Abstract
Background
Doxorubicin is a widely used chemotherapy drug for the treatment of a variety of cancers, however it also has serious side effects such as anaphylaxis and heart damage. Therefore, it’s very important to understand the downstream molecular pathways that are essential for Doxorubicin function in cancer treatment.
Methods
HeLa S3 cells were treated with different concentrations of Doxorubicin for 24 hours. Then, the mRNA levels of Notch pathway components in the Doxorubicin treated cells were determined by Real-Time qRT-PCR. Lentiviral transfection was used to up-regulate and down-regulate HES1 expression. Cell proliferation and apoptosis were measured with MTT assay and flow cytometry. Finally, immunofluorescence was used to detect protein subcellular location.
Result
Doxorubicin treatment strongly increases the expression of multiple Notch pathway components in cancer cells. The Notch target HES1 is activated by Doxorubicin and is required for the Doxorubicin driven apoptosis. In addition, over-expression of HES1 can further enhances Doxorubicin’s role in promoting apoptosis. Mechanistically, HES1 activates PARP1 and regulates the subcellular location of AIF to mediate the apoptosis response under Doxorubicin treatment.
Conclusion
Our results provided novel insights into the downstream molecular pathways underlying Doxorubicin treatment and suggested that manipulation of Notch signaling pathway could have synergistic effect with Doxorubicin for cancer treatment.
Keywords: Doxorubicin, Notch, HES1, PARP1, apoptosis, cancer treatment
Introduction
Doxorubicin is an anthracycline antibiotic that has a strong anticancer activity and has been widely used as an efficient broad-spectrum anticancer drug.1,2 Doxorubicin integrates into the DNA double-strand helix and forms a stable complex structure with DNA, which destroys the DNA tertiary structure and, therefore, prevents DNA replication and RNA synthesis, resulting in cellular apoptosis. Because of these DNA interfering mechanisms, although Doxorubicin preferentially targets cancer cells, it can also damage the DNA in the normal cells and result in some side effects such as congestive heart failure.3,4 Therefore, understanding of the downstream molecular pathways that are essential for Doxorubicin-driven apoptosis is crucial for reducing the side effects of Doxorubicin, resulting in better cancer treatment.
Notch signaling pathway is a highly conserved pathway that regulates cell fate determination and is frequently misregulated in cancers. For example, >50% of the T-cell acute lymphoblastic leukemia patients have the Notch overactive mutations. Importantly, these Notch mutations can induce T-cell acute lymphoblastic leukemia initiation and progression in mouse models. In addition, Notch activation promotes the growth and invasion of other cancers such as ovarian cancer5 and eye cancer,6 supporting the essential role of Notch as an oncogene in promoting carcinogenesis. On the other hand, the Notch pathway can also function as a tumor suppressor to inhibit tumor growth in other tissues, suggesting the tissue-specific multifunctioning of Notch pathway in regulation of cancers.7 Given the important function of Notch signaling pathway in cancers, the Notch pathway is a promising therapeutic target for cancer treatment.8–12 As an example, the Notch inhibitors such as DAPT or Compound E have been widely used to treat cancers with Notch gain-of-function mutations,7 while for cancers mediated by Notch loss-of-function mutations, activation of Notch signaling pathway is a promising therapeutic strategy.8–12
Notch signaling is mediated by ligand receptor interaction through cell contact. Binding of ligands (DLL1, DLL3, DLL4, JAGGED1 and JAGGED2) to the Notch receptors (NOTCH1, NOTCH2, NOTCH3 and NOTCH4) induces the activation of Notch signaling and cleavage of Notch receptors. The intracellular domain of Notch then enters into the nucleus, binds to the transcription factor CSL and recruits coactivators including MAML1–3 and P300 to activate the transcription of Notch targets including HES (hairy/enhancer of split) and HEY (HEY – hairy/enhancer-of-split related with YRPW motif family members) family genes.13,14 HES1 is a transcriptional repressor with three conserved domains: the bHLH, Orange and WRPW domains.15 As a key Notch target, HES1 is essential for the cancer stem cells’ maintenance, cancer metastasis and chemoresistance.15 The Notch-HES1 signaling axis regulates the functioning of essential genes in cellular proliferation and apoptosis regulation; therefore, the Notch signaling pathway is one of the key downstream pathways of cancer therapeutic drugs.8,10,16,17 However, the function and the underlying mechanisms of Notch pathway in Doxorubicin-induced apoptosis remain inclusive.12
Here, we studied the functional role of Notch signaling pathway in Doxorubicin-induced apoptosis of cancer cells. We found that Doxorubicin treatment strongly increased the expression of multiple Notch pathway components. The Notch target HES1 mediated Doxorubicin-driven apoptosis through activation of poly(ADP-ribose) polymerase-1 (PARP1) and regulation of apoptosis-inducing factor (AIF) subcellular location, suggesting the important role of Notch pathway in mediating Doxorubicin-driven apoptosis in cancer cells.
