Abstract
Over the last few decades, the incidence of oral cancer has gradually increased, due to the negative influence of environmental factors and also abnormalities within the genome. The main issues in oral cancer treatment consist in surpassing resistance and recurrence. However, continuous discovery of altered signaling pathways in these tumors provides valuable information for the identification of novel gene candidates targeted in personalized therapy. RNA interference (RNAi) is a natural mechanism that involves small interfering RNA (siRNA); this can be exploited in biomedical research by using natural or synthetic constructs for activation of the mechanism. Synthetic siRNA transcripts were developed as a versatile class of molecular tools that have a diverse range of programmable roles, being involved in the regulation of several biological processes, thereby providing the perspective of an alternative option to classical treatment. In this review, we summarize the latest information related to the application of siRNA in oral malignancy together with molecular aspects of the technology and also the perspective upon the delivery system. Also, the emergence of newer technologies such as clustered regularly interspaced short palindromic repeats/Cas9 or transcription activator-like effector nucleases in comparison with the RNAi approach is discussed in this paper.
Keywords: RNA interference, oral cancer, therapeutic strategy, mechanistic and biochemical insights
Introduction
A group of tumors on the lip, tongue, mouth, and oropharynx are grouped under the name of oral cancer.1 Global epidemiologic data of oral cancer reflect a 20-fold geographic variation, with almost two-thirds of all newly diagnosed cases being reported in developing countries.2,3 Betel chewing, a traditional practice in South-Eastern Asia and in the Pacific Ocean, represents the greatest risk factor, followed by tobacco smoking, human papillomavirus (HPV) infection, alcohol consumption, and nutrient deficiency.4 On a global scale, oral cancer could be considered a deadly disease, considering that out of the 300,000 newly diagnosed cases, 145,000 deaths were reported in 2012.3 The major challenge in the treatment of oral cancer remains the emergence of drug resistance and also recurrence episodes; therefore, improved therapeutic options that surpass these issues are necessary.5,6
In the past, molecular biology studies were centered around protein coding genes, supported by the central dogma of biology, which postulates that messenger RNA (mRNA) only acts as a mediator between a gene and its encoded protein.7–10 The noncoding regions of the genome were considered as nonfunctional entities and classified as “junk DNA” or “dark-matter.”11–15 However, genome sequencing performed over the last 2 decades brought scientists to the conclusion that the phenotypes are generated not only by proteins and their coding genes but also by noncoding regulatory elements that modulate gene expression programs.13,16–18 Noncoding RNAs (ncRNAs) have the common feature of not being translated into proteins, hence their name.19–21 As a very diverse class, these transcripts are classified based on the sequence length, in: long ncRNAs (with >200 nt) and short ncRNAs (with <200 nt), as presented in Figure 1. The short ncRNAs group contains transcripts, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs.9,19,22,23
Figure 1.
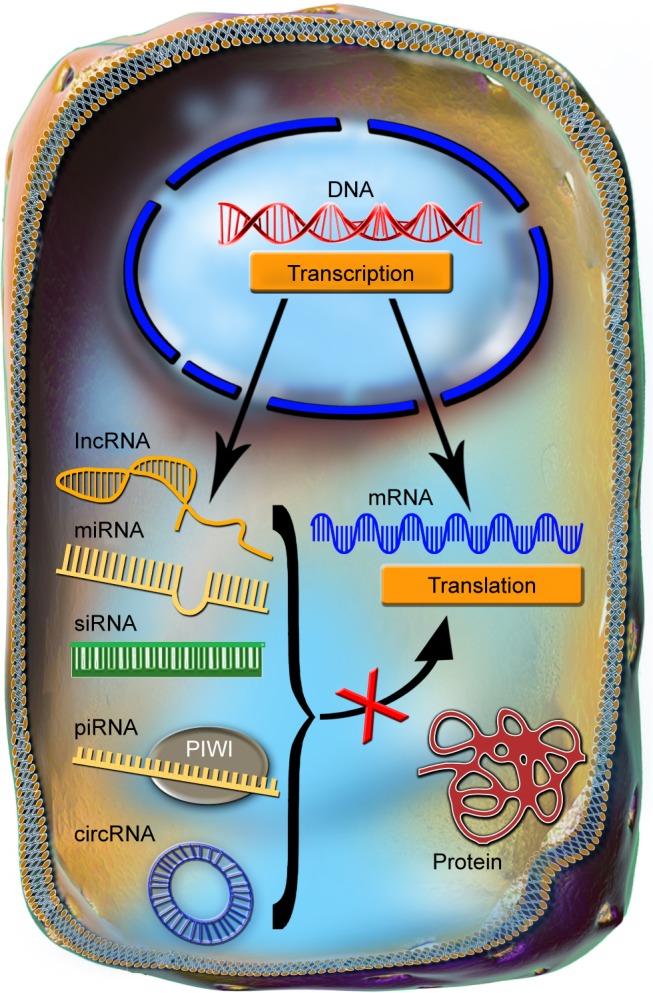
The common features of ncRNAs.
Notes: The ncRNAs are transcribed from the DNA and through multiple processing steps are exported into the cytoplasm, in a manner that bears a higher or lower degree of similarity with the processing of mRNA. In the cytoplasm, the main function of the ncRNAs is to repress the translation of mRNAs into proteins.
Abbreviations: circRNA, circular RNA; lncRNA, long noncoding RNAs; miRNA, microRNAs; mRNA, messenger RNA; ncRNAs, noncoding RNAs; piRNA, piwi-interacting RNA; siRNA, small interfering RNA.
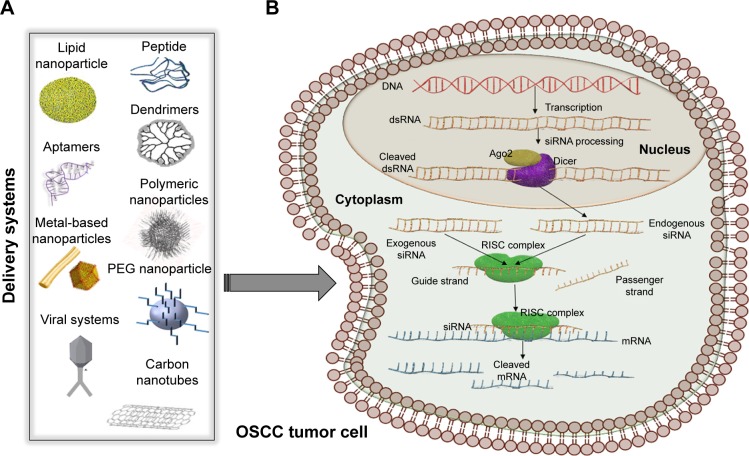
RNA interference (RNAi) was first considered as an exogenous RNA in plants.24,25 It was later discovered that siRNA can have an endogenous origin in many types of cells, human cells included, and that its expression is dysregulated during the development and progression of various pathologies, including cancer.25,26 Both siRNAs and miRNAs are processed by the same proteins: Dicer and RNA-induced silencing complex (RISC). Dicer cleaves the double-stranded precursor into a 23–25 nucleotide double-stranded RNA (dsRNA), with a two-nucleotide overhang at each end, while RISC has two functions: keeping only the guide strand of the dsRNA, while discarding the passenger strand and cleaving the mRNA between its 10th and 11th base, after siRNA full-complementarily binds to the 3′ end (Figure 2).25,27 The exogenous therapeutic siRNA can be delivered as dsRNA ready to be integrated into the RISC complex or as short-hairpin RNA (shRNA), a precursor of siRNA that is cleaved by Dicer and then loaded into the RISC complex. The dsRNA, referred from now on simply as siRNA, is the most commonly used. One example is represented by siRNAs targeting the epidermal growth factor receptor (EGFR) gene that is overexpressed in oral cancer cells.28
Figure 2.
siRNA therapeutic implication.
Notes: (A) Some relevant examples of nanosystems used for siRNA application, comprising a great variety of viral/nonviral delivery systems, some of which are used in oral cancer: lipid nanoparticles, nanorods, peptides, dendrimers, PEG nanoparticles, or adeno-associated viruses with application in oral cancer. (B) Schematic view of siRNA mechanism. siRNAs can have an endogenous origin, being transcribed from the genome and processed by Dicer or it can be delivered by artificial nanosystems. Both the exogenous and the endogenous siRNAs are loaded in the RISC complex, with the help of which only the guide strand is kept for further interaction with its corresponding mRNA.
