ABSTRACT
Macroautophagy, a highly conserved process in eukaryotic cells, is initiated in response to stress, especially nutrient starvation. Macroautophagy helps cells survive by engulfing proteins and organelles into an unusual double-membraned structure called the autophagosome, which then fuses with the lysosome. Upon degradation of the engulfed contents, the building blocks are recycled for synthesis of new macromolecules. Recent work has demonstrated that construction of the autophagosome requires a variety of small GTPases in variations of their normal roles in membrane traffic. In this Commentary, we review our own recent findings with respect to 2 different GTPases, Arl1, a member of the Arf/Arl/Sar family, and Ypt6, a member of the Rab family, in the yeast S. cerevisiae in light of other information from the literature and discuss future directions for further discerning the roles of small GTPases in autophagy.
KEYWORDS: Arl1, autophagosome, GTPase-activating protein (GAP), guanine nucleotide exchange factor (GEF), macroautophagy, membrane traffic, Saccharomyces cerevisiae, Ypt6
Introduction
Macroautophagy is a process by which defective proteins or organelles are packaged and transported for breakdown in lysosomes (called vacuoles in the yeast Saccharomyces cerevisiae) so that building blocks (amino acids, lipids, etc.) can be recycled for reuse, especially under stress conditions such that induced by nitrogen starvation. Activation of this pathway, initiated by inhibition of the Tor complex results in the construction of an unusual double-membraned structure called the autophagosome, which grows from a structure called the phagophore at the phagophore assembly site (PAS). As macroautophagy proceeds, the autophagosomes fuse with the lysosome/vacuole then the inner membrane as well as the engulfed contents are broken down by degradative enzymes contained in the lysosome/vacuole.1,2
Packaging of material into autophagosomes is a complex process requiring membranes from a number of different organelles, including the ER, Golgi apparatus, plasma membrane and mitochondria.3 Construction and then consumption of autophagosomes require a number of proteins specific to autophagy, the Atg proteins. Finally, small GTPases of the Arf/Arl/Sar and Rab families are required for both construction of the autophagosome and fusion of the autophagosome with the lysosome (or vacuole in yeast) in a variation of their roles as membrane traffic regulators for the secretory pathway and endocytosis.4-7 In this Commentary, we will describe our recent work documenting roles for 2 GTPases in macroautophagy, Arl1, a member of the Arf/Arl/Sar family of small GTPases, and Ypt6, a member of the Rab family, in S. cerevisiae,8 describe how these data fit into a larger understanding of the roles of membrane traffic in macroautophagy, then discuss future directions. We will focus primarily on what has been learned from studies in yeast, but note that this process is highly conserved across eukaryotes, including higher plants and animals.
The roles of yeast ARL1 and YPT6 in macroautophagy
Arl1, highly conserved in eukaryotes, is involved in membrane traffic in the secretory and endocytic pathways.9,10 Arl1 is also a mediator of K+ homeostasis in yeast,11-13 although it is unknown whether Arl1 plays a similar role in other eukaryotes.
Our interest in exploring a potential role for Arl1 in macroautophagy was initially sparked by results describing a role for ARL1 in autophagic cell death in S. cerevisiae,14 specifically, that a mutant allele of ARL1, ARL1[D151G] extended the viability of a cdc28 mutant. By using specific autophagy assays, the GFP-Atg8 assay,15 which measures transfer of a key regulator of autophagy, Atg8 to the vacuole by examining whether free GFP is produced; and the Pho8Δ60 assay,16 which measures arrival of phosphatase activity in the vacuole by autophagy, we found that an arl1Δ mutant was unable to perform autophagy under certain conditions. Specifically the defective autophagy phenotype was only observed at the restrictive temperature of 37oC; autophagy proceeded normally at the permissive temperature, 30°C. In addition, the phenotype was fully reversible upon reincubation of the cells at 30°C.8
Because YPT6 exhibits synthetic lethality with ARL1,17 we also explored the potential role of YPT6 in macroautophagy, and found a similar phenotype: ypt6Δ strains are unable to complete autophagy at 37°C, yet the phenotype is reversible upon reincubation at 30°C.8 By using protein degron technology18 to construct a degradable version of Arl1, we temporarily induced loss of Arl1 in a ypt6Δ background and found that the cells now showed an autophagy defect at 30°C, suggesting that Arl1 and Ypt6 function reciprocally in autophagy.