Materials and methods
Plasmids and transfection
The following shRNA lentiviral constructs targeting the HES1 were obtained from Thermo Fisher Scientific. The hairpin sequence numbers are TRCN0000018989, TRCN0000018990, TRCN0000018991, TRCN0000018992 and TRCN0000018993. A vector for mammalian overexpression of HES1 was generated by cloning HES1 CDS (Genbank No NM_005524.3) into the multiple cloning site of the pOZ-FH-N vector via XhoI and BamHI restriction sites. The primers were as follows: HES1 forward 5′-GCCTCGAGCCAGCTGATATAATGGAGAA-3′ and reverse 5′-TGGATCCTCAGTTCCGCCACGGCCT-3′. Integrity of the construct was verified by sequencing. Lenti-viral transfection was performed as described previously.18 Briefly, 50% confluent 293T cells seeded in 100 mm plates were transfected with lentiviral vector targets HES1 (HES1-shRNA1-5) and HES1 overexpression lentiviral, by using Effectene Transfection Reagent (Qiagen, Hilden, Germany). HeLa cells were plated at 40%–50% confluence in 100 mm plates and subsequently infected with viral supernatant for three times with 2.0 μg/mL Polybere (Sigma-Aldrich, St Louis, MO, USA). Finally, the cells were replaced with a complete medium containing puromycin (1.0 μg/mL; Sigma-Aldrich) for 1 week.
Cell culture
Human HeLa S3 cell line (HeLa) was obtained from American Type Culture Collection. The cells were cultured in DMEM (HyClone; GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin and 100 ng/mL streptomycin at 37°C in a humidified atmosphere of 95% air/5% CO2. Doxorubicin (sc-280681) was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). Rucaparib was purchased from Selleck (AG-014699). For Doxorubicin treatment, cells were cultured in fresh culture medium with Doxorubicin (0, 100, 200, 300, 500 and 750 μM) for 24 hours. For PARP1 inhibitor treatment, cells were cultured in fresh culture medium with Rucaparib (1 nM) for 7 days.
Real-time reverse transcriptase polymerase chain reaction (RT-PCR)
Real-time RT-PCR was performed as described previously.19 Total RNA was extracted from HeLa S3 cells using Trizol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. By using the PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa, Tokyo, Japan), total RNA was subjected to reverse transcription. Real-time RT-PCR analysis was performed on LightCycler 96 (Hoffman-La Roche Ltd., Basel, Switzerland) using the SYBR Premix Ex TaqTM Kit (TaKaRa). GAPDH was used as an internal control. The primers used are listed in Table 1.
Table 1.
List of primers used in this study
| Name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| GAPDH | GAGTCAACGGATTTGGTCGT | GACAAGCTTCCCGTTCTCAG |
| HES1 | ATTTTTGGAGTTCTTCACGAAA | GAATCCCCCGTCTACCTCTC |
| HES5 | CCCAAAGAGAAAAACCGACTGC | ATGTCGGCCTTCTCCAGCTT |
| HES7 | ACGGCCCCAAGATGCTCAA | TTTCTCCAGCTTCGGGTTCC |
| NOTCH1 | GATGACCTGGGCAAGTCCG | TGCGCTCCTGTGCGATGT |
| NOTCH2 | AGGTGTCAGAATGGAGGGGT | GTGCAGAACTGTCCTGTCCA |
| NOTCH3 | TGTGAACTCCTCTCCCCCTG | CGTTCAGGCATGGGTCTTG |
| NOTCH4 | CCCTTGTCCTCCCTCCTTCT | CTGCTCACCTGTCCATCCAG |
| JAG1 | ACGACCCCCTGTGAAGTGAT | TCCCGACTGACTCTTGCACT |
| JAG2 | CGCCAATGAGTGTGAAGGGA | TCGTTGACGTTGATATGGCAGT |
| DLL1 | ACCTCGCAACAGAAAACCCA | GTGTTCGTCACACACGAAGC |
| DLL3 | GGTCCGAGCTCGTCCGTAG | GAAAAGGGGCGTCGCTACC |
| DLL4 | ATCAGCGATATGCTCCCCCA | TGCCTTATACCTCCGTGGCA |
| MAML1 | GCCACGCATCTTCATGATACAG | CCATTGGAAGAGATGGCAACTC |
| MAML2 | CTGATTGCGCTCCAGGGTTC | TCCACAAAGCCATTGGGTCG |
| MAML3 | ACCACACGCTGATCATGCTAC | GCACCATTCTGCTGGTCTCC |
| MYC | GGTAGTGGAAAACCAGCAGCC | TTCTCCTCCTCGTCGCAGTA |
| HEY1 | GCAGGTAATGGAGCAAGGATCT | AACCTTTCCCTCCTGCCGTA |
| HEY2 | GCAACAGGGGGTAAAGGCTAC | ACCGCGCAACTTCTGTTAGG |
| HEYL | GTCCCCACTGCCTTTGAGAA | TCGGGCATCAAAGAATCCTGT |
| CSL | CAACAGCGAGAGAGAAGGGG | GCTTCTTCCTGCAGCCATTG |
Immunofluorescence
Cells were grown on glass chamber slides and treated with Doxorubicin and PARP1 inhibitor Rucaparib for 24 hours. After treatment, the cells were fixed with 4% formaldehyde for 10 minutes and then blocked with 5% bovine serum albumin for 1 hour to reduce the background.20 The cells were then incubated with the antibody of AIF (Santa Cruz Biotechnology Inc.) at 5 μM overnight at 4°C. The secondary antibody (green) was goat anti-mouse antibody (Santa Cruz Biotechnology Inc.) used at 1/250 dilution for 1 hour. DAPI was used to stain the cell nuclei (blue) at concentration of 1.43 μM.