Abbreviations: dsRNA, double-stranded RNA; mRNA, messenger RNA; OSCC, oral squamous cell carcinoma; PEG, polyethylene glycol; RISC, RNA-induced silencing complex; siRNA, small interfering RNA.
In this review, we analyze the main challenges associated with siRNA therapy, as a modern and effective way of reducing cell proliferation, invasion, metastasis, increasing immune response targeting invasion/migration, preventing metastasis, or even sensitizing cancer cells to chemotherapy. The siRNA therapeutic approach is summarized in two main categories: classical Lipofectamine-transfected siRNAs and nanoparticle-loaded siRNAs as single or multiple drug delivery.
Overcoming the challenges of siRNA therapy
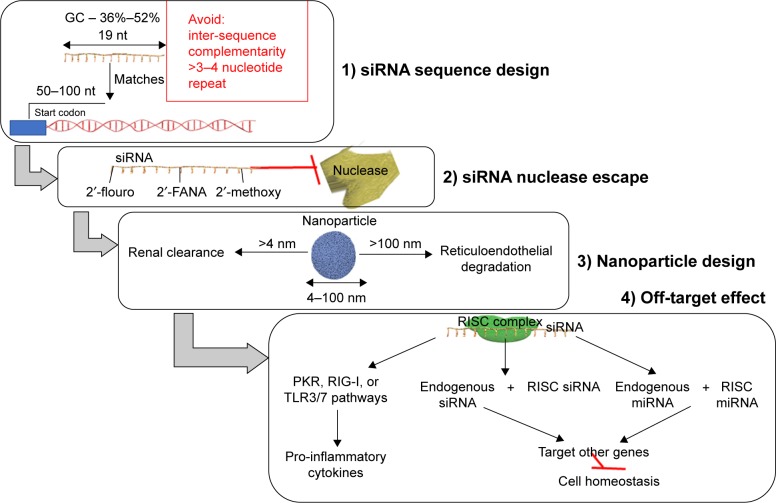
The main challenges associated with siRNA therapy are represented by the sequence design, the high instability of RNAs, the danger of nuclease degradation, endosomal entrapment, and also off-target effects,29 aspects summarized in Figure 3.
Figure 3.
An efficient therapeutic siRNA design has to consider: 1) the siRNA sequence design; 2) modification made to the sequence as to escape the blood nuclease; 3) an efficient nanoparticle size range essential for efficient siRNA delivery; and 4) the off-target effects exogenous siRNA.
Abbreviations: 2′-FANA, 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid; miRNA, microRNAs; PKR, protein kinase R; RIG-I, retinoic acid-inducible gene I; RISC, RNA-induced silencing complex; siRNA, small interfering RNA; TLR, Toll-like receptor.
When designing an siRNA sequence, the first step consists in the selection of the targeted gene. The targeted region is composed of 50–100 nucleotides upstream from the start codon, and the single nucleotide polymorphism containing areas are excluded. The recommended length of the siRNA sequence is 19 nucleotides and the guanine–cytosine content should be between 36% and 52%.29,30 The antisense strand has to be enriched in A/U nucleotides at the 5′ end, while the sense strand is advised to contain more G/C nucleotides at the 5′ end.29,31 Inter-sequence complementarity and >3 or 4 repeats of the same nucleotide should be avoided, as it causes changes in the secondary structure of siRNA.30,31
In the blood, there are nucleases that degrade nucleic acids and are characterized by an increased serum half-life.32 The chemical structure of siRNA undergoes some modification in order to be protected particularly from the action of RNA nucleases. Some examples of these modifications are: the insertion of ribonucleic analogs – 2′-flouro, 2′-FANA, 2′-methoxy; 5′-/3′-terminal inverted abasic end caps – or algorithms predicting the nuclease cleaved sites and modifying these with 2′-methoxy.33
Particle size control in delivery represents an important issue. If the nanoparticles are smaller than 4 nm, they are rapidly cleared from the blood via renal filtration and urinary excretion.34 At the same time, if they are larger than 100 nm in diameter, they will be degraded in the reticuloendothelial system and are difficult to be internalized.35 Lipid nanosystems have a long circulation time in the blood and can easily modify their surface, but sometimes they can cause pseudo-allergic reactions. Neutral liposomes have a size of 30–40 nm and are the most commonly used lipid-based nanosystems.36
siRNA therapy is well-tolerated, but in some cases, particularly for the case of transcripts with >23 nucleotides, interferon responses can be activated together with the induction of cell death within in vitro systems. The immune reaction is different, dependent of the type of cells used, and it is hard to predict the in vivo responses.37
Off-target effects are very frequently retrieved in practice. The siRNAs are loaded on the RISC complex and can sometimes target other genes than the intended ones, by an imperfect match. The siRNAs can also be incorporated into dendritic cells and initiate an immune response,38 by stimulating the protein kinase R, retinoic acid-inducible gene I, or Toll-like receptor 3 or 7 pathways for production of pro-inflammatory cytokines.39 Through the off-target effect, the therapeutic siRNA can also impair the endogenous siRNA and the endogenous miRNAs to exert their normal function, ultimately, interfering with the homeostatic state of the cell.38
The trafficking of siRNAs from endosomes into the cytoplasm constitutes a main rate-limiting issue for many delivery approaches.40 siRNA is recognized by surface proteins and internalized into an early endosome that fuses with a sorting endosome and transfers its contents into a late endosome. The late endosome interacts with the lysosome, the pH drops to 4.5, and the siRNA sequence is degraded. The siRNA molecules can form a complex with a special class of lipids and then form a micelle-like structure (called phosphatidylethanolamines [PEs]). The lipoplex fuses rapidly with the anionic membranes, and the content of the endosome is released in the cytoplasm, before lysosomal interaction.40 Lipofectamine, a cationic liposome, used very often in siRNA transfection studies in vitro is a PE-based liposome (Table 1).41 Another class of proteins associated with siRNAs are the viroporins, proteins of viral origin that have the ability of forming pores in a lipid bilayer.40 The influenza virus peptide, termed 599, combined with cancerous inhibitor of protein phosphatase 2A (CIP2A) siRNA proved to be able to overcome endosomal entrapment and effectively silenced the CIP2A gene in vitro in the CAL 27 cell line.42
Table 1.
Studies using siRNA-targeted delivery as treatment strategies in oral cancer
| Targeted gene | Expression | Delivery system | Cell line | Animal model | Methods | Effect on | Reference |
|---|---|---|---|---|---|---|---|
| CIP2A | Up | 599 peptide (viroporins) + Lipofectamine | CAL 27 and SCC-25 | – | Agarose gel shift assay, fluorescence microscopy, RT-PCR, Western blot, CellTiter kit for cell proliferation, CytoSelect 24-well cell invasion assay kit, anchorage-independent growth assay | Regulate anchorage- independent growth and inhibition of invasion | 42 |
| Cip2A | Up | EGFR-binding peptide and endosome-disruptive peptide | CAL 27 and SCC-15 | Nude mice | Cell proliferation assay, hydrodynamic diameter and zeta potential measurement, BIO-RAD ChemiDoc XRS system | Prevent invasion and tumor growth | 28 |
| CxcR4 | Up | Lentiviral vectors | SCC-9, SCC-4, and Tca8113 cell lines | Nude mice | RT-PCR, Western blot, propidium iodide staining for cell cycle, Annexin V for apoptosis, CCK8, IHC, and microarray | Regulate cell cycle and proliferation | 67 |
| VEGF | Up | Folic acid-decorated polyamidoamine dendrimer G4 | HN-12 | Female athymic nude mice | ELISA, qRT-PCR, tumor size, H&E staining | Inhibit angiogenesis | 68 |
| MDR1 and MDR2 | Up | Adeno-associated viral vector | KB | No | RT-PCR, Western blot, flow cytometry, MTT assay | Regulate MDR | 51 |
| MDR1 | Up | Polymer polyethylenimine | Doxorubicin | KBV cells, BALB/c nude mice | Cytotoxicity assays in vitro, drug loading rate and release assays, agarose gel retardation assay, qRT-PCR, apoptosis by flow cytometry | Regulate MDR | 69 |
Abbreviations: CCK8, cell counting kit 8; EGFR, epidermal growth factor receptor; ELISA, enzyme-linked immunosorbent assay; IHC, immunohistochemistry; MDR, multidrug resistance; KBV, oral squamous cells; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; siRNA, small interfering RNA.
