were determined for all PHE masses tested and are listed in Table S5. The zero-order PHE transformation rates in PHE-exposed cells increased significantly as the PHE mass increased (p-value < 0.01) (Table S5). The zero-order PHE transformation rates in 1-octyne controls were not statistically different for any of the PHE masses tested (p-value > 0.05). The zero-order rates for this study were within the same order of magnitude as other Mycobacterium sp. strains, including and PYRGCK, JS19b1, czh-3, and czh-117 (−0.0020 to −0.019 µmol hr−1) and A1-PYR (−0.019 µmol hr−1).60,62
PHE transformation by ELW1 followed first-order kinetics, as indicated by a straight regression line when the natural logarithm of PHE concentration was plotted over time (Figure S3). Non-normalized first-order PHE transformation rates (hr−1), kPHE, and half-lives (hr), t1/2, were calculated for PHE-exposed cells and 1-octyne controls, and are listed in Table S6. The non-normalized first-order rates of PHE transformation in this study (−0.16 to −0.51 hr−1) were much faster than those previously reported for Mycobacterium sp. PYR-1 (−0.0015 hr−1) and Pasteurella spp. was (−0.0005 hr−1).63
Average biomass-normalized zero-order rates of OHPHE metabolites formation increased with increasing PHE mass for both PHE-exposed cells and 1-octyne controls (Table S5). Rates of formation for each OHPHE, in both PHE-exposed cells and 1-octyne controls, significantly increased (p-value < 0.04) as the initial PHE mass increased. For several of the OHPHEs measured in the 1-octyne controls, zero-order rates could not be determined because of limitations due to instrumental detection limits and/or poor chromatography (Table S7).
Mass balances were assessed by comparing zero-order PHE transformation rates to the sum of the zero-order OHPHE formation rates (Table S7). In PHE-exposed cells, the rate of PHE transformation was in good agreement with the OHPHE formation rates, with percent differences ranging 6–46%. There were higher percent differences for the 1-octyne controls because OHPHE concentrations were close to the instrumental detection limits.
Toxicity of PHE and OHPHE Metabolites
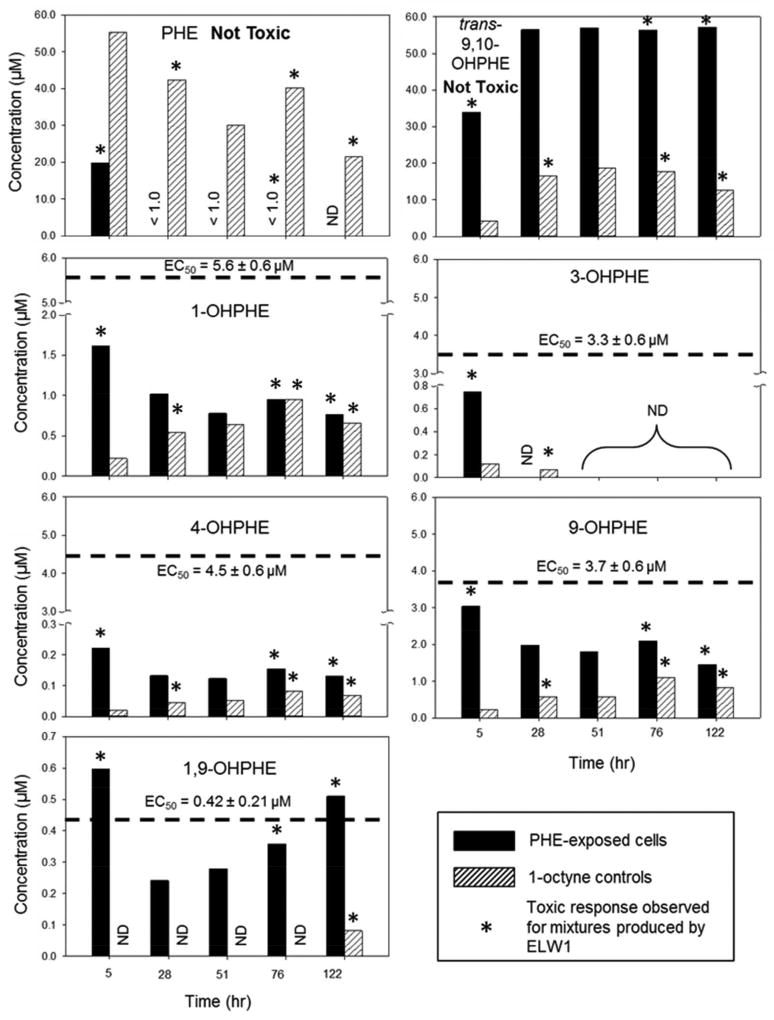
The developmental toxicity of PHE and OHPHE standards were tested using the embryonic zebrafish model, at nominal concentrations ranging from 0.01 – 60 µM. These concentrations were not measured in the 96-well plates, but previously published studies indicate that sorptive losses in this test system for lower molecular weight PAHs, such as phenanthrene, are less than 10%.64 PHE and trans-9,10-OHPHE were not toxic to zebrafish at the range of concentrations tested. The relative order of the mean effective concentrations at which any negative effect was observed in half the embryonic zebrafish (EC50), at 120 hpf, were 1,9-OHPHE (most toxic) < 3-OHPHE < 9-OHPHE < 4-OHPHE < 1-OHPHE (least toxic) and are shown in Figure 3 as the dashed lines for each standard.