The GTP-restricted allele of Arl1, ARL1[Q72L], complements defects in membrane traffic,9,10 while a nucleotide-free version of the protein, encoded by ARL1[N127I] complements defects in K+ homeostasis.12,19 We therefore investigated which Arl1 alleles complemented the autophagy phenotype and found only wild type and the GTP-restricted allele, ARL1[Q72L] were able to do so, supporting the hypothesis that Arl1s role in the process is as a membrane traffic regulator. Interestingly, the ARL1 allele, ARL1[D151G], despite the fact that this allele appeared to extend lifespan in a cdc28 mutant14 was not able to complement the phenotypes we measured. However, similar to Arl1, the GTP-restricted allele of YPT6, YPT6[Q69L], complemented the phenotype whereas a GDP-restricted allele, YPT6[T24N] did not.8
Arl1 and Ypt6 are necessary for the construction of the autophagosome, and are required for the anterograde traffic of the sole transmembrane protein known to be involved in autophagy, Atg9, to this structure. Moreover, the 2 GTPases are required for at least delivery of membrane components from the Golgi apparatus to the PAS, but whether they also are required for delivery of membrane components from other membranes (ER, mitochondria, etc.) remains an open question. Finally, based on previous data showing both Arl1 and Ypt6 interact with the Golgi-associated retrograde protein (GARP) complex (specifically, Arl1 binds to the Vps53 subunit20 and Ypt6 binds to the Vps52 subunit21 of the GARP complex) and that the GARP complex is necessary for some forms of autophagy,22 we examined the colocalization of Arl1 and Ypt6 with GARP complex subunits Vps52 and Vps53 at the PAS upon induction of autophagy,8 which resulted in the following model for the roles of these 2 small GTPases in macroautophagy (Fig. 1).
Figure 1.
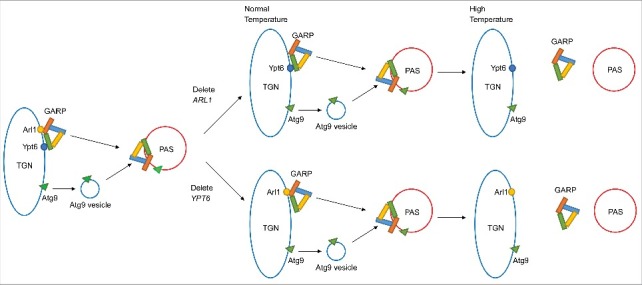
Current model for the reciprocal roles of Arl1 and Ypt6 in macroautophagy. Data shown in ref. 8 demonstrate that Arl1 and Ypt6 in S. cerevisiae function to deliver Atg9-containing vesicles from the Golgi apparatus to the growing phagophore at the phagophore assembly site (PAS) to make the autophagosome by virtue of their interactions with the Golgi-associated retromer complex (GARP). In mutants lacking either ARL1 or YPT6, autophagy proceeds normally at the permissive temperature of 30°C because one of the 2 proteins is sufficient to bind to the GARP comples. However, in mutants lacking either of the genes, autophagy is inhibited at the restrictive temperature of 37°C presumably because the strength of the interaction with a single small GTPase is insufficient to retain GARP on the membrane at this temperature. A conditional mutant lacking both small GTPases is unable to perform autophagy at the permissive temperature.
Future directions
What is the complete set of small GTPase proteins required for macroautophagy?