In order to overcome all these challenges related to siRNAs, several databases were developed. This is the case of siRNAdb, a complex database that furnishes a gene-centric view of siRNA experimental data, providing information related to the siRNA sequence, thermodynamic characteristics, and potential off-target effects.43 siDRM is one of the most promising tools for the design of effective siRNAs.44 RFRCDB-siRNA is another online tool for the design of siRNAs based on the random forest regression model connected with database searching.45 Without any doubt, RNAcentral is the most complex database that has joined 12 new resources and started the importing of new types of data for ncRNAs,46 such as nucleotides modification furnished from MODOMICS and Protein Data Bank, offering also free access.44
Nano-delivery systems for siRNA
The plasma membrane represents a physical barrier for siRNA uptake, due to the high hydrophobicity and negative charge, blocking the crossing of the biological barrier. The delivery methods need to counterwork this limitation and to assist cellular uptake.40
Naked siRNA have a very reduced internalization rate; therefore, in order to increase the therapeutic efficacy, often siRNA is loaded on a carrier alone or in tandem with other therapeutic systems. Also, siRNA may be conjugated with fluorescein isothiocyanate, in order to emit fluorescence, being easily monitored for cellular localization.47
An effective delivery system should be stable at the body temperature and pH variations, have an endocytosis promoting shape, should not be toxic, should exhibit high siRNA loading abilities, and have a size that avoids rapid renal and hepatic clearance.48
All the delivery systems developed for gene therapy can be adapted for siRNA delivery.49 siRNA delivery strategies that use viral particles have a constitutive effect while different nanostructures have only a transitory effect, implying multiple administration doses.36
The viral delivery of siRNA is composed of two main strategies: siRNA is either chemically synthesized and loaded into a viral capsule or it can be expressed from the DNA of a recombinant virus. The major disadvantages of viral siRNA delivery are lower targeted delivery of a specific cell in vivo, the host immune reaction, and the danger of oncogenic transformation of the virus.50 The multidrug resistance in oral cancer can be surpassed through the delivery of siRNA against the multidrug resistance (MDR) gene in adeno-associated viral vector. AAV-siMDR25mer and AAV-siMDR28mer inhibited the MDR1 and MDR2 gene expression and reduced the level of P-glycoprotein, expressed by the MDR1 gene, in KB cells.51 The viral delivery of interference RNA, although highly efficient, presents many challenges when delivered to a living organism, because it causes an inflammatory response following the recognition of the dsRNA longer than 30 nucleotides as nonself.52
Nonviral delivery systems for siRNA are far more diverse and are proposed more often in recent studies.52 These contain several subtypes, among which are liposomes, peptides, immunoliposomes, purified collagen,53 dendrimers,54 gold nanorods,55 carbon nanotubes,56,57 and RNAi microsponges.58 In general, the nanoparticles are PEGylated to mask the unspecific immune response, although it has been proven that the immune system can produce polyethylene glycol (PEG) antibodies.59 The most frequently tested nonviral carriers are liposomes, which have again presented the major challenge of immune reaction.53 The main problems related to siRNA-based drugs are represented by high toxicity.60 The comprehension and elimination of these issues can be considered a major constituent in the progress of safe and effective siRNA therapeutic systems.61
For effective delivery of therapeutic siRNA, the transcripts can be conjugated with cholesterol reaching better tumor retention in vivo. However, the siRNA-cholesterol complex is effective when administered in the tumor proximity; hence, it is unsuitable for systemic delivery.62
Aptamers are oligonucleotides or peptides that target specific molecules and by being bound to siRNA enhance their specificity.48 A nucleotide aptamer selected with the use of Systematic Evolution of Ligands by Exponential Enrichment technology was internalized in the HPV16-infected tonsil epithelial cells more efficiently than in the noninfected cells;63 it was proposed to be conjugated with E6 oncogene targeting siRNA.64 The cationic cell penetrating peptides facilitate endosomal escape or mediate endosomal-free entry into the cell without any need of modifying the siRNA structure.65
The polymers are positively charged; when they are conjugated with the negatively charged nucleic acids, they cause the nucleic acids to condensate and be protected.48 A polymeric nanoparticle galactose-modified trimethyl chitosan-cysteine loaded with vascular endothelial growth factor (VEGF)-targeted siRNA was used for increased specificity and efficiency in inhibiting oral cancer cell proliferation in vitro and tumor growth in vivo.49 Dendrimers are also artificial polymers that have a positive charge and are highly branched at the end-chains.
Polyamidoamine (PAMAM) is a dendrimer that, when conjugated with siRNA, can have both nuclear and perinuclear localization.48 The shRNA-mediated silencing of human telomerase reverse transcriptase loaded on PAMAM is a more potent knockdown method with an increased therapeutic efficacy.66
The targeted delivery of a complex composed of CIP2A siRNA, EGFR binding-peptide, and an endosome-disruptive peptide was proven to silence more efficiently the CIP2A oncogene, in vitro in the oral squamous cell carcinoma (OSCC) cell lines CAL 27 and squamous cell carcinoma (SCC)-15 and in vivo in xenografted mouse tumors of transfected CAL 27 cells.28 Recently, it was proven that the co-delivery of doxorubicin and MDR1 siRNA by using polymer polyethylenimine was related to an increased response to treatment, demonstrated both in vitro and in vivo. More details over the targeted delivery of siRNA as a therapeutic option in oral cancer are provided in Table 1.
Therapeutic assessment of siRNA in oral cancer
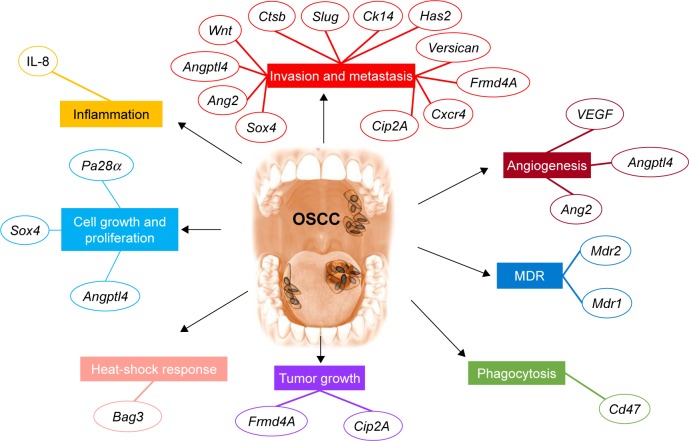
The essential characteristic of siRNA is its highly specific targeting of a single gene transcript. The various studies that use siRNA in cancer focus on the identification of a particular gene function or on the development of new therapies by silencing the overexpressed genes with oncogenic role (Figure 4).
Figure 4.
The main pathway targeted using siRNA delivery systems in oral cancer.
Notes: Examples of siRNA therapeutics alter multiple hallmarks applied for oral cancer. Cell growth and proliferation are impaired by siRNA for Pa28α, Sox4, or Angptl4. Local inflammation is reduced by IL-8. Invasion and metastasis are decreased by siRNA for Sox4, Ang2, Angptl4, Wnt, Ctsb, Slug, Ck14, Has2, Versican, Frmd4A, Cxcr4, and Cip2A. Angiogenesis is inhibited by siRNA targeting VEGF, Angptl4, and Ang2. MDR is inhibited by siRNA for Mdr1 and Mdr2. Phagocytosis is stimulated by siRNA for Cd47. Tumor growth is reduced in the presence of siRNA for Cip2A and Frmd4A. The heat-shock response is decreased by siRNA for Bag3.
Abbreviations: IL-8, interleukin-8; MDR, multidrug resistance; siRNA, small interfering RNA; VEGF, vascular endothelial growth factor.