Intracellular apoptosis measurement
HeLa S3 cells were harvested and washed twice in PBS before the analyses of apoptosis. The percentages of apoptosis were examined using Multicaspase Kit (EMD Millipore). According to the manufacturer’s instructions, multicaspase was added to each sample for 30 minutes at 37°C in dark and then incubated with 7-aminoactinomycin D (EMD Millipore) for 5 minutes at room temperature in dark. After incubation, the events for live, dead early and late apoptotic cells were counted with the Muse Cell Analyzer.21
Cell viability
The MTT assay was used to detect cell viability. Briefly, cells were seeded onto 96-well plates at a density of 2000 cells per well. Cell proliferation was measured with MTT assay at different time points. Briefly, 20 μL MTT (5 mg/mL; Sigma-Aldrich) solution was added to the wells and then the 96-well plates were incubated at 37°C for 2–4 hours. Then the medium was removed and 200 μL of dimethyl sulfoxide was added per well. After 10 minutes of shaking, the wavelength at 490 nm was measured by a microplate reader (BioTek Instruments, Winooski, VT, USA).
Western blotting
Western blot was performed as described previously.22 The antibodies used in the study were HES1 (Santa Cruz Biotechnology Inc.) and alpha-tubulin (Affinity, Cincinnati, OH, USA). α-Tubulin was used as an internal control.
Statistical analysis
All assays were performed in triplicate. The results were expressed as the means ± SD. Statistical analyses were performed with Student’s t-test for comparison of two groups (GraphPad Prism 5 software). Significant values were shown as 0.01<P<0.05 (*), P<0.01 (**) and P>0.05 (#).
Results
Role of Doxorubicin in regulation of apoptosis
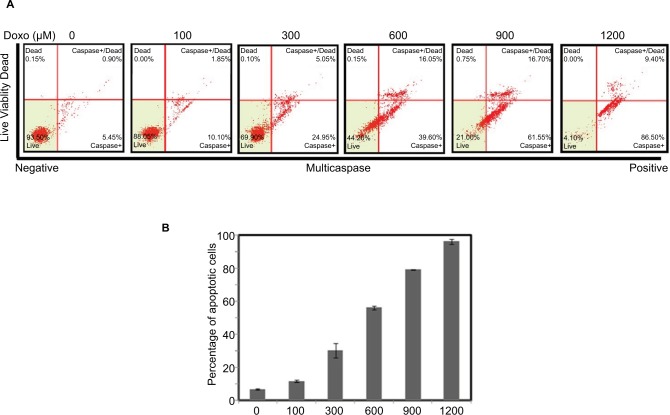
To study the role of Notch oncogenic pathway in Doxorubicin-driven apoptosis, we first established the apoptosis model by treating the HeLa S3 cells with different concentrations (0, 100, 300, 600, 900 and 1200 μM) of Doxorubicin for 24 hours. As shown in Figure 1A and B, Doxorubicin treatment induced apoptosis in a dose-dependent manner, confirming the role of Doxorubicin in killing cancer cells.
Figure 1.
Doxorubicin affects Hela S3 cell apoptosis.
Notes: The HeLa S3 cells were treated with Doxorubicin (0, 100, 300, 600, 900 and 1200 μM) for 24 hours. (A) The percentage of apoptosis cells is shown. (B) Bars represent mean of three independent experiments performed in triplicate.