Figure 3.
PHE and identified OHPHE metabolite concentrations in mixtures formed by ELW1 in the PHE-exposed cells (solid bars) and 1-octyne controls (dashed bars) and used for zebrafish testing. Mean EC50s (with standard error, n = 32) for individual OHPHE standards are shown in dotted lines. The initial PHE mass was 1.8 µmol in the reactors. Mixtures that elicited a toxic response after 120 hpf in PHE-exposed cells were harvested at 5, 76, and 122 hr and in 1-octyne controls at 28, 76, and 122 hr, as indicated by the asterisk (*) above the corresponding bars. ND: Compounds that were not detected.
To investigate the influence of physical-chemical properties on the developmental toxicity of PHE and the OHPHEs, their log Kow values were estimated (Table S2). PHE had the highest log Kow and was not developmentally toxic, while the OHPHEs had lower log Kow values than PHE and were toxic. However, trans-9,10-OHPHE had the lowest log Kow value of all of the compounds tested and was not toxic. This suggests that log Kow alone does not account for the measured developmental toxicity.
Figure 3 was created to determine if the measured toxicity of the metabolite mixtures formed by ELW1 could be explained by the concentrations of the individual OHPHEs identified in the mixtures, as evidenced by their concentrations in the mixture exceeding their individual EC50 values. The data shown in Figure 3 shows the highest concentration of the PHE and OHPHE metabolite mixtures formed by ELW1 that the zebrafish were exposed to (~60 µM), in both the PHE-exposed cells and 1-octyne controls. Toxicity was observed for the PHE-exposed cell mixtures at 5, 76, and 122 hr and for the 1-octyne control mixtures at 28, 76, and 122 hr (indicated by asterisks in Figure 3). PHE and OHPHE metabolites were not detected in extracts from the cells-only controls and these extracts were not toxic to embryonic zebrafish (data not shown). In the PHE-exposed cells, only the 1,9-OHPHE concentrations in the mixtures (at 5 and 122 hr) were higher than the EC50 value for the individual 1,9-OHPHE standard (Figure 3), suggesting that the measured toxicity at these time points may have been caused by 1,9-OHPHE. In the 1-octyne controls and in the remaining PHE-exposed cell time points, the PHE and OHPHE concentrations in the mixtures did not exceed the EC50 values for any individual standards, suggesting that measured toxicity could not be explained by any one identified compound (Figure 3).
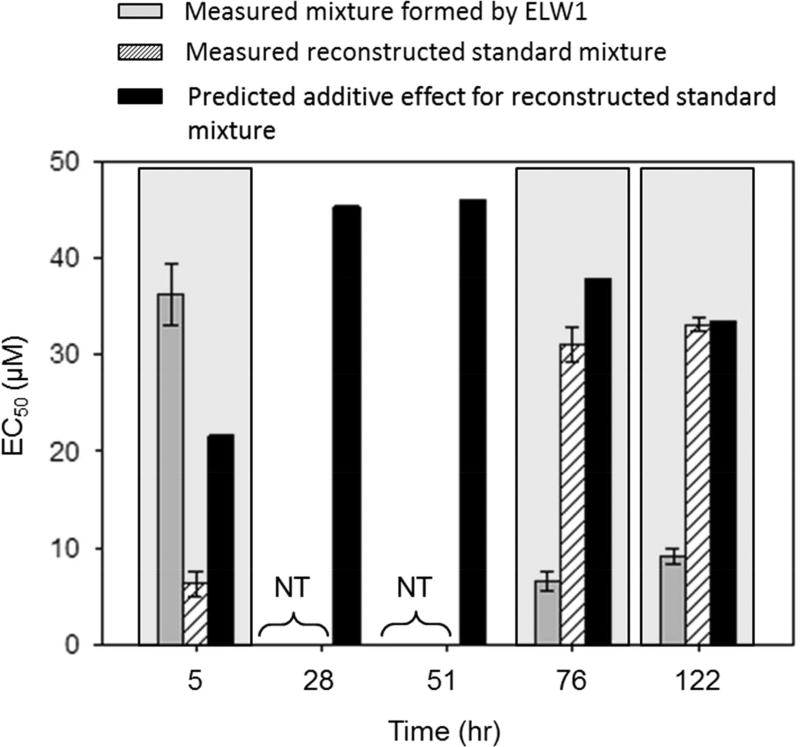
In order to evaluate, and account for, the potential for mixture toxicity of the OHPHE metabolites identified in the ELW1 produced mixtures, we reconstructed the mixture using single PHE and OHPHE standards of the identified metabolites (prepared in the same PHE and OHPHE ratios as the mixture extracts formed by ELW1) and measured the developmental toxicity of this reconstructed standard mixture. EC50 values were measured for both the reconstructed standard mixtures of the identified OHPHE metabolites and the original mixtures formed by ELW1 and are shown in Figure 4. Additionally, predicted EC50 values for the reconstructed standard mixtures of the identified OHPHE metabolites were calculated assuming additive