Recently, the identification of subcellular targets in oral cancer cells guided the well-founded progression in the field of “siRNA-targeted therapy.” These newly designed siRNA systems are directed to target cellular components involved in signal transduction pathways that control epigenetics effectors, cell proliferation, cell cycle, apoptosis, immune response, or angiogenesis.20,70–72
The balance of histone acetylation and deacetylation is an important regulator of epigenetic pathways, affecting the mechanisms of gene expression regulation.73 Alteration or mutation within the histone deacetylase 8 (HDAC8) is associated with tumor progression caused by induction of aberrant transcription factors that regulate important cellular function.73 HDAC8 was found to be overexpressed in OSCC cancer cells. By targeting HDAC8 with siRNA, the expression of this enzyme was reduced, leading to reduced proliferation and increased apoptosis within the cells.74 After silencing the Frmd4A gene, the CAL 27 cells remained in the G0 phase with impaired proliferative, invasive, and migration capacities.75
Molecular analysis showed that a number of genes were affected by the treatment and that these genes were involved in the cell cycle, in the following signaling pathways: JAK/STAT, p53, and extracellular matrix proteins.67
SOX4 is an important regulator of the oncogenic PI3K/AKT and MAPK signaling pathways, which sustains cell proliferation and differention.76,77 SOX4 functions as an oncogene in the OSCC event described after siRNA treatment, followed by a decline in the proliferation, invasion, and migration of UM1 cancer cells.78 A reduction in the mutant p53 expression caused by its corresponding siRNA increased the apoptotic and necrotic SCC-4 cells and also lowered their metastasis capability.79
Previous studies reveal the complex effects of immune response mechanisms in OSCC.80–83 IL-8 is an important pro-inflammatory interleukin, involved in head and neck SCC progression, since its knockdown by an analog antisense ncRNA affected the cellular viability and the colony formation capacity mediated by NOD signaling pathway.81 IL-8 was also found to be overexpressed in the saliva of OSCC patients, when compared to healthy subjects and patients with precancerous lesions.82
siRNA knockdown of C-X-C chemokine receptor type 4 (Cxcr4) in OSCC decreased cell proliferation, while increasing the apoptosis rate and causing cell cycle arrest. Also, Cxcr4 supports epithelial to mesenchymal transition (EMT) progress and lymph node metastasis in tongue squamous cell carcinoma (TSCC); thus, the silencing of this gene by its corresponding siRNA led to the regression of these events in vitro as well as in xenografted tumors in mice.83
Cytokeratin 14 (Ck14) is a cytoskeletal component of the epithelial tissue and an important promoter of local invasion and metastasis. The Ck14 siRNA resulted in a 80% drop in the collective invasion of cells belonging to a three-dimensional salivary adenoid cystic carcinoma (SACC) culture.84 Hyaluronic synthases 2 (Has2) is overexpressed only in cancer-associated fibroblasts (CAFs) and not in normal fibroblasts. When this gene is knocked down by one of its two specific siRNAs in CAFs, and then the CAFs co-cultured with oral OSCC cells, the cancer invasion and migration abilities were decreased. This process is mediated by increased expression level of matrix metalloproteinase (MMP)-1 and MMP-3 and decreased expression of tissue inhibitor of metalloproteinase 1 and MMP-10.85
In some cases, multiple siRNAs can be used as a way of following the interaction of multiple elements from a signaling pathway. The regulation of EMT, a key process due to the fact that oral cancer cells are characterized by high epithelial plasticity, can be exploited to develop novel targeted therapies. EMT is related to inhibition/loss of E-cadherin expression, reduction of cellular adhesion, a fact that promotes invasion/migration, and finally leads to metastatic disease.86 For instance, by knocking down the MMP-10 and the Slug gene, it was proven that transforming growth factor β induces EMT and invasion of human oral squamous cell carcinoma (HSC)-4 cells, through Slug-dependent activation of MMP-10.87
Sometimes, siRNA can be used to study the role of a gene in OSCC pathology. The knocking down of Cdh11 gene enhanced the proliferation and invasion of UM-SCC-29 and UM-SCC-47 cells, thus providing proof of its function as a tumor suppressor.88 The methods by which the above-mentioned siRNA effects were determined are listed in Table 2.
Table 2.
Relevant studies of siRNA as a silencing mechanism in OSCC using commercial delivery systems used to furnish new mechanistic insights
| Targeted gene | Expression | Delivery system | Type of cell line/animal model | Methods | Effect on | Reference |
|---|---|---|---|---|---|---|
| Sox4 | Up | Lipofectamine 3000 | UM1 cell line | MTT assay, cell counting kit, matrigel invasion assay | Reduce proliferation and invasion | 78 |
| Pa28α | Up | Lipofectamine™ 2000 | CAL 27 and HSC-3 cell line BALB/c nude mice | Western blot, MTT assay, plate clonogenic assay, Edu assay, TUNEL assay, single scratch wound assay, tumor size measurement, immunohistochemical analysis of PCNA and CD34 | Cell growth and proliferation | 93 |
| Cd47 | Up | Lipofectamine RNAiMAX | HSC-3 cell line cocultured with CD68+ and CD163+ macrophage | Flow cytometry CFSE+ CD33+- engulfed cells | Anti-phagocytic | 94 |
| IL-8 | Up | Lipofectamine RNAiMAX | SCC-4, SCC-9, SCC-25 cell lines | qRT-PCR, Western blot, immunohistochemical staining, MTT assay, clonogenic assay, Lipofectamine 2000 siRNA transfection | Reduce proliferation and colony formation | 81 |
| Ck14 | Up | Lipofectamine 2000 | 3D cultures from fresh tissues of patients with salivary adenoid cystic carcinomas | Collective invasion in an ex vivo 3D culture assay, Western blot, micrographs of the border | Reduce proliferation rate and metastatic capacity | 84 |
| Cxcr4 | Up | Lipofectamine 2000 | TSCC cell line | Wound healing assay, transwell assay, Western blot, Annexin V-FITC apoptosis detection kit, histopathological analysis, immunohistochemistry analysis | Regulate EMT and prevent metastasis | 83 |
| Frmd4A | Up | Lipofectamine RNAiMAX | CAL 27 cell line | Western blot, propidium iodide staining and flow cytometry, cell counting kit, wound healing assay, matrigel invasion assay | Reduce tumor progression and metastasis | 75 |
| Has2 | Up | Lipofectamine 2000 | Primary fibroblasts from 48 patients – NF and CAF, the CAL 27 cell line | Western blot, immunofluorescence, wound healing and matrigel invasion assay, bromodeoxyuridine assay, RayBio antibody array | Regulate EMT and reduce invasion | 85 |
| Versican | Up | Lipofectamine 2000 | SCC-15 cell line | Western blot, transwell assay, matrigel invasion assay | Reduce migration, invasion, and tumor progression | 95 |
| Angptl4 | Up in late- stages | Lipofectamine 3000 | TSCC cells | Immunohistochemistry staining, qRT-PCR, Western blot, transwell plate invasion, colony-forming assay, cell counting kit | Reduce cell growth, angiogenesis, and metastasis | 89 |
| Ctsb | Up | Not specified | OC2 and CAL 27 cell lines | Western blot, transwell migration assays | Prevent metastasis | 96 |
| MMP-10 and Slug | TGFβ-induced expression | Lipofectamine RNAiMAX | HSC-4 cell line | qRT-PCR, Western blot, Boyden chamber assay | Regulate TGFβ-induced invasion | 87 |
| Ang2 | Up | FuGENE HD transfection reagent | TCA8113 cell line, BALB/c nude mice | Western blot, MTS assay, fluorescence channel analysis, flow cytometry, transwell chamber, ELISA, tumor volume measurement, TUNEL assay, immunohistochemical and quantification analysis of CD31 protein | Regulate EMT, prevent angiogenesis, invasion, and metastasis Inhibit apoptosis | 90 |
Abbreviations: ELISA, enzyme-linked immunosorbent assay; EMT, epithelial to mesenchymal transition; MTS, 3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; siRNA, small interfering RNA; TGFβ, transforming growth factor β; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
siRNA was used to target angiopoietin-like protein 4 mRNA in TSCC cells, as a way of inhibiting its role in cellular growth and colony formation, with the perspective of improving the survival rate of patients.89 Following siRNA silencing of Ang2, the expression of Vimentin, Snail, and Twist genes was downregulated at the same time as E-cadherin was upregulated. Thereby, the TCA8113 cells would present a reduction in their capacity to invade and migrate through the EMT process.90 The same pattern was observed in the case of siRNA inhibition of superoxide dismutase 2 in SACC-LM that led to E-cadherin increased levels as well as the decreased expression of migration-associated genes Vimentin, Slug, Snail, p-ERK, and MMP-2.91
Aberrant Notch4 expression has an essential function in OSCC tumorigenesis and promotes metastasis by perineural spread,92 becoming an important prognosis marker and therapeutic target.92 Notch4 siRNA caused the 1.4-fold drop in proliferation and 1.9-fold drop in cell motility, after 48-hour post-transfection in an oral cancer cell line, HSC-3.92
Combination of siRNA with other therapies
Nano-delivery of WNT siRNA in KB cells interrupted the WNT, β-catenin, and Vimentin pathway, hindering mesenchymal transition and by including photodynamic therapy, it could advance into a novel combinatorial therapeutic strategy.97 The combination of plasmonic photothermal therapy (PPTT) and heat-shock protein (HSP) siRNA is another instance of this kind of approach. By removing the protective role of Hsp70, which opposes heat-induced aggregation of proteins, thereby preventing cell death, these ncRNAs enhance the beneficial effects of PPTT.98 An important remark is the fact that a few years ago it was discovered that the KB cells have probably been contaminated with the HeLa cell line of cervical cancer, since it shows high genetic similarity with it; the controversy still remains around this cell line, because phenotypically it maintained the same morphology as the originally isolated cells of OSCC.99 Hence, studies using the KB cell line as oral cancer model should be regarded with a rational trace of doubt regarding its application in oral cancer.