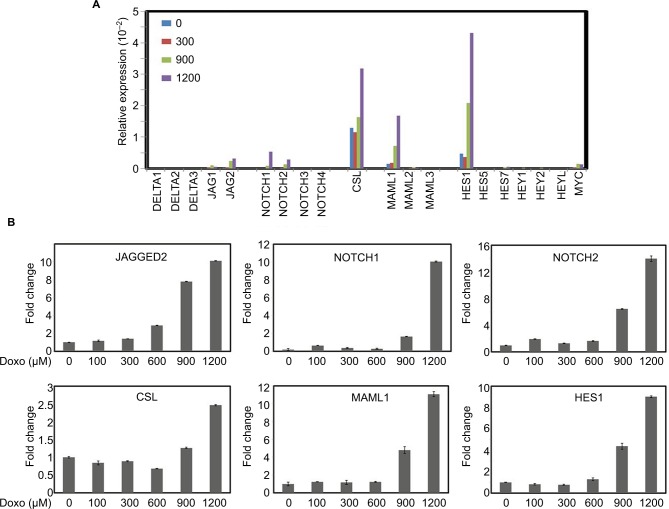
Role of Doxorubicin in Notch pathway gene expression
We then studied the role of Doxorubicin in regulation of expression of Notch pathway genes. The mRNA levels of Notch pathway ligands (DELTA1, DELTA3, DELTA4 and JAGGED1, JAGGED2), receptors (NOTCH1, NOTCH2, NOTCH3 and NOTCH4), transcription factor CSL, coactivators (MAML1, MAML2 and MAML3) and downstream targets (HES1, HES5, HES7, HEY1, HEY2, HEYL and MYC) in the Doxorubicin-treated cells were determined by real-time quantitative RT-PCR. As shown in Figure 2, Doxorubicin treatment increased the expression of NOTCH pathway components JAGGED2, NOTCH1 and NOTCH2, CSL, MAML1 and HES1 in a dose-dependent manner (Figure 2B), indicating that Doxorubicin treatment regulates the JAGGED2-NOTCH1/2-CSL-MAML1-HES1 axis in cancer cells.
Figure 2.
Effects of different concentrations of Doxorubicin on the mRNA expression of Notch pathway members.
Notes: (A) HeLa S3 cells were treated with Doxorubicin (0, 300, 900 and 1200 μM) for 24 hours. The mRNA expression levels of Notch pathway components and targets were examined by real-time RT-PCR. (B) Selective representation of Doxorubicin-responsive Notch pathway genes. GAPDH was used as an internal control for the normalization of gene expression.
Abbreviation: RT-PCR, reverse transcriptase polymerase chain reaction.
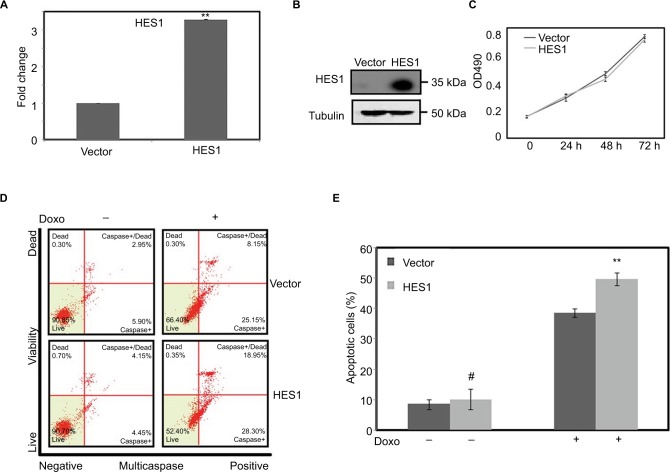
HES1 overexpression promotes Doxorubicin-driven apoptosis
Given that HES1 is the only NOTCH target gene that is highly expressed in the HeLa S3 cells and is strongly induced under Doxorubicin treatment (Figure 2), we next determined the potential role of HES1 in Doxorubicin-induced apoptosis in cancer cells. We first established a HES1 overexpressing stable cell line and confirmed the HES1 mRNA and protein levels by real-time RT-PCR (Figure 3A) and Western blotting (Figure 3B). Overexpression of HES1 in HeLa S3 cells had little effect on cancer cell proliferation (Figure 3C), but modestly increased cellular apoptosis (Figure 3D, E). Interestingly, when we treated the HES1-overexpressing and the control cells with Doxorubicin, we found that HES1 overexpression promoted the cellular apoptosis induced by Doxorubicin treatment. Overall, the above mentioned data suggest that HES1 acts as a downstream target of Doxorubicin in regulation of apoptosis.
Figure 3.
Upregulation of HES1 expression could promote Doxorubicin-induced apoptosis.