effects of the individual toxic OHPHE metabolites identified (see equation 1) and are shown in Figure 4.51 The measured EC50 values for the mixtures formed by ELW1 were lower (more toxic) than the measured EC50 values for the reconstructed standard mixtures of the identified OHPHE metabolites at the 76 and 122 hr time points (Figure 4), suggesting that the mixtures formed by ELW1 may contain additional unidentified toxic metabolites that contribute to the toxicity of the complex mixture. In addition, the measured EC50 values for the reconstructed standard mixtures of the identified OHPHE metabolites were in good agreement with the predicted EC50 values of the identified metabolites for PHE-exposed cells at 76 and 122 hr (Figure 4). Together, these data suggest that an additive toxic effect to embryonic zebrafish was observed at these time points for the identified metabolites and that the presence of yet unidentified metabolites may have contributed to an increase in toxicity in the mixtures formed by ELW1, at these time points. EC50 values for the mixtures formed by ELW1 and the reconstructed standard mixtures of the identified OHPHE metabolites at 28 and 51 hr were not measured because these mixtures did not elicit a toxic response at the concentrations tested. The predicted EC50 values at 28 and 51 hr indicated that higher concentrations of the mixtures were needed to elicit a toxic response (Figure 4).
Figure 4.
Measured EC50s (with standard error bars, n = 32) for PHE and OHPHE mixtures formed by ELW1 (solid grey bar) and the reconstructed standard mixtures of the identified OHPHE metabolites (dashed bar), as well as the predicted EC50s for the reconstructed standard mixture using equation 1 (black bar) (n = 1), over time (hr), for PHE-exposed cells. Mixtures at 5, 76, and 122 hours elicited a toxic response after 120 hpf and are highlighted with a light grey background. NT: Not Toxic at the concentrations tested on embryonic zebrafish.
Unidentified Metabolites
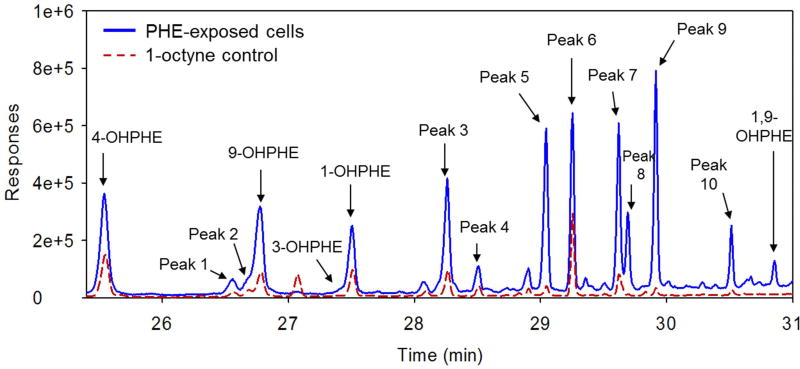
The mixtures formed by ELW1 were derivatized and analyzed in full scan mode to identify the presence of unknown metabolites. Ten peaks, corresponding to yet unidentified metabolites, were observed in the chromatograms for both the PHE-exposed cells and 1-octyne controls, with retention times between ~26–31 min (Figure 5). Chromatograms for each of the time points from PHE-exposed cells and 1-octyne controls are shown in Figure S4 and Figure S5, respectively. The peak retention times, primary ions, and tentative identifications are listed in Table S9 and may include other mono-OHPHE and di-OHPHE metabolites that we were not able to identify because of the lack of commercially available standards. The unidentified metabolites in both PHE-exposed cells and 1-octyne controls may have contributed to the toxicity of the metabolite mixture formed by ELW1. Several of the chromatogram peaks (Peak 3, 7, 8, 10) increased in area with the time of cell exposure (up to 122 hr) (Figure S4), which corresponds to the increase in measured toxicity with time. For example, Peak 7, was tentatively identified as a di-OHPHE, which could be produced via the transformation of initial mono-OHPHE compounds.