There are several dozen small GTPases found in S. cerevisiae. The number found in multicellular eukaryotes is even larger, especially with respect to the Rab protein family (equivalent to the Ypt family in S. cerevisiae). No systematic study has been undertaken of all the GTPase proteins in even a simple unicellular organism like S. cerevisiae, although it is clear several members of the Arf/Arl/Sar and Ypt/Rab families are required for construction of the autophagosome and for fusion of the autophagosome with the vacuole.4,23,24 Members of the Rac/Rho/Cdc42 family, proteins generally viewed as regulators of cell polarity and cytoskeletal function, also appear to have signaling roles in autophagy.25,26 Interestingly, RhoA along with its downstream effector, ROCK1 appears to mediate switching between autophagy and apoptosis via control of Beclin-1 (the ortholog of Atg6 in S. cerevisiae) levels in mammalian cells.24 Ras proteins appear to be involved in initiation of autophagy via regulation of TORC1.27 In contrast, the GTPase complex made of Gtr1 and Gtr2 (equivalent to RagA and RagB in mammals) appears to stimulate TORC1.28,29 At present, there is no evidence that Ran proteins, which regulate movement of molecules in and out of the nucleus via nuclear pores, have a role, but this question appears not to have been explored to date.
How does regulation of nucleotide binding on small GTPases affect macroautophagy?
The GTP-restricted versions of Arl1 and Ypt6 are required for autophagy.8 Other GTPases in autophagy also function in the GTP-bound state; examples include Ypt1,30,31 Ypt31/32,32 and Ypt7,33-35 suggesting that guanine nucleotide exchange factors (GEFs) are also important for autophagy. Indeed, the Mon1/Ccz1 GEF for Ypt733-35 and the Trs130 protein,32 part of the complex that regulates Ypt31/32, have been shown to be necessary for autophagy. However, it may be challenging to determine which GEF is the relevant one for autophagy for a given small GTPase, including Arl1 and Ypt6, given that many small GTPases are turned on by several different GEFs and that many GEFs activate several different GTPases. For example, a network of GEFs and GTPases appear to work together for Arf and Arl proteins.36,37 In addition, GEF proteins can be regulated spatially and temporally by the addition of protein subunits. As an example, the TRAPP complex which serves as a GEF for Ypt1 is found in 3 different forms, TRAPP I, which regulates traffic from the ER to the cis-Golgi; TRAPP II, which regulates intra-Golgi traffic; and TRAPP III, which is specific for autophagy share a number of subunits, but TRAPP II and TRAPP III have more subunits than TRAPP I.30,31,38-41
Recently, it has been demonstrated that Syt1, a GEF for Arl1, is phosphorylated upon induction of the unfolded protein response, resulting in increased activation of Arl1.42 Similarly, the Rab12 GEF, DENND3 is phosphorylated by ULK1 (the ortholog of Atg1 in S. cerevisiae) which then promotes autophagy.43,44 It is conceivable that other GEF post-translational modifications might be important for activation of small GTPases in their roles as modulators of autophagy.
By similar reasoning, GTPase activating proteins (GAPs) would also be expected to be important for regulation of autophagy, since they would be responsible for terminating the signals transmitted by GTPases in the GTP-bound state. This appears to be the case, at least for Rab proteins, where it has been shown that several Rab GAPs coordinate autophagy and “normal” functions of Rab proteins in the secretory pathway and endocytosis.45-47 While the issue of networks of GAPs and GTPases may make this a challenging problem to investigate,36,37 recent work has elucidated some of the details with respect to Rabs and RabGAPs in particular in different forms of autophagy, including in xenophagy, the process by which autophagy is induced in response to bacteria or viruses.6
Which effectors are required for macroautophagy?
Arl1 and Ypt6 appear to direct the GARP tethering complex to the growing autophagosome.8 Other elements of the membrane traffic apparatus appear to be co-opted by the autophagy machinery in order to grow the autophagosome and then fuse the autophagosome with the vacuole/lysosome, including other tethering complexes, such as the HOPS complex downstream of Ypt7;48 SNAREs;49 coat proteins such as COPII, downstream of Ypt1;50 and membrane deformation proteins such as Ivy1, an I-BAR protein downstream of Ypt7.48 Likely other downstream effectors, including other modulators of membrane traffic will be revealed to have roles in autophagy.