Notes: Cells were stably transfected with HES1 overexpression plasmids. Cells transfected with empty pOZ vector were used as the control group. (A) The overexpression efficiency of HES1 was measured by real-time RT-PCR. (B) The levels of the overexpressed protein were detected by Western blot. (C) The cell viability of overexpressed cell line was detected by MTT. (D) The overexpressed cells were stained with Multicaspase/7-AAD for quantitative measurement by using flow cytometry. (E) Bars represent the mean of three independent experiments performed in triplicate of (D). #P>0.05, **P<0.01. The cells were treated with Doxorubicin (300 μM) for 24 hours.
Abbreviations: 7-AAD, 7-aminoactinomycin D; RT-PCR, reverse transcriptase polymerase chain reaction.
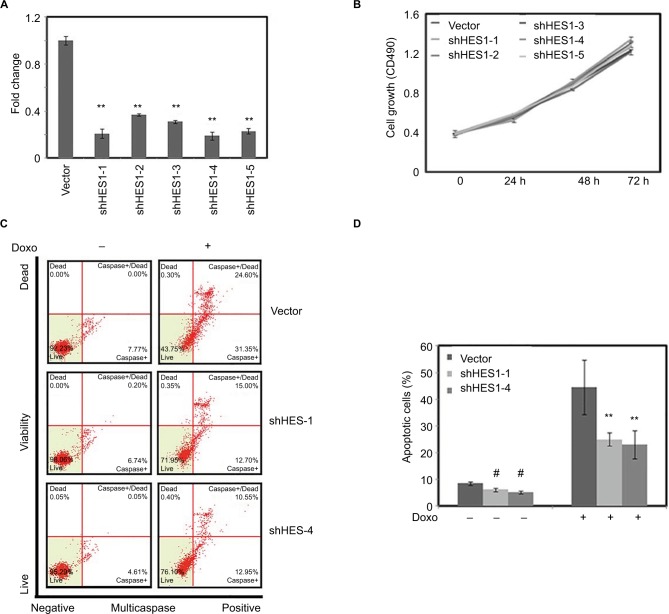
Depletion of HES1 suppresses Doxorubicin-induced apoptosis
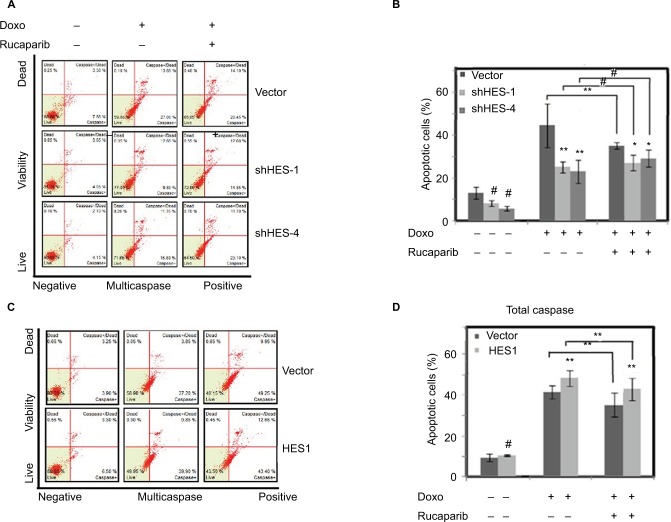
We next studied the role of HES1 depletion in regulation of Doxorubicin-induced apoptosis. Five different shRNAs were used to knockdown HES1 expression in HeLa cells (Figure 4A). As shown in Figure 4B and C, knockdown of HES1 has little effect in cellular proliferation and apoptosis. We further determined the role of HES1 depletion in cellular apoptosis under the treatment of Doxorubicin. We found that Doxorubicin could strongly promote the apoptosis of the control cells, while in the HES1-depleted cells, the effect of Doxorubicin in promoting apoptosis was significantly lower than in the control cells (Figure 4C and D), suggesting that Doxorubicin HES1 is an essential factor that mediates Doxorubicin’s function in inducing apoptosis.
Figure 4.
Downregulation of HES1 expression could inhibit Doxorubicin-induced apoptosis.
Notes: Cells were stably transfected with HES1 shRNA plasmids. Cells transfected with empty pLko.1 vector were used as the control group. (A) The knockdown efficiency of HES1 was measured by real-time RT-PCR. (B) The cell viability of knockdown cell lines was detected by MTT. (C) The knockdown and control cells were treated with Doxorubicin (300 μM) for 24 hours and then stained with Multicaspase/7-AAD for the quantitative measurement by using flow cytometry. (D) Bars represent the mean of three independent experiments performed in triplicate of (C). **P<0.01. #P>0.05.
Abbreviations: 7-AAD, 7-aminoactinomycin D; RT-PCR, reverse transcriptase polymerase chain reaction.