Nanoparticles composed of PEG, chlorine e6, and polyethylenimine loaded with WNT siRNA decrease the in vitro viability and invasion capacity of KB cells and the effects were even more pronounced when gene silencing was combined with photodynamic therapy, thus proving the fact that combinatorial therapy is still the best option.100 Similar results were obtained in xenografted TSCC cell lines, SCC-4 and SAS treated with calcium phosphate lipid nanoparticles containing VEGF siRNA, in which the tumor growth was reduced due to efficient silencing of the pro-angiogenic factor VEGF.101 The same type of nanoparticles combined with photodynamic therapy and loaded with HIF1α siRNA again proved to be more effective than single therapy in SCC-4 and SAS cells in vitro and in xenografted tumors in vivo.102 siRNA-mediated silencing of Bag3 is more efficient in vivo in xenografted tumors of CAL 27 cells, by combining ncRNA with gold nanorods and applying photodynamic therapy.55 A summary of the data and details are provided in Table 3.
Table 3.
Studies using siRNA in combination with other therapies in oral cancer
| Targeted gene | Expression | Delivery system | Combination | Animal model, cell line | Methods | Target mechanism | Reference |
|---|---|---|---|---|---|---|---|
| Hsp70 | Up | Gold nanorods | Plasmonic photothermal therapy | HSC (oral) | XTT assay, PI staining, Annexin- V-FITC, immunofluorescence, immunoblot | Inhibition of proliferation and activation of apoptosis | 98 |
| WNT3 | Up | Nanoparticles of PEG, Ce6, and PEI | Photodynamic therapy | KB | Singlet oxygen generation measurement, particle size determination with TEM, MTT, Annexin V/PI staining, qRT-PCR, Western blot | Regulate EMT and prevent invasion | 97, 100 |
| VEGF | Up | Calcium phosphate lipid nanoparticles | Photodynamic therapy | SCC-4 and SAS cell lines, C57BL/6 mice | Western blot, tumor measurement, hematoxylin and eosin, IHC staining for Ki-67, cleaved caspase-3, α-SMA, CD31, VEGF, TUNEL, liver and kidney function assessment via blood measurement of ASP, ALT, and urea nitrogen | Inhibit angiogenesis | 101 |
| Hif1a | Up | Calcium phosphate lipid nanoparticles | Photodynamic therapy | SCC-4 and SAS cell lines, female nude mice | Western blot, Texas Red-DNA oligos, MTT, qRT-PCR, IHC, TUNEL assay, DAB detection kit, liver toxicity with ALT, AST, total bilirubin, kidney toxicity with creatinine, blood urea nitrogen, blood total albumin, protein, globulin, LDH, creatine kinase | Inhibit angiogenesis via hypoxic-related pathways | 102 |
| Bag3 | Up | Gold nanorods | Photodynamic therapy | CAL 27 cell line, BALB/c nude mice | TEM, MTT, RT-PCR, Western blot, fluorescence microscopy, flow cytometry, IHC, TUNEL assay | Implicated in heat- shock response | 55 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; EMT, epithelial to mesenchymal transition; FITC, fluorescein isothiocyanate; LDH, lactate dehydrogenase; PEG, polyethylene glycol; PEI, polyethylenimine; PI, propidium iodine; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; siRNA, small interfering RNA; TEM, transmission electron microscope; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; VEGF, vascular endothelial growth factor; α-SMA, α-smooth muscle actin; XTT, 2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide.
Clinical trial in oral cancer using siRNA therapy
The main challenge for siRNA delivery is the proper choice of a delivery route. In the clinical trials listed for therapeutic delivery systems in oral cancer, the majority of them (three out of four) use the adenovirus injected locally as a carrier of the therapeutic agent, while a single clinical trial loaded lipid nanoparticles with the siRNA for p53, which are again injected locally.
A way of preventing transformation of premalignant oral lesions may be the local delivery of the adenoviral containing wild-type p53 gene, the Ad5CMV-p53 gene. In a clinical trial first proposed in 2006, the injection of oral lesion of Ad5CMV-p53 was followed by a rinse of the mouth. It was demonstrated that Ad5CMV-p53 is safe and can be repetitively administered for improvement of the therapeutic efficacy, lacking signs of severe toxicity. Its presence in the circulation of the adenovirus at 24 hours posttreatment, and the presence of p53 transgene in the tumor tissue were demonstrated. This certifies the utility of this therapeutic system for the mutated p53 gene,103 being well-tolerated by patients with oral cancers.104
The study was terminated and another was performed.105,106 siRNA delivery was carried out alone, before surgical tumor resection or in combination with standard chemotherapy, in order to observe how it would affect the survival rate of patients with advanced oral or maxillofacial malignancies.107 The antisense DNA of EGFR gene was delivered in lipid nanoparticles, in a clinical trial for advanced SCC of the head and neck,108,109 being well-tolerated, showing only reduced inflammation at the injection site.109
siRNA therapies versus novel genomic editing tools
RNAi is a natural mechanism that has the capacity to regulate gene expression via an endogenous systematic system with effects on the expression level of a particular phenotype. It still remains an important technology for the study of gene function despite several disadvantages related to the hypomorphic phenotypes, some inconsistencies in complete loss-of-function and also due to genetic mutation.110
A basically distinct method to regulate gene function was developed based on the comprehension of the mechanism of action of the programmable DNA-binding proteins, retrieved in the literature as transcription activator-like effectors nucleases (TALEN).110 Another important tool for genome editing is represented by the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system.110–112 CRISPR/Cas9 is currently the most popular system used and is based on a programmable DNA nuclease similar to TALEN.112
In contrast to RNAi, CRISPR or TALEN seem to be able to have a consistent and robust knockdown effect upon the targeted DNA,110 but the main problem related to these two approaches consists in the possible mutagenic effect achieved during procedure, an effect that is maintained persistently in the DNA sequence.