Figure 5.
Full scan chromatogram between ~26 – 31 min for derivatized PHE-exposed cells and 1-octyne controls collected at 122 hr.
This study has shown that ELW1 can rapidly co-metabolize PHE in resting cell tests. Using commercially available standards, OHPHE metabolites were identified and quantified. The primary metabolite formed by ELW1 was trans-9,10-OHPHE, which was not further transformed. Developmental toxicity of individual metabolites, and metabolite mixtures, was assessed using embryonic zebrafish and showed PHE and trans-9,10-OHPHE were not toxic to zebrafish, but all other identified metabolites (1-, 3-, 4-, 9-, and 1,9-OHPHE) were toxic. ELW1 formed toxic metabolites through the co-metabolism of PHE and the mixtures of these metabolites were also toxic to zebrafish. The increased toxicity observed from the metabolite mixtures formed by ELW1, relative to the reconstructed standard mixtures of the identified OHPHE metabolites, was likely caused by unidentified metabolites in the extract, since a mixture of the known metabolites exerted less toxicity than the metabolic mixture. Future research should include an integrated approach for identifying these toxic transformation products.65 The time dependent nature of the toxicity of the ELW1 formed metabolites indicates initial products may have been co-metabolized to form more toxic secondary products, which warrants further study. Based on these results, future studies in bioremediation of PAHs should consider the possibility that more toxic metabolites may be formed during bioremediation, as well as investigating the metabolites formed by more recalcitrant PAHs.
Supplementary Material
Acknowledgments
This publication was made possible in part by grant number P42 ES016465 and P30-ES00210 from the National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NIEHS, NIH. The authors would also like to thank Sinnhuber Aquatic Research Laboratory (SARL) for performing the toxicity testing and Dr. Michael Hyman from North Carolina State University for providing the EWL1 culture and helpful comments.
Footnotes
Supporting Information. Tables S1–S11 and Figures S1–S8 provide additional information on the chemistry, biodegration and toxicity experiments described in the manuscript. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Registry, A. f. T. S. a. D. Public Health Statement: Polycyclic Aromatic Hydrocarbons (Pahs) D. o. H. a. H, editor. Services. 1995:1–6. www.atsdr.cdc.gov.
- 2.Kriech AJ, Kurek JT, Osborn LV, Wissel HL, Sweeney BJ. Determination of Polycyclic Aromatic Compounds in Asphalt and in Corresponding Leachate Water. Polycyclic Aromatic Compounds. 2002;22(3–4):517–535. [Google Scholar]
- 3.Neff JM, Stout SA, Gunster DG. Ecological Risk Assessment of Polycyclic Aromatic Hydrocarbons in Sediments: Identifying Sources and Ecological Hazard. Integrated Environmental Assessment and Management. 2005;1(1):22–33. doi: 10.1897/ieam_2004a-016.1. [DOI] [PubMed] [Google Scholar]
- 4.Fetzer JC, Kershaw JR. Identification of Large Polycyclic Aromatic-Hydrocarbons in a Coal-Tar Pitch. Fuel. 1995;74(10):1533–1536. [Google Scholar]
- 5.Ledesma EB, Kalish MA, Nelson PF, Wornat MJ, Mackie JC. Formation and Fate of Pah During the Pyrolysis and Fuel-Rich Combustion of Coal Primary Tar. Fuel. 2000;79(14):1801–1814. [Google Scholar]
- 6.Schubert P, Schantz MM, Sander LC, Wise SA. Determination of Polycyclic Aromatic Hydrocarbons with Molecular Weight 300 and 302 in Environmental-Matrix Standard Reference Materials by Gas Chromatography/Mass Spectrometry. Analytical Chemistry. 2003;75(2):234–246. doi: 10.1021/ac0259111. [DOI] [PubMed] [Google Scholar]
- 7.Gevao B, Jones KC. Kinetics and Potential Significance of Polycyclic Aromatic Hydrocarbon Desorption from Creosote-Treated Wood. Environmental Science & Technology. 1998;32(5):640–646. [Google Scholar]
- 8.Kohler M, Kunniger T, Schmid P, Gujer E, Crockett R, Wolfensberger M. Inventory and Emission Factors of Creosote, Polycyclic Aromatic Hydrocarbons (Pah), and Phenols from Railroad Ties Treated with Creosote. Environmental Science & Technology. 2000;34(22):4766–4772. [Google Scholar]
- 9.Cancer), I. I. A. f. R. o. Some Non-Heterocyclic Polycyclic Aromatic Hydrocarbons and Some Related Exposures. World Health Organization; Lyon, France: 2010. Iarc Monographs on the Evaluation of Carcinogenic Risks to Humans. [PMC free article] [PubMed] [Google Scholar]
- 10.Agency, U. S. E. P. [July 10, 2016];Integrated Risk Information System (Iris) https://www.epa.gov/iris.