HES1 induces apoptosis through PARP1 activation
We next determined the molecular mechanisms of HES1 in regulation of Doxorubicin-driven apoptosis. A previous study reported that HES1 interacts with PARP1 and induces the activation of PARP1-mediated proapoptotic signal pathway in a cell type–specific manner;23 therefore, we explored the potential interplay between HES1 and PARP1 in Doxorubicin-induced apoptosis. Treatment of HES1-depleted and control cells with Doxorubicin and the combination of Doxorubicin with the PARP1 inhibitor Rucaparib revealed that Rucaparib could significantly reduce Doxorubicin-driven apoptosis in the control cells, while in the HES1-depleted cells, Rucaparib had little effect on the cellular apoptosis induced by Doxorubicin treatment (Figure 5A and B). This suggests that HES1 depletion reduces PARP1 activity; as a result, the HES1-depleted cells are irresponsive to PARP1 inhibition by Rucaparib. We further tested the role of HES1 overexpression in the regulation of PARP1 activity. As shown in Figure 5C and D, Rucaparib treatment resulted in increased apoptosis in the HES1-overexpressing cells compared to the control cells, suggesting that overexpression of HES1 activates PARP1 and, therefore, sensitizes the cells to PARP1 inhibition.
Figure 5.
HES1 regulates apoptosis by activating PARP1.
Notes: HeLa cells were pretreated with PARP1 inhibitor Rucaparib (1 nM) for 7 days, and then, the cells were treated with Doxorubicin (300 μM) for 24 hours. (A) Cell apoptosis of HES1-knockdown cell lines was determined by flow cytometry. (B) Bars represent the mean of three independent experiments performed in triplicate of (A). (C) Cell apoptosis of HES1 overexpressing cell line was determined by flow cytometry. (D) Bars represent the mean of three independent experiments performed in triplicate of (C). **P<0.01, *P<0.05, #P>0.05.
Abbreviation: PARP1, poly(ADP-ribose) polymerase-1.
HES1-PARP1 regulates AIF subcellular location upon Doxorubicin treatment
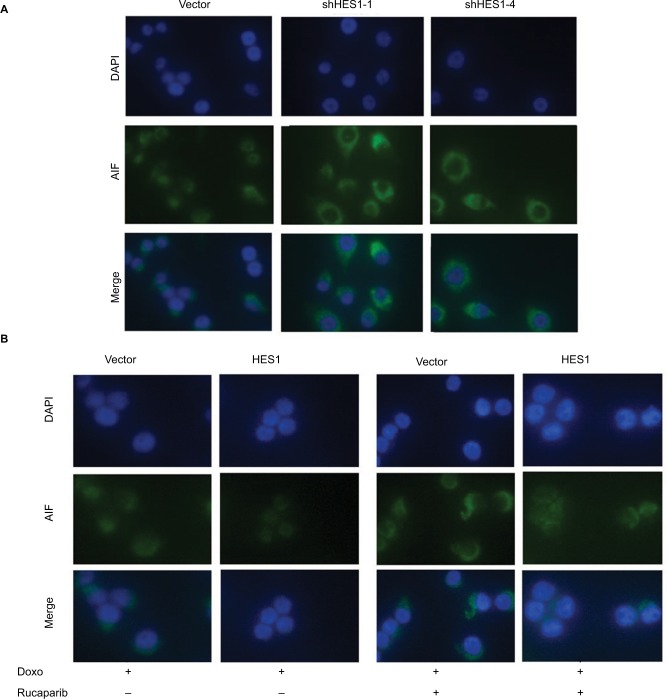
In order to further explore the mechanism of HES1 in the regulation of Doxorubicin-driven apoptosis, we used immunofluorescence to detect changes in the location of AIF. As can be seen from Figure 6A, when the cells were treated with Doxorubicin alone, most of the AIF in the control group had been transferred from the mitochondria into the nucleus, thereby inducing apoptosis. While in the HES1-depleted cells, the fluorescence intensity of AIF in the nucleus strongly decreased, indicating that inhibition of HES1 expression suppressed the transfer of AIF from the mitochondria into the nucleus. Treatment of HES1 overexpressing cells with Doxorubicin resulted in further migration of AIP into the nucleus (Figure 6B). Interestingly, the PARP1 inhibitor Rucaparib could prevent the Doxorubicin-induced nucleus accumulation of AIP, similar to the effect of HES1 depletion. Overall, our results indicated that the HES1-PARP1 axis regulates the subcellular location of AIF to facilitate the proapoptotic effect of Doxorubicin in cancer cells.
Figure 6.
HES1 interplays with PARP1 to regulate AIF subcellular location.