The subject regarding the advantages and disadvantages between the newly implemented technologies, and RNAi is currently under major debate; however, the RNAi strategy has several advantages that make this approach a strong contender in different experimental settings. One of these advantages consists in the simplicity of the method, in that RNAi does not require additional elements for the generation of knockdowns. The fact that the majority of the targeted cells already contain the RNAi silencing machinery, that is actually the full equipment necessary, makes the implementation of such strategies easier and also more cost efficient. Moreover, RNAi targets the transcriptomic landscape and not the genomic DNA sequences, such as CRISPR or TALEN; therefore, the accessibility to the targeted sequence is not hampered by the chromatin state.110 Also, RNAi does not target the transcription start site (TSS), an act that facilitates the use of the technology in organisms without annotated TSSs.113
Conclusion and perspectives
siRNA therapeutic development in oral cancer has to overcome some major challenges related especially to the delivery methods and potential off-target effects. Also, the danger of being degraded by bloodstream nucleases, loaded onto the reticuloendothelial system, endosomal escape, avoidance of the immune response, and RISC interaction are important aspects to be considered in the future.
The siRNA delivery issue can be overcome through chemical modification of the exogenous sequence, loading of the particle in a viral capsule, or loading siRNA on different types of nanoparticles.
Often, siRNA is used as a means of evaluating the function of a particular gene, for the study of particular molecular mechanisms. During the first evaluations of the therapeutic potential of siRNA against certain oral cancer tumor promoting genes: CIP2A, CXCR4, VEGF, MDR1, and MDR2, specific cancer hallmarks were targeted. One step further in the development of siRNA therapeutics is the concept of combining siRNA therapy with other therapies such as photodynamic therapy or PPTT. Lipid nanoparticles targeting EGFR were proved to have a significant therapeutic potential, along with a clinical trial for the silencing of the p53 gene exploiting RNAi mechanisms. These two clinical trials have demonstrated only minimal drawbacks of using a more targeted approach on oral cancer malignancies and opened the path for a brighter future of oral cancer therapies.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.D’Souza G, McNeel TS, Fakhry C. Understanding personal risk of oropharyngeal cancer: risk-groups for oncogenic oral HPV infection and oropharyngeal cancer. Ann Oncol. 2017;28(12):3065–3069. doi: 10.1093/annonc/mdx535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Petti S. Lifestyle risk factors for oral cancer. Oral Oncol. 2009;45(4–5):340–350. doi: 10.1016/j.oraloncology.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Irimie AI, Braicu C, Cojocneanu-Petric R, Berindan-Neagoe I, Campian RS. Novel technologies for oral squamous carcinoma biomarkers in diagnostics and prognostics. Acta Odontol Scand. 2015;73(3):161–168. doi: 10.3109/00016357.2014.986754. [DOI] [PubMed] [Google Scholar]
- 6.Pileczki V, Braicu C, Gherman CD, Berindan-Neagoe I. TNF-alpha gene knockout in triple negative breast cancer cell line induces apoptosis. Int J Mol Sci. 2012;14(1):411–420. doi: 10.3390/ijms14010411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setoyama T, Ling H, Natsugoe S, Calin GA. Non-coding RNAs for medical practice in oncology. Keio J Med. 2011;60(4):106–113. doi: 10.2302/kjm.60.106. [DOI] [PubMed] [Google Scholar]
- 9.Berindan-Neagoe I, Calin GA. Molecular pathways: microRNAs, cancer cells, and microenvironment. Clin Cancer Res. 2014;20(24):6247–6253. doi: 10.1158/1078-0432.CCR-13-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64(5):311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. 2010;11(8):559–571. doi: 10.1038/nrg2814. [DOI] [PubMed] [Google Scholar]
- 12.Francastel C, Hubé F, Mani S, Santulli G, Taube J, Szweykowska-Kulinska Z. The non-coding RNA journal club: highlights on recent papers – 2. Noncoding RNA. 2015;1(2):167. doi: 10.3390/ncrna1020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin G, Vazquez F. Welcome to the new journal Non-Coding RNA! Noncoding RNA. 2015;1(1):1–3. doi: 10.3390/ncrna1010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaharie F, Muresan MS, Petrushev B, et al. Exosome-carried microRNA-375 inhibits cell progression and dissemination via Bcl-2 blocking in colon cancer. J Gastrointestin Liver Dis. 2015;24(4):435–443. doi: 10.15403/jgld.2014.1121.244.375. [DOI] [PubMed] [Google Scholar]
- 16.Ferracin M, Gautheret D, Hubé F, et al. The non-coding RNA journal club: highlights on recent papers. Noncoding RNA. 2015;1(1):87. doi: 10.3390/ncrna1010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7–8):454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 18.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121(4):141–158. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 19.Catana CS, Pichler M, Giannelli G, Mader RM, Berindan-Neagoe I. Non-coding RNAs, the Trojan horse in two-way communication between tumor and stroma in colorectal and hepatocellular carcinoma. Oncotarget. 2017;8(17):29519–29534. doi: 10.18632/oncotarget.15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braicu C, Catana C, Calin GA, Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr Pharm Des. 2014;20(42):6565–6574. doi: 10.2174/1381612820666140826153529. [DOI] [PubMed] [Google Scholar]
- 21.Pop-Bica C, Gulei D, Cojocneanu-Petric R, Braicu C, Petrut B, Berindan-Neagoe I. Understanding the role of non-coding RNAs in bladder cancer: from dark matter to valuable therapeutic targets. Int J Mol Sci. 2017;18(7):E1514. doi: 10.3390/ijms18071514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braicu C, Cojocneanu-Petric R, Chira S, et al. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine. 2015;10:791–800. doi: 10.2147/IJN.S72904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimie AI, Braicu C, Sonea L, et al. A looking-glass of non-coding RNAs in oral cancer. Int J Mol Sci. 2017;18(12):E2620. doi: 10.3390/ijms18122620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 25.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dana H, Chalbatani GM, Mahmoodzadeh H, et al. Molecular mechanisms and biological functions of siRNA. Int J Biomed Sci. 2017;13(2):48–57. [PMC free article] [PubMed] [Google Scholar]
- 27.Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander-Bryant AA, Zhang H, Attaway CC, et al. Dual peptide-mediated targeted delivery of bioactive siRNAs to oral cancer cells in vivo. Oral Oncol. 2017;72:123–131. doi: 10.1016/j.oraloncology.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fakhr E, Zare F, Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 2016;23(4):73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- 30.Tilesi F, Fradiani P, Socci V, Willems D, Ascenzioni F. Design and validation of siRNAs and shRNAs. Curr Opin Mol Ther. 2009;11(2):156–164. [PubMed] [Google Scholar]
- 31.Tafer H. Bioinformatics of siRNA design. Methods Mol Biol. 2014;1097:477–490. doi: 10.1007/978-1-62703-709-9_22. [DOI] [PubMed] [Google Scholar]
- 32.Hickerson RP, Vlassov AV, Wang Q, et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides. 2008;18(4):345–354. doi: 10.1089/oli.2008.0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chernolovskaya EL, Zenkova MA. Chemical modification of siRNA. Curr Opin Mol Ther. 2010;12(2):158–167. [PubMed] [Google Scholar]