- 11.Gan S, Lau EV, Ng HK. Remediation of Soils Contaminated with Polycyclic Aromatic Hydrocarbons (Pahs) Journal of Hazardous Materials. 2009;172(2–3):532–549. doi: 10.1016/j.jhazmat.2009.07.118. [DOI] [PubMed] [Google Scholar]
- 12.Woo SH, Park JM. Microbial Degradation and Enhanced Bioremediation of Polycyclic Aromatic Hydrocarbons. Journal of Industrial and Engineering Chemistry. 2004;10(1):16–23. [Google Scholar]
- 13.Bastida F, Jehmlich N, Lima K, Morris BEL, Richnow HH, Hernandez T, von Bergen M, Garcia C. The Ecological and Physiological Responses of the Microbial Community from a Semiarid Soil to Hydrocarbon Contamination and Its Bioremediation Using Compost Amendment. Journal of Proteomics. 2016;135:162–169. doi: 10.1016/j.jprot.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Festa S, Coppotelli BM, Morelli IS. Comparative Bioaugmentation with a Consortium and a Single Strain in a Phenanthrene-Contaminated Soil: Impact on the Bacterial Community and Biodegradation. Applied Soil Ecology. 2016;98:8–19. [Google Scholar]
- 15.Nzila A. Update on the Cometabolism of Organic Pollutants by Bacteria. Environmental Pollution. 2013;178:474–482. doi: 10.1016/j.envpol.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 16.Mackay D, Shiu WY, Ma K-C, Lee SC. Handbook of Physical-Chemical Properties and Environmental Fate for Organic Chemicals: Introduction and Hydrocarbons. 2. Vol. 1 Taylor & Francis Group; Boca Raton, FL: 2006. [Google Scholar]
- 17.Chauhan A, Fazlurrahman, Oakeshott JG, Jain RK. Bacterial Metabolism of Polycyclic Aromatic Hydrocarbons: Strategies for Bioremediation. Indian Journal of Microbiology. 2008;48(1):95–113. doi: 10.1007/s12088-008-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallick S, Chakraborty J, Dutta TK. Role of Oxygenases in Guiding Diverse Metabolic Pathways in the Bacterial Degradation of Low-Molecular-Weight Polycyclic Aromatic Hydrocarbons: A Review. Critical Reviews in Microbiology. 2011;37(1):64–90. doi: 10.3109/1040841X.2010.512268. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa K, Suenaga H, Goto M. Biphenyl Dioxygenases: Functional Versatilities and Directed Evolution. J. Bacteriol. 2004;186(16):5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferraro DJ, Okerlund AL, Mowers JC, Ramaswamy S. Structural Basis for Regioselectivity and Stereoselectivity of Product Formation by Naphthalene 1,2-Dioxygenase. J. Bacteriol. 2006;188(19):6986–6994. doi: 10.1128/JB.00707-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resnick SM, Lee K, Gibson DT. Diverse Reactions Catalyzed by Naphthalene Dioxygenase from Pseudomonas Sp Strain Ncib 9816. Journal of Industrial Microbiology & Biotechnology. 1996;17(5–6):438–457. [Google Scholar]
- 22.Selifonov SA, Grifoll M, Eaton RW, Chapman PJ. Oxidation of Naphthenoaromatic and Methyl-Substituted Aromatic Compounds by Naphthalene 1,2-Dioxygenase. Applied and Environmental Microbiology. 1996;62(2):507–514. doi: 10.1128/aem.62.2.507-514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vila J, Lopez Z, Sabate J, Minguillon C, Solanas AM, Grifoll M. Identification of a Novel Metabolite in the Degradation of Pyrene by Mycobacterium Sp Strain Ap1: Actions of the Isolate on Two- and Three-Ring Polycyclic Aromatic Hydrocarbons. Applied and Environmental Microbiology. 2001;67(12):5497–5505. doi: 10.1128/AEM.67.12.5497-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jouanneau Y, Meyer C, Jakoncic J, Stojanoff V, Gaillard J. Characterization of a Naphthalene Dioxygenase Endowed with an Exceptionally Broad Substrate Specificity toward Polycyclic Aromatic Hydrocarbons. Biochemistry. 2006;45(40):12380–12391. doi: 10.1021/bi0611311. [DOI] [PubMed] [Google Scholar]
- 25.Schuler L, Jouanneau Y, Chadhain SMN, Meyer C, Pouli M, Zylstra GJ, Hols P, Agathos SN. Characterization of a Ring-Hydroxylating Dioxygenase from Phenanthrene-Degrading Sphingomonas Sp Strain Lh128 Able to Oxidize Benz a Anthracene. Applied Microbiology and Biotechnology. 2009;83(3):465–475. doi: 10.1007/s00253-009-1858-2. [DOI] [PubMed] [Google Scholar]