Are small GTPases required for other forms of autophagy?
The preceding discussion focuses on macroautophagy, that induced by starvation. However, less is known about the roles of small GTPases in other forms of autophagy. We demonstrated that Arl1 and Ypt6 have modest roles in the cytosol-to-vacuole (CVT) pathway, a constitutive process responsible for delivery of several enzymes to the vacuole in yeast.8 However, we have not yet investigated whether these 2 GTPases have roles in other forms of autophagy, including mitophagy, induced to recycle defective mitochondria; pexophagy, for elimination of unwanted peroxisomes; ER-phagy, for elimination of excess ER; etc. These pathways, which have been shown to involve other small GTPases, will provide interesting avenues for future research into the roles of Arl1 and Ypt6 in cellular functions.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931-7; PMID:17712358; https://doi.org/ 10.1038/nrm2245 [DOI] [PubMed] [Google Scholar]
- [2].Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861-73; PMID:18006683; https://doi.org/ 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- [3].Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol 2012; 22:R29-34; PMID:22240478; https://doi.org/ 10.1016/j.cub.2011.11.034 [DOI] [PubMed] [Google Scholar]
- [4].Yang S, Rosenwald AG. The roles of monomeric GTP-Binding Proteins in Macroautophagy in saccharomyces cerevisiae. Int J Mol Sci 2014; 15:18084-101; PMID:25302616; https://doi.org/ 10.3390/ijms151018084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bento CF, Puri C, Moreau K, Rubinsztein DC. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci 2013; 126:1059-69; PMID:23620509; https://doi.org/ 10.1242/jcs.123075 [DOI] [PubMed] [Google Scholar]
- [6].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ 2014; 21:348-58; PMID:24440914; https://doi.org/ 10.1038/cdd.2013.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Szatmari Z, Sass M. The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy 2014; 10:1154-66; PMID:24915298; https://doi.org/ 10.4161/auto.29395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang S, Rosenwald AG. Autophagy in Saccharomyces cerevisiae requires the monomeric GTP-binding proteins, Arl1 and Ypt6. Autophagy 2016; 12(10):1-17; https://doi.org/10.1080/15548627.2016.1196316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Burd CG, Strochlic TI, Gangi Setty SR. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol 2004; 14:687-94; PMID:15564045; https://doi.org/ 10.1016/j.tcb.2004.10.004 [DOI] [PubMed] [Google Scholar]
- [10].Munro S. The Arf-like GTPase Arl1 and its role in membrane traffic. Biochem Soc Trans 2005; 33:601-5; PMID:16042553; https://doi.org/ 10.1042/BST0330601 [DOI] [PubMed] [Google Scholar]
- [11].Fell GL, Munson AM, Croston MA, Rosenwald AG. Identification of yeast genes involved in K+ homeostasis: Membrane traffic genes affect K+ uptake. G3: Genes, Genomes, Genetics 2011; 1:43-56; https://doi.org/full_text [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Munson AM, Haydon DH, Love SL, Fell GL, Palanivel VR, Rosenwald AG. Yeast ARL1 encodes a regulator of K+ influx. J Cell Sci 2004; 117:2309-20; PMID:15126631; https://doi.org/ 10.1242/jcs.01050 [DOI] [PubMed] [Google Scholar]
- [13].Maresova L, Vydareny T, Sychrova H. Comparison of the influence of small GTPases Arl1 and Ypt6 on yeast cells' tolerance to various stress factors. FEMS Yeast Res 2012; 12:332-40; PMID:22188384; https://doi.org/ 10.1111/j.1567-1364.2011.00780.x [DOI] [PubMed] [Google Scholar]
- [14].Abudugupur A, Mitsui K, Yokota S, Tsurugi K. An ARL1 mutation affected autophagic cell death in yeast, causing a defect in central vacuole formation. Cell Death Differ 2002; 9:158-68; PMID:11840166; https://doi.org/ 10.1038/sj.cdd.4400942 [DOI] [PubMed] [Google Scholar]