- 34.Choi HS, Liu W, Misra P, et al. Renal clearance of nanoparticles. Nat Biotechnol. 2007;25(10):1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miele E, Spinelli GP, Miele E, et al. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irimie AI, Sonea L, Jurj A, et al. Future trends and emerging issues for nanodelivery systems in oral and oropharyngeal cancer. Int J Nanomedicine. 2017;12:4593–4606. doi: 10.2147/IJN.S133219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85(2):187–200. [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 39.Meng Z, Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123(Pt 8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 41.Dalby B, Cates S, Harris A, et al. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33(2):95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 42.Cantini L, Attaway CC, Butler B, Andino LM, Sokolosky ML, Jakymiw A. Fusogenic-oligoarginine peptide-mediated delivery of siRNAs targeting the CIP2A oncogene into oral cancer cells. PLoS One. 2013;8(9):e73348. doi: 10.1371/journal.pone.0073348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalk AM, Warfinge RE, Georgii-Hemming P, Sonnhammer ELL. siRNAdb: a database of siRNA sequences. Nucleic Acids Res. 2005;33(Database issue):D131–D134. doi: 10.1093/nar/gki136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong W, Ren Y, Zhou H, Wang Y, Kang S, Li T. siDRM: an effective and generally applicable online siRNA design tool. Bioinformatics. 2008;24(20):2405–2406. doi: 10.1093/bioinformatics/btn442. [DOI] [PubMed] [Google Scholar]
- 45.Jiang P, Wu H, Da Y, et al. RFRCDB-siRNA: improved design of siRNAs by random forest regression model coupled with database searching. Comput Methods Programs Biomed. 2007;87(3):230–238. doi: 10.1016/j.cmpb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 46.RNAcentral Consortium RNAcentral: an international database of ncRNA sequences. Nucleic Acids Res. 2015;43(Database issue):D123–D129. doi: 10.1093/nar/gku991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocker M, Neureiter D, Lueders M, et al. Variants of bcl-2 specific siRNA for silencing antiapoptotic bcl-2 in pancreatic cancer. Gut. 2005;54(9):1298–1308. doi: 10.1136/gut.2004.056192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials (Basel) 2017;7(4):E77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han L, Tang C, Yin C. Oral delivery of shRNA and siRNA via multifunctional polymeric nanoparticles for synergistic cancer therapy. Biomaterials. 2014;35(15):4589–4600. doi: 10.1016/j.biomaterials.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira S, Storm G, Schiffelers RM. Targeted delivery of siRNA. J Biomed Biotechnol. 2006;2006(4):63675. doi: 10.1155/JBB/2006/63675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu D, McCarty D, Fernandes A, Fisher M, Samulski RJ, Juliano RL. Delivery of MDR1 small interfering RNA by self-complementary recombinant adeno-associated virus vector. Mol Ther. 2005;11(4):523–530. doi: 10.1016/j.ymthe.2004.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akhtar S, Benter IF. Nonviral delivery of synthetic siRNAs in vivo. J Clin Invest. 2007;117(12):3623–3632. doi: 10.1172/JCI33494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13(18):1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 54.Biswas S, Torchilin VP. Dendrimers for siRNA delivery. Pharmaceuticals (Basel) 2013;6(2):161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang BK, Yu XF, Wang JH, et al. Gold-nanorods-siRNA nanoplex for improved photothermal therapy by gene silencing. Biomaterials. 2016;78:27–39. doi: 10.1016/j.biomaterials.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 56.Neagoe IB, Braicu C, Matea C, et al. Efficient siRNA delivery system using carboxilated single-wall carbon nanotubes in cancer treatment. J Biomed Nanotechnol. 2012;8(4):567–574. doi: 10.1166/jbn.2012.1411. [DOI] [PubMed] [Google Scholar]
- 57.Gherman C, Tudor MC, Constantin B, et al. Pharmacokinetics evaluation of carbon nanotubes using FTIR analysis and histological analysis. J Nanosci Nanotechnol. 2015;15(4):2865–2869. doi: 10.1166/jnn.2015.9845. [DOI] [PubMed] [Google Scholar]
- 58.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 2012;11(4):316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Q, Lai SK. Anti-PEG immunity: emergence, characteristics, and unaddressed questions. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2015;7(5):655–677. doi: 10.1002/wnan.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Human Gene Ther. 2008;19(2):111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 61.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 62.Chernikov IV, Gladkikh DV, Meschaninova MI, et al. Cholesterol-containing nuclease-resistant siRNA accumulates in tumors in a carrier-free mode and silences MDR1 gene. Mol Ther Nucleic Acids. 2017;6:209–220. doi: 10.1016/j.omtn.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gourronc FA, Rockey WM, Thiel WH, Giangrande PH, Klingelhutz AJ. Identification of RNA aptamers that internalize into HPV-16 E6/E7 transformed tonsillar epithelial cells. Virology. 2013;446(1–2):325–333. doi: 10.1016/j.virol.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe-Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus-positive cancer cells. Oncogene. 2003;22(38):5938–5945. doi: 10.1038/sj.onc.1206894. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X, Huang H, Wang J, et al. Dendrimers-delivered short hairpin RNA targeting hTERT inhibits oral cancer cell growth in vitro and in vivo. Biochem Pharmacol. 2011;82(1):17–23. doi: 10.1016/j.bcp.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 67.Yu T, Wu Y, Huang Y, et al. RNAi targeting CXCR4 inhibits tumor growth through inducing cell cycle arrest and apoptosis. Mol Ther. 2012;20(2):398–407. doi: 10.1038/mt.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu L, Yeudall WA, Yang H. Folic acid-decorated polyamidoamine dendrimer exhibits high tumor uptake and sustained highly localized retention in solid tumors: its utility for local siRNA delivery. Acta Biomater. 2017;57:251–261. doi: 10.1016/j.actbio.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D, Xu X, Zhang K, et al. Codelivery of doxorubicin and MDR1-siRNA by mesoporous silica nanoparticles-polymerpolyethylenimine to improve oral squamous carcinoma treatment. Int J Nanomedicine. 2018;13:187–198. doi: 10.2147/IJN.S150610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braicu C, Pileczki V, Irimie A, Berindan-Neagoe I. p53siRNA therapy reduces cell proliferation, migration and induces apoptosis in triple negative breast cancer cells. Mol Cell Biochem. 2013;381(1–2):61–68. doi: 10.1007/s11010-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 71.Aldea M, Craciun L, Tomuleasa C, et al. Repositioning metformin in cancer: genetics, drug targets, and new ways of delivery. Tumour Biol. 2014;35(6):5101–5110. doi: 10.1007/s13277-014-1676-8. [DOI] [PubMed] [Google Scholar]
- 72.Aldea MD, Petrushev B, Soritau O, et al. Metformin plus sorafenib highly impacts temozolomide resistant glioblastoma stem-like cells. J BUON. 2014;19(2):502–511. [PubMed] [Google Scholar]
- 73.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1(1):19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahn MY, Yoon JH. Histone deacetylase 8 as a novel therapeutic target in oral squamous cell carcinoma. Oncol Rep. 2017;37(1):540–546. doi: 10.3892/or.2016.5280. [DOI] [PubMed] [Google Scholar]
- 75.Zheng X, Jia B, Lin X, et al. FRMD4A: a potential therapeutic target for the treatment of tongue squamous cell carcinoma. Int J Mol Med. 2016;38(5):1443–1449. doi: 10.3892/ijmm.2016.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramezani-Rad P, Geng H, Hurtz C, et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood. 2013;121(1):148–155. doi: 10.1182/blood-2012-05-428938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Cui L, Huang J, et al. SOX4 promotes progression in OLP-associated squamous cell carcinoma. J Cancer. 2016;7(11):1534–1540. doi: 10.7150/jca.15689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Georgakopoulou EA, Troupis TG, Troupis G, Gorgoulis VG. Update of the cancer-associated molecular mechanisms in oral lichen planus, a disease with possible premalignant nature. J Buon. 2011;16(4):613–616. [PubMed] [Google Scholar]
- 79.Irimie AI, Braicu C, Pileczki V, et al. Knocking down of p53 triggers apoptosis and autophagy, concomitantly with inhibition of migration on SSC-4 oral squamous carcinoma cells. Mol Cell Biochem. 2016;419(1–2):75–82. doi: 10.1007/s11010-016-2751-9. [DOI] [PubMed] [Google Scholar]