- 26.Van de Wiele T, Vanhaecke L, Boeckaert C, Peru K, Headley J, Verstraete W, Siciliano S. Human Colon Microbiota Transform Polycyclic Aromatic Hydrocarbons to Estrogenic Metabolites. Environmental Health Perspectives. 2005;113(1):6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chibwe L, Geier MC, Nakamura J, Tanguay RL, Aitken MD, Simonich SLM. Aerobic Bioremediation of Pah Contaminated Soil Results in Increased Genotoxicity and Developmental Toxicity. Environmental Science & Technology. 2015;49(23):13889–13898. doi: 10.1021/acs.est.5b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaiswal PK, Gupta J, Shahni S, Thakur IS. Nadph Oxidase-Mediated Superoxide Production by Intermediary Bacterial Metabolites of Dibenzofuran: A Potential Cause for Trans-Mitochondrial Membrane Potential (Delta Im) Collapse in Human Hepatoma Cells. Toxicol. Sci. 2015;147(1):17–27. doi: 10.1093/toxsci/kfv107. [DOI] [PubMed] [Google Scholar]
- 29.Moorthy B, Chu C, Carlin DJ. Polycyclic Aromatic Hydrocarbons: From Metabolism to Lung Cancer. Toxicol. Sci. 2015;145(1):5–15. doi: 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubrovskaya EV, Pozdnyakova NN, Muratova AY, Turkovskaya OV. Changes in Phytotoxicity of Polycyclic Aromatic Hydrocarbons in the Course of Microbial Degradation. Russian Journal of Plant Physiology. 2016;63(1):172–179. [Google Scholar]
- 31.Lewtas J, Walsh D, Williams R, Dobiáš L. Air Pollution Exposure–DNA Adduct Dosimetry in Humans and Rodents: Evidence for Non-Linearity at High Doses. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1997;378(1–2):51–63. doi: 10.1016/s0027-5107(97)00097-3. [DOI] [PubMed] [Google Scholar]
- 32.White PA. The Genotoxicity of Priority Polycyclic Aromatic Hydrocarbons in Complex Mixtures. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2002;515(1–2):85–98. doi: 10.1016/s1383-5718(02)00017-7. [DOI] [PubMed] [Google Scholar]
- 33.Libalová H, Krčková S, Uhlířová K, Milcová A, Schmuczerová J, Ciganek M, Kléma J, Machala M, Šrám RJ, Topinka J. Genotoxicity but Not the Ahr-Mediated Activity of Pahs Is Inhibited by Other Components of Complex Mixtures of Ambient Air Pollutants. Toxicology Letters. 2014;225(3):350–357. doi: 10.1016/j.toxlet.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 34.Søfteland L, Kirwan JA, Hori TSF, Storseth TR, Sommer U, Berntssen MHG, Viant MR, Rise ML, Waagbø R, Torstensen BE, Booman M, Olsvik PA. Toxicological Effect of Single Contaminants and Contaminant Mixtures Associated with Plant Ingredients in Novel Salmon Feeds. Food and Chemical Toxicology. 2014;73:157–174. doi: 10.1016/j.fct.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Lu S, Liu G, Zhou Y, Lv Y, She J, Fan R. Co-Exposure to Polycyclic Aromatic Hydrocarbons, Benzene and Toluene and Their Dose–Effects on Oxidative Stress Damage in Kindergarten-Aged Children in Guangzhou, China. Science of The Total Environment. 2015;524–525:74–80. doi: 10.1016/j.scitotenv.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Martins M, Santos JM, Diniz MS, Ferreira AM, Costa MH, Costa PM. Effects of Carcinogenic Versus Non-Carcinogenic Ahr-Active Pahs and Their Mixtures: Lessons from Ecological Relevance. Environmental Research. 2015;138:101–111. doi: 10.1016/j.envres.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 37.Martins M, Santos JM, Costa MH, Costa PM. Applying Quantitative and Semi-Quantitative Histopathology to Address the Interaction between Sediment-Bound Polycyclic Aromatic Hydrocarbons in Fish Gills. Ecotoxicology and Environmental Safety. 2016;131:164–171. doi: 10.1016/j.ecoenv.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 38.Michalec F-G, Holzner M, Souissi A, Stancheva S, Barras A, Boukherroub R, Souissi S. Lipid Nanocapsules for Behavioural Testing in Aquatic Toxicology: Time–Response of Eurytemora Affinis to Environmental Concentrations of Pahs and Pcb. Aquatic Toxicology. 2016;170:310–322. doi: 10.1016/j.aquatox.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Bucker M, Glatt HR, Platt KL, Avnir D, Ittah Y, Blum J, Oesch F. Mutagenicity of Phenanthrene and Phenanthrene K-Region Derivatives. Mutation Research. 1979;66(4):337–348. doi: 10.1016/0165-1218(79)90044-2. [DOI] [PubMed] [Google Scholar]