- [15].Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy 2011; 7:1546-50; PMID:22108003; https://doi.org/ 10.4161/auto.7.12.18424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; PMID:19185711; https://doi.org/ 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- [17].Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al.. The genetic landscape of a cell. Science 2010; 327:425-31; PMID:20093466; https://doi.org/ 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 2009; 6:917-22; PMID:19915560; https://doi.org/ 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- [19].Manlandro CM, Palanivel VR, Schorr EB, Mihatov N, Antony AA, Rosenwald AG. Mon2 is a negative regulator of the monomeric G protein, Arl1. FEMS Yeast Res 2012; 12:637-50; PMID:22594927; https://doi.org/ 10.1111/j.1567-1364.2012.00814.x [DOI] [PubMed] [Google Scholar]
- [20].Panic B, Whyte JR, Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol 2003; 13:405-10; PMID:12620189; https://doi.org/ 10.1016/S0960-9822(03)00091-5 [DOI] [PubMed] [Google Scholar]
- [21].Siniossoglou S. Affinity purification of Ypt6 effectors and identification of TMF/ARA160 as a Rab6 interactor. Methods Enzymol 2005; 403:599-607; PMID:16473623; https://doi.org/ 10.1016/S0076-6879(05)03052-1 [DOI] [PubMed] [Google Scholar]
- [22].Reggiori F, Klionsky DJ. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J Cell Sci 2006; 119:2903-11; PMID:16787937; https://doi.org/ 10.1242/jcs.03047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Amaya C, Fader CM, Colombo MI. Autophagy and proteins involved in vesicular trafficking. FEBS Lett 2015; 589:3343-53; PMID:26450776; https://doi.org/ 10.1016/j.febslet.2015.09.021 [DOI] [PubMed] [Google Scholar]
- [24].Lin PY, Chang CD, Chen YC, Shih WL. RhoA/ROCK1 regulates Avian Reovirus S1133-induced switch from autophagy to apoptosis. BMC Vet Res 2015; 11:103; PMID:25944062; https://doi.org/ 10.1186/s12917-015-0417-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, et al.. A protein interaction map for cell polarity development. J Cell Biol 2001; 154:549-71; PMID:11489916; https://doi.org/ 10.1083/jcb.200104057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Till A, Saito R, Merkurjev D, Liu JJ, Syed GH, Kolnik M, Siddiqui A, Glas M, Scheffler B, Ideker T, et al.. Evolutionary trends and functional anatomy of the human expanded autophagy network. Autophagy 2015; 11:1652-67; PMID:26103419; https://doi.org/ 10.1080/15548627.2015.1059558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 2004; 279:20663-71; PMID:15016820; https://doi.org/ 10.1074/jbc.M400272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino Acids Stimulate TORC1 through Lst4-Lst7, a GTPase-Activating protein complex for the rag family GTPase Gtr2. Cell Rep 2015; 13:1-7; PMID:26387955; https://doi.org/ 10.1016/j.celrep.2015.08.059 [DOI] [PubMed] [Google Scholar]
- [29].Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal 2013; 6:ra42; PMID:23716719; https://doi.org/ 10.1126/scisignal.2004112 [DOI] [PubMed] [Google Scholar]
- [30].Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, Brumell JH, Ferro-Novick S, Klionsky DJ. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A 2010; 107:7811-6; PMID:20375281; https://doi.org/ 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, Ferro-Novick S. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A 2013; 110:9800-5; PMID:23716696; https://doi.org/ 10.1073/pnas.1302337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zou S, Chen Y, Liu Y, Segev N, Yu S, Liu Y, Min G, Ye M, Zeng Y, Zhu X, et al.. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic 2013; 14:233-46; PMID:23078654; https://doi.org/ 10.1111/tra.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu X, Klionsky DJ. The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion. Autophagy 2016; 12:894-5; PMID:26986547; https://doi.org/ 10.1080/15548627.2016.1162364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Piekarska I, Kucharczyk R, Mickowska B, Rytka J, Rempola B. Mutants of the Saccharomyces cerevisiae VPS genes CCZ1 and YPT7 are blocked in different stages of sporulation. Eur J Cell Biol 2010; 89:780-7; PMID:20709422; https://doi.org/ 10.1016/j.ejcb.2010.06.009 [DOI] [PubMed] [Google Scholar]