Notes: HeLa cells were pretreated with PARP1 inhibitor Rucaparib (1 nM) for 7 days, and then, the cells were treated with Doxorubicin (300 μM) for 24 hours. (A) Immunofluorescence of AIF (green) in the HES1-knockdown and control cells. DAPI staining (blue) marks the nuclei. Merged images are also shown. (B) Immunofluorescence of AIF (green) in the HES1-overexpressing and control cells. The picture shows the location of AIF (green) in cells. DAPI staining (blue) marks the nuclei. Merged images are also shown.
Abbreviations: AIF, apoptosis-inducing factor; PARP1, poly(ADP-ribose) polymerase-1.
Discussion
Doxorubicin is commonly used for cancer chemotherapy, but it also causes strong side effects such as low white blood cell count, vomiting and heart problems.4,11 Therefore, a better understanding of the molecular pathways underlying Doxorubicin-induced apoptosis in cancer cells is essential for effective and safer cancer treatment.12,24 In this study, we revealed that Notch signaling pathway is activated upon Doxorubicin treatment. Doxorubicin increased the expression of multiple Notch pathway components and Notch downstream target gene HES1, which is required for the Doxorubicin-induced apoptosis of cancer cells. Similarly, a previous study revealed that, in osteosarcoma, Doxorubicin treatment activated the Notch pathway and siRNA knockdown of Notch reduced the effects of Doxorubicin on the viability and apoptosis of osteosarcoma cells.25 Our result confirmed the essential role of Notch pathway in Doxorubicin-driven apoptosis of cancer cells and further specified that the molecular event of Doxorubicin-mediated apoptosis is through the upregulation of the key Notch target HES1. Mechanistically, HES1 activates PARP1 and regulates the subcellular location of AIF to mediate the apoptosis response under Doxorubicin treatment.
PARP1 is a major member of the poly ADP-ribose polymerase family and is a substrate for the apoptotic core members of caspase.26–29 PARP1 plays an important role in DNA damage repair and apoptosis.27,29,30 Studies have shown that PARP1 can directly regulate AIF release and lead to apoptosis.31,32 In addition, HES1 and PARP1 can interact in B-cell acute lymphoblastic leukemia cell lines to regulate PARP1 and induce apoptosis.23 Our results suggest that inhibition of PARP1 activity can reduce the apoptosis caused by Doxorubicin-induced DNA damage. In addition, HES1 can induce apoptosis by altering the location of AIF in DNA damage. This suggests that HES1 regulates apoptosis induced by Doxorubicin by promoting the migration of AIF to the nucleus through PARP1. Though previous studies have revealed that Doxorubicin regulates Notch and HES1 regulates PARP1-AIF in other cell lines, our study provided evidence that specifically, HES1 is the key Notch target that is essential for the Doxorubicin-driven apoptosis in cancer cells. In addition, we provided direct evidence that Doxorubicin regulates PARP1-AIF through the Notch target HES1.
Overall, our results suggest that the Notch target HES1 promotes Doxorubicin-induced apoptosis through PARP1 interactions, suggesting that Notch signaling pathway is essential for the Doxorubicin-mediated apoptosis of cancer cells. Given that HES expression could enhance Doxorubi-cin’s function in promoting apoptosis in cancer cells, modulation of Notch pathway could have synergistic effect with Doxorubicin and, therefore, could reduce the side effects of Doxorubicin in cancer treatment.