- 80.Winck FV, Prado Ribeiro AC, Ramos Domingues R, et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep. 2015;5:16305. doi: 10.1038/srep16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan LP, Wang LF, Chiang FY, Lee KW, Kuo PL, Liang CH. IL-8 promotes HNSCC progression on CXCR1/2-mediated NOD1/RIP2 signaling pathway. Oncotarget. 2016;7(38):61820–61831. doi: 10.18632/oncotarget.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Punyani SR, Sathawane RS. Salivary level of interleukin-8 in oral precancer and oral squamous cell carcinoma. Clin Oral Investig. 2013;17(2):517–524. doi: 10.1007/s00784-012-0723-3. [DOI] [PubMed] [Google Scholar]
- 83.Duan Y, Zhang S, Wang L, et al. Targeted silencing of CXCR4 inhibits epithelial–mesenchymal transition in oral squamous cell carcinoma. Oncol Lett. 2016;12(3):2055–2061. doi: 10.3892/ol.2016.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao XL, Wu JS, Cao MX, et al. Cytokeratin-14 contributes to collective invasion of salivary adenoid cystic carcinoma. PLoS One. 2017;12(2):e0171341. doi: 10.1371/journal.pone.0171341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Z, Tao D, Zhang P, et al. Hyaluronan synthase 2 expressed by cancer-associated fibroblasts promotes oral cancer invasion. J Exp Clin Cancer Res. 2016;35(1):181. doi: 10.1186/s13046-016-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patil S, Rao RS, Ganavi BS. Mesenchymal–epithelial transition in oral cancer. J Int Oral Health. 2015;7(9):i–ii. [PMC free article] [PubMed] [Google Scholar]
- 87.Hino M, Kamo M, Saito D, et al. Transforming growth factor-beta1 induces invasion ability of HSC-4 human oral squamous cell carcinoma cells through the Slug/Wnt-5b/MMP-10 signalling axis. J Biochem. 2016;159(6):631–640. doi: 10.1093/jb/mvw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Piao S, Inglehart RC, Scanlon CS, Russo N, Banerjee R, D’Silva NJ. CDH11 inhibits proliferation and invasion in head and neck cancer. J Oral Pathol Med. 2017;46(2):89–97. doi: 10.1111/jop.12471. [DOI] [PubMed] [Google Scholar]
- 89.Huang Z, Xie J, Lin S, et al. The downregulation of ANGPTL4 inhibits the migration and proliferation of tongue squamous cell carcinoma. Arch Oral Biol. 2016;71:144–149. doi: 10.1016/j.archoralbio.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 90.Li C, Li Q, Cai Y, et al. Overexpression of angiopoietin 2 promotes the formation of oral squamous cell carcinoma by increasing epithelial–mesenchymal transition-induced angiogenesis. Cancer Gene Ther. 2016;23(9):295–302. doi: 10.1038/cgt.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang B, Yang H, Jiao Y, et al. SOD2 deregulation enhances migration, invasion and has poor prognosis in salivary adenoid cystic carcinoma. Sci Rep. 2016;6:25918. doi: 10.1038/srep25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mk H, Prince S, Mohan AM, Krishnan KV, Devi A. Association of Notch4 with metastasis in human oral squamous cell carcinoma. Life Sci. 2016;156:38–46. doi: 10.1016/j.lfs.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 93.Feng X, Jiang Y, Xie L, et al. Overexpression of proteasomal activator PA28α serves as a prognostic factor in oral squamous cell carcinoma. J Exp Clin Cancer Res. 2016;35:35. doi: 10.1186/s13046-016-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakakura K, Takahashi H, Kaira K, et al. Relationship between tumor-associated macrophage subsets and CD47 expression in squamous cell carcinoma of the head and neck in the tumor microenvironment. Lab Invest. 2016;96(9):994–1003. doi: 10.1038/labinvest.2016.70. [DOI] [PubMed] [Google Scholar]
- 95.Wei T, Cong X, Wang XT, et al. Interleukin-17A promotes tongue squamous cell carcinoma metastasis through activating miR-23b/versican pathway. Oncotarget. 2017;8(4):6663–6680. doi: 10.18632/oncotarget.14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang WE, Ho CC, Yang SF, et al. Cathepsin B expression and the correlation with clinical aspects of oral squamous cell carcinoma. PLoS One. 2016;11(3):e0152165. doi: 10.1371/journal.pone.0152165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma C, Shi L, Huang Y, et al. Nanoparticle delivery of Wnt-1 siRNA enhances photodynamic therapy by inhibiting epithelial–mesenchymal transition for oral cancer. Biomater Sci. 2017;5(3):494–501. doi: 10.1039/c6bm00833j. [DOI] [PubMed] [Google Scholar]
- 98.Ali MR, Ali HR, Rankin CR, El-Sayed MA. Targeting heat shock protein 70 using gold nanorods enhances cancer cell apoptosis in low dose plasmonic photothermal therapy. Biomaterials. 2016;102:1–8. doi: 10.1016/j.biomaterials.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 99.Jiang L, Zeng X, Wang Z, Chen Q. Cell line cross-contamination: KB is not an oral squamous cell carcinoma cell line. Eur J Oral Sci. 2009;117(1):90–91. doi: 10.1111/j.1600-0722.2008.00599.x. [DOI] [PubMed] [Google Scholar]
- 100.Ghahhari NM, Babashah S. Interplay between microRNAs and WNT/β-catenin signalling pathway regulates epithelial-mesenchymal transition in cancer. Eur J Cancer. 2015;51(12):1638–1649. doi: 10.1016/j.ejca.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 101.Lecaros RL, Huang L, Lee TC, Hsu YC. Nanoparticle delivered VEGF-A siRNA enhances photodynamic therapy for head and neck cancer treatment. Mol Ther. 2016;24(1):106–116. doi: 10.1038/mt.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen WH, Lecaros RL, Tseng YC, Huang L, Hsu YC. Nanoparticle delivery of HIF1α siRNA combined with photodynamic therapy as a potential treatment strategy for head-and-neck cancer. Cancer Lett. 2015;359(1):65–74. doi: 10.1016/j.canlet.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tolcher AW, Hao D, de Bono J, et al. Phase I, pharmacokinetic, and pharmacodynamic study of intravenously administered Ad5CMV-p53, an adenoviral vector containing the wild-type p53 gene, in patients with advanced cancer. J Clin Oncol. 2006;24(13):2052–2058. doi: 10.1200/JCO.2005.03.6756. [DOI] [PubMed] [Google Scholar]
- 104.INGN 201: Ad-p53, Ad5CMV-p53, adenoviral p53, p53 gene therapy-introgen, RPR/INGN 201. Drugs R D. 2007;8(3):176–187. doi: 10.2165/00126839-200708030-00005. [DOI] [PubMed] [Google Scholar]
- 105.M.D. Anderson Cancer Center Adenovirus for Oral Premalignancies. [Accessed January 10, 2018]. Available from: https://clinicaltrials.gov/ct2/show/NCT00410865?term=p53&cond=oral+cancer&draw=1&rank=2. NLM identifier: NCT00410865.
- 106.National Cancer Institute (NCI) Gene Therapy in Preventing Cancer in Patients With Premalignant Carcinoma of the Oral Cavity or Pharynx. [Accessed January 10, 2018]. Available from: https://clinicaltrials.gov/ct2/show/study/NCT00064103?term=p53&cond=oral+cancer&draw=1&rank=3. NLM identifier: NCT00064103.
- 107.Shenzhen SiBiono GeneTech Co., Ltd rAd-p53 Gene Therapy for Advanced Oral and Maxillofacial Malignant Tumors (rAd-p53) [Accessed January 10, 2018]. Available from: https://clinicaltrials.gov/ct2/show/study/NCT00902083?term=p53&cond=oral+cancer&draw=1&rank=6. NLM identifier: NCT00902083.
- 108.University of Pittsburgh Gene Therapy in Treating Patients With Advanced Head and Neck Cancer. [Accessed January 10, 2018]. Available from: https://clinicaltri-als.gov/ct2/show/study/NCT00009841?term=gene&cond=Oral+Cancer&draw=3. NLM identifier: NCT00009841.
- 109.Zeng Q, Kanter PM, Dhir R, Gooding WE, Huang L, Grandis JR. Lack of toxicity of EGFR antisense gene therapy. J Exp Ther Oncol. 2002;2(3):174–186. doi: 10.1046/j.1359-4117.2002.01033.x. [DOI] [PubMed] [Google Scholar]
- 110.Boettcher M, McManus MT. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell. 2015;58(4):575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chira S, Gulei D, Hajitou A, Berindan-Neagoe I. Restoring the p53 ‘guardian’ phenotype in p53-deficient tumor cells with CRISPR/Cas9. Trends Biotechnol. 2018 Feb 22; doi: 10.1016/j.tibtech.2018.01.014. Epub. [DOI] [PubMed] [Google Scholar]
- 112.Gulei D, Berindan-Neagoe I. CRISPR/Cas9: a potential life-saving tool. What’s next? Mol Ther Nucleic Acids. 2017;9:333–336. doi: 10.1016/j.omtn.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Carninci P, Sandelin A, Lenhard B, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38(6):626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]