- 40.Moody JD, Freeman JP, Doerge DR, Cerniglia CE. Degradation of Phenanthrene and Anthracene by Cell Suspensions of Mycobacterium Sp Strain Pyr-1. Applied and Environmental Microbiology. 2001;67(4):1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moscoso F, Teijiz I, Deive FJ, Sanroman MA. Efficient Pahs Biodegradation by a Bacterial Consortium at Flask and Bioreactor Scale. Bioresource Technology. 2012;119:270–276. doi: 10.1016/j.biortech.2012.05.095. [DOI] [PubMed] [Google Scholar]
- 42.Muratova A, Pozdnyakova N, Makarov O, Baboshin M, Baskunov B, Myasoedova N, Golovleva L, Turkovskaya O. Degradation of Phenanthrene by the Rhizobacterium Ensifer Meliloti. Biodegradation. 2014;25(6):787–795. doi: 10.1007/s10532-014-9699-9. [DOI] [PubMed] [Google Scholar]
- 43.Peng LB, Deng DY, Ye FT. Efficient Oxidation of High Levels of Soil-Sorbed Phenanthrene by Microwave-Activated Persulfate: Implication for in Situ Subsurface Remediation Engineering. J. Soils Sediments. 2016;16(1):28–37. [Google Scholar]
- 44.Kottegoda S, Waligora E, Hyman M. Metabolism of 2-Methylpropene (Isobutylene) by the Aerobic Bacterium Mycobacterium Sp. Strain Elw1. Applied and Environmental Microbiology. 2015;81(6):1966–1976. doi: 10.1128/AEM.03103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motorykin O, Schrlau J, Jia Y, Harper B, Harris S, Harding A, Stone D, Kile M, Sudakin D, Massey Simonich SL. Determination of Parent and Hydroxy Pahs in Personal Pm2.5 and Urine Samples Collected During Native American Fish Smoking Activities. Science of The Total Environment. 2015;505:694–703. doi: 10.1016/j.scitotenv.2014.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schummer C, Delhomme O, Appenzeller BMR, Wennig R, Millet M. Comparison of Mtbstfa and Bstfa in Derivatization Reactions of Polar Compounds Prior to Gc/Ms Analysis. Talanta. 2009;77(4):1473–1482. doi: 10.1016/j.talanta.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 47.EPA Method - 8280a Epa. The Analysis of of Polychlorinated Dibenzo-P-Dioxins and Polychlorinated Dibenzofurans by High-Resolution Gas Chromatography/Low Resolution Mass Spectrometry (Hrgc/Lrms) [Google Scholar]
- 48.Truong L, Bugel SM, Chlebowski A, Usenko CY, Simonich MT, Simonich SLM, Tanguay RL. Optimizing Multi-Dimensional High Throughput Screening Using Zebrafish. Reproductive Toxicology. 2016;65:139–147. doi: 10.1016/j.reprotox.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Truong L, Harper SL, Tanguay RL. Evaluation of Embryotoxicity Using the Zebralfish Model. Methods in Molecular Biology. 2011;691:271–279. doi: 10.1007/978-1-60761-849-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loewe S, Muischnek H. Über Kombinationswirkungen. Naunyn-Schmiedebergs Archiv für experimentelle Pathologie und Pharmakologie. 1926;114(5):313–326. [Google Scholar]
- 51.Backhaus T, Scholze M, Grimme LH. The Single Substance and Mixture Toxicity of Quinolones to the Bioluminescent Bacterium Vibrio Fischeri. Aquatic Toxicology. 2000;49(1–2):49–61. doi: 10.1016/s0166-445x(99)00069-7. [DOI] [PubMed] [Google Scholar]
- 52.Narro ML, Cerniglia CE, Vanbaalen C, Gibson DT. Metabolism of Phenanthrene by the Marine Cyanobacterium Agmenellum-Quadruplicatum Pr-6. Applied and Environmental Microbiology. 1992;58(4):1351–1359. doi: 10.1128/aem.58.4.1351-1359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seo J-S, Keum Y-S, Hu Y, Lee S-E, Li QX. Phenanthrene Degradation in Arthrobacter Sp. P1-1: Initial 1,2-, 3,4- and 9,10-Dioxygenation, and Meta- and Ortho-Cleavages of Naphthalene-1,2-Diol after Its Formation from Naphthalene-1,2-Dicarboxylic Acid and Hydroxyl Naphthoic Acids. Chemosphere. 2006;65(11):2388–2394. doi: 10.1016/j.chemosphere.2006.04.067. [DOI] [PubMed] [Google Scholar]