Acknowledgments
This work was sponsored by the Fundamental Research Funds of the State Key Laboratory of Ophthalmology and the National Natural Science Foundation of China (81670874 and 81772999).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.O’Bryan RM, Baker LH, Gottlieb JE, et al. Dose response evaluation of adriamycin in human neoplasia. Cancer. 1977;39(5):1940–1948. doi: 10.1002/1097-0142(197705)39:5<1940::aid-cncr2820390505>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 2.Blum RH, Carter SK. Adriamycin. A new anticancer drug with significant clinical activity. Ann Intern Med. 1974;80(2):249–259. doi: 10.7326/0003-4819-80-2-249. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee K, Zhang J, Honbo N, Karliner JS. Doxorubicin cardiomyopathy. Cardiology. 2010;115(2):155–162. doi: 10.1159/000265166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minow RA, Benjamin RS, Lee ET, Gottlieb JA. Adriamycin cardiomyopathy-risk factors. Cancer. 1977;39(4):1397–1402. doi: 10.1002/1097-0142(197704)39:4<1397::aid-cncr2820390407>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 5.Chiaramonte R, Colombo M, Bulfamante G, et al. Notch pathway promotes ovarian cancer growth and migration via CXCR4/SDF1alpha chemokine system. Int J Biochem Cell Biol. 2015;66:134–140. doi: 10.1016/j.biocel.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Asnaghi L, Ebrahimi KB, Schreck KC, et al. Notch signaling promotes growth and invasion in uveal melanoma. Clin Cancer Res. 2012;18(3):654–665. doi: 10.1158/1078-0432.CCR-11-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aster JC, Pear WS, Blacklow SC. The varied roles of Notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S, Tian L, Shen H, et al. DDX5 is a positive regulator of oncogenic NOTCH1 signaling in T cell acute lymphoblastic leukemia. Oncogene. 2013;32(40):4845–4853. doi: 10.1038/onc.2012.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Previs RA, Coleman RL, Harris AL, Sood AK. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. 2015;21(5):955–961. doi: 10.1158/1078-0432.CCR-14-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menietti E, Xu XY, Ostano P, Joseph JM, Lefort K, Dotto GP. Negative control of CSL gene transcription by stress/DNA damage response and p53. Cell Cycle. 2016;15(13):1767–1778. doi: 10.1080/15384101.2016.1186317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaish V, Kim J, Shim M. Jagged-2 (JAG2) enhances tumorigenicity and chemoresistance of colorectal cancer cells. Oncotarget. 2017;8(32):53262–53275. doi: 10.18632/oncotarget.18391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Shen H, Jin B, et al. Brief report: blockade of Notch signaling in muscle stem cells causes muscular dystrophic phenotype and impaired muscle regeneration. Stem Cells. 2013;31(4):823–828. doi: 10.1002/stem.1319. [DOI] [PubMed] [Google Scholar]
- 14.Ehebauer M, Hayward P, Martinez-Arias A. Notch signaling pathway. Sci STKE. 2006;2006(364):cm7. doi: 10.1126/stke.3642006cm7. [DOI] [PubMed] [Google Scholar]
- 15.Liu ZH, Dai XM, Du B. Hes1: a key role in stemness, metastasis and multidrug resistance. Cancer Biol Ther. 2015;16(3):353–359. doi: 10.1080/15384047.2015.1016662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamstrup MR, Biskup E, Manfe V, et al. Chemotherapeutic treatment is associated with Notch1 induction in cutaneous T-cell lymphoma. Leuk Lymphoma. 2017;58(1):171–178. doi: 10.1080/10428194.2016.1180681. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Guo WC, Wang L, et al. Notch signaling is important for epithelial-mesenchymal transition induced by low concentrations of doxo-rubicin in osteosarcoma cell lines. Oncol Lett. 2017;13(4):2260–2268. doi: 10.3892/ol.2017.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Tang Y, Wu L, et al. The hepatocyte-specific HNF4alpha/miR-122 pathway contributes to iron overload-mediated hepatic inflammation. Blood. 2017;130(8):1041–1051. doi: 10.1182/blood-2016-12-755967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mondal J, Samadder A, Khuda-Bukhsh AR. Psorinum 6 × triggers apoptosis signals in human lung cancer cells. J Integr Med. 2016;14(2):143–153. doi: 10.1016/S2095-4964(16)60230-3. [DOI] [PubMed] [Google Scholar]
- 21.Khan A, Gillis K, Clor J, Tyagarajan K. Simplified evaluation of apoptosis using the Muse cell analyzer. Postepy Biochem. 2012;58(4):492–496. [PubMed] [Google Scholar]
- 22.Shen H, McElhinny AS, Cao Y, et al. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20(6):675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannan S, Fang W, Song G, et al. Notch/HES1-mediated PARP1 activation: a cell type-specific mechanism for tumor suppression. Blood. 2011;117(10):2891–2900. doi: 10.1182/blood-2009-12-253419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borges KS, Andrade AF, Silveira VS, et al. The aurora kinase inhibitor AMG 900 increases apoptosis and induces chemosensitivity to anticancer drugs in the NCI-H295 adrenocortical carcinoma cell line. Anticancer Drugs. 2017;28(6):634–644. doi: 10.1097/CAD.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 25.Ji P, Yu L, Guo WC, et al. Doxorubicin inhibits proliferation of osteosarcoma cells through upregulation of the Notch signaling pathway. Oncol Res. 2014;22(4):185–191. doi: 10.3727/096504015X14343704124340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu SW, Wang H, Poitras MF, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 27.Chiarugi A, Moskowitz MA. Cell biology. PARP-1-a perpetrator of apoptotic cell death? Science. 2002;297(5579):200–201. doi: 10.1126/science.1074592. [DOI] [PubMed] [Google Scholar]
- 28.Schiewer MJ, Goodwin JF, Han S, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2(12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto M, Takii R, Takaki E, et al. The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun. 2017;8(1):1638. doi: 10.1038/s41467-017-01807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31(6):446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 31.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25(5):259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23(16):2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]