- [35].Polupanov AS, Nazarko VY, Sibirny AA. CCZ1, MON1 and YPT7 genes are involved in pexophagy, the Cvt pathway and non-specific macroautophagy in the methylotrophic yeast Pichia pastoris. Cell Biol Int 2011; 35:311-9; PMID:21155714; https://doi.org/ 10.1042/CBI20100547 [DOI] [PubMed] [Google Scholar]
- [36].Shin HW, Nakayama K. Guanine nucleotide-exchange factors for arf GTPases: their diverse functions in membrane traffic. J Biochem 2004; 136:761-7; PMID:15671486; https://doi.org/ 10.1093/jb/mvh185 [DOI] [PubMed] [Google Scholar]
- [37].Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol 2000; 10:60-7; PMID:10652516; https://doi.org/ 10.1016/S0962-8924(99)01699-2 [DOI] [PubMed] [Google Scholar]
- [38].Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol 2010; 11:759-63; PMID:20966969; https://doi.org/ 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- [39].Kim JJ, Lipatova Z, Segev N. TRAPP Complexes in Secretion and Autophagy. Front Cell Dev Biol 2016; 4:20; PMID:27066478; https://doi.org/ 10.3389/fcell.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lipatova Z, Kim JJ, Segev N. Ypt1 and TRAPP interactions: optimization of multicolor bimolecular fluorescence complementation in yeast. Methods Mol Biol 2015; 1298:107-16; PMID:25800836; https://doi.org/ 10.1007/978-1-4939-2569-8_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Taussig D, Lipatova Z, Segev N. Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic 2014; 15:327-37; PMID:24329977; https://doi.org/ 10.1111/tra.12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hsu JW, Tang PH, Wang IH, Liu CL, Chen WH, Tsai PC, Chen KY, Chen KJ, Yu CJ, Lee FJ. Unfolded protein response regulates yeast small GTPase Arl1p activation at late Golgi via phosphorylation of Arf GEF Syt1p. Proc Natl Acad Sci U S A 2016; 113:E1683-90; PMID:26966233; https://doi.org/ 10.1073/pnas.1518260113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Xu J, McPherson PS. DENND3: a signaling/trafficking interface in autophagy. Cell Cycle 2015; 14:2717-8; PMID:26177209; https://doi.org/ 10.1080/15384101.2015.1071136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Xu J, Fotouhi M, McPherson PS. Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep 2015; 16:709-18; PMID:25925668; https://doi.org/ 10.15252/embr.201440006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 2016; 35:281-301; PMID:26711178; https://doi.org/ 10.15252/embj.201592695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kern A, Dikic I, Behl C. The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 2015; 11:2393-7; PMID:26565612; https://doi.org/ 10.1080/15548627.2015.1110668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Spang N, Feldmann A, Huesmann H, Bekbulat F, Schmitt V, Hiebel C, Koziollek-Drechsler I, Clement AM, Moosmann B, Jung J, et al.. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy 2014; 10:2297-309; PMID:25495476; https://doi.org/ 10.4161/15548627.2014.994359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Numrich J, Peli-Gulli MP, Arlt H, Sardu A, Griffith J, Levine T, Engelbrecht-Vandre S, Reggiori F, De Virgilio C, Ungermann C. The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J Cell Sci 2015; 128:2278-92; PMID:25999476; https://doi.org/ 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- [49].Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al.. SNARE proteins are required for macroautophagy. Cell 2011; 146:290-302; PMID:21784249; https://doi.org/ 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Davis S, Ferro-Novick S. Ypt1 and COPII vesicles act in autophagosome biogenesis and the early secretory pathway. Biochem Soc Transactions 2015; 43:92-6; PMID:25619251; https://doi.org/ 10.1042/BST20140247 [DOI] [PubMed] [Google Scholar]