- 54.Tongpim S, Pickard MA. Cometabolic Oxidation of Phenanthrene to Phenanthrene Trans-9,10-Dihydrodiol by Mycobacterium Strain S1 Growing on Anthracene in the Presence of Phenanthrene. Canadian Journal of Microbiology. 1999;45(5):369–376. [PubMed] [Google Scholar]
- 55.Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. Identification of Pyrene-Induced Proteins in Mycobacterium Sp. Strain 6py1: Evidence for Two Ring-Hydroxylating Dioxygenases. J. Bacteriol. 2003;185(13):3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinyakong O, Habe H, Kouzuma A, Nojiri H, Yamane H, Omori T. Isolation and Characterization of Genes Encoding Polycyclic Aromatic Hydrocarbon Dioxygenase from Acenaphthene and Acenaphthylene Degrading Sphingomonas Sp. Strain A4. FEMS Microbiology Letters. 2004;238(2):297–305. doi: 10.1016/j.femsle.2004.07.048. [DOI] [PubMed] [Google Scholar]
- 57.Kim SJ, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE. Molecular Cloning and Expression of Genes Encoding a Novel Dioxygenase Involved in Low- and High-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation in Mycobacterium Vanbaalenii Pyr-1. Applied and Environmental Microbiology. 2006;72(2):1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kweon O, Kim SJ, Freeman JP, Song J, Baek S, Cerniglia CE. Substrate Specificity and Structural Characteristics of the Novel Rieske Nonheme Iron Aromatic Ring-Hydroxylating Oxygenases Nidab and Nida3b3 from Mycobacterium Vanbaalenii Pyr-1. Mbio. 2010;1(2) doi: 10.1128/mBio.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seo J-S, Keum Y-S, Li QX. Mycobacterium Aromativorans Js19b1t Degrades Phenanthrene through C-1,2, C-3,4 and C-9,10 Dioxygenation Pathways. International Biodeterioration & Biodegradation. 2012;70:96–103. doi: 10.1016/j.ibiod.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hennessee CT, Li QX. Effects of Polycyclic Aromatic Hydrocarbon Mixtures on Degradation, Gene Expression, and Metabolite Production in Four Mycobacterium Species. Applied and Environmental Microbiology. 2016;82(11):3357–3369. doi: 10.1128/AEM.00100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinyakong O, Habe H, Supaka N, Pinpanichkarn P, Juntongjin K, Yoshida T, Furihata K, Nojiri H, Yamane H, Omori T. Identification of Novel Metabolites in the Degradation of Phenanthrene by Sphingomonas Sp. Strain P2. FEMS Microbiology Letters. 2000;191(1):115–121. doi: 10.1111/j.1574-6968.2000.tb09327.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhong Y, Luan T, Lin L, Liu H, Tam NFY. Production of Metabolites in the Biodegradation of Phenanthrene, Fluoranthene and Pyrene by the Mixed Culture of Mycobacterium Sp. And Sphingomonas Sp. Bioresource Technology. 2011;102(3):2965–2972. doi: 10.1016/j.biortech.2010.09.113. [DOI] [PubMed] [Google Scholar]
- 63.Sepic E, Bricelj M, Leskovsek H. Biodegradation Studies of Polyaromatic Hydrocarbons in Aqueous Media. Journal of Applied Microbiology. 1997;83(5):561–568. doi: 10.1046/j.1365-2672.1997.00261.x. [DOI] [PubMed] [Google Scholar]
- 64.Chlebowski AC, Tanguay RL, Simonich SLM. Quantitation and Prediction of Sorptive Losses During Toxicity Testing of Polycyclic Aromatic Hydrocarbon (Pah) and Nitrated Pah (Npah) Using Polystyrene 96-Well Plates. Neurotoxicology and Teratology. 2016;57:30–38. doi: 10.1016/j.ntt.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chibwe L, Titaley IA, Hoh E, Simonich SLM. Integrated Framework for Identifying Toxic Transformation Products in Complex Environmental Mixtures. Environmental Science & Technology Letters. 2017 doi: 10.1021/acs.estlett.6b00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.